Abstract
Assisted reproductive technologies are an emerging field in equine reproduction, with species-dependent peculiarities, such as the low success rate of conventional IVF. Here, the ‘cumulome’ was related to the developmental capacity of its corresponding oocyte. Cumulus–oocyte complexes collected from slaughterhouse ovaries were individually matured, fertilized by ICSI, and cultured. After maturation, the cumulus was collected for proteomics analysis using label-free mass spectrometry (MS)-based protein profiling by nano-HPLC MS/MS and metabolomics analysis by UPLC-nanoESI MS. Overall, a total of 1671 proteins and 612 metabolites were included in the quantifiable ‘cumulome’. According to the development of the corresponding oocytes, three groups were compared with each other: not matured (NM; n = 18), cleaved (CV; n = 15), and blastocyst (BL; n = 19). CV and BL were also analyzed together as the matured group (M; n = 34). The dataset revealed a closer connection within the two M groups and a more distinct separation from the NM group. Overrepresentation analysis detected enrichments related to energy metabolism as well as vesicular transport in the M group. Functional enrichment analysis found only the KEGG pathway ‘oxidative phosphorylation’ as significantly enriched in the NM group. A compound attributed to ATP was observed with significantly higher concentrations in the BL group compared with the NM group. Finally, in the NM group, proteins related to degradation of glycosaminoglycans were lower and components of cumulus extracellular matrix were higher compared to the other groups. In summary, the study revealed novel pathways associated with the maturational and developmental competence of oocytes.
Keywords: equine, oocyte, cumulus cells, proteomics, metabolomics, biomarker, developmental competence, maturation, IVM, IVF
Graphical Abstract
Graphical Abstract.
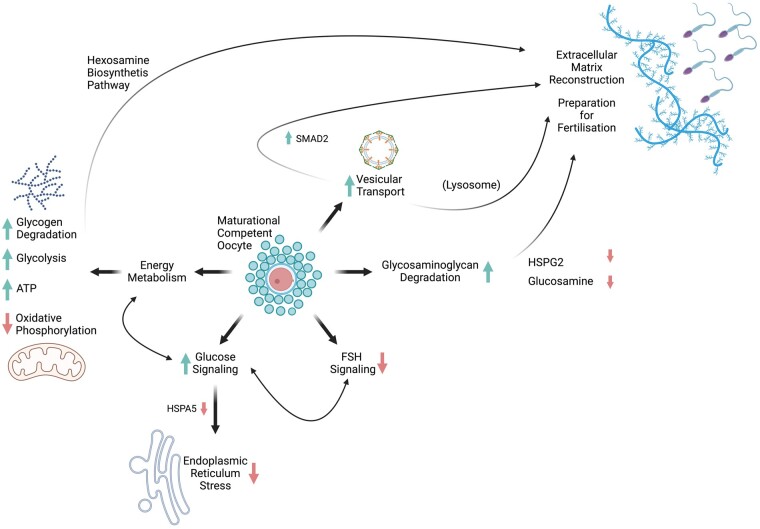
Cumulus signaling associated with oocyte developmental competence. HSPA5, endoplasmic reticulum chaperone BiP; HSPG2, basement membrane-specific heparan sulfate proteoglycan core protein; SMAD2, mothers against decapentaplegic homolog 2.
Introduction
In a broad variety of species, assisted reproductive technologies are applied to optimize reproductive efficiency or to overcome fertility issues. Amongst these, the horse is tainted with some peculiarities regarding reproductive biology. The response to hormonal induction of multiple ovulations is limited (Squires and McCue, 2007; Blanco et al., 2009). Therefore, superovulation of mares for embryo transfer or collection of mature oocytes for IVF is mostly irrelevant in clinical programs. Instead, immature oocytes are collected by oocyte retrieval and subjected to IVM (Galli et al., 2001). Unfortunately, IVM is one of the major challenges in ART across species (Rouhollahi Varnosfaderani et al., 2020), as it results in an impaired developmental capacity of the oocyte. In particular, the maturational competence of equine oocytes seems to be affected by processing factors, such as the time until the oocytes are recovered from the ovaries after slaughter or the duration in the maturation medium (Hinrichs, 2010). Maturation rates of equine oocytes in vitro can be up to 71%, especially with prolonged maturation times (36–38 h) (Lazzari et al., 2020). Rates for expanded cumulus–oocyte complexes (COCs) are usually higher (63%) than for compact COCs (23%) (Galli et al., 2007; Hinrichs, 2010). However, a direct comparison of the developmental rates of oocytes matured in vitro with oocytes matured in vivo, which were all transferred back to the fallopian tube of inseminated recipient mares, resulted in lower development of embryonic vesicles on Day 16 for the in vitro group (7% vs 82%) (Scott et al., 2001). Similar results have also been described when ICSI was used, but if the dominant stimulated follicles start to mature in vivo, a blastocyst (BL) rate of 70% can be achieved, which is twice as high as that of immature oocytes (Foss et al., 2013).
For a long time, standard IVF in horses, resulting in live offspring, was only successful with oocytes that had matured in vivo (Palmer et al., 1991) and therefore, IVM was considered unsuitable for embryo production (Leemans et al., 2016). The explanation for this IVF failure in the horse is not fully elucidated. Besides inadequate sperm capacitation, hardening of the zona pellucida after IVM that prevents sperm penetration of the oocyte has been discussed (Dell’Aquila et al., 1999; Hinrichs et al., 2002; Leemans et al., 2016; Lazzari et al., 2020). To overcome this fertilization issue, the technical achievements of ICSI from human ART were transferred into equine reproduction and contributed to satisfactory protocols for the laboratory production of equine embryos (Palermo et al., 1992; Squires et al., 1996; Lazzari et al., 2020; Briski and Salamone, 2022). Only recently, a standard IVF using an adapted system using extended incubation with semen, with 22 h sperm pre-incubation and 3 h COC co-incubation, was described, and this led to a satisfactory BL rate (Felix et al., 2022). Nevertheless, there is a great demand to increase the efficiency of equine ART and to understand the relevant processes.
The metabolic profile of equine oocytes during maturation differs significantly from other species and depends on the age of the mare (Lewis et al., 2020a; Catandi et al., 2023). Therefore, investigating metabolism during this critical period of maturation can help to elucidate factors associated with the developmental competence of the oocyte. A unique cell source to examine metabolism non-invasively for the oocyte is cumulus cells (CCs). They hold a close bidirectional communication with their oocyte and accompany the oocyte throughout the maturation process (Russell et al., 2016). And, they are shed naturally after fertilization or removed artificially after maturation to perform ICSI. This brought CCs into the focus of biomarker research for the prediction of oocyte development, with the rationale to find additional prognostic tools to select embryos for intra-uterine transfer (Sirait et al., 2021).
Especially for bovine and human reproduction, a variety of gene-expression studies on pooled cumulus samples are available, and have identified numerous genes linked to maturation status (Assou et al., 2006; Ouandaogo et al., 2011, 2012; Bunel et al., 2014; Li et al., 2014, 2015; Wyse et al., 2020) or post-fertilization development (Anderson et al., 2009; Adriaenssens et al., 2010; Assou et al., 2010; Assidi et al., 2011; Wathlet et al., 2011, 2012, 2013; Bunel et al., 2014; Scarica et al., 2019). Global transcriptomic studies are usually performed on pooled samples, for reasons of simplicity and cost. Pooling of samples in biomarker research deals with negative effects such as a reduction of biological variance or masking of individual candidates (Diz et al., 2009; Telaar et al., 2010). With the increasing sensitivity of the techniques, studies requiring no or only moderate pooling (n = 5) of cumulus samples from individual cultured COCs for transcriptomic analysis have arisen (Bunel et al., 2015; Borup et al., 2016; Artini et al., 2017; Green et al., 2018; Demiray et al., 2019; Aguila et al., 2020; El-Maarri et al., 2021). Analyzing the proteome or metabolome at the single COC level is more challenging, however, as this approach includes no enrichment steps. Nevertheless, proteomic analysis of single human and bovine oocytes has already been successful (Virant-Klun et al., 2016; Labas et al., 2017). Also, metabolomic profiling of glucose metabolism and lipid fingerprinting has been successful at the level of single oocytes (González-Serrano et al., 2013; Sessions-Bresnahan et al., 2016; Anderson et al., 2022; Catandi et al., 2022). However, the metabolomics analysis of human CCs in moderately pooled samples (n = 12) has provided only poor predictive value for the developmental competence of the enclosed oocytes (Martinez-Moro et al., 2021).
In previous ‘cumulomics’ studies, the metabolome and proteome of bovine and equine CCs (‘cumulome’) corresponding to single oocytes were analyzed. These studies elucidated important metabolic aberrations between in vivo maturation and IVM, using an ultrasensitive mass spectrometry (MS)-based workflow (Fortes et al., 2014; Walter et al., 2019, 2020). Now, this approach provides the unique opportunity to correlate the ‘cumulome’ directly to the developmental competence of the corresponding oocyte to find markers for the developmental competence of the oocytes (Catandi et al., 2022, 2023). The use of a multi-omics approach gives unprecedented possibilities to enhance the current understanding of metabolism during IVM (Jendoubi, 2021). Up to now, the available studies have failed to identify universal markers that predict the developmental potential of oocytes (El-Maarri et al., 2021). Single marker genes were not homogenous across studies, but the differentially expressed genes, proteins, or metabolites can lead to pathways that might be able to distinguish between oocytes of good or poor quality (El-Maarri et al., 2021). With the identification of global molecular signatures determining the oocytes’ developmental competence, novel approaches to overcome the limitations in equine IVF can evolve. Therefore, this study related the ‘cumulome’ to the maturational and developmental potential of the corresponding individual oocytes. These ‘cumulomic signatures’ identified novel paths for augmenting oocyte quality. As the horse can serve as a good model for human reproduction, the obtained data might also contribute to overcome similar challenges in human IVM (Carnevale, 2008; Dell’Aquila et al., 2012; Virant-Klun and Krijgsveld, 2014).
Materials and methods
Experimental design
For the correlation of the ‘cumulome’ with the developmental competence of the oocyte, cumulus samples of individual COCs originating from three different groups were analyzed. One group contained cumulus of oocytes that failed to mature (NM: not matured, n = 18). Oocytes of the other groups matured successfully (M: matured, n = 34). The matured group included oocytes that arrested their development after cleavage (CV: cleaved, n = 15) and oocytes that developed further to the BL stage (BL, n = 19). Figure 1 illustrates the experimental design with the different developmental groups.
Figure 1.
Experimental design with sample distribution on developmental groups.
COC collection and maturation
Oocytes were collected from ovaries of slaughtered mares of mixed ages, breed, and reproductive status. Since the ovaries collection was performed in the northern hemisphere in April–May, most of the mares were cycling. A total of 80 ovaries in two different replicates were used in this study. The COCs were collected by follicular scraping and flushing to obtain enough CCs. During the time of collection (around 4 h), oocytes were held at room temperature in HEPES-buffered synthetic oviductal fluid (H-SOF), then the whole procedure from oocyte maturation to embryo culture was performed as a single culture protocol, in which each oocyte was matured and decumulated. All successfully matured oocytes were fertilized by ICSI and in vitro cultured individually, to keep track of their developmental competence. The in vitro production (IVP) of embryos was performed according to Lazzari et al. (2020). In brief, single COC IVM was performed in individual wells of four-well plates containing 500 μl of a mixture of DMEM and Ham’s F12 with 10% fetal calf serum (Gibco, Thermo Fisher, Segrate, Italy), supplemented with insulin (5 mg/l), transferrin (5 mg/l), sodium selenite (5 mg/l), epidermal growth factor (50 ng/ml) (PeproTech, London, UK), 1 mmol/l sodium pyruvate and the gonadotropins, follicle-stimulating hormone (0.1 IU/ml), and luteinizing hormone (0.1 IU/ml) (Menopur, Ferring, Milan, Italy). Only suppliers of reagents not purchased from Sigma Aldrich (Merck, Milan, Italy) are specially indicated. IVM was performed in a humified atmosphere at 38°C with 20% O2 and 5% CO2 for on average of 27 h. Before maturation, each oocyte was classified as ‘compact’ or ‘expanded’ based on the morphology and consistency of the CCs.
Cumulus sampling and preparation
After maturation, each COC was individually transferred into H-SOF. After four washing steps in 100 μl drops of PBS with bovine serum albumin (BSA), glucose, and pyruvate (PBS-BSA), each oocyte was manually pipetted after a short incubation in trypsin. Most of the CCs were recovered in a volume of 3 μl wash media and immediately snap-frozen in liquid nitrogen. Then, each oocyte was fully denuded in H-SOF supplemented with hyaluronidase by pipetting, washed once, and transferred back to its maturation well until ICSI.
Fertilization by ICSI and embryo culture
The maturation status was documented and only oocytes that extruded the first polar body were selected for fertilization. Oocytes without polar body extrusion were assigned to the group of NM COCs, indicated in the ‘cumulome’ analysis as ‘NM’ (NM, n = 18). COCs of the ‘NM’ group included only cumulus of oocytes with normal morphology; CCs of degenerated or broken oocytes after maturation were not used for this experiment. ICSI was performed in two different sessions using semen from the same frozen-thawed batch of a stallion of proven fertility. The procedure was also summarized in the review by Lazzari et al. (2020). Briefly, the semen was prepared using a 90–45% discontinuous density gradient centrifugation (Redigrad, Amersham Biosciences, Freiburg i. Br., Germany). Piezo pulses were used to immobilize sperm and to penetrate the zona pellucida of the oocyte. Injected oocytes were individually cultured in 20 μl drops covered with mineral oil of modified synthetic oviductal fluid: in vitro culture (SOF-IVC) medium with BSA and amino acids (Lazzari et al., 2002). The zygotes/embryos were regularly monitored to document further development until Day 9. On Day 2 after ICSI, they were checked for a first cleavage division. The single COC-IVP-protocol achieved consistent and satisfactory developmental rates (maturation rate 59.7%; BL rate per COC 25.9%). A total of 52 COCs were used for the ‘cumulome’ analysis. CCs from sperm-injected oocytes that developed to the BL stage until Day 9 were assigned to the BL group (BL, n = 19), whereas the ones that cleaved but did not develop to the BL stage were assigned to the cleavage group (CV, n = 15). The matured group (M, n = 34) represented both group CV and group BL.
Bottom-up proteomics
For the protein extraction, the protocol developed for the ‘Cumulomics’ project was applied as previously documented (Fortes et al., 2014; Walter et al., 2019). The remaining pellet of individual cumuli after methanol extraction was treated using a sonoreactor for cell lysis (Rial-Otero et al., 2007) followed by a filter-aided sample preparation (Wiśniewski et al., 2009). Trypsin digestion was performed overnight on the filter unit. Desalting using C18 solid phase extraction columns (Finisterre, Teknokroma, Barcelona, Spain) preceded the final resolubilization in LC-MS solution. The MS analysis was performed using reverse-phase LC-MS/MS on an Orbitrap Fusion mass spectrometer (Thermo Scientific, Monza, Italy) in data-dependent acquisition mode. The instrument was coupled to a nano-HPLC system (EASY-nLC 1000, Thermo Scientific, Monza, Italy). A gradient of 120 min (1–35% acetonitrile) was used for the elution of peptides. Precursor ions charged between +2 and +6 with a minimum signal intensity of 1e4 were fragmentated. A pool containing an aliquot from each sample was analyzed as an aligning reference in each analysis run.
The quantification was conducted label-free in Progenesis QI for proteomics software version 3.0 (Nonlinear Dynamics, Waters, Milford, MA, USA). For protein quantification, the average of the normalized abundance of the three most intense peptide ions of each protein group was calculated for each sample individually. This resulted in the normalized quantitative protein abundance (Grossmann et al., 2010). Spectra were identified against the NCBI database for horses (NCBI Taxonomy ID 9796, release date 20170523). The false discovery rate was estimated using reversed sequences (Nesvizhskii et al., 2003; Käll et al., 2008). Database searching was performed on Mascot Server v.2.5.1.3. (Matrix Science, London, UK). Settings were 10 ppm tolerance for the monoisotopic precursor masses, 0.5 Da for fragment ion tolerance with a maximum of two allowed missed cleavages, and the value for number of 13C was set to 1. Specified as fixed modification was carbamidomethylation of cysteine; selected for variable modifications were oxidation of methionine and deamidation from glutamine and asparagine. In Scaffold v4.1.1 (Proteome Software, Portland, OR, USA), protein probabilities were analyzed (Käll et al., 2008). A filtered scaffold spectrum report (protein false discovery rate 10%; peptide false discovery rate 5%) was loaded to Progenesis QI to assign peptide and protein data to the MS1 spectra. The false discovery rate for the proteins (with at least two peptides) in all biological samples was <0.4% (Käll et al., 2008). Differences between the developmental groups were analyzed in Progenesis QI software using two group comparisons, which calculated fold changes (linear values) and P-values (t-test on arcsinh values). These pairwise comparisons were used to generate sets of candidates with relation to developmental competence that were afterward used for downstream analysis. A separate Progenesis experiment was set up for the comparison between the NM (n = 18) and matured (n = 34) COCs to obtain fold changes and P-values for separate downstream analysis.
Untargeted metabolomics
In the first step, methanol extraction of the metabolome was conducted for individual cumuli as already documented in detail by Walter et al. (2019). Separation of metabolites was achieved by liquid chromatography on a nanoAcquity UPLC column (Waters), which was coupled by a nano-ESI source to a Synapt HDMS G2 mass spectrometer (Waters). The nano-ESI source was operated in negative polarization mode. The LC–MS dataset was analyzed using Progenesis QI software (Waters). Pooled samples and reference compound mixtures were used for quality control, to check the technical accuracy and stability. Detected ions were identified based on accurate mass, detected adduct patterns, and isotope patterns by comparing them with entries in the Human Metabolome Data Base. A mass accuracy tolerance of 5mDa was set for the searches. Fragmentation patterns were considered for the identification of metabolites.
Normalized abundances were exported together with the retention time (RT) and mass-to-charge ratio (m/z) to an Excel table (Windows) where, for pairwise comparisons (CV-NM, BL-NM, BL-CV), log2-fold changes, and P-values based on two-sided t-tests and log10-transformed intensity values were calculated for each detected ion. As ATP is known to break down in ADP during analysis, the compounds 6.25_426.0214m/z (ADP) and 6.25_506.9957n (ATP) were summed up for further analysis. The list of metabolites was filtered for a fold change threshold level of 1.2 and a P-value <0.05 in at least one of the pairwise comparisons.
Downstream analysis and data visualization
All data were analyzed in pairwise comparisons. The group with the higher developmental potential was always analyzed in comparison to the group with the lower developmental potential (CV/NM, BL/NM, BL/CV). This resulted in negative log2-fold changes for proteins with higher expression in CCs of oocytes with lower developmental potential. Between-group analysis (BGA) for both datasets were run in R software using the package made4 (Bioconductor repository) (http://www.bioconductor.org/packages/release/bioc/html/made4.html).
The equine NCBI database for horses (NCBI Taxonomy ID 9796, release date 20170523) was blasted to the reviewed sequences of the human uniprot database to reveal orthologous proteins (canonical UniProt database: Tax ID: 9606, 20161209: file: fgcz_9606_reviewed_cnl_contaminantNoHumanCont_20161209.fasta) to access tools for downstream analysis with the human IDs (orthologues were accepted when the blastp e-value was 1E-15 or smaller). The individual group comparisons were screened for overlaps between both matured groups (CV, BL) against the NM group and these identified candidates for maturational competence were analyzed for enriched pathways and protein clusters using enrichment analysis in STRING-DB (http://string-db.org) (Szklarczyk et al., 2021), as well as gene ontology (GO) grouping using ClueGO plugin of Cytoscape (NIGMS/NIH). Data of the progenesis analysis combining CV and BL (M) were analyzed in a functional enrichment analysis against the whole protein-encoding genome in STRING-DB (http://string-db.org), using all proteins ranked according to their log2-fold change (M/NM). Boxplots were generated in Prism 9 (GraphPad Software, Boston, MA, USA) based on the hyperbolic arcsine of normalized abundances. Integration of proteomic and metabolomic results was achieved by the use of KEGG Mapper v3.1 (release date 1 October 2017; http://www.genome.jp/kegg/mapper.html), as well as Metacore (Cortellis, Clarivate, London, UK). Pathways were illustrated using the BioRender App (Biorender.com). Proteomics data were deposited in the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier 033551 (Perez-Riverol et al., 2019). Data tables with the complete results of proteomics and metabolomics data are available in Supplementary Tables S1 and S2.
Results
Detected proteins and metabolites
Over the three biological groups, a total of 1671 proteins with NCBI accessions were identified and considered as quantifiable (i.e. ≥two peptides per protein); 40 of these were linked to more than one orthologous human Uniprot accession. Human Uniprot IDs linked to more than one equine NCBI accession are marked in a column in the full results table (Supplementary Table S1). The metabolomic analysis quantified a total of 612 metabolic compounds. For 12 compounds, a putative metabolite ID was assigned.
Between-group analyses
The BGA for the proteomics and metabolomics analysis illustrates a shift of the cluster centers along the horizontal axis between the two matured groups (CV, BL) and the NM group (Fig. 2).
Figure 2.
Results of the between-group analysis (BGA). The groups show some overlaps but considering the shift of the cluster centers along the horizontal axis 1, there is a clear separation, especially between the not matured group (NM, blue) and the two matured groups: cleaved (CV, green) and blastocyst (BL, red). Left: Metabolomics data; Right: Proteomics data; grid line distance d = 20.
Pairwise comparisons of groups
Individual two-group comparisons
This section describes the results of the pairwise comparisons between all three developmental groups. The data for these three comparisons (FC and P-values) were generated in Progenesis QI software (Nonlinear Dynamics). The individual comparisons were also screened for overlaps between both matured groups (CV, BL) against the NM group. These identified candidates for maturational competence were analyzed for enriched pathways and protein clusters using enrichment analysis in STRING-DB (http://string-db.org), as well as gene ontology (GO) grouping using the ClueGO plugin of Cytoscape (NIGMS/NIH).
The two-group comparisons revealed a result similar to the BGA analysis (Fig. 2), with less difference between the two matured groups CV and BL than between these groups and the NM group (Table 1).
Table 1.
Overview on proteomics and metabolomics data.
| Quantifiable | CV>NM | NM>CV | BL>NM | NM>BL | BL>CV | CV>BL | |
|---|---|---|---|---|---|---|---|
| Proteins | 1671 | 208 | 7 | 148 | 16 | 4 | 14 |
| Metabolites | 612 (12) | 61 (2) | 30 | 100 (4) | 59 (6) | 10 | 35 |
Listed are the overall quantifiable compounds as well as the number of significant candidates in the two group comparisons (FC > 1.2; P < 0.05). The number of metabolomic compounds with putative metabolite IDs are listed in brackets.
CV, oocytes that arrested their development after cleavage; BL, oocytes developing to blastocyst stage; NM, oocytes that failed to mature.
Comparison of CV versus NM groups
The comparison of the group that cleaved but did not develop to the BL stage (CV) with the group that failed to mature (NM) revealed a total of 215 proteins with significantly different expression (FC > 1.2, P < 0.05). Of these, 208 proteins were more abundant in the CV group, whereas only seven proteins were overrepresented in the NM group.
The metabolomic data revealed 91 compounds with significant differences (FC > 1.2, P < 0.05). Of these, 61 showed a higher abundance in the CV group, including two compounds with a putative metabolite ID (oxalacetic acid, oxoglutaric acid) while 30 compounds had a higher abundance in the NM group.
Comparison of BL versus NM groups
The comparison of the group with final development to the BL stage (BL) against the group that failed to mature (NM) identified 164 significantly different proteins. Again, the majority was more abundant in BL samples (148) compared to NM samples (16).
This group comparison revealed the highest number of significant differences in the metabolomic dataset (159). Higher abundances in the BL group were detected for 100 compounds: 4 with putative metabolite ID (oxalacetic acid, oxoglutaric acid, oxoadipic acid, ATP (ATP+ADP)). The results of the compounds 6.25_426.0214m/z (ADP) and 6.25_506.9957n (ATP) were both increasing from NM over CV to BL with significant differences between NM and BL (ADP: P < 0.01; ATP: P < 0.05). Individual results are listed in Supplementary Table S2. In the NM group, 59 compounds showed significantly higher abundances, of which six compounds were attributed to a metabolite ID (purine, succinic acid, hydroxyglutaric acid, glucosamine, malic acid, glyceric acid).
Comparison of BL versus CV groups
The comparison of the two groups that matured successfully detected only minor differences with a total of 18 significantly different proteins. The majority were overexpressed in the CV group (14) and only four proteins were more abundant in the BL group.
The metabolomic data showed only minor differences between the CV and BL groups, with only 45 compounds showing significant differences (10 higher in BL, 35 higher in CV). None of them could be attributed to a putative metabolite ID.
Comparison of NM versus CV and BL (matured) groups
As previously described, the data of the individual two-group comparison (Table 1) reflect the result of the BGA analysis (Fig. 2), where larger deviations were noticed between the NM group compared to the CV and BL groups. To extrapolate the intersection in proteins and metabolites with significant alterations between the matured (M) groups (CV and BL) and the NM group, the results were filtered for proteins and metabolites with both fold changes >1.2 and at least one P-value <0.05. This procedure identified a total of 243 proteins and 102 metabolic compounds which showed higher expression in M against NM (CV/BL>NM). Vice versa, 16 proteins and 56 metabolic compounds were more highly expressed in the NM group compared to CV and BL (NM>CV/BL) group. Full result lists are available in Supplementary Tables S1 and S2.
Enrichment analysis of the proteins with significant higher abundances in the CV and BL groups revealed various significant enrichments. The complete enrichment analysis can be accessed online on STRING-DB (https://version-11-5.string-db.org/cgi/network?networkId=bxhDLO88HwDV). Figure 3 illustrates a selection of enriched KEGG pathways (Supplementary Table S3), which were selected for detailed presentation in the section on biological categories. This includes the pathways ‘glycolysis/gluconeogenesis’ (hsa00010), ‘fructose and mannose metabolism’ (hsa00051, FDR<0.01), ‘starch and sucrose metabolism’ (hsa00500, FDR<0.05), ‘endocytosis’ (hsa04144), and ‘protein processing in endoplasmic reticulum’ (hsa04141).
Figure 3.
Enrichment analysis using STRING-DB. The analysis was conducted on 243 proteins (241 matched with database, minimum required interaction score 0.7) with significant higher expression (both FC>1.2, one P<0.05) in the cleaved (CV) or blastotyst (BL) groups compared to the not matured group (NM). Highlighted are proteins in the overrepresented KEGG groups ‘glycolysis/gluconeogenesis’ (hsa00010, FDR 0.01, orange nodes), ‘fructose and mannose metabolism’ (hsa00051, FDR<0.01, yellow nodes), and ‘starch and sucrose metabolism’ (hsa00500, FDR<0.05, green nodes). Also, proteins overrepresented in the pathways ‘endocytosis’ (hsa04144, FDR <1E−07, pink nodes) and ‘protein processing in endoplasmic reticulum’ (hsa04141, FDR=0.001, turquoise nodes) are indicated. The complete enrichment analysis can be accessed online on STRING-DB (https://version-11-5.string-db.org/cgi/network?networkId=bxhDLO88HwDV). Enriched KEGG pathways are listed in Supplementary Table S3.
Furthermore, an analysis of enriched Gene Ontology groups for biological processes was performed using the ClueGO plugin in Cytoscape (NIGMS/NIH). The same proteins overexpressed in the M group against the NM group (n = 243) were considered. This analysis included similar terms as the enriched KEGG pathways in the STRING-DB analysis: vesicle, carbohydrate catabolic process, or endoplasmic reticulum (ER) (Supplementary Fig. S1A and B).
Combined analysis of matured (M=CV&BL) versus NM groups
As the BGA revealed a closer relation between the CV and BL groups, a separate Progenesis QI (Nonlinear Dynamics) analysis of the proteomic data, where these groups were directly combined to a matured (M) group was run. This two group-analysis of all NM and matured (M) samples provided the opportunity to run functional enrichment analysis on the full-ranked protein lists. This list was ranked along log2-fold change and analyzed using the functional enrichment tool in STRING-DB. The functional enrichment analysis was performed against the whole protein-encoding genome. The result identified a significantly enriched KEGG pathway, ‘oxidative phosphorylation’ (hsa00190), which was shifted toward the NM group (enrichment score 0.6, false discovery rate <0.01) (Szklarczyk et al., 2021). The analysis assigned 41 proteins of the input list to this pathway (Supplementary Fig. S2). The complete functional enrichment analysis can be accessed online on STRING-DB (https://version-11-5.string-db.org/cgi/globalenrichment?networkId=bboFFIjSDO2L). The same ranked list was analyzed using the Core Analysis with default settings in Ingenuity Pathway Analysis (Qiagen IPA) software (filtered for P < 0.05). The results of the canonical pathway analysis are listed in Table 2 (filtered for pathways with negative or positive z-score and a −log(P-value) >2). The pathway with the most significant result and a positive z-score was ‘remodeling of epithelial adherence junctions’. Related to energy metabolism, the pathway ‘glycogen degradation’ also revealed a significant result with a positive z-score. The upstream regulator analysis (Table 3) identified potential upstream regulators that were inhibited (negative z-score) or activated (positive z-score). Results were filtered for a P-value <5.00E-05. Amongst the activated regulators were glucose, beta-estradiol, and estrogen receptor 2, whereas the activation z-score for FSH was negative.
Table 2.
Results of the canonical pathway analysis.
| Ingenuity canonical pathways | −log(P-value) | Ratio | z-score | Molecules |
|---|---|---|---|---|
| Remodeling of epithelial adherens junctions | 1.11E+01 | 1.76E−01 | 2.24 | ACTA2, ACTG1, ACTR2, ACTR3, ARPC2, ARPC3, CTNNA2, RAB7A, T UBB, TUBB2A, TUBB6, VCL |
| Huntington’s disease signaling | 9.61E+00 | 6.76E−02 | 0.45 | AP2A2, BAX, CAPN1, CAPNS1, DCTN1, GLS, HSPA1A/HSPA1B, NA PA, PSMA3, PSMB10, PSMB3, PSMB8, PSMC2, PSMD10, PSMD13, PSMD6, PSMD7, PSME1, RPS27A |
| Inhibition of ARE-mediated mRNA degradation pathway | 7.66E+00 | 8.07E−02 | 2.00 | PRKACA, PRKAR1A, PSMA3, PSMB10, PSMB3, PSMB8, PSMC2, PS MD10, PSMD13, PSMD6, PSMD7, PSME1, YWHAE |
| EIF2 signaling | 7.64E+00 | 6.70E−02 | 1.13 | ACTA2, EIF3D, EIF3H, RPL13, RPL26, RPL27A, RPL35, RPL35A, RPS 23, RPS27A, RPS27L, RPS28, RPS6, RPS8, WARS1 |
| Integrin signaling | 7.07E+00 | 6.57E−02 | 3.05 | ACTA2, ACTG1, ACTR2, ACTR3, ARF3, ARPC2, ARPC3, CAPN1, CAP NS1, GSN, RAC1, RHOG, TLN1, VCL |
| Synaptogenesis signaling pathway | 5.76E+00 | 4.78E−02 | 3.74 | ACTR2, ACTR3, AP2A2, AP2B1, AP2S1, ARPC2, ARPC3, CAMK2D, N AP1L1, NAPA, PRKACA, PRKAR1A, RAC1, THBS1, TLN1 |
| Coronavirus pathogenesis pathway | 5.66E+00 | 5.91E−02 | −1.16 | ATP6V1A, BAX, DDX58, EEF1A1, RAB7A, RPS23, RPS27A, RPS27L, RPS28, RPS6, RPS8, SERPINE1 |
| Glycogen degradation II | 5.22E+00 | 3.33E−01 | 2.00 | PGM1, PYGB, PYGL, TYMP |
| Fcγ receptor-mediated phagocytosis in macrophages and monocytes | 5.10E+00 | 8.51E−02 | 2.83 | ACTA2, ACTG1, ACTR2, ACTR3, ARPC2, ARPC3, RAC1, TLN1 |
| Glycogen degradation III | 4.92E+00 | 2.86E−01 | 2.00 | PGM1, PYGB, PYGL, TYMP |
| Actin cytoskeleton signaling | 4.82E+00 | 4.90E−02 | 2.11 | ACTA2, ACTG1, ACTR2, ACTR3, ARPC2, ARPC3, GSN, MYH1, NCKA P1, RAC1, TLN1, VCL |
| HIF1α signaling | 4.77E+00 | 5.29E−02 | 1.90 | CAMK2D, COPS5, ELOC, GPI, HK1, HSPA1A/HSPA1B, PKM, RAC1, R PS6, SERPINE1, VIM |
| Regulation of actin-based motility by Rho | 4.43E+00 | 6.90E−02 | 2.65 | ACTA2, ACTR2, ACTR3, ARPC2, ARPC3, GSN, RAC1, RHOG |
| Coronavirus replication pathway | 3.92E+00 | 1.11E−01 | 2.24 | COPG2, DDX1, TUBB, TUBB2A, TUBB6 |
| Dilated cardiomyopathy signaling pathway | 3.68E+00 | 5.41E−02 | 2.45 | ACTA2, ACTG1, BAX, CAMK2D, LMNA, MYH1, PRKACA, PRKAR1A |
| Epithelial adherens junction signaling | 3.51E+00 | 5.10E−02 | 2.83 | ACTR2, ACTR3, ARPC2, ARPC3, CTNNA2, RAC1, VCL, YWHAE |
| RHOA signaling | 3.40E+00 | 5.65E−02 | 2.65 | ACTA2, ACTG1, ACTR2, ACTR3, ARPC2, ARPC3, SEPTIN7 |
| Reelin signaling neurons | 3.36E+00 | 5.56E−02 | 2.45 | ACTR2, ACTR3, ARPC2, ARPC3, CAMK2D, PAFAH1B3, RAC1 |
| Actin nucleation by ARP-WASP complex | 3.29E+00 | 6.45E−02 | 2.45 | ACTR2, ACTR3, ARPC2, ARPC3, RAC1, RHOG |
| RHOGDI signaling | 3.25E+00 | 4.19E−02 | −2.83 | ACTA2, ACTG1, ACTR2, ACTR3, ARPC2, ARPC3, MYH1, RAC1, RHOG |
| Signaling by Rho family GTPases | 3.16E+00 | 3.73E−02 | 3.16 | ACTA2, ACTG1, ACTR2, ACTR3, ARPC2, ARPC3, RAC1, RHOG, SEPT IN7, VIM |
| tRNA charging | 3.10E+00 | 1.03E−01 | 2.00 | DARS1, FARSB, QARS1, WARS1 |
| Xenobiotic metabolism PXR signaling pathway | 2.92E+00 | 4.15E−02 | 1.41 | ALDH1A1, ALDH9A1, CAMK2D, GSTA3, GSTM3, PPP1R7, PRKACA, PRKAR1A |
| PFKFB4 signaling pathway | 2.83E+00 | 8.70E−02 | 1.00 | GPI, HK1, PRKACA, PRKAR1A |
| Calcium signaling | 2.61E+00 | 3.70E−02 | 2.24 | ACTA2, ATP2B1, ATP2B4, CAMK2D, MYH1, PRKACA, PRKAR1A, TP M2 |
| Ferroptosis signaling pathway | 2.51E+00 | 4.55E−02 | −0.45 | ARF3, CTSB, GCLC, H2AC21, HSPB1, MACROH2A1 |
| Death receptor signaling | 2.42E+00 | 5.21E−02 | 1.34 | ACTA2, ACTG1, HSPB1, LMNA, SPTAN1 |
| RAC signaling | 2.41E+00 | 4.35E−02 | 2.45 | ACTR2, ACTR3, ARPC2, ARPC3, NCKAP1, RAC1 |
| Apoptosis signaling | 2.27E+00 | 4.81E−02 | 2.24 | BAX, CAPN1, CAPNS1, LMNA, SPTAN1 |
| ILK signaling | 2.24E+00 | 3.54E−02 | 0.45 | ACTA2, ACTG1, MYH1, RAC1, RHOG, VCL, VIM |
| Semaphorin neuronal repulsive signaling pathway | 2.23E+00 | 3.97E−02 | −0.82 | DPYSL2, DPYSL3, PRKACA, PRKAR1A, RAC1, SMC3 |
| Paxillin signaling | 2.20E+00 | 4.63E−02 | 2.24 | ACTA2, ACTG1, RAC1, TLN1, VCL |
Output gene sets are filtered for pathways with negative or positive z-score and a −log(P-value) >2.
Table 3.
Results of the upstream regulator analysis.
| Upstream regulator | Expr. log ratio | Molecule type | Predicted activation state | Activation z-score | P-value of overlap | Target molecules in dataset |
|---|---|---|---|---|---|---|
| MYC | Transcription regulator | Activated | 3.04 | 2.52E-14 | ABCE1, AHCY, ANXA5, ARF3, BAX, CAPNS1, CTSB, CTSV, DBI, ECM1, E IF3D, GCLC, GDI1, GLS, GOLGA2, GPI, HNRNPAB, HNRNPD, HSPB1, LXN, METAP2, NCKAP1, NDRG1, NOP56, NUDT21, PFKFB3, PKM, PLS3, PSMB8, RBP1, RPL13, RPL26, RPL27A, RPL35, RPL35A, RPS23, RPS27A, RPS27L, RPS28, RPS6, RPS8, SAE1, SERPINE1, THBS1, TLN1, VI M | |
| APP | 0.45 | Other | Activated | 2.17 | 6.40E-13 | ACO2, ACTA2, ACTG1, ANXA5, ATP6V1A, BAX, CAMK2D, CAP1, CAPN 1, CD68, CTSB, CTSV, DBI, DDAH1, DDX58, DPYSL2, DSTN, GPI, GSTM 3, HK1, HMGB1, HSPB1, HSPG2, IGF2R, PFKFB3, PKM, PRDX5, PRKAC A, PSME1, RPS6, SEPTIN7, SERPINE1, SPTAN1, TAGLN, TPP1, TUBB, TUBB2A, TUBB6, UBE2K, YWHAE |
| TP53 | Transcription regulator | Activated | 2.70 | 5.16E-12 | ACADVL, ACO2, ACTA2, AHCY, AKR1B1, ALDH1A1, ALDH9A1, ATL3, B AX, BCAP31, CAMK2D, CAP1, CAPNS1, CTSB, DBI, DCTN2, DSTN, EEF1A1, ENO3, FBXO22, FKBP4, GPI, GSN, HSPB1, HSPG2, HSPH1, KPNA2, MACROH2A1, MPI, MVP, MYO6, NAP1L1, NDRG1, PDCD6IP, PDLIM5, PKM, PSAP, RPS27L, SEC23B, SEC61B, SERPINB9, SERPINE1, ST 13, THBS1, TLN1, TPP1, TRIM28, UBA1, VAPA, VCL, VIM, XPO5 | |
| LARP1 | Translation regulator | Inhibited | −3.06 | 1.16E-11 | EEF1A1, EIF3H, RPL13, RPL26, RPL27A, RPL35, RPL35A, RPS23, RPS27A, RPS28, RPS6, RPS8, VIM | |
| YAP1 | Transcription regulator | Activated | 2.80 | 1.31E-11 | ACTA2, ACTG1, BAX, DDAH1, LMNA, LMNB1, RPL13, RPL26, RPL27A, RPL35, RPL35A, RPS23, RPS27A, RPS27L, RPS28, RPS6, RPS8, SERPINE1, TAGLN, THBS1, TUBB, VCL | |
| RICTOR | Other | Inhibited | −3.16 | 7.50E-10 | ATP6V1A, BAX, PFKFB3, PSMA3, PSMB10, PSMB3, PSMB8, PSMC2, PSMD13, PSMD6, PSMD7, PSME1, RAC1, RPL26, RPL35A, RPS23, RPS27A, RPS6, RPS8 | |
| TGFB1 | Growth factor | Activated | 2.90 | 1.11E-08 | ABCE1, ACTA2, ARPC2, BAX, CAP1, CAPNS1, CD68, COL12A1, CTPS1, CTSB, DNPEP, DPYSL2, DPYSL3, ECM1, ENO3, FSCN1, GCLC, GFPT2 GLS, GNS, GPI, GSN, GUSB, HEXA, HK1, HNRNPAB, HSPA1A/HSPA1 B, HSPB1, HSPG2, PDLIM5, PKM, PLOD1, PLS3, PPT1, PRDX4, RAC1, SEC61B, SERPINE1, TAGLN, THBS1, TPM2, TUBB2A, TYMP, UBE2K, V CL, VIM, YWHAE | |
| MLXIPL | Transcription regulator | Activated | 2.89 | 1.21E-07 | PYGL, RPL13, RPL26, RPL27A, RPL35, RPL35A, RPS23, RPS27A, RPS27L, RPS28, RPS6, RPS8 | |
| MKNK1 | Kinase | Activated | 3.16 | 1.57E-07 | ANXA5, DCTN1, DPYSL3, GPS1, KLC1, MYO6, PDLIM5, TAGLN, TPP1, VIM | |
| GABA | Chemical—endogenous mammalian | Inhibited | −3.44 | 4.46E-07 | AP2A2, ATP2B4, CD63, EEF1A1, HEXA, MTHFD1, PRKAR1A, PRMT5, PSAP, RAC1, RPL13, RPL35, RPL35A, RPS28, RPS8, SEC24A, UBA1, UBE2M, UBE2N | |
| D-glucose | Chemical—endogenous mammalian | Activated | 4.31 | 4.58E-07 | ACO2, ACTA2, AKR1B1, ARF3, ATP2B1, ATP2B4, BAX, CAP1, CTSB, CT SV, DBI, GPI, GSN, HMGB1, HSPB1, PFKFB3, PGM1, PKM, PON2, SERPINE1, TAGLN, THBS1, VCL | |
| Beta-estradiol | Chemical—endogenous mammalian | Activated | 4.48 | 1.20E-06 | ACADVL, ACTA2, ACTG1, ARSA, ATP2B1, BAX, CD68, CRYZ, CTNNA2, CTPS1, CTSB, DDAH1, EIF3D, ELOC, FARSB, GCLC, GLS, GNS, GSN, GUSB, HNRNPD, HSD17B12, HSPB1, HSPH1, IGF2R, KPNA2, LMNA, ND RG1, NOP56, PKM, PLIN2, PON2, PPT1, PRDX4, PSAP, RAB7A, RAC1, RBP1, S100A13, SERPINE1, SFRP4, THBS1, VAPA, VCL, VIM, YWHAE | |
| SRF | Transcription regulator | Activated | 2.75 | 7.99E-06 | ACTA2, ACTG1, CAP1, DSTN, GLRX3, GSN, HSPG2, MYH1, PDCD10, PGM1, PTGR1, TAGLN, TLN1, VCL | |
| ESR2 | Ligand-dependent nuclear receptor | Activated | 2.57 | 1.31E-05 | ABCE1, CTPS1, DDAH1, DPYSL3, GCLC, HPRT1, KPNA2, LMNA, LMNB1, MYH1, NASP, PLEC, PSMD7, PSME1, SFRP4, VIM | |
| CAT | Enzyme | Inhibited | −2.43 | 2.95E-05 | BAX, GCLC, SERPINE1, TAGLN, THBS1, VIM | |
| UBA1 | 0.469 | Enzyme | Inhibited | −2.00 | 3.43E-05 | ANXA5, HSPB1, TAGLN, VIM |
| NFASC | Other | Activated | 2.00 | 4.37E-05 | DCTN1, DYNC1LI1, DYNLT3, KLC1 | |
| FSH | Complex | Inhibited | −2.04 | 4.70E-05 | ACTA2, ACTR2, AMH, ATP2B1, COPS5, CTSV, GCLC, NPC2, PKM, PRKAR1A, SERPINE1, THBS1, TLN1, TPM2, VCL |
Genes are filtered for negative (inhibition) or positive (activation) z-score and a P-value <5.00E−05.
Proteins and metabolites decreasing or increasing with developmental competence (NM>CV>BL or BL>CV>NM)
A total of 13 proteins showed increased concentrations with developmental competence (all fold changes ³1.2 and at least one P-value <0.05), with the lowest means in NM and the highest means in BL groups. Decreasing concentrations with developmental competence were found for a total of seven proteins (Supplementary Table S1). Amongst these seven proteins were NADH dehydrogenase (ubiquinone) iron-sulfur protein 8, mitochondrial (NDUFS8) of the KEGG pathway ‘oxidative phosphorylation’ (Fig. 4), and basement membrane-specific heparan sulfate proteoglycan core protein (HSPG2). Regarding metabolites (Supplementary Table S2), 33 compounds showed increased concentrations with developmental competence (criteria according to proteomics results), of which only two were attributed to a metabolite (ATP, ADP). There were 25 compounds found to be decreasing with developmental competence of which three were attributed to a metabolite (purine, succinate, glucosamine).
Figure 4.
Energy metabolism. Visualized are the affected pathways of glycogen degradation, glycolysis, tricarboxylic acid cycle, and oxidative phosphorylation. The illustration includes 15 proteins (blue labels) and 6 metabolites (green labels) with significant differences in abundance between not matured (NM) and/or cleaved (CV)/blastocyst (BL) groups that belong to the KEGG pathways ‘starch and sucrose metabolism’ (hsa00500), ‘glycolysis/gluconeogenesis’ (hsa00010), and ‘fructose and mannose metabolism’ (hsa00051). Box plots were generated on ASINH normalized abundances (Whiskers from min to max; red: NM, orange: CV, green: BL). Significant differences in the pairwise comparison are marked with a bar. For ATP the sum of compounds 6.25_426.0214m/z and 6.25_506.9957n is illustrated. Adapted from ‘Warburg Effect’ template, using BioRender.com (2021). ACO1, Cytoplasmic aconitate hydratase; AKR1B1, Aldo-keto reductase family 1 member B1; ALDH9A1, 4-trimethylaminobutyraldehyde dehydrogenase; ALDOC, Fructose-bisphosphate aldolase C; ENO3, Beta-enolase; FBP1, Fructose-Bisphosphatase 1; GLUT, Glucose Transporter; GPI, Glucose-6-phosphate isomerase; GYS1, Glycogen Synthase 1; HK1, Hexokinase-1; MCT, Monocarboxylate Transporter; MPI, Mannose-6-phosphate isomerase; NDUFS8, NADH dehydrogenase (ubiquinone) iron-sulfur protein 8, mitochondrial; PFKFB3, 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3; PGLS, 6-phosphogluconolactonase; PGM1, Phosphoglucomutase-1; PKM, Pyruvate kinase PKM; PYGB, Glycogen phosphorylase, brain form; PYGL, Glycogen phosphorylase, liver form.
Biological categories
Energy metabolism
The enrichment analysis revealed a significant overrepresentation of proteins that belong to the KEGG groups ‘glycolysis/gluconeogenesis’ (hsa00010, FDR 0.01), ‘fructose and mannose metabolism’ (hsa00051, FDR<0.01), and ‘starch and sucrose metabolism’ (hsa00500, FDR<0.05) (Fig. 3). Therefore, an integrated illustration including 15 proteins and six metabolites related to cellular energy metabolism was drawn (Fig. 4). Proteins involved in glycolysis and glycogen utilization showed a homogenous lower expression in the NM group. Instead, proteins involved in oxidative phosphorylation were more highly expressed in the NM group. This finding was supported by the result of the functional enrichment analysis in STRING-DB, where the KEGG pathway ‘oxidative phosphorylation’ (hsa00190) was significantly enriched (Supplementary Fig. S2).
Finally, an important result of this study is that the highest concentrations of ATP/ADP were detected in the cumulus samples of oocytes that developed to the BL stage.
Vesicular transport
The enrichment analysis revealed a significant overrepresentation of proteins involved in the pathways ‘endocytosis’ (hsa04144, FDR 7e−07) and ‘protein processing in the ER’ (hsa04141, FDR 0.001) in both matured groups (Fig. 3). Therefore, an integrated illustration of proteins of these KEGG pathways was drawn (Fig. 5). The 28 proteins illustrated in Fig. 5 had significantly lower expression in the NM group compared to either of the matured groups (CV and/or BL).
Figure 5.
Vesicular transport with endocytic and secretory pathways. The illustration includes 28 proteins (blue labels) with significant differences in the abundance between the not matured (NM) and/or cleaved (CV)/blastocyst (BL) groups and belonging to the KEGG pathways ‘endocytosis’ (hsa04144) or ‘protein processing in endoplasmic reticulum’ (hsa04141). Box plots were generated on ASINH normalized abundances (Whiskers from min to max; red: NM, orange: CV, green: BL). Significant differences in the pairwise comparison are marked with a bar. Adapted from the ‘Intracellular Transport’ template using BioRender.com (2021). AP1G1, AP-1 complex subunit gamma-1; BAX, Apoptosis regulator BAX; BCAP31, B-cell receptor-associated protein 31; CAPN1, Calpain-1 catalytic subunit; HSPA5, Endoplasmic reticulum chaperone BiP; HSPH1, Heat shock protein 105 kDa; IGF2R, Cation-independent mannose-6-phosphate receptor; RAB7A, Ras-related protein Rab-7a; SEC23A, Protein transport protein Sec23A; SEC23B, Protein transport protein Sec23B; SEC24A, Protein transport protein Sec24A; SEC24C, Protein transport protein Sec24C; SEC24D, Protein transport protein Sec24D; SEC61A1, Protein transport protein Sec61 subunit alpha isoform 1.
Glycosaminoglycan metabolism
The KEGG pathway ‘glycosaminoglycan (GAG) degradation’ was not significantly enriched in the StringDB analysis of proteins that matured successfully. However, three proteins involved in GAG degradation showed significant higher abundances in cumulus complexes that matured successfully: β-glucuronidase (GUSB), N-acetylglucosamine-6-sufatase (GNS), and hexosaminidase (HEXA). Heparan sulfate proteoglycan (HSPG2, PGBM, Perlecan) was significantly higher in NM compared to CV and BL cumulus complexes. This protein belongs to the group of seven proteins that decreased with increasing developmental potential, showing the lowest abundance in the BL group. In the metabolomics analysis, one compound that showed significant lower abundances in the BL group (4.58_178.0716 m/z) was putatively assigned to the substance glucosamine. Figure 6 illustrates the GAG degradation and the measured compounds in this pathway.
Figure 6.
Glycosaminoglycan degradation. The illustration includes four proteins (blue labels) and one metabolite (green label) with significant differences in the abundance between not matured (NM) and/or cleaved (CV)/blastocyst (BL) groups and belonging to the KEGG pathway ‘glycosaminoglycan degradation’ (hsa00531). Box plots were generated on ASINH normalized abundances (Whiskers from min to max; red: NM, orange: CV, green: BL). Significant differences in the pairwise comparison are marked with a bar. Created with BioRender.com. GNS, N-acetylglucosamine-6-sulfatase; GUSB, Beta-glucuronidase; HEXA, Beta-hexosaminidase subunit alpha; HSPG2, Basement membrane-specific heparan sulfate proteoglycan core protein; HYAL, Hyaluronidase.
Discussion
General discussion
With a total of 1671 proteins and 612 metabolic compounds identified, a similar range of detectable candidates was observed as in the previously published comparison of the equine ‘cumulome’ after in vivo maturation and IVM (1811 proteins and 906 metabolic compounds) (Walter et al., 2019). The BGA analysis revealed a more precise separation of the two matured groups (M: CV and BL) from the NM group (Fig. 2). This closer relation between the matured groups is also reflected by the limited number of candidates with significant difference between these two groups (proteomics 18, metabolomics 45, Table 1). Thus, the concept of predicting the developmental competence of the oocyte through analysis of the ‘cumulome’ has some limitations. The results of this study nicely illustrate wide-ranging differences in the ‘cumulome’ of oocytes with and without maturational competence, although the cause of a failure to mature could not be determined using this approach. Nevertheless, the application of this ‘cumulomics’ approach to discriminate between oocytes with the potential to develop to the BL stage and oocytes that arrest their development after the first cleavage division is difficult. Considering the biological function of the CC, it is not surprising that the largest differences were detected between the NM and matured groups. CCs have a close bidirectional communication with their oocytes throughout the maturation process through gap junctional communication, paracrine signaling or vesicular trafficking of large cargo like mRNA or long noncoding RNA (Macaulay et al., 2014; Russell et al., 2016). In the horse, the coupling between CCs and oocytes is interrupted after the oocytes are incubated in maturation medium for at least 18 h (Colleoni et al., 2004). Even after maturation and ovulation, the cumulus complex participates in capturing the COCs to the fallopian tube and guiding the sperm during fertilization. After fertilization, the oocytes shed their cumulus vesture. This disconnection between cumulus and oocyte is reflected in the broad similarities of the ‘cumulome’ of oocytes that matured successfully and were only different regarding their post-fertilization developmental capacity. Another reason for these similarities might be the heterogeneity in the CV group, which included all zygotes that arrested their development between the cleavage and the BL stage. Nevertheless, with 18 proteins (1.1%) and 45 metabolites (7.4%), a couple of significantly different candidates were also identified between CV and BL (Table 1). A similarly designed human study, but on a limited number of pooled CCs (n = 10 per group), found only six equally expressed proteins for the different developmental groups in a total of 87 detected proteins (Braga et al., 2016). A study comparing CCs of single bovine oocytes before and after maturation, using intact cell MALDI-TOF MS, identified 20 differentially expressed compounds as potential markers for successful maturation (Labas et al., 2017).
Pathway analyses of differentially expressed proteins between successfully matured and failing-to-mature COCs identified potential upstream regulators that might be responsible for the detected alterations in the ‘cumulome’. Amongst these, FSH appears to have an influence on the expression of various molecules found in the data, such as AMH or actin-related protein 2 (Tables 2 and 3). Thus, it is a promising candidate for optimization of maturation media, which was inhibited in the matured group. FSH and LH are gonadotrophins traditionally supplemented in maturation media with important roles in the orchestration of the maturation process in bovine COCs (Khan et al., 2015). Even though FSHR protein expression was detected in equine oocytes (Scarlet et al., 2015), the influence of FSH on oocyte quality seems to be mostly mediated by CCs (Buratini et al., 2021). The available literature gives reason to assume that excessive FSH availability may result in impaired developmental competence of the oocyte. Molecular pathways can link increased FSH signaling to impaired communication through transzonal processes between CCs and the oocyte (Buratini et al., 2021). The optimal gonadotrophin concentrations during IVM as well as their signaling mechanisms to achieve oocytes with the best developmental potential are still under investigation. In human IVM, concentrations >70 IU/l improved the nuclear maturation of human oocytes by downregulating FSHR and upregulating LHCGR in CCs (Cadenas et al., 2021). For murine oocytes, a shorter incubation time with FSH (2 h) during maturation was beneficial for fertilization and BL rates (Lin et al., 2011). Besides FSH concentration and exposure time, the ratio between FSH and LH can also influence the maturation success. In human oocytes, a ratio of FSH:LH at 1:10 resulted in significantly improved embryo development beyond Day 2 compared to a 1:1 ratio. In bovine oocytes, the development to the BL stage was not significantly different for the two ratios, but still higher using the 1:10 ratio than the 1:1 (Anderiesz et al., 2000). Gene-expression analysis of CCs of individual human oocytes also identified FSH as an important upstream regulator for developmental competence. Interestingly, lower developmental potential can be associated with over- or under-stimulation by FSH in comparison to samples from oocytes with high developmental (pregnancy-positive) potential (El-Maarri et al., 2021). This result also indicates that too much FSH (too high concentration or too long exposure) as well as too little FSH might hinder the utilization of the oocyte’s full intrinsic maturational potential. This concept of just the right concentration or stimulation was described by Leese et al. (2016) for the right amount of embryonic metabolic activity in preimplantation embryos that results in optimal developmental competence: called the ‘Goldilocks Principle’. This hypothesis was later modified and renamed the ‘Goldilocks zone’, as it could be demonstrated that there is an optimal area of metabolic activity, within which oocytes and embryos with maximum development potential are located (Leese et al., 2022).
Energy metabolism
The dependence of the oocyte on the delivery of glycolytic products from the surrounding CCs was elucidated already half a century ago (Biggers et al., 1967). The essential role of glucose metabolism in COCs for the acquisition of developmentally competent oocytes is highlighted in numerous studies and reviews (Sugiura et al., 2005; Sutton-McDowall et al., 2010; González-Fernández et al., 2018). The oocytes depend on CCs, especially for the utilization of glucose (Richani et al., 2021). Consumption of glucose in CCs was estimated to be 23 times higher than in oocytes (Thompson et al., 2007). The comparison of the ‘cumulome’ between maturation in vivo and in vitro already revealed a shift toward glycolysis under in vitro conditions. For the interpretation of these results, the experimental condition needs to be considered. With a concentration of 17.5 mmol/l, DMEM-F12-based maturation media contain about four times more glucose than follicular fluid in the physiological environment (Walter et al., 2019; Fernández-Hernández et al., 2020). With these high-glucose media, significantly improved cleavage and BL rates were achieved in the horse (Galli et al., 2007). Nevertheless, maturation in TCM199-based medium containing 5.6 mM glucose also resulted in satisfactory results (Hinrichs et al., 2005; Brom-de-Luna et al., 2019). A recent study, comparing physiological (5.6 mM) and supraphysiological (17 mM) concentrations of glucose, found no differences in maturation or BL rate, but altered mitochondrial function (Lewis et al., 2020b). For this ‘cumulomics’ study, a high-glucose DMEM/F12-based maturation medium was used, and glucose was identified as an upstream regulator in the proteomics dataset (Table 3).
One of the most important roles of CCs is to supply pyruvate to the oocyte for ATP production via tricarboxylic acid (TCA) cycle and oxidative phosphorylation (Sutton-McDowall et al., 2010; Richani et al., 2021). The cumulomics data showed that KEGG pathways related to carbohydrate metabolism are significantly overrepresented in the proteins that are overexpressed in CV and BL cumulus (Fig. 3). In particular, numerous enzymes involved in glycolysis were significantly higher expressed in cumulus of maturational competent oocytes (CV/BL) (Fig. 4). These results demonstrate that glycolysis, generating pyruvate/lactate, is as important for the oocyte’s energy supply in the horse as in other species (Downs et al., 2002; Johnson et al., 2007; Bresnahan et al., 2024). A comprehensive analysis of metabolism during maturation of equine COCs in vitro revealed that glucose consumption and lactate production did not differ between degenerated and matured COCs nor between matured COCs that cleaved or failed to cleave (Lewis et al., 2020a,b). However, the glucose consumption of developmental competent COCs was close to the mean of all COCs, which supports again the ‘Goldilocks principle’ (Leese et al., 2016; Lewis et al., 2020b). Metabolism during IVM under high-glucose/high oxygen conditions also directs energy production toward glycolysis compared to maturation in vivo (Walter et al., 2019).
The glycolysis product pyruvate may either be transferred to the oocyte or metabolized directly in the mitochondria of CCs by TCA cycle and oxidative phosphorylation (Sutton-McDowall et al., 2010; Richani et al., 2021). In contrast to glycolysis, higher expression of the proteins or metabolites involved in these metabolic pathways was not consistently observed in maturing COCs (Fig. 4). On the contrary, the enzymes aconitase (ACON, TCA cycle) and mitochondrial NADH dehydrogenase iron-sulfur protein 8 (Ndufs8, oxidative phosphorylation) showed significantly lower expression in cumulus of maturational competent oocytes. The same distribution was observed for the TCA cycle metabolites succinate and malate. Succinate is a crucial metabolite linking several metabolic pathways. Succinate dehydrogenase couples the TCA cycle and the respiratory chain in mitochondria. It is also involved in the formation and elimination of reactive oxygen species (Tretter et al., 2016). For bovine COCs, TCA metabolites decrease in CCs over the time course of maturation but were secreted into the medium. This may indicate an overall reduced pyruvate utilization in CCs (Uhde et al., 2018). The TCA cycle metabolites oxalacetate and α-ketoglutarate showed different distribution patterns (Fig. 4) with increased expression in maturation-competent oocytes. This is not necessarily contradictory as the levels can be filled directly from outside the TCA cycle: oxalacetate from pyruvate (by pyruvate carboxylase) and α-ketoglutarate from glutamate (by glutamate dehydrogenase).
With the used global metabolomics approach, it is not possible to finely discriminate between ATP and ADP. Nevertheless, both compounds showed the same pattern between developmental groups, with the highest level in cumulus of oocytes that developed to the BL stage and the lowest in the ones that failed to mature. In combination with the other alterations in energy metabolism (Fig. 4), this is one of the most intriguing candidates in this study, which promotes the hypothesis that CCs not only provide glycolytic products to the oocyte for energy production but may also directly fuel the oocyte with ATP. Further studies are necessary to consolidate this theory. Still, it is already supported by the available literature on other species. In bovine germinal vesicle COCs, oxidative phosphorylation was the most prominent metabolic pathway in CCs, but not in oocytes. Additionally, a higher expression of core proteins involved in oxidative phosphorylation was found in CCs compared to oocytes (Peddinti et al., 2010). Measurement of ATP during IVM in single mouse COCs revealed higher levels of ATP in COCs accompanied by their cumulus vesture (Dalton et al., 2014; Scantland et al., 2014). This observation was abolished by the inhibition of gap junction communication (Dalton et al., 2014). In the NM group, the functional enrichment analysis using unfiltered ranked data showed that the KEGG pathway ‘oxidative phosphorylation’ (hsa00190) was significantly enriched (Supplementary Fig. S2). On the individual protein level, NDUFS8, which catalyzes the electron transfer from NADH in the respiratory chain, was significantly higher in the NM group, with the lowest value in the BL group (Fig. 4). In COCs that failed to mature, the cumulus seems to counter-steer this ATP/energy shortage by increasing oxidative phosphorylation. These results raise the hypothesis that the high ATP content of developmentally competent COCs might come from other pathways than oxidative phosphorylation. One option for this might be the adenosine salvage pathway, which phosphorylates AMP in two steps to ATP (Richani et al., 2021). ATP amounts of this pathway (from isotopic-labeled AMP) were higher in intact COCs than in denuded oocytes and lower in pharmacological uncoupled COC gap junctions (Richani et al., 2019). These results also indicate that CCs provide not only energy substrates but also ATP directly to their oocyte through gap junctions (Richani et al., 2019, 2021).
The positive correlation of ATP content in the oocyte with its developmental competence was documented for a variety of species (Van Blerkom, 2011; Richani et al., 2021). Bioassays investigating mitochondrial function in human CCs of IVF patients also found correlations with maturity of the oocytes (Dumesic et al., 2016; Anderson et al., 2018). Interestingly, this effect seems to be negatively influenced by higher FSH concentrations during stimulation (Dumesic et al., 2016). Finally, lower ATP levels and impaired mitochondrial function in CCs were found in diabetic mice as well as in human IVF patients with endometriosis (Wang et al., 2010; Hsu et al., 2015). The present study adds data regarding the horse by describing a positive correlation of ATP content in CCs with developmental competence of the oocyte. Metabolic flux analysis in equine COCs during maturation suggests that the major metabolic fate of glucose is the release of lactate. Measurements of the oxygen consumption rate revealed that ∼87% of ATP comes from oxidative phosphorylation and only 13% came from glycolytic production of lactate. Therefore, the equine COC also seems to use other substrates, like lipids, amino acids, or glycogen, for oxidative ATP production (Lewis et al., 2020a). Here, the ‘cumulomics’ results add novel data regarding utilization of glycogen in CCs. Several enzymes involved in glycogen degradation were significantly overexpressed in the cumulus of maturational competent oocytes (glycogen phosphorylase (PYGL/B) and phosphoglucomutase-1 (PGM1); Fig. 4). In addition, IPA analysis also identified glycogen degradation as an activated canonical pathway in the cumulus of maturational competent oocytes (Table 2). Literature on glycogen utilization in COCs during maturation is very limited. Some data are available on the storage of glycogen in the female gamete or early embryo of model organisms. A study in mouse embryos found glycogen in large amounts in cleavage-stage oocytes. This glycogen seemed to be utilized only from the BL stage onward (Thomson and Brinster, 1966). Similar results were detected in a teleost (Yurowitzky and Milman, 1972). In the shrew, an induced ovulator, the cumulus oophorus of MII oocytes is rich in glycogen (Kaneko et al., 2003). Also, in oocytes from diabetic mice, glycogen storage was significantly increased (Ratchford et al., 2007). A study on Drosophila oogenesis elucidated that mitochondria enter a reversible low respiratory state in late oogenesis, establishing oocyte quiescence. This decrease in oxidative metabolism results in the accumulation of glycolytic and TCA cycle intermediates and leads to glycogen storage (Sieber et al., 2016). In the macaque, the mRNA content of glycogen phosphorylase in mural granulosa cells increased after hCG administration (Brogan et al., 2011) and decreased outside the breeding season (VandeVoort et al., 2015). Glycogen breakdown in CCs increases the availability of glucose and may therefore be associated with a higher developmental potential of the oocyte. In human IVF patients, mitochondrial utilization of glycogen in granulosa cells was significantly correlated with higher fertilization rates (Kordus et al., 2020). This directly reflects the results that were obtained in this study for the horse, with higher expression of proteins involved in glycogen degradation. Based on the detection in the pre-ovulatory follicular fluid of glucose-1-phosphate, which is derived from glycogen, Fernández-Hernández et al. (2020) have suggested that the equine oocyte also relies on glycogen metabolism as proposed in previous reports (Lewis et al., 2020a). In in vitro-matured bovine oocytes, the consumption of serum-derived glycogen was mediated by follicle-stimulating hormone (Cantanhêde et al., 2021).
In summary, the metabolic signature in the cumulus of maturational competent oocytes seems to be characterized by increased glycogen degradation and glycolysis as well as a decrease in TCA cycle/oxidative phosphorylation. The cumulus ATP (ATP/ADP) content was identified as an important determinant of the oocyte’s developmental competence.
Vesicular transport
Analysis of the overrepresentation of proteins with higher expression in oocytes that have the potential to mature (CV&BL>NM) also identified the pathway of ‘endocytosis’ (hsa04144) and ‘protein processing in endoplasmic reticulum’ (hsa04141), with many proteins related to clathrin-mediated endocytosis, early endosome, multivesicular body formation or COPII vesicle coating (Fig. 3). These pathways are involved in intracellular membrane trafficking, which enables the cells to communicate with their environment (Alberts et al., 2008). These processes are illustrated in Fig. 5, which shows boxplots for the proteins with relevant expression differences between the developmental groups of this study. Transport vesicles for endo- and exocytosis are coated by proteins, which drive vesicle budding and select the vesicle cargo (Robinson, 1997). Three different types of coated vesicles are distinguished by their coat proteins: clathrin-coated, COPI-coated, and COPII-coated. Clathrin-coated vesicles are involved in endocytosis, carrying vesicles from the plasma membrane to the endosome or Golgi compartment. COP-coated vesicles are involved in secretory pathways, with COPI-coated vesicles budding from the Golgi complex and COPII-coated vesicles budding from the ER (Alberts et al., 2008). Different adaptor proteins are responsible for cargo selection into the vesicles. The AP-2 adaptor links the clathrin coat with proteins to be endocytosed (Pearse et al., 2000). All four subunits showed a significant higher expression in maturational competent proteins. The same pattern was observed for SEC23/SEC24, the adaptor proteins for COPII vesicles at the ER (Barlowe et al., 1994). ADP-ribosylation factors (ARFs) are small coat-recruitment GTPases that trigger assembly and disassembly of coated carrier vesicles (Alberts et al., 2008; Kondo et al., 2012). ADP-ribosylation factor 3 (ARF3) was significantly higher in CV and BL groups compared to NM (Fig. 5).
A synapse-like communication between CCs and oocytes was demonstrated at the tip of transzonal processes, through which CCs can deliver large cargo like RNA to the oocytes (Allworth and Albertini, 1993; Macaulay et al., 2014, 2016). Canonical pathway analysis revealed synaptogenesis signaling as an activated pathway in the cumulus of maturational-competent oocytes (Table 2). This pathway includes proteins of clathrin-mediated endocytosis, which is the predominant mechanism in synaptic vesicle recycling (Gan and Watanabe, 2018). Clathrin-mediated endocytosis is also an important route for internalization of extracellular vesicles (Gurung et al., 2021). In other species, supplementation of extracellular vesicles to the maturation medium already improved developmental competence of the oocytes (da Silveira et al., 2017; de Avila et al., 2020; Tesfaye et al., 2020; Javadi et al., 2021).
The WASH complex is a nucleation-promoting factor that activates the actin nucleator ARP2/3 complex (ARP2/3) and is responsible for actin assembly in the cell (Duleh and Welch, 2010; Buratini et al., 2021). Assembly of actin filaments plays a central role in endocytosis, especially for clathrin-coated vesicles (Galletta and Cooper, 2009). A recent study showed that the uncoupling of the somatic cells from the oocytes is an active retraction of the transzonal processes into the body of CCs. This retraction is initiated by LH (in vivo) or EGF (in vitro) signaling and involves two proteins with higher expression in the cumulus of maturational competent oocytes: calpain1 catalytic subunit (CAPN1) triggers the loss of adhesion between transzonal processes, and actin-related protein 2/3 complex (ARP2/3) which is responsible for the retraction of transzonal projections (TZPs) into the CCs (Abbassi et al., 2021). Both proteins were significantly more abundant in the cumulus of maturational competent oocytes. This loss of the TZPs during maturation might also be a reason for the detected accumulation of ATP in cumulus of developmental competent oocytes. However, the release of ATP from cells does not only occur through gap junction channels in transzonal processes. ATP storage in secretory granules with release through exocytosis is well documented and might be another mechanism by which CCs nurse their oocytes with ATP (Lazarowski, 2012).
Another overrepresented pathway in maturational competent COCs involved in vesicular transport was ‘protein processing in endoplasmic reticulum’ (hsa04141) (Fig. 3). Besides five proteins of the adaptor complex of COPII vesicles (SEC23/24), particularly channel proteins connecting the ER lumen with the cytoplasm (BCAP31, SEC61A1, CAPN1) were enriched. ER stress, together with inflammation and oxidative stress, results in the unfolding protein response (Ron and Walter, 2007). Conditions leading to ER stress are glucose deprivation, inflammation, oxidative stress, elevated free fatty acids, impaired Ca2+ homeostasis, or hypoxia (Huang et al., 2017; Brenjian et al., 2020). Therapeutic strategies targeting ER stress (resveratrol and salubrinal) were found to improve the developmental potential of COCs (Sutton-McDowall et al., 2016; Brenjian et al., 2020).
Finally, mothers against decapentablegic homolog 2 protein (SMAD2) expression was higher in developmental-competent COCs. The available literature already highlights the role of SMAD signaling pathways in mediating TGFB1 signaling in cumulus and granulosa cells, leading to hyaluronan (HA) synthesis and cumulus expansion (Li et al., 2008; Wang et al., 2019).
GAG metabolism
During maturation, the COC undergoes significant alterations in the composition of the extracellular matrix. The gonadotrophin surge stimulates CCs to produce an HA-rich matrix, which is mainly responsible for the expansion of the cumulus complex (Nagyova, 2018). In addition to HA, the CCs secrete proteoglycans and proteins (Zhuo and Kimata, 2001). Basement membrane-specific heparan sulfate proteoglycan core protein (PGBM, HSPG2, Perlecan) was detected at a significantly lower level in developmental-competent cumulus complexes. This is in accordance with recent findings, where lower HSPG2 mRNA levels were related to human oocyte quality (Ma et al., 2020). A metabolic compound that was attributed to glucosamine (4.58_178.0716 m/z) was also detected at the highest concentrations in the NM group and lowest in the BL group. Glucosamine is part of hyaluronic acid (HA), a polymer of N-acetyl-D-glucosamine (GlcNAc): an amide between glucosamine, acetic acid, and D-glucuronic acid. Glucosamine and acetylglucosamine in culture media are incorporated in hyaluronic acid and support the expansion of cumulus (Ball et al., 1982; Salustri et al., 1989; Chen et al., 1990). HA is generated by CCs via the hexosamine biosynthesis pathway (HBP). Increased activity can lead to an alternate fate of UDP-N-acetylglucosamine toward O-linked glycosylation, which results in a reduction of the developmental competence of bovine oocytes (Thompson et al., 2007).
The low concentrations of these compounds in the cumulus of developmental-competent oocytes might reflect the successful maturation of the extracellular matrix in preparation for fertilization. This hypothesis is supported by the increase of three proteins related to GAG degradation in the maturational competent groups: GUSB, GNS, and HEXA. Figure 6 illustrates GAG degradation and the measured players in this pathway. The involved proteins are located in lysosomes. Literature on the function of these proteins in CCs is limited. Degradation of HA is important for fertilization and the sperm acrosome contains specific isoforms of hyaluronidase, the first enzyme involved in cumulus degradation, which enables the sperm to cross the cumulus mass (Fouladi-Nashta et al., 2017). The specific role of these proteins in CCs needs further investigation, but their overexpression in the cumulus of developmental competent oocytes gives reason for the assumption that the COC itself might also play an important role in regulating its own reconstruction in preparation of sperm penetration.
Conclusion
In summary, the analysis of the ‘cumulome’ in relation to the developmental competence of the oocytes revealed novel mechanisms that might be related to the triggering of the developmental competence of oocytes. The importance of energy metabolism was highlighted by the strong correlation between ATP content in CCs and the developmental competence of the oocyte. The data also revealed an emerging role of vesicular transport within and between CCs, which seems to be important for the CCs to modulate their own extracellular environment.
Supplementary Material
Contributor Information
Jasmin Walter, Clinic of Reproductive Medicine, Department for Farm Animals, Vetsuisse Faculty, University of Zurich, Zurich, Switzerland.
Silvia Colleoni, Avantea srl, Laboratory of Reproductive Technologies, Cremona, Italy.
Giovanna Lazzari, Avantea srl, Laboratory of Reproductive Technologies, Cremona, Italy.
Claudia Fortes, Functional Genomics Centre Zurich, University and ETH Zurich, Zurich, Switzerland.
Jonas Grossmann, Functional Genomics Centre Zurich, University and ETH Zurich, Zurich, Switzerland; Swiss Institute of Bioinformatics (SIB), Zurich, Switzerland.
Bernd Roschitzki, Functional Genomics Centre Zurich, University and ETH Zurich, Zurich, Switzerland.
Endre Laczko, Functional Genomics Centre Zurich, University and ETH Zurich, Zurich, Switzerland.
Hanspeter Naegeli, Institute of Pharmacology and Toxicology, Vetsuisse Faculty, University of Zurich, Zurich, Switzerland.
Ulrich Bleul, Clinic of Reproductive Medicine, Department for Farm Animals, Vetsuisse Faculty, University of Zurich, Zurich, Switzerland.
Cesare Galli, Avantea srl, Laboratory of Reproductive Technologies, Cremona, Italy.
Supplementary data
Supplementary data are available at Molecular Human Reproduction online.
Data availability
All proteomic data are available in PRIDE-Archive (EMBL-EBI) with the dataset identifier PXD033551 (https://www.ebi.ac.uk/pride/archive/).
Authors’ roles
All authors contributed to the concept and experimental design of this study. All IVF lab work was conducted at Avantea srl. by S.C., G.L., and C.G. J.W. contributed to IVF lab work, analyzed, and interpreted data, designed the illustrations, and wrote the manuscript. Proteomics analysis was performed at the Functional Genomics Centre Zurich by C.F. and B.R. J.G. analyzed the proteomics data. Metabolomics analysis as well as data analysis was conducted by E.L. J.W., U.B., and H.N. were involved in drafting the manuscript and revised the article for critically important intellectual content. The final version of the manuscript was approved by all authors, and they agreed to be accountable for all aspects of the work.
Funding
‘Forschungskredit’ of the University of Zurich (‘Clinomics-Project’) (grant number FK-13-062).
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Abbassi L, El-Hayek S, Carvalho KF, Wang W, Yang Q, Granados-Aparici S, Mondadori R, Bordignon V, Clarke HJ.. Epidermal growth factor receptor signaling uncouples germ cells from the somatic follicular compartment at ovulation. Nat Commun 2021;12:1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adriaenssens T, Wathlet S, Segers I, Verheyen G, De Vos A, Van der Elst J, Coucke W, Devroey P, Smitz J.. Cumulus cell gene expression is associated with oocyte developmental quality and influenced by patient and treatment characteristics. Hum Reprod 2010;25:1259–1270. [DOI] [PubMed] [Google Scholar]
- Aguila L, Treulen F, Therrien J, Felmer R, Valdivia M, Smith LC.. Oocyte selection for in vitro embryo production in bovine species: noninvasive approaches for new challenges of oocyte competence. Animals 2020;10:2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts B, Johnson A, Lewis J, Morgan D, Raff M, Roberts K, Walter P.. Molecular Biology of the Cell, 6th edn. New York, NY, USA: Garland Science, 2008. [Google Scholar]
- Allworth AE, Albertini DF.. Meiotic maturation in cultured bovine oocytes is accompanied by remodeling of the cumulus cell cytoskeleton. Dev Biol 1993;158:101–112. [DOI] [PubMed] [Google Scholar]
- Anderiesz C, Ferraretti A, Magli C, Fiorentino A, Fortini D, Gianaroli L, Jones GM, Trounson AO.. Effect of recombinant human gonadotrophins on human, bovine and murine oocyte meiosis, fertilization and embryonic development in vitro. Hum Reprod 2000;15:1140–1148. [DOI] [PubMed] [Google Scholar]
- Anderson RA, Sciorio R, Kinnell H, Bayne RAL, Thong KJ, De Sousa PA, Pickering S.. Cumulus gene expression as a predictor of human oocyte fertilisation, embryo development and competence to establish a pregnancy. Reproduction 2009;138:629–637. [DOI] [PubMed] [Google Scholar]
- Anderson S, Xu P, Frey AJ, Goodspeed JR, Doan MT, Orris JJ, Clements N, Glassner MJ, Snyder NW.. Cumulus cell acetyl-CoA metabolism from acetate is associated with maternal age but only partially with oocyte maturity. Syst Biol Reprod Med 2022;68:36–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson SH, Glassner MJ, Melnikov A, Friedman G, Orynbayeva Z.. Respirometric reserve capacity of cumulus cell mitochondria correlates with oocyte maturity. J Assist Reprod Genet 2018;35:1821–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artini PG, Tatone C, Sperduti S, D’Aurora M, Franchi S, Di Emidio G, Ciriminna R, Vento M, Di Pietro C, Stuppia L. et al. ; Italian Society of embryology, Reproduction and Research (SIERR). Cumulus cells surrounding oocytes with high developmental competence exhibit down-regulation of phosphoinositol 1,3 kinase/protein kinase B (PI3K/AKT) signalling genes involved in proliferation and survival. Hum Reprod 2017;32:2474–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assidi M, Montag M, Van der Ven K, Sirard M-A.. Biomarkers of human oocyte developmental competence expressed in cumulus cells before ICSI: a preliminary study. J Assist Reprod Genet 2011;28:173–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assou S, Anahory T, Pantesco V, Le Carrour T, Pellestor F, Klein B, Reyftmann L, Dechaud H, De Vos J, Hamamah S.. The human cumulus–oocyte complex gene-expression profile. Hum Reprod 2006;21:1705–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assou S, Haouzi D, De Vos J, Hamamah S.. Human cumulus cells as biomarkers for embryo and pregnancy outcomes. Mol Hum Reprod 2010;16:531–538. [DOI] [PubMed] [Google Scholar]
- Ball GD, Bellin ME, Ax RL, First NL.. Glycosaminoglycans in bovine cumulus–oocyte complexes: morphology and chemistry. Mol Cell Endocrinol 1982;28:113–122. [DOI] [PubMed] [Google Scholar]
- Barlowe C, Orci L, Yeung T, Hosobuchi M, Hamamoto S, Salama N, Rexach MF, Ravazzola M, Amherdt M, Schekman R.. COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell 1994;77:895–907. [DOI] [PubMed] [Google Scholar]
- Biggers JD, Whittingham DG, Donahue RP.. The pattern of energy metabolism in the mouse oocyte and zygote. Proc Natl Acad Sci USA 1967;58:560–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco IDP, Devito LG, Ferreira HN, Araujo GHM, Fernandes CB, Alvarenga MA, Landim-Alvarenga FC.. Collection rates and morphology of equine oocytes obtained from immature follicles after treatment with equine pituitary extract (EPE) and human chorionic gonadotropin. J Equine Vet Sci 2009;29:613–617. [Google Scholar]
- Borup R, Thuesen LL, Andersen CY, Nyboe-Andersen A, Ziebe S, Winther O, Grondahl ML.. Competence classification of cumulus and granulosa cell transcriptome in embryos matched by morphology and female age. PLoS One 2016;11:e0153562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braga DPAF, Setti AS, Lo Turco EG, Cordeiro FB, Cabral EC, Cortezzi SS, Ono E, Figueira RCS, Eberlin MN, Borges E.. Protein expression in human cumulus cells as an indicator of blastocyst formation and pregnancy success. J Assist Reprod Genet 2016;33:1571–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenjian S, Moini A, Yamini N, Kashani L, Faridmojtahedi M, Bahramrezaie M, Khodarahmian M, Amidi F.. Resveratrol treatment in patients with polycystic ovary syndrome decreased pro-inflammatory and endoplasmic reticulum stress markers. Am J Reprod Immunol 2020;83:e13186. [DOI] [PubMed] [Google Scholar]
- Bresnahan DR, Catandi GD, Peters SO, Maclellan LJ, Broeckling CD, Carnevale EM.. Maturation and culture affect the metabolomic profile of oocytes and follicular cells in young and old mares. Front Cell Develop Biol 2024;11:1280998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briski O, Salamone DF.. Past, present and future of ICSI in livestock species. Anim Reprod Sci 2022;246:106925. [DOI] [PubMed] [Google Scholar]
- Brogan RS, MacGibeny M, Mix S, Thompson C, Puttabyatappa M, VandeVoort CA, Chaffin CL.. Dynamics of intra-follicular glucose during luteinization of macaque ovarian follicles. Mol Cell Endocrinol 2011;332:189–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brom-de-Luna JG, Salgado RM, Canesin HS, Diaw M, Hinrichs K.. Equine blastocyst production under different incubation temperatures and different CO2 concentrations during early cleavage. Reprod Fertil Dev 2019;31:1823–1829. [DOI] [PubMed] [Google Scholar]
- Bunel A, Jorssen EP, Merckx E, Leroy JL, Bols PE, Sirard MA.. Individual bovine in vitro embryo production and cumulus cell transcriptomic analysis to distinguish cumulus–oocyte complexes with high or low developmental potential. Theriogenology 2015;83:228–237. [DOI] [PubMed] [Google Scholar]
- Bunel A, Nivet AL, Blondin P, Vigneault C, Richard FJ, Sirard MA.. Cumulus cell gene expression associated with pre-ovulatory acquisition of developmental competence in bovine oocytes. Reprod Fertil Dev 2014;26:855–865. [DOI] [PubMed] [Google Scholar]
- Buratini J, Dellaqua TT, Dal Canto M, La Marca A, Carone D, Mignini Renzini M, Webb R.. The putative roles of FSH and AMH in the regulation of oocyte developmental competence: from fertility prognosis to mechanisms underlying age-related subfertility. Hum Reprod Update 2021;28:232–254. [DOI] [PubMed] [Google Scholar]
- Cadenas J, Nikiforov D, Pors SE, Zuniga LA, Wakimoto Y, Ghezelayagh Z, Mamsen LS, Kristensen SG, Andersen CY.. A threshold concentration of FSH is needed during IVM of ex vivo collected human oocytes. J Assist Reprod Genet 2021;38:1341–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantanhêde LF, Santos-Silva CT, Moura MT, Ferreira-Silva JC, Oliveira JMB, Gonçalves DNA, Teixeira ÁAC, Wanderley-Teixeira V, Oliveira MAL.. Follicle-stimulating hormone mediates the consumption of serum-derived glycogen by bovine cumulus–oocyte complexes during in vitro maturation. Vet World 2021;14:2512–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnevale EM. The mare model for follicular maturation and reproductive aging in the woman. Theriogenology 2008;69:23–30. [DOI] [PubMed] [Google Scholar]
- Catandi GD, Bresnahan DR, Peters SO, Fresa KJ, Maclellan LJ, Broeckling CD, Carnevale EM.. Equine maternal aging affects the metabolomic profile of oocytes and follicular cells during different maturation time points. Front Cell Dev Biol 2023;11:1239154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catandi GD, LiPuma L, Obeidat YM, Maclellan LJ, Broeckling CD, Chen T, Chicco AJ, Carnevale EM.. Oocyte metabolic function, lipid composition, and developmental potential are altered by diet in older mares. Reproduction 2022;163:183–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Wert SE, Hendrix EM, Russell PT, Cannon M, Larsen WJ.. Hyaluronic acid synthesis and gap junction endocytosis are necessary for normal expansion of the cumulus mass. Mol Reprod Dev 1990;26:236–247. [DOI] [PubMed] [Google Scholar]
- Colleoni S, Luciano AM, Gandolfi F.. Cumulus-oocyte communications in the horse: role of the breeding season and of the maturation medium. Reprod Domest Anim 2004;39:70–75. [DOI] [PubMed] [Google Scholar]
- da Silveira JC, Andrade GM, Del Collado M, Sampaio RV, Sangalli JR, Silva LA, Pinaffi FVL, Jardim IB, Cesar MC, Nogueira MFG. et al. Supplementation with small-extracellular vesicles from ovarian follicular fluid during in vitro production modulates bovine embryo development. PLoS One 2017;12:e0179451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton CM, Szabadkai G, Carroll J.. Measurement of ATP in single oocytes: impact of maturation and cumulus cells on levels and consumption. J Cell Physiol 2014;229:353–361. [DOI] [PubMed] [Google Scholar]
- de Avila A, Bridi A, Andrade GM, Del Collado M, Sangalli JR, Nociti RP, da Silva Junior WA, Bastien A, Robert C, Meirelles FV. et al. Estrous cycle impacts microRNA content in extracellular vesicles that modulate bovine cumulus cell transcripts during in vitro maturationdagger. Biol Reprod 2020;102:362–375. [DOI] [PubMed] [Google Scholar]
- Dell’Aquila ME, Cho YS, Martino NA, Uranio MF, Rutigliano L, Hinrichs K.. OMICS for the identification of biomarkers for oocyte competence, with special reference to the mare as a prospective model for human reproductive medicine. In: Swan A (ed). Meiosis—Molecular Mechanisms and Cytogenetic Diversity. London, UK: IntechOpen Ltd., 2012, 257–282. 10.5772/33270. https://www.intechopen.com/chapters/30377 [DOI] [Google Scholar]
- Dell’Aquila ME, De Felici M, Massari S, Maritato F, Minoia P.. Effects of fetuin on zona pellucida hardening and fertilizability of equine oocytes matured in vitro. Biol Reprod 1999;61:533–540. [DOI] [PubMed] [Google Scholar]
- Demiray SB, Goker ENT, Tavmergen E, Yilmaz O, Calimlioglu N, Soykam HO, Oktem G, Sezerman U.. Differential gene expression analysis of human cumulus cells. Clin Exp Reprod Med 2019;46:76–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diz AP, Truebano M, Skibinski DOF.. The consequences of sample pooling in proteomics: an empirical study. Electrophoresis 2009;30:2967–2975. [DOI] [PubMed] [Google Scholar]
- Downs SM, Humpherson PG, Leese HJ.. Pyruvate utilization by mouse oocytes is influenced by meiotic status and the cumulus oophorus. Mol Reprod Dev 2002;62:113–123. [DOI] [PubMed] [Google Scholar]
- Duleh SN, Welch MD.. WASH and the Arp2/3 complex regulate endosome shape and trafficking. Cytoskeleton (Hoboken) 2010;67:193–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumesic DA, Guedikian AA, Madrigal VK, Phan JD, Hill DL, Alvarez JP, Chazenbalk GD.. Cumulus cell mitochondrial resistance to stress in vitro predicts oocyte development during assisted reproduction. J Clin Endocrinol Metab 2016;101:2235–2245. [DOI] [PubMed] [Google Scholar]
- El-Maarri O, Jamil MA, Koster M, Nusgen N, Oldenburg J, Montag M, van der Ven H, van der Ven K.. Stratifying cumulus cell samples based on molecular profiling to help resolve biomarker discrepancies and to predict oocyte developmental competence. Int J Mol Sci 2021;22:6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix MR, Turner RM, Dobbie T, Hinrichs K.. Successful in vitro fertilization in the horse: production of blastocysts and birth of foals after prolonged sperm incubation for capacitation†. Biol Reprod 2022;107:1551–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Hernández P, Sánchez-Calabuig MJ, García-Marín LJ, Bragado MJ, Gutiérrez-Adán A, Millet Ó, Bruzzone C, González-Fernández L, Macías-García B.. Study of the metabolomics of equine preovulatory follicular fluid: a way to improve current in vitro maturation media. Animals 2020;10:883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortes C, Roschitzki B, Walter J, Schlapbach R.. Cumulomics—optimization of sample preparation for low amount cumulus samples. Annual Conference of the American Society for Mass Spectrometry, Baltimore, Maryland, USA, 2014. [Google Scholar]
- Foss R, Ortis H, Hinrichs K.. Effect of potential oocyte transport protocols on blastocyst rates after intracytoplasmic sperm injection in the horse. Equine Vet J Suppl 2013;(45):39–43. [DOI] [PubMed] [Google Scholar]
- Fouladi-Nashta AA, Raheem KA, Marei WF, Ghafari F, Hartshorne GM.. Regulation and roles of the hyaluronan system in mammalian reproduction. Reproduction 2017;153:R43–R58. [DOI] [PubMed] [Google Scholar]
- Galletta BJ, Cooper JA.. Actin and endocytosis: mechanisms and phylogeny. Curr Opin Cell Biol 2009;21:20–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli C, Colleoni S, Duchi R, Lagutina I, Lazzari G.. Developmental competence of equine oocytes and embryos obtained by in vitro procedures ranging from in vitro maturation and ICSI to embryo culture, cryopreservation and somatic cell nuclear transfer. Anim Reprod Sci 2007;98:39–55. [DOI] [PubMed] [Google Scholar]
- Galli C, Crotti G, Notari C, Turini P, Duchi R, Lazzari G.. Embryo production by ovum pick up from live donors. Theriogenology 2001;55:1341–1357. [DOI] [PubMed] [Google Scholar]
- Gan Q, Watanabe S.. Synaptic vesicle endocytosis in different model systems. Front Cell Neurosci 2018;12:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Fernández L, Sánchez-Calabuig MJ, Alves MG, Oliveira PF, Macedo S, Gutiérrez-Adán A, Rocha A, Macías-García B.. Expanded equine cumulus–oocyte complexes exhibit higher meiotic competence and lower glucose consumption than compact cumulus–oocyte complexes. Reprod Fertil Dev 2018;30:297–306. [DOI] [PubMed] [Google Scholar]
- González-Serrano AF, Pirro V, Ferreira CR, Oliveri P, Eberlin LS, Heinzmann J, Lucas-Hahn A, Niemann H, Cooks RG.. Desorption electrospray ionization mass spectrometry reveals lipid metabolism of individual oocytes and embryos. PLoS One 2013;8:e74981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green KA, Franasiak JM, Werner MD, Tao X, Landis JN, Scott RT Jr, Treff NR.. Cumulus cell transcriptome profiling is not predictive of live birth after in vitro fertilization: a paired analysis of euploid sibling blastocysts. Fertil Steril 2018;109:460–466.e2. [DOI] [PubMed] [Google Scholar]
- Grossmann J, Roschitzki B, Panse C, Fortes C, Barkow-Oesterreicher S, Rutishauser D, Schlapbach R.. Implementation and evaluation of relative and absolute quantification in shotgun proteomics with label-free methods. J Proteomics 2010;73:1740–1746. [DOI] [PubMed] [Google Scholar]
- Gurung S, Perocheau D, Touramanidou L, Baruteau J.. The exosome journey: from biogenesis to uptake and intracellular signalling. Cell Commun Signal 2021;19:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinrichs K. The equine oocyte: factors affecting meiotic and developmental competence. Mol Reprod Dev 2010;77:651–661. [DOI] [PubMed] [Google Scholar]
- Hinrichs K, Choi YH, Love LB, Varner DD, Love CC, Walckenaer BE.. Chromatin configuration within the germinal vesicle of horse oocytes: changes post mortem and relationship to meiotic and developmental competence. Biol Reprod 2005;72:1142–1150. [DOI] [PubMed] [Google Scholar]
- Hinrichs K, Love CC, Brinsko SP, Choi YH, Varner DD.. In vitro fertilization of in vitro-matured equine oocytes: effect of maturation medium, duration of maturation, and sperm calcium ionophore treatment, and comparison with rates of fertilization in vivo after oviductal transfer. Biol Reprod 2002;67:256–262. [DOI] [PubMed] [Google Scholar]
- Hsu AL, Townsend PM, Oehninger S, Castora FJ.. Endometriosis may be associated with mitochondrial dysfunction in cumulus cells from subjects undergoing in vitro fertilization-intracytoplasmic sperm injection, as reflected by decreased adenosine triphosphate production. Fertil Steril 2015;103:347–352.e1. [DOI] [PubMed] [Google Scholar]
- Huang N, Yu Y, Qiao J.. Dual role for the unfolded protein response in the ovary: adaption and apoptosis. Protein Cell 2017;8:14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javadi M, Soleimani Rad J, Pashaiasl M, Farashah MSG, Roshangar L.. The effects of plasma-derived extracellular vesicles on cumulus expansion and oocyte maturation in mice. Reprod Biol 2021;22:100593. [DOI] [PubMed] [Google Scholar]
- Jendoubi T. Approaches to integrating metabolomics and multi-omics data: a primer. Metabolites 2021;11:184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MT, Freeman EA, Gardner DK, Hunt PA.. Oxidative metabolism of pyruvate is required for meiotic maturation of murine oocytes in vivo. Biol Reprod 2007;77:2–8. [DOI] [PubMed] [Google Scholar]
- Käll L, Storey JD, MacCoss MJ, Noble WS.. Assigning significance to peptides identified by tandem mass spectrometry using decoy databases. J Proteome Res 2008;7:29–34. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Iida H, Bedford JM, Oda S, Mōri T.. Mating-induced cumulus-oocyte maturation in the shrew, Suncus murinus. Reproduction 2003;126:817–826. [DOI] [PubMed] [Google Scholar]
- Khan DR, Guillemette C, Sirard MA, Richard FJ.. Characterization of FSH signalling networks in bovine cumulus cells: a perspective on oocyte competence acquisition. Mol Hum Reprod 2015;21:688–701. [DOI] [PubMed] [Google Scholar]
- Kondo Y, Hanai A, Nakai W, Katoh Y, Nakayama K, Shin HW.. ARF1 and ARF3 are required for the integrity of recycling endosomes and the recycling pathway. Cell Struct Funct 2012;37:141–154. [DOI] [PubMed] [Google Scholar]
- Kordus RJ, Hossain A, Malter HE, LaVoie HA.. Mitochondrial metabolic substrate utilization in granulosa cells reflects body mass index and total follicle stimulating hormone dosage in in vitro fertilization patients. J Assist Reprod Genet 2020;37:2743–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labas V, Teixeira-Gomes A-P, Bouguereau L, Gargaros A, Spina L, Marestaing A, Uzbekova S.. Intact cell MALDI-TOF mass spectrometry on single bovine oocyte and follicular cells combined with top-down proteomics: a novel approach to characterise markers of oocyte maturation. J Proteomics 2017;175:56–74. [DOI] [PubMed] [Google Scholar]
- Lazarowski ER. Vesicular and conductive mechanisms of nucleotide release. Purinergic Signal 2012;8:359–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzari G, Colleoni S, Crotti G, Turini P, Fiorini G, Barandalla M, Landriscina L, Dolci G, Benedetti M, Duchi R. et al. Laboratory production of equine embryos. J Equine Vet Sci 2020;89:103097. [DOI] [PubMed] [Google Scholar]
- Lazzari G, Wrenzycki C, Herrmann D, Duchi R, Kruip T, Niemann H, Galli C.. Cellular and molecular deviations in bovine in vitro-produced embryos are related to the large offspring syndrome. Biol Reprod 2002;67:767–775. [DOI] [PubMed] [Google Scholar]
- Leemans B, Gadella BM, Stout TA, De Schauwer C, Nelis H, Hoogewijs M, Van Soom A.. Why doesn’t conventional IVF work in the horse? The equine oviduct as a microenvironment for capacitation/fertilization. Reproduction 2016;152:R233–R245. [DOI] [PubMed] [Google Scholar]
- Leese HJ, Brison DR, Sturmey RG.. The quiet embryo hypothesis: 20 years on. Front Physiol 2022;13:899485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leese HJ, Guerif F, Allgar V, Brison DR, Lundin K, Sturmey RG.. Biological optimization, the Goldilocks principle, and how much is lagom in the preimplantation embryo. Mol Reprod Dev 2016;83:748–754. [DOI] [PubMed] [Google Scholar]
- Lewis N, Hinrichs K, Leese HJ, Mc GAC, Brison DR, Sturmey R.. Energy metabolism of the equine cumulus oocyte complex during in vitro maturation. Sci Rep 2020a;10:3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis N, Hinrichs K, Leese HJ, McGregor Argo C, Brison DR, Sturmey RG.. Glucose concentration during equine in vitro maturation alters mitochondrial function. Reproduction 2020b;160:227–237. [DOI] [PubMed] [Google Scholar]
- Li Q, Pangas SA, Jorgez CJ, Graff JM, Weinstein M, Matzuk MM.. Redundant roles of SMAD2 and SMAD3 in ovarian granulosa cells in vivo. Mol Cell Biol 2008;28:7001–7011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S-H, Lin M-H, Hwu Y-M, Lu C-H, Yeh L-Y, Chen Y-J, Lee RK-K.. Correlation of cumulus gene expression of GJA1, PRSS35, PTX3, and SERPINE2 with oocyte maturation, fertilization, and embryo development. Reprod Biol Endocrinol 2015;13:93–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Li RQ, Ou SB, Zhang NF, Ren L, Wei LN, Zhang QX, Yang DZ.. Increased GDF9 and BMP15 mRNA levels in cumulus granulosa cells correlate with oocyte maturation, fertilization, and embryo quality in humans. Reprod Biol Endocrinol 2014;12:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YH, Hwang JL, Seow KM, Huang LW, Hsieh BC, Chen HJ, Tzeng CR, Bai CH.. Effect of incubation with different concentrations and durations of FSH for in-vitro maturation of murine oocytes. Reprod Biomed Online 2011;23:111–117. [DOI] [PubMed] [Google Scholar]
- Ma Y, Jin J, Tong X, Yang W, Ren P, Dai Y, Pan Y, Zhang Y, Zhang S.. ADAMTS1 and HSPG2 mRNA levels in cumulus cells are related to human oocyte quality and controlled ovarian hyperstimulation outcomes. J Assist Reprod Genet 2020;37:657–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay AD, Gilbert I, Caballero J, Barreto R, Fournier E, Tossou P, Sirard M-A, Clarke HJ, Khandjian ÉW, Richard FJ. et al. The gametic synapse: RNA transfer to the bovine oocyte. Biol Reprod 2014;91:90–90. [DOI] [PubMed] [Google Scholar]
- Macaulay AD, Gilbert I, Scantland S, Fournier E, Ashkar F, Bastien A, Saadi HAS, Gagné D, Sirard M-A, Khandjian ÉW. et al. Cumulus cell transcripts transit to the bovine oocyte in preparation for maturation. Biol Reprod 2016;94:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Moro A, Lamas-Toranzo I, Bermejo-Alvarez P.. Metabolomics analysis of human cumulus cells from oocytes exhibiting different developmental competence. Reprod Fertil Dev 2021;33:172. [Google Scholar]
- Nagyova E. The biological role of hyaluronan-rich oocyte-cumulus extracellular matrix in female reproduction. Int J Mol Sci 2018;19:283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R.. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 2003;75:4646–4658. [DOI] [PubMed] [Google Scholar]
- Ouandaogo ZG, Frydman N, Hesters L, Assou S, Haouzi D, Dechaud H, Frydman R, Hamamah S.. Differences in transcriptomic profiles of human cumulus cells isolated from oocytes at GV, MI and MII stages after in vivo and in vitro oocyte maturation. Hum Reprod 2012;27:2438–2447. [DOI] [PubMed] [Google Scholar]
- Ouandaogo ZG, Haouzi D, Assou S, Dechaud H, Kadoch IJ, De Vos J, Hamamah S.. Human cumulus cells molecular signature in relation to oocyte nuclear maturity stage. PLoS One 2011;6:e27179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palermo G, Joris H, Devroey P, Van Steirteghem AC.. Pregnancies after intracytoplasmic injection of single spermatozoon into an oocyte. Lancet 1992;340:17–18. [DOI] [PubMed] [Google Scholar]
- Palmer E, Bezard J, Magistrini M, Duchamp G.. In-vitro fertilization in the horse—a retrospective study. J Reprod Fertil 1991;44:375–384. [PubMed] [Google Scholar]
- Pearse BM, Smith CJ, Owen DJ.. Clathrin coat construction in endocytosis. Curr Opin Struct Biol 2000;10:220–228. [DOI] [PubMed] [Google Scholar]
- Peddinti D, Memili E, Burgess SC.. Proteomics-based systems biology modeling of bovine germinal vesicle stage oocyte and cumulus cell interaction. PLoS One 2010;5:e11240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M. et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 2019;47:D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratchford AM, Chang AS, Chi MM, Sheridan R, Moley KH.. Maternal diabetes adversely affects AMP-activated protein kinase activity and cellular metabolism in murine oocytes. Am J Physiol Endocrinol Metab 2007;293:E1198–1206. [DOI] [PubMed] [Google Scholar]
- Rial-Otero R, Carreira RJ, Cordeiro FM, Moro AJ, Fernandes L, Moura I, Capelo JL.. Sonoreactor-based technology for fast high-throughput proteolytic digestion of proteins. J Proteome Res 2007;6:909–912. [DOI] [PubMed] [Google Scholar]
- Richani D, Dunning KR, Thompson JG, Gilchrist RB.. Metabolic co-dependence of the oocyte and cumulus cells: essential role in determining oocyte developmental competence. Hum Reprod Update 2021;27:27–47. [DOI] [PubMed] [Google Scholar]
- Richani D, Lavea CF, Kanakkaparambil R, Riepsamen AH, Bertoldo MJ, Bustamante S, Gilchrist RB.. Participation of the adenosine salvage pathway and cyclic AMP modulation in oocyte energy metabolism. Sci Rep 2019;9:18395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MS. Coats and vesicle budding. Trends Cell Biol 1997;7:99–102. [DOI] [PubMed] [Google Scholar]
- Ron D, Walter P.. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 2007;8:519–529. [DOI] [PubMed] [Google Scholar]
- Rouhollahi Varnosfaderani S, Hajian M, Jafarpour F, Ghazvini Zadegan F, Nasr-Esfahani MH.. Granulosa secreted factors improve the developmental competence of cumulus oocyte complexes from small antral follicles in sheep. PLoS One 2020;15:e0229043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell DL, Gilchrist RB, Brown HM, Thompson JG.. Bidirectional communication between cumulus cells and the oocyte: old hands and new players? Theriogenology 2016;86:62–68. [DOI] [PubMed] [Google Scholar]
- Salustri A, Yanagishita M, Hascall VC.. Synthesis and accumulation of hyaluronic acid and proteoglycans in the mouse cumulus cell–oocyte complex during follicle-stimulating hormone-induced mucification. J Biol Chem 1989;264:13840–13847. [PubMed] [Google Scholar]
- Scantland S, Tessaro I, Macabelli CH, Macaulay AD, Cagnone G, Fournier E, Luciano AM, Robert C.. The adenosine salvage pathway as an alternative to mitochondrial production of ATP in maturing mammalian oocytes. Biol Reprod 2014;91:75. [DOI] [PubMed] [Google Scholar]
- Scarica C, Cimadomo D, Dovere L, Giancani A, Stoppa M, Capalbo A, Ubaldi FM, Rienzi L, Canipari R.. An integrated investigation of oocyte developmental competence: expression of key genes in human cumulus cells, morphokinetics of early divisions, blastulation, and euploidy. J Assist Reprod Genet 2019;36:875–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarlet D, Walter I, Hlavaty J, Aurich C.. Expression and immunolocalisation of follicle-stimulating hormone receptors in gonads of newborn and adult female horses. Reprod Fert Dev 2015;28:1340–1348. [DOI] [PubMed] [Google Scholar]
- Scott TJ, Carnevale EM, Maclellan LJ, Scoggin CF, Squires EL.. Embryo development rates after transfer of oocytes matured in vivo, in vitro, or within oviducts of mares. Theriogenology 2001;55:705–715. [DOI] [PubMed] [Google Scholar]
- Sessions-Bresnahan DR, Schauer KL, Heuberger AL, Carnevale EM.. Effect of obesity on the preovulatory follicle and lipid fingerprint of equine oocytes1. Biol Reprod 2016;94:15, 11–12. [DOI] [PubMed] [Google Scholar]
- Sieber MH, Thomsen MB, Spradling AC.. Electron transport chain remodeling by GSK3 during oogenesis connects nutrient state to reproduction. Cell 2016;164:420–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirait B, Wiweko B, Jusuf AA, Iftitah D, Muharam R.. Oocyte competence biomarkers associated with oocyte maturation: a review. Front Cell Dev Biol 2021;9:710292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires EL, McCue PM.. Superovulation in mares. Anim Reprod Sci 2007;99:1–8. [DOI] [PubMed] [Google Scholar]
- Squires EL, Wilson JM, Kato H, Blaszczyk A.. A pregnancy after intracytoplasmic sperm injection into equine oocytes matured in vitro. Theriogenology 1996;45:306. [Google Scholar]
- Sugiura K, Pendola FL, Eppig JJ.. Oocyte control of metabolic cooperativity between oocytes and companion granulosa cells: energy metabolism. Dev Biol 2005;279:20–30. [DOI] [PubMed] [Google Scholar]
- Sutton-McDowall ML, Gilchrist RB, Thompson JG.. The pivotal role of glucose metabolism in determining oocyte developmental competence. Reproduction 2010;139:685–695. [DOI] [PubMed] [Google Scholar]
- Sutton-McDowall ML, Wu LL, Purdey M, Abell AD, Goldys EM, MacMillan KL, Thompson JG, Robker RL.. Nonesterified fatty acid-induced endoplasmic reticulum stress in cattle cumulus oocyte complexes alters cell metabolism and developmental competence. Biol Reprod 2016;94:23. [DOI] [PubMed] [Google Scholar]
- Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P. et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res 2021;49:D605–D612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telaar A, Nürnberg G, Repsilber D.. Finding biomarker signatures in pooled sample designs: a simulation framework for methodological comparisons. Adv Bioinformatics 2010;2010:318573–318578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesfaye D, Hailay T, Salilew-Wondim D, Hoelker M, Bitseha S, Gebremedhn S.. Extracellular vesicle mediated molecular signaling in ovarian follicle: Implication for oocyte developmental competence. Theriogenology 2020;150:70–74. [DOI] [PubMed] [Google Scholar]
- Thompson JG, Lane M, Gilchrist RB.. Metabolism of the bovine cumulus–oocyte complex and influence on subsequent developmental competence. Soc Reprod Fertil Suppl 2007;64:179–190. [DOI] [PubMed] [Google Scholar]
- Thomson JL, Brinster RL.. Glycogen content of preimplantation mouse embryos. Anat Rec 1966;155:97–102. [DOI] [PubMed] [Google Scholar]
- Tretter L, Patocs A, Chinopoulos C.. Succinate, an intermediate in metabolism, signal transduction, ROS, hypoxia, and tumorigenesis. Biochim Biophys Acta 2016;1857:1086–1101. [DOI] [PubMed] [Google Scholar]
- Uhde K, van Tol HTA, Stout TAE, Roelen BAJ.. Metabolomic profiles of bovine cumulus cells and cumulus-oocyte-complex-conditioned medium during maturation in vitro. Sci Rep 2018;8:9477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Blerkom J. Mitochondrial function in the human oocyte and embryo and their role in developmental competence. Mitochondrion 2011;11:797–813. [DOI] [PubMed] [Google Scholar]
- VandeVoort CA, Mtango NR, Midic U, Latham KE.. Disruptions in follicle cell functions in the ovaries of rhesus monkeys during summer. Physiol Genomics 2015;47:102–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virant-Klun I, Krijgsveld J.. Proteomes of animal oocytes: what can we learn for human oocytes in the in vitro fertilization programme? Biomed Res Int 2014;2014:856907–856911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virant-Klun I, Leicht S, Hughes C, Krijgsveld J.. Identification of maturation-specific proteins by single-cell proteomics of human oocytes. Mol Cell Proteomics 2016;15:2616–2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, Huwiler F, Fortes C, Grossmann J, Roschitzki B, Hu J, Naegeli H, Laczko E, Bleul U.. Analysis of the equine “cumulome” reveals major metabolic aberrations after maturation in vitro. BMC Genomics 2019;20:588–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, Monthoux C, Fortes C, Grossmann J, Roschitzki B, Meili T, Riond B, Hofmann-Lehmann R, Naegeli H, Bleul U.. The bovine cumulus proteome is influenced by maturation condition and maturational competence of the oocyte. Sci Rep 2020;10:9880–9815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Chang HM, Yi Y, Li H, Leung PCK.. TGF-beta1 promotes hyaluronan synthesis by upregulating hyaluronan synthase 2 expression in human granulosa-lutein cells. Cell Signal 2019;63:109392. [DOI] [PubMed] [Google Scholar]
- Wang Q, Frolova AI, Purcell S, Adastra K, Schoeller E, Chi MM, Schedl T, Moley KH.. Mitochondrial dysfunction and apoptosis in cumulus cells of type I diabetic mice. PLoS One 2010;5:e15901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wathlet S, Adriaenssens T, Segers I, Verheyen G, Janssens R, Coucke W, Devroey P, Smitz J.. New candidate genes to predict pregnancy outcome in single embryo transfer cycles when using cumulus cell gene expression. Fertil Steril 2012;98:432–439.e1–4. [DOI] [PubMed] [Google Scholar]
- Wathlet S, Adriaenssens T, Segers I, Verheyen G, Van de Velde H, Coucke W, El RR, Devroey P, Smitz J.. Cumulus cell gene expression predicts better cleavage-stage embryo or blastocyst development and pregnancy for ICSI patients. Hum Reprod 2011;26:1035–1051. [DOI] [PubMed] [Google Scholar]
- Wathlet S, Adriaenssens T, Segers I, Verheyen G, Van Landuyt L, Coucke W, Devroey P, Smitz J.. Pregnancy prediction in single embryo transfer cycles after ICSI using QPCR: validation in oocytes from the same cohort. PLoS One 2013;8:e54226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiśniewski JR, Zougman A, Nagaraj N, Mann M.. Universal sample preparation method for proteome analysis. Nat Methods 2009;6:359–362. [DOI] [PubMed] [Google Scholar]
- Wyse BA, Fuchs Weizman N, Kadish S, Balakier H, Sangaralingam M, Librach CL.. Transcriptomics of cumulus cells—a window into oocyte maturation in humans. J Ovarian Res 2020;13:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurowitzky YG, Milman LS.. Changes in enzyme activity of glycogen and hexose metabolism during oocyte maturation in a teleost, Misgurnus fossilis L. Wilhelm Roux Arch Entwickl Mech Org 1972;171:48–54. [DOI] [PubMed] [Google Scholar]
- Zhuo L, Kimata K.. Cumulus oophorus extracellular matrix: its construction and regulation. Cell Struct Funct 2001;26:189–196. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All proteomic data are available in PRIDE-Archive (EMBL-EBI) with the dataset identifier PXD033551 (https://www.ebi.ac.uk/pride/archive/).