Abstract
Several dozen Mendelian mutants have been discovered in axolotl (Ambystoma mexicanum) populations, including several that affect pigmentation. Four recessive mutants have been described in the scientific literature and genes for three of these have been identified. Here we describe and genetically dissect copper, a mutant with an albino-like phenotype known only from the pet trade. We performed a cross segregating copper and wildtype color phenotypes and used bulked segregant RNA-Seq to identify a region on chromosome 6 that was enriched for single-nucleotide polymorphisms (SNPs) between the color phenotypes. This region included Tyrosinase-like Protein 1 (Tyrp1), a melanin synthesis protein that when mutated, is associated with lighter than black melanin coloration in animal models and oculocutaneous albinism in humans. Inspection of RNA-Seq reads identified a single nucleotide deletion that is predicted to change the coding frame, introduce a premature stop codon in exon 6 and yield a truncated Tyrp1 protein in copper individuals. Using CRISPR-Cas9 editing, we show that wildtype Tyrp1 crispants exhibit copper pigmentation, thus confirming Tyrp1 as the copper locus. Our results suggest that commercial and hobbyist axolotl populations may harbor useful mutants for biological research.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-73283-1.
Subject terms: Mutation, Disease model, Herpetology
Introduction
The axolotl (Ambystoma mexicanum) has a long and storied history in biological research. Axolotls provide models for studies of embryonic and post-embryonic development, including most famously the study of whole organ regeneration1. The primary stock center for axolotl research is a captive bred population that has been maintained for almost 100 years2,3. Many mutants have been maintained in this population over time, including four different color mutants that are determined by single genes: white, melanoid, albino, and axanthic. Each of these mutants has an interesting history. The white mutant was presumably collected from Mexico in 1862 along with 33 wildtype individuals4. These axolotls were transported to Paris to establish the first laboratory axolotl population. The white mutant is caused by a Edn3 splicing defect3. The melanoid mutant was originally identified from laboratory crosses made using wild-caught axolotls from Mexico, and like white, melanoid is presumably also a natural color variant5. Subsequent analyses revealed that melanoid is genetically associated with Ltk6. Similarly, albino was originally identified in a wild captured tiger salamander and crossed into axolotl stocks7. In contrast, axanthic appears to have arisen spontaneously within a laboratory axolotl strain8. Although the gene for axanthic has not been identified, Woodcock et al.3 showed that albino is caused by a deletion in the Tyr coding sequence.
For as long as axolotls have been studied in research, they have also been highly prized as aquarium pets. All axolotl color mutants known to biological research, and even transgenic axolotls, are available in the pet trade. Additionally, pet breeders have identified color variants that have not received biological study, including a recessive Mendelian mutant called copper. Pet breeders describe copper axolotls as having copper-colored bodies with yellow xanthophores, brownish melanophores, and iridescent iridophores. The brownish and not black color of melanophores suggests copper is a form of albinism, perhaps involving mutation of a melanin synthesis protein. Given the importance of animal models in the study of albinism diseases, we used bulked segregant RNA-Seq (BSR-Seq)9 and CRISPR-Cas910 to identify Tyrp1as the copper locus. Our results suggest that commercial and private axolotl collections may harbor genetic variants that may prove useful as animal models in biological research.
Results
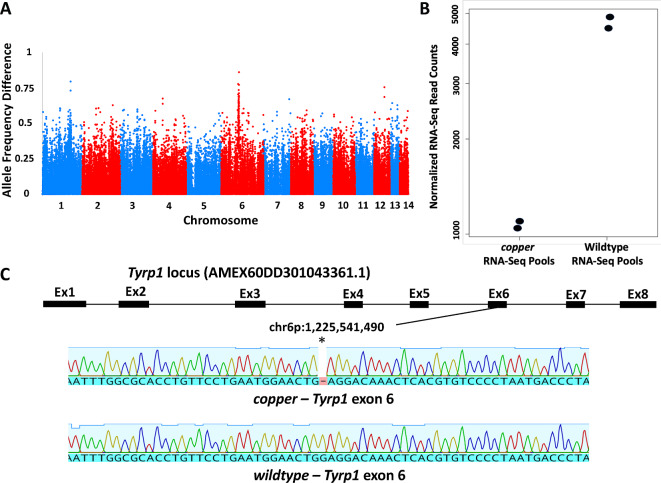
BSR-Seq9 was used to identify SNPs linked to the copper locus. Embryos from a cross that segregated copper and wildtype individuals were used to create two copper and two wildtype RNA pools (N = 36 and 27 for copper and wildtype respectively, per pool) that were each subjected to Illumina short-read RNA sequencing to identify single nucleotide polymorphisms (SNPs). Analysis of SNPs across the axolotl genome revealed a region on chromosome 6p with dissimilar frequencies between the pools (Fisher’s exact test F = 686, p = 2.93E−123 at peak association) (Fig. 1a; Supplementary File 1). Examination of genes within this region identified Tyrosinase-like protein 1 (Tyrp1) as a candidate gene. Tyrp1 encodes an enzyme that functions in melanin synthesis. Tyrp1 mutations generate lighter than black melanin shades of color in animal models11–13 and Oculocutaneous Albinism 3 (OCA3) in humans14. Additionally, significantly more Tyrp1 transcripts were identified from the wild-type vs. copper RNA-Seq pools (Dseq2 Wald Test = 20.03, adjusted P-value = 1.06E-84) (Fig. 1b). The axolotl Tyrp1 locus (AMEX60DD301043361.1) is distributed across 8 exons and the 1663 bp coding sequence encodes a 524 amino acid protein. A single nucleotide deletion (chr6p:1,225,541,490) was identified in RNA-Seq reads generated for the copper RNA pool (Fig. 1c; Supplementary File 2). This deletion, in exon 6, was confirmed by sequencing genomic DNA isolated from two copper and two wildtype individuals that were used to construct the RNA BSR-Seq pools (Fig. 1c). The deletion in the copper Tyrp1 sequence is predicted to change the reading frame and introduce a premature stop codon at amino acid position 416 (Supplementary File 3).
Fig. 1.
Identification of Tyrp1 as a candidate gene for copper. (A) Plot showing genome wide SNP (allele) frequency differences between copper and wildtype BSA-RNA-Seq pools. (B) Transcript counts identified for Tyrp1 between copper and wildtype BSA-RNA-Seq pools. The difference between pools is statistically significant (W = 20.03, adjusted P-value = 1.06E-84). (C) Genomic map of the axolotl Tyrp1 locus and deletion detected in exon 6 between copper and wildtype alleles.
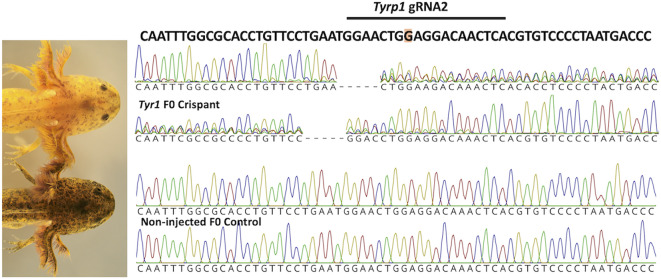
CRISPR-CAS910 was performed to determine if disruption of the Tyrp1 coding sequence would yield individuals that presented copper pigmentation. Two guide RNAs targeting exons 2 and 6 of the Tyrp1 coding sequence were injected into 100, one-cell stage wildtype embryos and visually assessed for color at developmental stage 4215. Crispant individuals were characterized by having fewer melanophores than non-injected controls and presented a yellowish overall color, as is typical of copper larvae in the pet trade. PCR and DNA sequencing of Tyrp1 regions targeted by CRISPR-Cas9 confirmed that crispant embryos (N = 12) were edited at both gRNA-target sites (Fig. 2). Five Tyrp1 crispants (that were edited at both gRNA1 and gRNA2) were reared to approximately 17 cm total body length (1.2 years old) and photographed to document pigmentation. Relative to the yellowish color observed during the larval stage, all five Tyrp1 crispants presented a darker copper skin color that was very similar to the color of adult copper axolotls (Fig. 3). The pigmentation phenotypes resulting from CRISPR-Cas9 genome editing strongly implicate Tyrp1 as the gene for copper.
Fig. 2.
CRISPR-CAS9 knockdown of Tyrp1. Melanin pigmentation was greatly reduced in F0 Tyrp1 crispant larvae relative to F0 non-injected control siblings. F0 Tyrp1 crispant electropherograms showed overlapping sequence traces at the gRNA target site for forward and reverse DNA sequencing reactions, consistent with genome editing. F0 non-injected control DNA sequence does not show evidence of genome editing. The gRNA target sequence (underlined) overlaps the deleted nucleotide in the copper Tyrp1 allele.
Fig. 3.

Images of F0 Tyrp1 crispants relative to a wildtype axolotl and copper mutant. Salamanders are 15–18 cm total body length and 1-1.2 years old. A USA copper penny (1.9 cm diameter) is provided as a color and size reference.
Discussion
Several different axolotl pigmentation variants are present in laboratories and households around the world. Several of these variants, including white, albino and melanoid, have received considerable attention in biological research. Previously, we identified genes for these variants to increase their value as research models3,5. In this study we identified a new gene in an axolotl pigmentation variant known only from the pet trade. Specifically, we show that copper, an axolotl mutant with copper coloration, is likely determined by a single nucleotide deletion in the Tyrp1 coding sequence. Tyrp1 encodes an enzyme that functions in melanophores to produce a black pigment called melanin. Mutations in Tyrp1 are associated with decreased production of melanin and structural alterations that result in lighter than expected coloration, for example brown coat color in mice11 and blonde hair in Melanesian humans16. copper axolotls in the larval stage have fewer dark pigmented melanophores than those observed in wildtype animals, and in absence of melanophores pigmentation is primarily determined by yellow xanthophores. As copper axolotls age, a brownish pigment emerges and individuals developed a uniform copper skin color, as is typical of animal models with Tyrp1 mutations. To functionally validate Tyrp1 for copper, we generated copper-like pigmentation in wildtype individuals using CRISPR-Cas9 genome editing of Tyrp1. The combination of SNP genome association data with CRISPR-Cas9 functional genomics results strongly implicates Tyrp1 as the copper locus.
The copper deletion is predicted to alter the Tyrp1 coding sequence and introduce a premature stop codon in exon 6, yielding a truncated protein with an altered function. We note that the exon 6 deletion in copper Tyrp1 parallels an exon 6 deletion identified in the first human case report of OCA314, with both deletions leading to a premature stop codon. No Tyrp1 mRNA or protein was detected in the human case while Tyrp1 transcripts were recovered by BSR-Seq in copper axolotls, albeit at lower transcript abundances. We speculate that Tyrp1 transcript abundances are lower in copper axolotls because the premature stop codon causes nonsense-mediated mRNA decay. Although it remains to be determined if a copper Tyrp1 protein is generated in axolotl, a nonsense mutation in the catalytic, tyrosinase-like domain of Tyrp1 is expected to affect protein function and melanogenesis17.
Domestic plant and animal populations have long served as reservoirs for phenotypes of commercial, biological, and biomedical relevance18–20. While the pet trade has gained access to axolotls from research labs, including for example GFP transgenics, axolotl researchers have not assessed commercial and domestic populations for new research models. Now that Tyrp1 has been identified as the causative gene for copper, this new axolotl model can be used to study molecular functions underlying OCA3 phenotypes. As is observed in humans with OCA3, copper is characterized by a reduction in melanin. A reduction in melanin could trace to many different cellular mechanisms as Tyrp1 functions in multiple pathways that directly or indirectly regulate melanin biosynthesis, as well as affecting melanophore cell division and death21. In addition to copper, other axolotl pigment variants in the pet trade include hypomelanistic, which is characterized by a reduction in melanin and UV-light excitable face/cranium fluorescence, and “starburst” which presents increased numbers of iridophores in albino axolotls. These and other phenotypic variants in the axolotl pet trade may provide useful models for biological research.
Materials and methods
Approval for animal experiments
Animal care procedures were approved under University of Kentucky IACUC protocol 2017–2580 and performed in accordance with ARRIVE22 and AAALAC23 guidelines and standards.
Animal procedures
Axolotls used in this study were obtained from a commercial population (Strohls Herptiles: wildtype and copper sibling larvae) and the Ambystoma Genetic Stock Center (AGSC RRID: SCR_006372; wildtype axolotl embryos RRID: AGSC_100E). All experiments were performed using either 50% (pre-hatching) or 100% (post-hatching) axolotl rearing water (ARW: 1.75 g NaCl, 100 mg MgSO4, 50 mg CaCl2, and 25 mg KCl per liter, buffered with NaHCO3 to pH 7.3–7.5) in a room maintained at 17–18 °C. Larvae were housed in 1 L glass or 2 L polypropylene bowls at either low density (8–10 per bowl) or one per container. After larvae reached 5 cm they were transferred to 9 L boxes on an Aquatic Enterprises recirculating system. Larvae were initially fed newly hatched brine shrimp until 3 cm total body length and then transitioned to California Black worms (J&R Enterprises) until large enough to be fed fish pellets (Rangen). Animals were anesthetized using a 0.02% benzocaine solution. Benzocaine was first dissolved in 4 ml 100% EtOH and then the chemical and solvent were diluted in 1 L of ARW.
Bulked segregant RNA-Seq
A total of 36 copper and 27 wildtype siblings were sampled from a cross between a homozygous copper male and a heterozygous female carrier and reared to approximately 3 cm. Tail tissue was collected from each while under benzocaine anesthesia. The resulting tail tips were pooled into separate 1.7 ml plastic tubes and flash frozen with dry ice to create copper and wildtype bulk segregant pools for RNA isolation. Tail tissue samples were dissociated with 23- and 26-gauge needles, and RNA was isolated with TRIzol and then further purified using a QIAGEN RNeasy Mini Kit with DNase treatment. The resulting RNA pools were used to generate outsourced Poly(A) RNA-seq libraries that were sequenced on an Illumina NovaSeq X Plus (150 bp paired end reads) by Novogene. Reads from each pool of copper or wildtype individuals were mapped to the axolotl genome assembly24 using HiSat225 v. 2.2.0 (http://daehwankimlab.github.io/hisat2). SNPs were identified using BCFtools26 v. 1.12-57-g0c2765b (https://samtools.github.io/bcftools) and SNP frequencies at polymorphic sites were compared between copper and wildtype pools using Popoolation227 v. 1.201 (https://sourceforge.net/projects/popoolation2). Additionally, DEseq228 v. 1.44.0 (https://www.bioconductor.org/packages/release/bioc/html/DESeq2.html) was used to compare Tyrp1 transcript abundances between cooper and wildtype pools. RNA sequence data will be published in the Short Read Archive at the National Center for Biotechnology Information.
Two guide RNAs (gRNAs) (CTGGCCACTGCGGAGAGCCT, TTTGTCCTCCAGTTCCATTC) were designed to target the 2nd and 6th exons, respectively, of axolotl Tyrp1 (TYRP1|AMEX60DD301043361.1). To generate targeted mutations, gRNAs were first duplexed with Alt-R tracrRNA, aliquoted and stored at -80⁰ C. All RNA products were synthesized by Integrated DNA Technologies. Guide RNAs and Alt-R tracrRNA were mixed with Cas9 to generate ribonucleoproteins that were injected into 100, 1-cell stage wildtype AGSC embryos as described previously 29. A total of 12 injected embryos were reared to approximately 3 cm and tail tissue was collected from each while under benzocaine anesthesia. During animal rearing, larvae were fed brine shrimp. The resulting tail tips were placed into separate 1.7 ml plastic tubes and placed on ice for DNA isolation. DNA isolations were performed using a New England Biolabs (NEB) Monarch genomic DNA isolation kit. DNA concentrations were determined using a Nanodrop (Thermo Scientific) and all samples were diluted to 30 ng / ul for PCR. PCR primers were designed to amplify DNA amplicons spanning gRNA target sites. The flanking PCR primers for gRNA1: Tyrp1_gRNA1_5. 1 TTGGTTTTATAGTTCCAGGTCCCG and Tyrp1_gRNA1_3.1 AGACAGAAGCCATTCATCGACTG. The flanking PCR primers for gRNA2: Tyrp1_gRNA2_5.1 AAGGTGGTTGAATCTTGTCTCCTT and Tyrp1_gRNA2_3.1 TTTTAAGACAGGTTACCCCCGAG. PCR amplicons were treated with NEB Exo-CIP and shipped to Eurofins for Sanger sequencing. The resulting sequences were compared using Geneious Prime software to evaluate CRISPR editing.
Homozygous copper and wildtype (axolotls not carrying copper alleles) larvae were sampled from two separate spawns. PCR was performed using Tyrp1_gRNA2_5.1 and Tyrp1_gRNA2_3.1 primers to amplify Tyrp1 exon 6 genomic sequence from 4 individuals (2 copper, 2, wildtype) and amplicons were sequenced as described above. The resulting sequences were aligned using Geneious Prime software and searched for polymorphisms.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was funded by the Office of Infrastructure Programs at the National Institutes of Health (R24OD010435, P40OD019794).
Author contributions
R.F.C. performed animal care procedures, collected tissues for RNA and DNA isolation, isolated RNA and DNA, designed PCR primers and gRNAs for CRISPR-CAS9, performed PCR, prepared samples for RNA and DNA sequencing, analyzed and summarized results from PCR and DNA sequencing, and contributed to writing of the manuscript. L.S. contributed to study design, supplied copper and wildtype larvae for RNA-Seq, provided information about axolotl stocks in the pet trade, assessed candidate genes for association to copper, and contributed to writing of the manuscript. M.T. and J.S. performed animal care procedures and took pictures of salamanders. N.T. developed and applied methods and pipelines for bioinformatic analyses and contributed to writing of the manuscript. J.J.S. contributed to study design, developed and applied methods and pipelines for bioinformatic analyses, summarized results from bioinformatic analyses, and contributed to writing of the manuscript. S.R.V. contributed to study design, performed animal care procedures, collected tissues for RNA and DNA isolation, performed embryo microinjections, photographed salamanders, analyzed, and summarized results from the study, and drafted the original manuscript.
Data availability
Raw DNA sequence reads and transcript abundance estimates may be found under GEO GSE269079.
Supplementary files
Competing interests
The authors declare no competing interests.
Supplementary file 1
Manhattan plots showing the degree of association of segregating genotypes with copper phenotype vs. wildtype in BSR-Seq pools. (A) Values shown are -log10(p-values) from Fisher’s exact tests. (B) Values are Z-scores from statistical tests of allele frequency difference.
Supplementary file 2
Nucleotide sequence for wildtype Tyrp1 showing where a nucleotide deletion (nonsense mutation at 1156 bp) occurs in the copper Tyrp1 allele.
Supplementary file 3
Predicted amino acid sequence for wildtype Tyrp1 showing where the copper amino acid sequence is altered by a nonsense mutation that results in a stop codon at amino acid residue 415.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Voss, S. R., Epperlein, H. H. & Tanaka, E. M. Ambystoma mexicanum, the axolotl: a versatile amphibian model for regeneration, development, and evolution studies. Cold Spring Harbor Protoc.8, artpdbemo128 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Voss, S. R., Woodcock, M. R. & Zambrano, L. A tale of two axolotls. Bioscience. 65, 1134–1140 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woodcock, M. R. et al. Identification of mutant genes and introgressed tiger salamander DNA in the laboratory axolotl. Ambystoma mexicanum Sci. Rep.7, 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith, H. M. Discovery of the axolotl and its early history in biological research. In: Developmental Biology of the Axolotl (eds (eds Armstrong, J. B. & Malacinski, G. M.) 3–12 (Oxford University Press (1989).
- 5.Humphrey, R. R. & Bagnara, J. T. A color variant in the Mexican axolotl. J. Hered. 58, 251–256 (1967). [Google Scholar]
- 6.Kabangu, M. et al. Leukocyte tyrosine kiltke (Ltk) is the mendelian determinant of the axolotl melanoid color variant. Genes (Basil). 14, 904 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Humphrey, R. R. Albino axolotls from an albino tiger salamander through hybridization. J. Hered. 58, 95–101 (1967). [DOI] [PubMed] [Google Scholar]
- 8.Lyerla, T. A. & Dalton, H. C. Genetic and developmental characteristics of a new color variant, axanthic, in the Mexican axolotl, Ambystoma mexicanum Shaw. Dev. Biol.24, 1–18 (1971). [DOI] [PubMed] [Google Scholar]
- 9.Liu, S., Yeh, C-T., Tang, H. M., Nettleton, D. & Schnable, P. S. Gene mapping via bulked segregant RNA-Seq (BSR-Seq). PLoS ONE. 7, e36406 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doudna, J. A. & Charpentier, E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 346, 1258096 (2014). [DOI] [PubMed] [Google Scholar]
- 11.J Jackson, I. A cDNA encoding tyrosinase-related protein maps to the brown locus in mouse. Proc. Nat. Acad. Sci. U S A. 85, 4392–4396 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmidt-Küntzel, A., Eizirik, E., O’Brien, S. J. & Menotti-Raymond, M. Tyrosinase and tyrosinase related protein 1 alleles specify domestic cat coat color phenotypes of the albino and brown loci. J. Hered. 96, 289–301 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Peterson, S. M. et al. Genetic variants in melanogenesis proteins TYRP1 and TYR are associated with the golden rhesus macaque phenotype. G3 13, jkad168 (2023). [DOI] [PMC free article] [PubMed]
- 14.Boissy, R. E. et al. Mutation in and lack of expression of tyrosinase-related protein-1 (TRP-1) in melanocytes from an individual with brown oculocutaneous albinism: a new subtype of albinism classified as OCA3. Am. J. Hum. Genet.58, 1145–1156 (1996). [PMC free article] [PubMed] [Google Scholar]
- 15.Bordzilovskaya, N. P., Dettlaff, T. A., Duhon, S. T. & Malacinski, G. M. Developmental stage series of axolotl embryos. In: Developmental Biology of the Axolotl (eds (eds Armstrong, J. B. & Malacinski, G. M.) 201–209 (Oxford University Press (1989).
- 16.Kenny, E. E. et al. Melanesian blond hair is caused by an amino acid change in TYRP1. Science. 336, 554 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lai, X., Wichers, H. J., Soler-Lopez, M. & Dijkstra, B. W. Structure of human tyrosinase related protein 1 reveals a binuclear zinc active site important for Melanogenesis. Angew Chem. Int. Ed. Engl.56, 9812–9815 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bennett, D. C. & Lamoreux, M. L. The color loci of mice–a genetic century. Pigment Cell. Res.16, 333–344 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Andersson, L. Domestic animals as models for biomedical research. Upsala J. Med. Sci.121, 1–11 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kantar, M. B., Nashoba, A. R., Anderson, J. E., Blackman, B. K. & Rieseberg, L. H. The genetics and genomics of plant domestication. BioScience. 67, 971–982 (2017). [Google Scholar]
- 21.Kamaraj, B. & Purohit, R. Mutational analysis of oculocutaneous albinism: a compact review. Biomed. Res. Int. 905472 (2014). (2014). [DOI] [PMC free article] [PubMed]
- 22.Percie du Sert. The ARRIVE guidelines 2.0: updated guidelines for reporting animal research. PLoS Biol.18, e3000410 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.National Research Council. Guide for the Care and Use of Laboratory Animals: Eighth Edition (National Academies, 2011). 10.17226/12910
- 24.Schloissnig, S. et al. The giant axolotl genome uncovers the evolution, scaling and transcriptional control of complex gene loci. Proc. Nat. Acad. Sci. U.S.A. 118, e2017176118 (2021). [DOI] [PMC free article] [PubMed]
- 25.Kim, D. et al. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol.37, 907–915 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 27, 2987–2993 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kofler, R., Pandey, R. V. & Schlötterer, C. PoPoolation2: identifying differentiation between populations using sequencing of pooled DNA samples (Pool-Seq). Bioinformatics. 27, 3435–3436 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Love, M. I., Huber, W. & Anders, S. Moderated estimation of Fold change and dispersion for RNA-seq data with DESeq2. Genome Biol.15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trofka, A. et al. Genetic basis for an evolutionary shift from ancestral preaxial to postaxial limb polarity in non-urodele vertebrates. Curr. Biol.31, 4923–4934 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw DNA sequence reads and transcript abundance estimates may be found under GEO GSE269079.