Abstract
The Sendai virus (SeV) C gene codes for a nested set of four C proteins that carry out several functions, including the modulation of viral RNA synthesis and countering of the cellular antiviral response. Using mutant C genes (and in particular a C gene with a deletion of six amino acids present only in the larger pair of C proteins) and recombinant SeV carrying these mutant C genes, we find that the nested set of C proteins carry out a nested set of functions. All of the C proteins interdict interferon (IFN) signaling to IFN-stimulated genes (ISGs) and prevent pY701-Stat1 formation. However, only the larger C proteins can induce STAT1 instability, prevent IFN from inducing an antiviral state, or prevent programmed cell death. Remarkably, interdiction of IFN signaling to ISGs and the absence of pY701-Stat1 formation did not prevent IFN-α from inducing an anti-Vesicular stomatitis virus (VSV) state. It is possible that IFN-α signaling to induce an anti-VSV state can occur independently of the well-established Jak/Stat/ISGF3 pathway and that it is this parallel pathway that is targeted by the longer C proteins.
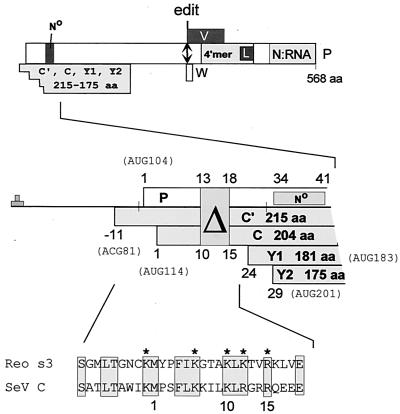
Paramyxovirus P genes are remarkable for the complexity of their genetic organization and expression. This gene, named for the phosphoprotein that is an essential component of the P/L viral RNA polymerase (vRNAP), contains additional open reading frames (ORFs) that overlap the beginning and the middle of the P protein ORF (the C and V ORFs, respectively) (Fig. 1). The C and V overlapping ORFs are accessed by a variety of unusual mechanisms of ribosomal choice (reviewed in references 9 and 31) and mRNA editing (39, 41), respectively. The overlapping ORFs are also referred to as “accessory” genes (1) because in each case there is at least one virus within the subfamily Paramyxovirinae that does not contain (or express) the V or C genes. For Sendai virus (SeV), a nested set of 2 longer C proteins (C′ACG81 and CAUG114) and 2 shorter C proteins (Y1AUG183 and Y2AUG201) are expressed (Fig. 1). The two longer C proteins (along with the P protein) are initiated by scanning ribosomes, whereas the two shorter C proteins are initiated by a ribosomal shunt (31).
FIG. 1.
ORF organization and expression of the SeV P gene. The ORFs expressed as protein (P, C, V, and W) are shown as horizontal boxes, drawn roughly to scale (above). Several domains of the P protein, that which chaperones unassembled N protein (N°), its tetramerization domain including the L protein binding site, and that which binds to the N:RNA nucleocapsdid, are indicated. The double-headed arrow shows the editing site (codon 317), where G residues are added cotranscriptionally, to access the V and W ORFs. A blowup of the 5′ end of the mRNA is shown in the middle, and the five ribosomal start sites in this region are indicated. The numbers refer to the codon positions of the P and C ORFs. For the C ORF, the AUG114-initiated C protein is the point of reference; hence, the ACG81-initiated C′ protein begins at position −11. All four C proteins terminate at codon 205. The 18-nucleotide deletion that eliminates codons 13 to 18 of P and codons 10 to 15 of C is shown, along with the N chaperone site of P. An alignment of residues −9 to 19 of the C ORF with residues 273 to 300 of the reovirus ς3 protein (part of the dsRNA binding motif) is shown at the bottom. The five basic residues essential for dsRNA binding are indicated with asterisks.
The first property of the multifunctional C proteins noted was their ability to modulate viral RNA (vRNA) synthesis. When vRNA synthesis is reconstituted in vitro with purified N:RNA nucleocapsids and the products of the P/C and L genes (expressed in transfected cell extracts), specific ablation of C gene expression (or its replacement with mutant CF170S genes) increases vRNAP specific activity ∼8-fold (5). The longer C′ or C proteins are equally active in this respect, but the shorter Y proteins are inactive. The inhibitory effects of C expression are promoter specific, in that genomic promoters are considerably more sensitive to inhibition than antigenomic promoters (1, 38). Remarkably, the negative effects of C on vRNA synthesis are accompanied by an equivalent increase in promoter fidelity, in that minigenomes with promoter mutations (including those which destroy hexamer genome length) that are lethal in the presence of C gene expression can replicate in its absence (34, 38).
C was initially thought to be a nonstructural virus protein, but it is present in SeV particles at relatively low levels (ca. 40 molecules/genome [29, 43]). Its relative abundance increases greatly during intracellular replication (C also increases ca. 20-fold relative to the P and L proteins during the infection), and the inhibitory effects of C are concentration dependent (38). As C accumulates during infection, it is thought to first shift vRNA synthesis from that of mRNAs and antigenomes toward minus-strand genomes and then to shut down mRNA synthesis entirely in preparation for assembling minus-strand genome nucleocapsids into virions. The C protein(s) also plays a more direct role in virus assembly, since progeny virions from infections missing all four C proteins are poorly infectious and have altered morphology. The absence of C interactions with the M protein is thought to be responsible for this defect in virus assembly (20, 28). Moreover, progeny virions from infections missing only the two larger C proteins have less virion RNAP activity in vitro (unpublished), and there is a significant delay in the onset of intracellular virus amplification during infection (30). In this respect, the essentially nonstructural C proteins also act as an infectivity factor similar to the human immunodeficiency virus type 1 vif protein, another accessory (nonstructural) virus protein which counteracts the innate antiviral activity of cells (36, 40).
The SeV CF170S mutation was uncovered as one of two mutations (along with LE2050A) in a highly virulent strain (SeVM; 50% lethal dose [LD50] of 40) that became avirulent (SeVMVC; LD50 of >800,000) on adaptation for growth in LLC-MK2 cells (21). Recombinant SeV (rSeV) carrying the CMVC gene in an otherwise wild-type strain Z background (rSeVZ-CMVC) is also avirulent in mice, whereas rSeV strains carrying the CM gene have not lost virulence (15). SeV C gene activity is thus clearly required for virulence in mice. Although rSeVZ-CMVC was avirulent, it grew normally in the mouse respiratory tract at first, but virus was then quickly cleared. The rapidity of this antiviral response in immunologically naive mice, which mirrored that found in rSeV-[V-minus] infections (23), suggested that some aspect of innate immunity was involved (17). One such function has recently been identified. Infection of interferon (IFN)-competent cells with SeV or simian virus 5 (SV5) does not induce an antiviral state. These paramyxovirus infections do not affect IFN-α/β secretion, but somehow interfere with its receptor-mediated signaling since they also prevent added IFN from inducing an antiviral state (12, 17, 19). For SeV, the C gene (Fig. 1) is clearly involved, since viruses which carry the CF170S mutation, or those which do not express any of the four C proteins, do not interfere with IFN action (17, 28). Moreover, rSeV which cannot specifically express CAUG114 (rSeVZ-[C-minus]) does not prevent the IFN-induced antiviral state, even though this virus normally expresses C′ACG81 and Y1, and overexpresses Y2. SeVZ-[C-minus] is also avirulent in mice. The SeV CAUG114 protein is thus specifically required for the full range of IFN-mediated antiviral action and virulence in mice (18, 28, 30). Human parainfluenza virus type 1, like SeV, expresses four C proteins, but most paramyxoviruses are thought to express a single C protein, equivalent to SeV CAUG114. For the rubulavirus SV5, which does not express a C protein, it is the V protein that apparently functions to interdict IFN signaling (12, 13).
The SeV C proteins appear to be multifunctional proteins, playing roles in such diverse processes as modulating vRNA synthesis, interdicting IFN action, delaying CPE (antiapoptosis), and ensuring proper virion assembly and infectivity. We describe here experiments designed to elucidate the role of the various C proteins in some of these multiple functions.
MATERIALS AND METHODS
Cells and viruses.
Murine BF cells (cloned from a primary cell culture of a BALB/c mouse embryo) and mouse embryo fibroblasts (MEFs) (44) were grown as monolayers in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum. The generation of rSeV expressing alternate C (and P) proteins has been described elsewhere (10, 14, 15, 30). All SeV stocks were grown in the allantoic cavity of 10-day-old embryonated chicken eggs. Virus present in the allantoic fluids was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie blue staining after virus pelleting. Virus titers were determined by plaque formation on LLC-MK2 cells. The vesicular stomatitis virus (VSV) stock (Mudd-Summers) was grown in BHK cells. Virus released into the culture medium was clarified by centrifugation to remove cell debris, and the titer was determined by plaque formation on LLC-MK2 cells.
Plasmid DNAs.
The IFN-α/β responsive reporter plasmid, p(9-27)4tk(−39)lucter (24, 25), referred to here as pISRE(f)luc, contains four tandem repeats of the IFN-inducible gene 9-27 ISRE fused to the firefly luciferase gene. pTK-r.luc., used as a transfection standard, contains the herpes simplex virus TK promoter region upstream of the Renilla luciferase gene (Promega).
Transient transfections.
Monolayers of BF cells in 5.5-cm-diameter plates (at 50% confluence) were transfected with a total of 2 μg of DNA and 6 μl of Effectene (Qiagen) according to the manufacturer's instructions. At 24 h posttransfection, the cells were (or were not) infected with 20 PFU of SeV per cell and treated with 1,000 IU of recombinant IFN-α2/α1 (42) per ml at 48 h posttransfection. At 4 to 12 h after IFN treatment, cells were harvested and assayed for firefly and Renilla luciferase activity (dual-luciferase reporter assay system; Promega). Relative expression levels were calculated by dividing the firefly luciferase values by those of the Renilla luciferase.
Immunoblotting.
Proteins were separated by SDS-PAGE and transferred to Immunobilon-P membranes by semidry transfer. The primary antibodies used included a rabbit polyclonal antiserum to VSV P protein (provided by J. Perrault and D. Summers), a rabbit polyclonal antiserum to SeV P protein isolated from an SDS gel (anti-PSDS; L. Roux, Geneva, Switzerland), a mouse monoclonal antibody to SeV N (N877 [33]), a rabbit polyclonal antiserum to SeV C protein (provided by Y. Nagai, Tokyo, Japan), a mouse monoclonal antibody to Stat1 C terminus (Transduction Laboratories S21120), a rabbit polyclonal antiserum to Phospho-Stat (Y701) (Upstate Biotechnology 06-657), and a rabbit polyclonal antiserum to actin (provided by G. Gabbiani, Geneva, Switzerland). The secondary antibodies used were alkaline phosphatase-conjugated goat antibodies specific for either rabbit or mouse immunoglobulin G (Bio-Rad). The immobilized proteins were detected by light-enhanced chemiluminescence (Bio-Rad).
Measurement of phosphatidylserine exposure by Annexin-V fluorescence.
HeLa cells from 5.5-cm petri dishes were harvested and analyzed by flow cytometry using Annexin-V-Alexa 568 (Roche) and BOBO-1 (Molecular Probes) at 48 h postinfection, according to the manufacturer's instructions.
In vitro RNA synthesis.
RNA synthesis in vitro was performed essentially as described by Curran et al. (6) with the modifications outlined in Curran (8). N:RNA nondefective templates were isolated from infected egg allantoic fluid (strain Z) by banding twice on 20 to 40% CsCl gradients. Templates were resuspended at a concentration of ca. 250 ng/μl in TE (10 mM Tris [pH 7.4], 1 mM EDTA) containing 1 mM dithiothreitol–10% glycerol and stored at −80°C. A549 cells (5-cm-diameter petri dish) were transfected with 5 μg of pGEM-P/C (or mutant P/C clone) and 1 μg of pGEM-L. In vitro RNA synthesis was generally carried out in 100-μl reaction mixtures containing 5 μl of the template, 90 μl of vaccinia virus-T7 (VV-T7)-infected A549 cell extract, 30 μCi of [32P]GTP, and 20 μg of actinomycin D per ml at 30°C for 3 h. After the reaction, 500 μl of lysis buffer (150 mM NaCl, 50 mM Tris [pH 7.4], 10 mM EDTA, 0.6% NP-40) was added, and the RNA was recovered by pelleting through 5.7 M CsCl. Products were analyzed directly on 1.5% agarose-HCHO gels.
RESULTS
C gene inhibition of vRNA synthesis and rSeV recovery are separate events.
The coexpression of either of the two larger C proteins during SeV vRNAP complex formation in transfected cells strongly reduces the specific activity of the P/L vRNAP (P/Cwt versus P/Cstop, Fig. 2), but the coexpression of Y1 and/or Y2 is not inhibitory (5). This capacity of the longer C proteins to inhibit vRNA synthesis thus appears to depend on the 23 NH2-terminal residues (or C1–23) of CAUG114 (Fig. 1). Moreover, virtually the entire C ORF upstream of Y1AUG183 can be aligned with a small portion of the double-stranded RNA (dsRNA) binding domain of the reovirus ς3 protein (ς3 also functions in preventing the host cell's innate antiviral response, presumably by sequestering dsRNA and preventing PKR activation [11]). We were therefore interested in further defining the function of the C1–23 domain (the 23 NH2-terminal residues of CAUG114) and, if possible, within a rSeV so that its properties in cellular and animal infections could be determined.
FIG. 2.
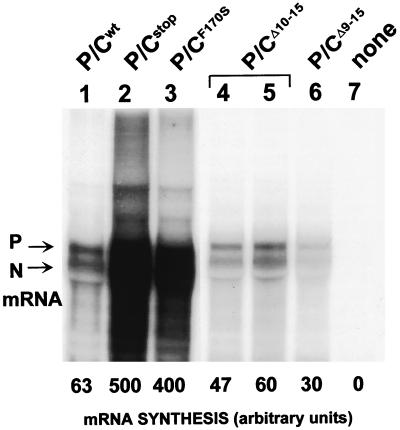
C protein expression inhibits SeV mRNA synthesis in transfected cell extracts. Viral mRNA synthesis was carried out with purified N:RNA nucleocapsids and cytoplasmic extracts of vTF7-3 infected A549 cells that had been transfected with pGEM-L and various pGEM-P/C plasmids (as indicated above), in the presence of actinomycin D and [α-32P]GTP (see Materials and Methods). P/Cstop contains a stop codon just downstream of AUG201 (Y2) and does not express any of the C proteins; none, control (no pGEM-P/C). The products of the reaction were purified by Trizol extraction and separated on a 1.5% formaldehyde-agarose gel. The N and P mRNA bands were quantified in a PhosphorImager, and their relative intensities are shown below.
It has not yet been possible to separate the P and C gene ORFs into individual transcription units in rSeV. rSeV with deletions within C1–23 of the C ORF will also carry the corresponding deletions in the P ORF, and both deletions could potentially affect rSeV viability. We had originally introduced a series of eight short deletions within the first 69 codons of the SeV P ORF (to locate a P protein domain (N°, Fig. 1) that chaperones unassembled N protein during the nascent-chain assembly step of genome replication [7]). Three of these deletions, CΔ2-8/PΔ5-11, CΔ9-15/PΔ12-18, and CΔ16-22/PΔ19-25, were located within C1–23. In spite of the importance of the C gene in many aspects of SeV replication, its coexpression from the pGEM-P/C support plasmid during rSeV recovery from DNA in the VV-infected and plasmid-transfected (VV-T7) cell system is highly deleterious and needs to be suppressed (1, 14, 15). Our VV-T7 recovery system can also be highly recombinogenic, and rSeV carrying P/C gene mutations which lead to a selective disadvantage vs rSeVwt are generally lost in the recovery unless the pGEM-P/C support plasmid also carries the same mutation (see below). However, when these three deleted pGEM-P/C support plasmids were examined for their ability to rescue rSeVwt from DNA, we were surprised to find that, whereas CΔ2-8 and CΔ16-22 prevented rSeV recovery as had Cwt, CΔ9-15 had lost this property (data not shown). pGEM-PΔ12-18/CΔ9-15 could then be used (if necessary) to rescue rSeV without inactivating its C protein expression.
We had originally assumed that the capacities of the C proteins to inhibit rSeV recovery and vRNA synthesis were related, because C is a powerful inhibitor of minigenome replication in the VV-T7 system and CF170S genes were null mutants for both properties (1, 38). We therefore expected that at least one of these three deletions (including CΔ9-15) would further identify a domain required for CAUG114 to inhibit vRNA synthesis. However, when CΔ2-8, CΔ9-15, and CΔ16-22 were examined for their ability to inhibit RNA synthesis, we were further surprised to find that all were wild type in this respect (Fig. 2 and data not shown; see Discussion). Thus, although the properties of inhibiting vRNA synthesis and rSeV recovery are linked by their requirement for F170, they are also separate events (see Fig. 7). Moreover, we have uncovered other mutant C genes that have precisely this phenotype, i.e., their products inhibit vRNA synthesis normally but no longer prevent rSeV recovery (e.g., CG162A [data not shown]).
FIG. 7.
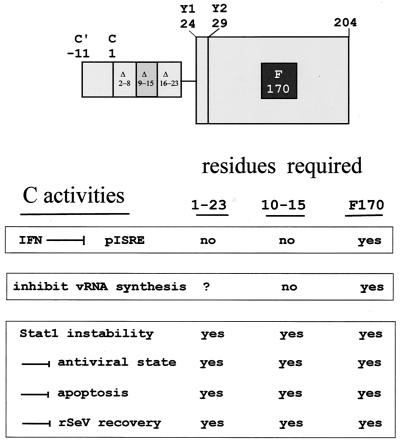
Schematic representation the SeV C proteins and their activities. The longer C proteins are shown as being composed of the Y protein module (large box) in which F170 is critical for all activities, plus an NH2-terminal extension of either 23 (CAUG114) or 34 (C′ACG81) residues. The deletions within C1–23 examined are indicated. The various activities of the C proteins are listed below in three groups distinguished by the phenotypes of the various mutant C genes examined. The numbers 1 to 23 and 10 to 15 refer to these residues within CAUG114. For further explanation, see the text. The values for residues 1 to 23 were determined from the activity of the Y1 protein.
rSeV-[CΔ10-15/PΔ13-18].
A rSeV containing a 21-nucleotide-deleted genome (encompassing C protein residues 9Leu-Lys-Leu-Arg-Gly-Arg-Arg15) is not expected to be viable (2). We therefore restricted ourselves to the six-amino-acid aa deletion of CΔ10-15 (and PΔ13-18), since CΔ10-15 also inhibits vRNA synthesis as had Cwt (Fig. 2, lanes 1, 4, and 5). Recovery of rSeV carrying this deletion was first attempted with our pGEM-P/Cstop support plasmid, but only rSeVwt was recovered under these conditions (presumably as the result of recombination between pGEM-P/Cstop and the full-length viral cDNA in the VV-T7 system). rSeV-[CΔ10-15/PΔ13-18] was only recovered when the P gene support plasmid contained the same deletion. Although rSeV-[CΔ10-15/PΔ13-18] is viable and produces normal plaques, this virus clearly cannot compete against the small amounts of SeVwt that are generated by recombination during virus recovery with pGEM-P/Cwt. rSeV-[PΔ13-18/CΔ10-15] is also avirulent in mice (Table 1). It is possible that the P protein deletion also contributes to the reduced fitness of rSeV-[CΔ10-15/PΔ13-18], even though the PΔ13-18 protein is wild type in all aspects of vRNA synthesis that can be reproduced in transfected cells and extracts (Fig. 2 and data not shown). rSeV-[PΔ13-18/CΔ10-15] grows as well as rSeVwt in MEFs (see Fig. 6, lower panel) and HeLa and BHK cells (data not shown). However, unlike rSeVwt rSeV-[PΔ13-18/CΔ10-15] is partially sensitive to IFN treatment (see Fig. 6 and below).
TABLE 1.
Pathogenicity of rSeV-[CΔ10-15]
| Inoculum size (CIU/mouse)a | No. of surviving mice/total no. of mice infected with:
|
|
|---|---|---|
| rSeVZ | rSeVZ-[CΔ10-15] | |
| 1.25 × 105 | 0/3 | 3/3 |
| 1.25 × 104 | 0/3 | 3/3 |
| 1.25 × 103 | 3/3 | 3/3 |
| 1.25 × 102 | 3/3 | 3/3 |
The LD50 values for rSeVZ and rSeVZ [CΔ10-15] were determined to be 6.9 × 103 and >6.9 × 105, respectively. CIU, cell infectious units.
FIG. 6.
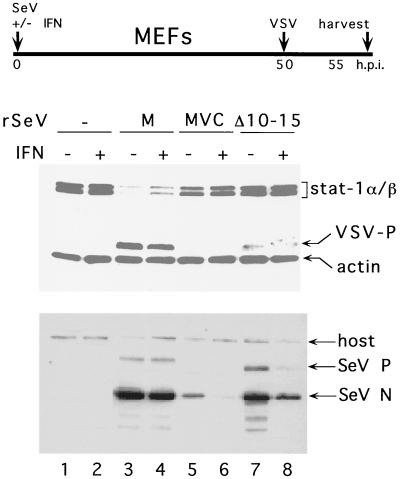
Effect of SeV infection on Stat1 levels and induction of an antiviral state. Parallel cultures of NIH 3T3 MEFs were infected with either SeVZ-CM (M), SeVZ-CMVC (MVC), or SeV-[CΔ10-15] or were not infected and then concomitantly treated (or not) with 1,000 U of IFN-α, per ml, as indicated. The cultures were superinfected with VSV (50 PFU/cell) at 50 h post-SeV infection and harvested at 55 hpi. The relative levels of the cellular Stat1 and VSV P proteins were determined by immunoblotting (upper panel). An anti-actin antibody was included to control for the amount of cellular material loaded onto each lane. Equal samples of the various cell extracts were used to determine the relative levels of the SeV N and P proteins present by immunoblotting (lower panel).
Characterization of rSeV-CΔ10-15/PΔ13-18, and IFN signaling through the Jak/Stat/ISGF3 pathway.
IFN-α/β molecules bind to a common IFN-α/β receptor, and this initiates a series of tyrosine phosphorylation events, including activation of the Janus tyrosine kinases Jak1 and Tyk2, which are found associated with the IFN-α/β receptor. Activation of Jak1 and Tyk2 results in the tyrosine phosphorylation of Stat1 and Stat2 and in the formation of IFN-stimulated gene factor 3 (ISGF3) composed of Stat1, Stat2, and p48. Nuclear translocation of ISGF3 and its subsequent binding to IFN-stimulated response elements (ISRE) leads to the activation of a variety of IFN-stimulated genes (ISG) which control the biological activity of IFN-α/β (reviewed in reference 37).
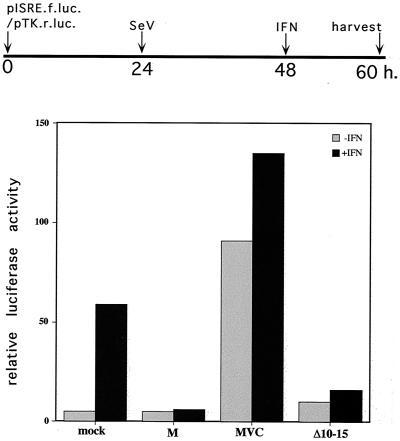
As shown previously (17), wild-type SeVM infection of IFN-competent BF cells that have been transfected with pISRE-luc (a luciferase reporter plasmid containing a basal promoter with a tandemized ISRE) (24, 25) induces only low levels of luciferase (Fig. 3). SeVM infection, moreover, also inhibits IFN signaling to pISRE-luc (+IFN, Fig. 3). Infection with SeVMVC, in contrast, induces high levels of luciferase even without IFN treatment, since SeVMVC infection induces strong IFN production without being able to interfere with its action (17). Infection with rSeV-[CΔ10-15/PΔ13-18] is almost as efficient as wild-type SeVM in suppressing IFN-induced luciferase activity (Fig. 3). Identical results were obtained by expressing CΔ10-15 independent of SeV infection in a transfected-cell assay (data not shown). These results are consistent with the previous demonstration that the Y1 and Y2 proteins are as effective as the longer C proteins at interdicting IFN signaling to pISRE-luc, either when expressed independent of, or within an rSeV infection (17, 18).
FIG. 3.
IFN signaling to pISRE-luciferase. BF cells were transfected with pISRE-(f)luc and pTK-(r)luc and then infected (or not) with 20 PFU of various rSeV types per cell as indicated on the x axis. Some of the cultures were treated with 1,000 U of IFN-α per ml at 48 h posttransfection. All of the cultures were harvested at 60 h, and the relative levels of Renilla and firefly luciferase activities were determined. A timeline of the experiment is shown at the top of the figure.
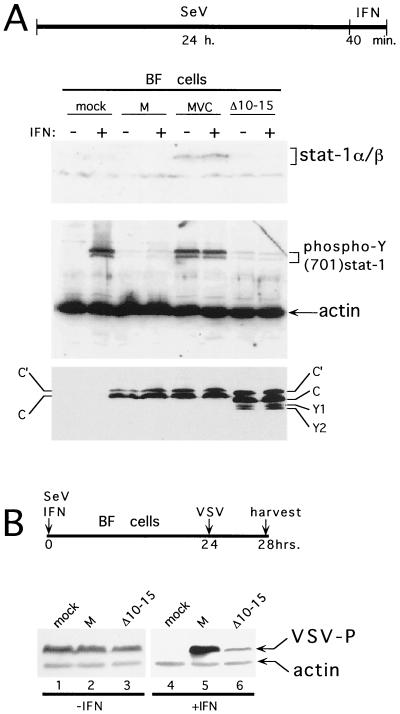
IFN-α signaling through the Jak/Stat pathway depends on the critical phosphorylation of Y701 of Stat1 (37) and SeV infection of HeLa cells prevents IFN-α-induced formation of pY701-Stat1 (19). To determine whether this block was due to the action of the C gene, we infected BF cells with SeV carrying either the wild-type CM, mutant CMVC (F170S), or CΔ10-15 genes. At 24 h postinfection (hpi), the cultures were treated (or not) with IFN and harvested 40 min later, and the relative levels of pY701-Stat1 present in whole-cell extracts were determined by immunoblotting. As shown in Fig. 4, 24 h of SeVMVC infection induces almost as much pY701-Stat1 as a 40-min treatment of mock-infected cells with 1,000 U of IFN-α. SeVM infection, in contrast, induces only barely detectable levels of pY701-Stat1 and, furthermore, prevents IFN treatment from inducing pY701-Stat1. Thus, the SeV block on IFN-induced pY701-Stat1 formation is indeed due to the action of its C gene, and the interdiction of IFN signaling to pISRE-luc can be explained, at least in large part, by its action at or upstream of pY701-Stat1 formation in this signaling pathway. rSeV-[CΔ10-15] infection was found to be as effective as that of wild-type rSeVM in preventing IFN-induced pY701-Stat1 formation. Moreover, identical results were obtained when pS727-Stat1 formation was examined (data not shown). Note that the two M strain viruses equally express C′ and C, but undetectable amounts of Y1 or Y2, the ribosomal shunt appears to be inoperative here (Fig. 4). Thus, the differences between SeVM and SeVMVC infections are not due to differences in the type and amount of C proteins. rSeV-CΔ10-15 (in the Z strain background), on the other hand, expresses significant amounts of all four C proteins, as did SeVZ-wt. However, the total amount of C proteins accumulated during these three SeV infections is roughly similar. Thus, interdiction of IFN signaling to pISRE-luc, including pY701-Stat1 and pS727-Stat1 formation, does not appear to require C1–23, but only a functional Y protein.
FIG. 4.
IFN induced formation of pY701-Stat1 and the antiviral (VSV) state. (A) BF cells were infected (or not) with 20 PFU of the various rSeVs per cell as indicated above and treated (or not) with 1,000 U of IFN-α per ml at 24 hpi, as indicated. Total cell extracts were prepared 40 min later, separated by SDS-PAGE, and immunoblotted with anti-Stat1 (top), anti-pY701-Stat1 and anti-actin (middle), and anti-C (bottom). A timeline of the experiment is shown at the top of the figure. (B) BF cells were infected (or not) with 20 PFU of the various rSeVs per cell as indicated above and treated (or not) with 1,000 U of IFN-α per ml at time zero, as indicated. All of the cultures were infected with 50 PFU of VSV per cell at 24 hpi. The cells were harvested at 28 hpi, and the levels of VSV P protein in cytoplasmic extracts were determined by immunoblotting. An anti-actin antibody was included as a loading control. A timeline of the experiment is shown above.
Remarkably, even though rSeV-[CΔ10-15] prevents IFN-induced pY701-Stat1 formation and activation of pISRE-luc, this virus is largely unable to prevent the IFN-induced anti-VSV state. As shown in Fig. 4B, BF cells are refractory to VSV replication upon 24 h of IFN treatment, but VSV replicates well in these cells if they are simultaneously infected with SeVM, as was previously found (17). However, if these IFN-treated cells are simultaneously infected with rSeV-[CΔ10-15], VSV replication is still largely restricted.
Cytopathic effects of rSeV.
SeVM infections induce programmed cell death (PCD) poorly either in LLC-MK2 cells or the mouse respiratory tract, whereas SeVMVC infections are highly cytopathic in both cases (21). Although SeVM and SeVMVC differ by two amino acids substitutions (CF170S and LE2050A [22]), further work with rSeVZ-CM and rSeVZ-CMVC has indicated that this difference in the cellular response to the infection is, at least in part, another property of the C gene (data not shown).
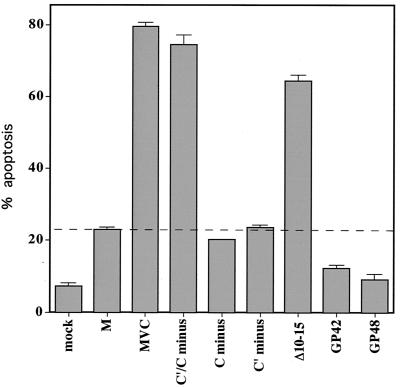
We have examined a panel of rSeV mutants to determine whether their infections are highly cytopathic (like SeVMVC) or relatively noncytopathic (like SeVM), by examining Annexin V staining (apoptosis) of rSeV-infected HeLa cells, one indicator of PCD (Fig. 5). Infection with rSeV that cannot specifically express either of the longer C proteins individually (rSeV-[C′-minus] or rSeV-[C-minus]) are as poorly cytopathic as SeVM, whereas rSeV-[C′/C-minus], the double mutant, is as cytopathic as SeVMVC. Thus, the two longer C proteins (but not Y1 and Y2) appear to actively suppress virus-induced apoptosis. This property specifically requires C10–15, since rSeV-CΔ10-15 is also highly apoptogenic. The NH2-terminal extensions of the C protein and, in particular, the highly basic residues C10–15, are thus critical in preventing or delaying at least one aspect of virus-induced cytopathic effects. In contrast, “copyback” rSeV that express wild-type C genes but that do not contain C-sensitive promoters (GP42 and GP48) are even less cytopathic than SeVM, as has been previously noted (16).
FIG. 5.
SeV-induced programmed cell death. HeLa cells were infected (or not) with 20 PFU of the various SeVs per ml as indicated below. At 48 hpi, the cells were stained with Annexin V and examined by fluorescence-activated cell sorting. The average percentage of cells considered apoptotic (from duplicate infections) is shown.
Antiviral state.
We have recently described a 3T3 MEF cell line which appears to be in an antiviral (VSV) state without IFN treatment, and this antiviral (VSV) state is associated with high basal Stat1 levels (18). When these MEFs are infected with SeV containing wild-type C genes (e.g., rSeVZ-CM, listed as “M” in lanes 3 and 4 in Fig. 6), Stat1 levels are strongly decreased, and VSV can now replicate in these cells. Moreover, exogenous IFN treatment of the cells does not decrease the level of rSeVZ-CM replication (lower panel, lane 4). When these MEFs are infected with rSeV-[CΔ10-15], they behave like those infected with rSeVZ-CMVC, namely, the Stat1 levels are not decreased and VSV replication continues to be largely restricted. Moreover, intracellular replication of rSeV-[CΔ10-15] is clearly reduced by IFN treatment, like that of rSeVZ-CMVC (lower panel). These results are similar to those obtained with rSeVZ-[C′/C-minus], whose infection is also unable to prevent the IFN-induced anti-VSV state (17). The entire CAUG114 protein and, in particular, the highly basic residues C10–15 are thus required to effectively counter the IFN-induced antiviral (VSV) state.
DISCUSSION
The SeV C proteins consist of a nested set of two longer (C′ and C) and two shorter (Y1 and Y2) proteins, which carry out multiple functions. We have investigated three (or more) of these functions, including (i) inhibiting or modulating vRNA synthesis, (ii) interdicting IFN signaling through the Jak/Stat/ISGF3 pathway, and (iii) a group of events that may be linked, namely, reducing Stat1 levels, preventing the IFN-induced antiviral (VSV) state, preventing programmed cell death (including apoptosis), and the enigmatic property of preventing rSeV recovery in the VV-T7 system. Our results indicate that the nested set of C proteins carry out a nested set of these functions, affecting both viral replication and the cellular antiviral response (Fig. 7). Because of these effects, rSeV strains which express only the shorter Y1 and Y2 proteins are highly debilitated, and those which do not express any of the four C proteins are even further debilitated (they are at the limit of rSeV recovery [28, 30]). Here, we report the properties of rSeV containing a deletion of a short, but notable, region of the C (and overlapping P) gene. rSeV-[CΔ10-15] is also highly debilitated (avirulent in mice and outgrown by rSeVwt in mixed infections), underscoring the importance of the first 23 residues of CAUG114 in virus replication. rSeV that cannot express the V or W proteins (two other accessory proteins of the P gene; Fig. 1), in contrast, replicates well in cell culture (10, 23).
The shorter Y proteins appear to be both necessary and sufficient to interdict IFN signaling to pISRE-luc (Fig. 3) or to prevent the critical formation of pY701-Stat1 required for this IFN signaling (Fig. 4). The interdiction of IFN signaling through the Jak/Stat/ISGF3 pathway is therefore one function that can be carried out by all of the C proteins (18). Since SeV infection prevents IFN-induced pY701-Stat1 formation within 2 hpi (26), this block on IFN signaling is a very early effect of the C gene, which occurs when very little of this essentially nonstructural protein is present intracellularly. Large amounts of the C proteins accumulate during infection, and these gene products also appear to be responsible for the dramatic turnover of Stat1 that occurs upon infection of some cells (Fig. 6). This effect on Stat1 stability, however, requires C10–15, and it is therefore unlikely to be due to the same C interaction that suppresses pY701-Stat1 formation (Fig. 7).
The C proteins also inhibit vRNA synthesis in a promoter-specific manner but, in contrast to their interdiction of the Jak/Stat/ISGF3 pathway, the shorter Y proteins are unable to carry out this task. This suggested that a domain within C1–23 was specifically required for this inhibition, but we were unable to map such a domain by serial deletions. The role of C1–23 in the inhibition of viral RNA synthesis is thus unclear. C1–23, however, is likely to play a specific role in SeV infection, because several properties of this gene (its ability to reduce Stat1 levels, to prevent the SeV infection from inducing a cellular antiviral state, or PCD, or to prevent rSeV recovery in the VV-T7 system [Fig. 7]) are all inactivated by the deletion of C10–15. Leaving aside the enigmatic rSeV recovery phenomenon, the remaining three properties may very well be linked. The IFN-induced antiviral state and PCD are complex cellular responses that integrate signals from different pathways, and Stat1 is prominent in both cellular responses. Eliminating Stat1 could act to prevent PCD (4, 27, 35), and there may be elements of IFN action that require basal levels of Stat1, independent of their phosphorylation status (3). If so, these three properties could be due to a common interaction of CAUG114 (but not Y1 or Y2) and Stat1, along with other cellular components.
The Y proteins appear to be as active as the longer C proteins in interdicting IFN signaling through the Jak/Stat pathway in both transfected cells and rSeV-infected cells (17, 18). We were therefore surprised to find that Y1 or Y2 expression alone, in the context of rSeV-[C′/C-minus] or rSeV-[CΔ10-15] infection, was unable to prevent IFN from inducing an effective anti-VSV state. We had assumed that the interdiction of this now well-established IFN signaling pathway would have been sufficient to prevent IFN action, but this seems not to be so in IFN-competent BF cells and MEFs. The IFN-induced antiviral state is known to be remarkably complex (37), and interdicting IFN signaling to ISRE-promoted ISGs is clearly not the same as preventing IFN from inducing an anti-VSV state. There is increasing evidence that while Jak/Stat pathways are essential, they are not necessarily sufficient for all aspects of the IFN-induced response. There may be another parallel pathway through which IFN acts to induce an antiviral (VSV) state, and which could require basal levels of Stat1 (independent of their phosphorylation). For example, Novick et al. (32) have recently described monoclonal antibodies to the IFNAR2 chain that neutralize IFN-α action but that neither prevent IFN-α from binding to its receptor, Jak/Stat tyrosine phosphorylation, or ISGF3 complex formation. It is perhaps this pathway that is targeted by the longer C proteins.
REFERENCES
- 1.Cadd T, Garcin D, Tapparel C, Itoh M, Homma M, Roux L, Curran J, Kolakofsky D. The Sendai paramyxovirus accessory C proteins inhibit viral genome amplification in a promoter-specific fashion. J Virol. 1996;70:5067–5074. doi: 10.1128/jvi.70.8.5067-5074.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calain P, Roux L. The rule of six: a basic feature for efficient replication of Sendai virus defective interfering RNA. J Virol. 1993;67:4822–4830. doi: 10.1128/jvi.67.8.4822-4830.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chatterjee-Kishore M, Wright K L, Ting J P, Stark G R. How Stat1 mediates constitutive gene expression: a complex of unphosphorylated Stat1 and IRF1 supports transcription of the LMP2 gene. EMBO J. 2000;19:4111–4122. doi: 10.1093/emboj/19.15.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chin Y E, Kitagawa M, Kuida K, Flavell R A, Fu X-Y. Activation of the STAT signaling pathway can cause expression of caspase 1 and apoptosis. Mol Cell Biol. 1997;17:5328–5337. doi: 10.1128/mcb.17.9.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Curran J, Marq J-B, Kolakofsky D. The Sendai virus nonstructural C proteins specifically inhibit viral mRNA synthesis. Virology. 1992;189:647–656. doi: 10.1016/0042-6822(92)90588-g. [DOI] [PubMed] [Google Scholar]
- 6.Curran J, Pelet T, Kolakofsky D. An acidic activation-like domain of the Sendai virus P protein is required for RNA synthesis and encapsidation. Virology. 1994;202:875–884. doi: 10.1006/viro.1994.1409. [DOI] [PubMed] [Google Scholar]
- 7.Curran J, Marq J-B, Kolakofsky D. An N-terminal domain of the Sendai virus P protein acts as a chaperone for NP protein during the nascent chain assembly step of genome replication. J Virol. 1995;69:849–855. doi: 10.1128/jvi.69.2.849-855.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Curran J. Reexamination of the Sendai virus P protein domains required for RNA synthesis: a possible supplemental role for the P protein. Virology. 1996;221:130–140. doi: 10.1006/viro.1996.0359. [DOI] [PubMed] [Google Scholar]
- 9.Curran J, Latorre P, Kolakofsky D. Translational gymnastics on the Sendai virus P/C mRNA. Semin Virol. 1998;8:351–357. [Google Scholar]
- 10.Delenda C, Hausmann S, Garcin D, Kolakofsky D. Normal cellular replication of Sendai without the trans-frame, nonstructural V protein. Virology. 1997;228:55–62. doi: 10.1006/viro.1996.8354. [DOI] [PubMed] [Google Scholar]
- 11.Denzler K L, Jacobs B L. Site-directed mutagenic analysis of reovirus sigma 3 protein binding to dsRNA. Virology. 1994;204:190–199. doi: 10.1006/viro.1994.1523. [DOI] [PubMed] [Google Scholar]
- 12.Didcock L, Young D F, Goodbourn S, Randall R E. The V protein of simian virus 5 inhibits interferon signaling by targeting STAT1 for proteasome-mediated degradation. J Virol. 1999;73:9928–9933. doi: 10.1128/jvi.73.12.9928-9933.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Didcock L, Young D F, Goodbourn S, Randall R E. Sendai virus and simian virus 5 block activation of interferon-responsive genes: Importance for virus pathogenesis. J Virol. 1999;73:3125–3133. doi: 10.1128/jvi.73.4.3125-3133.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcin D, Pelet T, Calain P, Roux L, Curran J, Kolakofsky D. A highly recombinogenic system for the recovery of infectious Sendai paramyxovirus from cDNA: generation of a novel copy-back nondefective interfering virus. EMBO J. 1995;14:6087–6094. doi: 10.1002/j.1460-2075.1995.tb00299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcin D, Itoh M, Kolakofsky D. A point mutation in the Sendai virus accessory C proteins attenuates virulence for mice, but not virus growth in cell culture. Virology. 1997;238:424–431. doi: 10.1006/viro.1997.8836. [DOI] [PubMed] [Google Scholar]
- 16.Garcin D, Taylor G, Tanebayashi K, Compans R, Kolakofsky D. The short Sendai virus leader region controls induction of programmed cell death. Virology. 1998;243:340–353. doi: 10.1006/viro.1998.9063. [DOI] [PubMed] [Google Scholar]
- 17.Garcin D, Latorre P, Kolakofsky D. Sendai virus C proteins counteract the interferon-mediated induction of an antiviral state. J Virol. 1999;73:6559–6565. doi: 10.1128/jvi.73.8.6559-6565.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcin D, Curran J, Kolakofsky D. Sendai virus C proteins must interact directly with cellular components to interfere with interferon action. J Virol. 2000;74:8823–8830. doi: 10.1128/jvi.74.19.8823-8830.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gotoh B, Takeuchi K, Komatsu T, Yokoo J, Kimura Y, Kurotani A, Kato A, Nagai Y. Knockout of the Sendai virus C gene eliminates the viral ability to prevent the interferon-alpha/beta-mediated responses. FEBS Lett. 1999;459:205–210. doi: 10.1016/s0014-5793(99)01241-7. [DOI] [PubMed] [Google Scholar]
- 20.Hasan M K, Kato A, Muranaka M, Yamaguchi R, Sakai Y, Hatano I, Tashiro M, Nagai Y. Versatility of the accessory C proteins of Sendai virus: contribution to virus assembly as an additional role. J Virol. 2000;74:5619–5628. doi: 10.1128/jvi.74.12.5619-5628.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Itoh M, Hotta H, Homma M. Increased induction of apoptosis by a Sendai virus mutant is associated with attenuation of mouse pathogenicity. J Virol. 1998;72:2927–2934. doi: 10.1128/jvi.72.4.2927-2934.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Itoh M, Isegawa Y, Hotta H, Homma M. Isolation of an avirulent mutant of Sendai virus with two amino acid mutations from a highly virulent field strain through adaptation to LLC-MK2 cells. J Gen Virol. 1997;78:3207–3215. doi: 10.1099/0022-1317-78-12-3207. [DOI] [PubMed] [Google Scholar]
- 23.Kato A, Kiyotani K, Sakai Y, Yoshida T, Nagai Y. The paramyxovirus Sendai virus V protein encodes a luxury function required for viral pathogenesis. EMBO J. 1997;16:678–587. doi: 10.1093/emboj/16.3.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.King P, Goodbourn S. The β-interferon promoter responds to priming through multiple independent regulatory elements. J Biol Chem. 1994;269:30609–30615. [PubMed] [Google Scholar]
- 25.King P, Goodbourn S. STAT1 is inactivated by a caspase. J Biol Chem. 1998;273:8699–8704. doi: 10.1074/jbc.273.15.8699. [DOI] [PubMed] [Google Scholar]
- 26.Komatsu T, Takeuchi K, Yokoo J, Tanaka Y, Gotoh B. Sendai virus blocks alpha interferon signaling to signal transducers and activators of transcription. J Virol. 2000;74:2477–2480. doi: 10.1128/jvi.74.5.2477-2480.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar A, Commane M, Flickinger T W, Horvath C M, Stark G R. Defective TNFα-induced apoptosis in STAT1-null cells due to low constitutive levels of caspases. Science. 1997;278:1630–1632. doi: 10.1126/science.278.5343.1630. [DOI] [PubMed] [Google Scholar]
- 28.Kurotani A, Kiyotani K, Kato A, Shioda T, Sakai Y, Mizumoto K, Yoshida T, Nagai Y. Sendai virus C proteins are categorically nonessential gene products but silencing their expression severely impairs viral replication and pathogenesis. Genes Cells. 1998;3:111–124. doi: 10.1046/j.1365-2443.1998.00170.x. [DOI] [PubMed] [Google Scholar]
- 29.Lamb R A, Mahy B W, Choppin P W. The synthesis of Sendai virus polypeptides in infected cells. Virology. 1996;69:116–131. doi: 10.1016/0042-6822(76)90199-9. [DOI] [PubMed] [Google Scholar]
- 30.Latorre P, Cadd T, Itoh M, Curran J, Kolakofsky D. The various Sendai virus C proteins are not functionally equivalent and exert both positive and negative effects on viral RNA accumulation during the course of infection. J Virol. 1998;72:5984–5993. doi: 10.1128/jvi.72.7.5984-5993.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Latorre P, Kolakofsky D, Curran J. Sendai virus Y proteins are initiated by a ribosomal shunt. Mol Cell Biol. 1998;18:5021–5031. doi: 10.1128/mcb.18.9.5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Novick D, Nabioullin R R, Ragsdale W, McKenna S, Weiser W, Garone L, Burkins C, Kim S H, Rubinstein M, Tepper M A, Arulanandam A R. The neutralization of type I IFN biologic actions by anti-IFNAR-2 monoclonal antibodies is not entirely due to inhibition of jak-stat tyrosine phosphorylation. J Interferon Cytokine Res. 2000;20:971–982. doi: 10.1089/10799900050198417. [DOI] [PubMed] [Google Scholar]
- 33.Orvell C, Grandien M. The effects of monoclonal antibodies on biological activities of structural proteins of Sendai virus. J Immunol. 1982;129:2779–2787. [PubMed] [Google Scholar]
- 34.Pelet T, Delenda C, Gubbay O, Garcin D, Kolakofsky D. Partial characterization of a Sendai virus replication promoter and the rule of six. Virology. 1996;224:405–414. doi: 10.1006/viro.1996.0547. [DOI] [PubMed] [Google Scholar]
- 35.Schindler C. STATs as activators of apoptosis. Trends Cell Biol. 1998;8:97–98. doi: 10.1016/s0962-8924(98)01233-1. [DOI] [PubMed] [Google Scholar]
- 36.Simon J H, Gaddis N C, Fouchier R A, Malim M H. Evidence for a newly discovered cellular anti-HIV-1 phenotype. Nat Med. 1998;4:1397–400. doi: 10.1038/3987. [DOI] [PubMed] [Google Scholar]
- 37.Stark G R, Kerr I M, Williams B R, Silverman R H, Schreiber R D. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 38.Tapparel C, Hausmann S, Pelet T, Curran J, Kolakofsky D, Roux L. Inhibition of Sendai virus genome replication due to promoter-increased selectivity: a possible role for the accessory C proteins. J Virol. 1997;71:9588–9599. doi: 10.1128/jvi.71.12.9588-9599.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomas S M, Lamb R A, Paterson R G. Two mRNAs that differ by two nontemplated nucleotides encode the amino coterminal proteins P and V of the parmyxovirus SV5. Cell. 1988;54:891–892. doi: 10.1016/S0092-8674(88)91285-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trono, D. When accessories turn out to be essential. Nat. Med. 4:1368–1369. [DOI] [PubMed]
- 41.Vidal S, Curran J, Kolakofsky D. A stuttering model for paramyxovirus P mRNA editing. EMBO J. 1990;9:2017–2022. doi: 10.1002/j.1460-2075.1990.tb08330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weber H, Valenzuela D, Luijber G, Gubler M, Weissmann C. Single amino acid changes that render human IFN-alpha 2 biologically active on mouse cells. EMBO J. 1987;6:591–598. doi: 10.1002/j.1460-2075.1987.tb04795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamada H, Hayata T, Omata-Yamada T, Taira H, Mizumoto K, Iwasaki K. Association of the Sendai virus C protein with nucleocapsids. Arch Virol. 1990;113:245–253. doi: 10.1007/BF01316677. [DOI] [PubMed] [Google Scholar]
- 44.Yang Y L, Reis L F, Pavlovic J, Aguzzi A, Schafer R, Kumar A, Williams B R, Aguet M, Weissmann C. Deficient signaling in mice devoid of double-stranded RNA-dependent protein kinase. EMBO J. 1995;14:6995–7006. doi: 10.1002/j.1460-2075.1995.tb00300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]