Abstract
Human herpesvirus 8 (HHV-8) is found in immunoblastic B cells of patients with multicentric Castleman's disease (MCD) and, predominantly in a latent form, in primary effusion lymphoma (PEL) cells and Kaposi's sarcoma (KS) spindle cells. Recent studies have shown that upon reactivation, HHV-8 expresses factors that downregulate major histocompatibility class I proteins and coactivation molecules and that may enable productively infected cells to escape cytotoxic T lymphocytes and natural killer cell responses. One of these viral factors is encoded by open reading frame (ORF) K3. Here we show that in PEL cells, ORF K3 is expressed through viral transcripts that are induced very early upon virus reactivation, including bicistronic RNA molecules containing coding sequences from viral ORFs K3 and 70. Specifically, we found that a bicistronic transcript was expressed in the absence of de novo protein synthesis, thereby identifying a novel HHV-8 immediate-early gene product. Several features of the RNA molecules encoding the K3 product, including multiple transcriptional start sites, multiple donor splicing sites, and potential alternative ATG usage, suggest that there exists a finely tuned modulation of ORF K3 expression. By contrast, ORF K3 transcripts are not detected in the majority of cells present in KS lesions that are latently infected by the virus, suggesting that there are other, as-yet-unknown mechanisms of immune evasion for infected KS spindle cells. Nevertheless, because HHV-8 viremia precedes the development of KS lesions and is associated with the recrudescence of MCD symptoms, the prompt expression of ORF K3 in productively infected circulating cells may be important for virus pathogenesis. Thus, molecules targeting host or viral factors that activate ORF K3 expression or inactivate the biological functions of the K3 product should be exploited for the prevention or treatment of HHV-8-associated diseases in at-risk individuals.
Human herpesvirus 8 (HHV-8) (8) is a novel gammaherpesvirus found in immunoblastic B cells from persons with multicentric Castleman's disease (MCD), primary effusion lymphoma (PEL) cells, and Kaposi's sarcoma (KS) spindle cells or endothelial cells lining normal vessels in KS lesions (5, 7, 17, 20, 60, 64). In PEL and KS, HHV-8 is generally present in a latent form (4, 11, 15, 17, 49, 61–64; M. Stürzl, G. Ascherl, C. Blasig, S. R. Opalenik, B. Ensoli, and P. J. Browning, Letter, AIDS 12:1105–1106, 1998).
Recent observations indicate that both reactivation of HHV-8 infection (28, 40, 50) and impaired immunological control of HHV-8-infected circulating cells (43, 48; P. Monini, M. C. Sirianni, M. Stürzl, M. Franco, L. Vincenzi, S. Topino, P. Leone, D. Goletti, P. Leone, M. Andreoni, O. Barduagni, G. Rezza, and B. Ensoli, Abstr. 2nd Int. Workshop KSHV/HHV-8 Relat. Agents, abstr. 30, 1999; M. C. Sirianni, L. Vincenzi, S. Topino, A. Giovannetti, F. Mazzetta, C. Alario, and B. Ensoli, Abstr. 2nd Int. Workshop KSHV/HHV-8 Relat. Agents, abstr. 29, 1999; M. C. Sirianni, C. Alario, F. Libi, D. Scaramuzzi, S. Topino, F. Ensoli, and P. Monini, Abstr. 3rd Int. Workshop Kaposi's Sarcoma-Associated Herpesvirus Relat. Agents, abstr. 105, 2000) lead to massive virus spreading that is associated with development of KS (3, 4, 9, 19, 23, 33, 42, 54, 55, 59, 71). These observations are supported by studies indicating that patients with AIDS-associated KS undergoing highly active antiretroviral therapy can show clearance of HHV-8 DNA from the circulation and that this is associated with KS regression (34; D. A. Rizzieri, J. Liu, S. T. Traweek, and G. D. Miralles, Letter, Lancet 349:775–776, 1997; M. C. Sirianni, L. Vincenzi, S. Topino, A. Giovannetti, F. Mazzetta, C. Alario, and B. Ensoli, Abstr. 2nd Int. Workshop KSHV/HHV-8 Relat. Agents, abstr. 29, 1999). In this context, our recent work has shown that latent HHV-8 is reactivated in monocytes and B cells from persons with KS or at risk of KS upon the exposure of peripheral blood mononuclear cells to specific Th-1-type inflammatory cytokines (ICs) that are found to be increased in persons with KS or at risk of KS (40).
There is evidence indicating that the lack of immunological control of HHV-8 infection may involve not only immune depression but also specific virus immune escape mechanisms. Recent studies, in fact, have shown that the gene products of HHV-8 open reading frames (ORFs) K3 and K5 are capable of downregulating major histocompatibility class I (MHC-I) proteins and coactivation molecules, thereby allowing HHV-8-infected cells to escape both cytotoxic T-lymphocyte and natural killer (NK) cell responses (10, 26, 29, 30). Viral mechanisms of immune evasion are likely to be important when immune depression is not yet apparent, and virus spreading and dissemination to tissues are essentially driven by virus reactivation upon IC stimulation. In addition, immune evasion is likely to be relevant also for other HHV-8-associated diseases, such as MCD, the exacerbation and worsening of which appear to be associated with increased HHV-8 viremic levels (25, 46).
To ensure a prompt evasion of the immune response by cells undergoing virus reactivation, ORFs K3 and K5 must be efficiently expressed with early kinetics. In this context, HHV-8 reactivation is known to depend on the induction of viral immediate-early (IE) genes whose transcription is driven by host factors and is independent of de novo viral protein synthesis (24, 35–37, 58, 67, 69, 74). However, there are few data on the transcription pattern of HHV-8 ORFs K3 and K5. In previous studies, both of these ORFs were shown to be expressed with early kinetics upon virus reactivation in PEL cell lines (36, 67). In addition, ORF K5 was shown to encode IE transcripts in an HHV-8-positive, Epstein-Barr virus (EBV)-negative PEL cell line (26), but not in PEL cells coinfected by HHV-8 and EBV (67, 74). However, the molecular pattern and kinetics of K3 transcription have not yet been elucidated, and there are no available data on K3 expression in KS tissue.
Here we show that K3 expression in EBV-negative PEL cells occurs through the synthesis of multiple transcripts containing coding sequences from viral ORFs K3 and 70. All transcripts appear to be expressed at very early times upon virus reactivation. In addition, resistance of the expression of a bicistronic transcript to the protein synthesis inhibitor cycloheximide indicates that the ORF K8 gene product is in part expressed through an IE RNA molecule. By contrast, ORF K3 transcripts were undetectable by in situ hybridization in the majority of latently infected spindle cells of KS lesions.
MATERIALS AND METHODS
Cell cultures.
BCBL-1 cells, obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health, were cultured in RPMI 1640 containing 10% fetal calf serum (FCS), 1 mM sodium pyruvate, and 50 μM β-mercaptoethanol. BC-1 cells (provided by Patrick Moore, Columbia University, New York, N.Y.) were cultured in RPMI 1640 containing 15% FCS. Cells were split at a density of 3 × 105/ml twice a week.
Induction of HHV-8 lytic replication in PEL cells.
Exponentially growing BCBL-1 cells (106/ml) were resuspended at a density of 5 × 105/ml and grown for an additional 24 h. HHV-8 lytic replication was then induced in these cells by treatment with 20 ng of phorbol 12-myristate 13-acetate (TPA; Sigma, Milan, Italy)/ml for various periods of time (53).
Determination of inhibition of protein synthesis by CHX.
Exponentially growing BCBL-1 cells (1.5 × 108) were collected, washed with phosphate-buffered saline (PBS), suspended in methionine-free growth medium (107/ml), and seeded in 24-well plates (3 × 106/well). Cells were incubated for 45 min at 37°C prior to the addition of [35S]methionine (25 μCi/well) and cycloheximide (CHX; 10 or 100 μg/ml). Protein synthesis inhibition was then determined after various periods of time by comparing the percentage of trichloroacetic acid-insoluble [35S]methionine from CHX-treated cultures with that from untreated cells.
Purification of HHV-8 genomic DNA.
BCBL-1 cells (about 3 × 108) were cultured for 6 days in the presence of TPA. Cell supernatants were cleared and filtered by using a 0.45-μm-pore-size membrane and then centrifuged for 2 h at 28,000 × g to collect the virus. The virus pellet was suspended in 400 μl of Tris-EDTA-NaCl buffer (0.1 M Tris-HCl [pH 8.0], 0.01 M EDTA [pH 8.0], 0.1 M NaCl) containing 20 mM MgCl2 and 250 IU of pancreatic DNase I (Boehringer GmbH, Mannheim, Germany)/ml and incubated at 37°C for 1 h. DNase digestion was repeated three times with addition of fresh DNase I on each occasion. At the end of the DNase digestion, EDTA was added to a final concentration of 50 mM and the virus suspension was rapidly heated to 95°C and incubated at this temperature for 10 min. The viral particles were then digested with proteinase K (Boehringer GmbH; 0.4 mg/ml) in the presence of 2% (wt/vol) sodium dodecyl sulfate at 37°C for 15 h. After proteinase K inactivation at 95°C for 10 min, viral DNA was extracted by using phenol and chloroform, ethanol precipitated, and digested with the XbaI restriction enzyme.
Recombinant plasmids.
HHV-8 nut-1/T1.1, ORF 12/kaposin, and 18S ribosomal cDNA recombinant plasmids used as probes for Northern blot analysis or for the screening of the suppression subtractive hybridization (SSH) cDNA library (see below) were described previously (39). Other probes used for Northern blotting, screening of the SSH library, or in situ hybridization were constructed by cloning PCR products amplified from HHV-8 phage or plasmid clones obtained from a classic KS lesion genomic library in the pCRII plasmid vector (Invitrogen, Carlsbad, Calif.). The nucleotide boundaries of the probes used in the study are as follows: ORF K3, nucleotides (nt) 18608 to 19609; ORF 70, nt 20137 to 21104; ORF 50, nt 72639 to 73299; ORF K5, nt 25871 to 26219; ORF 2, nt 17881 to 18598; and ORF K8, nt 74961 to 75243 (nucleotide enumeration as for GenBank accession no. KSU75698).
Northern blot hybridization.
Total RNA was extracted from BCBL-1 cells by using an RNeasy Mini Kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer's instructions. Equal amounts of total RNA (5 to 20 μg) were suspended in a buffer containing 40 mM morpholinepropanesulfonic acid (MOPS; pH 7.0), 10 mM sodium acetate (pH 4.5), 10 mM EDTA (pH 8.0), 50% (vol/vol) formamide, and 2.2 M formaldehyde; incubated at 65°C for 15 min; and chilled on ice. A 50% (vol/vol) glycerol solution containing 0.25% (wt/vol) bromophenol blue and ethidium bromide was then added to the samples, and the RNA was subjected to electrophoresis through formaldehyde-agarose gels, transferred onto nylon membranes (Hybond N; Amersham, Little Chalfont, United Kingdom), and hybridized to 32P-labeled DNA probes by standard procedures.
SSH.
BCBL-1 cells (5 × 108) were preincubated in the presence of CHX (10 μg/ml) for 30 min and induced for 4 h with 20 ng of TPA/ml in the continuous presence of CHX. Total RNA was then extracted and purified by using an RNeasy Midi Kit (Qiagen). Poly(A)+ RNA was isolated by using an Oligotex mRNA Midi Kit (Qiagen). cDNA synthesis and SSH (12, 13) were performed with a PCR-Select cDNA Subtraction Kit (Clontech, Palo Alto, Calif.) according to the instructions of the manufacturer. Prior to subtractive hybridization, cDNA from BCBL-1 cells (tester cDNA) was RsaI restricted and linked to oligonucleotide adapters. To minimize the subtraction of BCBL-1 IE cDNA, uninduced BC-1 cells were chosen as the source of driver (subtracting) cDNA because these cells are characterized by a very low frequency of spontaneous reactivation (reference 74 and data not shown). The first and second hybridization steps were performed with a fivefold excess of RsaI-restricted driver cDNA and a 1:1 ratio of RsaI-restricted tester and driver cDNAs, respectively. Nested-PCR products from the subtracted cDNA were used as 32P-labeled probes for Southern blot hybridization analysis of XbaI-digested HHV-8 DNA.
Construction and screening of the subtracted cDNA library.
SSH PCR products were cloned in the pCRII plasmid vector by using a TA cloning kit (Invitrogen) and employed to transform competent One Shot bacteria (Invitrogen). The titer of the resulting subtracted cDNA library was determined to be about 2,500 ampicillin-resistant bacterial colonies. A total of 315 randomly selected, individual colonies were picked and screened with 32P-labeled HHV-8 subgenomic DNA probes specific for viral ORF 50, ORF K12, ORF K5, ORF K8, ORF 2, or ORF K3 or with 32P-labeled HHV-8 DNA purified from BCBL-1 cell supernatants.
RACE.
The 5′ and 3′ boundaries of HHV-8 K3 transcripts were determined by rapid amplification of cDNA ends (RACE) (21, 22), using a Marathon cDNA amplification kit (Clontech), with cDNA from purified poly(A)+ RNA isolated from BCBL-1 cells induced with TPA in the presence of CHX. PCR primers internal to the SSH cDNA clone were as follows: SSH 1 (5′-AGAGATAGATCACGTCGCTG-3′, nt 19219 to 19200), SSH 2 (5′-TATTGCCTCGGCTGACTTAC-3′, nt 19398 to 19379), SSH 3 (5′-CTCGAGAACGTCCATAGAAG-3′, nt 19522 to 19503), and SSH 4 (5′-TTAGACTGGTGGACTACTGC-3′, nt 20146 to 20127) for amplification of 3′ cDNA ends and SSH I (5′-GCAGTAGTCCACCAGTCTAA-3′, nt 20127 to 20146), SSH III (5′-CGTAGTGGCTCTATATGCGT-3′, nt 20178 to 20197), and SSH IV (5′-GCCTGAGATACTGAAGTTCC-3′, nt 20927 to 20946) for amplification of 5′ cDNA ends (nucleotide enumeration as for GenBank accession no. KSU75698). PCR products were cloned into the pCRII plasmid vector (Invitrogen) and analyzed by DNA sequencing.
Amplification of sequences internal to the K3 transcripts.
Regions internal to the 5′ and 3′ RACE boundaries were amplified with primers K3ca (5′-TTAATGAAACATAAGGGCAGACG-3′, nt 18608 to 18630) and K3cb (5′-ATGTTTCCGTTTGTACC-3′, nt 21104 to 21088), spanning the ORF K3 stop codon and the ORF 70 ATG (underlined), or with primers SSH V (5′-CAGCGACGTGATCTATCTCT-3′, nt 19251 to 19270) and SSH 5 (5′-GGAACTTCAGTATCTCAGGC-3′, nt 20946 to 20927), which are located in more internal regions of ORFs K3 and ORF 70, respectively (nucleotide enumeration as for GenBank accession no. KSU75698). PCR amplification was performed with a GeneAmp XL PCR kit for long-range PCR (Perkin-Elmer, Branchburg, N.J.) according to the manufacturer's instructions.
DNA sequencing.
DNA sequencing was performed with an ABI 377 sequencer, using an ABI BigDye Terminator Cycle Sequencing Ready Reaction Kit (Perkin-Elmer).
Immunofluorescence assay.
BCBL-1 cells were spotted onto multitest slides (ICN, Aurora, Ohio) at a density of 5 × 104 per well, air dried, and fixed in 4% paraformaldehyde for 20 min at room temperature. After permeabilization with PBS containing 0.2% Triton X-100 for 10 min, the cells were incubated in 100 mM glycine for 10 min and then in PBS containing 10% FCS for 20 min. The cells were then incubated for 1 h with anti-viral-interleukin-6 (vIL-6) rabbit polyclonal antibody (Advanced Biotechnologies Inc., Columbia, Md.; 1:200 dilution) or control rabbit nonimmune serum at 37°C. After three washes (10 min each) with PBS, fluorescein isothiocyanate (FITC)-conjugated anti-rabbit immunoglobulin G antibody (Sigma) was added. After a 1-h incubation at 37°C, the cells were washed three times with PBS and once with PBS containing evans blue (Sigma) as a counterstaining dye. For quantitation of fluorescent cells, four microscopic fields (25× magnification) were randomly collected for each sample with a cooled charge-coupled device camera, and images were processed using OptiLab 2.6 software (Graphtek, Mirmande, France) to evaluate the percentage of positive (FITC-stained) cells present in each microscopic field.
In situ hybridization.
Bioptic samples from KS lesions were fixed in PBS-buffered 4% paraformaldehyde at 4°C, dehydrated, and paraffin embedded as previously described (65, 66). Thin tissue sections (5 to 10 μm thick) were hybridized to antisense 35S-radiolabeled K3 or K8 RNA probes or to a control β-actin RNA probe as previously described (39, 64). Briefly, the RNA probe solution (10 to 15 μl) was applied to the deparaffinized tissue sections at an adjusted activity of 50,000 cpm/μl in hybridization buffer (50% deionized formamide, 0.3 M NaCl, 20 mM Tris-HCl [pH 7.4], 5 mM EDTA, 10 mM NaPO4 [pH 8.0], 10% dextran sulfate, 1× Denhardt's solution, 50 μg of total yeast RNA/ml). Hybridization was carried out at 50°C for 16 h. At the end of the hybridization step, tissue sections were washed at 50°C in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing 10 mM dithiothreitol; stringently washed at 60°C in a solution containing 50% formamide, 2× SSC, and 0.1 M dithiothreitol; covered with a film emulsion; and incubated for 14 days.
Prediction of K3 splicing pattern and transcriptional promoters.
Analysis of the HHV-8 K3 locus for splicing donor-acceptor sites and transcriptional start sites was performed by the neural network method (45, 51, 52) with the software from the Berkeley Drosophila Genome Project (http://www.fruitfly.org/index.html).
Nucleotide sequence accession numbers.
The nucleotide sequences of the K3 transcript cDNAs have been submitted to GenBank and given the following accession numbers: T(lm)2, AF307519; T1, AF307581; T2, AF307517; and T3, AF307516.
RESULTS
Analysis of HHV-8 ORF K3 expression.
Herpesvirus IE gene transcription is driven by preexisting viral or host factors; therefore, it can proceed in the absence of de novo protein synthesis (32, 56). In contrast, efficient transcription of herpesvirus delayed-early (DE) or late viral genes requires de novo synthesis of viral transcriptional factors (32, 56).
The transcription pattern of HHV-8 ORF K3 was determined in latently HHV-8-infected PEL-derived BCBL-1 cells that had been subjected to TPA treatment, in the presence or absence of CHX, to induce replication of the virus (39, 53). Since PEL cells have been shown to be extremely sensitive to the toxic effects of CHX (36, 74), BCBL-1 cells were maintained for various periods of time in the presence or absence of a low (10 μg/ml) or high (100 μg/ml) concentration of CHX and analyzed for both inhibition of protein synthesis and cell survival. Cell protein synthesis was found to be blocked at both CHX concentrations within the first 30 min of treatment, as indicated by the very low percentage (<5%) of 35S incorporation by CHX-treated cells (data not shown). The percentage of viable (i.e., trypan blue-excluding) cells remained unchanged during the early hours of treatment; however, cell viability was found to be significantly decreased after 8 h (70% or fewer viable cells after 16 h). Since the loss of cell viability was less pronounced at the low CHX concentration, all further experiments were performed by treating BCBL-1 cells for up to 16 h in the presence or absence of CHX at a concentration of 10 μg/ml.
To analyze the expression pattern of the K3 gene product, BCBL-1 cells were cultured for various periods of time with or without TPA, in the presence or absence of CHX, and total RNA was analyzed by Northern blot hybridization with a DNA probe spanning HHV-8 ORF K3. To ensure a maximal blockade of protein synthesis upon virus reactivation, cells were induced with TPA 30 min after CHX addition.
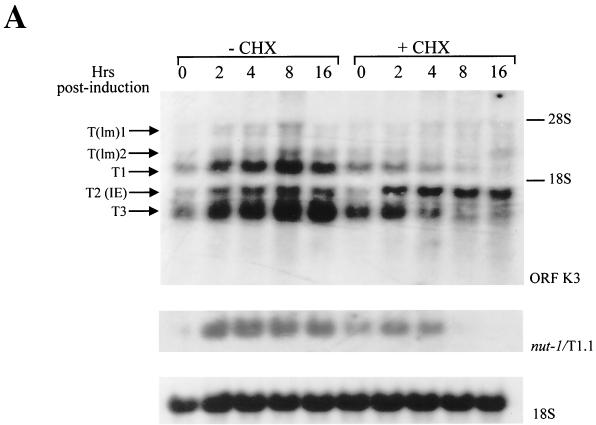
Under these conditions, Northern blot analysis identified three major K3 transcripts (T1, T2, and T3), with approximate sizes of 2.5, 1.5, and 1.3 kb, respectively, and two low-intensity hybridization bands of lower mobility [T(lm)1 and T(lm)2] (Fig. 1A). In the absence of CHX, the expression of all K3 transcripts appeared to be induced above background levels at early times after TPA induction and reached the maximal level of expression between 4 and 8 h postinduction (Fig. 1A). The expression of the majority of the K3 transcripts was clearly inhibited by CHX at all times post-TPA induction, indicating that the majority of K3 transcripts have an expression pattern typical of herpesvirus DE genes (Fig. 1A). However, the 1.5-kb transcript (T2) was consistently induced above background levels at all times postinduction, even in the presence of CHX (Fig. 1A), and thus a novel HHV-8 IE transcript containing K3 coding sequences was identified. As a control, hybridization of the same blot with a probe specific for the HHV-8 DE gene nut-1/T1.1 (39, 57, 68, 73) produced a band that, as predicted, was clearly sensitive to CHX treatment (Fig. 1A).
FIG. 1.
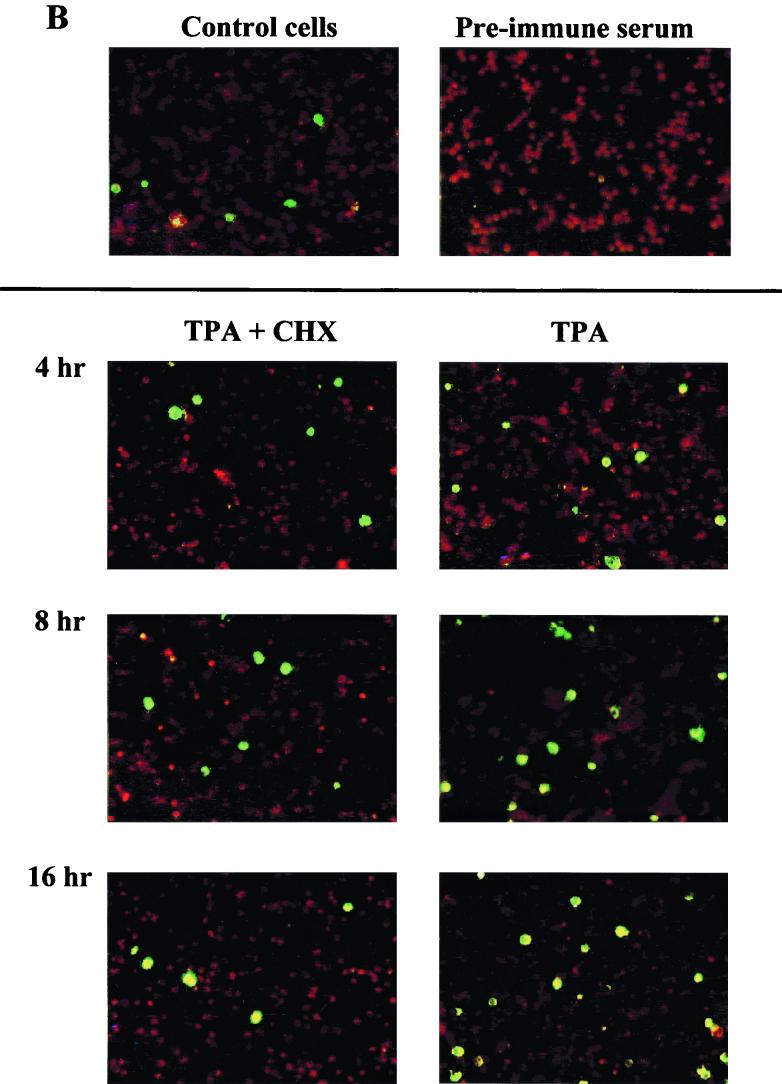
Expression of ORF K3, nut-1/T1.1, vIL-6, and 18S rRNA in TPA-induced BCBL-1 cells cultured in the presence or absence of CHX. (A) Kinetics of induction of ORF K3 transcripts, nut-1/T1.1 RNA, or 18S RNA by TPA in cells treated or not treated with CHX. CHX (10 μg/ml) was added to BCBL-1 cells, and after 30 min, cells were induced with TPA (20 ng/ml). Cell cultures were harvested at the indicated time points after TPA induction, and total RNA (10 μg) was analyzed by Northern blot hybridization with the pCRII-K3 probe. Two low-intensity bands, corresponding to low-mobility transcripts [T(lm)1 and T(lm)2], and three major transcripts of higher mobility (T1, T2, and T3) were identified. The membrane was stripped and rehybridized with probes specific for nut-1/T1.1, as a control for inhibition by CHX of HHV-8 DE gene induction, or 18S RNA, as a control for the amount of sample loaded on the gel. The positions of the 28S and 18S rRNAs, as molecular size markers, are indicated. (B) Kinetics of induction of vIL-6 by TPA in cells treated or not treated with CHX. (Upper panels) Left, uninduced cell cultures grown in the absence of CHX; right, uninduced cells incubated with a preimmune rabbit serum. (Lower panels) Left (TPA + CHX), cells induced with TPA in the presence of CHX; right (TPA), cells induced with TPA in the absence of CHX. Pictures are representative microscopic fields of FITC-stained cells counterstained with evans blue (magnification, 100×). In the absence of CHX, vIL-6 expression in TPA-induced cells continued to increase, and a significantly higher percentage of TPA-induced cells was found to be positive for vIL-6 expression at 48 h postinduction (data not shown). vIL-6 expression was quantified by analysis with a cooled charge-coupled device camera and OptiLab 2.6 software, confirming that viral protein synthesis was efficiently blocked in BCBL-1 cells induced with TPA in the presence of CHX at the indicated time points (data not shown).
To determine whether viral protein synthesis (including early viral transcriptional activators) was completely blocked by CHX in the cells analyzed for K3 gene expression, aliquots of the same cell cultures were analyzed for expression of vIL-6 (41, 44, 67) by an immunofluorescence assay. As shown in Fig. 1B, upon TPA induction, vIL-6 expression showed a clear increase to levels above the background, reaching a peak at 8 to 16 h postinduction; however, this upregulation was absent in the cells treated with CHX, confirming that de novo viral protein synthesis was completely inhibited by CHX. These experiments showed that HHV-8 ORF K3 transcription occurs through a complex program involving IE and DE viral transcripts.
Isolation of IE K3 cDNA sequences by SSH.
To identify and characterize the HHV-8 IE K3 transcript, total RNA from BCBL-1 cells was subjected to SSH, a PCR-based technique that allows isolation of cDNA sequences from genes that are differentially expressed on exposure to specific stimuli (12, 13). To this end, BCBL-1 cells were treated with CHX for 30 min and then induced with TPA for 4 h in the continuous presence of CHX. Poly(A)+ cDNA prepared from these cell cultures was digested with the RsaI restriction enzyme and subjected to subtractive hybridization with an excess of RsaI-restricted cDNA from uninduced cells (see Materials and Methods). To minimize subtraction of BCBL-1 IE cDNA sequences, the subtraction step was performed with cDNA from uninduced BC-1 cells, a PEL cell line characterized by very-low-level HHV-8 spontaneous reactivation (reference 74 and data not shown). These experimental conditions allowed the enrichment of cDNA RsaI restriction fragments from HHV-8 genes whose expression was induced upon virus reactivation in the presence of CHX. Under these conditions, SSH yielded a smear underlying several discrete DNA bands ranging in size from 0.3 to about 1 kb (data not shown).
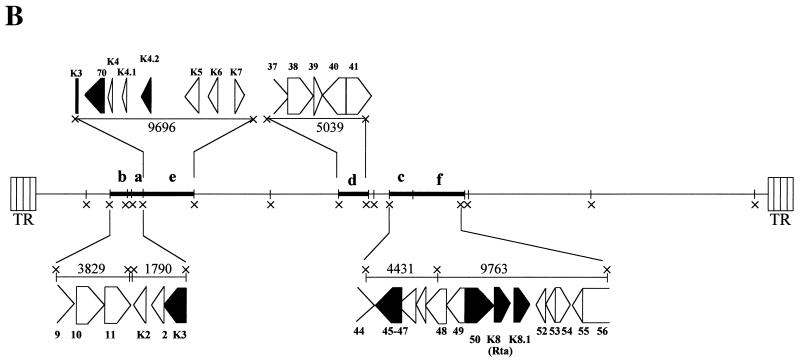
To verify that the subtracted cDNA contained viral sequences from HHV-8 ORF K3, genomic HHV-8 DNA was purified from TPA-induced BCBL-1 supernatants, digested with the XbaI restriction endonuclease, and hybridized to the SSH cDNA that was used as a probe. As expected, this analysis identified two hybridization bands whose sizes corresponded to the two XbaI HHV-8 DNA fragments containing sequences from ORF K3 (Fig. 2, bands a and e). In addition, other bands with sizes consistent with XbaI restriction fragments containing other viral IE genes, including ORFs 50, K8, K8.1, and K8.2 (which all encode the Rta transactivator) (24, 36, 37, 58, 67, 69, 74), ORF K4.2, ORF K5, and ORF 45 (26, 74), also hybridized with the SSH cDNA probe (Fig. 2).
FIG. 2.
Isolation and identification of HHV-8 ORF K3 IE cDNA by SSH. The SSH cDNA was used as a probe for hybridization of HHV-8 genomic DNA digested with the XbaI restriction endonuclease. (A) Southern blot hybridization of HHV-8 genomic DNA with the SSH cDNA probe. Viral genomic DNA extracted from TPA-induced BCBL-1 cell supernatants (1 μg) was extensively digested with XbaI, electrophoresed through a 0.8% agarose gel, transferred to a nylon membrane, and hybridized with the bulk of SSH cDNA labeled with 32P. Six bands (a to f) reacting with the SSH cDNA probe, with sizes ranging from 1.8 to 10 kbp, were identified. The 10-kbp band corresponds to a doublet of similarly sized restriction fragments (e and f) (see panel B). Viral ORFs contained in XbaI restriction fragments a to f (panel B) are indicated on the left side of the autoradiogram. Sequences from ORF K3 are contained in bands a and e. In addition, bands containing larger-sized fragments, possibly corresponding to the largest XbaI restriction fragments or to fragments containing viral terminal repeats, were also detected. HindIII-digested lambda phage DNA was used as a molecular size marker. The positions of the lambda DNA fragments are indicated by dashes; the arrows point to the hybridization bands a to f. The sizes of the DNA fragments are indicated in kilobase pairs. (B) Diagram of HHV-8 XbaI (×) restriction pattern. DNA fragments with sizes corresponding to the hybridization bands reacting with the SSH cDNA probe (a to f) are indicated in bold. The sizes of the reacting fragments (in kilobase pairs) and the encoded HHV-8 ORFs are shown. Restriction fragments a and e contain sequences from ORF K3. ORFs whose sequences are known to be present within HHV-8 IE transcripts are shown in black. Sequences from ORF 70 were determined to be encompassed in the IE ORF K3 transcript (see text for details). TR, terminal repeats.
To isolate the cDNA sequences hybridizing to ORF K3, the bulk of the SSH cDNA was cloned in a plasmid vector and a representative aliquot of the resulting subtracted cDNA library was screened with an HHV-8 DNA genomic probe. Four of the several HHV-8-positive colonies hybridized with an ORF K3-specific probe. Notably, several of the HHV-8-positive colonies hybridized with probes specific for other HHV-8 IE genes, including ORF 50 and ORF K5, and six colonies reacted with a probe specific for ORF K8, whose sequences have been shown to be present in IE transcripts encoding the viral transactivator Rta (24, 36, 37, 58, 69, 74). In contrast, none of the colonies hybridized with a probe for the HHV-8 DE ORF 2 viral dihydrofolate reductase (vDHFR) gene (which is adjacent to ORF K3 in the HHV-8 genome [Fig. 2B]) (67) or with the HHV-8 latency-associated ORF K12 (64, 73), showing that the SSH cDNA pool was indeed enriched in HHV-8 IE cDNA sequences.
Characterization of the IE ORF K3 SSH cDNA.
By DNA sequencing it was determined that all four SSH K3-positive recombinant clones carried the same cDNA fragment, spanning HHV-8 sequences between two RsaI restriction sites present at nt 19241 and 21092 in the viral genome (Fig. 3). The SSH cDNA insert contained sequences corresponding to both ORF K3 and ORF 70, which encodes the HHV-8 thymidylate synthase (TS) homologue (57). ORF 70 coding sequences from nt 20301 to 20842 were removed by in-frame splicing, and a large part of the noncoding sequence between ORFs 70 and K3 (nt 19612 to 20088) was also deleted by a splicing event resulting in a three-exon cDNA fragment (Fig. 3).
FIG. 3.
Molecular arrangement of the SSH K3 cDNA. The arrangement of the four SSH cDNA clones that were positive at screening with the ORF K3-specific probe is shown. The four clones were identical and spanned sequences between two RsaI sites present within ORFs K3 and 70. The nucleotide positions of exon-intron junctions (enumerated as for GenBank accession no. KSU75698) are indicated. Splicing donor and acceptor sites were detected by neural network analysis (52) at the exon-intron junctions (data not shown).
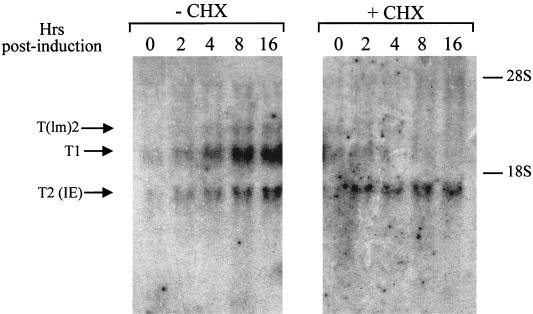
To confirm that the K3 IE transcript corresponded to the SSH cDNA clone, the RNA from the experiment shown in Fig. 1A was hybridized with a probe specific for ORF 70. This analysis proved that the IE 1.5-kb K3 transcript, T2, also contained sequence from ORF 70 (Fig. 4). In addition, the DE 2.5-kb transcripts, T(lm)2 and T1, also reacted with the ORF 70 probe (Fig. 4). The low sensitivity of this hybridization, however, did not allow the detection of the presence of ORF 70 sequences in the T3 transcript, which, as indicated by subsequent analysis, also contained stretches of sequences from HHV-8 ORF 70 (see below).
FIG. 4.
Detection of ORF 70 sequences in ORF K3 transcripts. The RNA from the experiment shown in Fig. 1A was hybridized to an ORF 70-specific DNA probe. Sequences reacting with the ORF 70 probe were detected in the IE transcript T2 and in the DE transcript T1. Similar results were obtained upon rehybridization of the Northern blot membrane shown in Fig. 1A (data not shown).
Mapping of 5′ and 3′ K3 mRNA boundaries by RACE.
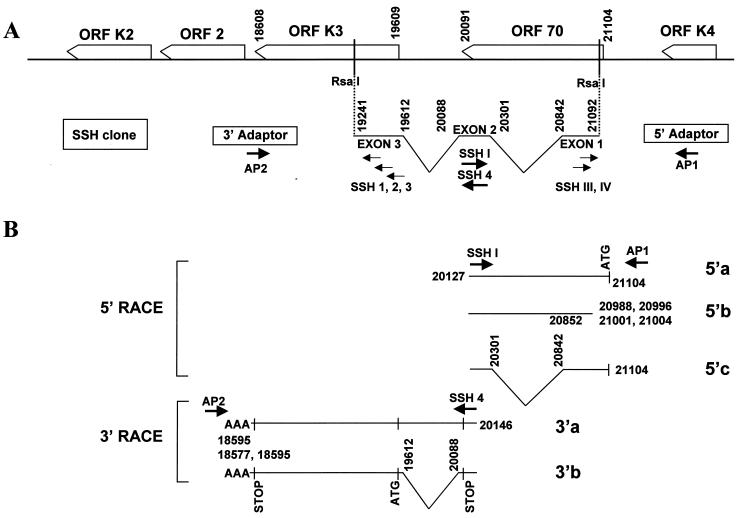
To determine the 5′ and 3′ ends of the HHV-8 K3 transcripts, cDNA from BCBL-1 cells induced for 4 h with TPA in the presence of CHX was subjected to RACE (21, 22). To this end, PCR primers designed against regions internal to the second exon of the SSH K3 cDNA clone were used in combination with primers specific for 5′ or 3′ RACE oligonucleotide adapters (Fig. 5A, primers SSH I, SSH 4, AP1, and AP2). RACE resulted in several 5′ and 3′ amplicons (Fig. 5B).
FIG. 5.
Diagram of the amplicons obtained by RACE. (A) The HHV-8 genomic region containing ORFs K3 and 70 and the structure of the SSH cDNA clone with the primers employed for RACE analysis are shown. (B) Molecular structure of the 5′ and 3′ RACE amplicons. Oligonucleotide primers specific for exon 2 (primers SSH I and SSH 4) and RACE adapters (primers AP1 and AP2) produced three 5′ amplicons (5′a, 5′b, and 5′c) and two 3′ amplicons (3′a and 3′b). The principal features of the 5′ and 3′ RACE products are depicted, including boundaries of 5′ or 3′ ends, splicing pattern, presence of ATG or stop codons from ORF K3 or ORF 70, and 3′ poly(A) sequences (AAA). Primers SSH 1 to 3 and SSH III and IV were also used in combination with adapter primers AP2 and AP1, respectively, producing unspliced amplicons exhibiting the same 5′ or 3′ RACE boundaries as the amplicons shown in this figure (data not shown). It is unlikely that unspliced amplicons were obtained upon amplification of contaminant DNA; in fact, (i) BCBL-1 cDNA was obtained from poly(A)+ purified RNA, (ii) the cDNA preparation was negative for amplification of genomic β-actin DNA sequences (data not shown), (iii) ligation of RACE adapters requires DNA blunt ends that are infrequently found at randomly nicked contaminant DNA termini, (iv) the unspliced 3′ RACE clone (3′a) has the typical mRNA poly(A) sequence that is not found on the HHV-8 genome, and (v) the 5′ RACE unspliced amplicon (5′b) shows several alternative and clustered 5′ ends that are usually found upon mapping of transcriptional start sites on cDNA molecules. (Nucleotide enumeration as for GenBank accession no. KSU75698.)
DNA sequencing of PCR products from 5′ RACE identified three different amplicons (Fig. 5B, amplicons 5′a, 5′b, and 5′c). Two of these amplicons (amplicons 5′a and 5′b) were unspliced, and one (amplicon 5′c) had the same splicing pattern as the IE SSH K3 cDNA clone (Fig. 5B). The 5′ boundary of one of the unspliced forms (amplicon 5′a) and of the IE spliced form (amplicon 5′c) overlapped the ORF 70 ATG at nt 21104, whereas the other unspliced amplicon (amplicon 5′b) corresponded to an mRNA molecule that was transcribed from a downstream promoter, internal to ORF 70, with four putative transcriptional start sites mapping from nt 20988 to 21004 (Fig. 5B). 3′ RACE analysis identified an unspliced polyadenylated amplicon as well as a polyadenylated amplicon exhibiting the same splicing pattern as the SSH cDNA (Fig. 5B, amplicons 3′a and 3′b, respectively). Amplicon 3′b showed two alternative polyadenylation sites, at nt 18577 and 18595, located immediately downstream of the stop codon of ORF K3.
As shown in Table 1, the most abundant amplicons obtained by RACE (amplicons 5′c and 3′b) had the same splicing pattern as the IE SSH K3 cDNA clone. Since RACE was performed with cDNA obtained in the presence of CHX, this further confirmed that the IE HHV-8 K3 transcript corresponded to the SSH K3 cDNA clone.
TABLE 1.
Percentages of 5′ and 3′ RACE clones obtained with cDNA from BCBL-1 cells induced with TPA for 4 h in the continuous presence of CHXa
| Amplicon | RACE boundary (nt) | Molecular structure | Clones (no. positive/ total) |
|---|---|---|---|
| 5′a | 21104 (ORF 70 ATG) | Unspliced | 1/16 |
| 5′b | 20988 to 21004 | Unspliced | 4/16 |
| 5′c | 21104 (ORF 70 ATG) | SSH K3 clone splicing pattern | 11/16 |
| 3′a | 18595 | Unspliced | 1/8 |
| 3′b | 18595, 18577 | SSH K3 clone splicing pattern | 7/8 |
5′ and 3′ RACE products were cloned in the plasmid PCRII (Invitrogen), and several randomly selected clones were analyzed by DNA sequencing.
Additional primers specific for exons 1 and 3 (Fig. 5A) yielded, as expected, unspliced amplicons with the same DNA ends as the amplicons obtained with primers internal to exon 2 (data not shown). All RACE amplicons were reamplified with nested-PCR primers, and the sequences of the nested-RACE fragments proved to be identical to those of the unnested-RACE products (data not shown).
Molecular arrangement of the K3 transcripts.
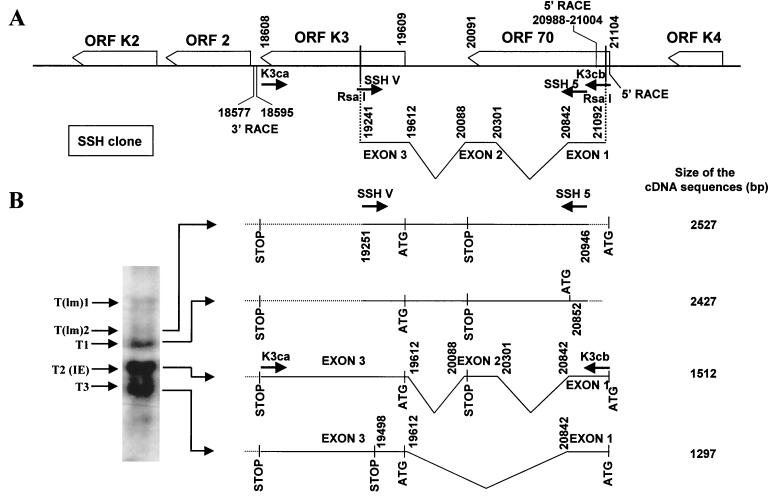
To characterize the molecular arrangement of the viral transcripts, amplification of the full-length K3 cDNAs was attained by long-range reverse transcription-PCR with primers spanning the ORF 70 ATG and the ORF K3 stop codons, i.e., with primers specific for the 5′ and 3′ RACE boundaries (primers K3ca and K3cb [Fig. 6]). This analysis, which was performed with cDNA from BCBL-1 cells that had been induced with TPA for 4 h in the presence of CHX, yielded two amplicons, corresponding to the IE doubly spliced SSH K3 cDNA clone (the most abundant transcript when CHX treatment was applied) and to a singly spliced form lacking exon 2 because of alternative splicing (Fig. 6B). These primers, however, did not allow amplification of cDNA from unspliced transcripts because these transcripts lacked sequences from the ORF 70 ATG (Fig. 5B) and/or were expressed at very low levels in the presence of CHX (Fig. 1A and Table 1). Amplification of cDNA sequences corresponding to the unspliced transcripts was, nevertheless, achieved by PCR with more-recessed primers (SSH V and SSH 5 [Fig. 6]). Overall, a total of four major transcripts with alternative splicing and/or transcriptional start sites, carrying sequences from both HHV-8 ORFs K3 and 70, were identified by combining data from RACE and long-range PCR analysis (Fig. 6B). The molecular sizes of these transcripts indicated that they corresponded to the low-mobility hybridization band T(lm)2, to the 2.5-kb band (T1), to the 1.5-kb IE transcript (T2), and to the 1.3-kb band (T3) (Fig. 6B).
FIG. 6.
Molecular structure of ORF K3 transcripts. (A) Diagram of the HHV-8 ORF K3-ORF 70 locus and of the SSH K3 cDNA clone. Primers K3cb and K3ca, which were designed to contain the ATG from ORF 70 and the stop codon from ORF K3, respectively, and primers SSH 5 and SSH V, designed in regions internal to the SSH clone, are indicated. The 5′ and 3′ RACE boundaries of the transcripts are shown. (B) Structure of the transcripts as deduced by combining data from RACE with that from PCR analysis with primers specific for RACE ends or internal to the SSH K3 cDNA clone. The major features of the transcripts are described; solid lines represent sequences amplified with primer pair K3ca-K3cb or SSH 5-SSH V, and broken lines represent sequences identified by RACE (see also Fig. 5B). The relative abundance of the transcripts obtained in the presence of CHX is depicted in the Northern blot inset (B, left); the various transcripts were assigned to the respective Northern blot bands on the basis of their molecular sizes. The sizes of the cDNA sequences are also shown (as calculated for the 3′ boundary present at nt 18577). (Nucleotide enumeration as for GenBank accession no. KSU75698.)
Two of these transcripts—namely, the T(lm)2 unspliced form and the IE doubly spliced transcript T2—were shown to contain the ATG and the stop codons from both ORFs 70 and K3; therefore, they have the molecular arrangement typical of bicistronic transcripts (Fig. 6B). Although these transcripts have the potential to express both of the ORFs, depending on the ATG usage, expression of a functional TS by the IE transcript is unlikely to occur because of extensive deletion of ORF 70 coding sequences by RNA splicing. The start site of the 2.4-kb unspliced DE transcript T1 was found to be located downstream of the ORF 70 ATG; however, this transcript potentially encodes a truncated TS homologue, starting at an internal in-frame ATG (nt 20852) (Fig. 6B), as well as a complete ORF K3 product. By contrast, the 1.3-kb singly spliced DE transcript, T3, contained both the ORF 70 and ORF K3 ATGs but lacked the stop codon from ORF 70. Depending on the ATG used, this transcript can encode the ORF K3 product and/or a fusion peptide; the latter, however, is truncated at nt 19498 because of a K3 out-of-frame translation pattern (Fig. 6B).
Notably, three of the four transcripts had a 5′ RACE end mapping to the ORF 70 ATG (Fig. 6B). However, as discussed below, further analysis indicated that the 5′ RACE end of these transcripts likely corresponds to a reverse transcriptase pausing site, the true transcriptional start sites being located further downstream in the HHV-8 genome.
Expression of ORF K3 in KS lesions.
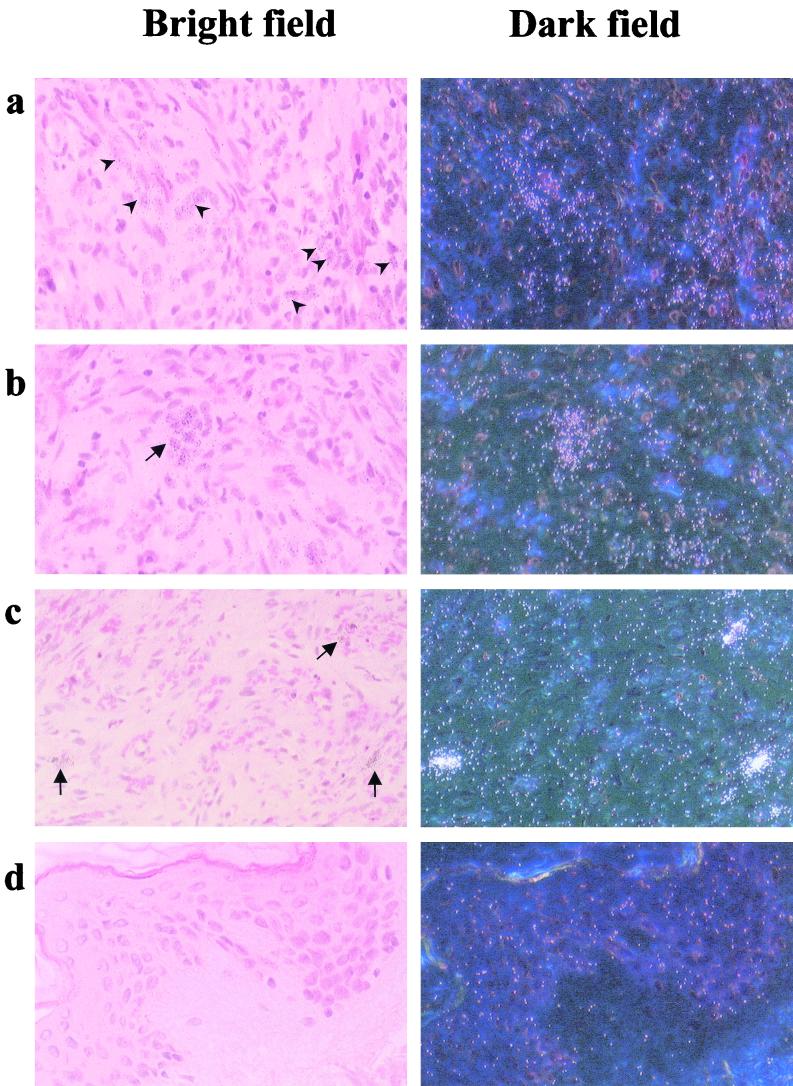
HHV-8 is present as a latent virus in the majority of infected KS spindle cells or endothelial cells lining lesional vessels, and it replicates productively in a small percentage of cells present in the lesions, including tissue-infiltrating lymphomonocytes (4, 47, 62–64; M. Stürzl, G. Ascherl, C. Blasig, S. R. Opalenik, B. Ensoli, and P. J. Browning, Letter, AIDS 12:1105–1106, 1998). Transcription of HHV-8 IE genes in KS lesions is, therefore, predicted to occur only in the small fraction of cells undergoing productive virus replication. To examine the expression pattern of HHV-8 ORF K3 in KS, seven KS lesions were analyzed by in situ hybridization (ISH) with an ORF K3-specific antisense RNA probe. K3 transcripts were found to be expressed by only a few cells in two of the seven lesions analyzed (Fig. 7a and b and data not shown), whereas the other five lesions were found to be negative (data not shown). A scattered pattern of expression by the HHV-8 ORF K8 (Fig. 7c), which was used as a control for HHV-8 early-gene expression, was also evident. In particular, sequences from this viral ORF are present in both IE transcripts (24, 36, 37, 58, 69, 74), which in BCBL-1 cells accumulate within 12 h of TPA induction (58), as well as DE transcripts encoding the viral transactivator K-bZIP (35, 58). ISH signals from ORF K3 were less frequent and less intense than those from ORF K8, suggesting that ORF K3 RNA is expressed at low levels in KS lesions (Fig. 7). By contrast, control (normal) skin above KS lesions was consistently negative for both ORF K3 and ORF K8 expression (Fig. 7d and data not shown).
FIG. 7.
Detection of transcripts from HHV-8 ORF K3 in a KS lesion by ISH. Microscopic bright fields (left panels) and corresponding dark fields (right panels) are shown (magnification, ×200). Upon hybridization with an HHV-8 ORF K3 antisense probe, low-intensity signals were detected in few cells that were scattered (a, arrowheads) or formed swirl-like structures within the lesion (b, arrow). Scattered cells with intense ISH signals (c, arrows) were detected in the same lesion upon hybridization with an antisense probe specific for the IE-DE HHV-8 ORF K8. No signals from HHV-8 ORF K3 or K8 were detected in normal skin above the KS lesion (panel d and data not shown). Consecutive sections were subjected to ISH with a control antisense β-actin RNA probe with positive results (data not shown).
DISCUSSION
In this study, we have shown that transcription of the MHC-I-downregulating HHV-8 ORF K3 in PEL cells is induced with kinetics typical of a viral early gene, confirming recent data pointing to virus-mediated downregulation of MHC-I molecules as an event associated with early phases of virus replication (26, 36, 67). Several early viral transcripts containing coding sequences from ORF K3 were identified, some of which showed a typical bicistronic structure. Although alternative ATG usage may enable bicistronic messengers to express the HHV-8 TS homologue, this is highly unlikely to occur for one of the bicistronic RNA molecules, from which a large part of the TS coding sequence from ORF 70 has been removed by RNA splicing. Indeed, this bicistronic molecule appears to play a key role in the transcriptional program leading to early induction of the K3 product; in fact, its expression was found to occur in the absence of de novo protein synthesis. By contrast, expression of all other K3 transcripts apparently requires newly synthesized factors. These data indicate that HHV-8 has evolved mechanisms to ensure a prompt and modulated expression of the K3 gene product through both IE and DE transcripts. Viral products downregulating MHC-I molecules are known to be encoded by early genes in other herpesviruses as well, including herpes simplex virus and cytomegalovirus (see references 27 and 31 and references cited therein). In fact, prompt immune evasion of productively infected cells is required for herpesvirus reactivation, as opposed to the efficient busting of cytotoxic T-lymphocyte responses by intermittent virus reactivation occurring throughout the host's life span (1, 6, 72). Notably, previous work and the present data indicate that HHV-8 ORFs K3 and K5 are expressed through both IE and DE transcripts in EBV-free PEL cell lines but only through DE transcripts in HHV-8- and EBV-coinfected cells (26, 67, 74), suggesting that EBV may interfere with the expression of some HHV-8 IE transcripts upon virus reactivation.
HHV-8 ORF K3 expression appears to occur through a complex transcriptional program that is likely to be modulated in many ways. In fact, several of the features of the HHV-8 K3 transcripts, including alternative donor splicing sites and alternative ATG usage, are possible targets of regulatory mechanisms tuning the expression of the K3 product. In addition, ORF K3 expression appears to be modulated through the activation of multiple transcriptional promoters. In fact, two different 5′ ends were identified, one mapping to a region internal to ORF 70 coding sequences and the other at the ORF 70 ATG. In particular, three of four K3 transcripts, including the IE doubly spliced RNA molecule, the DE spliced RNA molecules, and one of the unspliced transcripts, appeared to have been initiated at the first nucleotide of the ATG from ORF 70. This finding may suggest that these transcripts originate via the induction of a common IE large RNA precursor to undergo differential splicing by preexisting or de novo-synthesized splicing factors. This view, however, is in contrast with the DE kinetics of expression of the unspliced transcript. Indeed, neural network analysis of the HHV-8 K3 locus (45, 51) has identified in this region several additional transcriptional promoters, and preliminary reverse transcription-PCR analysis has indicated that these K3 transcripts are likely to be initiated within downstream sequences located between ORFs 70 and K4 (data not shown). Thus, the 5′ RACE ends of these transcripts may reflect the presence of a strong pausing site for reverse transcriptase, leading to premature cDNA termination. Additional studies are therefore required to fully elucidate the transcriptional patterns of these viral transcripts and their coding potentials.
The transcriptional factors responsible for inducing HHV-8 IE and DE K3 transcripts remain to be identified; however, by analogy with the other herpesviruses, it can be anticipated that activation of the IE transcript is dependent on housekeeping host genes, whereas the DE K3 transcripts are most likely induced by virus-encoded transcription factors. In this context, the transcription factor from the IE ORF 50, Rta, is known to activate several viral transcriptional promoters, including the K5 promoter (26), resulting in virus reactivation (24, 36, 37, 69). Since several studies indicate that active and prolonged HHV-8 reactivation is predictive of KS development (28, 40, 42, 54, 55, 71) or is associated with MCD recrudescence (25, 46), these viral factors, together with the ORF K3 product, may represent key factors in HHV-8 pathogenicity.
HHV-8 ORF K3 was found to be expressed at low levels only by rare cells present in KS lesions. Since only a few types of cells, including lymphomonocytes, are productively infected by HHV-8 in KS tissue (4, 47, 62; M. Stürzl, G. Ascherl, C. Blasig, S. R. Opalenik, B. Ensoli, and P. J. Browning, Letter, AIDS 12:1105–1106, 1998), these data are in agreement with the pattern of expression expected for a lytic IE or DE viral gene. However, since the vast majority of cells present in lesions were found to be negative for ORF K3 expression, these data also point to additional, as-yet-unknown mechanisms ensuring immune evasion to latently infected KS spindle cells. KS lesions are, in fact, infiltrated by activated (e.g., gamma-interferon-expressing) monocytes/macrophages and CD4+ and CD8+ T cells, as well as by NK cells; however, these effector cells are incapable of clearing infected cells from lesions (18, 19, 59). One of the possible mechanisms for immune evasion of latently infected KS spindle cells is related to the expression of the HHV-8 FLICE-inhibitory protein homologue (vFLIP) (2, 16, 70), which, in fact, is inversely correlated with cell apoptosis in KS lesions (63) and has been shown to inhibit FAS-mediated cell killing by CD8+ T cells in a murine model (16). Other observations, however, point to host mechanisms. In particular, our recent work indicates a decreased NK cell cytotoxic activity in KS patients (M. C. Sirianni, L. Vincenzi, S. Topino, A. Giovannetti, F. Mazzetta, C. Alario, and B. Ensoli, Abstr. 2nd Int. Workshop KSHV/HHV-8 Relat. Agents, abstr. 29, 1999) and the upregulation of killing-inhibitory receptors (KIR) both in a subset of CD8+ T cells and in NK cells from persons with KS or at risk of KS (M. C. Sirianni, C. Alario, F. Libi, D. Scaramuzzi, S. Topino, F. Ensoli, and P. Monini, Abstr. 3rd Int. Workshop Kaposi's Sarcoma-Associated Herpesvirus Relat. Agents, abstr. 105, 2000).
Early expression of HHV-8 ORFs K3 and K5, leading to prompt downregulation of MHC-I proteins and coactivation molecules (10, 26, 29, 30), may be crucial to KS development. First, this may represent a potent mechanism for immune evasion of lytically infected lymphomonocytes infiltrating KS lesions; these cells, in turn, may be required for virus transmission and long-lasting latent infection of KS spindle cells, as suggested by the loss of HHV-8 DNA from KS cells upon culture in vitro (14, 38; C. Lebbè, P. de Cremoux, M. Rybojad, C. Costa Da Cunha, P. Morel, and F. Calvo, Letter, Lancet 345:1180, 1995). Second, HHV-8 reactivation by the ICs present in KS (40) leads to peripheral blood viremia and virus dissemination in tissues that can precede both overt immunosuppression and KS development (3, 4, 9, 19, 33, 42, 59, 71). Thus, prompt and efficient immune evasion by cells undergoing virus reactivation may be a key mechanism for HHV-8 spreading during the inapparent phase that precedes KS development. Therefore, agents directed at blocking the expression of the HHV-8 K3 gene product or at inhibiting its biological function may be exploited for the prevention of HHV-8-associated diseases in infected individuals.
ACKNOWLEDGMENTS
This work was supported by grants from the AIDS Project of the Italian Ministry of Health to B. Ensoli, A. Caputo, and E. Cassai; by a grant from the “Associazione Italiana per la Ricerca sul Cancro” (AIRC, Milan) to B. Ensoli; and by the Biofuture program of the German Ministry of Education and Research (BMBF) and the Deutsche Krebshilfe (Mildred Scheel Stifung) to M. Stürzl. P. Rimessi was the recipient of a postdoctoral fellowship in E. Cassai's laboratory.
We thank A. Lippa and F. M. Regini for editorial assistance.
REFERENCES
- 1.Arvin A M. Varicella-zoster virus. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Vol. 2. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2547–2582. [Google Scholar]
- 2.Bertin J, Armstrong R C, Ottilie S, Martin D A, Wang Y, Banks S, Wang G H, Senkevich T G, Alnemri E S, Moss B, Lenardo M J, Tomaselli K J, Cohen J I. Death effector domain-containing herpesvirus and poxvirus proteins inhibit both Fas- and TNFR1-induced apoptosis. Proc Natl Acad Sci USA. 1997;94:1172–1176. doi: 10.1073/pnas.94.4.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blackbourn D J, Lennette E T, Ambroziak J, Mourich D V, Levy J A. Human herpesvirus 8 detection in nasal secretions and saliva. J Infect Dis. 1998;177:213–216. doi: 10.1086/517356. [DOI] [PubMed] [Google Scholar]
- 4.Blasig C, Zietz C, Haar B, Neipel F, Esser S, Brockmeyer N H, Tschachler E, Colombini S, Ensoli B, Stürzl M. Monocytes in Kaposi's sarcoma lesions are productively infected by human herpesvirus 8. J Virol. 1997;71:7963–7968. doi: 10.1128/jvi.71.10.7963-7968.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boshoff C, Schulz T F, Kennedy M M, Graham A K, Fisher C, Thomas A, McGee J O, Weiss R A, O'Leary J J. Kaposi's sarcoma-associated herpesvirus infects endothelial and spindle cells. Nat Med. 1995;1:1274–1278. doi: 10.1038/nm1295-1274. [DOI] [PubMed] [Google Scholar]
- 6.Britt W J, Alford C A. Cytomegalovirus. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Vol. 2. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2493–2523. [Google Scholar]
- 7.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 8.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 9.Corbellino M, Poirel L, Bestetti G, Pizzuto M, Aubin J T, Capra M, Bifulco C, Berti E, Agut H, Rizzardini G, Galli M, Parravicini C. Restricted tissue distribution of extralesional Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS patients with Kaposi's sarcoma. AIDS Res Hum Retrovir. 1996;12:651–657. doi: 10.1089/aid.1996.12.651. [DOI] [PubMed] [Google Scholar]
- 10.Coscoy L, Ganem D. Kaposi's sarcoma-associated herpesvirus encodes two proteins that block cell surface display of MHC class I chains by enhancing their endocytosis. Proc Natl Acad Sci USA. 2000;97:8051–8056. doi: 10.1073/pnas.140129797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis M A, Stürzl M A, Blasig C, Schreier A, Guo H G, Reitz M, Opalenik S R, Browning P J. Expression of human herpesvirus 8-encoded cyclin D in Kaposi's sarcoma spindle cells. J Natl Cancer Inst. 1997;89:1868–1874. doi: 10.1093/jnci/89.24.1868. [DOI] [PubMed] [Google Scholar]
- 12.Diatchenko L, Lukyanov S, Lau Y F, Siebert P D. Suppression subtractive hybridization: a versatile method for identifying differentially expressed genes. Methods Enzymol. 1999;303:349–380. doi: 10.1016/s0076-6879(99)03022-0. [DOI] [PubMed] [Google Scholar]
- 13.Diatchenko L, Lau Y F, Campbell A P, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov E D, Siebert P D. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci USA. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dictor M, Rambech E, Way D, Witte M, Bendsoe N. Human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus) DNA in Kaposi's sarcoma lesions, AIDS Kaposi's sarcoma cell lines, endothelial Kaposi's sarcoma simulators, and the skin of immunosuppressed patients. Am J Pathol. 1996;148:2009–2016. [PMC free article] [PubMed] [Google Scholar]
- 15.Dittmer D, Lagunoff M, Renne R, Staskus K, Haase A, Ganem D. A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus. J Virol. 1998;72:8309–8315. doi: 10.1128/jvi.72.10.8309-8315.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Djerbi M, Screpanti V, Catrina A I, Bogen B, Biberfeld P, Grandien A. The inhibitor of death receptor signaling, FLICE-inhibitory protein, defines a new class of tumor progression factors. J Exp Med. 1999;190:1025–1032. doi: 10.1084/jem.190.7.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dupin N, Fisher C, Kellam P, Ariad S, Tulliez M, Franck N, van Marck E, Salmon D, Gorin I, Escande J P, Weiss R A, Alitalo K, Boshoff C. Distribution of human herpesvirus-8 latently infected cells in Kaposi's sarcoma, multicentric Castleman's disease, and primary effusion lymphoma. Proc Natl Acad Sci USA. 1999;96:4546–4551. doi: 10.1073/pnas.96.8.4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ensoli, B., P. Monini, and M. Stürzl. Reactivation and role of HHV-8 in Kaposi's sarcoma initiation. Adv. Cancer Res., in press. [DOI] [PubMed]
- 19.Fiorelli V, Gendelman R, Sirianni M C, Chang H K, Colombini S, Markham P D, Monini P, Sonnabend J, Pintus A, Gallo R C, Ensoli B. γ-Interferon produced by CD8+ T cells infiltrating Kaposi's sarcoma induces spindle cells with angiogenic phenotype and synergy with human immunodeficiency virus-1 Tat protein: an immune response to human herpesvirus-8 infection? Blood. 1998;91:956–967. [PubMed] [Google Scholar]
- 20.Foreman K E, Bacon P E, Hsi E D, Nickoloff B J. In situ polymerase chain reaction-based localization studies support role of human herpesvirus-8 as the cause of two AIDS-related neoplasms: Kaposi's sarcoma and body cavity lymphoma. J Clin Investig. 1997;99:2971–2978. doi: 10.1172/JCI119492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frohman M A. Rapid amplification of complementary DNA ends for generation of full-length complementary DNAs: thermal RACE. Methods Enzymol. 1993;218:340–356. doi: 10.1016/0076-6879(93)18026-9. [DOI] [PubMed] [Google Scholar]
- 22.Frohman M A, Dush M K, Martin G R. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA. 1988;85:8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao S J, Kingsley L, Hoover D R, Spira T J, Rinaldo C R, Saah A, Phair J, Detels R, Parry P, Chang Y, Moore P S. Seroconversion to antibodies against Kaposi's sarcoma-associated herpesvirus-related latent nuclear antigens before the development of Kaposi's sarcoma. N Engl J Med. 1996;335:233–241. doi: 10.1056/NEJM199607253350403. [DOI] [PubMed] [Google Scholar]
- 24.Gradoville L, Gerlach J, Grogan E, Shedd D, Nikiforow S, Metroka C, Miller G. Kaposi's sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line. J Virol. 2000;74:6207–6212. doi: 10.1128/jvi.74.13.6207-6212.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grandadam M, Dupin N, Calvez V, Gorin I, Blum L, Kernbaum S, Sicard D, Buisson Y, Agut H, Escande J P, Huraux J M. Exacerbations of clinical symptoms in human immunodeficiency virus type 1-infected patients with multicentric Castleman's disease are associated with a high increase in Kaposi's sarcoma herpesvirus DNA load in peripheral blood mononuclear cells. J Infect Dis. 1997;175:1198–1201. doi: 10.1086/593567. [DOI] [PubMed] [Google Scholar]
- 26.Haque M, Chen J, Ueda K, Mori Y, Nakano K, Hirata Y, Kanamori S, Uchiyama Y, Inagi R, Okuno T, Yamanishi K. Identification and analysis of the K5 gene of Kaposi's sarcoma-associated herpesvirus. J Virol. 2000;74:2867–2875. doi: 10.1128/jvi.74.6.2867-2875.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hill A, Jugovic P, York I, Russ G, Bennink J, Yewdell J, Ploegh H, Johnson D. Herpes simplex virus turns off the TAP to evade host immunity. Nature. 1995;375:411–415. doi: 10.1038/375411a0. [DOI] [PubMed] [Google Scholar]
- 28.Hudnall S D, Rady P L, Tyring S K, Fish J C. Serologic and molecular evidence of human herpesvirus 8 activation in renal transplant recipients. J Infect Dis. 1998;178:1791–1794. doi: 10.1086/314482. [DOI] [PubMed] [Google Scholar]
- 29.Ishido S, Wang C, Lee B-S, Cohen G B, Jung J U. Downregulation of major histocompatibility complex class I molecules by Kaposi's sarcoma-associated herpesvirus K3 and K5 proteins. J Virol. 2000;74:5300–5309. doi: 10.1128/jvi.74.11.5300-5309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishido S, Choi J K, Lee B S, Wang C, DeMaria M, Johnson R P, Cohen G B, Jung J U. Inhibition of natural killer cell-mediated cytotoxicity by Kaposi's sarcoma-associated herpesvirus K5 protein. Immunity. 2000;13:365–374. doi: 10.1016/s1074-7613(00)00036-4. [DOI] [PubMed] [Google Scholar]
- 31.Jones T R, Wiertz E J, Sun L, Fish K N, Nelson J A, Ploegh H L. Human cytomegalovirus US3 impairs transport and maturation of major histocompatibility complex class I heavy chains. Proc Natl Acad Sci USA. 1996;93:11327–11333. doi: 10.1073/pnas.93.21.11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kieff E. Epstein-Barr virus and its replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Vol. 2. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2343–2353. [Google Scholar]
- 33.LaDuca J R, Love J L, Abbott L Z, Dube S, Freidman-Kien A E, Poiesz B J. Detection of human herpesvirus 8 DNA sequences in tissues and bodily fluids. J Infect Dis. 1998;178:1610–1615. doi: 10.1086/314514. [DOI] [PubMed] [Google Scholar]
- 34.Lebbè C, Blum L, Pellet C, Blanchard G, Verola O, Morel P, Danne O, Calvo F. Clinical and biological impact of antiretroviral therapy with protease inhibitors on HIV-related Kaposi's sarcoma. AIDS. 1998;12:F45–F49. doi: 10.1097/00002030-199807000-00002. [DOI] [PubMed] [Google Scholar]
- 35.Lin S-F, Robinson D R, Miller G, Kung H-J. Kaposi's sarcoma-associated herpesvirus encodes a bZIP protein with homology to BZLF1 of Epstein-Barr virus. J Virol. 1999;73:1909–1917. doi: 10.1128/jvi.73.3.1909-1917.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lukac D M, Kirshner J R, Ganem D. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J Virol. 1999;73:9348–9361. doi: 10.1128/jvi.73.11.9348-9361.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lukac D M, Renne R, Kirschner J R, Ganem D. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology. 1998;252:304–312. doi: 10.1006/viro.1998.9486. [DOI] [PubMed] [Google Scholar]
- 38.Monini P, Rotola A, de Lellis L, Corallini A, Secchiero P, Albini A, Benelli R, Parravicini C, Barbanti-Brodano G, Cassai E. Latent BK virus infection and Kaposi's sarcoma pathogenesis. Int J Cancer. 1996;66:717–722. doi: 10.1002/(SICI)1097-0215(19960611)66:6<717::AID-IJC1>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 39.Monini P, Carlini F, Stürzl M, Rimessi P, Superti F, Franco M, Melucci-Vigo G, Cafaro A, Goletti D, Sgadari C, Buttò S, Leone P, Leone P, Chiozzini C, Barresi C, Tinari A, Bonaccorsi A, Capobianchi M R, Giuliani M, Di Carlo A, Andreoni M, Rezza G, Ensoli B. Alpha interferon inhibits human herpesvirus 8 (HHV-8) reactivation in primary effusion lymphoma cells and reduces HHV-8 load in cultured peripheral blood mononuclear cells. J Virol. 1999;73:4029–4041. doi: 10.1128/jvi.73.5.4029-4041.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Monini P, Colombini S, Stürzl M, Goletti D, Cafaro A, Sgadari C, Buttò S, Franco M, Leone P, Fais S, Leone P, Melucci-Vigo G, Chiozzini C, Carlini F, Ascherl G, Cornali E, Zietz C, Ramazzotti E, Ensoli F, Andreoni M, Pezzotti P, Rezza G, Yarchoan R, Gallo R C, Ensoli B. Reactivation and persistence of human herpesvirus-8 infection in B cells and monocytes by Th-1 cytokines increased in Kaposi's sarcoma. Blood. 1999;93:4044–4058. [PubMed] [Google Scholar]
- 41.Moore P S, Boshoff C, Weiss R A, Chang Y. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science. 1996;274:1739–1744. doi: 10.1126/science.274.5293.1739. [DOI] [PubMed] [Google Scholar]
- 42.Moore P S, Kingsley L A, Holmberg S D, Spira T, Gupta P, Hoover D R, Parry J P, Conley L J, Jaffe H W, Chang Y. Kaposi's sarcoma-associated herpesvirus infection prior to onset of Kaposi's sarcoma. AIDS. 1996;10:175–180. doi: 10.1097/00002030-199602000-00007. [DOI] [PubMed] [Google Scholar]
- 43.Moosa M R, Treurnicht F K, van Rensburg E J, Schneider J W, Jordaan H F, Engelbrecht S. Detection and subtyping of human herpesvirus-8 in renal transplant patients before and after remission of Kaposi's sarcoma. Transplantation. 1998;66:214–218. doi: 10.1097/00007890-199807270-00013. [DOI] [PubMed] [Google Scholar]
- 44.Neipel F, Albrecht J-C, Ensser A, Huang Y-Q, Li J J, Friedman-Kien A E, Fleckenstein B. Human herpesvirus 8 encodes a homolog of interleukin-6. J Virol. 1997;71:839–842. doi: 10.1128/jvi.71.1.839-842.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ohler U, Harbeck S, Niemann H, Noth E, Reese M G. Interpolated Markov chains for eukaryotic promoter recognition. Bioinformatics. 1999;15:362–369. doi: 10.1093/bioinformatics/15.5.362. [DOI] [PubMed] [Google Scholar]
- 46.Oksenhendler E, Carcelain G, Aoki Y, Boulanger E, Maillard A, Clauvel J P, Agbalika F. High levels of human herpesvirus 8 viral load, human interleukin-6, interleukin-10, and C reactive protein correlate with exacerbation of multicentric Castleman's disease in HIV-infected patients. Blood. 2000;96:2069–2073. [PubMed] [Google Scholar]
- 47.Orenstein J M, Alkan S, Blauvelt A, Jeang K T, Weinstein M D, Ganem D, Herndier B. Visualization of human herpesvirus type 8 in Kaposi's sarcoma by light and transmission electron microscopy. AIDS. 1997;11:F35–F45. doi: 10.1097/00002030-199705000-00001. [DOI] [PubMed] [Google Scholar]
- 48.Osman M, Kubo T, Gill J, Neipel F, Becker M, Smith G, Weiss R, Gazzard B, Boshoff C, Gotch F. Identification of human herpesvirus 8-specific cytotoxic T-cell responses. J Virol. 1999;73:6136–6140. doi: 10.1128/jvi.73.7.6136-6140.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parravicini C, Chandran B, Corbellino M, Berti E, Paulli M, Moore P S, Chang Y. Differential viral protein expression in Kaposi's sarcoma-associated herpesvirus-infected diseases: Kaposi's sarcoma, primary effusion lymphoma, and multicentric Castleman's disease. Am J Pathol. 2000;156:743–749. doi: 10.1016/S0002-9440(10)64940-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parravicini C, Olsen S J, Capra M, Poli F, Sirchia G, Gao S J, Berti E, Nocera A, Rossi E, Bestetti G, Pizzuto M, Galli M, Moroni M, Moore P S, Corbellino M. Risk of Kaposi's sarcoma-associated herpes virus transmission from donor allografts among Italian posttransplant Kaposi's sarcoma patients. Blood. 1997;90:2826–2829. [PubMed] [Google Scholar]
- 51.Reese, M. G. Application of a time-delay neural network to the annotation of the Drosophila melanogaster genome. Comput. Chem., in press. [DOI] [PubMed]
- 52.Reese M G, Eeckman F H, Kulp D, Haussler D. Improved splice site detection in Genie. J Comput Biol. 1997;4:311–323. doi: 10.1089/cmb.1997.4.311. [DOI] [PubMed] [Google Scholar]
- 53.Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 54.Renwick N, Halaby T, Weverling G J, Dukers N H, Simpson G R, Coutinho R A, Lange J M, Schulz T F, Goudsmit J. Seroconversion for human herpesvirus 8 during HIV infection is highly predictive of Kaposi's sarcoma. AIDS. 1998;12:2481–2488. doi: 10.1097/00002030-199818000-00018. [DOI] [PubMed] [Google Scholar]
- 55.Rezza G, Andreoni M, Dorrucci M, Pezzotti P, Monini P, Zerboni R, Salassa B, Colangeli V, Sarmati L, Nicastri E, Barbanera M, Pristera R, Aiuti F, Ortona L, Ensoli B. Human herpesvirus-8 seropositivity and risk of Kaposi's sarcoma and other acquired immunodeficiency syndrome-related diseases. J Natl Cancer Inst. 1999;91:1468–1474. doi: 10.1093/jnci/91.17.1468. [DOI] [PubMed] [Google Scholar]
- 56.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Vol. 2. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2231–2241. [Google Scholar]
- 57.Russo J J, Bohenzky R A, Chien M C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV-8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Seaman W T, Ye D, Wang R X, Hale E E, Weisse M, Quinlivan E B. Gene expression from the ORF50/K8 region of Kaposi's sarcoma-associated herpesvirus. Virology. 1999;263:436–449. doi: 10.1006/viro.1999.9963. [DOI] [PubMed] [Google Scholar]
- 59.Sirianni M C, Vincenzi L, Fiorelli V, Topino S, Scala E, Uccini S, Angeloni A, Faggioni A, Cerimele D, Cottoni F, Aiuti F, Ensoli B. γ-Interferon production in peripheral blood mononuclear cells and tumor infiltrating lymphocytes from Kaposi's sarcoma patients: correlation with the presence of human herpesvirus-8 in peripheral blood mononuclear cells and lesional macrophages. Blood. 1998;91:968–976. [PubMed] [Google Scholar]
- 60.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d'Agay M F, Clauvel J P, Raphael M, Degos L. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 61.Staskus K A, Zhong W, Gebhard K, Herndier B, Wang H, Renne R, Beneke J, Pudney J, Anderson D J, Ganem D, Haase A T. Kaposi's sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J Virol. 1997;71:715–719. doi: 10.1128/jvi.71.1.715-719.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stürzl M, Wunderlich A, Ascherl G, Hohenadl C, Monini P, Zietz C, Browning P J, Neipel F, Biberfeld P, Ensoli B. Human herpesvirus-8 (HHV-8) gene expression in Kaposi's sarcoma (KS) primary lesions: an in situ hybridization study. Leukemia. 1999;13:110–112. doi: 10.1038/sj.leu.2401323. [DOI] [PubMed] [Google Scholar]
- 63.Stürzl M, Hohenadl C, Zietz C, Castanos-Velez E, Wunderlich A, Ascherl G, Biberfeld P, Monini P, Browning P J, Ensoli B. Expression of the K13/vFLIP gene of human herpesvirus 8 and apoptosis in Kaposi's sarcoma spindle cells. J Natl Cancer Inst. 1999;91:1725–1733. doi: 10.1093/jnci/91.20.1725. [DOI] [PubMed] [Google Scholar]
- 64.Stürzl M, Blasig C, Schreier A, Neipel F, Hohenadl C, Cornali E, Ascherl G, Esser S, Brockmeyer N H, Ekman M, Kaaya E E, Tschachler E, Biberfeld P. Expression of HHV-8 latency-associated T0.7 RNA in spindle cells and endothelial cells of AIDS-associated, classical and African Kaposi's sarcoma. Int J Cancer. 1997;72:68–71. doi: 10.1002/(sici)1097-0215(19970703)72:1<68::aid-ijc10>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 65.Stürzl M, Brandstetter H, Zietz C, Eisenburg B, Raivich G, Gearing D P, Brockmeyer N H, Hofschneider P H. Identification of interleukin-1 and platelet-derived growth factor-B as major mitogens for the spindle cells of Kaposi's sarcoma: a combined in vitro and in vivo analysis. Oncogene. 1995;10:2007–2016. [PubMed] [Google Scholar]
- 66.Stürzl M, Roth W K, Brockmeyer N H, Zietz C, Speiser B, Hofschneider P H. Expression of platelet-derived growth factor and its receptor in AIDS-related Kaposi sarcoma in vivo suggests paracrine and autocrine mechanisms of tumor maintenance. Proc Natl Acad Sci USA. 1992;89:7046–7050. doi: 10.1073/pnas.89.15.7046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sun R, Lin S-F, Staskus K, Gradoville L, Grogan E, Haase A, Miller G. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J Virol. 1999;73:2232–2242. doi: 10.1128/jvi.73.3.2232-2242.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sun R, Lin S-F, Gradoville L, Miller G. Polyadenylylated nuclear RNA encoded by Kaposi sarcoma-associated herpesvirus. Proc Natl Acad Sci USA. 1996;93:11883–11888. doi: 10.1073/pnas.93.21.11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sun R, Lin S-F, Gradoville L, Yuan Y, Zhu F, Miller G. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc Natl Acad Sci USA. 1998;95:10866–10871. doi: 10.1073/pnas.95.18.10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thome M, Schneider P, Hofmann K, Fickenscher H, Meinl E, Neipel F, Mattmann C, Burns K, Bodmer J L, Schroter M, Scaffidi C, Krammer P H, Peter M E, Tschopp J. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature. 1997;386:517–521. doi: 10.1038/386517a0. [DOI] [PubMed] [Google Scholar]
- 71.Whitby D, Howard M R, Tenant-Flowers M, Brink N S, Copas A, Boshoff C, Hatzioannou T, Suggett F E, Aldam D M, Denton A S, Miller R F, Weller I V D, Weiss R A, Tedder R S, Schulz T F. Detection of Kaposi sarcoma associated herpesvirus in peripheral blood of HIV-infected individuals and progression to Kaposi's sarcoma. Lancet. 1995;346:799–802. doi: 10.1016/s0140-6736(95)91619-9. [DOI] [PubMed] [Google Scholar]
- 72.Whitley R J. Herpes simplex viruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Vol. 2. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2297–2342. [Google Scholar]
- 73.Zhong W, Wang H, Herndier B, Ganem D. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc Natl Acad Sci USA. 1996;93:6641–6646. doi: 10.1073/pnas.93.13.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhu F X, Cusano T, Yuan Y. Identification of the immediate-early transcripts of Kaposi's sarcoma-associated herpesvirus. J Virol. 1999;73:5556–5567. doi: 10.1128/jvi.73.7.5556-5567.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]