Abstract
Oral potentially malignant disorders (OPMDs) with genomic alterations have a heightened risk of evolving into oral squamous cell carcinoma (OSCC). Currently, genomic data are typically obtained through invasive tissue biopsy. However, brush biopsy is a non-invasive method that has been utilized for identifying dysplastic cells in OPMD but its effectiveness in reflecting the genomic landscape of OPMDs remains uncertain. This pilot study investigates the potential of brush biopsy samples in accurately reconstructing the genomic profile and tumor evolution in a patient with both OPMD and OSCC. We analyzed single nucleotide variants (SNVs), copy number aberrations (CNAs), and subclonal architectures in paired tissue and brush biopsy samples. The results showed that brush biopsy effectively captured 90% of SNVs and had similar CNA profiles as those seen in its paired tissue biopsies in all lesions. It was specific, as normal buccal mucosa did not share these genomic alterations. Interestingly, brush biopsy revealed shared SNVs and CNAs between the distinct OPMD and OSCC lesions from the same patient, indicating a common ancestral origin. Subclonal reconstruction confirmed this shared ancestry, followed by divergent evolution of the lesions. These findings highlight the potential of brush biopsies in accurately representing the genomic profile of OPL and OSCC, proving insight into reconstructing tumor evolution.
Subject terms: Cancer, Oral cancer, Oncology, Cancer, Surgical oncology, Evolvability, Cancer genetics, Cancer genomics, Genomics, Mutation, Sequencing
Introduction
Oral cancer is a significant public health concern worldwide with more than 90% of the oral cavity malignancies being oral squamous cell carcinomas (OSCC)1. Some of these lesions develop from oral potentially malignant disorders (OPMDs). OPMDs are a group of conditions that possess the risk of malignant transformation to OSCC2,3. Risk of transformation varies based on the type of lesion and pathologic severity, with an overall transformation risk across all OPMDs being 7.9%4. Once transformed, patients with OSCC have an overall 5-year survival rate of 64.3%.
It is known that genomic alterations increase the risk of transformation from OPMD to OSCC5,6. Copy number alterations (CNAs) in specific regions of interest such as 3p, 9p, 17p loss as well as loss of heterozygosity (LOH) in genes associated with OSCC such as TP53, CASP8, and CDKN2A are predictive of malignant transformation of OPMD7–11.
However, the tumor evolution and the clonal selection of these genomic events are not as well understood as in other cancers. For example, in colorectal cancer, the transition from adenoma to carcinoma is characterized by the early accumulation of specific genetic alterations such as mutation in APC, KRAS, and TP53 genes12. Similarly, in breast cancer, the progression from ductal carcinoma in situ (DCIS) to invasive ductal carcinoma (IDC) involves a series of genetic and epigenetic changes, including clonal alterations in TP53, BRCA1, and PIK3CA13.
To study genomic alterations and tumor evolution, the gold standard is longitudinal tissue sampling. Obtaining tissue to study genomic alterations sometimes coincides with treatment for OPMDs, which can involve surgical biopsy and surgical resection and/or laser ablation. However, longitudinal serial biopsies and biopsies for only research purposes result in morbidity and is rarely acceptable for patients. As such, a non-invasive technique of obtaining tissue would be ideal to study tumor evolution. Cytological study of oral cells is a non-invasive technique that is well accepted by the patient for the diagnosis of dysplastic cells in OPMDs and could be an attractive option to study tumor evolution in OPMD14,15. Indeed, there is evidence that brush biopsy is able to capture genomic alterations in target tissue. Poell et al. demonstrated that brush biopsy was comparable to tissue biopsy in detecting mutations with a sensitivity and specificity of 85% and 86%, respectively, as well as comparable in detecting CNA events with sensitivity and specificity of 81% and 80%, respectively, using target-enriched deep sequencing in formalin-fixed paraffin-embedded (FFPE) samples16. Other studies5,10,17,18 have looked at the genomic characterizations of OPMDs (mainly oral leukoplakia) in the context of CNA and a panel of mutations using FFPE5,10 samples, transcriptomic data17 , or deep sequencing using liquid biopsy18. Nevertheless, no studies to date have utilized brush biopsy to study clonal evolution through genome-wide sequencing.
In this proof-of-concept pilot study, we compare whether brush biopsies of OPMD and OSCC samples can recapitulate the genome-wide alterations as well as clonal evolution found in their matched fresh tissue using whole exome sequencing. In this study, we found that brush biopsy was capable of faithfully recapitulating the mutations and CNAs of target tissue, thereby having the ability to reconstruct tumor evolution. By studying tumor evolution through brush biopsies, we provide evidence of unique tumor evolution, by demonstrating that a spatially distinct OPMD and OSCC from the same patient shared a common ancestor clone and subsequent divergent evolution.
Results and discussion
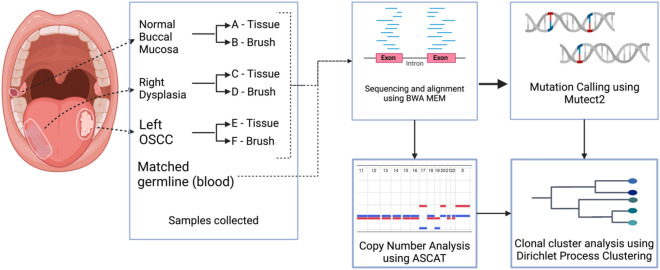
This pilot study enrolled a patient who had a clinical diagnosis of OSCC on the left lateral tongue and leukoplakia (OPMD) on the right lateral side, with these distinct lesions separated by normal epithelium (Fig. 1). Tissue and brush biopsy were collected from the normal buccal mucosa, OPMD, and OSCC lesions. OPMD and OSCC samples showed negative resection margins. High-coverage whole-exome sequencing (WES) was performed to a median coverage of 145X on the paired OPMD, OSCC, and normal buccal brush and tissue biopsy.
Fig. 1.
Workflow for subclonal reconstruction. Illustration of an oral potentially malignant disorder (OPMD) and oral squamous cell carcinoma (OSCC) lesions and normal buccal mucosa sampled from the same patient with both brush and tissue biopsies. The samples were aligned, and somatic mutations were called from the aligned reads. CNA profiles were constructed using ASCAT and SNV CCFs were then clustered to identify subclonal lineages in the samples and phylogenetic reconstruction was done.
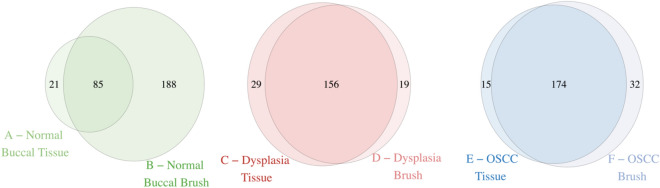
We first investigated whether the DNA of brush samples reflects the number of mutations compared to its paired tissue sample. We found that brush biopsies captured the majority of the single nucleotide variants (SNVs) found in its paired target tissue (Fig. 2, Supplementary Table 1). The paired brush sample captured 85 out of 106 (80.2%) SNVs found in the normal buccal oral tissue, 156 out of 185 (89%) in the dysplasia tissue, and 174 out of 189 (92%) SNVs in the OSCC tissue. In the case of the normal buccal mucosa and OSCC lesions, the brush biopsy sample detected more mutations compared to its paired tissue biopsy, with 167 SNVs (157%) and 17 SNVs (9.0%) more respectively. The substantial increase in mutations detected in the normal buccal brush sample may be attributed to brushing a broader, non-discrete area compared to the more focused targeted OSCC and OPMD lesion.
Fig. 2.
Mutations shared between the respective brush and tissue samples of each lesion.
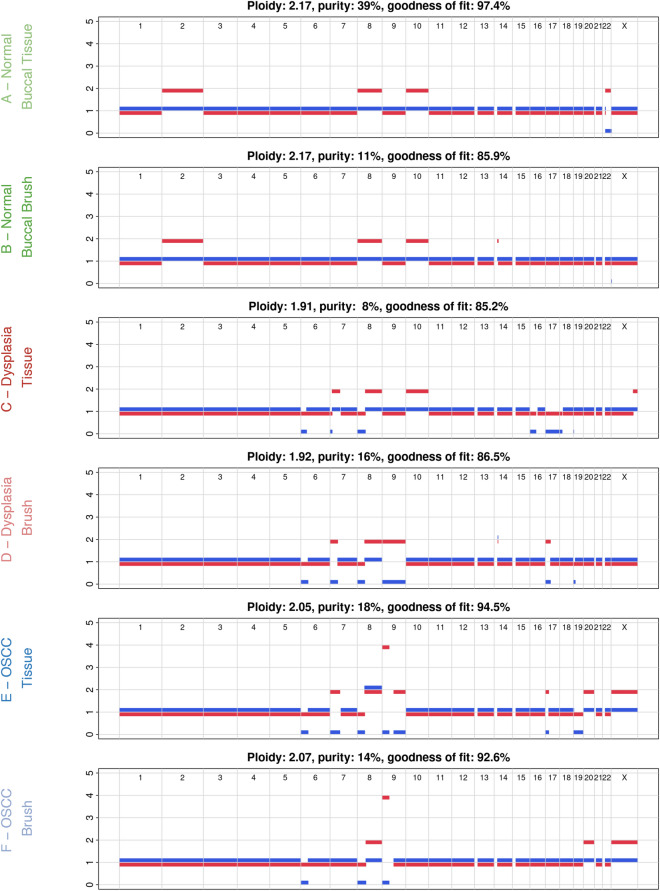
To identify CNAs we used Allele-Specific Copy Number Analysis of Tumors (ASCAT)19. Comparisons of CNA profiles between paired brush and tissue samples from each lesion revealed that brush biopsies accurately reflected the CNA of the corresponding tissue (Fig. 3). In normal buccal mucosa, dysplasia, and OSCC lesions, brush samples matched 98.7%, 81.6%, and 89.5% of the tissue samples’ allele-specific copy number calls, respectively. Significantly, brush biopsy captured CNAs found in dysplasia and OSCC tissue that play a known role in carcinogenesis, such as LOH of the short arm of chromosome 8 which houses tumor suppressor genes20–24 such DLC1 and NKX3.1, chromosome 9 which includes the CDKN2A locus25–27, chromosome 1728–31 which has the TP53 gene and is a significant marker for OSCC progression. Interestingly, brush biopsy also captured two TP53 mutations in conjunction with an LOH on chromosome 17 pointing to both mutations being present on one allele. However, it is crucial to note that the OSCC brush sample didn’t show any LOH with respect to chromosome 17 even with two TP53 mutations, which may be improved with a higher depth of sequencing. Given the shared CNAs between the dysplasia and OSCC sample, the minimum event distance32 between ASCAT profiles was calculated and a copy number-based phylogenetic tree was reconstructed using MEDICC232 (Supplementary Fig. 1). This analysis showed a clear separation between normal buccal epithelium and dysplasia/OSCC samples, with consistent clustering of the dysplasia and OSCC samples based on copy number events.
Fig. 3.
ASCAT profiles showing copy number alterations in each sample. Comparison of the tissue and brush samples show similar patterns in losses and gains in each lesion. Tumor purity is defined as fraction of cancer cells in the sample. The red and blue line represent the major and minor allele, respectively.
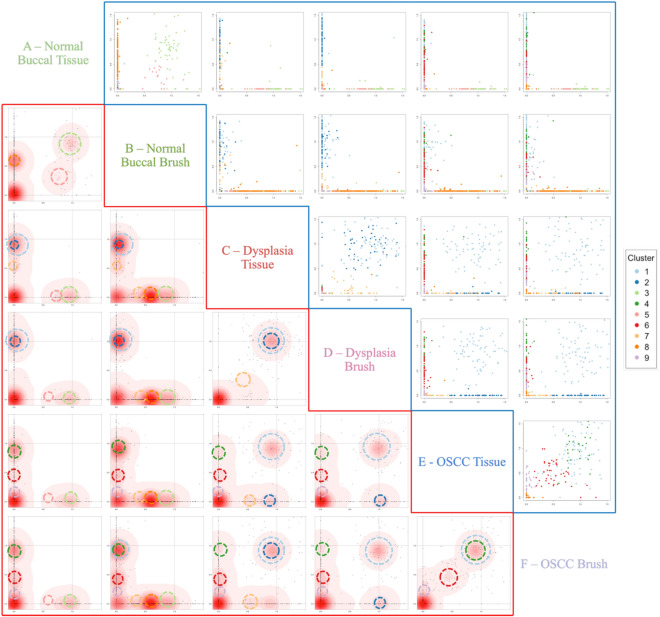
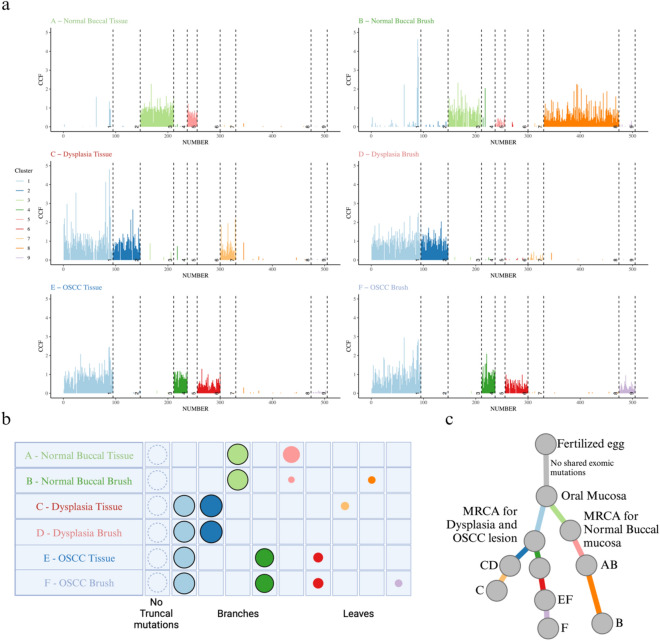
To identify clones and subclones, we used DPClust to cluster the SNVs based on their Cancer Cell Fraction (CCF) 33. CCF is the proportion of tumor cells that carry each SNV and is calculated using SNV variant allele frequencies (VAF) corrected for purity, ploidy, and copy number. Shared and/or unique subclones were identified (Figs. 4 and 5a). A consistent finding across all lesions was that brush biopsies captured all clonal and subclonal populations present in their corresponding tissue samples. Intriguingly, a unique subclonal population (cluster 8) was detected in the normal buccal brush sample that was absent in the paired tissue, likely due to the larger surface area covered by the brush biopsy, thereby picking up additional subclones. Surprisingly, paired samples from both dysplasia and OSCC showed a shared subclonal cluster, cluster 1, noting the presence of a common ancestral clone. Following the shared cluster 1 ancestor, the dysplasia and OSCC samples separate with unique subclones, demonstrating evolutionary divergence.
Fig. 4.
SNV Clustering using DPClust. Clusters are colored according to the legend provided. The bottom left red section shows the density of SNVs and the circles around the densities mark which color-coded cluster they belong to. The upper right blue section displays the distribution of the SNVs with each SNV colored according to the cluster it falls in.
Fig. 5.
Reconstruction of subclonal architecture. (a) Line plot of CCF in each mutation. The plot shows the number of mutations in each cluster, clusters present in each sample, and the number of shared and unique clusters between samples. Each line represents a mutation, and the length of the line corresponds to the CCF value of that mutation. (b) Each clone and subclone detected is represented as a set of color-coded circles across all samples. Each row represents a sample, and the area of the circle is proportional to the CCF of the corresponding subclone. Clonal and near-clonal populations are shown with solid borders. The circle plots are divided into three types: trunk (CCF = 1 in all samples, not seen in current samples), branches (present in more than one sample and either not found in all samples or subclonal in at least one), and leaf (specific to single samples). (c) Subclonal tree showing the relationship between subclones and the most common recent ancestor (MRCA) for the samples. The branch lengths of the phylogenetic tree are proportional to the number of mutations in each cluster and branches are annotated with samples in which they are present.
To evaluate the specificity of the brush biopsies, comparisons were made between brush samples from the dysplasia and OSCC lesions to the normal buccal brush and tissue samples. The absence of shared SNVs or clonal/subclonal populations with the normal buccal brush sample showed that the brush biopsies specifically captured the genomes of their target tissues (Fig. 4 and 5a). Interestingly there was a high mutational burden in the clinically normal buccal mucosa, which aligns with previous data from patients with significant smoking history34,35.
Finally, to assess the evolutionary history of the target lesions, a phylogenetic tree was constructed using SNV-based subclonal reconstruction (Fig. 5b and 5c). This identified three distinct branches in the tree with an initial joint branching of dysplasia and OSCC together from the normal buccal tissue through a shared common ancestor (branch AB and branch CDEF), consistent with the copy-number-based phylogenetic reconstruction (Supplementary Fig. 1). Following this, there was a divergence between OSCC and dysplasia with unique subclones (branches CD and EF) (Fig. 5c). It is important to note that the paired brush and tissue of each lesion consistently showed close phylogenetic relationships, denoting that brush biopsies can recapitulate the evolution of a tumor (Supplementary Fig. 2).
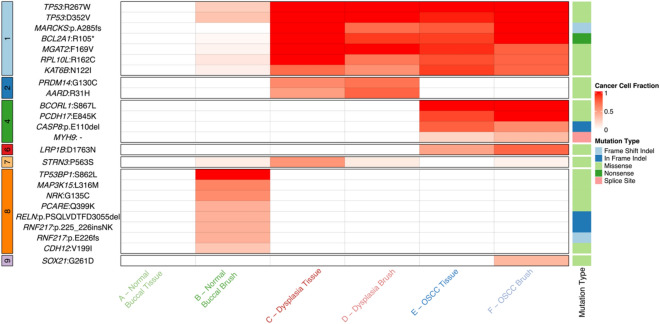
To assess putative driver mutations in our clonal populations, the catalog of mutated cancer genes from the Interactive OncoGenomics (IntOGen)36 was used (Fig. 6). We looked at all putative drivers present in our sample set and not just specific driver mutations in oral cancer to increase our comparisons. Most notably clonal cluster 1 had two TP53 mutations (R267W and D352V), cluster 4 had CASP837,38 and MYH939 mutations, which are associated with necrosis and radioresistance, and cluster 6 had a LRP1B mutation, which is associated with an unfavorable prognosis40–43. In addition to this, other potential driver mutations include BCL2A144, KAT6B45, and MGAT246 in cluster 1, PRDM1440 in cluster 2, and PCDH1747 in cluster 4. These mutations are involved in several important pathways, that play crucial roles in tumorigenesis, cell proliferation, differentiation, and survival. This shows that each subclone harbors additional mutations which may impart a selective advantage.
Fig. 6.
Oncoplot showing the presence of mutations in known driver genes in each sample, the cluster they fall in, and their CCF value. Genes and their protein change are denoted in each row. CCF values of the driver mutations are shown on a gradient scale from white to red for 0 to 1 CCF values. CCF of 0 (white) shows the absence of the mutation in the sample. The mutations are also denoted according to the cluster they fall in (left of the plot) and the type of mutation (right side of the plot).
In summary, this pilot proof-of-concept study demonstrated that brush biopsies are effective in accurately replicating SNVs, CNAs, and phylogenetic relationships, in comparison to their paired tumor tissue. Moreover, our study confirmed the specificity of brush biopsies to their target tissue, by demonstrating no contamination from normal buccal SNVs or CNAs. These results highlight the potential of brush biopsy as a minimally invasive method of analyzing genomic alterations in OPMDs. This strategy could provide valuable insight into oral carcinogenesis by affording the capacity for longitudinal serial analysis, the gold standard for studying tumor evolution, with the long-term goal of using this data for risk stratification.
Methods
Patient samples
This study followed the principles of the Helsinki Declaration and the institutional review board at the University of Texas MD Anderson Cancer Center at Houston reviewed, approved, and monitored this study. Data and biospecimens were collected with voluntary informed consent. For this pilot study, a 69-year-old female patient with an 11.5-pack-year smoking with no history of alcohol or smokeless tobacco use seen at MD Anderson Cancer Center, Houston, USA was selected for the study. She was a prime candidate for the study as she presented with an OSCC on the left lateral side of the tongue and leukoplakia on the right lateral side of the tongue. Fresh tissue biopsies were collected from both lesions for diagnosis. Brush biopsy samples of these lesions were obtained using soft Rovers Orcellex brushes (Rovers Medical Devices B.V., Oss, the Netherlands). Additionally, fresh tissue and brush biopsies were also collected from normal buccal mucosa for the study. Blood sample was used as germline sample to filter germline variants. Brush biopsy was collected immediately prior to tissue collection, and both were performed on the same day.
DNA extraction and quality control
gDNA was extracted by QIAamp® DNA Mini Kit (Qiagen) and then quantified using Quant-iT™ PicoGreen™ dsDNA Assay Kit (ThermoFisher SCIENTIFIC) and quality was accessed using Genomic DNA ScreenTape and Reagents on the Tapestation 4200 (Agilent Technologies).
Library preparation
Up to 200 ng of each gDNA sample based on the PicoGreen quantification was sheared (mechanically fragmented) using the E220 Focused-ultrasonicator Covaris (Covaris). To ensure the proper fragment size, samples were examined on TapeStation 4200 using the DNA High Sensitivity kit (Agilent Technologies).
The sheared DNA was proceeded to library preparation using Agilent SureSelect XT_HS2 DNA Reagent Kit with 384 unique dual sample indexing (UDI) and dual Molecular Barcodes to better suppress false positives and more accurately detect low variant allele frequency (VAF). The quality and quantity of the prepared libraries were evaluated using TapeStation 4200 and the DNA High Sensitivity kit (Agilent Technologies) to verify correct fragment size and to ensure complete removal of primer dimers.
Subsequently, the prepared libraries were individually hybridized to Agilent SureSelect Human All Exon v.4 probes (Agilent Technologies). The hybridization steps were automated on the Sciclone G3 NGSx Workstation (PerkinElmer, Inc.). Agilent captures were hybridized as single sample reactions using 500—1000 ng of prepared library as input. All Hybridization and Post-hybridization capture & washes were performed according to Agilent’s protocol.
Sequencing
The captured libraries were sequenced on Illumina NovaSeq 6000 platform for 2 × 150 paired end reads with an 8nt read for indexes using Cycle Sequencing v3 reagents (Illumina). The Demultiplexing was performed using Illumina’s bcl2fastq or BCL Convert software to generate paired end reads based on the dual indexes and remove sequences with incorrectly paired P5 and P7 indexes. Agilent Genomics NextGen Toolkit (AGeNT) was used for molecular barcode extraction and trimming.
Data analysis
Read mapping and quality control. We aligned the whole exome sequencing (WES) capture deep-sequencing data to human reference assembly hg38 (GRCh38) using BWA (v.0.7.17)48 and removed duplicated reads using Picard (v.2.23.8)49 with default parameter as described in the DNA-Seq analysis Pipeline in NCI-GDC Documentation50. Quality control was performed using FastQC (v.0.11.5), SamTools (v.1.15)51, Picard (v.2.23.8)49, and Genome Analysis Toolkit (GATK, v.4.2.4.0)52. The files were recalibrated using BaseRecalibrator and ApplyBQSR as part of the GATK pipeline.
Somatic mutation calling. All samples were analyzed against the matched normal blood sample. Single Nucleotide Variants (SNVs) were called using Mutect252,53 in tumor-normal mode. SNVs were filtered using the following cutoffs: (1) mapping quality score 50, (2) > 2 tumor reads supporting variant, (3) tumor sequence depth at locus 10. Mutations were further visualized using the Integrative Genome Viewer54 for false positives. The filtered variants were annotated using Funcotator (FUNCtional annOTATOR) as part of the GATK pipeline.
Mutation signature analysis. We used SigFit55 to identify the linear combination of predefined Head and Neck Cancer signatures in COSMIC56 that most accurately reconstructed the mutational profile of the cohort. For the cohort signature analysis, signatures with contributions of less than 5% were assumed to be absent.
Copy number alteration calling. Segmental copy number information was derived for each sample using ASCAT (v3.1.2)19, as previously described, from which gains, losses, copy number-neutral events, and loss of heterozygosity (LOH) can accurately be determined. In addition, the ASCAT profiles reveal differences in aberrant tumor cell fraction, ploidy, gains, losses, LOH, and copy number-neutral events between the samples. The purity and ploidy estimates were cross-checked and if necessary refitted after DPClust clustering of mutation data and review of B allele frequency and LogR profiles. Minimum event distance (MED) based copy number phylogeny was constructed using MEDICC232 to quantify the distance between copy number profiles.
Subclonal reconstruction. To model the subclonal structure of the 6 oral samples sequenced, we employed DPClust57,58 in 6 dimensions. Indels were not considered in the clustering process, as their variant allele frequencies (VAF) are over-dispersed and biased towards the reference allele, which would add noise to the clustering. However, they were assigned to clusters post hoc based on their CCF values across the samples. Phylogenetic relationships between identified subclones were reconstructed following the pigeonhole principle33. 215 SNVs were excluded from the DPClust analysis since they had very low CCF in all samples (median CCF = 0.0063), which is reminiscent of false positive artifacts.
Supplementary Information
Acknowledgements
This work was in part funded by the Pre-Cancerous Atlas (PCA) at The University of Texas MD Anderson Cancer Center. Pre-sequencing processing work was carried out in the Cancer Genomics Laboratory, The University of Texas MD Anderson Cancer Center Sheikh Khalifa Bin Zayed Al Nahyan Institute for Personalized Cancer Therapy” and supported by the MD Anderson Moon Shot programs. T.M.B. is supported by a PhD fellowship from Boehringer Ingelheim Fonds. P.V.L., T.L., and T.M.B. were supported by the Francis Crick Institute which receives its core funding from Cancer Research UK (CC2008), the UK Medical Research Council (CC2008), and the Wellcome Trust (CC2008). P.V.L. is a CPRIT Scholar in Cancer Research and acknowledges CPRIT grant support (RR210006).
Author contributions
E.J. carried out the majority of the analyses with assistance from T.L. and supervision from X.Z. and P.V.L.; E.J., X.Z., and P.V.L. wrote the manuscript with comments and contributions from T.L., M.T., and J.R.W.; M.T. and T.M.B. provided bioinformatics assistance. A.G. provided the clinical samples and developed the clinical protocols. T.M.B., M.T., A.G., and J.R.W. provided comments and edited the manuscript. All authors have read and approved the final manuscript.
Data availability
The sequencing data has been uploaded to European Genome Archive (EGA) and are is available upon request.
Competing interests
The authors declare no competing interests.
Ethics declarations
The collection and analysis of tissues was approved at the UT MD Anderson Cancer Center under IRB protocols 2021–0129 and PA17-0050.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Peter Van Loo and Xiao Zhao these authors jointly supervised the work.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-72946-3.
References
- 1.Conway, D. I., Purkayastha, M. & Chestnutt, I. G. The changing epidemiology of oral cancer: definitions, trends, and risk factors. Br Dent J225, 867–873. 10.1038/sj.bdj.2018.922 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Warnakulasuriya, S. Oral potentially malignant disorders: A comprehensive review on clinical aspects and management. Oral Oncol102, 104550. 10.1016/j.oraloncology.2019.104550 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Ray, J. G. Oral potentially malignant disorders: Revisited. J Oral Maxillofac Pathol21, 326–327. 10.4103/jomfp.JOMFP_224_17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iocca, O. et al.. Potentially malignant disorders of the oral cavity and oral dysplasia: A systematic review and meta-analysis of malignant transformation rate by subtype. Head Neck42, 539–555. 10.1002/hed.26006 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Wils, L. J. et al. Elucidating the Genetic Landscape of Oral Leukoplakia to Predict Malignant Transformation. Clin Cancer Res29, 602–613. 10.1158/1078-0432.CCR-22-2210 (2023). [DOI] [PubMed] [Google Scholar]
- 6.Wils, L. J. et al. The role of differentiated dysplasia in the prediction of malignant transformation of oral leukoplakia. J Oral Pathol Med52, 930–938. 10.1111/jop.13483 (2023). [DOI] [PubMed] [Google Scholar]
- 7.Zhou, G., Liu, Z. & Myers, J. N. TP53 Mutations in Head and Neck Squamous Cell Carcinoma and Their Impact on Disease Progression and Treatment Response. J Cell Biochem117, 2682–2692. 10.1002/jcb.25592 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Califano, J. et al. Genetic progression model for head and neck cancer: implications for field cancerization. Cancer Res56, 2488–2492 (1996). [PubMed] [Google Scholar]
- 9.Zhang, L. et al. Loss of heterozygosity (LOH) profiles–validated risk predictors for progression to oral cancer. Cancer Prev Res (Phila)5, 1081–1089. 10.1158/1940-6207.CAPR-12-0173 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farah, C. S. et al. Exome sequencing of oral leukoplakia and oral squamous cell carcinoma implicates DNA damage repair gene defects in malignant transformation. Oral Oncol96, 42–50. 10.1016/j.oraloncology.2019.07.005 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Yoo, W. J. et al. Loss of heterozygosity on chromosomes 3p,8p,9p and 17p in the progression of squamous cell carcinoma of the larynx. J Korean Med Sci19, 345–351. 10.3346/jkms.2004.19.3.345 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Armaghany, T., Wilson, J. D., Chu, Q. & Mills, G. Genetic alterations in colorectal cancer. Gastrointest Cancer Res5, 19–27 (2012). [PMC free article] [PubMed] [Google Scholar]
- 13.Wang, J. et al. Progression from ductal carcinoma in situ to invasive breast cancer: molecular features and clinical significance. Signal Transduct Target Ther9, 83. 10.1038/s41392-024-01779-3 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Babshet, M., Nandimath, K., Pervatikar, S. & Naikmasur, V. Efficacy of oral brush cytology in the evaluation of the oral premalignant and malignant lesions. J Cytol28, 165–172. 10.4103/0970-9371.86342 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mehrotra, R., Mishra, S. & Singh, M. The efficacy of oral brush biopsy with computer-assisted analysis in identifying precancerous and cancerous lesions. Head Neck Oncol3, 39. 10.1186/1758-3284-3-39 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poell, J. B. et al. Oral cancer prediction by noninvasive genetic screening. Int J Cancer152, 227–238. 10.1002/ijc.34277 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghosh, A. et al. Integrative analysis of genomic and transcriptomic data of normal, tumour, and co-occurring leukoplakia tissue triads drawn from patients with gingivobuccal oral cancer identifies signatures of tumour initiation and progression. J Pathol257, 593–606. 10.1002/path.5900 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Errazquin, R. et al. Early Diagnosis of Oral Cancer and Lesions in Fanconi Anemia Patients: A Prospective and Longitudinal Study Using Saliva and Plasma. Cancers (Basel)15, 10.3390/cancers15061871 (2023). [DOI] [PMC free article] [PubMed]
- 19.Van Loo, P. et al. Allele-specific copy number analysis of tumors. Proc Natl Acad Sci U S A107, 16910–16915. 10.1073/pnas.1009843107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu, C. L. et al. Deletion mapping defines three discrete areas of allelic imbalance on chromosome arm 8p in oral and oropharyngeal squamous cell carcinomas. Genes Chromosomes Cancer20, 347–353. 10.1002/(sici)1098-2264(199712)20:4%3c347::aid-gcc5%3e3.0.co;2-1 (1997). [DOI] [PubMed] [Google Scholar]
- 21.El-Naggar, A. K. et al. Localization of chromosome 8p regions involved in early tumorigenesis of oral and laryngeal squamous carcinoma. Oncogene16, 2983–2987. 10.1038/sj.onc.1201808 (1998). [DOI] [PubMed] [Google Scholar]
- 22.Zhou, X. et al. Allelic imbalance analysis of oral tongue squamous cell carcinoma by high-density single nucleotide polymorphism arrays using whole-genome amplified DNA. Hum Genet118, 504–507. 10.1007/s00439-005-0069-x (2005). [DOI] [PubMed] [Google Scholar]
- 23.Pathare, S., Schäffer, A. A., Beerenwinkel, N. & Mahimkar, M. Construction of oncogenetic tree models reveals multiple pathways of oral cancer progression. Int J Cancer124, 2864–2871. 10.1002/ijc.24267 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshioka, S. et al. Genomic profiling of oral squamous cell carcinoma by array-based comparative genomic hybridization. PLoS One8, e56165. 10.1371/journal.pone.0056165 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosin, M. P. et al. Use of allelic loss to predict malignant risk for low-grade oral epithelial dysplasia. Clin Cancer Res6, 357–362 (2000). [PubMed] [Google Scholar]
- 26.Zhang, L. & Rosin, M. P. Loss of heterozygosity: a potential tool in management of oral premalignant lesions?. J Oral Pathol Med30, 513–520. 10.1034/j.1600-0714.2001.300901.x (2001). [DOI] [PubMed] [Google Scholar]
- 27.van der Riet, P. et al. Frequent loss of chromosome 9p21-22 early in head and neck cancer progression. Cancer Res54, 1156–1158 (1994). [PubMed] [Google Scholar]
- 28.Miller, C. et al. Human p53 gene localized to short arm of chromosome 17. Nature319, 783–784. 10.1038/319783a0 (1986). [DOI] [PubMed] [Google Scholar]
- 29.McBride, O. W., Merry, D. & Givol, D. The gene for human p53 cellular tumor antigen is located on chromosome 17 short arm (17p13). Proc Natl Acad Sci U S A83, 130–134. 10.1073/pnas.83.1.130 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martín-Ezquerra, G. et al. Multiple genetic copy number alterations in oral squamous cell carcinoma: study of MYC, TP53, CCDN1, EGFR and ERBB2 status in primary and metastatic tumours. Br J Dermatol163, 1028–1035. 10.1111/j.1365-2133.2010.09947.x (2010). [DOI] [PubMed] [Google Scholar]
- 31.Choi, K. Y. et al. Loss of heterozygosity in mammary serine protease inhibitor (maspin) and p53 at chromosome 17 and 18 in oral cavity squamous cell carcinoma. Head Neck37, 1239–1245. 10.1002/hed.23741 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Kaufmann, T. L. et al. MEDICC2: whole-genome doubling aware copy-number phylogenies for cancer evolution. Genome Biol23, 241. 10.1186/s13059-022-02794-9 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tarabichi, M. et al. A practical guide to cancer subclonal reconstruction from DNA sequencing. Nat Methods18, 144–155. 10.1038/s41592-020-01013-2 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.South, A. P. et al. Mutation signature analysis identifies increased mutation caused by tobacco smoke associated DNA adducts in larynx squamous cell carcinoma compared with oral cavity and oropharynx. Sci Rep9, 19256. 10.1038/s41598-019-55352-y (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shani, T. et al. Chromosomal numerical aberrations in apparently normal oral mucosa of heavy smokers affected by lung cancer. Oral Oncol46, 96–99. 10.1016/j.oraloncology.2009.10.010 (2010). [DOI] [PubMed] [Google Scholar]
- 36.Martínez-Jiménez, F. et al. A compendium of mutational cancer driver genes. Nat Rev Cancer20, 555–572. 10.1038/s41568-020-0290-x (2020). [DOI] [PubMed] [Google Scholar]
- 37.Uzunparmak, B. et al. Caspase-8 loss radiosensitizes head and neck squamous cell carcinoma to SMAC mimetic-induced necroptosis. JCI Insight5, 10.1172/jci.insight.139837 (2020). [DOI] [PMC free article] [PubMed]
- 38.Singh, R. et al. Study of Caspase 8 mutation in oral cancer and adjacent precancer tissues and implication in progression. PLoS One15, e0233058. 10.1371/journal.pone.0233058 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yabe, T. E. et al. MYH9 mutation and squamous cell cancer of the tongue in a young adult: a novel case report. Diagn Pathol17, 23. 10.1186/s13000-022-01210-x (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu, Y., Jiang, E., Shao, Z. & Shang, Z. Long Noncoding RNAs in the Metastasis of Oral Squamous Cell Carcinoma. Front Oncol10, 616717. 10.3389/fonc.2020.616717 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cao, C. H. et al. mutation is associated with tumor HPV status and promotes poor disease outcomes with a higher mutation count in HPV-related cervical carcinoma and head & neck squamous cell carcinoma. Int J Biol Sci17, 1744–1756. 10.7150/ijbs.56970 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim, S. Y. et al. Aberrantly hypermethylated tumor suppressor genes were identified in oral squamous cell carcinoma (OSCC). Clin Epigenetics11, 116. 10.1186/s13148-019-0715-0 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakagawa, T. et al. Genetic or epigenetic silencing of low density lipoprotein receptor-related protein 1B expression in oral squamous cell carcinoma. Cancer Sci97, 1070–1074. 10.1111/j.1349-7006.2006.00283.x (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng, Q., Gan, G., Gao, X., Luo, Q. & Chen, F. Targeting the IDO-BCL2A1-Cytochrome c Pathway Promotes Apoptosis in Oral Squamous Cell Carcinoma. Onco Targets Ther14, 1673–1687. 10.2147/OTT.S288692 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.You, J. S. & Jones, P. A. Cancer genetics and epigenetics: two sides of the same coin?. Cancer Cell22, 9–20. 10.1016/j.ccr.2012.06.008 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taniguchi, N. & Kizuka, Y. Glycans and cancer: role of N-glycans in cancer biomarker, progression and metastasis, and therapeutics. Adv Cancer Res126, 11–51. 10.1016/bs.acr.2014.11.001 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Haruki, S. et al. Frequent silencing of protocadherin 17, a candidate tumour suppressor for esophageal squamous cell carcinoma. Carcinogenesis31, 1027–1036. 10.1093/carcin/bgq053 (2010). [DOI] [PubMed] [Google Scholar]
- 48.Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics25, 1754–1760. 10.1093/bioinformatics/btp324 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DePristo, M. A. et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet43, 491-+, 10.1038/Ng.806 (2011). [DOI] [PMC free article] [PubMed]
- 50.Institute, N. C. DNA-Seq Analysis Pipeline, <https://docs.gdc.cancer.gov/Data/Bioinformatics_Pipelines/DNA_Seq_Variant_Calling_Pipeline/> (
- 51.Danecek, P. et al. Twelve years of SAMtools and BCFtools. Gigascience10, 10.1093/gigascience/giab008 (2021). [DOI] [PMC free article] [PubMed]
- 52.McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res20, 1297–1303. 10.1101/gr.107524.110 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cibulskis, K. et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol31, 213–219. 10.1038/nbt.2514 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Robinson, J. T. et al. Integrative genomics viewer. Nat Biotechnol29, 24–26. 10.1038/nbt.1754 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gori, K. & Baez-Ortega, A. sigfit: flexible Bayesian inference of mutational signatures. bioRxiv, 372896, 10.1101/372896 (2020).
- 56.Alexandrov, L. B. et al. The repertoire of mutational signatures in human cancer. Nature578, 94–101. 10.1038/s41586-020-1943-3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nik-Zainal, S. et al. The life history of 21 breast cancers. Cell149, 994–1007. 10.1016/j.cell.2012.04.023 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bolli, N. et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat Commun5, 2997. 10.1038/ncomms3997 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data has been uploaded to European Genome Archive (EGA) and are is available upon request.