Abstract
Many RNA viruses have genetically diverse populations known as quasispecies. Important biological characteristics may be related to the levels of diversity in the quasispecies (quasispecies cloud size), including adaptability and host range. Previous work using Tobacco mosaic virus and Cucumber mosaic virus indicated that evolutionarily related viruses have very different levels of diversity in a common host. The quasispecies cloud size for these viruses remained constant throughout serial passages. Inoculation of these viruses on a number of hosts demonstrated that quasispecies cloud size is not constant for these viruses but appears to be dependent on the host. The quasispecies cloud size remained constant as long as the viruses were maintained on a given host. Shifting the virus between hosts resulted in a change in cloud size to levels associated with the new host. Quasispecies cloud size for these viruses is related to host-virus interactions, and understanding these interactions may facilitate the prediction and prevention of emerging viral diseases.
Genetic diversity is the essential component in a population that allows a species to evolve in an ever-changing environment with shifting selection pressures. Viruses have extreme evolutionary capacities that have allowed them to adapt to parasitize all known groups of organisms and, in many cases, to rapidly adapt to numerous host species within a kingdom. Considerable effort has been made to demonstrate and understand the population structure and evolutionary capacities of viruses such as Human immunodeficiency virus, Vesicular stomatitis virus, and Hepatitis C virus (1, 2, 15, 18, 25). These viruses, with RNA genomes or pregenomes, are associated with error-prone replication and short generation times that result in large, highly diverse replicating populations (7). When such a replicating population reaches equilibrium in its levels of diversity, it is termed a quasispecies. The level of genetic diversity in a viral population, termed the quasispecies cloud size, is an intrinsic property of the quasispecies.
The theoretical advantage of maintaining a diverse quasispecies is that, when the virus is shifted to a new environmental niche or selective regimen, a variant may already be present in the population which will be more fit in the new environment. However, excessive diversity can create problems if the virus is subjected to repeated bottlenecks. Since most mutations are deleterious, frequent bottlenecks can result in the rapid loss of fitness known as Muller's ratchet (3, 12, 13, 28). In order to survive, a virus must be diverse enough to adapt rapidly to changing environments without losing fitness during passage from host to host. Understanding the forces that control viral quasispecies may lead to novel ways of predicting the emergence of new viral pathogens and resistance-breaking variants, as well as lend insight into the population structures of other organisms.
Plant viruses are a convenient model system for studying viral evolution, because they allow for the use of infectious in vitro-generated viral RNAs as an inoculum for host organisms and permit controlled studies in multiple, genetically similar, intact hosts. The Alpha-like group of RNA viruses includes a number of plant and animal viruses with similarities in their genome organizations and nonstructural proteins (6). In a previous study, three Alpha-like plant viruses, Tobacco mosaic virus (TMV), Cucumber mosaic virus (CMV), and Cowpea chlorotic mottle virus (CCMV), were chosen for a direct comparison of quasispecies cloud size in a common host, Nicotiana benthamiana. Similar genome organizations and conserved elements in their replication proteins indicate that TMV, CMV, and CCMV evolved from a common ancestral virus. Despite their similarities, these three viruses differ greatly in host range size: CMV infects about a thousand plant species, TMV infects 80 to 100 plant species, and CCMV infects only a few plant species. In N. benthamiana, CMV, TMV, and CCMV had differences in the levels of population variation that correlated with their relative host range sizes (21). The comparison of the three viruses was the first controlled experimental evidence of a correlation between quasispecies cloud size and host range, an important biological implication of quasispecies theory. Additionally, serial passages resulted in no significant change in diversity levels through 10 consecutive passages, consistent with the portion of quasispecies theory which predicts that, when the replicating population reaches equilibrium, all individuals in the population will have a similar average genetic distance from the consensus sequence (4).
While the previous experiments were useful in understanding the population structures of plant viruses, they were limited to a single host for direct comparison. It was still unknown whether the quasispecies cloud size was constant for these viruses in all environments or whether it varied. It also remained to be seen if the phenomenon of constant cloud size throughout passages was universal for all hosts or was an artifact associated with N. benthamiana. Using various plant hosts as changing environments, we demonstrate here that the virus quasispecies reaches an equilibrium of diversity that is maintained through several passages and that the level of diversity in the quasispecies cloud is a property of both the virus and the host. Identifying the factors affecting the diversity levels of viral populations is the first step toward controlling quasispecies cloud size, which could contribute significantly to the understanding and eventual control of the viral disease evolution.
MATERIALS AND METHODS
Plants and viruses.
Infections with CMV and TMV were initiated either with infectious in vitro transcripts from cDNA clones (CMV) (14) or with infectious transcripts generated in vivo from 35S promoter-driven constructs (22) as previously described (21). Plant hosts included zucchini squash (Cucurbita pepo cv. Elite), tomato (Lycopersicon esculentum cv. Rutgers), pepper (Capsicum annuum cv. Morengo), tobacco (Nicotiana tabacum cv. Xanthi nc Cornell or Xanthi nn), and N. benthamiana. Protoplasts were generated from BY2 tobacco suspension cell cultures (24) and inoculated with in vitro transcripts or in vivo transcript-generating clones (10). Passage experiments were done by sap inoculation as previously described (21). Briefly, two plants were inoculated with transcript (passage 0), and sap samples from both plants were pooled to inoculate two more plants (passage 1), continuing through passage 10.
Cloning and sequencing of viral populations.
The viral RNAs from individual plants from each treatment were cloned separately and treated as unique populations. Total RNA was isolated from plants 2 weeks postinoculation and from protoplasts 24 h after inoculation as previously described (11). The total RNAs from whole plants and protoplasts were used as templates for high-fidelity reverse transcription-PCR (RT-PCR) as previously described (21). RT reactions were primed with primers 4144 (AACCTACTGCAGTGAGTCCGAGGATTA) for CMV and 4145 (TAATCGGAATTCAGGAAACAGCTATGACCCGCGCGATCCAGACAC) for TMV. The PCR primers used (4139 [AACCTACTGCAGTGAGTCCGAGGATTA] and 4144 for CMV and 4143 [AACCTACTGCAGCGGGTTTCTGTCCGC] and 4145 for TMV) amplified a region of the viral genomes including the 3′ ends of the movement protein genes, the CMV intercistronic region, the coat protein genes, and a portion of the 3′ nontranslated regions (Fig. 1). Thermal cycling reactions were carried out for 15 cycles (94°C denaturation for 30 s, 50°C annealing for 1 min, and 72°C extension for 1 min) and included a polymerase with proofreading capability (Pfu; Stratagene). The PCR product was cloned, and the sequence was determined by automated sequencing using an ABI 377 sequencer according to the manufacturer's protocols. Sequencing was performed with flanking T7 and T3 plasmid primers and one internal primer for TMV (positioned from bases 5672 to 5691) or two internal primers for CMV (positioned from bases 1270 to 1290 and bases 1570 to 1590). From 11 to 15 clones were analyzed for each population (a minimum of 11,000 bases/population). The consensus sequence for each population was determined, the sequence of each clone was compared to the consensus sequence, and changes were recorded. The mutation frequency (total number of changes/total number of bases sequenced) and the percentage of mutated clones were used as indicators of genetic diversity.
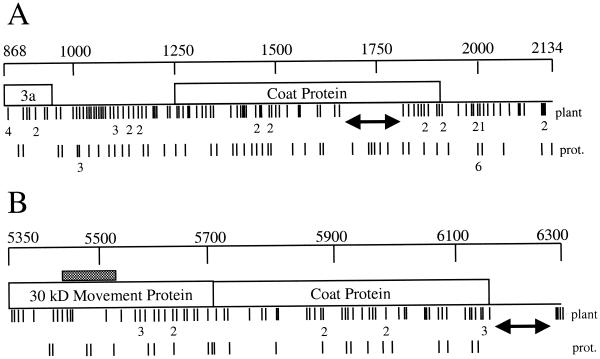
FIG. 1.
Distribution of mutations in cloned regions of CMV and TMV. Maps of the cloned regions of CMV (A) and TMV (B) are shown, with nucleotide positions marked above. The locations of mutations from cloned populations are shown as lines directly below the maps. The mutations from whole-plant populations are directly underneath the maps, with mutations from protoplast (prot.) populations below them. Mutation-free regions from whole-plant populations are marked with a double-headed arrow. In instances where multiple mutations have occurred at the same site, the site is marked with a single line with the number of mutations indicated directly below. The proposed origin-of-assembly region in TMV is denoted by the shaded bar over the movement protein gene. None of the changes in the origin-of-assembly region dramatically affected the proposed structure.
Statistical analysis.
Comparisons between viral populations in different hosts were tested for statistical significance using the ANOVA (analysis of variance) test from the statistical package MStat (Michigan State University, Lansing) to determine least significant differences. A standard runs test was used to assess the randomness of mutation distribution. The significance of mutation-free zones was tested by generating 50 computer-simulated mutation distributions for both TMV and CMV with a number of mutations equal to the combined total of all observed populations. A map was generated for each of the 50 simulated populations, and the largest mutation-free zone of each computer simulation was recorded. The mutation-free zones from the 50 simulated distributions are assumed to represent what could be expected if the mutations occurred completely by chance. The mean values and standard deviations were determined for the largest mutation-free zones from the 50 simulated populations, and these values were compared to the values for the observed mutation-free zones from whole-plant populations of TMV and CMV. The mean values for simulated mutation-free zones were subtracted from the observed mutation-free zone size, and this value was divided by the standard deviation to determine the number of standard deviations by which the observed value varied from the randomly generated mean. Because the randomly generated values form a normal distribution, the number of standard deviations can be used to estimate the likelihood of the observed mutation-free zone occurring by chance.
RESULTS
To investigate the effects of changes in the environment on quasispecies cloud size, TMV and CMV were inoculated onto several additional hosts using a genetically identical source (Table 1). Young plants were inoculated mechanically, and 14 days postinoculation, total RNA was extracted from noninoculated, systemically infected leaves. This RNA was used as a template for RT, and the resulting cDNA was PCR amplified with virus-specific primers. The amplified products (approximately 1 kb in length including the coat protein genes and some flanking sequences) were cloned, and 11 to 16 clones were analyzed for each virus population.
TABLE 1.
Quasispecies variation in CMV and TMV populations in different host species
| Virus and host | % Mutated clones | Total mutations/bases sequenced | Mutation frequency (103)a |
|---|---|---|---|
| CMV | |||
| N. benthamiana | 43 (6/14) | 8/16,454 | 0.5A |
| Tobacco | 70 (7/10) | 12/11,197 | 1.0B |
| Squash | 30 (3/10) | 8/11,193 | 0.7A |
| Tomato | 64 (7/11) | 9/13,786 | 0.7A |
| Pepper | 100 (11/11) | 23/13,820 | 1.8C |
| Tobacco protoplast | 100 (11/11) | 34/13,358 | 2.5D |
| TMV | |||
| N. benthamiana | 29 (4/14) | 7/13,227 | 0.5F |
| Tobacco | 46 (6/13) | 11/11,850 | 0.9G |
| Tomato | 8 (1/12) | 2/11,353 | 0.2E |
| Pepper | 77 (10/13) | 18/12,471 | 1.5B |
| Tobacco protoplast | 100 (12/12) | 23/11,358 | 2.0I |
Substitutions, insertions, and deletions are all counted equally in determining mutation frequency. Statistically significant differences in diversity levels are denoted by capital letters next to mutation frequencies. Mutation frequencies with the same letter are not statistically different. Least significant differences were determined using the ANOVA test (P < 0.05). The levels are specific to each virus, i.e., CMV populations are compared to only other CMV populations.
Each clone represented a unique viral RNA, and comparing the sequences of the viral clones to the consensus sequence (in most cases, identical to the sequence of the progenitor cDNA clone) provided a snapshot of the genetic diversity generated within a given viral population. Using these clones as a representative sample of viral populations, mutation frequency and the percentage of mutated viral clones were used as indicators of population diversity. Mutation frequency is the number of bases that differ from the consensus sequence divided by the total number of bases sequenced. It is not a measure of mutation rate (i.e., misincorporation by the polymerase) but an indication of diversity, as described by convention (2). At least two plants were sampled for each virus-host combination, and the viral populations were cloned separately. Cloning and sequencing two RT-PCR sets for each plant treatment would detect potential differences in levels of error introduced in independent RT-PCRs. There were no significant differences between the sampled plants of any given host-virus combination, even in subsequent passage experiments where multiple plants from a single host species were used, and so the data from clones for each virus-host combination were pooled. Comparisons between treatments were done using ANOVA tests to determine least significant differences. In all cases, care was taken to minimize the level of error introduced by the cloning process. The number of PCR cycles was limited (15 cycles), and high-fidelity polymerase was used as previously described. Control experiments using in vitro transcripts as template RNA indicated that the level of variability introduced by the experimental procedure (4.5 × 10−5) was significantly lower than levels observed in the viral populations (21).
No bias for synonymous mutations was observed in any of the populations (Tables 2 and 3), consistent with the observation that selection in these experiments is acting on the RNA level (21). Thus, synonymous and nonsynonymous mutations, as well as substitutions, insertions, and deletions, were treated alike in terms of counting mutations and determining diversity levels. Occasionally, multiple bases were mutated close to one another. In these cases, each individual mutated base was counted as a mutation, even though the mutations may have arisen from a single mutational event. However, if these mutations are grouped and counted as a single mutation, it does not affect the statistical comparisons significantly (data not shown).
TABLE 2.
Types of mutations observed in CMV and TMV populations in whole plants
| Virus | No. of type of mutationa
|
|||||
|---|---|---|---|---|---|---|
| Translated
|
Nontrans. | Subs. | Ins. | Del. | ||
| Syn. | Nonsyn. | |||||
| CMV | 16 | 38 | 85 | 114 | 12 | 13 |
| TMV | 23 | 51 | 15 | 65 | 8 | 16 |
Abbreviations: Syn., synonymous; Nonsyn., nonsynonymous; Nontrans., nontranslated; Subs., substitution; Ins., insertion; Del., deletion.
TABLE 3.
Substitution types
| Virus | Original base | No. of substitutions at mutated base
|
|||
|---|---|---|---|---|---|
| G | A | U | C | ||
| CMV | G | 41 | 5 | 1 | |
| A | 11 | 4 | 2 | ||
| U | 7 | 8 | 17 | ||
| C | 2 | 0 | 16 | ||
| TMV | G | 14 | 2 | 2 | |
| A | 12 | 3 | 1 | ||
| U | 2 | 3 | 12 | ||
| C | 0 | 3 | 11 | ||
The initial experiments demonstrated that the quasispecies cloud size for CMV and TMV varied with changing environments (i.e., changing hosts). Both CMV and TMV showed marked differences in variability depending on the host (Table 1). Even on hosts in the same genus (tobacco and N. benthamiana), TMV and CMV generated viral populations with significantly different cloud sizes. The range of cloud sizes was quite large: for example, TMV populations had very little detectable variation in tomato, while populations in pepper had high percentages of mutated clones and high mutation frequencies. CMV also had host-dependent changes in quasispecies variation, particularly in the percentage of mutated clones. Clearly, the size of the quasispecies cloud for a particular virus was not constant; rather, it depended on the host and/or the environment, and certain virus-host combinations or environmental effects appeared to contribute to increases in viral variation capacities.
CMV populations from pepper (high diversity), TMV populations from pepper (high diversity), and TMV populations from tomato (low diversity) were passaged 10 times by plant sap inoculation to test the stability of quasispecies cloud size in these hosts. In all cases, there was little adjustment in the level of population diversity over the course of serial passage (Table 4). After 10 passages, TMV populations from pepper were transferred to tomato, TMV populations from tomato were transferred to pepper, and CMV populations from pepper were transferred to N. benthamiana. By 14 days after transfer to the new host, the TMV and CMV populations developed variation levels that were equivalent to those of populations that had been passaged on the new host 10 times (Table 4). There were no changes to the consensus sequence of the sequenced regions associated with shifting between high- and low-diversity hosts, and so the changes in quasispecies cloud size were not directly associated with adaptation of the coat protein or flanking sequences to new hosts. Selection appears to act on the diversity levels of the viral populations. This demonstrates that virus populations rapidly reach and maintain diversity levels that are specific to individual host species, and viral RNA populations are selected for levels of diversity in addition to overall fitness.
TABLE 4.
Quasispecies variation in CMV and TMV populations during serial passages on tomato and pepper
| Virus and host | Passage | % Mutated clones | Total mutations/bases sequenced | Mutation frequency (103)a |
|---|---|---|---|---|
| CMV | ||||
| Pepper | 0 | 100 (11/11) | 23/13,820 | 1.8B |
| 1 | 100 (12/12) | 23/15,160 | 1.5B | |
| 10 | 93 (13/14) | 25/17,606 | 1.4B | |
| N. benthamiana (after passage 10) | 36 (4/11) | 5/13,770 | 0.4A | |
| TMV | ||||
| Pepper | 0 | 77 (10/13) | 18/12,471 | 1.5D |
| 1 | 64 (9/14) | 12/13,466 | 0.9D | |
| 10 | 60 (9/15) | 15/14,121 | 1.1D | |
| Tomato (after passage 10) | 22 (2/13) | 2/12,265 | 0.2C | |
| Tomato | 0 | 8 (1/12) | 2/11,353 | 0.2C |
| 1 | 23 (3/13) | 3/12,511 | 0.2C | |
| 10 | 0 (0/14) | 0/13,151 | 0.0C | |
| Pepper (after passage 10) | 69 (9/13) | 12/11,875 | 1.0D |
Substitutions, insertions, and deletions are all counted equally in determining mutation frequency. Statistically significant differences in diversity levels are denoted by capital letters next to mutation frequencies. Mutation frequencies with the same letter are not statistically different. Least significant differences were determined using the ANOVA test (P < 0.05).
To examine the rate of quasispecies cloud expansion, TMV and CMV populations were cloned following infection and 24 h of incubation in tobacco protoplasts, where selection for cell-to-cell and long-distance movement is removed. In protoplast infections, the virus is limited to a single inoculated protoplast, and the selection pressures on viral populations are primarily for replication and RNA stability. In just 24 h, both TMV and CMV generated very diverse populations, with larger cloud sizes than those that were observed after 2 weeks in intact tobacco plants (Table 1). Clearly, these viruses are capable of generating tremendous amounts of diversity very rapidly when selective pressures or other factors are removed.
Mutations became fixed in viral populations in only two cases; most populations maintained the original consensus sequence. One fixed mutation was found in the tobacco populations of CMV (Table 1), a change from C to U at position 1486 in the coat protein gene. Another fixed mutation was observed in the passaged TMV tomato populations (Table 4), a G-to-A transition at position 5514 in the movement protein gene. The fixed mutation in the passage 10 TMV tomato populations resulted in a new consensus sequence. As such, no mutations from the consensus sequence are recorded for the passage 10 TMV populations in tomato, nor is the fixed mutation from CMV tobacco populations represented in the final mutation frequency. It is impossible to say if the fixed mutations were truly adaptive or if they became fixed in the population by chance, although both fixed mutations were nonsynonymous. In general, the lack of fixed mutations emphasizes the rare nature of adaptive mutations.
As noted previously (21), there is a bias for transition mutations in both TMV and CMV populations (Tables 2 and 3). CMV in particular demonstrated a strong bias for G-to-A substitutions. This is consistent with transition biases noted in other viral systems (9, 23). There are very few C-to-A and A-to-C transversions in CMV populations, a trend that is also consistent with phylogenetic analyses of CMV strains (17). A runs test indicated that neither the CMV nor the TMV mutations were clustered. However, both TMV and CMV demonstrated an uneven distribution of mutations in the sequenced regions (Fig. 1). In CMV, there were mutational hot spots, such as the area around base 2000 in the 3′ nontranslated region where numerous mutations were recovered from all CMV populations, both in whole plants and in protoplasts. This is probably related to the sequence in this region, where there is a stretch of six uracil residues. The most common mutations in this region were deletions or additions in the poly(U) stretch or G-to-A substitutions of the base immediately preceding the poly(U) stretch (data not shown). There were no such mutational hot spots observed in the TMV populations.
In contrast to the mutational hot spot in CMV, there were zones in both TMV and CMV cloned regions where mutations were not recovered. In CMV, there was a 149-base region in the coat protein gene where no mutations were observed in any of the whole-plant populations. Interestingly, a number of mutations did occur in this region in tobacco protoplast populations (Fig. 1). A similar 110-base mutation-free zone was observed in the 3′ nontranslated region of TMV. This region also remained mutation free in protoplast populations (Fig. 1). To statistically test the significance of such mutation-free regions, 50 simulated mutation distributions were constructed by randomly generating an equal number of mutation locations. The simulated mutations were mapped and compared to the actual mutation distribution observed for TMV and CMV in whole plants. None of the 50 randomly generated mutation distributions had a mutation-free zone equal in size to the ones observed for TMV and CMV (data not shown). The average of the largest mutation-free zones from the randomly generated distributions was 75.5 bases for CMV (standard deviation = 21.5) and 70.4 bases for TMV (standard deviation = 16.6). The observed mutation-free zones for both TMV and CMV were more than 2 standard deviations away from the mean mutation-free zones for the randomly generated distributions, indicating that the probability of such a mutation-free zone occurring by chance in viral populations from whole plants is less than 5%.
DISCUSSION
It is difficult to imagine a mechanism that would select a specific level of diversity for each host-virus relationship. Two possible scenarios could explain this observation. First, in certain hosts the viral replicase may make fewer errors and thus generate less diversity. The host contributes components to the replicase complex in addition to controlling concentrations of available nucleotides, pH, and other soluble components such as divalent cations, any of which could affect fidelity. Alternatively, these viruses may be capable of generating equivalent levels of diversity in different hosts, but some selection pressure specific to a particular host acts as a cap, limiting the accumulation of diversity. The levels of diversity observed in protoplasts after just 24 h were higher than those observed in any intact plant species (including whole tobacco plants) where the population was sampled 2 weeks postinoculation (Table 1). This suggests that the quasispecies is quite capable of generating diversity rapidly, and other factors limit the eventual accumulation of variation in whole plants.
The observed mutation frequency in whole plants is surprisingly low. At the highest level in tobacco protoplasts, the CMV mutation frequency is 2.5 × 10−3 in just 24 h. Very little information is available about the generation time for plant viruses, although TMV begins to produce progeny virus in 35 to 40 min (26). To account for the mutation frequency observed in tobacco protoplasts with an error rate of 10−4, there must have been approximately 20 to 25 generations in the protoplasts, or a generation time of about 1 h. This estimate of generations is probably low, because it assumes that the mutation frequency is completely due to random mutations and that there is no negative selection on deleterious mutations. In protoplasts, there may be little selection for the normal functions of the movement protein or coat protein; however, there is undoubtedly selection against mutations that would render the RNA unstable or unable to replicate. In whole plants, the cytoplasm is a contiguous system between cells connected by plasmodesmata, making estimations of generation numbers difficult. Regardless, assuming that the CMV replicase error rate is similar to other RNA-dependent RNA polymerase error rates, we can crudely estimate at least one generation per hour in single infected cells.
In infected plants, generation time may fluctuate with the metabolic state of the plant. In addition, viral replication may not be continuous in the plant. However, one generation per hour in the plant is probably not an unreasonable estimate. Hence, in 14 days, there should be about 336 generations, and a theoretical mutation frequency of 3.4 × 10−2. Clearly, the observed mutation frequencies are much lower than this. Two factors potentially are decreasing the mutation frequency: selection (predominantly negative) and bottlenecks. It is clear that selection plays an important role, because although mutations are not biased for synonymous changes (Table 1), they are not evenly distributed (Fig. 1). However, even in the most densely mutated regions we do not approach the theoretical mutation frequency, and so we must assume that bottlenecks associated with long-distance or cell-to-cell movement also are limiting the diversity.
It is important to note that the cloned viral populations are being selected for at the RNA level. There are more than enough nonmutated copies of genes to produce all the necessary coat proteins in trans in even the most highly mutated populations. The coat protein must fulfill its primary function of encapsidation in trans, because multiple proteins encapsidate a single viral RNA. Even the most mutated populations are capable of efficient systemic infection, and these diverse populations do not lose the ability to be passaged successfully, further indicating that there are sufficient levels of wild-type virus proteins to complete the infection cycle. Thus, the presence of regions where mutations are not recovered clearly indicates the presence of selection for RNA sequence. Both TMV and CMV have such regions, and computer simulations with randomized distributions of mutations indicated that these regions are not occurring by chance. Undoubtedly, mutations do occur in these regions, but viral RNAs with such mutations are rapidly selected out of the quasispecies cloud.
The mutation-free region in the 3′ nontranslated region of TMV is easily explained, since this region acts as the promoter for minus-strand synthesis. Mutations in this region would potentially debilitate the ability of a viral RNA to be replicated. It is not surprising that mutations did not occur in these areas in protoplast populations as well. Fewer mutations were also recovered in another highly selectable region of TMV, the origin of assembly (bases 5436 to 5532), which controls the encapsidation of individual RNAs (20). Although the gaps between mutations in this region were not larger than expected by chance, there are clearly fewer mutations in this region than in flanking sequences on either side (Fig. 1). In addition, two of the five mutations in this region did not affect the proposed stem-loop required for efficient encapsidation (20), and none of the mutations affected the end of the stem-loop. This pattern of mutation-free regions also occurred in protoplast populations, where all mutations in the origin of assembly did not affect secondary structure, and no mutations occurred in the 3′ nontranslated region.
In contrast, CMV populations demonstrate patterns of mutations in whole plants different from those in protoplasts. The different distribution of mutations seems to indicate that there are selection pressures working on the RNA level that are present in whole plants but absent in protoplasts. The most obvious selection pressures on viral RNAs are replication capacity and stability (encapsidation would prevent viral RNA degradation by host nucleases), but these selection pressures are present in both environments. Clearly, viruses are required to move both cell to cell and systemically in whole plants, a portion of the life cycle that is absent in protoplasts. Perhaps, the mutation-free region in the coat protein gene of whole-plant CMV populations is related to movement of viral RNAs. Examining the sequence of the mutation-free region in the CMV coat protein gene does not reveal any obvious selectable traits. The possibility that this region is a cis-acting replication signal not necessary in protoplasts but required in whole plants is unlikely but cannot be ruled out completely. Unusual distributions of mutations resulting from passage bottlenecks have been observed with human immunodeficiency virus (27). Possibly, bottlenecks associated with viral movement in plants are reflected in the differences in mutation distributions for CMV populations between whole plants and protoplasts.
There are two particularly significant findings in this work. First, the quasispecies cloud sizes for these viruses are not constant; rather, they vary depending on the host environment. These differences may be due to differences in the ability of TMV and CMV to generate diversity through replicase error and recombination (16), but these experiments suggest that the differences are due to selection or bottlenecks imposed by the host-virus infection cycle. Second, this work clearly demonstrates that the viral quasispecies is selected for a particular level of variation specific to the virus-host combination. Although this possibility had been suggested in early descriptions of quasispecies theory, this is the first experimental evidence to indicate that such selection occurs. This suggests the possibility that different hosts may accelerate or decelerate the rate of viral evolution by permitting or denying high levels of diversity in viral populations. Diversity in viral quasispecies has been described previously as a mechanism to avoid host resistance responses (5, 8) or a reservoir to maintain variants with selective advantages in other environments (19) and has been correlated with the ability to infect numerous hosts (21). If the factors that contribute to permitting viral diversity can be identified and manipulated, a whole new set of tools will become available to prevent the development of new diseases. Perhaps, the very resistance mechanisms that we use to combat viruses are in fact generating high-diversity quasispecies that act as a source of new pathogenic variants. A greater understanding of the forces that drive and control the levels of virus genetic variation may help us one day predict and possibly prevent the scenarios that lead to the evolution of emerging viral diseases.
ACKNOWLEDGMENTS
This work was supported by the Samuel Roberts Noble Foundation.
We acknowledge Yiming Bao for assistance with tobacco protoplasts and Richard Dixon, Wayne Versaw, and Ulrich Melcher for critical review of the manuscript.
REFERENCES
- 1.Domingo E, Escarmís C, Sevilla N, Moya A, Elena S F, Quer J, Novella I, Holland J J. Basic concepts in RNA virus evolution. FASEB J. 1996;10:859–864. doi: 10.1096/fasebj.10.8.8666162. [DOI] [PubMed] [Google Scholar]
- 2.Domingo E, Holland J J. RNA virus mutations and fitness for survival. Annu Rev Microbiol. 1997;51:151–178. doi: 10.1146/annurev.micro.51.1.151. [DOI] [PubMed] [Google Scholar]
- 3.Duarte E, Clarke D, Moya A, Domingo E, Holland J. Rapid fitness losses in mammalian RNA virus clones due to Muller's ratchet. Proc Natl Acad Sci USA. 1992;89:6015–6019. doi: 10.1073/pnas.89.13.6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eigen M. The physics of molecular evolution. Chem Scr. 1986;26B:13–16. [Google Scholar]
- 5.Feuer R, Boone J D, Netski D, Morzunov S P, St. Jeor S C. Temporal and spatial analysis of Sin Nombre virus quasispecies in naturally infected rodents. J Virol. 1999;73:9544–9554. doi: 10.1128/jvi.73.11.9544-9554.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goldbach R, deHaan P. RNA viral supergroups and the evolution of RNA viruses. In: Morse S S, editor. The evolutionary biology of viruses. New York, N.Y: Raven Press; 1994. pp. 105–120. [Google Scholar]
- 7.Holland J, Spindler K, Horodyski F, Grabau E, Nichol S, VandePol S. Rapid evolution of RNA genomes. Science. 1982;215:1577–1585. doi: 10.1126/science.7041255. [DOI] [PubMed] [Google Scholar]
- 8.Lech W J, Wang G, Yang Y L, Chee Y, Dorman K, McCrae D, Lazzeroni L C, Erickson J W, Sinsheimer J S, Kaplan A H. In vivo sequence diversity of the protease of human immunodeficiency virus type 1: presence of protease inhibitor-resistant variants in untreated subjects. J Virol. 1996;70:2038–2043. doi: 10.1128/jvi.70.3.2038-2043.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mansky L M, Temin H M. Lower in vivo mutation rate of human immunodeficiency virus type 1 than that predicted from the fidelity of purified reverse transcriptase. J Virol. 1995;69:5087–5094. doi: 10.1128/jvi.69.8.5087-5094.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maule A J, Boulton M I, Wood K R. An improved method for the infection of cucumber leaf protoplasts with cucumber mosaic virus. Phytopathol Z. 1980;97:118–126. [Google Scholar]
- 11.Mirkov T E, Kurath G, Mathews D M, Elliott K, Dodds J A, Fitzmaurice L. Factors affecting efficient infection of tobacco with in vitro RNA transcripts from cloned cDNAs of satellite tobacco mosaic virus. Virology. 1990;179:395–402. doi: 10.1016/0042-6822(90)90307-d. [DOI] [PubMed] [Google Scholar]
- 12.Novella I S, Elena S F, Moya A, Domingo E, Holland J J. Size of genetic bottlenecks leading to virus fitness loss is determined by mean initial population fitness. J Virol. 1995;69:2869–2872. doi: 10.1128/jvi.69.5.2869-2872.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Novella I S, Quer J, Domingo E, Holland J J. Exponential fitness gains of RNA virus populations are limited by bottleneck effects. J Virol. 1999;73:1668–1671. doi: 10.1128/jvi.73.2.1668-1671.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rizzo T M, Palukaitis P. Construction of full-length cDNA clones of cucumber mosaic virus RNAs 1, 2 and 3: generation of infectious RNA transcripts. Mol Gen Genet. 1990;222:249–256. doi: 10.1007/BF00633825. [DOI] [PubMed] [Google Scholar]
- 15.Rodrigo A G. HIV evolutionary genetics. Proc Natl Acad Sci USA. 1999;96:10559–10561. doi: 10.1073/pnas.96.19.10559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roossinck M J. Mechanisms of plant virus evolution. Annu Rev Phytopathol. 1997;35:191–209. doi: 10.1146/annurev.phyto.35.1.191. [DOI] [PubMed] [Google Scholar]
- 17.Roossinck M J, Zhang L, Hellwald K-H. Rearrangements in the 5′ nontranslated region and phylogenetic analyses of cucumber mosaic virus RNA 3 indicate radial evolution of three subgroups. J Virol. 1999;73:6752–6758. doi: 10.1128/jvi.73.8.6752-6758.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rouzine I M, Coffin J M. Interplay between experiment and theory in development of a working model for HIV1 population dynamics. In: Domingo E, Webster R, Holland J, editors. Origin and evolution of viruses. London, United Kingdom: Academic Press; 1999. pp. 225–262. [Google Scholar]
- 19.Ruiz-Jarabo C M, Arias A, Baranowski E, Escarmís C, Domingo E. Memory in viral quasispecies. J Virol. 2000;74:3543–3547. doi: 10.1128/jvi.74.8.3543-3547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saito T, Yamanaka K, Okada Y. Long-distance movement and viral assembly of tobacco mosaic virus mutants. Virology. 1990;176:329–336. doi: 10.1016/0042-6822(90)90002-9. [DOI] [PubMed] [Google Scholar]
- 21.Schneider W L, Roossinck M J. Evolutionarily related Sindbis-like plant viruses maintain different levels of population diversity in a common host. J Virol. 2000;74:3130–3134. doi: 10.1128/jvi.74.7.3130-3134.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shintaku M H, Carter S A, Bao Y, Nelson R S. Mapping nucleotides in the 126-kDa protein gene that controls the differential symptoms induced by two strains of tobacco mosaic virus. Virology. 1996;221:218–225. doi: 10.1006/viro.1996.0368. [DOI] [PubMed] [Google Scholar]
- 23.Vartanian J-P, Plikat U, Henry M, Mahieux R, Guillemot L, Meyerhans A, Wain-Hobson S. HIV genetic variation is directed and restricted by DNA precursor availability. J Mol Biol. 1997;270:139–151. doi: 10.1006/jmbi.1997.1104. [DOI] [PubMed] [Google Scholar]
- 24.Watanabe Y, Meshi T, Okada Y. Infection of tobacco protoplasts with in vitro transcribed tobacco mosaic virus RNA using an improved electroporation method. FEBS Lett. 1987;219:65–69. [Google Scholar]
- 25.Wei X, Ghosh S K, Taylor M E, Johnson V A, Emini E A, Deutsch P, Lifson J D, Bonhoeffer S, Nowak M A, Hahn B H, Saag M S, Shaw G M. Viral dynamics in human immunodeficiency virus type 1 infection. Nature. 1995;373:117–122. doi: 10.1038/373117a0. [DOI] [PubMed] [Google Scholar]
- 26.Wu X, Xu Z, Shaw J G. Uncoating of tobacco mosaic virus RNA in protoplasts. Virology. 1994;200:256–262. doi: 10.1006/viro.1994.1183. [DOI] [PubMed] [Google Scholar]
- 27.Yuste E, Lopez-Galindez C, Domingo E. Unusual distribution of mutations associated with serial bottleneck passages of human immunodeficiency virus type 1. J Virol. 2000;74:9546–9552. doi: 10.1128/jvi.74.20.9546-9552.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuste E, Sanchez-Palomino S, Casado C, Domingo E, Lopez-Galindez C. Drastic fitness loss in human immunodeficiency virus type 1 upon serial bottleneck events. J Virol. 1999;73:2745–2751. doi: 10.1128/jvi.73.4.2745-2751.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]