Abstract
Background
Genetic variants in COL4A2 are less common than those of COL4A1 and their fetal clinical phenotype has not been well described to date. We present a fetus from China with an intronic variant in COL4A2 associated with a prenatal diagnosis of severe cerebral encephalomalacia and subdural hemorrhage.
Methods
Whole exome sequencing (WES) was applied to screen potential genetic causes. Bioinformatic analysis was performed to predict the pathogenicity of the variant. In in vitro experiment, the minigene assays were performed to assess the variant’s effect.
Results
In this proband, we observed ventriculomegaly, subdural hemorrhage, and extensive encephalomalacia that initially suggested cerebral hypoxic-ischemic and/or hemorrhagic lesions. WES identified a de novo heterozygous variant c.549 + 5G > A in COL4A2 gene. This novel variant leads to the skipping of exon 8, which induces the loss of 24 native amino acids, resulting in a shortened COL4A2 protein (p.Pro161_Gly184del).
Conclusion
Our study demonstrated that c.549 + 5G > A in COL4A2 gene is a disease-causing variant by aberrant splicing. This finding enriches the variant spectrum of COL4A2 gene, which not only improves the understanding of the fetal neurological disorders associated with hypoxic-ischemic and hemorrhagic lesions from a clinical perspective but also provides guidance on genetic diagnosis and counseling.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12920-024-02012-4.
Keywords: COL4A2, Heterozygous intronic variant, Minigene splicing assays, Encephalomalacia
Introduction
Collagen IV, a major component of basement membranes and extracellular matrix, comprises six alpha chains encoded by distinct and tandemly arranged genes. These genes include COL4A1 and COL4A2 (located on chromosomes 13q), COL4A3 and COL4A4 (located on chromosomes 2q), COL4A5 and COL4A6 (located on chromosomes xq), respectively. COL4A2 gene (Collagen Type IV Alpha 2 Chain) (OMIM:120090), encodes one of the six subunits of type IV collagen which composes a heterotrimeric helix with COL4A1 in a constant 1:2 ratio (α1ɑ1ɑ2). Variants in COL4A3 to COL4A5 were associated with benign familial hematuria and Alport syndrome [1–3], and COL4A6 variants were associated with hearing loss [4]. COL4A1 and COL4A2 are most ubiquitously expressed among these six genes and constitute the major structural component of basement membranes, they are transcribed from a shared, bidirectional promoter [5]. This explains why the variants in COL4A1/COL4A2 affect the stability and integrity of the vascular basement membranes and then cause a wide spectrum of disorders characterized by the presence of cerebrovascular diseases such as intracerebral hemorrhage, porencephaly, leukoencephalopathy, hydrocephalus, and developmental defects [6–9]. COL4A2 and COL4A1 pathogenic variants were considered the most frequent genetic cause of fetal intracranial hemorrhage [10]. The variants in COL4A1/COL4A2 increase the risk of germinal matrix hemorrhage in the last trimester or around birth [11]. The variation spectrum of COL4A1 is more extensive than that of COL4A2 to date. The phenotypic manifestation of COL4A1/COL4A2 variants can be quite different within and among families, marked by clinical heterogeneity and variable expressivity [12–14]. The onset can range from early fetal development to the postnatal period, even in late adulthood [13].
Generally, individuals diagnosed with a gene-related disorder may have been affected by parents, in a study including 194 fetuses with intracerebral hemorrhage fetuses identified 35 variants in COL4A1/COL4A2 gene and reported that 70% of probands carried de novo variant [10]. These data suggested that COL4A1/COL4A2 de novo variants were in a high frequency. Among the reported variants, missense mutations were the most and other variants such as deletion mutations, repetitive mutations, and splice mutations had previously been reported [15–18]. There are few reports of splice variants and the functional characterizations of splice–junction variants are rarely explained [19].
In this study, we observed severe encephalomalacia and subdural hemorrhage in a fetus from China, tests for abnormalities in coagulation, metabolism, and infectious diseases ruled out other potential sources of intracranial hemorrhage. A rare, previously undescribed COL4A2 intronic variant c.549 + 5G > A was discovered. The genetic and molecular characterization of COL4A2 spliceogenic variants was corroborated by detection through in silico algorithm, which lead to the skipping of exon 8 (c.478_549del) in COL4A2 gene, this misspliced transcript did not create a downstream frameshift or a premature termination codon in the COL4A2 mRNA sequence, only leading to the loss of 24aa inside the protein (p.Pro161_Gly184del). This discovery broadens the range of COL4A2 gene variants, which not only advances clinical knowledge of the fetal neurological disorders associated with hypoxic-ischemic and hemorrhagic lesions but also provides guidance on genetic diagnosis and counseling.
Materials and methods
Participants
The proband was a fetus with ventriculomegaly, subdural hemorrhage, and extensive encephalomalacia. He was admitted to the neonatal intensive care unit (NICU) after birth. He did not have any siblings and no relevant family history was recorded. Symptoms such as physical dysfunction, mental retardation and language disorder are important in family history. We have excluded the above symptoms of the proband’s parents, whose phenotype is normal.
We further performed a physical examination, ultrasonic examinations, and brain magnetic resonance imaging (MRI), and measured the biochemical parameters of the proband and whole exome sequencing was further performed in this family.
Ethical compliance and consent to Participate
This study was approved by the ethics committee of The First Affiliated Hospital of Wenzhou Medical University. Informed consent was obtained before commencing the study.
Whole exome sequencing (WES) and Sanger sequencing
Genomic DNA was extracted, purified, fragmented into random segments, and then captured using the Agilent Sure Select Human All Exome V6 Kit (Agilent Technologies, USA). WES was performed by in-solution hybridization followed by high-throughput paired-end sequencing on the NovaSeq 6000 platform (Illumina, Inc., San Diego, CA, USA), with a reading length of 150 bp. Sequence alignment was performed, then annotated and filtered gene variants. The main reference databases we used in this study included population (dbSNP, 1000G, and gnomAD) and disease databases (HGMD, OMIM, UCSC, ClinVar, and DECIPHER). Sanger sequencing was performed to validate suspected variants. Primers were designed to amplify exons and intron boundaries using polymerase chain reaction (PCR). The products were sequenced directly using a 3500xL Genetic Analyzer (Applied Biosystems, Waltham, MA, USA).
Copy number variation (CNV) detection
Copy number variation (CNV) is caused by genome rearrangement, generally referring to an increase or decrease in the copy number of large fragments of the genome with a length of 50 bp or more, mainly manifested as deletion and duplication at the submicroscopic level. The peripheral blood of the proband was collected. Add EDTA and Tiangen stabilizer to the sample. The sample was sent to Beijing Zhiyin Oriental Translational Medicine Research Center, CNV-seq was performed to detect CNV. CNV-seq is the most comprehensive CNV information obtained through whole genome sequencing, which can compensate for the shortcomings of whole exome sequencing in detecting structural variations, the detection accuracy of CNVs greater than 100k can reach 99%. The detection of DNA copy number variation (CNV) in our case was negative.
In silico assay
The splice mutation site was analyzed with Human Splicing Finder (HSF) version 3.0 (https://www.umd.be/HSF/), varSEAK (https://varseak.bio/), SpliceAI (https://spliceailookup.broadinstitute.org/), and RDCCSC (https://rddc.tsinghua-gd.org/). The protein domains of COL4A2 were interpreted using the online tool InterProScan (http://www.ebi.ac.uk/interpro/).
Minigene constructs and mutagenesis
In vitro minigene assay was carried out to examine the target gene regions covering COL4A2 exon 7–9, and intron 7 and 8, which were amplified from the cDNA of the proband (Fig. 4a). Using genomic DNA as a template, the wt COL4A2 gene fragment was obtained by nested PCR with 120,011-COL4A2-F/123,097-COL4A2-R and 120,334-COL4A2-F/122,754-COL4A2-R (Table 1) as primers. Three pairs of primers (pcDNA3.1-COL4A2-KpnI-F/COL4A2-mut-R, COL4A2-mut-F/pcDNA3.1-COL4A2-EcoRI-R, and pcDNA3.1-COL4A2-KpnI-F/pcDNA3.1-COL4A2-EcoRI-R; Table 1) were developed to amplify the heterozygous c.549 + 5G > A variant site from the product of nested PCR by seamless cloning (Vazyme Biotech Co., Ltd., Nanjing, China). The amplified DNA products were then recombined and cloned into the two digestion sites (HIKpnI/EcoRI) of the pcDNA3.1 vector (Hitrobio Biotechnology Co., Ltd., Beijing, China). Furthermore, the recombinant plasmids pcDNA3.1-COL4A2-wt (wild-type) and pcDNA3.1-COL4A2-mut (c.549 + 5G > A) were validated by Sanger sequencing. To substantiate the conclusion drawn in this study, the assay was conducted repeatedly in another pcMINI-N vector using the same experimental method described above.
Table 1.
Primer sequences used to analyze the variant of the COL4A2 gene and vector pcDNA3.1
| Primer name | Primer sequence (5` − 3`) |
|---|---|
| 120,011-COL4A2-F | CGTTGCGGGTGTGTCTTCTC |
| 120,334-COL4A2-F | CCCAGGTGAAGCAGAACTTT |
| 122,754-COL4A2-R | GGATGACCGGACGATGGTCC |
| 123,097-COL4A2-R | ATATGCCTGCTAGCATGGGT |
| pcDNA3.1-COL4A2-KpnI-F | GCTTGGTACCATGGGACACCCGGGGCAAGGTGG |
| COL4A2-mut-F | ACAGATATCGGGTACATTTGCAAGAGATGGG |
| COL4A2-mut-R | CCCATCTCTTGCAAATGTACCCGATATCTGT |
| pcDNA3.1-COL4A2-EcoRI-R | TGCAGAATTCCTGGAAACCGACCAATCCAG |
| pcDNA3.1-F | CTAGAGAACCCACTGCTTAC |
| pcDNA3.1-R(BGH-R) | TAGAAGGCACAGTCGAGG |
Cell culture and transfection
Human embryonic kidney (HEK293) and HeLa cell lines (Cell Resource Center of the Chinese Academy of Medical Science, Beijing, China) were maintained at 37 °C in a humidified 5% CO2 environment in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. COL4A2-wt and variant cDNAs were inserted into the pcDNA3.1 vector and then transfected into HEK293T and HeLa cells using Lipo3000 Transfection Reagent (Glpbio Technology Inc, Montclair, CA, USA). The transfected cells were incubated for 48 h before RNA extraction.
RNA extraction, PCR, and sequencing
Total RNA from HEK293T and HeLa cells transfected with minigene plasmids was extracted using the GenEluteMammalian Total RNA Kit (Sigma–Aldrich), following the manufacturer’s recommendations. DNA was degraded on a column with DNase I (Qiagen, Valencia, CA, USA). Extracted total RNAs were reverse-transcribed into cDNA, and reverse transcription polymerase chain reaction (RT-PCR) was performed using the primer pair pcDNA3.1-F/pcDNA3.1-R (Table 1). Next, the cDNA products were examined by 1% agarose gel electrophoresis and further confirmed by Sanger sequencing. The length of wild-type cDNA products was determined to be 410 bp in the pcDNA3.1-COL4A2 vector and 392 bp in the pcMINI-N-COL4A2 vector.
Results
Clinical data
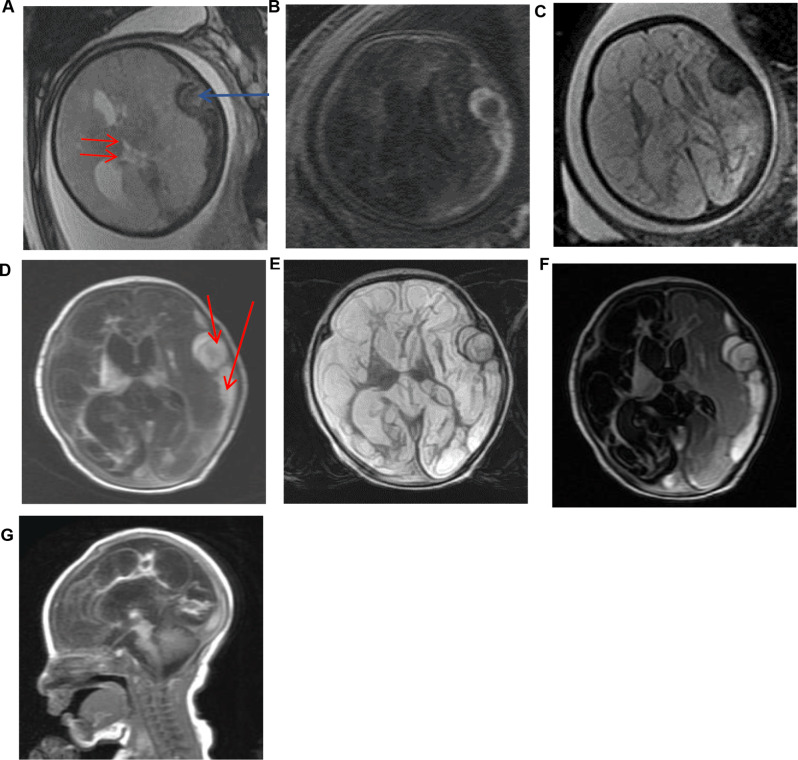
The proband was a boy and the first symptoms appeared prenatally. Ultrasound examination at 33 gestational weeks found a fusiform echogenic mass measuring 22 × 13 cm in the left side of the cerebrum close to the skull and suspected porencephalia. Fetal brain MRI at 34 weeks confirmed the left subdural hemorrhage and lateral displacement of the cerebrum (Fig. 1a). At 35 weeks, MRI displayed lateral ventricle dilation and diffuse abnormal intensities in cerebrum, suggesting extensive encephalomalacia (Fig. 1b and c).
Fig. 1.
Brain magnetic resonance imaging (MRI) of the proband. Fetal brain MRI performed at 34 weeks: (a) T2*-weighted axial image showing hypointensities (blue arrow) due to subdural hemorrhage in the left. Lateral displacement of the brain (red arrows). Brain MRI performed at 35 weeks: (b) T1-weighted axial image showing diffuse hypointensities in cerebrum parenchyma and left subdural hemorrhage. (c)T2-weighted axial image showing diffuse hypperintensities in cerebrum parenchyma and left subdural hemorrhage. Brain MRI on the Second day after birth: (d-g) Axial and sagittal images displaying extensive cerebrumencephalomalacia, dilation of lateral ventricle, and subdural hemorrhage in left
The proband was delivered vaginally at 37+ 6 weeks. At birth, the infant had a weight of 2,470 g (-1 SD), a length of 45 cm (-1 SD), and a head circumference of 32.8 cm (0 SD). Apgar scores at 1 and 5 min were 8 and 10, respectively. Postnatal imaging performed on day 2 revealed the left subdural hemorrhage and extensive encephalomalacia which implied an overall reduction of parenchyma in cerebrum. T2-Flair found that the intensity of the right cerebrum was equal to cerebrospinal fluid, suggesting a porencephalic lesion in the right part (Fig. 1d-g). After birth, the proband boy exhibited a weakness in sucking, necessitating nasal feeding. Approximately 25 h post-birth, he was administered phenobarbital to manage seizures. Subsequent follow-ups revealed significant developmental delay and cognitive impairment. By the age of 18 months, he was unable to balance his head, laugh, babble, turn over, or sit unassisted. Tragically, the patient passed away at the age of two. The parents exhibited no clinical or radiological abnormalities, indicating good health. During the process of obtaining the proband’s medical history, the family member reported an absence of any incidents of abdominal trauma during the pregnancy. And the family history did not present any noteworthy findings.
Variant detection and prediction
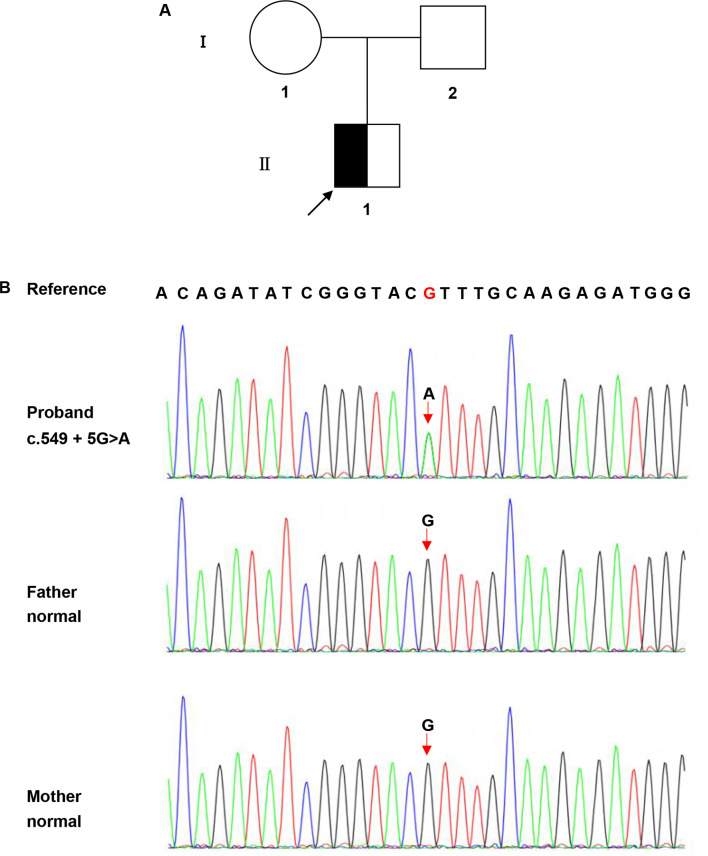
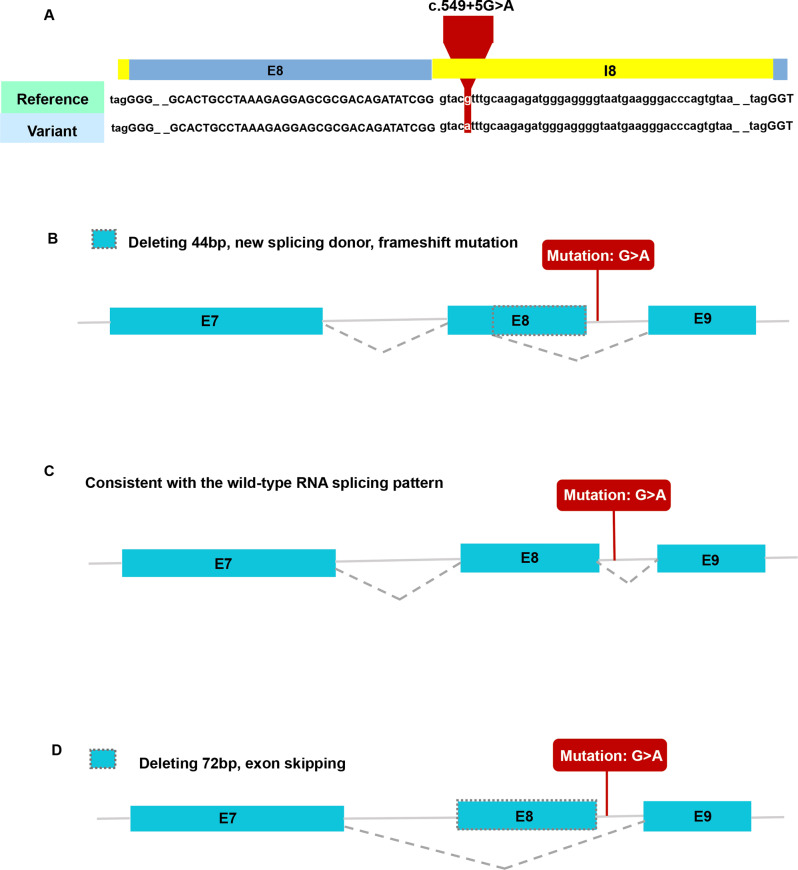
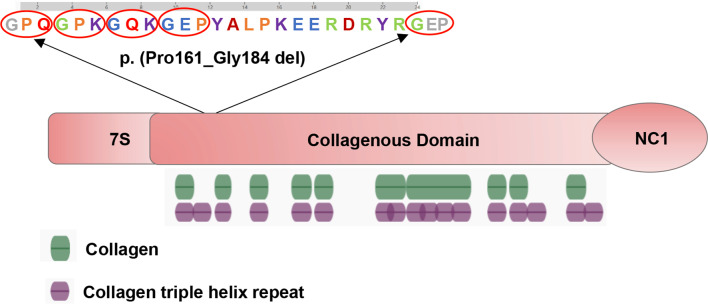
A novel heterozygous point variant in the intron 8 splice donor site of COL4A2 (c.549 + 5G > A; NM_001846.4) was identified in the proband (II-1; Fig. 2a) by PCR-based Sanger sequencing (Fig. 2b). Analysis of the DNA from his parents (I-1 and I-2) was normal, showing that the variant might be de novo in the proband (paternity/maternity testing was not performed). The results of the HSF indicated that this variant destroyed the original donor site and most probably affected splicing (Table 2). The results were validated using varSEAK (Fig. 3a). The Splice AI algorithm resulted in a high value (Δ score = 0.79) for c.549 + 5G > A. The meaning of the value is the probability of affecting splicing, the closer to 1 the greater the probability of affecting splicing. The variant was predicted to be pathogenic, with three patterns of RNA splicing prediction (Fig. 3b-d). Predicted protein domains showed the loss of 24aa (p.Pro161_ Gly184del) located in the collagenous domain including five (Gly-X-Y) repeats within the triple helical domain (Fig. 5). Therefore, all these predictions indicated that this variant affects mRNA splicing.
Fig. 2.
Pedigree of the studied family and the COL4A2 sequencing results. (a) Pedigree of the family. ◪ represents the affected male; □ represents the normal male; ○ represents the normal female. Arrow indicates the proband. (b) The sequencing result of the family. The rectangular symbol indicates the variant site. The mutation is might a de novo variant
Table 2.
Results of the prediction of the c.549 + 5G > A COL4A2 variant in HSF
| Mutation | HGVS | Predict Impact |
|---|---|---|
| 13 110,429,962 G > A | ENST00000360467.7: c.549 + 5G > A |
λ Broken WT Donor Site: Alteration of the WT Donor site, most probably affecting splicing (HSF) λ Broken WT Donor site: Alteration of the WT Donor site, most probably affecting splicing (MaxEnt) |
Fig. 3.
Results of in silico assay. (a) Result of the prediction in varSEAK online consists of these in silico assays by Human Splicing Finder (HSF). (b-d) Three patterns of RNA splicing prediction. b. Pattern one: deleting 44 bp, new splicing donor, frameshift mutation, premature termination. c. Pattern two: consistent with the wild-type RNA splicing pattern. d. Pattern three: deleting 72 bp, exon skipping. The minigene results showed that our variant of interest was consistent with pattern three. E7: exon 7; E8: exon 8; E9: exon 9; I8: intron 8
Minigene assay
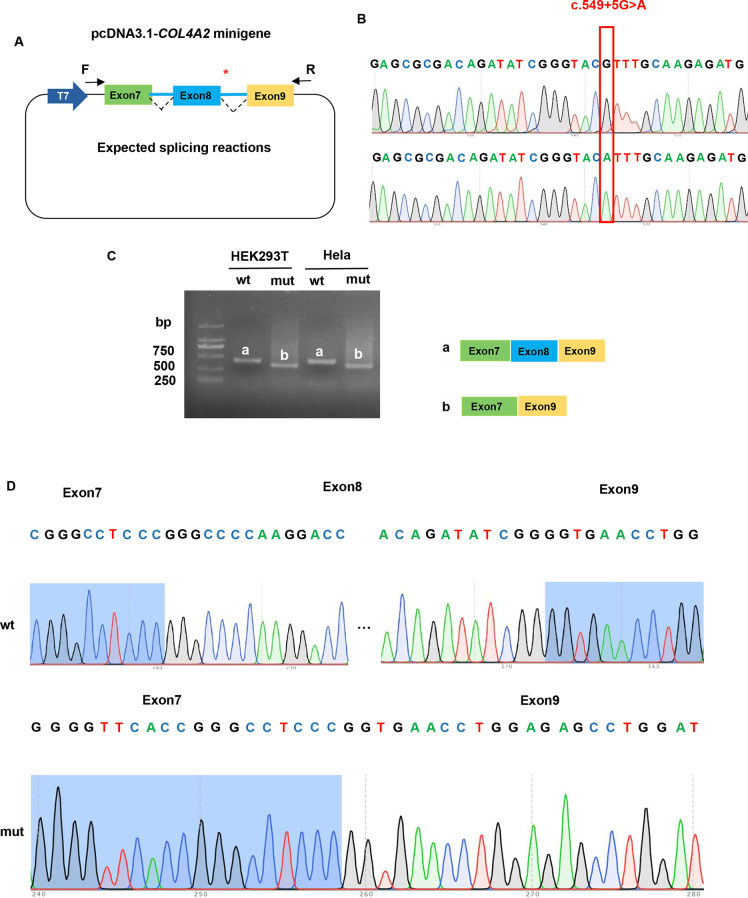
To analyze the effect of the c.549 + 5G > A variant (Fig. 4b) on COL4A2 mRNA splicing, an in vitro transcription assay was conducted. The pcDNA3.1-COL4A2-wt minigene was 1970 bp long and covered DNA regions including exon 7 (117 bp), intron 7 (1301 bp), exon 8 (72 bp), intron 8 (444 bp), and exon 9 (36 bp; Fig. 4a). A full-length 410 bp RT-PCR product of the expected normal size (partial plasmid sequence 185 bp and target gene 225 bp) was detected in HEK293T and HeLa cells transfected with pcDNA3.1-COL4A2-wt minigene as the most abundant transcript (Fig. 4c). Sanger sequencing confirmed that the 410-bp band corresponded to normal COL4A2 mRNA, with exon 7-8-9 ligated and intervening intron 7–8 removed (Fig. 4d). The minigene construct containing the c.549 + 5G > A variant in the pcDNA3.1-COL4A2-mut minigene was transfected into HeLa and HEK293T cell lines. In sharp contrast, normal COL4A2 mRNA was not detected in the mutant (mut) lanes, except for one smaller band (338 bp; Fig. 4c). In the mut lanes, the shorter band of 338-bp corresponds to a misspliced transcript of COL4A2 with complete skipping of exon 8; it which only included exons 7 and 9. The Sanger DNA chromatogram of the misspliced transcript of COL4A2 is shown in Fig. 4d. We also used the pcMINI-N vector to confirm these results, which contains the general MCS-Intron B-Exon B by the same experimental procedure. Full-length sequence translation analysis of misspliced transcripts identified by minigene analyses indicated that this variant induces complete skipping of exon 8 including 72 bp nucleotides (c.478_549del), resulting in the loss of 24aa inside the shorter protein (p.Pro161_Gly184del). These results were consistent with the prediction of in silico assays, and the pattern of RNA splicing was validated to be consistent with the prediction of splicing pattern three (Fig. 3d).
Fig. 4.
Functional analysis of the COL4A2 variant effect on mRNA splicing. (a) Construction strategy of pcDNA3.1 minigene vector, * representative of the variant site. (b) The sequencing of wildtype COL4A2 gene vector and c.549 + 5G > A COL4A2 variant gene vector. (c) Gel electrophoresis of reverse transcription polymerase chain reaction products displayed a single band a (estimated 410 bp) from the wild type (wt) and a smaller band b (estimated 338 bp) in the mutant type (mut). (d) Illustration of the sequencing of band a (wild type in HEK293T and Hela cells) and band b (variant c.549 + 5G > A in HEK293T and Hela cells) products lead to a shorter transcript with deletion of exon 8 including 72 bp
Discussion
In our study, we observed ventriculomegaly, subdural hemorrhage, and extensive encephalomalacia in a fetus. Genetic analysis revealed a novel heterozygous intronic variant c.549 + 5G > A in COL4A2, which has not been previously reported. In silico and in vitro minigene splicing assay indicated that c.549 + 5G > A was a spliceogenic variant resulting in complete skipping of exon 8 (c.478_ 549del) in COL4A2 gene. The misspliced transcript did not create a downstream frameshift or a premature termination codon in the COL4A2 mRNA sequence, only leading to the loss of 24aa inside the protein (p.Pro161_Gly184del).
We performed toxoplasmosis, rubella, cytomegalovirus, herpes, parvovirus B19, assays for immune and alloimmune thrombocytopenia, platelet count, prothrombin time, partial thromboplastin time, plasminogen, von Willebrand factor, factor V Leiden, anticoagulant protein S, activated protein C, and antiplatelet antibodies on the proband, all of these were normal. So we excluded the diseases including congenital infectious disease, fetal and neonatal alloimmune thrombocytopenia (FNAIT), thrombocytopenic disease, coagulation system diseases, inherited bleeding diseases like Vascular hemophilia, anticoagulant system diseases such as genetic protein C deficiency. After excluding other causes, a novel heterozygous intronic variant c.549 + 5G > A in COL4A2 was identified in the proband. The COL4A2 protein contains three major domains: an amino-terminal 7 S domain, a central triple-helix-forming (collagenous) domain, and a carboxyterminal non-collagenous (NC1) domain with each domain possessing different functions. The 7 S domain is involved in macromolecular organization and intermolecular cross-linking. The majority of the protein is made up of the triple helical domain, which consists of long (Gly-X-Y)n repeats with varied X and Y amino acids. Globular NC1 domains are responsible for the assembly of heterotrimers [20]. The majority of variants occurred at conserved Gly position in (Gly-X-Y)n repeats within the triple helical domain, resulting in alteration of the heterotrimeric helix (α1α1α2) [5, 6]. Triple helical structure has cardinal significance. Within the endoplasmic reticulum (ER), NC1 domains initiate heterotrimer assembly and then proceed by the progressive winding of the triple helical domains. The pathogenicity of COL4A1 and COL4A2 variants is typically attributed to heterotrimeric helix (α1ɑ1ɑ2) misfolding and defective secretion. Important pathogenic events have been proposed for both intracellular retention and extracellular deficiency of (α1ɑ1ɑ2) [21–25]. The variant c.549 + 5G > A resulted in the loss of 24aa (p.Pro161_Gly184del) located in the collagenous domain including five (Gly-X-Y) repeats within the triple helical domain (Fig. 5). It is likely to affect the stability and flexibility of COL4A2 protein.
Fig. 5.
Predicted COL4A2 protein domains showed the loss of 24aa (p.Pro161_ Gly184del) located in collagenous domain including five (Gly-X-Y) repeats within the triple helical domain. Green denotes collagen, purple denotes collagen triple helix repeat
COL4A2 variants can cause cytotoxicity and impair the secretion of COL4A1 and COL4A2 [24]. The occurrence of cerebral injury was due to variants in COL4A1/COL4A2 affecting the stability of the vascular basement membranes. In a study of Col4a1 mutation mouse model, inconsistent density and uneven edges of vascular basement membranes were displayed [26]. COL4A2-related diseases are typically AD inheritance. However, recent work reported that the homozygous variant (p.G1158R) in COL4A2 exhibits autosomal recessive inheritance, with intellectual disability, epilepsy, and spastic cerebral palsy in probands, suggesting that the COL4A2 diseases exist in both dominant and recessive forms [15]. Individuals with the same variant have different clinical manifestations, this may be due to incomplete penetrance, which may be contributed to the combined effects of genetic context and environmental factors [27, 28]. Allelic heterogeneity has been demonstrated to influence the severity and penetrance of diseases [23].
The chromosome analysis of the proband revealed normal 46, XY. A phenotype of cerebral injury was found during his fetal period. The father and mother were validated as negative by sequencing analysis. Therefore, this novel variant was deduced to be de novo, rather than inherited (paternity/maternity testing was not performed). As this variant COL4A2 c.549 + 5G > A has might occurred de novo, the recurrence risk to any siblings of the patient is low, although there remains a small risk of recurrence due to the possibility of parental germline mosaicism. Investigating splicing abnormalities in transcripts from the samples of affected proband will provide the evidence to confirm splicing events in vivo. However, patients are unwilling to provide more samples, and transcriptional analysis cannot be performed, which is a limitation of our study. Therefore, multiple online splicing pattern prediction tools combined with in vitro minigene detection may be a feasible alternative method.
In summary, we describe a deleterious COL4A2 splice variant c.549 + 5G > A for the first time, that caused aberrant splicing within intron 8. We predicted the impact of this variant using different algorithms and arrived at the same conclusion with minigene assay. The in vitro minigene splicing assay indicated that the variant promotes skipping of exon 8 (c.478_549del) in COL4A2, leading to the loss of 24aa inside the protein (p.Pro161_Gly184del), this probably results in the loss function and structural instability of COL4A2 protein. Therefore, this novel splice-site variant is considered pathogenic. Our findings expand the variant spectrum of the COL4A2 gene in the etiological investigation of the fetal neurological disorders associated with hypoxic-ischemic and hemorrhagic lesions.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The authors are grateful to the patient and his family for their understanding and cooperation, and to express gratitude to Berry Genomics Co., for the technical support received.
Author contributions
RS and YX drafted the article. DW and CL conceived the study and revised the manuscript. SH, HX, JP, and MZ collected clinical data. RS, TZ, and YG investigated the data. QH and YL analyzed the data. RS and TZ performed the bioinformatic analysis. HW, JS, MZ, SH, and YG performed the experiments. All authors contributed to revising the manuscript and read through and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (82171701), the Medical Science and Technology project of Zhejiang Province (2022YK839), the Social Programs of Wenzhou Technology Bureau (2023Y0329).
Data availability
The datasets generated and/or analyzed during the current study are available from the corresponding author upon reasonable request. The novel variant revealed in the study were submitted to ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/) under Accession Numbers SCV004041824.
Declarations
Ethical compliance and consent to Participate
The study was conducted in accordance with the Declaration of Helsinki and approved by the ethics committee of The First Affiliated Hospital of Wenzhou Medical University (No. YS2022-798). Informed consent to participate was obtained from all of the participants and parents of the minor participant in the study.
Consent for publication
Informed consent for publication of identifying images or other personal or clinical details was obtained from all of the participants and from parents for minor participant.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rong-Yue Sun, Yue Xu and Qing-Qing Huang contributed equally to this work.
Contributor Information
Chao-Sheng Lu, Email: lucs363@163.com.
Dan Wang, Email: wd608044@wmu.edu.cn.
References
- 1.Yuan X, et al. Genetic variants of the COL4A3, COL4A4, and COL4A5 genes contribute to thinned glomerular basement membrane lesions in sporadic IgA nephropathy patients. J Am Soc Nephrol. 2023;34(1):132–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gibson JT, et al. Genotype-phenotype correlations for COL4A3-COL4A5 variants resulting in gly substitutions in Alport syndrome. Sci Rep. 2022;12(1):2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oohashi T, et al. Clonal overgrowth of esophageal smooth muscle cells in diffuse leiomyomatosis-Alport syndrome caused by partial deletion in COL4A5 and COL4A6 genes. Matrix Biol. 2011;30(1):3–8. [DOI] [PubMed] [Google Scholar]
- 4.O’Brien A, et al. Confirmation of COL4A6 variants in X-linked nonsyndromic hearing loss and its clinical implications. Eur J Hum Genet. 2022;30(1):7–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khoshnoodi J, Pedchenko V, Hudson BG. Mammalian Collagen IV Microsc Res Tech. 2008;71(5):357–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yoneda Y, et al. De novo and inherited mutations in COL4A2, encoding the type IV collagen α2 chain cause porencephaly. Am J Hum Genet. 2012;90(1):86–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jeanne M, Gould DB. Genotype-phenotype correlations in pathology caused by collagen type IV alpha 1 and 2 mutations. Matrix Biol, 2017. 57–8: pp. 29–44. [DOI] [PMC free article] [PubMed]
- 8.Meuwissen ME, et al. Sporadic COL4A1 mutations with extensive prenatal porencephaly resembling hydranencephaly. Neurology. 2011;76(9):844–6. [DOI] [PubMed] [Google Scholar]
- 9.Decio A, et al. A novel mutation in COL4A1 gene: a possible cause of early postnatal cerebrovascular events. Am J Med Genet A. 2015;167a(4):810–5. [DOI] [PubMed] [Google Scholar]
- 10.Coste T, et al. COL4A1/COL4A2 and inherited platelet disorder gene variants in fetuses showing intracranial hemorrhage. Prenat Diagn. 2022;42(5):601–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verbeek E, et al. COL4A2 mutation associated with familial porencephaly and small-vessel disease. Eur J Hum Genet. 2012;20(8):844–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Plaisier E, et al. COL4A1 mutations and hereditary angiopathy, nephropathy, aneurysms, and muscle cramps. N Engl J Med. 2007;357(26):2687–95. [DOI] [PubMed] [Google Scholar]
- 13.van der Knaap MS, et al. Neonatal porencephaly and adult stroke related to mutations in collagen IV A1. Ann Neurol. 2006;59(3):504–11. [DOI] [PubMed] [Google Scholar]
- 14.Rannikmäe K, et al. Common variation in COL4A1/COL4A2 is associated with sporadic cerebral small vessel disease. Neurology. 2015;84(9):918–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bakhtiari S, et al. Recessive COL4A2 mutation leads to intellectual disability, Epilepsy, and spastic cerebral palsy. Neurol Genet. 2021;7(3):e583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hausman-Kedem M, et al. Deletion in COL4A2 is associated with a three-generation variable phenotype: from fetal to adult manifestations. Eur J Hum Genet. 2021;29(11):1654–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koene S, et al. Intracerebral hemorrhage in a neonate with an intragenic COL4A2 duplication. Am J Med Genet A. 2021;185(2):571–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meuwissen ME, et al. The expanding phenotype of COL4A1 and COL4A2 mutations: clinical data on 13 newly identified families and a review of the literature. Genet Med. 2015;17(11):843–53. [DOI] [PubMed] [Google Scholar]
- 19.Stein S, Bahrami-Samani E, Xing Y. Using RNA-Seq to Discover genetic polymorphisms that produce hidden splice variants. Methods Mol Biol. 2017;1648:129–42. [DOI] [PubMed] [Google Scholar]
- 20.Murray LS, et al. Chemical chaperone treatment reduces intracellular accumulation of mutant collagen IV and ameliorates the cellular phenotype of a COL4A2 mutation that causes haemorrhagic stroke. Hum Mol Genet. 2014;23(2):283–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayashi G, Labelle-Dumais C, Gould DB. Use of sodium 4-phenylbutyrate to define therapeutic parameters for reducing intracerebral hemorrhage and myopathy in Col4a1 mutant mice. Dis Model Mech, 2018. 11(7). [DOI] [PMC free article] [PubMed]
- 22.Mayne R, et al. Monoclonal antibodies against chicken type IV and V collagens: electron microscopic mapping of the epitopes after rotary shadowing. J Cell Biol. 1984;98(5):1637–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuo DS, et al. Allelic heterogeneity contributes to variability in ocular dysgenesis, myopathy and brain malformations caused by Col4a1 and Col4a2 mutations. Hum Mol Genet. 2014;23(7):1709–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeanne M, et al. COL4A2 mutations impair COL4A1 and COL4A2 secretion and cause hemorrhagic stroke. Am J Hum Genet. 2012;90(1):91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gould DB, et al. Col4a1 mutation causes endoplasmic reticulum stress and genetically modifiable ocular dysgenesis. Hum Mol Genet. 2007;16(7):798–807. [DOI] [PubMed] [Google Scholar]
- 26.Gould DB, et al. Mutations in Col4a1 cause perinatal cerebral hemorrhage and porencephaly. Science. 2005;308(5725):1167–71. [DOI] [PubMed] [Google Scholar]
- 27.Jeanne M, Jorgensen J, Gould DB. Molecular and genetic analyses of collagen type IV mutant mouse models of spontaneous intracerebral hemorrhage identify mechanisms for Stroke Prevention. Circulation. 2015;131(18):1555–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mao M, et al. Type IV collagens and basement membrane diseases: Cell Biology and pathogenic mechanisms. Curr Top Membr. 2015;76:61–116. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are available from the corresponding author upon reasonable request. The novel variant revealed in the study were submitted to ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar/) under Accession Numbers SCV004041824.