Abstract
Purpose
Structural mosaicism has been previously implicated in developmental disorders. We aimed to identify rare mosaic chromosomal alterations (MCAs) in probands with severe undiagnosed developmental disorders.
Methods
We identified MCAs in genotyping array data from 12,530 probands in the Deciphering Developmental Disorders study using mosaic chromosome alterations caller (MoChA).
Results
We found 61 MCAs in 57 probands, many of these were tissue specific. In 23 of 26 (88.5%) cases for which the MCA was detected in saliva in which blood was also available for analysis, the MCA could not be detected in blood. The MCAs included 20 polysomies, comprising either 1 arm of a chromosome or a whole chromosome, for which we were able to show the timing of the error (25% mitosis, 40% meiosis I, and 35% meiosis II). Only 2 of 57 (3.5%) of the probands in whom we found MCAs had another likely genetic diagnosis identified by exome sequencing, despite an overall diagnostic yield of ∼40% across the cohort.
Conclusion
Our results show that identification of MCAs provides candidate diagnoses for previously undiagnosed patients with developmental disorders, potentially explaining ∼0.45% of cases in the Deciphering Developmental Disorders study. Nearly 90% of these MCAs would have remained undetected by analyzing DNA from blood and no other tissue.
Keywords: Anueploidy, Chromosomal alterations, Developmental disorders, Mosaic, Uniparental disomy
Introduction
Genetic mosaicism is the presence of 2 or more genetically distinct lineages of cells in 1 individual, arising from post-zygotic variants. Mosaic variation can consist of single-nucleotide variants (SNVs) and indels, or it may involve larger stretches of the genome, including copy number variants (CNVs) and aneuploidies. Mosaicism has been associated with diseases including neurodevelopmental disorders.1, 2, 3, 4 The clinical consequences of mosaicism vary according to the nature of the mosaic event, the stage of development at which this event occurs, and the tissue types in which this event is present.5
Very large chromosomal abnormalities, such as complete autosomal aneuploidy, are generally incompatible with life, with the exception of trisomy 21. However, mosaic aneuploidies are better tolerated and have been identified in many autosomes, including chromosomes 7, 8, 9, 14, 16, 17, 19, and 22.6, 7, 8 Moreover, children with mosaic trisomies of chromosomes 13 and 18 live for much longer than those with constitutive trisomies, with 80% and 70% of patients with mosaic trisomy 13 and 18, respectively, surviving for at least a year compared with 8% with non-mosaic trisomies.9,10 Additionally, mosaic uniparental disomy (UPD, in which 2 copies of 1 chromosome are inherited from 1 parent) has also been associated with several developmental disorders.5,11, 12, 13, 14
Mosaic chromosomal alterations (MCAs) can be detected in SNP genotyping array data by identifying differences from the expected log R ratio (LRR) and B-allele frequency (BAF). LRR gives a measure of the intensity at any given position on the array and deviations from the expected LRR indicate an abnormal copy number. BAF is a normalized measure of the intensity ratio of 2 alleles (A and B), such that a BAF of 1 or 0 indicates the complete absence of 1 of the 2 alleles (eg, homozygous AA or BB), and a BAF of 0.5 indicates the equal presence of both alleles (eg, heterozygous AB). Deviations in BAF can indicate the presence of CNVs or UPD.
There are several tools that use LRR and BAF to detect MCAs from genotyping array data. Mosaic alteration detection (MAD) uses the genome alteration detection algorithm to detect mosaic CNVs and UPDs.15,16 Parent-of-origin-based detection in trios (triPOD) uses an overlapping window approach to detect mosaic CNVs and UPDs in parent-offspring trios, but the absence of parental data makes this tool unsuitable for many cohorts.17 MONTAGE is a recently developed tool using a sliding window approach for rapid detection of mosaic CNVs; however, it is unable to detect mosaic UPDs.18 MoChA is a bcftools plugin, which identifies mosaic CNVs and UPDs in array data using a hidden Markov model to detect imbalances in phased BAF and LRR.19,20 We chose to use MoChA because it is quick to run, sensitive, does not require trio information, and is able to detect both mosaic CNVs and UPDs.
The Deciphering Developmental Disorders study (DDD) is a cohort of 13,612 children with severe developmental disorders.21 The DDD study recruited patients from 2011 to 2015 who remained undiagnosed after expert review by a clinical geneticist and completion of routine genetic testing. These patients had neurodevelopmental disorders, congenital anomalies, abnormal growth parameters, dysmorphic features, and genetic disorders of significant impact for which the molecular basis was unknown. 58.4% of these patients were male and the median age at recruitment was 7 years (range 0-63 years). Approximately 85% had undergone array analysis (CMA) before recruitment, often supplemented by phenotype-targeted gene sequencing, but remained without a molecular genetic diagnosis to explain their phenotypes. MCAs have previously been investigated in this cohort using exome sequencing (ES) from 4,911 probands using MrMosaic and additionally from SNP genotyping array data for 1,303 of these probands using MAD and triPOD.3,4 However, the majority of DDD probands have not been analyzed systematically for the presence of MCAs. Here, we used MoChA to detect MCAs across all 12,530 probands with SNP genotyping array data in the DDD study.
Materials and Methods
Patient cohort
A total of 13,612 probands with developmental disorders, and their parents, were recruited to the DDD study. Blood-extracted DNA and/or saliva samples were collected from all probands, and, where possible, saliva samples were collected from parents. SNP genotyping array data were generated for 12,530 of the probands in this study. Probands were systematically phenotyped by consultant clinical geneticists using the Human Phenotype Ontology22 and a structured questionnaire in DECIPHER (www.deciphergenomics.org).23
MCA detection from array
Samples from 1465 probands were genotyped on the Illumina HumanOmniExpress chip, and samples from 11,065 probands were genotyped on the Illumina HumanCoreExome chip. Intensity data were converted into VCF (variant call format), including BAF and LRR, using gtc2vcf.24
MCAs were detected using MoChA.19,20 The output was filtered to remove samples with the following: BAF phase concordance across phased heterozygous sites underlying the call of >0.51, calls <100 kbp, calls with a LOD score of <10 for the model based on BAF and genotype phase, calls flagged by MoChA as likely germline CNVs, and calls with an estimated cell fraction of >50%. More stringent filters were subsequently applied to identify rare MCAs of likely clinical significance: events that occur in more than 1% of the cohort were removed, events overlapping CNVs previously identified in the cohort were removed, and events that were <1 Mb in length were removed unless they overlapped genes known to cause developmental disorders (https://www.ebi.ac.uk/gene2phenotype).25 All MCAs remaining after these filters were manually reviewed to evaluate data quality; events with low deviation in BAF, events in regions where the genotyping array had sparse SNPs and events in samples with noisy data were removed.
Results
Potentially clinically relevant MCAs were identified in 57 probands with developmental disorders
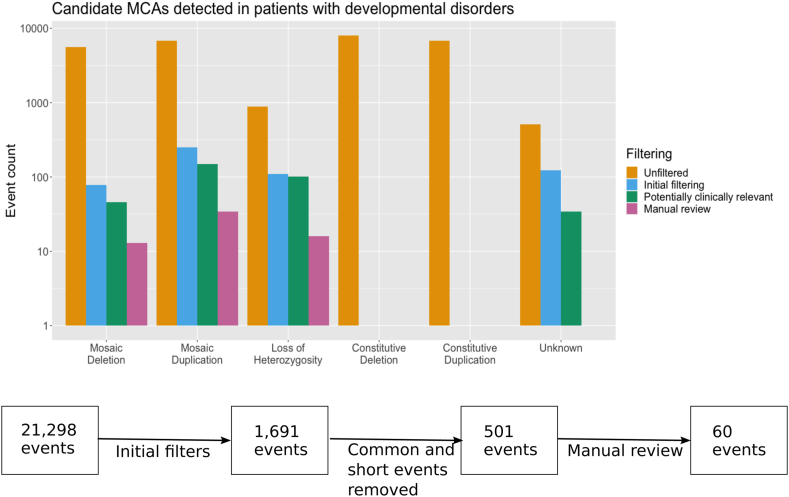
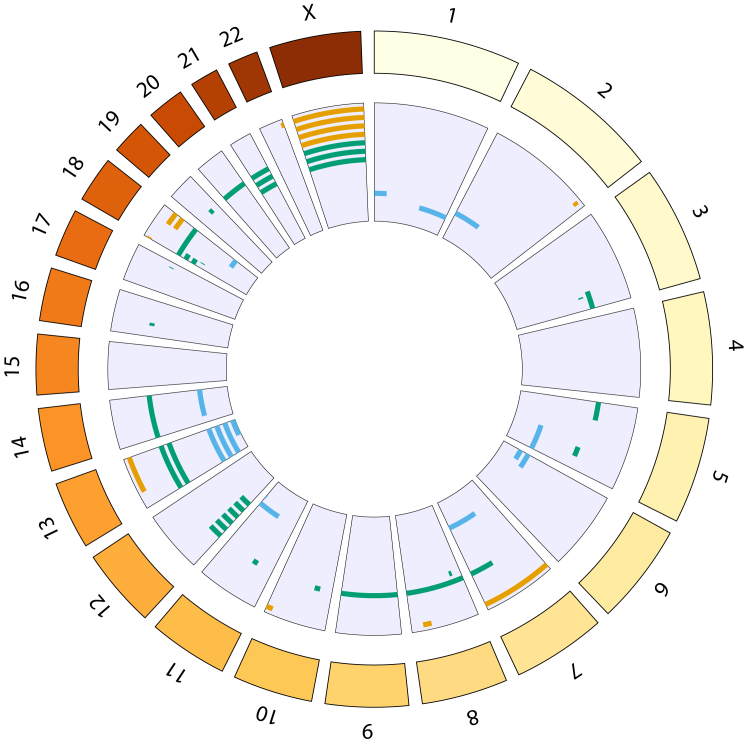
A total of 28,864 candidate MCAs were identified by MoChA in the 12,530 probands studied. It was our intention to identify potentially clinically relevant MCAs and our stringent filtering will inevitably have discarded some true positive MCAs. Besides the filtering recommended by the authors of the MoChA software, we filtered on size, frequency, and visibility to the naked eye of deviation on plots of BAF and LRR. Short MCAs, common MCAs, and those invisible to the naked eye because of low deviation in BAF are less likely to be pathogenic. After initial filtering (Methods), 558 candidate MCAs remained. These events comprised 249 duplications, 78 deletions, 109 copy number neutral loss-of-heterozygosity events, and 122 events in which the type could not be determined. After further filtering to identify events of potential clinical relevance, 330 events were reviewed manually to evaluate data quality, and, subsequently, 61 events from 57 probands were identified for clinical evaluation (Figure 1, Supplemental Table 1). These MCAs represent a potential diagnostic yield of 0.45% in our cohort. These 61 events comprise 33 duplications, 12 deletions, 14 copy number neutral loss-of-heterozygosity events, a deletion flanked by copy number neutral loss of heterozygosity, and a duplication followed by UPD of the majority of the q-arm of chromosome 1. The 33 duplications affect 18 different chromosomes, with the most frequently affected being chromosome 12 (6 events), the 12 deletions affect 7 different chromosomes, of which the most frequently affected is chromosome X (4 events), and the UPDs affect 9 different chromosomes, of which the most frequently affected is chromosome 13 (4 events) (Figure 2). All of these 57 probands had previously undergone ES, but pathogenic or likely pathogenic variants had only been identified in 2 of these individuals,26 indicative that these MCAs are likely diagnostic findings for the developmental disorders in these individuals because no alternative diagnoses were evident.
Figure 1.
Workflow used to identify potentially clinically relevant MCAs. The flowchart shows the different filtering stages and the total number of events remaining at each stage. The bar plot shows the number of events of each type remaining at each stage.
Figure 2.
The distribution of MCAs in the genome. Each bar represents 1 event; deletions are shown in orange, duplications in green, and loss of heterozygosity in blue.
Tissue specificity was observed for the majority of clinically relevant MCAs
A total of 42 MCAs were detected in saliva from 38 probands (Supplemental Table 1). For 9 of the probands in which a total of 11 MCAs were found in saliva, we also had genotyping array data from blood; the MCA was also detected in only 3 of these. In an additional 14 probands that 15 MCAs were detected in saliva, we also had ES and/or array comparative genomic hybridization (aCGH) data from blood; there was no evidence of the MCA in any of these. For the remaining 16 MCAs from 15 probands, no other tissue type was available for testing. A total of 22 MCAs were detected in blood from 22 probands. There were genotyping array data available from saliva in 3 of these, and, in all 3 cases, the MCA was also detected in saliva at a cell fraction higher than in blood, eg, mosaic deletion in ID 259003 is present in saliva at 53% and blood at 32% (see Table 1).
Table 1.
Pathogenic and likely pathogenic MCAs in DDD patients
| Chr. | Event type | Start-end (GRCh37) | Size (Mb) | Blood Saliva % % |
DECIPHER | Syndrome | Key Phenotypes Shown by Patients with Recurrent MCAs |
|---|---|---|---|---|---|---|---|
| 1 | dup+upd | 165589535-249250621 (q) | 83.66 | nd 39 | 296586 | Mosaic likely pathogenic duplication - ACMG score 0.90 (G1A, G3C) | |
| 2 | del | 223873590-232804522 | 8.93 | 0 22 | 265112 | Mosaic pathogenic deletion - ACMG score 1.90 (L1A, L2A, L3C) | |
| 3 | dup | 153567441-198022430 | 44.46 | 0 54 | 258956 | Mosaic pathogenic partial trisomy 3q23-ter | |
| 5 | dup | 1-46174864 | 46.18 | nd 36 | 305868 | Mosaic trisomy 5p | |
| 5 | dup | 123851734-148651711 | 24.80 | 0 34 | 261240 | Mosaic likely pathogenic duplication - ACMG score 0.90 (G1A, G3C) | |
| 7 | del | 1-159138663 (w) | 159.14 | 58 nd | 285424 | MIRAGE syndrome due to SAMD9 variant | |
| 7 | upd | 64864800-159138663 (q) | 94.27 | 7 0 | 275728 | MIRAGE syndrome due to SAMD9 variant | |
| 7 | dup | 99227172-159138663 | 59.91 | 0 43 | 283385 | Mosaic pathogenic partial trisomy 7q21.11-ter | |
| 8 | dup | 1-146364022 (w) | 146.36 | 45 nd | 275705 | Mosaic trisomy 8 | |
| 8 | dup | 22487087-29344462 | 6.85 | 25 nd | 263580 | Mosaic pathogenic duplication - ACMG score 1.35 (G1A, G2H, G3C, G5A) | |
| 8 | del | 101011612-120215645 | 19.20 | nd 33 | 290927 | Langer Gideon syndrome (includes EXT1) | |
| 9 | dup | 1-141213431 (w) | 141.21 | 19 nd | 295318 | Mosaic trisomy 9 | |
| 10 | del | 121375181-135534747 | 14.16 | 0 43 | 274013 | Mosaic pathogenic deletion - ACMG score 1.35 (L1A, L2C, L5A) | |
| 11 | dup | 41887927-47877800 | 5.99 | 0 22 | 259029 | Mosaic pathogenic duplication - ACMG score 1.35 (G1A G3C, G5A) | |
| 12 | dup | 1-26749137 (p) | 26.75 | 0 33 | 280908 | Pallister-Killian syndrome | ) ) |
| 12 | dup | 1-34523378 (p) | 34.52 | 0 34 | 283911 | Pallister-Killian syndrome | ) |
| 12 | dup | 1-34758266 (p) | 34.76 | 0 63 | 299715 | Pallister-Killian syndrome | GDD, irregular pigmentation, sparse scalp hair |
| 12 | dup | 1-34781187 (p) | 34.78 | 0 44 | 261373 | Pallister-Killian syndrome | ) |
| 12 | dup | 1-34801271 (p) | 34.80 | 0 25 | 286521 | Pallister-Killian syndrome | ) |
| 12 | dup | 1-34826574 (p) | 34.83 | 0 63 | 265800 | Pallister-Killian syndrome | ) |
| 13 | dup | 1-115169878 (w) | 115.17 | 0 11 | 264072 | Mosaic trisomy 13 | ) Congenital heart disease and GDD |
| 13 | dup | 1-115169878 (w) | 115.17 | 16 nd | 283167 | Mosaic trisomy 13 | ) |
| 13 | del flanked by loh | 33986219-115169878 | 81.18 | 23 nd | 293046 | Mosaic pathogenic deletion - ACMG score 2.35 (L1A, L2A, L3C, L5A) | |
| 14 | dup | 1-107349540 (w) | 107.35 | nd 45 | 306061 | Mosaic trisomy 14 | |
| 14 | upd (pat) | 1-107349540 (w) | 85.62 | nd 36 | 303525 | Mosaic Kagami-Ogata syndrome | |
| 16 | dup | 27183151-34747045 | 7.56 | 0 31 | 263654 | Mosaic pathogenic duplication - ACMG score 1.35 (G1A, G2H, G3C, G5A) | |
| 18 | dup | 1-14084928 (p) | 14.09 | nd 16 | 266471 | Mosaic pathogenic duplication - ACMG score 1.35 (G1A, G2H, G3C, G5A) | ) GDD ,hypotonia |
| 18 | dup | 1-14988113 | 14.99 | 0 25 | 273553 | Mosaic pathogenic duplication - ACMG score 1.35 (G1A, G2H, G3C, G5A) | ) |
| 18 | dup | 1-78077248 (p) | 78.08 | 14 nd | 260037 | Mosaic partial trisomy 18 | |
| 18 | del | 48368256-78077248 | 29.71 | 0 45 | 260462 | Mosaic partial monosomy 18q12.3-ter | ) Microcephaly, hypotonia |
| 18 | del | 49315539-78077248 | 28.76 | 0 49 | 274600 | Mosaic partial monosomy 18q12.3-ter | ) |
| 19 | dup | 31740744-41525952 | 9.79 | nd 29 | 290927 | Mosaic pathogenic duplication - ACMG score 1.35 (G1A, G2H, G3C, G5A) | |
| 20 | dup | 1-63025520 (w) | 63.03 | 0 22 | 258190 | Mosaic trisomy 20 | |
| 21 | dup | 1-48129895 (w) | 48.13 | nd 12 | 294112 | Mosaic trisomy 21 | ) |
| 21 | dup | 1-48129895 (w) | 48.13 | nd 40 | 301048 | Mosaic trisomy 21 | GDD, delayed speech and language, sandal gap |
| 21 | dup | 1-48129895 (w) | 48.13 | nd 30 | 306282 | Mosaic trisomy 21 | ) |
| 22 | del | 45311891-51304566 | 5.99 | 32 53 | 259003 | Mosaic pathogenic deletion - ACMG score 1.45 (L1A, L2A, L3A, L5A) | |
| X | dup | 1-155270560 (w) | 155.27 | 3 nd | 287504 | Mosaic XXXY/XXXXY syndrome | |
| X | dup | 38541235-41749282 | 3.21 | 19 nd | 280407 | Mosaic triple X syndrome | ) Cognitive impairment |
| X | dup | 38557085-41742515 | 3.19 | 15 nd | 277716 | Mosaic triple X syndrome | ) |
| X | del | 1-155270560 (w) | 155.27 | 0 43 | 283385 | Mosaic Turner syndrome | ) |
| X | del | 1-155270560 (w) | 155.27 | nd 55(p) 24(q) |
291029 | Mosaic Turner syndrome | ) Short stature |
| X | del | 1-155270560 (w) | 155.27 | 25 nd | 291198 | Mosaic Turner syndrome | ) |
| X | del | 1-155270560 (w) | 155.27 | nd 28 | 300814 | Mosaic Turner syndrome | ) |
Chr., chromosome; del, deletion; dup, duplication; GDD, global developmental delay; MCA, mosaic chromosomal alterations; nd, not done; p, p-arm; q, q-arm; upd, uniparental disomy; upd (pat), paternal uniparental disomy; w, whole chromosome.
Mosaic aneuploidy can originate in mitosis, meiosis I or meiosis II
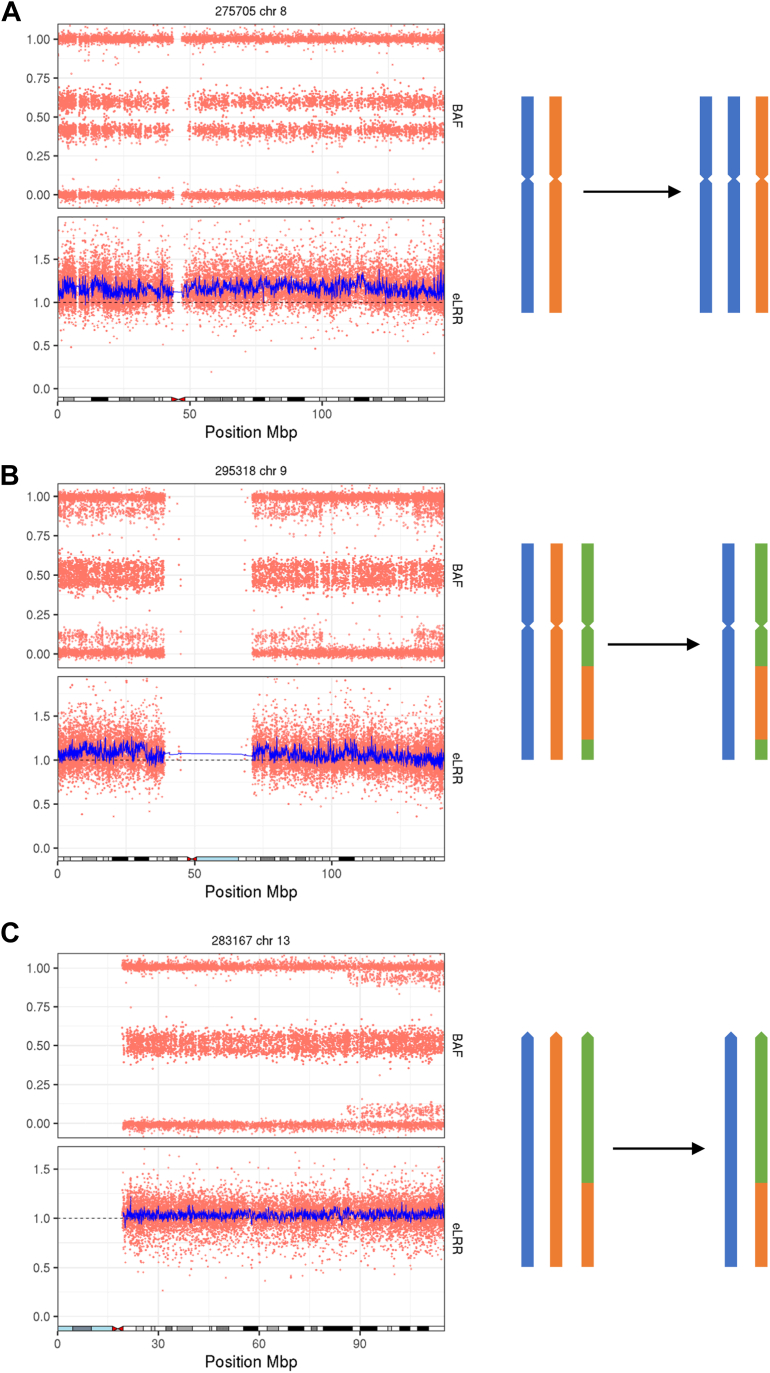
The 33 observed duplications include 20 polysomies, 11 of which affect a whole chromosome and 9 of which consist of the p-arm only (Supplemental Table 1). Ten different chromosomes were affected by these polysomies (5, 8, 9, 12, 13, 14, 18, 21, 20, and X). The origin of a trisomy can be determined by examination of the BAF pattern.6 The absence of a third haplotype indicates that 5 of these events (3 in chromosome 12 p-arm, 1 in chromosome 8, and 1 in chromosome X) have a mitotic origin. Eight events (1 in chromosome 5 p-arm, 1 in each of chromosomes 9, 13, 14, 18, and 20, and 2 in chromosome 21) have a BAF pattern consistent with occurrence in meiosis I, where 3 haplotypes are observed near to the centromere. A pattern consistent with occurrence in meiosis II, with additional haplotypes present at the telomeres but not at the centromere, is observed in the remaining 7 cases (3 chromosome 12 p-arm, 2 chromosome 18 p-arm, 1 chromosome 13, and 1 chromosome 21) (Figure 3).
Figure 3.
Origin of mosaic trisomies. A. A mosaic trisomy that has occurred during mitosis. B. A mosaic trisomy that has occurred during meiosis I. C. A mosaic trisomy that has occurred during meiosis II.
The 12 observed deletions include 4 mosaic monosomies, 1 in chromosome 7, and 3 in the X chromosome. One of the X chromosome monosomies has mosaic monosomy of the p-arm in 50% of cells and mosaic monosomy of the q-arm in 25 % of cells (Supplemental Figure 1). In all of the observed monosomies, the BAF patterns are consistent with origination via mitotic nondisjunction, with 2 distinct haplotypes observed, rather than monosomy rescue.6
The 14 mosaic copy number neutral loss-of-heterozygosity events identified include 5 mosaic UPDs that comprise all or most of 1 arm of a chromosome (1q, 2p, 6p, 7q, and 11q), 4 mosaic UPDs that affect an entire chromosome (3 in chromosome 13 and 1 in chromosome 14), and 5 smaller loss-of-heterozygosity events. In 1 case, the UPD in the p-arm of chromosome 6 shows 2 different clonalities and is therefore likely to have arisen as 2 different events (Supplemental Figure 2). Furthermore, we detected complex chromosomal events in several probands, including the following: deletion flanked by copy number neutral loss-of-heterozygosity, spanning a total of 80.9 Mb in chromosome 13 (Supplemental Figure 3A), a duplication followed by UPD of the majority of chromosome 1 q-arm (Supplemental Figure 3B), and a patient with a 59.9 Mb duplication in chromosome 7, mosaic polysomy of the first 95 Mb of chromosome X and non-mosaic polysomy of the remainder of chromosome X (Supplemental Figure 3C). We were unable to determine the origin of these events.
Discussion
Using these filters, we have identified MCAs of interest using genotyping array data in 57 of 12,530 (0.45%) probands with severe developmental disorders. Fifty-four patients had single events, 2 had 2 independent events, and 1 had 3 MCA events. Our findings are consistent with Sherman et al in which 46 mosaic CNVs were identified in 12,077 probands with autism spectrum disorder (ASD).27 After ES, only 2 (3.5%) patients with potentially clinically significant MCAs identified here have previously identified pathogenic or likely pathogenic SNVs, indels, or CNVs. Compared with the cohort-wide diagnostic yield in the DDD study of ∼40%,28 the observed enrichment of undiagnosed patients in this group suggests that most of these MCAs are diagnostic.
Clinical evaluation of the phenotypic and genomic data by an experienced clinical geneticist resulted in 44 diagnoses of MCAs that were either well-established pathogenic variants, eg, Mosaic tetrasomy 12p in Pallister-Killian syndrome, or where the chromosomal anomaly was classified as Pathogenic or Likely Pathogenic using the ACMGTM CNV classifier29 (Table 1). These comprised mosaic polysomies of chromosomes 12p, 18p, and 20, mosaic duplications of chromosomes 5, 8, 11, and 17, mosaic deletions of chromosomes 2 and 22, mosaic loss of heterozygosity in chromosome 5, and a mosaic deletion-duplication-deletion in chromosome 18. Clinical features indicative of a mosaic event, including abnormalities of skin pigmentation, syndactyly, and/or asymmetry, were observed in only 7 of the 44 probands with a diagnostic finding. The remainder of the MCAs were interpreted to be variants of uncertain significance. Recruitment to DDD was by ∼200 experienced consultant clinical geneticists. The fact that apparently recognizable disorders, such as Pallister-Killian syndrome or mosaic trisomy 13 or 21, have been identified in this study demonstrates that clinical assessment of mosaic disorders is not entirely reliable, and it is easy for them to be overlooked in clinic.
MoChA is unable to distinguish between mosaic trisomies, mosaic tetrasomies, or other mosaic polysomies. The 6 mosaic polysomies involving chromosome 12p are likely to be Pallister-Killian syndrome, in which an isochromosome comprising 2 copies of chromosome 12p is present.30 We also identify a case that is likely to be mosaic tetrasomy 5p. Only 5 cases of mosaic tetrasomy 5p, in which an isochromosome consisting of 2 copies of the p-arm of chromosome is present, have been reported to date.31 In addition a case of an isochromosome consisting of 2 partial copies of 5p has been reported.32 A small number of live-born cases of mosaic isochromosome 18p have been reported in the literature;33, 34, 35 we identify a likely mosaic tetrasomy 18p.
Mosaic trisomy can occur by meiotic non-disjunction in the oocyte or sperm followed by trisomy rescue or by mitotic nondisjunction at a later stage of development. Using genotyping array data, we were able to distinguish between mosaic polysomies occurring via nondisjunction at mitosis or meiosis, and 15 of 20 trisomies detected (75%) were meiotic in origin. The timing of the event has implications for counseling families because some women have a higher rate of meiotic nondisjunction and therefore a greater recurrence risk.36,37 This estimate is somewhat higher than Conlin et al, who found that 10 of 20 (50%) of trisomies had a meiotic origin6 and may reflect ascertainment differences between the cohorts.
The mosaic monosomies we detected all arose by mitotic nondisjunction, rather than monosomy rescue, which would result in homozygosity. This finding has important implications for recurrence risk because for the former this is negligible, whereas the latter raises the potential for gonadal mosaicism. We were unable to detect monosomy rescue because the method used is phase-based and therefore cannot detect events in runs of homozygosity;20 however, Conlin et al also only reported mitotic events.6 Similarly, our study can only detect mosaic UPD arising from trisomy rescue and resulting in heterodisomy because any events arising from monosomy rescue will result in isodisomy and lack heterozygous regions.
Two of the MCAs described here, a mosaic monosomy and a mosaic UPD, are in chromosome 7 and include the SAMD9 gene. In both patients pathogenic SAMD9 variants have previously been reported. Loss of chromosome 7 and UPD of 7q have previously been described in patients with MIRAGE syndrome (MIM #617053), this is believed to be an adaptation to the growth-suppressing effect of the SAMD9 variants.38,39
We found MoChA to be a highly effective tool for detecting clinically relevant MCAs. Smaller subsets of the DDD cohort have previously been analyzed for MCAs using alternative methods. Previously published analysis of structural mosaicism in genotyping arrays from 1303 DDD probands using MAD and triPOD described MCAs in 12 probands.3 However, neither MAD or triPOD detected all 12 of these events, and it was shown that a combination of algorithms was necessary to maximize diagnostic yield. We tested 11 of these probands and found 9 of the previously reported events. MoChA identifies events found by MAD and missed by triPOD and vice versa. One of the events missing in our filtered MoChA data set was a genome-wide paternal UPD. This event was found by MoChA; however, the sample was removed by the MoChA default filters designed to exclude samples that are either contaminated or low quality DNA based on high-phased BAF auto-correlation. The second event that is not found by MoChA was a UPD of chromosome 14 present in around two-thirds of cells; it is not clear why this event was not found by MoChA; however, no mosaic UPDs with a cell fraction of >0.4 were detected. Additionally, a duplication in chromosome 17 not previously identified using MAD and triPOD was detected using MoChA. Furthermore, the previously published analysis of structural mosaicism in ES data from 4911 DDD probands using MrMosaic described MCAs in 9 probands, all of which were detected using MoChA.4 In the same 4911 probands, an additional 5 events were detected using MoChA that were not detected using MrMosaic, including the following: 2 mosaic polysomies of chromosome 18p, 1 mosaic polysomy of chromosome 8, 1 mosaic UPD of chromosome 13, and 1 mosaic UPD of chromosome 2p. These results show that using more than 1 tool will increase the number of MCAs detected; however, if only a single tool is to be used (for example from a cost-benefit perspective), then MoChA is a good choice because of its high sensitivity, rapid run time, and ability to detect both mosaic CNVs and UPDs.
Importantly, 23 of 26 (88.5%) of MCAs detected from saliva in which blood was also available for testing could not be detected in blood-derived DNA. This result contrasts with our previous observation that mosaic de novo SNVs were observed at similar variant allele fractions in both blood and saliva40 and may suggest stronger negative selection against MCAs within blood lineages. One limitation of our study is that we only have data from 2 tissues, blood, and saliva. Although study of saliva yields more mosaic events than blood, variants occurring later in embryonic development are likely to be present in a narrower range of tissues and we may therefore miss potentially diagnostic events by not having more tissue types available to study. Nonetheless, our observations highlight the importance of testing saliva (or other tissues) where possible to avoid missing mosaic structural events.
There is currently a paucity of large-scale studies of MCAs in disease cohorts. Our results are comparable in both size and yield to those of Sherman et al, who report 46 mosaic CNVs in a cohort of 12,077 patients with autism spectrum disorder (0.38%),27 our MCAs included mosaic CNVs in 43 of our 12,530 patients (0.34%). Study of mosaic aneuploidies and UPDs by Conlin et al in a cohort of 2019 patients referred to the Children's Hospital of Philadelphia Clinical CytoGenomics laboratory had a higher yield than our study (30/2019, 1.5%).6 Our results add to this body of literature but are likely to be an underestimate of the true diagnostic yield from MCAs in developmental disorders because of under-ascertainment in the DDD study of cases who would have been previously diagnosed using prior clinical genetic testing (such as karyotyping and microarray analysis).41
Our results show that rare MCAs are an important source of diagnoses in severe developmental disorders. The meiotic or mitotic origin of the variant can often be determined through careful analysis of genotyping array data and has important implications for recurrence risk. This work suggests that routinely analyzing SNP genotyping array data could provide potential diagnoses that are currently difficult to detect via ES and that diagnostic yield will be increased by the analysis of saliva samples. We recommend that clinical teams consider the use of saliva-derived DNA for genotyping array analysis for the investigation of neurodevelopmental disorders to complement genome-wide sequencing using blood-derived DNA.
Data Availability
Diagnostic variants and phenotypes for probands included in this study are available via the DECIPHER database (https://deciphergenomics.org/). Genotype array data are available in EGA.
Conflict of Interest
M.E.H. is a co-founder and non-executive director of Congenica Ltd and an advisor to AstraZeneca. All other authors declare no conflicts of interest.
Acknowledgments
We thank the DDD participants and their families for their participation in this study. We thank Giulio Genovese for his assistance when running MoChA. We thank the Sanger Institute genotyping facility. The DDD study presents independent research commissioned by the Health Innovation Challenge Fund (grant number HICF-1009-003). This study makes use of DECIPHER (http://decipher.genomics.org), which is funded by Wellcome. See Nature PMID: 25533962 or www.ddduk.org/access.html for full acknowledgment. This research was funded in part by the Wellcome grant (206194). H.F. receives support from Wellcome grant 200990/A/16/Z and from the NIHR Cambridge Biomedical Research Centre (NIHR203312). The views expressed are those of the authors and not necessarily those of the NIHR or the Department of Health and Social Care. For the purpose of open access, the author has applied a CC BY public copyright license to any Author Accepted Manuscript version arising from this submission.
Author Information
Conceptualization: M.E.H.; Data Curation: R.Y.E.; Formal Analysis: R.Y.E., H.V.F.; Funding Acquisition: M.E.H.; Investigation: R.Y.E.; Visualization: R.Y.E.; Writing-original draft: R.Y.E.; Writing-review and editing: R.Y.E., H.V.F., C.F.W., M.E.H., D.R.F.
Ethics Declaration
The study has UK Research Ethics Committee approval (10/H0305/83, granted by the Cambridge South Research Ethics Committee and GEN/284/12, granted by the Republic of Ireland Research Ethics Committee). Informed consent for participation in the DDD study was obtained from all participants as required by the REC.
Footnotes
The Article Publishing Charge (APC) for this article was paid by Dr Ruth Eberhardt, Wellcome Sanger Institute, Hinxton, Cambs, UK.
Additional Information
The online version of this article (https://doi.org/10.1016/j.gimo.2023.100836) contains supplemental material, which is available to authorized users.
Additional Information
References
- 1.Lupski J.R. Genetics. Genome mosaicism—one human, multiple genomes. Science. 2013;341(6144):358–359. doi: 10.1126/science.1239503. [DOI] [PubMed] [Google Scholar]
- 2.Poduri A., Evrony G.D., Cai X., Walsh C.A. Somatic mutation, genomic variation, and neurological disease. Science. 2013;341(6141) doi: 10.1126/science.1237758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.King D.A., Jones W.D., Crow Y.J., et al. Mosaic structural variation in children with developmental disorders. Hum Mol Genet. 2015;24(10):2733–2745. doi: 10.1093/hmg/ddv033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.King D.A., Sifrim A., Fitzgerald T.W., et al. Detection of structural mosaicism from targeted and whole-genome sequencing data. Genome Res. 2017;27(10):1704–1714. doi: 10.1101/gr.212373.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forsberg L.A., Gisselsson D., Dumanski J.P. Mosaicism in health and disease-clones picking up speed. In:. Nat Rev Genet: Nature Publishing Group. 2017;18(2):128–142. doi: 10.1038/nrg.2016.145. [DOI] [PubMed] [Google Scholar]
- 6.Conlin L.K., Thiel B.D., Bonnemann C.G., et al. Mechanisms of mosaicism, chimerism and uniparental disomy identified by single nucleotide polymorphism array analysis. Hum Mol Genet. 2010;19(7):1263–1275. doi: 10.1093/hmg/ddq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yong P.J., Barrett I.J., Kalousek D.K., Robinson W.P. Clinical aspects, prenatal diagnosis, and pathogenesis of trisomy 16 mosaicism. J Med Genet. 2003;40(3):175–182. doi: 10.1136/jmg.40.3.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biesecker L.G., Spinner N.B. A genomic view of mosaicism and human disease. Nat Rev Genet. 2013;14(5):307–320. doi: 10.1038/nrg3424. [DOI] [PubMed] [Google Scholar]
- 9.Goel N., Morris J.K., Tucker D., et al. Trisomy 13 and 18-Prevalence and mortality-A multi-registry population based analysis. Am J Med Genet A. 2019;179(12):2382–2392. doi: 10.1002/ajmg.a.61365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu J., Springett A., Morris J.K. Survival of trisomy 18 (Edwards syndrome) and trisomy 13 (Patau syndrome) in England and Wales: 2004-2011. Am J Med Genet A. 2013;161(10):2512–2518. doi: 10.1002/ajmg.a.36127. [DOI] [PubMed] [Google Scholar]
- 11.Ahram D.F., Stambouli D., Syrogianni A., et al. Mosaic partial pericentromeric trisomy 8 and maternal uniparental disomy in a male patient with autism spectrum disorder. Clin Case Rep. 2016;4(12):1125–1131. doi: 10.1002/ccr3.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKinlay Gardner R.J., Amor D.J. Vol. 1. Oxford University Press; Oxford, United Kingdom: 2018. Uniparental Disomy and Disorders of Imprinting. [Google Scholar]
- 13.Myers K.A., Bennett M.F., Chow C.W., et al. Mosaic uniparental disomy results in GM1 gangliosidosis with normal enzyme assay. Am J Med Genet A. 2018;176(1):230–234. doi: 10.1002/ajmg.a.38549. [DOI] [PubMed] [Google Scholar]
- 14.Haug M.G., Brendehaug A., Houge G., Kagami M., Ogata T. Mosaic upd(14)pat in a patient with mild features of Kagami-Ogata syndrome. Clin Case Rep. 2018;6(1):91–95. doi: 10.1002/ccr3.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.González J.R., Rodríguez-Santiago B., Cáceres A., et al. A fast and accurate method to detect allelic genomic imbalances underlying mosaic rearrangements using SNP array data. BMC Bioinformatics. 2011;12:166. doi: 10.1186/1471-2105-12-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pique-Regi R., Monso-Varona J., Ortega A., Seeger R.C., Triche T.J., Asgharzadeh S. Sparse representation and Bayesian detection of genome copy number alterations from microarray data. Bioinformatics. 2008;24(3):309–318. doi: 10.1093/bioinformatics/btm601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baugher J.D., Baugher B.D., Shirley M.D., Pevsner J. Sensitive and specific detection of mosaic chromosomal abnormalities using the Parent-of-Origin-based Detection (POD) method. BMC Genomics. 2013;14(1):367. doi: 10.1186/1471-2164-14-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glessner J.T., Chang X., Liu Y., et al. MONTAGE: a new tool for high-throughput detection of mosaic copy number variation. BMC Genomics. 2021;22(1):133. doi: 10.1186/s12864-021-07395-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loh P.R., Genovese G., McCarroll S.A. Monogenic and polygenic inheritance become instruments for clonal selection. Nature. 2020;584(7819):136–141. doi: 10.1038/s41586-020-2430-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loh P.R., Genovese G., Handsaker R.E., et al. Insights into clonal haematopoiesis from 8,342 mosaic chromosomal alterations. Nature. 2018;559(7714):350–355. doi: 10.1038/s41586-018-0321-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Firth H.V., Wright C.F., DDD Study The Deciphering Developmental Disorders (DDD) study. Dev Med Child Neurol. 2011;53(8):702–703. doi: 10.1111/j.1469-8749.2011.04032.x. [DOI] [PubMed] [Google Scholar]
- 22.Köhler S., Carmody L., Vasilevsky N., et al. Expansion of the Human Phenotype Ontology (HPO) knowledge base and resources. Nucleic Acids Res. 2019;47(D1):D1018–D1027. doi: 10.1093/nar/gky1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Firth H.V., Richards S.M., Bevan A.P., et al. DECIPHER: database of chromosomal imbalance and phenotype in humans using Ensembl resources. Am J Hum Genet. 2009;84(4):524–533. doi: 10.1016/j.ajhg.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Genovese G. gtc2vcf. Github. https://github.com/freeseek/gtc2vcf Accessed July 2, 2020.
- 25.Thormann A., Halachev M., McLaren W., et al. Flexible and scalable diagnostic filtering of genomic variants using G2P with Ensembl VEP. Nat Commun. 2019;10(1):2373. doi: 10.1038/s41467-019-10016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wright C.F., Fitzgerald T.W., Jones W.D., et al. Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. Lancet. 2015;385(9975):1305–1314. doi: 10.1016/S0140-6736(14)61705-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sherman M.A., Rodin R.E., Genovese G., et al. Large mosaic copy number variations confer autism risk. Nat Neurosci. 2021;24(2):197–203. doi: 10.1038/s41593-020-00766-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wright C.F., McRae J.F., Clayton S., et al. Making new genetic diagnoses with old data: iterative reanalysis and reporting from genome-wide data in 1,133 families with developmental disorders. Genet Med. 2018;20(10):1216–1223. doi: 10.1038/gim.2017.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Riggs E.R., Andersen E.F., Cherry A.M., et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen) Genet Med. 2020;22(2):245–257. doi: 10.1038/s41436-019-0686-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Izumi K., Krantz I.D. Pallister-Killian syndrome. Am J Med Genet C Semin Med Genet. 2014;166C(4):406–413. doi: 10.1002/ajmg.c.31423. [DOI] [PubMed] [Google Scholar]
- 31.Brock J.-A.K., Dyack S., Ludman M., Dumas N., Gaudet M., Morash B. Mosaic tetrasomy 5p resulting from an isochromosome 5p marker chromosome: case report and review of literature. Am J Med Genet A. 2012;158a(2):406–411. doi: 10.1002/ajmg.a.34272. [DOI] [PubMed] [Google Scholar]
- 32.Roulet-Coudrier F., Rouibi A., Thuillier C., et al. Unusual isochromosome 5p marker chromosome. Am J Med Genet A. 2015;167A(2):455–459. doi: 10.1002/ajmg.a.36843. [DOI] [PubMed] [Google Scholar]
- 33.Chen C.P., Ko T.M., Chen Y.Y., et al. Prenatal diagnosis and molecular cytogenetic characterization of low-level mosaicism for tetrasomy 18p at amniocentesis in a pregnancy with a favorable outcome. Taiwan J Obstet Gynecol. 2017;56(6):836–839. doi: 10.1016/j.tjog.2017.10.024. [DOI] [PubMed] [Google Scholar]
- 34.Bai J.L., Jin Y.W., Qu Y.J., Wang H., Cao Y.Y., Song F. Mosaicism of tetrasomy 18p: clinical and cytogenetic findings in a female child. Chin Med J (Engl) 2017;130(6):744–746. doi: 10.4103/0366-6999.201604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Plaiasu V., Ochiana D., Motei G., Georgescu A. A rare chromosomal disorder – isochromosome 18p syndrome. Maedica. 2011;6(2):132–136. [PMC free article] [PubMed] [Google Scholar]
- 36.De Souza E., Halliday J., Chan A., Bower C., Morris J.K. Recurrence risks for trisomies 13, 18, and 21. Am J Med Genet A. 2009;149A(12):2716–2722. doi: 10.1002/ajmg.a.33099. 12. [DOI] [PubMed] [Google Scholar]
- 37.Warburton D., Dallaire L., Thangavelu M., Ross L., Levin B., Kline J. Trisomy recurrence: a reconsideration based on North American data. Am J Hum Genet. 2004;75(3):376–385. doi: 10.1086/423331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Narumi S., Amano N., Ishii T., et al. SAMD9 mutations cause a novel multisystem disorder, Mirage syndrome, and are associated with loss of chromosome 7. Nat Genet. 2016;48(7):792–797. doi: 10.1038/ng.3569. [DOI] [PubMed] [Google Scholar]
- 39.Wong J.C., Bryant V., Lamprecht T., et al. Germline SAMD9 and SAMD9L mutations are associated with extensive genetic evolution and diverse hematologic outcomes. JCI Insight. 2018;3(14) doi: 10.1172/jci.insight.121086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wright C.F., Prigmore E., Rajan D., et al. Clinically relevant postzygotic mosaicism in parents and children with developmental disorders in trio exome sequencing data. Nat Commun. 2019;10(1):2985. doi: 10.1038/s41467-019-11059-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wright C.F., Campbell P., Eberhardt R.Y., et al. Genomic diagnosis of rare pediatric disease in the United Kingdom and Ireland. N Engl J Med. 2023;388(17):1559–1571. doi: 10.1056/NEJMoa2209046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Diagnostic variants and phenotypes for probands included in this study are available via the DECIPHER database (https://deciphergenomics.org/). Genotype array data are available in EGA.