Abstract
DEV is an obligatory lytic Pseudomonas phage of the N4-like genus, recently reclassified as Schitoviridae. The DEV genome encodes 91 ORFs, including a 3398 amino acid virion-associated RNA polymerase (vRNAP). Here, we describe the complete architecture of DEV, determined using a combination of cryo-electron microscopy localized reconstruction, biochemical methods, and genetic knockouts. We built de novo structures of all capsid factors and tail components involved in host attachment. We demonstrate that DEV long tail fibers are essential for infection of Pseudomonas aeruginosa but dispensable for infecting mutants with a truncated lipopolysaccharide devoid of the O-antigen. We determine that DEV vRNAP is part of a three-gene operon conserved in 191 Schitoviridae genomes. We propose these three proteins are ejected into the host to form a genome ejection motor spanning the cell envelope. We posit that the design principles of the DEV ejection apparatus are conserved in all Schitoviridae.
Subject terms: Bacteriophages, Cryoelectron microscopy, Phage biology, Molecular biology
Lokareddy et al. determine the complete structure of DEV, a lytic virus that infects Pseudomonas aeruginosa, using a combination of cryo-electron microscopy, biochemical methods, and genetic knockouts. They propose that the virion-associated RNA polymerase may be part of a genome ejection motor.
Introduction
The Escherichia coli phage N4 and the growing list of N4-like bacteriophages represent some of biology’s most understudied bacterial viruses. Long classified as Podoviridae for the relatively small size of the tail apparatus (based on low-resolution negative staining electron microscopy analysis), N4-like phages are genetically and structurally profoundly different from classical Podoviridae. In 2020, a large-scale bioinformatics analysis revealed 115 N4-like viruses, referred to as Schitoviridae, after Gian Carlo Schito, the scientist who first isolated the Escherichia phage N4 in 1966 from sewers in Genoa1. Schitoviridae have grown to include eight subfamilies and numerous new genera. The number of N4-like phages characterized in recent years continues to grow, from just 33 members in 2015 to 115 viruses in 20201, with many more reported in the last three years (e.g., phage AM.P22, PL143, VL14, ΦImVa-15). The genomic complexity of Schitoviridae is surprisingly vast and consistent with a genome size of ~75 kb, nearly twice that of most Podoviridae like P22 or T7, that encode about 90 ORFs, including three genes encoding two RNA polymerases, one of which is a massive virion-associated RNA polymerase (vRNAP) unique to this family of bacterial viruses.
Phage N4 is the best-studied member of the family Schitoviridae. A medium-resolution asymmetric reconstruction of the N4 mature virion elucidated the basic organization of this virus6 that has a ~700 Å icosahedral head connected to a short, non-contractile tail comprising just ten structural gene products (gp). The phage encodes three distinct RNAPs that regulate the temporal expression of its genome7. The massive 3500 residue vRNAP8,9 is ejected into the host upon infection7. This enzyme is responsible for an early burst of RNA synthesis observed immediately after N4 infection, even after inhibiting the host RNA pol10. N4 vRNAP initiates transcription of early genes gp1 and gp2 encoding cofactors for the heterodimeric polymerase N4 RNAPII (gp15:gp16) required to transcribe middle genes7. Among these transcripts, N4SSB redirects the host RNA pol to late promoters, mediating the expression of late genes involved in virion assembly, DNA replication, packaging, and host lysis. Interestingly, N4 vRNAP transcribes efficiently only denatured N4 DNA in vitro but is inactive on native N4 DNA, suggesting the transcription of early promoters requires host DNA gyrases to introduce negative supercoils into the phage genome. In stark contrast to the N4 transcriptional program that has been studied in depth7, many fundamental aspects of N4-like phage biology, such as capsid assembly, tail morphogenesis, and genome ejection/packaging, are poorly understood. A scaffolding protein has not been identified in the N4 genome, and it is also unclear if N4 is a cos or pac packager11, given that the virus packaging strategy is entirely unknown. N4 putative large (TerL) and small (TerS) terminase subunits were identified as gp68 and gp69, respectively, using comparative genomics analysis12.
This study focuses on the Pseudomonas phage DEV, an obligatory lytic phage of the N4-like genus. DEV is part of the CKϕ4 phage cocktail that eradicates P. aeruginosa infections in Galleria mellonella (wax moth) larvae and vertebrate models13,14. The DEV genome ( ~ 72.7 kb) encodes 91 ORFs, including a 3,398 amino acid vRNAP. DEV is slightly larger than N4 ( ~ 70.2 kb), which encodes 72 ORFs. DEV and N4 similarity is limited to 30% of phage proteomes, and even essential DEV proteins like the portal or tail fibers are undetectable using conventional bioinformatic analysis. DEV receptor has not been identified yet. Most Pseudomonas phages studied so far exploit either the lipopolysaccharide (LPS) or the Type IV pilus (T4P) as receptors for adsorption15. DEV does not require T4P to infect bacteria, and LPS’s role in infection is unclear16. The phage cannot adsorb PAO1 mutants lacking the O-antigen polymerase Wzy, which accumulate a lipooligosaccharide (LOS) decorated with a single O-antigen repeat (C + 1 LOS). On the contrary, loss-of-function galU, algC, or wapH mutants, which produce LOS species with a truncated core, are susceptible to DEV infection15,16.
Leveraging the power of cryogenic Electron Microscopy (cryo-EM), single particle analysis (SPA), and localized reconstruction17, in this study, we decipher building blocks and provide a complete annotation of all structural open reading frames (ORFs) conserved in the Pseudomonas phage DEV. The structural, biochemical, and functional results presented here elucidate fundamental aspects of DEV biology with general applicability to Schitoviridae and shed light on the existence of a genome ejection motor formed by ejection proteins after expulsion into the bacterial cell envelope.
Results
Overview of DEV
We purified the Pseudomonas phage vB_PaeP_DEV (Taxonomy ID: 2034344), abbreviated as DEV13, from PAO1 cells using cesium chloride ultracentrifugation18. SDS-PAGE of proteins extracted from infectious DEV virions revealed a band pattern (Supplementary Fig. 1A) very similar to that of LIT1 Litunavirus19, whose genome is 93.4% identical to DEV’s over 98% of its length. DEV virions were analyzed by nano-Liquid Chromatography High Resolution Mass Spectrometry (n-LC HRMS) to identify DEV virion proteins. We found that nine out of 14 DEV proteins are orthologues of LIT1 virion proteins, including the giant virion-encapsidated vRNAP (e.g., gp71)1 (Supplementary Table S1). The same preparation used for n-LC HRMS yielded infectious virions partially contaminated with cellular debris (Supplementary Fig. 1B) that we used for extended data collection on a Titan Krios 300 kV transmission electron microscope equipped with a Gatan K3 detector (Table 1). 2D classification found that the specimen contained two populations of ~8000 and ~16,000 particles, which we will refer to as half-filled (HF) and fully-filled (FF), respectively (Supplementary Fig. 1C). Both HF and FF capsids were mature and fully angular with a small tail apparatus (Supplementary Fig. 1D, E). However, HF particles lacked bulk DNA inside the capsid, whereas FF virions were filled with DNA. We computed icosahedral reconstructions for either species by applying I3 symmetry and identified the unique 5-fold vertex containing the tail region using localized reconstruction and C12 averaging of the tail region. We also applied C5 symmetry at the rotation center of the unique penton to obtain a high-resolution reconstruction of the proteins forming the capsid shell. A C5-C12 aligned asymmetric virion map was then used as a reference to generate asymmetric (C1) reconstructions of FF virions (Fig. 1A) and HF particles (Fig. 1B) using a tight mask.
Table 1.
Map and model refinement statistics
| Data Collection Statistics | |||||
|---|---|---|---|---|---|
| Specimen | Pseudomonas Phage DEV Virion | DEV gp72:gp73 Ejection Proteins | |||
| Facility/Microscope | NCEF / Titan Krios | SLAC-Stanford / Titan Krios | |||
| Detector | Gatan K3 | Falcon 4 | |||
| Imaging Software | SerialEM | EPU | |||
| Magnification | 81,000 x | 150,000 x | |||
| Voltage (kV) | 300 | 300 | |||
| Exposure (e-/Å2) | 50 | 50 | |||
| Exposure Time (sec) | 3.2 | 12.9 | |||
| Defocus range/step (µm) | -0.8 to -1.6 (0.2 increments) | -0.8 to -2.2 (0.2 increments) | |||
| Pixel size (Å) | 1.12 (0.56) | 0.56 (1.12) | |||
| Total movies (frames/movie) | 17,245 (40) | 4897 (40) | |||
| Total dose (e-/Å2) | 50 | 50 | |||
| Refinement Statistics | |||||
|---|---|---|---|---|---|
| Entry | Major Capsid protein | Portal Protein/ HT-adapter/ tail tube complex |
gp72 pre-ejection conformation |
gp53 Long Tail Fiber N-term Barrel |
gp72:gp73 post-ejection conformation |
| PDB / EMDB entry |
9BGN/ EMD-44518 |
9BGM/ EMD-44517 |
9BGO/ EMD-44519 |
9COD/ EMD-45776 |
8VXQ/ EMD-43629 |
| Initial particles number | 109,000 | 310,000 | |||
| Final particles number | 19,000 | 16,000 | 16,000 | 8,000 | 61,000 |
| Map Resolution (Å) at FSC 0.143 | 3.3 | 3.1 | 4.0 | 4.7 | 3.15 |
| Map Symmetry | C5 | C12 | C12 | C15 | C9 |
| Initial Model | de novo | de novo | de novo | Alphafold | de novo |
| Chains / Residues | 9 / 3,591 | 36 / 13,212 | 12 / 4,740 | 15 / 4,740 | 18 / 4,140 |
| Model-to-Map Correlation Coefficient (CC)a | 0.89 | 0.89 | 0.70 | 0.70 | 0.78 |
| MolProbity / Clash | 1.7 / 5.1 | 2.0 / 10.9 | 2.0 / 13.2 | 2.0 / 13.2 | 1.6 / 3.2 |
|
R.M.S. deviations Bond Length (Å) / Angles (°) |
0.002 (1) / 0.6 (15) |
0.007 (0) / 0.7 (46) |
0.002(0) / 0.6(0) |
0.002(0) / 0.6(0) |
0.004(3) / 0.7(23) |
| Rotamer outliers (%) | 0.1 | 0.9 | 0.0 | 0.0 | 0.4 |
| Ramachandran (%) Fav / Allow / Outlier | 91.5 / 8.4 / 0.1 | 92.2 / 7.6 / 0.2 | 94.6 / 5.4 / 0.0 | 94.6 / 5.4 / 0.0 | 93.2 / 6.7 / 0.1 |
aThe model-to-map correlation coefficient CC is calculated in the map region around the model.
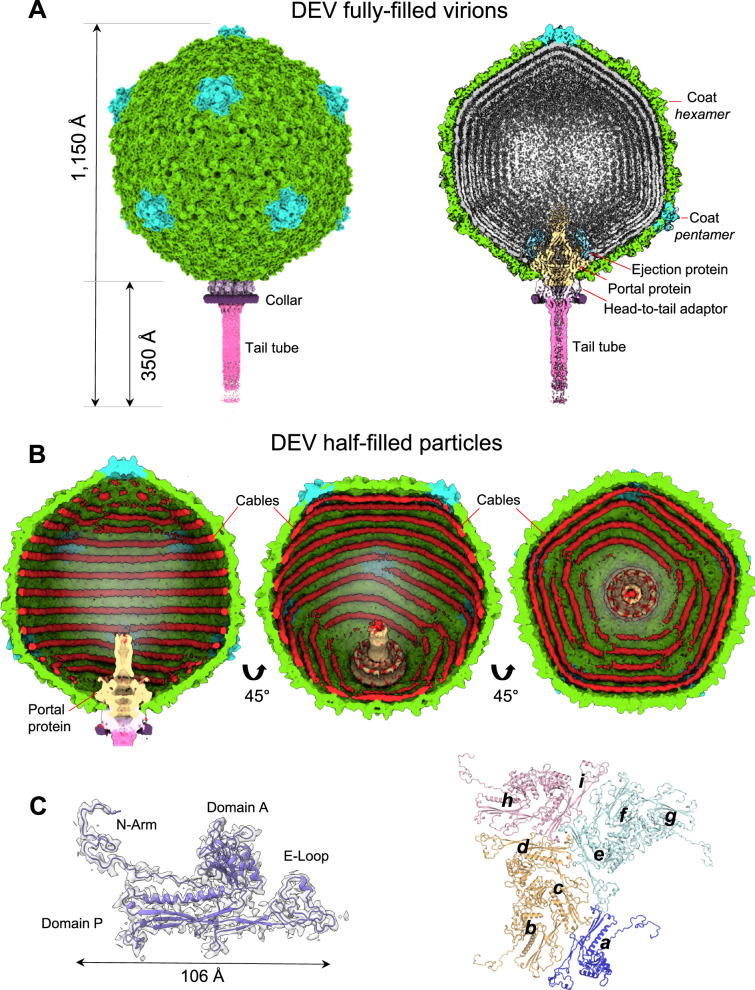
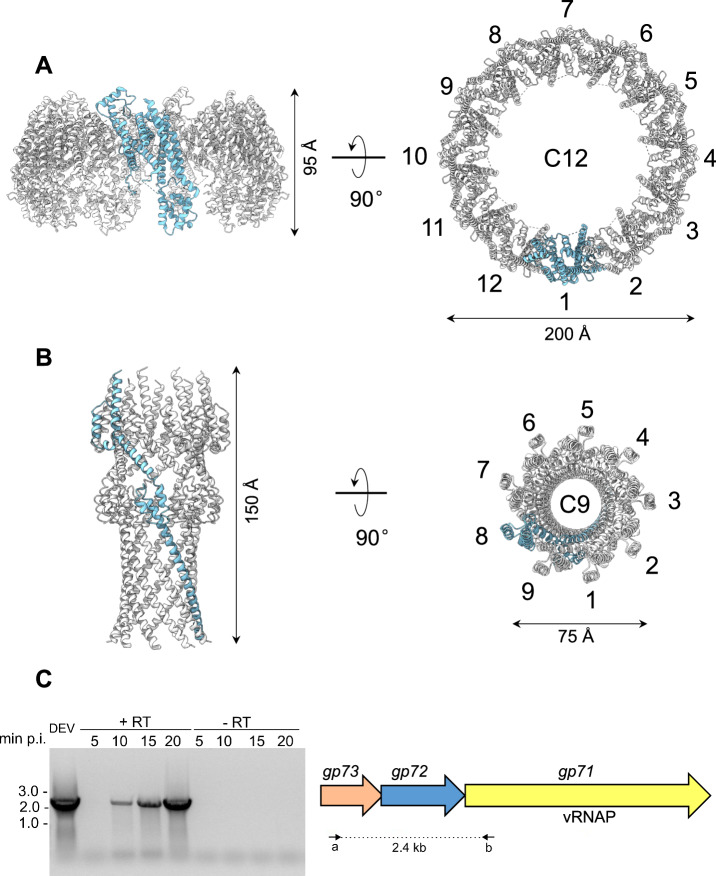
Fig. 1. Cryo-EM analysis of the Pseudomonas phage DEV.
A Asymmetric cryo-EM reconstruction of DEV FF virion in a side (left) and cutout (right) view. The T = 9 icosahedral shell is colored light green (hexamers) and cyan (pentons). B Asymmetric cryo-EM reconstructions of DEV HF particle. From left to right, three cutout views of the capsid are shown rotated in 45 Å increments. The cable density assigned to dsDNA is colored red. C (Left) DEV coat protein gp77 tertiary structure overlaid to a 3.3 Å C5-averaged localized reconstruction of the mature head map contoured at 5σ above background. (Right) Overview of DEV T = 9 icosahedral asymmetric unit comprising nine coat proteins labeled (a-i).
Phage DEV is ~1,200 Å in length, consisting of an ~800 Å icosahedral capsid with triangulation number T = 9 and a thin ~400 Å tail (Fig. 1A). The DEV major coat protein was built de novo in a 3.3 Å C5 map (Fig. 1C, Supplementary Fig. 2A) and identified as the gene product of ORF77 (gp77, 399 aa) (Supplementary Fig. 3). The coat protein, refined to a model-to-map Correlation Coefficient (CC) of 0.89 (Table 1), adopts a classic HK97 fold with an elongated N-terminal arm (res. 1-46) present in the mature virion. DEV capsid is built by 535 copies of the major coat protein (T = 9) (Fig. 1C), with one penton replaced by the dodecameric portal protein. Double-stranded DNA (dsDNA) fills the interior of FF virions, forming an 8-layer core (Fig. 1A). However, a medium-resolution map displayed at low contours revealed bulk density inside the capsid, looming over the portal vertex. Unfortunately, this density did not align with the capsid or portal axis and could not be improved. DEV FF virions also displayed a helical density surrounding the portal perimeter, contacting the outer two layers of dsDNA.
DEV half-filled particles contain DNA cables
DEV reconstructions obtained from HF particles and FF virions were identical from the outside, with superimposable coat protein structures. However, HF particles had peculiar rings of density laying against the capsid interior, which we termed cables, but lacked bulk dsDNA inside the capsid and the protein density surrounding the portal perimeter (Fig. 1B, Supplementary Fig. 4A). DEV cables most likely represent dsDNA that we estimated to account for ~6-6.2 kbp, equivalent to less than 10% of the DEV genome. Similar cable-like structures lining the interior of a capsid were observed in a recent cryo-EM reconstruction of the E. coli phage SU1020 capsid. SU10 is a Podoviridae of the Kuravirus genus characterized by a short non-contractile tail, a prolate head, and a large genome of 77,327 base pairs21. Cable-like structures inside the SU10 prolate capsid are concentric to the portal protein and were hypothesized to be remnants of the genome left in the capsid after genome ejection20. In our reconstructions, DNA cables are not readily distinguishable in DEV FF virions due to the averaging effect of DNA. However, an overlay of the HF and FF reconstructions reveals that cables of equal size and shape also exist in the mature virion. DEV cables appear concentric in the C1 reconstruction (Supplementary Fig. 4A), which is nonetheless biased by the 5-fold symmetry used during icosahedral reconstruction. To eliminate this bias, we calculated a focused reconstruction using a mask covering one of the five angles of the unique five-fold (Supplementary Fig. 4B). This revealed two different cable arrangements inside the mask: in one case, two cables run along the sides of a capsid hexon with a spacing of ~58 Å; in the other arrangement, one cable ends before encountering a penton, generating an asymmetric Y-shaped pattern (Supplementary Fig. 4C). Unfortunately, due to the low number of HF particles ( ~8,000), we were not able to generate a high-resolution map and follow the complete trajectory of cables that, in the general reconstruction, remain biased by 5-fold symmetry, except in the region covered by the mask. In addition, HF particles purified by sucrose gradient had an absorbance 260/A280 ratio of ~1.04, suggesting the presence of nucleic acids but did not reveal a predominant low molecular weight DNA/RNA species by agarose gel electrophoresis. This suggests the cables may comprise spurious DNA fragments of different sizes, arranged in a defined pattern inside the virion due to electrostatic contacts with the mildly basic capsid protein interior (Supplementary Fig. 4C).
The architecture of phage DEV neck and tail
The C12 localized reconstructions of the unique vertex for FF virions and HF particles yielded 3.1 Å and 3.5 Å resolution (Supplementary Fig. 2A). We used these maps to build de novo models of the dodecameric portal protein gp80 (726 aa), twelve copies of the head-to-tail adapter gp83 (244 aa), and twelve copies of the tail tube gp75 (321 aa) (Fig. 2A and Supplementary Fig. 3). In total, we built 13,212 amino acids, distributed in 36 chains (Fig. 2A), which were real-space refined to a final Correlation Coefficient (CC) of 0.89, indicating an excellent model-to-map fit (Table 1). The tail complexes reconstructed from HF particles (Supplementary Fig. 5A) and FF virions (Fig. 2A) are structurally identical, except that the former lacks bulk DNA inside the capsid and the helical density surrounding the portal perimeter, described below.
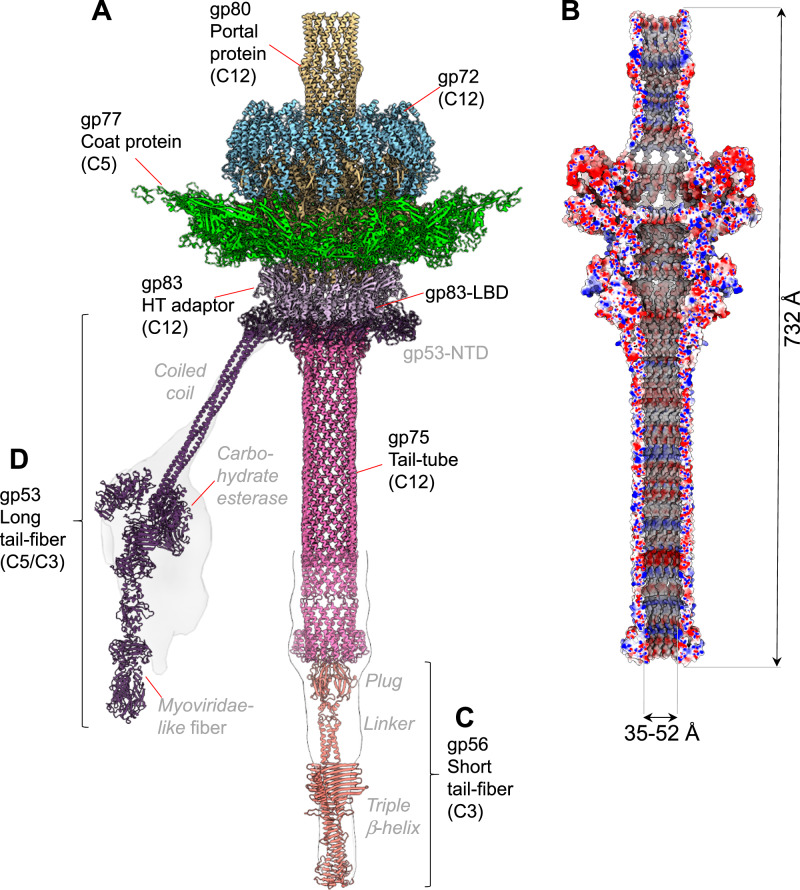
Fig. 2. DEV tail apparatus.
A Composite ribbon diagram of DEV tail reconstructed from FF virions. Tail factors identified de novo in the C12 localized reconstruction include the portal protein gp80 (yellow), the ejection protein gp72 (blue), the HT-adapter gp83 (light purple), and the tail tube gp75 (magenta). B Cross section of an electrostatic surface representation of the DEV tail channel. Red, blue, and white represent negative, positive, and neutral charges near the surface. C–D AlphaFold models for the short-tail fiber gp56 and long-tail fiber gp53 overlaid to low-resolution localized reconstructions shown as semitransparent surfaces. Individual tail factors are color-coded, as in panel (A).
DEV portal protein, gp80 adopts a conserved portal fold22 consisting of five regions: the wing (res. 160-237 and 272-351), clip (res. 408-450), stem (res. 376-407 and 451-476), crown (res. 564-614), and C-terminal barrel (res. 620-721) (Supplementary Fig. 3). The inner diameter of the portal ranges between ~26 Å (T490) and ~64 Å (E566) at the bottom of the crown domain, while the narrowest diameter of the inner barrel is ~28 Å. A DALI23 search identified the related bacteriophage SPP1 portal protein (PDB:2JES) as the most similar candidate to the DEV portal protomer. The RMSD between these two proteins is 6.7 Å, and the sequence identity is only 12%. A ring of dsDNA surrounds the portal perimeter24,25, making direct contact with the N-arm of the portal. In the FF virion reconstruction, the portal wing interacts with dsDNA through an additional factor we identified de novo as gp72 (Fig. 2A) (see next section). However, the RMSD between the portal in FF versus HF particles that lack density for gp72 is less than 0.05 Å, suggesting the latter does not affect the portal structure. Unexpectedly, DEV portal barrel26 was well-resolved in FF virions (Fig. 2A) and HF particles (Supplementary Fig. 5A), indicating the barrel is stably folded as a dodecamer without DNA. The interface between two neighboring barrel helices (res. 620-721) is stabilized by one salt bridge, 6 H-bonds, and 180 non-bonded contacts over 78 residues, significantly more bonds than in phage Sf627 and P2228 where the portal barrel is folded only in the presence of DNA24 but collapses without surrounding DNA.
We also identified and de novo built residues 2 to 244 of DEV head-to-tail adapter gp83 (HT-adapter) (Fig. 2A and Supplementary Fig. 3). DEV gp83 consists of an N-terminal four-helix bundle extending into a C-terminal arm that inserts at the binding interface between two DEV portal protomers29,30. DEV gp83 helical bundle contains a 101 amino acid lectin-binding domain (LBD) (res. 56-156) insertion between α-helices H2 and H3 (Supplementary Fig. 3), facing outward in the tail apparatus (Fig. 2A). Gp83 helical body extends the portal protein, providing an attachment point for the tail tube. The α-helical core and C-terminal arm are structurally similar to P22 HT-adapter gp4 (RMSD ~ 3.1 Å)28,30, although the sequence identity is only 10% (with a coverage of 106 residues of 142 total). Gp83 LBD, instead, is unique to DEV.
DEV non-contractile tail tube is 12-fold symmetric
DEV tail tube, the gene product of ORF75 (gp75, 321 aa), is the last tail factor we built in the C12 localized reconstruction map (Fig. 2A, Supplementary Fig. 3). DEV tail tube has no structural, sequence or even topological similarity to the tail tube of long-tailed Siphoviridae and Myoviridae phages31: it looks like an upside-down version of the portal protein barrel. It attaches to the HT-adapter and remains dodecameric without the typical 12:6 symmetry reduction seen in Podoviridae27,28. In the 3.1 Å C12 map (Supplementary Fig. 2A), the tail tube can only be visualized between residues 2–157, while the remaining residues 158–321 were modeled in a lower-resolution C1 map using an AlphaFold32 model (Fig. 2A). DEV gp75 is dodecameric with an outer diameter of 63 Å, an inner diameter of 36 Å, and approximately 330 Å in length (Fig. 2B, Supplementary Fig. 3). A short helix-turn-helix domain (res. 34–46) at the N-terminus of the tail tube is the only domain that contacts the HT-adapter. The total length of the DEV tail machine from the top of the barrel to the bottom of the tail tube is ~732 Å, with an internal diameter between ~35–52 Å (Fig. 2B). A Coulombic electrostatic potential surface reveals that the DNA channel is mildly basic, especially in the barrel region and bottom part of the tail tube but contains no physical gate or constriction to prevent DNA from exiting the capsid. A low-resolution C1 density of the whole virion revealed density inside the tail, possibly dsDNA that is stronger in the top portion of the portal but tapers down toward the bottom (Supplementary Fig. 5B). At low contour, a density is visible at the end of the tail channel, like a plug (Fig. 2A). However, it was impossible to define the molecular identity of this DEV gene product from the electron density. The lack of physical constriction and the continuous DNA density inside the tail tube suggest that the DEV tail is sealed by a plug at the distal tip relative to the capsid. We hypothesize that this plug could be analogous to the tail needle of podophages P2228 and Sf627, which seals the tail after genome packaging, stabilizing encapsidated DNA33.
DEV contains two flexible fibers: the long fiber gp53 and the short fiber gp56
Both low-resolution C1 density (Supplementary Fig. 5B) and high-resolution C12 localized reconstruction of DEV FF (Fig. 2A) had minimal density for two components of the DEV tail: the fiber emanating from the tail neck, known as the appendage in N46, and a putative plug at the tail tube tip. Both structural components of the DEV tail can be seen in aligned micrographs and 2D-class averages (Supplementary Figs. 6A, B) but are smeared in 3D reconstructions. The appendage density was especially weak and could not be aligned in a C1 map, underscoring its flexibility. Coincidentally, two ORFs were left unassigned in the DEV genome: ORF53, which encodes the 1090 aa gp53 protein, and ORF56, which encodes a smaller, 429 aa protein (Supplementary Fig. 3), both identified in the DEV mature virion using MS (Supplementary Table S1). We generated AlphaFold234 models for both ORFs that are predicted to fold into trimeric fibers. We named gp53 the long fiber and gp56 the short fiber. To validate the existence of these fibers in DEV FF virions, we computed low-resolution focused reconstructions of the DEV tail using masks of different shapes and sizes, which revealed smeared density around the neck and at the tail tip (Fig. 2C, D, Supplementary Fig. 6C, D). The trimeric short fiber gp56 was docked at the tail tube tip, sealing the dodecameric tube channel (Fig. 2C, Supplementary Figs. 6C, D). Here, a 12:3 symmetry mismatch is plausible, as the predicted gp56 N-terminal knob fits snugly inside the tail tube. This interaction resembles the Podoviridae tail needle35,36 that seals the tail hub channel in P2228 and Sf627, preventing DNA leakage.
DEV long-tail fiber gp53 assembles to the tail via a 15:12 symmetry mismatch
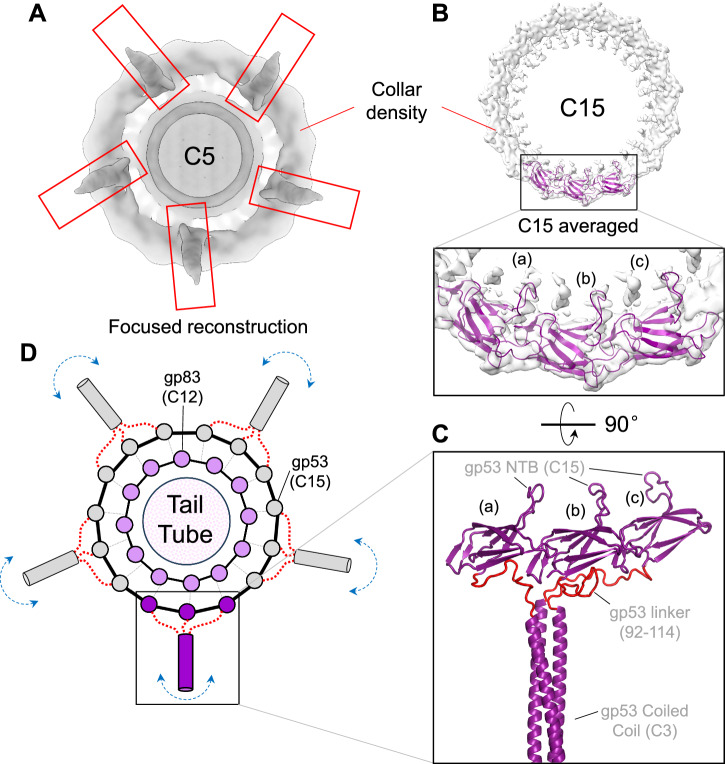
Deciphering how the long-tail fiber gp53 attaches to the dodecameric tail tube was challenging. Gp53 is DEV’s equivalent to phage N4 appendages that follow the tail C12 symmetry in this phage6. However, several lines of evidence suggest that DEV is not likely to contain 12 copies of gp53. First, the C12 localized map used to build the portal, HT-adapter, and tail tube had a smeared, collar-shaped density, ~174 Å in diameter and ~27 Å thick, bonding the HT-adapter (Fig. 1A). This density, visible at the same contour as the HT-adapter, is featureless in our C12 map, suggesting the gp53-collar is either misaligned relative to the tail in the reconstruction or averaged out by applying incorrect rotational symmetry. Second, a localized reconstruction of DEV putative appendages revealed five elongated densities emanating outward from the tail (Fig. 3A, Supplementary Fig. 6C). Third, AlphaFold234 predicts gp53 to fold into a trimeric fiber containing an N-terminal β-barrel (NTB, res. 1-91), flexibly connected to a trimeric coiled-coil domain (res. 115-250), a carbohydrate esterase-like domain (res. 265-656) and a Myoviridae-like fiber (res. 694-1090) (Supplementary Fig. 6C). We then hypothesized that the DEV gp53-collar has higher symmetry than the dodecameric tail. To test this idea, we expanded C12-aligned particles along the Z-axis to C24 and C30, followed by an asymmetric search without sampling with a donut-like tight mask. The 3D classification was not well resolved for C12 expanded particles but sufficient to identify 15 repeated density blobs. One class from the C30 symmetry expansion consisting of 8,000 particles after removing duplicates could be refined, yielding maps of the C15 symmetrized and asymmetric (C1) collar to 4.7 Å (Fig. 3B, Table 1) and 6 Å resolution (Supplementary Fig. 7), respectively.
Fig. 3. Topology and composition of DEV collar.
A A low-resolution C5 map of the DEV mature virion visualized from the bottom of the tail apparatus shows density for five appendages protruding from the collar and assigned to the tail fibers gp53. B A C15 symmetrized density of the DEV collar is visualized at a high contour (5σ). Fifteen copies of gp53-NTB were real-space refined against the C15 density. Only three NTB subunits (labeled ‘a-c’) are shown in the zoom-in panel. C A 90° rotated view of the three gp53-NTBs is shown in panel (B), which also includes an AlphaFold model of the gp53 flexible linker (res. 92-114), colored red, and the first part of the coiled-coil domain. D Schematic diagram of the proposed 12:15:pseudo-5 symmetry mismatch between DEV dodecameric HT-adapter gp83 (light pink circles) and 15 copies of gp53 NTB (gray/purple circles). A flexible linker in gp53 spanning residues 92–114 (shown as a red dotted line) allows three neighboring gp53 protomers to assemble into five trimeric long tail fibers that project outward. The boxed gp53 trimer at the bottom of panel (D) is shown in the same orientation as the AlphaFold model in panel (C).
To model the long-tail fiber, we docked 15 copies of gp53 NTB (Supplementary Fig. 3) into the C15 and C1 collar densities and real space refined them to a final CC = 0.7 and 0.82, respectively (Fig. 3B). The fitting is convincing, with 15 β-barrels (res. 1–91) making head-to-tail contacts and encircling the dodecameric HT-adapter. Notably, gp53 trimeric coiled-coil (res. 115–250) fits remarkably well with the elongated density emanating from the DEV collar (Fig. 3A), which positions the flexible esterase-like domain and Myoviridae-like fiber in the smeared density pointing away from the tail (Fig. 2D, Supplementary Fig. 6C). Together, these data are consistent with an assembly model whereby 15 copies of gp53-NTB form a collar around the dodecameric HT-adapter, while the rest of gp53 trimerizes via the coiled-coil domain to form five trimeric appendages (Fig. 3C, D). The proposed 12:15:pseudo-5 symmetry mismatch is topologically possible because of a 22-residue flexible linker in gp53 (res. 92-114) that connects NTBs in the C15 collar to five trimeric long fibers (pseudo-C5). Unfortunately, due to the weak and smeared density emanating from the DEV collar (Supplementary Fig. 6C), it is currently impossible to align DEV long tail fibers in a C1 map. Thus, our model remains hypothetical. However, a 12:15:pseudo-5 symmetry mismatch similar to that proposed for DEV (Figs. 3C, D) was recently observed in the high-resolution cryo-EM reconstruction of the podophage GP437. In this phage, which infects Ralstonia solanacearum, the dodecameric HT-adapter is encircled by 15 copies of the ‘tail-fiber adapter’, a 93 amino acids immunoglobulin-like protein, topologically superimposable to DEV gp53 NTBs. Notably, the GP4 tail-fiber adapter forms a 12:15 symmetry mismatch with the phage tail. It also mediates the attachment to three sets of fibers, including two 5-fold symmetric fibers I and II, generating a global 12:15:5 symmetry mismatch similar to that proposed for the DEV tail (Fig. 3D).
The long fiber gp53 is necessary but not sufficient to infect P. aeruginosa
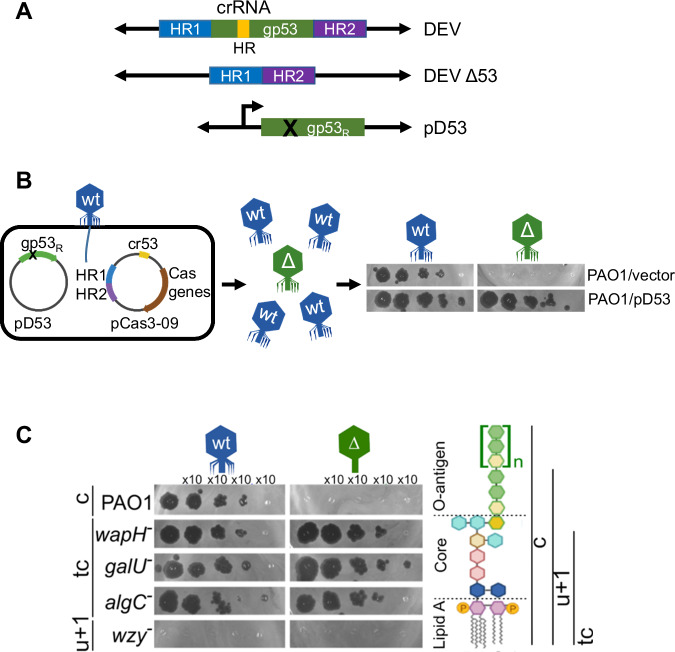
To characterize the function of DEV gp53 long-tail fiber and discern its role in phage adsorption, we generated a DEV gp53 deletion mutant (DEV Δ53; Fig. 4A). To obtain the mutant phage, we exploited a minimal Cascade-Cas3 system (Type I-C), generating large deletions whose boundaries can be specified by a homology-directed repair (HDR) template38. DEV genome editing was performed in P. aeruginosa PAO1 strain containing the plasmids pCas3-09 and pD53 (editing strain; Fig. 4B and Supplementary Table S2). pCas3-09 is a pCas3cRh derivative expressing all components of the Cascade-Cas3 (Type I-C) system (e.g., cas3, cas5, cas8, and cas738) and a gp53-specific gRNA (cr-RNA53). pCas3-09 also carried the HDR template, e.g., two fused DNA fragments corresponding to the gp53 flanking regions in the DEV genome. As expected, DEV did not grow on PAO1 expressing the cr-RNA53 Cascade complex (Supplementary Fig. 8A). pD53 contained the gp53 gene with silent mutations in the region recognized by the cr-RNA53 (gp53R allele; Fig. 4A and B) that made pD53 resistant to cr-RNA53-targeted Cas3 digestion. Expression of gp53R from pD53 complemented the Δ53 mutation so that even phage genomes lacking the gp53 gene were packaged into complete virions. The editing strain was infected with DEV, and the lysate was plated on the permissive PAO1/pD53 strain. Almost all phages unable to reproduce without pD53 (Fig. 4B) contained the expected gp53 deletion in their genomes (Supplementary Fig. 8B). The DEV Δ53 mutant did not reproduce in PAO1 but surprisingly, it grew in algC, galU, and wapH mutant strains, which all make uncapped LOS species lacking the O-antigen16,39 (Fig. 4C). Thus, gp53 is essential for PAO1 infection in the presence of smooth-type, O-antigen capped LPS but dispensable if rough-type, uncapped LPS is produced. Both DEV and DEV Δ53 mutant did not grow in the wzy mutant (Fig. 4C), which accumulates an LPS form capped with a single O-antigen repeat16,39, suggesting that in the wzy mutant, no receptor is present/accessible to the phage.
Fig. 4. Role of DEV long fiber gp53 in host attachment.
A Structure of the gp53 locus in DEV and DEV Δ53 phages and the pD53 plasmid. crRNA HR, region targeted by cr-RNA53 expressed by pCas3-09; HR1 and HR2, homology regions cloned in pCas3-09 plasmid; gp53R, cr-RNA53 resistant gp53 allele cloned in pD53. B Outline of DEV mutagenesis. Infection with DEV of PAO1 carrying pCas3-09 and pD53 produces a genetically mixed phage progeny with wt (in blue) and Δ53 (in green) virions. Unlike gp53+ DEV, Δgp53 mutants grow on PAO1 carrying pD53 (pD53) but not on PAO1 containing the empty vector pGM931 (vector). Source data are provided as a Source Data file. C DEV Δgp53 growth on mutants with LPS defects. Serial dilutions (x 10) of DEV or Δgp53 were replicated on PAO1 and the indicated PAO1 mutants with defective LPS. On the right is the structure of PAO1 LPS (c, capped) and the LPS portions present in the mutant LPS variants. tc, truncated core; u + 1, uncapped LPS + one O-antigen repeat. Source data are provided as a Source Data file.
An ejection protein surrounds the DEV portal protein
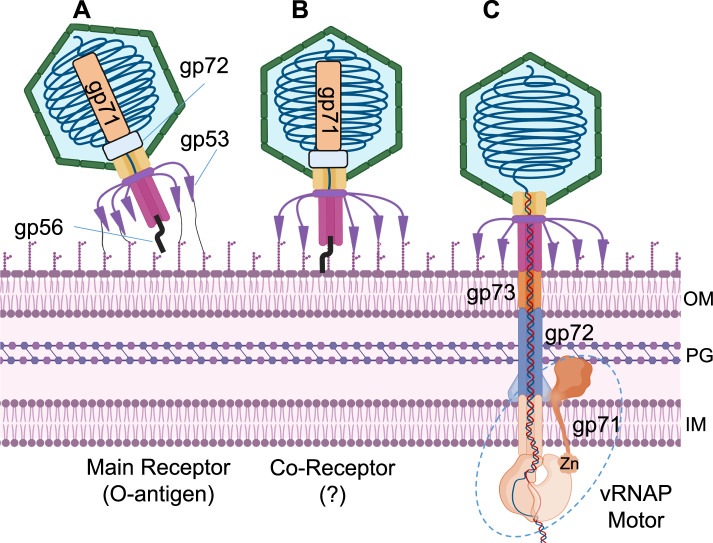
The initial localized reconstruction of the FF virion revealed tubular density surrounding the portal wings (Fig. 1A, right). To improve the unknown density, we computed a focused reconstruction using a tight mask, which only covers the helical region, and applied C12 rotational symmetry, yielding a 4.0 Å map. We built this density de novo and identified the protein surrounding the DEV portal as the gene product of ORF72 (gp72, 521 aa) (Fig. 5A). The modeled structure (res. 116-495) lacks residues 1–115 and 496–521, which are likely too flexible to be aligned. Applying 12-fold symmetry, we generated a dodecameric model of gp72, consisting of 4740 residues, which we real-space refined to a final CC = 0.70 (Table 1). Twelve copies of gp72 form a 200 Å-wide dodecameric ring, concentric to the portal protein. Bioinformatic analysis revealed that DEV gp72 has 7% identity and 15% similarity to phage T7 ejection protein gp15 that forms a periplasmic tunnel (PT) in the E. coli cell envelope after ejection40,41.
Fig. 5. Quaternary structures of DEV ejection protein gp72 pre- and post-ejection.
A The quaternary structure of DEV gp72 from FF virions determined in situ. Twelve gp72 subunits surround the portal protein, generating a ~ 200 Å wide ring. B Cryo-EM structure of the recombinant nonameric gp72 determined at 3.65 Å resolution in the post-ejection conformation. In panels (A and B) only one protomer is colored in cyan, whereas all other subunits are light gray. (C) DEV gp71, gp72, and gp73 genes are co-transcribed as an operon. (Left panel) Agarose gel electrophoresis of RT-PCR products. RNA samples extracted from PAO1 cultures at different time points post-infection (p.i.) with DEV (e.g., 5, 10, 15, 20 minutes) were reverse-transcribed ( + RT) or not (negative control, –RT) and used as templates for PCR amplification. Migration of MW (kb) markers is shown on the left. The assay was repeated three times with similar result. (Right panel) Schematic diagram of DEV ORFs encoding gp71, gp72, and gp73. Arrows represent the position of oligonucleotides used for amplification, yielding a 2.4 kb long amplification product. Source data are provided as a Source Data file.
The hallmark of ejection proteins is that they exist in a pre-ejection conformation in the virion, before genome ejection, and a post-ejection conformation inside the host cell envelope: we referred to these proteins as conformational gymnasts in a recent review42. Previous work on T7 ejection protein gp15 revealed that the recombinant protein expressed in bacteria43 adopts a post-ejection conformation, distinct from the metastable conformation seen in the mature virion before ejection (pre-ejection conformation)44. To test the hypothesis that DEV gp72 is an ejection protein, we cloned, purified, and solved a cryo-EM structure of the recombinant gp72 protein (Supplementary Fig. 9A–C). We found that recombinant gp72 assembles into a nonameric elongated channel, 80 Å in width, containing a 25 Å wide lumen large enough to fit dsDNA (Fig. 5B). Thus, like T7 ejection protein gp15, DEV gp72 adopts two drastically distinct conformations inside the virion (Fig. 5A) and when expressed recombinantly (Fig. 5B). This structural reorganization involves a tertiary structure refolding and a change in oligomerization stoichiometry from 12 to 9 subunits in DEV and 9 to 6 in T742, respectively. DEV gp72 and T7 gp15 have different sizes (e.g., 521 and 747 residues, respectively) and have limited sequence identity and similarity (14.6% and 28.4%, respectively, limited coverage due to the size difference). Despite differences in the primary, tertiary, and quaternary structures, both proteins are entirely α-helical and form a tube-like tunnel wide enough to accommodate dsDNA (Supplementary Fig. 10).
In phage T7 and many Podoviridae studied to date42, the ejection protein gp15 is neighbored by a smaller gene encoding the membrane protein gp14 and a much larger gene encoding gp16 (> 1300 residues), implicated in genome ejection45. T7 genes encoding gp14, gp15, and gp16 form an operon. We then asked if the two ORFs in DEV surrounding gp72, known as gp71 and gp73, also encode ejection proteins and whether the three proteins are expressed together as part of an operon42. RT-PCR of DEV-infected cells revealed that gp72 and the flanking gp71 and gp73 are expressed as a polycistronic mRNA ~10 minutes after DEV infection of PAO1 (Fig. 5C). Remarkably, DEV gp71 happens to be the giant vRNAP that in the related phage N4 is known to be injected in E. coli during infection7.
Structure of DEV ejection protein gp72:gp73 complex
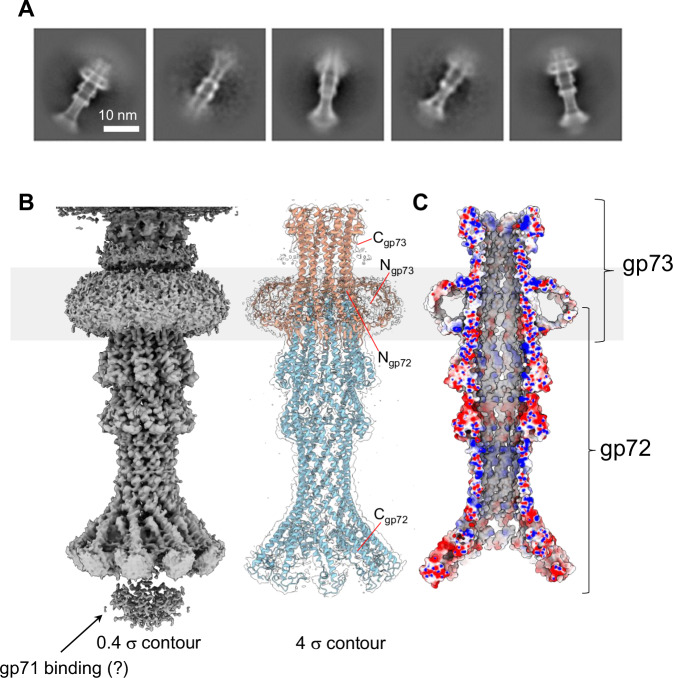
Phage ejection proteins form a channel in the bacterial cell envelope spanning the outer membrane (OM), periplasm, and inner membrane (IM) projecting into the host cytoplasm42. We have established that DEV gp72 resembles a PT, so we reasoned that gp73 may form an OM pore (OMP) like T7 gp14, which has pore-forming activity in lipid bilayers40, or P22 gp7 that partitions into lipid nanodiscs in vitro28. This hypothesis was bolstered by transmembrane prediction servers, MemBrain46 and DAS47, which predict a transmembrane α-helix spanning gp73 residues 77-91. In vitro, recombinant gp73 (M.W. 17.7 kDa) was completely insoluble but could be purified from an E. coli membrane fraction using sarcosine and n-dodecyl β-D-maltoside (DDM) (Supplementary Fig. 9A). To test if detergent-solubilized gp73 was active, we mixed it with gp72, which is water-soluble. Size exclusion chromatography (SEC) indicated that the two recombinant proteins form a complex (Supplementary Fig. 9A) that we successfully vitrified. A large dataset collected on a 300 kV Krios electron microscope allowed us to reconstruct the 3D structure of DEV gp72:gp73 using single particle analysis (Fig. 6A, B) at a resolution between 3.15–6 Å (Table 1, Supplementary Fig. 2C). The structure revealed a striking 9-fold symmetry that allowed us to dramatically improve the experimental density by applying C9 rotational symmetry. Overall, we built de novo residues 1–155 of gp73 and residues 25–331 of gp72, which were real-space refined to a final Correlation Coefficient (CC) of 0.78, indicating an excellent model-to-map fit (Table 1).
Fig. 6. DEV ejection proteins gp72 and gp73 form a tube-shaped complex.
A Representative 2D class averages of the gp72:gp73 complex. B 3D reconstruction of the gp72:gp73 complex visualized at low (left) and high (right) contours. The atomic models of gp73 and gp72 are overlaid to a semitransparent density calculated at 3.15 Å resolution. In gray is the putative position of the bacterium’s outer membrane. C The cross-section of an electrostatic surface representation of gp72:gp73 shows the lumen and surface charge inside the channel. Red, blue, and white represent negative, positive, and neutral charges near the surface.
The membrane protein gp73 forms a cap onto which gp72 inserts (Fig. 6B), generating a trumpet-shaped complex ~300 Å in length with an internal diameter of 35 Å (Fig. 6C). Gp73 and gp72 main chains run antiparallel, with the N-termini of the two proteins located in relative proximity and the C-termini displaced by more than 200 Å (Fig. 6B). Extensive intermolecular interactions between gp73 residues 64-93 and gp72 N-terminal helix (res. 25–54) stabilize the gp72:gp73 nonameric interface ( ~67,944 Å2), comprising 24 hydrogen bonds and 723 non-bonded interactions (Supplementary Table S4). Strikingly, the gp73 long α-helix spanning residues 77–137 makes up the inner channel and the region facing the bacterial outer membrane, which is very positively charged (Fig. 7A). We speculate that a positive charge may favor insertion in the negatively charged lipid A building the OM48. The soft density around the gp73 N-termini (Fig. 6B) likely comprises the disordered N-terminal residues 1–52, and DDM used for solubilization. Additional studies in lipid nanodiscs are required to determine the nature of the gp73 membrane-insertion domain. On the opposite tip, gp72 flowers like a trumpet to provide a binding domain for gp71. Notably, the gp73-bound structure of gp72 has an additional 20 residues visible at the N-terminus and 90 residues at the C-terminus relative to the isolated gp72 (e.g., the last residues visible in the electron density are D224 and E331 for gp72 alone and gp72:gp73 complex, respectively) (Supplementary Fig. 10). This suggests that gp73 binding stabilizes gp72 quaternary structure, which spans ~230 Å versus just 150 Å for the unbound conformation.
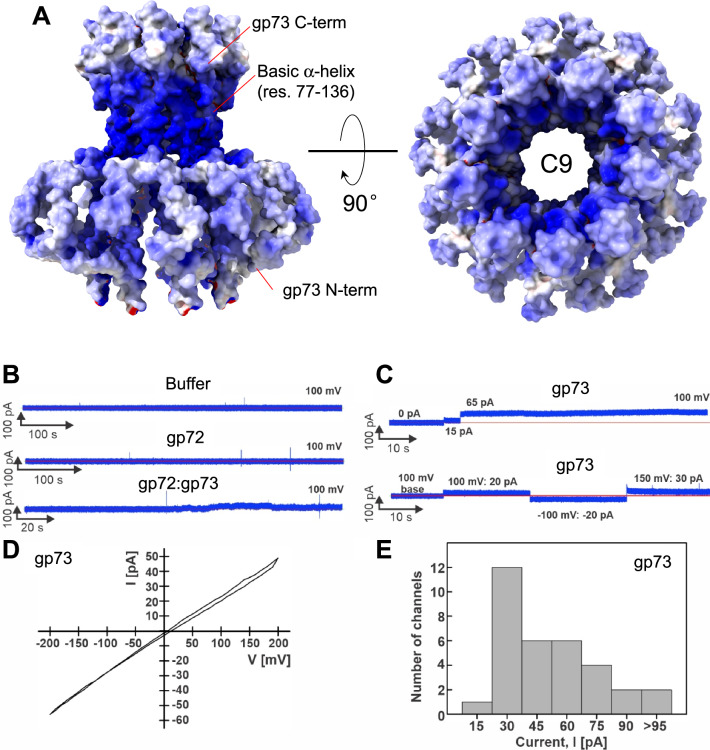
Fig. 7. Lipid bilayer experiments with purified DEV ejection protein gp73.
A The electrostatic surface representation of nonameric gp73 reveals a significant positive charge, mainly in the α-helical core. B,C Lipid bilayer experiments were performed at 100 mV applied potential in diphytanoylphosphotidylcholine (DPhPC) membranes bathed in 1 M KCl, 10 mM HEPES, pH 7.4 electrolyte. The protein samples were added to the grounded trans side of the cuvette, which had 100 µm SU-8 aperture. B Representative current traces. Top: 15 µl protein buffer in the cuvette. Six membranes were recorded with 1 – 15 µl of the protein buffer, and no activity of the buffer was observed. Middle: gp72 current trace. Seven membranes with up to 24 µg of gp72 in the cuvette were recorded, and no channel activity was observed. Bottom: gp72:gp73 complex. 19 membranes were recorded, and only one shown here had 10 – 20 pA fluctuations around the baseline when 10 µg of protein sample was in the cuvette. C Representative current traces of gp73. Top: Two insertions of gp73 (750 ng protein in the cuvette) with amplitudes of 15 pA, and 65 pA. Bottom: Continuous current trace of a single gp73 insertion (900 ng in the cuvette) at indicated voltages. D Current-voltage curve of one gp73 pore inserted in the DPhPC membrane at a voltage range of −200 to 200 mV. E Histogram of single-channel current amplitudes of gp73 at 100 mV. A total of 33 channels were observed with a mode current of 30 pA.
Lipid bilayer experiments reveal that DEV gp73 is a membrane-spanning pore protein
A phage ejectosome requires a continuous channel from the phage head to the bacterium cytoplasm. The cryo-EM structure of gp73 revealed a hollow nonameric channel with an internal diameter wide enough to accommodate DNA ( ~ 25 Å) and hydrophobic helices flowering outbound to gp72 and extensive basic residues, possibly involved in binding lipid A phosphate groups in the bacterial outer membrane (Fig. 7A). To examine the channel-forming properties of DEV ejection proteins gp72 and gp73, we performed lipid bilayer experiments as described previously40,43,49. Lipid bilayer measurements are a sensitive, well-established single-molecule method that measures ionic current across a lipid membrane bathed in a 1 M potassium chloride solution. Since the membrane is not permeable to ions, no current is generated when voltage is applied, and a baseline current is observed. To exclude buffer components or contaminations in the system that disrupt the membrane or form pores, a negative control is essential, namely, the buffer used to solubilize gp73, which contains 0.3 mM DDM. This buffer did not change the baseline current in several lipid bilayer experiments (Fig. 7B, top panel; Supplementary Table S5), demonstrating that the detergent alone does not perturb the lipid membrane. Similar to the buffer control, purified gp72 (Fig. 7B, middle panel) did not show any channel activity in our experiments. When 10 µg of gp72:gp73 complex was added, we observed only one membrane with current fluctuations between 10 and 20 pA in 19 experiments (Fig. 7B, bottom panel; Supplementary Table S5). These results are inconsistent with a predominant pore-forming structure, suggesting that the gp72:gp73 complex does not form a stable pore in the membrane. In contrast, the addition of purified gp73 protein in amounts ranging from 0.3 µg to 2.6 µg reproducibly resulted in a stepwise current increase indicative of insertions of open, water-filled channels into the lipid bilayer (Fig. 7C, D and Supplementary Figs. 11A, B). We analyzed 33 gp73 channels in 17 different membranes using different protein fractions after gel filtration from two different protein preparations and observed a distribution of open pore currents ranging from 15 to 95 pA at 100 mV (Fig. 7E, Supplementary Table S5; Supplementary Fig. 11C-F). The mode value of the open pore current is 30 pA, corresponding to a conductance of 300 pS for the gp73 channel (Fig. 7E). This gp73 conductance is within the range of other pore-forming proteins such as the E. coli proteins Tsx (10 pS)50, LamB (155 pS)51 and OmpF (1500 pS)52. In conclusion, these experiments demonstrate that gp73 forms water-filled channels in lipid membranes. The fact that we did not observe channels of the gp72:gp73 complex in our lipid bilayer experiments might be caused by a structural change of gp73 in complex with gp72, which may close the channel to prevent premature loss of the phage DNA. DNA ejection by the phage after binding to the cell surface of P. aeruginosa would open the gp72:gp73 channel, but this signal is missing in our lipid bilayer system.
Domain analysis of DEV ejection protein gp71
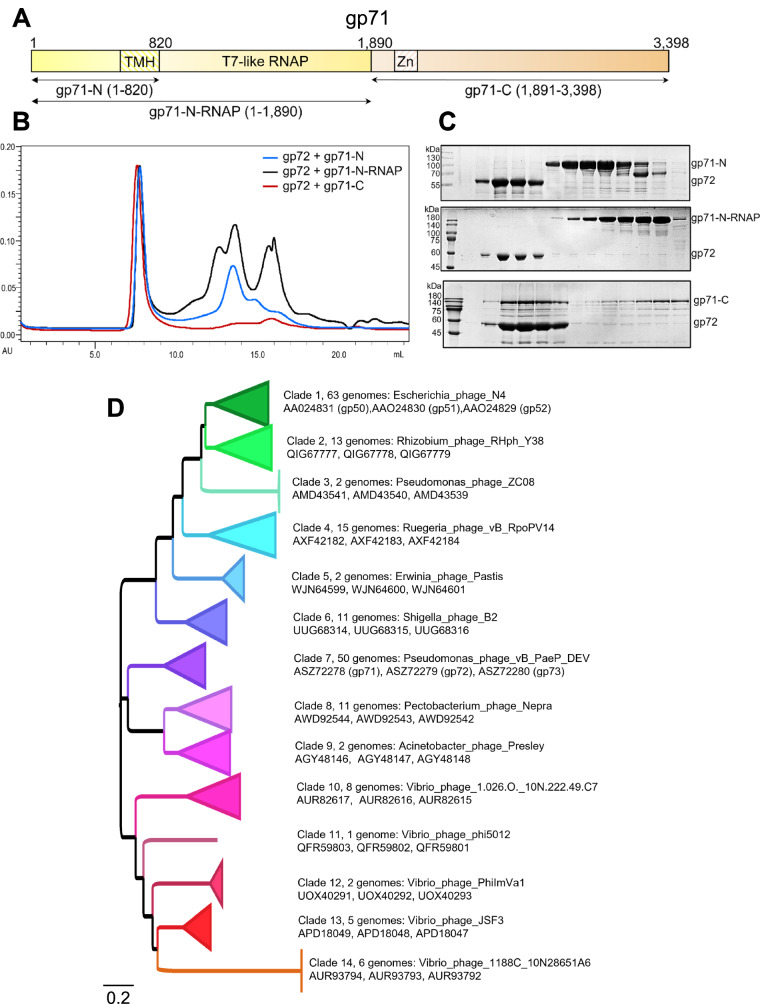
DEV’s largest gene, gp71, is part of the same operon as gp72 and gp73 (Fig. 5C) and encodes a 3398 vRNAP, homologous to phage N4 p50. MS analysis of purified DEV virions (Supplementary Table S1) confirmed that gp71 is present in the specimen used for cryo-EM SPA but is invisible in the reconstruction, possibly due to failure to align a non-icosahedral feature inside a DNA-filled capsid. The sequence identity/similarity between DEV gp71 and N4 gp50 is modest, just 20.8% and 36.9%, respectively (with 25.6% gaps). Nevertheless, the two proteins share a similar domain organization, containing a ~1000-residue single-unit RNAP core53, approximately 800 amino acids away from the start codon (Fig. 8A). Bioinformatic and AlphaFold analysis suggests that the DEV RNAP core is flanked by two regions enriched in α-helices: gp71-N (res. 1–820), which contains putative transmembrane-spanning helices (TMH) between residues 572–820, and a large gp71-C (res. 1900–3398), also predicted to harbor a putative zinc-binding motif between residues 2100-2150. Like N4 vRNAP gp50, which is ejected into the host upon infection7 and associates with bacterial membranes7,54–56, DEV gp71 also lacks cysteine residues.
Fig. 8. Domain mapping and conservation of DEV gp71.
A Schematic diagram of gp71 predicted domains. TMH = transmembrane helixes; Zn = zinc-binding domain. B SEC co-migration assay. Overlay of chromatograms obtained by running gp72 with gp71-N (blue line), gp71-N-RNAP (black line), and gp71-C (red line). C SDS-PAGE of fractions eluted from the co-migration assay in panel (B). Gp72 = 59 kDa; gp71-N = 88 kDa, gp71-N-RNAP = 205 kDa and gp71-C = 170 kDa. Each assay was repeated at least three times. Source data are provided as a Source Data file. D Phylogenetic relationships of Schitovirus DEV gp71/ N4 gp50 homologs inferred by maximum-likelihood. 191 homologs of DEV gp71/ N4 gp50 were identified by similarity searches. Various clades have collapsed, and the members of each clade can be found in Supplementary Table S6. Clade refers to groups of Schitoviridae to which the gp71/gp50 homolog was assigned. The longer the branch in the horizontal dimension, the larger the amount of change. The branch length units are amino acid substitutions per position (e.g., the number of amino acid changes divided by the length of the sequence). For each clade, an exemplar genome sequence is indicated, followed by the accession codes for the gp71/gp50 homolog, the inferred gp72/gp51 homolog, and the gp73/gp52 homolog found clustered in the expected sequence on the genome. The tree is arbitrarily rooted.
To identify the region of gp71 that is associated with gp72, we subcloned gp71-N (res. 1–820), gp71-N-RNAP (res. 1–1890), and gp71-C (res. 1891–3398) coding regions (Fig. 8A) and purified the three fragments from soluble bacterial lysates. In a co-migration assay by size exclusion chromatography (SEC), we found that only gp71-C co-migrated with gp72. In contrast, neither N-terminal construct had a detectable association with the periplasmic tunnel (Fig. 8B, C). Densitometric analysis of SDS-PAGE bands from SEC suggested ~2.7 ± 1 copies of gp71-C associated with a gp72 nonamer (Fig. 6C). This is close to the 4 ± 1 copies of N4 gp50 estimated to be inside the mature virion6. Thus, DEV’s largest ejection protein, gp71, is associated with the periplasmic tunnel gp72 via a large C-terminal domain. We speculate that gp72 C-terminal residues 332-521, disordered in the gp72:gp73 complex reconstruction (Fig. 6B), become structured upon binding to gp71.
Phylogenetic origin of the Schitoviridae ejection apparatus
To evaluate the possible conservation of the phage ejection machinery described here for DEV, we performed bioinformatics analyzes using the existing annotations from 234 schitovirus genomes available in Genbank as of April 23, 2024. We selected genomes that were assembled as a single contig and for which the best BlastP hit for DEV gp71 and DEV gp73 or N4 gp52 were encoded on the same strand and with an intergenic interval of 2500 nt or less. We subsequently focused on 191 genomes (Supplementary Table S6), where an open reading frame of between 900 and 2500 nt was annotated on the same strand between the DEV gp71 and DEV gp73/N4 gp52 matches (probable operons). Notably, the restricted dataset (191 phage genomes containing putative correctly annotated ejection operons) represented the entire taxonomic host range covered by the unfiltered dataset (members of the alpha-, beta- and gamma-proteobacteria). Next, we focused on individual putative ejection proteins.
Gp71 homologs are highly divergent, with multiple short blocks of conserved sequence and many large (10-50 aa) indels between clusters of gene products. Around 500 amino acid positions were identified as unambiguously aligned and subjected to phylogenetic analysis under the maximum likelihood criterion (LG substitution model with one invariable and four gamma-distributed variable substitution rate categories). The recovered topology consistently, but not universally, clusters gp71 homologs according to bacterial host annotations (at both the genus and class levels) (Fig. 8D), likely reflecting ancestral adaptation to the different cell envelopes through which these proteins form a transenvelope channel. Notably, extensive sequence diversity is observed among the gp71 homologs of vibrio pages and between vibrio and non-vibrio phage gp71 homologs (Fig. 8D). This observation is also reflected throughout the gp71 alignment, including regions not employed in the construction of the phylogenetic tree as well as in sequences of gp72/73 equivalents. Strikingly, while the RNAP core, located in the middle of gp71 (Fig. 8A), exhibits multiple short alignable sequence motifs, the C-terminal region of the protein, which we found to form physical associations with gp72 (Fig. 8B, C), shows higher levels of overall conservation.
Gp73 homologs (recovered by similarity to DEV gp73/N4 gp52) show high overall levels of sequence diversity but high levels of conservation within the groups defined by the gp71 tree, consistent with vertical coinheritance with gp71. As for gp71, the C-terminal region, which is proximal to gp72 and exposed above the OM in our model (Fig. 6B), is more conserved than the N-terminal region, where multiple insertions and deletions render confident alignment impossible.
Finally, putative gp72 equivalents are the most divergent of the three ejection proteins. For about 30% of the genomes, candidate gp72 homologs could not be identified by similarity with DEV gp72/N4 gp51. However, a relatively long ORF is consistently annotated between gp71 and gp73 homologs. As for other members of the putative ejection operon, levels of sequence divergence were extremely high, although conservation was evident within clusters defined by the virion RNA polymerase tree. However, it was not possible to obtain convincing evidence of homology between all these proteins from sequence alignment. Accordingly, we generated Alphafold2 structure predictions for representatives of each cluster within which homology was evident. As seen for the experimentally determined T7 gp15 and DEV gp72 (Supplementary Fig. 10), all predicted structures were extremely rich in α-helical secondary structure elements. We subsequently performed an all-against-all structural similarity search employing Foldseek57 to evaluate the structural similarity between the candidate gp72 equivalents, considering the probability of homology value for significant (e < = 10-5) structural matches. A summary of comparisons showing a ≥ 90% probability of homology is provided in Supplementary Fig. 12. It illustrates a robust network of predictions sustaining the overall homology of the sequences identified by similarity to DEV gp72/ N4 gp51 and those identified by positional conservation.
It has previously been suggested that a lack of cysteine residues in certain phage proteins might reflect the requirement for passage through the host periplasm, which contains proteins that catalyze disulfide bond formation58. The observed frequency of cysteine residues across the annotated proteomes of the 191 viruses in our survey was 0.865%. For the gp71 homologs, the equivalent value was 0.00075% (mean length 3475.75 + /- 175.65 amino acids), while for candidate gp72 homologs, the observed frequency of cysteines was 0.0055% (mean length 566.9 + /- 126.24 amino acids). Notably, all 6 of the cysteine residues found in candidate gp72 homologs were situated in regions lacking conservation even between otherwise closely related proteins or close to the end of apparently prematurely truncated protein predictions, consistent with annotation errors resulting from genomic sequence errors. No cysteines were observed in DEV gp73 / N4 gp52 homologs, although these proteins are relatively short (mean length 153.70 + /- 19.93 amino acids). Thus, as previously noted for N4 gp5059, suppression of cysteine residues is a conserved feature of proteins encoded by the putatively conserved genome ejection operon of Schitoviridae.
Discussion
Infections caused by the Gram-negative pathogen P. aeruginosa are a leading cause of morbidity and mortality worldwide. Phage therapy against P. aeruginosa has gained attention as a promising therapeutic weapon, especially in the fight against cystic fibrosis-related infections60,61. The N4-like phage DEV is part of an experimental phage cocktail to eradicate P. aeruginosa infections in vivo13,14. In this study, we determined a complete structural atlas of all phage DEV structural factors and elucidated fundamental aspects of DEV biology. We resolve a new symmetry mismatch in the tail apparatus, identify two putative fibers, and decipher three ejection proteins. As Schitoviridae genomes are largely unannotated and many ORFs have unknown functions, our work paves the way for the facile identification of structural components when a new Schitoviridae phage is discovered.
The mechanisms of Schitoviridae attachment to bacteria are poorly understood. N4 binds the E. coli membrane protein NfrA62 via gp65 (e.g., tail sheath)63, which surrounds the tail tube. However, this attachment mechanism is unlikely for DEV, which lacks a homologous factor surrounding the tail tube. Also, P. aeruginosa lacks a nfrA (and nfrB) orthologue. Based on all data presented here, we hypothesize that gp53 and gp56 mediate DEV adsorption to the P. aeruginosa surface. We posit that the long fiber gp53 binds the highly abundant O-antigen that serves as a primary receptor. The short fiber gp56 contacts a secondary receptor in the OM that triggers its detachment from the tail tube, prompting genome ejection.
In previous work, we postulated that DEV adsorption to P. aeruginosa PAO1 and other strains with LPS of the same serotype might involve two receptors: the O-antigen and another unidentified receptor64. Phage N4 can also rely on two receptors to infect its E. coli host: the outer membrane protein NfrA47 and an N-acetylmannosamine-based surface carbohydrate produced and exported to the cell surface of E. coli in a c-di-GMP-dependent manner57,58. DEV’s second receptor may be the inner core of the LPS or an outer membrane protein, like for N4. The lack of PAO1 infection of the DEV Δ53 is consistent with the hypothesis that gp53 is the receptor-binding protein involved in O-antigen recognition. The DEV phage may exploit gp53-dependent O-antigen binding to approach the second receptor when infecting PAO1. Conversely, gp53 is dispensable for adsorption to PAO1 mutants with a truncated LPS core, suggesting that long-tail fibers are not needed if the LPS does not mask the second receptor. Thus, DEV can bind directly to the secondary receptor when accessible using another receptor-binding protein. This is likely the short-tail fiber gp56, as short-tail fibers are known to bind receptors in other phages65. It remains unclear why the wzy mutant, which produces a LOS form capped with a single O-antigen repeat16,39, is resistant to both wild-type DEV and Δ53-DEV. We speculate that in wzy, the minimal O-antigen, which is reduced to a single unit repeat, is too short to allow gp53-dependent DEV binding but long enough to mask the second receptor.
Schitoviridae have large DNA genomes, but how these viruses eject their DNA into bacteria is unknown. In small Podoviridae like T7 and P22, ejection proteins form a DNA-ejectosome40,43 that extends the short tail, allowing DNA passage through the cell envelope41,42,66. However, ejection proteins have not been mapped in N4 or other members of the Schitoviridae family1. In this work, we found that the DEV portal is surrounded by 12 copies of a helical protein that we built de novo and identified as gp72. The position in the virion and helical fold of DEV gp72 is similar to the recently characterized gp45 cargo protein C1 found in the CrAssphage ΦcrAss00167. The discovery of gp72 led us to identify an operon, of which gp72 is also part, containing the membrane protein gp73 and the giant vRNAP. This highly divergent operon can be identified in the genomes of over 190 Schitoviridae members. Cryo-EM reconstructions revealed that the recombinant DEV gp72 and gp73 proteins assemble into a long channel large enough to accommodate dsDNA and span the bacterium periplasm. Lipid bilayer experiments demonstrated that gp73 can insert into membranes and form stable pores. Biochemical mapping studies found that the C-terminal domain of DEV gp71 associates with gp72, while a putative transmembrane region is predicted N-terminal of the RNAP core. Altogether, comparative genomics, protein structure determination and prediction, and biochemical analyses are highly consistent with the hypothesis that gp73, gp72, and gp71 are DEV ejection proteins functionally equivalent to phage T7 gp14, gp15, and gp1642. We propose that DEV ejection proteins assemble into a specialized ejection apparatus that uses the RNAP domain of gp71 as a motor for genome ejection, and the design principles of this molecular machine are conserved across the Schitoviridae family. Since DEV utilizes three RNA polymerases, we propose that the DEV genome is actively pulled into the host by the combined action of three sub-motors: first, the ejected vRNAP, gp71, required for early gene transcription, possibly aided by host gyrases that introduce negative supercoils into the phage genome; second, the transcription activity by DEV second RNA polymerase, RNAPII; lastly, the host RNA polymerase.
The results presented in this paper allow us to formulate a model for how DEV attachment to the Pseudomonas surface triggers the ejection of the phage genome through the tail tube68. We envision three steps in this infection process.
Step 1 (Fig. 9A). DEV long-tail fibers are flexible, stochastically fluctuating to enhance the chance of encountering LPS. The association of gp53 with the host O-antigen, possibly mediated by the carbohydrate esterase-like domain (res. 265-656) and Myoviridae-like fiber (res. 694-1090), tethers the phage closer to the host surface, likely inducing the sequential attachment of all five long-tail fibers. We did not detect LPS hydrolase activity in isolated DEV virions, suggesting the long-tail fibers only function by adsorbing the virion to the cell surface instead of shaving off LPS as in P2269. The association of multiple long-tail fibers with the LPS may reorient the virion to land perpendicular to the OM, as observed for phage P2266.
Fig. 9. Proposed model for DEV absorption onto P. aeruginosa surface and genome ejection.
Three proposed steps of infection are shown: each step is accompanied by distinct conformations of the long and short-tail fibers. A DEV interacts with the host O-antigen through flexible long-tail fibers (gp53), possibly reorienting the virion to land perpendicular to the OM. B The short-tail fiber gp56 interacts with a secondary receptor in the bacterium OM, triggering a conformational change that releases the short fiber. C The ejection proteins gp73, gp72, and gp71 are expelled into the bacterium cell envelope where gp73 forms an OM pore, gp72 spans the periplasm, and gp71 crosses the IM, projecting a large vRNAP motor into the bacterial cytoplasm, that begins pulling the viral genome inside the host. PG = peptidoglycan. Figure 9, created with BioRender.com, released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license.
Step 2 (Fig. 9B). After the virion has attached to the host surface, the short-tail fiber, which also functions as a tail plug, encounters a secondary receptor, likely an outer membrane protein shielded by LPS and thus inaccessible from the outside. We hypothesize that the interaction of gp56 with the secondary receptor transmits a mechanical signal that releases the short-tail fiber. Analogous to the tail needle gp26 in P22-like phages36, the short-tail fiber could be ejected into the host or removed upon association with the receptor.
Step 3 (Fig. 9C). Following the release of gp56, the ejection proteins gp73, gp72, and gp71 are expelled into the bacterial cell envelope where gp73 forms an outer membrane pore, analogous to P22 gp7 and T7 gp14. Gp72 spans the periplasm, like the periplasmic tunnel gp15 in T7, and gp71 crosses the IM, projecting a large RNA pol motor into the bacterial cytoplasm. As observed for phage T7, we determined a reduction of the gp72 oligomeric state from dodecameric inside the virion to nonameric after ejection40. In T7, the periplasmic tunnel gp15 is octameric in capsid and hexameric after ejection. Thus, a loss of ejection protein subunits accounts for the challenging refolding-coupled assembly of the DNA-ejectosome in Schitoviridae, as suggested for small Podoviridae70. However, the major difference is that Schitoviridae have a much larger genome ( ~ 75 kbs) and carry a vRNAP that must become transcriptionally active to promote successful infection7. This function is exerted by the host RNA polymerase in small Podoviridae that rely chiefly on the host transcription machinery to eject their smaller genomes.
In summary, we have deciphered the architecture and design principles of a prototypical N4-like phage used in an experimental phage therapy cocktail. We propose that the structural principles elucidated in this work are conserved in other Schitoviridae of the widespread N4-like family1. It is also possible that the anatomy of DEV ejection proteins, the interplay with virion-associated and encoded RNAPs, and their role in genome delivery will be conserved in crAss-like phages, the most abundant viruses in the human gut67. We anticipate that the 3D-atlas of DEV structural proteins described in this paper will allow the mapping of resistance mutations and facilitate the identification of ORFs in related Pseudomonas phages used in phage therapy cocktails.
Methods
Bacteria, bacteriophages and plasmids
P. aeruginosa strains, bacteriophages, and plasmids are listed in Supplementary Table S2; oligonucleotides are in Supplementary Table S3. P. aeruginosa genome coordinates refer to PAO1 strain, NCBI RefSeq NC_002516.2. Plasmids were constructed in Escherichia coli, and their relevant portions were sequenced before they were transferred into P. aeruginosa by transformation. pCas3cRh38 is a pHERD30T derivative and was purchased from Addgene. pCas3-01 was obtained by cloning the annealed 3697 and 3698 primers in the pCas3cRh BsaI restriction site. pCas3-09 and pD53 were constructed by assembling DNA fragments using the NEBuilder HiFi DNA Assembly Master Mix (New England Biolabs). pCas3-09 derives from the assembly of pCas3-01 linearized with BstZ17I with two fragments obtained by PCR on DEV DNA with primers 3807-3840 and 3808-3841. pGM2148 carries the DEV gp53 gene under the control of araBp promoter and was obtained by assembling pGM93171 digested with KpnI and the gp53 gene amplified by PCR on DEV DNA with primers 3804-3805. pD53, which carries crRNA53-resistant gp53, was obtained by PCR amplification of pGM2148 with primers 3859-3955 and ligation of the amplicon with the KLD Enzyme Mix (New England Biolabs). The genes encoding gp72 and gp73 were cloned by PCR from DEV DNA between restriction sites NdeI and XhoI in pET30b and pET22b, respectively. gp73 was cloned with C-terminal 6x His tag, while gp72 was untagged. Later, gp72 was cloned in pET28a, having N-terminal 6x His tag between NdeI and XhoI. Gp71-N (1–820 residues), gp71-N-RNAP (1–1890 residues), and gp71-C (1891–3398 residues) coding regions were cloned in pET-16b between NdeI/XhoI sites with N-terminal 9x His tag. Bacterial cultures were grown in Lysogeny Broth (LB) at 37 °C. Cultures of bacterial strains carrying plasmids were supplemented with 100 µg/ml ampicillin, 50 µg/ml gentamicin, 300 µg/ml carbenicillin, 50 µg/ml kanamycin, 0.2% arabinose (w/v), and 0.1% rhamnose (w/v) when needed.
Purification of DEV virions for cryo-EM
DEV was prepared for cryo-EM analysis as described18 with minor modifications (standard protocol) or upon chloroform extraction (modified protocol). In brief, P. aeruginosa strain PAO1 was grown at 37 °C to OD600 = 0.5, corresponding to about 2.5 × 107 cfu/ml in LB and infected with DEV phage13 (GenBank MF490238.1) at a multiplicity of infection (m.o.i.) of 0.001. Growth was continued until cell lysis was detected as a drop in the OD600. The lysate was incubated 30 min at 37 °C with DNase I and RNase A (1 µg/ml each) and centrifuged 20 min at 5000 × g. After supernatant filtration through a 0.45 µm filter, 58 g l-1 NaCl and 105 g l-1 polyethylene glycol (PEG) MW 6 K were dissolved in the supernatant. The solution was incubated 16 hours at 4 °C, and the phage particles recovered by centrifugation at 4 °C and 20,000 × g for 30 min. The pellet was resuspended in TN buffer (10 mM Tris-HCl, 150 mM NaCl pH 8), and phages were purified according to either standard or modified protocol. In standard protocol, the mixture was stratified on the top of a cesium chloride step gradient from 1.3 to 1.6 gm/cc cesium chloride, top to bottom, formed in polyallomer ultracentrifuge tubes for Beckman rotor SW41, and centrifuged at 100,000 × g for 120 minutes at 4 °C in a Beckmann Optima XE-90 ultracentrifuge using a SW41 rotor. The phage bands, which usually sediment at the 1.4 gm/cc step, were transferred into polyallomer tubes for SW41 Beckman rotor. The tubes were filled with a solution of cesium chloride 1.4 gm/cc in TN buffer and centrifuged 16 hours at 150,000 × g. After centrifugation, the phage bands were collected, dialyzed 2 x for 20 minutes against water and 16 hours against TN buffer, filtered through 0.22 µm filters and stored at 4 °C. In the modified protocol, the mixture was mixed with 0.5 volumes of chloroform and centrifuged for 15 min at 4500 × g. The upper phase was recovered, centrifuged on the 1.3 to 1.6 gm/cc cesium chloride step gradient, and dialyzed against water and TN buffer as described before. The phage mixture was layered onto a 10–40% (w/v) sucrose gradient in TN buffer and centrifuged 1 h at 210,000 × g at 4 °C using a SW41 rotor. After centrifugation, 0.4 ml fractions were collected, and their OD260, OD280, and OD320 were measured and plotted. The fractions corresponding to absorbance peaks were dialyzed, filtered through 0.22 µm filters, and stored at 4 °C. Both protocols yielded HF and FF particles; phages purified on sucrose gradients tend to lose the tails over time.
SDS-PAGE analysis of DEV virion proteins
To extract the proteins from phage particles, 0.5 ml of phages purified on double CsCl2 gradients was mixed with 0.5 ml of methanol and 0.375 ml of chloroform. After vigorous mixing, the sample was centrifuged for 5 min at 16,873 × g in a microfuge, and the upper fraction was discarded. The lower fraction and the interface were mixed with 0.38 ml of methanol and centrifuged as above. The protein pellet was dried and resuspended in Tris-HCl 10 mM pH 7.4, 6 M urea. The viral proteins were analyzed using 15 % SDS-PAGE and Coomassie staining.
Mass spectrometry analysis of DEV virion composition
20 µg of DEV virion proteins extracted as described above from phages purified on CsCl2-sucrose gradients were analyzed at UNITECH OMICs (University of Milano, Italy) using Dionex Ultimate 3000 nano-LC system (Sunnyvale CA, USA) connected to Orbitrap Fusion™ Tribrid™ Mass Spectrometer (Thermo Scientific, Bremen, Germany) equipped with nano electrospray ion source. The samples resuspended in 6 M urea were mixed 1:5 with 50 mM ammonium hydrogen carbonate (AMBIC) and incubated 30 min at 55 °C with 5 mM DTT. After adding 15 mM Iodoacetamide, samples were kept 20 min in the dark. Digestion was performed by overnight incubation with trypsin (final concentration, 0.007 mg/ml) at 37 °C. Peptide mixtures were pre-concentrated onto an Acclaim PepMap 100–100 µm × 2 cm C18 (Thermo Scientific) and separated on EASY Spray column ES900, 15 cm×75 µm ID packed with Thermo Scientific Acclaim PepMap RSLC C18, 3 µm, 100 Å using mobile phase A (0,1 % formic acid in water) and mobile phase B (0,1% formic acid in acetonitrile 20/80, v/v) at a flow rate of 0.300 µL/min. The temperature was set to 35 °C, and 2 µl samples were injected in triplicates. MS spectra were collected over an m/z range of 375–1500 Da at 120,000 resolutions, operating in the data-dependent mode, with a cycle time of 3 sec between master scans. HCD was performed with collision energy set at 35 eV and positive polarity. Data were processed using Proteome Discoverer 2.5 software (Thermo Scientific, USA), implemented with the SEQUEST algorithm, with the search database set as Pseudomonas phage vB_PaeP_DEV (NCBI: txid2034344) and trypsin as the digestion enzyme, with a minimal cross-correlation ≥ 2.2.
DEV mutagenesis
A culture of P. aeruginosa PAO1 carrying plasmids pCas3-09 and pD53 was grown in LB supplemented with gentamicin, carbenicillin, arabinose, and rhamnose to OD600 = 0.1. The two plasmids are maintained in the same cell, although they share the same replicon, in the presence of gentamycin and carbenicillin. 1 ml was infected with DEV phage at a m.o.i. of 10 and incubated at 37 °C for 5 min static and 40 min with agitation. The mixture was serially diluted and plated to obtain single plaques using PAO1/pD53. After overnight incubation at 30 °C, some plaques were analyzed by replica plating as described64 on PAO1/pD53 (permissive strain) and PAO1/pGM931 (non-permissive strain). In three replicate experiments, we found that 18, 8, and 7 out of 76, 84, and 78 analyzed plaques were formed by phages unable to reproduce without pD53. Decreasing the m.o.i. to 1 did not improve editing efficiency. The plaques formed by pD53-dependent phages (5 for each replicate experiment) were controlled by PCR with oligonucleotides 3806 and 3809 to confirm the presence of the gp53 deletion, which was present in 13/15 plaques (see Supplementary Fig. 8B).
RT-PCR analysis of DEV RNA
P. aeruginosa PAO1 cultures were grown in LB at 37 °C up to OD600 = 0.8 and infected with DEV at a m.o.i. of 5–6. 3 ml samples were collected at 0-, 10-, 15- and 20-minutes post-infection for RNA extraction. RNA extraction was performed by phenol-chloroform treatment of cell lysates as described72. After digestion with Turbo DNase (Ambion), 2 mg of RNA was retrotranscribed with Superscript III Reverse Transcriptase (Invitrogen) in 10 µl (final volume). A 0.5 µl aliquot of the reaction was PCR-amplified with primers 3969 and 3970 mapping within gp71 and gp73, respectively. Mock reverse transcription reactions (e.g., without Reverse transcriptase) were also PCR-amplified as a control for DNA contamination.
Expression and purification of recombinant gp1, gp72 and gp73
The constructs were expressed in the LOBSTR E. coli expression strain (Kerafast) supplemented with either 30 μg/mL kanamycin for gp72-pET30b(+) or 50 μg/mL ampicillin for gp73-pET22b(+). The cultures were grown in LB medium at 37 °C until OD600 = ~ 0.3 when the temperature was dropped to 28 °C until an OD600 = ~ 0.6 and were induced with 0.5 mM IPTG for 2-4 h. For gp73, cell pellets were lysed by sonication in Lysis buffer (20 mM Tris-HCl pH 8.0, 300 mM NaCl, 4 mM MgCl2, 1% glycerol, 2 mM EDTA, 0.1% Triton X-100, 1 mM PMSF, 20 μg/mL DNase). After centrifugation at 26,500 × g for 30 min, 4 °C, the insoluble fraction containing the protein was solubilized with rotation in Extraction buffer (20 mM Tris-HCl pH 8.0, 200 mM NaCl, 0.25-0.35% N-lauroylsarcosine, 1% glycerol, 20 μg/mL DNase) at room temperature for 1.5–2 h. After centrifugation at 26,500 × g for 30 min, 4 °C, the resulting supernatant was incubated with Nickel Agarose beads (GoldBio) for 2 h with rotation at 4 °C. The beads were washed with Wash buffer (20 mM Tris-HCl pH 8.0, 200 mM NaCl, 0.025% DDM, 2 mM MgCl2, 1% glycerol, 1 mM PMSF, 5 mM imidazole) and eluted with Wash buffer containing 20 - 320 mM imidazole. The protein fractions were dialyzed against Dialysis buffer (20 mM Tris-HCl pH 8.0, 100 mM NaCl, 0.025% DDM, 2 mM MgCl2, 1% glycerol, 1 mM PMSF). The gp73 was further polished by SEC using an in-house packed Superose 12 16/60 column equilibrated with degassed Dialysis buffer. For gp72, the cell pellet was lysed by sonication in Lysis buffer (20 mM Tris-HCl pH 8.0, 25 mM NaCl, 4 mM MgCl2, 1% glycerol, 2 mM EDTA, 0.1% Triton X-100, 1 mM PMSF, 20 μg/mL DNase). The soluble portion, after centrifugation at 26,500 × g for 30 min, 4 °C was passed through Heparin (GE) column. The unbound fraction was collected and subjected to MonoQ (GE) and eluted with a buffer containing 1 M NaCl. Enriched fractions containing gp72 eluted at ~550 nM NaCl were further purified by SEC using Superdex 200 16/600 (Cytiva). The protein used for Cryo-EM was further polished over Superose 6 10/300 GL (Cytiva). To form the gp72:gp73 complex, 90 µM gp72 was incubated with 180 µM gp73 in a total volume of ~1.2 mL for 3 h at room temperature. The volume was concentrated to 0.5 mL using 100 kDa MWCO (Vivaspin® 6, Sartorius) concentrator and subjected to Superose 6 10/300 GL. Gp72, gp71-N (1–820 residues), gp71-N-RNAP (1-1890 residues), and gp71-C (1891–3398 residues) were expressed in the NiCo21(DE3) E. coli expression strain (NEB) supplemented with either 50 µg/ml kanamycin or 100 µg/ml ampicillin. The growth conditions listed above were followed. Gp72 constructs were purified by metal affinity chromatography on low-density nickel agarose beads (GoldBio). The beads were washed with Wash buffer (20 mM Tris-HCl pH 8.0, 200 mM NaCl, 0.015% DDM, 2 mM MgCl2, 1% glycerol, 1 mM PMSF, 10-20 mM imidazole) and eluted with Wash buffer containing 50–400 mM imidazole. The protein elutions were injected directly on an in-house packed Superose 6 16/60 column equilibrated with degassed gel filtration buffer (20 mM Tris-HCl pH 8.0, 75 mM NaCl, 2 mM MgCl2, 1% (v/v) glycerol, 0.1 mM PMSF). Eluted protein fractions were concentrated using either 50 or 100 kDa MWCO (Vivaspin® 6, Sartorius) concentrator. To form a gp72:gp71 complex, 25 µM gp72 was incubated with 1.25-1.5 molar excess of gp71 constructs in a total volume of 500 µL for 3 h at room temperature and subjected to Superose 6 10/300 GL.
Vitrification and data collection
2.5 µl of DEV mature virions at 1 × 1012 phages/ml were applied to a 200-mesh copper Quantifoil R 2/1 holey carbon grid (EMS) previously glow-discharged negatively for 60 sec at 15 mA using an easiGlow (PELCO). The grid was blotted for 7.5 sec at blot force 2 and vitrified immediately in liquid ethane using a Vitrobot Mark IV (FEI). Micrographs were screened on 200 kV Glacios equipped with a Falcon4 detector. EPU software was used for data collection using accurate positioning mode. For high-resolution data collection, micrographs were collected on a Titan Krios microscope operated at 300 kV and equipped with a K3 direct electron detector camera (Gatan) at the National Cryo-EM Facility at the Frederick National Laboratory, MD. Micrographs were collected in super-resolution mode plus energy filter at 20 eV with an image pixel size of 0.56 Å at 81,000x magnification, a nominal total dose of 50 e/Å2, 40 frames, and a defocus range -0.8 to -1.6 μm. Further collection parameters are in Table 1.
Cryo-EM single particle analysis
All steps of SPA were carried out using RELION 3.1.273,74 and cryoSPARC75 software on a dual GPU Linux workstation. To reconstruct phage DEV, two sets of micrographs were combined after motion correction with MotionCor276, yielding 17,245 micrographs. RELION’s implementation of motion correction was applied to the micrographs with options of ‘dose-weighted averaged micrographs’ and ‘sum of non-dose weighted power spectra’ every 4 e-/Å2. CTF (Contrast Transfer Function) estimation was carried out using CTFFIND477. After 2D classification, symmetry-free low-res reconstruction, 3D classification, and 3D refinement in I3 symmetry, reference-free picked 109,000 particles were processed into two groups, HF capsids, and FF virions, with 8000 and 16,000 particles, respectively. The particles of both groups were then expanded according to I3 symmetry using RELION’s relion_particle_symmetry_expand function to obtain expanded particles of 60 different orientations. A cylindrical mask (r = 200 Å, l = 300 Å) was created using SCIPION 3.078 and then resampled onto the reference map from 3D refinement in I3 symmetry, covering the five-fold vertex on z-axis in Chimera79. The cylindrical mask was then used for symmetry-free sampling-free 3D classification to search for the tail. Locally aligned sub-particles were selected, extracted with z-axis shifted, and duplicates removed. The initial localized reference map was reconstructed directly from the selected particles using RELION’s relion_reconstruct routine. Selected particles were auto-refined using C5 symmetry, followed by 5-fold symmetry particle expansion. The 5-fold expanded particles were subjected to another round of sampling-free symmetry-free 3D classification aims for aligning ejecting components, portal, HT-adapter, and tail tube at the unique 5-fold vertex. The capsid-portal aligned particles were then 3D refined with limited initial angular, CTF refined, and polished to generate localized maps with and without C12 symmetry applied. The same concept of localized reconstruction was applied to searching for scaffolding protein and the whisker ring (appendage), as well as improving coat protein resolution from 3.7 Å (I1 symmetry) to 3.3 Å (local C5 averaged). The final densities were sharpened using phenix.auto_sharpen80. Electron density maps were displayed using ChimeraX81. To reconstruct the structure of recombinant gp72 and the gp72:gp73 complex, all steps of SPA were carried out using cryoSPARC75 imposing C9 symmetry.
De novo model building, AlphaFold modeling and refinement
All de novo atomic models presented in this paper were built using Coot82 or Chimera79. We used five different maps for model building: (i) a 3.3 Å C5-averaged localized reconstruction of the mature head (Supplementary Fig. 2A) that revealed the atomic structure of the DEV coat protein (res. 1–399). (ii) a 3.1 Å C12-averaged localized reconstruction of the FF virion (Supplementary Fig. 2A) was used to build a model of dodecameric portal protein (res. 21–721) bound to twelve copies of the head-to-tail adapter (res. 2-244), which also bound to twelve copies of the tail tube protein (2-158). The portal protein has no visible density of residues 1–21 in the C12 averaged map, but we were able to trace from residue #8 in the C5-C12 aligned asymmetric map; also, residues 722–726 of the C-terminal end of portal protein were not built due to missing densities. The HT-adapter was fully modeled except for the first methionine. The C-terminal of the tail tube was not visible at the 3.1 Å resolution map after residue #158. The tail tube residues 159–321 were modeled in a low-resolution localized reconstruction of the full-length tail. (iii) a 4.0 Å C12-averaged localized reconstruction of the portal protein from the FF virion (Supplementary Fig. 2A) was used to identify and model gp72 residues 95–503 in the pre-ejection conformation. (iv) a 3.65 Å C9-averaged reconstruction (Supplementary Fig. 2B) was used to model recombinant gp72 (res. 46–224) in the post-ejection conformation. (v) A 3.15 Å C9-averaged reconstruction (Supplementary Fig. 2C) was used to build recombinant gp72 (res. 25–331) bound to gp73 (res. 1–155) in the post-ejection conformation. The first 52 residues of gp73 are poorly ordered, and the density is very noisy, likely due to the presence of DDM. All atomic models were refined using several rounds of rigid-body, real-space, and B-factor refinement using phenix.real_space_refinement83 and validated using MolProbity84. Refinement statistics are in Table 1. The long- and short-tail fiber gp53 and gp56 prediction models were generated using AlphaFold32 and AlphaFold234.
Phylogenetic analysis of Schitoviridae ejection proteins
Genome sequences labeled schitovirus, as well as associated off annotations and protein sequences, were downloaded from Genbank (23/04/2024) using the NCBI datasets API (https://www.ncbi.nlm.nih.gov/datasets/). Genomes assembled as more than one contig, where full-length candidate gp71 (vRNAP) predictions were absent, or where low stringency local searches with BlastP85 failed to recover candidate homologs of DEV gp73 / N4 gp52 products encoded on the same strand and within 2500 nt downstream of the best match to DEV gp71 were discarded. Candidate gp72 homologs were identified either as best matches to DEV gp72 / N4 gp51 sequences (using a low stringency BlastP search) or as proteins of > 300 amino acids derived from annotated open reading frames situated between gp71 and gp73 candidates. Protein sequences were aligned using Muscle86 and edited in SeaView87. Conserved blocks for phylogenetic analyzes were identified with GBlocks (relaxed parameters as set in SeaView)88, and phylogenetic reconstruction was performed using PhyML89 under the LG substitution model90 with one invariable and four variable gamma-distributed rate categories. Protein structure prediction was performed with AlphaFold232 as implemented on the colab notebook (https://colab.research.google.com/github/deepmind/alphafold/blob/main/notebooks/AlphaFold.ipynb), recovering five alternative models for each selected protein. Structure similarity searches were performed for each alternative model against the collection of all generated models with Foldseek57, invoking the prob option to estimate the probability of homology between significant matches, and homology probability for the best match between any model for each template was recorded if ≥ 0.9. Counts of cysteine frequencies and other data manipulation operations were performed with custom scripts prepared in Python.
Structure analysis and bioinformatics tools
All renderings of protein structures and map surface representations were generated using ChimeraX81. Model analysis and inspection were carried out in PyMol91. Structural comparison and identification were done using the DALI server23. Binding interfaces were analyzed using PISA92 and PDBsum93. RMSD between superimposed PDBs was calculated using SuperPose Version 1.0 (superpose.wishartlab.com)94. The Coulombic Electrostatic Potential was calculated and displayed with surface coloring using ChimeraX81. The identity of Pseudomonas phage vB_PaeP_DEV protein sequences (Taxonomy ID: 2034344, NCBI:txid2034344) was analyzed using bioinformatics tools, including BlastP95 and searches in graphics mode of the NCBI Nucleotide section (GenBank MF490238.1). Sequence alignment was carried out in ebi.ac.uk/services96, including CLUSTAL O (1.2.4) for sequence alignment, EMBOSS Seqret for converting alignment format, and HMMER for homology search, and an in-house version of AlphaFold32 database for protein structure prediction. Sequence identity and similarity were carried out at Sequence Manipulation Suite (bioinformatics.org)97.
Lipid Bilayer Measurements
Lipid bilayer experiments were performed using a silicon chip with a SU-8 aperture of 100 µm diameter98. The chip was placed in a cis part (grounded) of a custom-made cuvette. Each side of a SU-8 aperture was pre-treated with 0.5 µl of 5 mg/ml diphytanoylphosphatidylcholine (DPhPC; Avanti Polar Lipids) in hexane. After hexane evaporation, the cis part was mounted on a trans part of the cuvette, and both parts were filled with 600 µl of an electrolyte solution containing 1 M KCl, 10 mM HEPES (pH 7.4). Lipid bilayers were painted across the aperture with a solution of 20 mg/ml of DPhPC in n-decane. After membrane formation, a protein sample was added to the cis part of the cuvette. Baseline and detergent-containing buffers were examined to exclude contamination and detergent interference. Experiments were performed at +100 mV potential. A pair of Ag/AgCl electrodes were inserted into the cuvette chambers and connected to an Axopatch 200 B patch-clamp amplifier (Molecular Devices). Currents were recorded at a sampling rate of 250 kHz, and low-pass filtered at 10 kHz. Data analysis was performed using Pythion, a custom software developed for analyzing current signals98. Pythion and SigmaPlot 11 (Systat Software) were used to generate the graphs shown in this study.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
We thank the staff at NCEF and Stanford-SLAC CryoEM Center (S2C2) for their assistance in cryo-EM data collection. We also thank Dr. Pramod Kumar for helping to draw a cartoon model. This work was supported by the National Institutes of Health grants R01 GM100888, R35 GM140733, and S10 OD030457 to GC, and by the Fondazione per la ricerca sulla Fibrosi cistica-Associazione Trentina Fibrosi Cistica ODV In ricordo di Pio Nicolini grant FFC#15/2021 to FB. A portion of this work was carried out at NCEF (supported by contract 75N91019D00024); Stanford-SLAC CryoEM Center (S2C2) is supported by the NIH Common Fund Transformative High-Resolution Cryo-Electron Microscopy program (U24 GM129541). Part of this work was carried out at UNITECH OMICs mass spectrometry platform of the Università degli Studi di Milano.
Author contributions
R.K.L, C-F.D.H., S.M.I., F.L., and G.C. performed biochemical analysis, cryo-EM, structural analysis, deposition of atomic coordinates and maps, and figure preparation. F.F. and F.B. purified the DEV virion, extracted DEV virion proteins, constructed and analyzed the DEV deletion mutant, and performed RT-PCR transcription analysis of the ejection operon. M.P. and M.N. carried out all steps of lipid bilayer experiments, including data collection and interpretation. D.S.H. performed the evolutionary analyzes. G.C. and F.B. supervised the project and wrote the paper. All authors contributed to the writing and editing of the manuscript.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Data availability
Atomic coordinates for phage DEV coat protein (C5), portal:HT-adapter:tail tube complex (C12), pre-ejection gp72 (C12), gp53 NTB (C15), and gp72:gp73 complex (C9) have been deposited in the Protein Data Bank with accession codes 9BGN, 9BGM, 9BGO, 9COD and 8VXQ. The relative cryo-EM density maps have been deposited in the Electron Microscopy Data Bank with accession codes EMD-44518, EMD-44517, EMD-44519, EMD-45776 and EMD-43629. Source data are provided in this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Federica Briani, Email: federica.briani@unimi.it.
Gino Cingolani, Email: gcingola@uab.edu.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-52752-1.
References
- 1.Wittmann, J. et al. From orphan phage to a proposed new family-the diversity of N4-Like viruses. Antibiotics (Basel)9, 663 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Menon, N. D. et al. A novel N4-like bacteriophage isolated from a wastewater source in south india with activity against several multidrug-resistant clinical pseudomonas aeruginosa isolates. mSphere6, e01215–e01220 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi, X. et al. Characterization and complete genome analysis of pseudomonas aeruginosa bacteriophage vB_PaeP_LP14 belonging to genus litunavirus. Curr. Microbiol77, 2465–2474 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Lerdsittikul, V. et al. A novel virulent Litunavirus phage possesses therapeutic value against multidrug resistant Pseudomonas aeruginosa. Sci. Rep.12, 21193 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tajuddin, S. et al. Genomic analysis and biological characterization of a novel Schitoviridae phage infecting Vibrio alginolyticus. Appl Microbiol Biotechnol.107, 749–768 (2023). [DOI] [PubMed] [Google Scholar]
- 6.Choi, K. H. et al. Insight into DNA and protein transport in double-stranded DNA viruses: the structure of bacteriophage N4. J. Mol. Biol.378, 726–736 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lenneman, B. R. & Rothman-Denes, L. B. Structural and biochemical investigation of bacteriophage N4-encoded RNA polymerases. Biomolecules5, 647–667 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Falco, S. C., Zehring, W. & Rothman-Denes, L. B. DNA-dependent RNA polymerase from bacteriophage N4 virions. Purification and characterization. J. Biol. Chem.255, 4339–4347 (1980). [PubMed] [Google Scholar]
- 9.Falco, S. C., Laan, K. V. & Rothman-Denes, L. B. Virion-associated RNA polymerase required for bacteriophage N4 development. Proc. Natl Acad. Sci. USA74, 520–523 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rothman-Denes, L. B. & Schito, G. C. Novel transcribing activities in N4-infected Escherichia coli. Virology60, 65–72 (1974). [DOI] [PubMed] [Google Scholar]
- 11.Casjens, S. R. & Gilcrease, E. B. Determining DNA packaging strategy by analysis of the termini of the chromosomes in tailed-bacteriophage virions. Methods Mol. Biol.502, 91–111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan, J. Z., Millard, A. D., Mann, N. H. & Schafer, H. Comparative genomics defines the core genome of the growing N4-like phage genus and identifies N4-like Roseophage specific genes. Front Microbiol5, 506 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forti, F. et al. Design of a Broad-range bacteriophage cocktail that reduces pseudomonas aeruginosa biofilms and treats acute infections in two animal models. Antimicrob Agents Chemother.62, e02573–17(2018). [DOI] [PMC free article] [PubMed]
- 14.Cafora, M. et al. Phage therapy against Pseudomonas aeruginosa infections in a cystic fibrosis zebrafish model. Sci. Rep.9, 1527 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wright, R. C. T., Friman, V. P., Smith, M. C. M. & Brockhurst, M. A. Cross-resistance is modular in bacteria-phage interactions. PLoS Biol.16, e2006057 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Forti, F. et al. Identification and impact on Pseudomonas aeruginosa virulence of mutations conferring resistance to a phage cocktail for phage therapy. Microbiol Spectr.11, e0147723 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hou, C. D., Li, F., Iglesias, S. & Cingolani, G. Use of localized reconstruction to visualize the shigella phage Sf6 tail apparatus. Methods Mol. Biol.2738, 215–228 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, F. et al. High-resolution cryo-EM structure of the Pseudomonas bacteriophage E217. Nat. Commun.14, 4052 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ceyssens, P. J. et al. Molecular and physiological analysis of three Pseudomonas aeruginosa phages belonging to the “N4-like viruses. Virology405, 26–30 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siborova, M. et al. Tail proteins of phage SU10 reorganize into the nozzle for genome delivery. Nat. Commun.13, 5622 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khan Mirzaei, M., Eriksson, H., Kasuga, K., Haggard-Ljungquist, E. & Nilsson, A. S. Genomic, proteomic, morphological, and phylogenetic analyses of vB_EcoP_SU10, a podoviridae phage with C3 morphology. PLoS One9, e116294 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dedeo, C. L., Cingolani, G. & Teschke, C. M. Portal protein: the orchestrator of capsid assembly for the dsDNA Tailed bacteriophages and herpesviruses. Annu Rev. Virol.6, 141–160 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holm, L. & Rosenstrom, P. Dali server: conservation mapping in 3D. Nucleic Acids Res38, W545–W549 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lokareddy, R. K. et al. Portal protein functions akin to a DNA-sensor that couples genome-packaging to icosahedral capsid maturation. Nat. Commun.8, 14310 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lander, G. C. et al. The structure of an infectious p22 virion shows the signal for headful DNA packaging. Science312, 1791-5 (2006). [DOI] [PubMed]
- 26.Tang, J. et al. Peering down the barrel of a bacteriophage portal: the genome packaging and release valve in p22. Structure19, 496–502 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, F. et al. High-resolution cryo-EM structure of the Shigella virus Sf6 genome delivery tail machine. Sci. Adv.8, eadc9641 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iglesias, S. M. et al. Molecular architecture of Salmonella typhimurium virus P22 genome ejection machinery. J. Mol. Biol.435, 168365 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olia, A. S. et al. Binding-induced stabilization and assembly of the phage P22 tail accessory factor gp4. J. Mol. Biol.363, 558–576 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Olia, A. S., Prevelige, P. E. Jr., Johnson, J. E. & Cingolani, G. Three-dimensional structure of a viral genome-delivery portal vertex. Nat. Struct. Mol. Biol.18, 597–603 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hardy, J. M., Dunstan, R. A., Lithgow, T. & Coulibaly, F. Tall tails: cryo-electron microscopy of phage tail DNA ejection conduits. Biochem Soc. Trans.50, 459–22W (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature596, 583–589 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strauss, H. & King, J. Steps in the stabilization of newly packaged DNA during phage P22 morphogenesis. J. Mol. Biol.172, 523–543 (1984). [DOI] [PubMed] [Google Scholar]
- 34.Skolnick, J., Gao, M., Zhou, H. & Singh, S. AlphaFold 2: why it works and its implications for understanding the relationships of protein sequence, structure, and function. J. Chem. Inf. Model61, 4827–4831 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olia, A. S., Casjens, S. & Cingolani, G. Structure of phage P22 cell envelope-penetrating needle. Nat. Struct. Mol. Biol.14, 1221–1226 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Bhardwaj, A., Walker-Kopp, N., Casjens, S. R. & Cingolani, G. An evolutionarily conserved family of virion tail needles related to bacteriophage P22 gp26: correlation between structural stability and length of the alpha-helical trimeric coiled coil. J. Mol. Biol.391, 227–245 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng, J. et al. Asymmetric structure of podophage GP4 reveals a novel architecture of three types of tail fibers. J. Mol. Biol.435, 168258 (2023). [DOI] [PubMed] [Google Scholar]
- 38.Csorgo, B. et al. A compact Cascade-Cas3 system for targeted genome engineering. Nat. Methods17, 1183–1190 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.King, J. D., Kocincova, D., Westman, E. L. & Lam, J. S. Review: lipopolysaccharide biosynthesis in Pseudomonas aeruginosa. Innate Immun.15, 261–312 (2009). [DOI] [PubMed] [Google Scholar]
- 40.Swanson, N. A. et al. Cryo-EM structure of the periplasmic tunnel of T7 DNA-ejectosome at 2.7 A resolution. Mol. Cell81, 3145–3159.e7 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perez-Ruiz, M. et al. Assisted assembly of bacteriophage T7 core components for genome translocation across the bacterial envelope. Proc. Natl Acad. Sci. USA118, e2026719118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swanson, N. A., Hou, C. D. & Cingolani, G. Viral ejection proteins: mosaically conserved, conformational gymnasts. Microorganisms10, 504 (2022). [DOI] [PMC free article] [PubMed]
- 43.Swanson, N. A. et al. Expression and purification of phage T7 ejection proteins for cryo-EM analysis. STAR Protoc.2, 100960 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen, W. et al. Structural changes in bacteriophage T7 upon receptor-induced genome ejection. Proc. Natl. Acad. Sci. USA.118, e2102003118 (2021). [DOI] [PMC free article] [PubMed]
- 45.Chang, C. Y., Kemp, P. & Molineux, I. J. Gp15 and gp16 cooperate in translocating bacteriophage T7 DNA into the infected cell. Virology398, 176–186 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yin, X., Yang, J., Xiao, F., Yang, Y. & Shen, H. B. MemBrain: an easy-to-use online webserver for transmembrane protein structure prediction. Nanomicro Lett.10, 2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cserzo, M., Wallin, E., Simon, I., von Heijne, G. & Elofsson, A. Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng.10, 673–676 (1997). [DOI] [PubMed] [Google Scholar]
- 48.Adams, P. G., Lamoureux, L., Swingle, K. L., Mukundan, H. & Montano, G. A. Lipopolysaccharide-induced dynamic lipid membrane reorganization: tubules, perforations, and stacks. Biophys. J.106, 2395–2407 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heinz, C. & Niederweis, M. Selective extraction and purification of a mycobacterial outer membrane protein. Anal. Biochem285, 113–120 (2000). [DOI] [PubMed] [Google Scholar]
- 50.Benz, R., Schmid, A., Maier, C. & Bremer, E. Characterization of the nucleoside-binding site inside the Tsx channel of Escherichia coli outer membrane. Reconstitution experiments with lipid bilayer membranes. Eur. J. Biochem176, 699–705 (1988). [DOI] [PubMed] [Google Scholar]
- 51.Benz, R., Schmid, A., Nakae, T. & Vos-Scheperkeuter, G. H. Pore formation by LamB of Escherichia coli in lipid bilayer membranes. J. Bacteriol.165, 978–986 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Benz, R., Janko, K., Boos, W. & Lauger, P. Formation of large, ion-permeable membrane channels by the matrix protein (porin) of Escherichia coli. Biochim Biophys. Acta511, 305–319 (1978). [DOI] [PubMed] [Google Scholar]
- 53.Murakami, K. S., Davydova, E. K. & Rothman-Denes, L. B. X-ray crystal structure of the polymerase domain of the bacteriophage N4 virion RNA polymerase. Proc. Natl Acad. Sci. USA105, 5046–5051 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Falco, S. C. & Rothman-Denes, L. B. Bacteriophage N4-induced transcribing activities in Escherichia coli. II. association of the N4 transcriptional apparatus with the cytoplasmic membrane. Virology95, 466–475 (1979). [DOI] [PubMed] [Google Scholar]
- 55.Falco, S. C. & Rothman-Denes, L. B. Bacteriophage N4-induced transcribing activities in Escherichia coli. I. detection and characterization in cell extracts. Virology95, 454–465 (1979). [DOI] [PubMed] [Google Scholar]
- 56.Kiino, D. R., Singer, M. S. & Rothman-Denes, L. B. Two overlapping genes encoding membrane proteins required for bacteriophage N4 adsorption. J. Bacteriol.175, 7081–7085 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Kempen, M. et al. Fast and accurate protein structure search with foldseek. Nat. Biotechnol.42, 243–246 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ritz, D. & Beckwith, J. Roles of thiol-redox pathways in bacteria. Annu Rev. Microbiol55, 21–48 (2001). [DOI] [PubMed] [Google Scholar]
- 59.Kazmierczak, K. M., Davydova, E. K., Mustaev, A. A. & Rothman-Denes, L. B. The phage N4 virion RNA polymerase catalytic domain is related to single-subunit RNA polymerases. EMBO J.21, 5815–5823 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Svoboda, E. Bacteria-eating viruses could provide a route to stability in cystic fibrosis. Nature583, S8–S9 (2020). [DOI] [PubMed] [Google Scholar]
- 61.Dedrick, R. M. et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med25, 730–733 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kiino, D. R., Licudine, R., Wilt, K., Yang, D. H. & Rothman-Denes, L. B. A cytoplasmic protein, NfrC, is required for bacteriophage N4 adsorption. J. Bacteriol.175, 7074–80 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McPartland, J. & Rothman-Denes, L. B. The tail sheath of bacteriophage N4 interacts with the Escherichia coli receptor. J. Bacteriol.191, 525–32 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Forti, F. et al. Identification and impact on Pseudomonas aeruginosa virulence of mutations conferring resistance to a phage cocktail for phage therapy. Microbiology Spectrum (2023). [DOI] [PMC free article] [PubMed]
- 65.Golomidova, A. K. et al. Branched lateral tail fiber organization in T5-like bacteriophages DT57C and DT571/2 is revealed by genetic and functional analysis. Viruses8, 26(2016). [DOI] [PMC free article] [PubMed]
- 66.Wang, C., Tu, J., Liu, J. & Molineux, I. J. Structural dynamics of bacteriophage P22 infection initiation revealed by cryo-electron tomography. Nat. Microbiol4, 1049–1056 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bayfield, O. W. et al. Structural atlas of a human gut crassvirus. Nature617, 409–416 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yap, M. L. et al. Role of bacteriophage T4 baseplate in regulating assembly and infection. Proc. Natl Acad. Sci. USA113, 2654–9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Andres, D. et al. Tailspike interactions with lipopolysaccharide effect DNA ejection from phage P22 particles in vitro. J. Biol. Chem.285, 36768–75 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lupo, D. et al. The T7 ejection nanomachine components gp15-gp16 form a spiral ring complex that binds DNA and a lipid membrane. Virology486, 263–71 (2016). [DOI] [PubMed] [Google Scholar]
- 71.Delvillani, F. et al. Tet-Trap, a genetic approach to the identification of bacterial RNA thermometers: application to Pseudomonas aeruginosa. RNA20, 1963–76 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Deho, G., Zangrossi, S., Sabbattini, P., Sironi, G. & Ghisotti, D. Bacteriophage P4 immunity controlled by small RNAs via transcription termination. Mol. Microbiol6, 3415–25 (1992). [DOI] [PubMed] [Google Scholar]
- 73.Scheres, S. H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol.180, 519–30 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife7, e42166(2018). [DOI] [PMC free article] [PubMed]
- 75.Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 76.Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rohou, A. & Grigorieff, N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol.192, 216–21 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Conesa Mingo, P. et al. Scipion web tools: Easy to use cryo-EM image processing over the web. Protein Sci.27, 269–275 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pettersen, E. F. et al. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput Chem.25, 1605–12 (2004). [DOI] [PubMed] [Google Scholar]
- 80.Terwilliger, T. C., Sobolev, O. V., Afonine, P. V. & Adams, P. D. Automated map sharpening by maximization of detail and connectivity. Acta Crystallogr D. Struct. Biol.74, 545–559 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Goddard, T. D. et al. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci.27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr D. Biol. Crystallogr60, 2126–32 (2004). [DOI] [PubMed] [Google Scholar]
- 83.Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr D. Struct. Biol.74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Davis, I. W. et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res35, W375–83 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol.215, 403–10 (1990). [DOI] [PubMed] [Google Scholar]
- 86.Edgar, R. C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinforma.5, 113 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gouy, M., Tannier, E., Comte, N. & Parsons, D. P. Seaview version 5: a multiplatform software for multiple sequence alignment, molecular phylogenetic analyses, and tree reconciliation. Methods Mol. Biol.2231, 241–260 (2021). [DOI] [PubMed] [Google Scholar]
- 88.Talavera, G. & Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol.56, 564–77 (2007). [DOI] [PubMed] [Google Scholar]
- 89.Guindon, S. et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol.59, 307–21 (2010). [DOI] [PubMed] [Google Scholar]
- 90.Le, S. Q. & Gascuel, O. An improved general amino acid replacement matrix. Mol. Biol. Evol.25, 1307–20 (2008). [DOI] [PubMed] [Google Scholar]
- 91.DeLano, W. L. et al. The PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC. (2002).
- 92.Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol.372, 774–97 (2007). [DOI] [PubMed] [Google Scholar]
- 93.Laskowski, R. A. PDBsum new things. Nucleic Acids Res37, D355–9 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Maiti, R., Van Domselaar, G. H., Zhang, H. & Wishart, D. S. SuperPose: a simple server for sophisticated structural superposition. Nucleic Acids Res32, W590–4 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Altschul, S. F. & Koonin, E. V. Iterated profile searches with PSI-BLAST-a tool for discovery in protein databases. Trends Biochem Sci.23, 444–7 (1998). [DOI] [PubMed] [Google Scholar]
- 96.Madeira, F. et al. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res50, W276–W279 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Stothard, P. The sequence manipulation suite: javascript programs for analyzing and formatting protein and DNA sequences. Biotechniques28, 1104 (2000). [DOI] [PubMed] [Google Scholar]
- 98.Kang, X., Alibakhshi, M. A. & Wanunu, M. One-pot species release and nanopore detection in a voltage-stable lipid bilayer platform. Nano Lett.19, 9145–9153 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates for phage DEV coat protein (C5), portal:HT-adapter:tail tube complex (C12), pre-ejection gp72 (C12), gp53 NTB (C15), and gp72:gp73 complex (C9) have been deposited in the Protein Data Bank with accession codes 9BGN, 9BGM, 9BGO, 9COD and 8VXQ. The relative cryo-EM density maps have been deposited in the Electron Microscopy Data Bank with accession codes EMD-44518, EMD-44517, EMD-44519, EMD-45776 and EMD-43629. Source data are provided in this paper.