Abstract
Cytopathic, T-cell-tropic feline leukemia viruses (FeLV-T) evolve from FeLV-A in infected animals and demonstrate host cell specificities that are distinct from those of their parent viruses. We recently identified two cellular proteins, FeLIX and Pit1, required for productive infection by these immunodeficiency-inducing FeLV-T variants (M. M. Anderson, A. S. Lauring, C. C. Burns, and J. Overbaugh, Science 287:1828–1830, 2000). FeLV-T is the first example of a naturally occurring type C retrovirus that requires two proteins to gain entry into target cells. FeLIX is an endogenous protein that is highly related to the N-terminal portion of the FeLV envelope protein, which includes the receptor-binding domain. Pit1 is a multiple-transmembrane phosphate transport protein that also functions as a receptor for FeLV-B. The FeLV-B envelope gene is derived by recombination with endogenous FeLV-like sequences, and its product can functionally substitute for FeLIX in facilitating entry through the Pit1 receptor. In the present study, we tested other retrovirus envelope surface units (SUs) with their cognate receptors to determine whether they also could mediate infection by FeLV-T. Cells were engineered to coexpress the transmembrane form of the envelope proteins and their cognate receptors, or SU protein was added as a soluble protein to cells expressing the receptor. Of the FeLV, murine leukemia virus, and gibbon ape leukemia virus envelopes tested, we found that only those with receptor-binding domains derived from endogenous FeLV could render cells permissive for FeLV-T. We also found that there is a strong preference for Pit1 as the transmembrane receptor. Specifically, FeLV-B SUs could efficiently mediate infection of cells expressing the Pit1 receptor but could only inefficiently mediate infection of cells expressing the Pit2 receptor, even though these SUs are able to bind to Pit2. Expression analysis of feline Pit1 and FeLIX suggests that FeLIX is likely the primary determinant of FeLV-T tropism. These results are discussed in terms of current models for retrovirus entry and the interrelationship among FeLV variants that evolve in vivo.
Genetic variation is one of the hallmarks of a retrovirus infection. This genetic plasticity allows the transmitted strain to respond to selective pressures and persist within an infected host. For example, changes in the gene coding for the envelope protein may facilitate immune system escape or broaden the cell tropism of the virus by altering envelope receptor recognition. Viral variation also facilitates the evolution of pathogenic viruses that cause disease in the host. For retroviruses that cause immunodeficiency, disease onset is correlated with the evolution of more cytopathic, T-cell-tropic variants. This progression has been observed not only in lentiviruses, such as the human and simian immunodeficiency viruses (24, 41), but also in the emergence of T-cell-tropic feline leukemia virus (FeLV-T) variants in infected cats (39). For FeLV-T, T-cell tropism is the result of changes in the viral envelope protein, and the envelope has been shown to be the major pathogenic determinant for immunodeficiency-inducing FeLV-T variants (12, 32, 33).
FeLV was originally classified into three receptor interference subgroups (A, B, and C) (42). FeLV-A is considered to be the ecotropic, transmissible form of FeLV, and it is not acutely pathogenic (40). FeLV-B arises in vivo through recombination between FeLV-A and endogenous FeLV-like sequences (enFeLV) (9, 34, 43). Acquisition of enFeLV sequences encoding portions of the envelope surface unit (SU) leads to changes in cell tropism that result from a change in receptor specificity (8, 44). This reflects the fact that the region in the FeLV-B envelope SU that is thought to be the receptor binding domain (RBD) is encoded by enFeLV in these recombinant retrovirus genomes (8). All FeLV-Bs use the phosphate transport protein, Pit1, as a receptor, but recent work indicates that subtle differences in the lengths and compositions of the enFeLV-derived sequences allow certain variants to efficiently use a related protein, Pit2, as a receptor (8; 43a; M. M. Anderson, A. S. Lauring, S. Roberston, C. Dirks, and J. Overbaugh, unpublished data).
The T-cell-tropic variants appear to constitute a distinct subgroup (29). The sequence of the FeLV-T envelope is most closely related to FeLV-A, and FeLV-T variants evolve from FeLV-A during the course of an in vivo infection (38, 39). The envelope gene of the pathogenic FeLV-T molecular clone, 61C, encodes an N-terminal 6-amino-acid deletion, a C-terminal 6-amino-acid insertion, and 11 scattered amino acid changes compared to the envelope gene of the relatively nonpathogenic FeLV-A-61E (32). Of these changes, the region encompassing the insertion and one or more of the N-terminal changes are the major determinants of T-cell tropism and cytopathicity (12, 16). Despite this similarity in envelope gene sequence, data from superinfection interference assays suggest that FeLV-A-61E and FeLV-T-61C use distinct cell surface receptors to gain entry into target cells (29). We recently reported the identification of a cellular cofactor that is necessary for infection by FeLV-T (2). This protein, FeLIX (for feline leukemia virus infection “x-cessory” factor), is expressed from endogenous FeLV-like sequences and corresponds to a truncated version of the FeLV SU. Consistent with the recombinatorial origin of FeLV-B envelopes, FeLIX is nearly identical (95%) in sequence to the RBD of the FeLV-B envelope. FeLIX is necessary but not sufficient for FeLV-T infection, acting in concert with Pit1, the FeLV-B receptor, to permit infection by FeLV-T (2).
Because of its requirement for two proteins to infect cells, FeLV-T may provide a new model for entry by simple, type C retroviruses. For most simple retroviruses, interactions between the viral envelope protein and transmembrane cellular receptor are thought to be both necessary and sufficient for entry (17). The SU of the envelope specifically binds the cell surface receptor. Receptor binding triggers conformational changes in the envelope that lead to activation of the fusion machinery. While the transmembrane (TM) subunit of the envelope contains the fusion peptide and mediates the actual process of fusion, recent work has identified amino acids in the N terminus and C-terminal half of retrovirus SUs that are necessary for postbinding events (4, 21, 22, 51). Interestingly, FeLV-T-61C has two substitutions in the N-terminal domain and an insertion in the C-terminal domain relative to FeLV-A-61E (32).
To better understand the mechanism of FeLV-T entry, we have attempted to define the cofactor and receptor requirements for FeLV-T infection. Specifically, we determined whether other retrovirus envelope SUs could facilitate FeLV-T infection when bound to their cognate receptors. Here we show that only FeLIX and the FeLV-B SU can efficiently function as cofactors for FeLV-T infection. Similarly, we also found that FeLV-T has a strong preference for Pit1 as a receptor because related receptors do not efficiently mediate FeLV-T infection even when their cognate envelopes are supplied as cofactors. Together, our data indicate that FeLV-T entry is a specific process, mediated by particular receptor and cofactor combinations.
MATERIALS AND METHODS
Cell culture and viruses.
Molecular clones FeLV-A-61E, FeLV-B-90Z [called EE(Z1-5)E in reference 8], FeLV-B-90ZRBD [called EE(Z1-4)E in reference 8], and FeLV-T-61C have been described previously (8, 9, 32, 43a). AH927 feline fibroblasts and 293T human embryonic kidney fibroblasts were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 100 U of penicillin per ml, 100 μg of streptomycin per ml, and 0.25 μg of amphotericin B per ml (complete DMEM). The construction of MDTF cell lines expressing human Pit1 (HuPit1), HuPit2, feline Pit1 (FePit1), and FePit2 is described elsewhere (13; Anderson et al., unpublished data). MDTF-Pit cell lines were maintained in complete DMEM containing 0.6 mg of G418/ml.
Construction of MLV genomes containing retrovirus envelope genes.
Envelope genes were cloned into murine retrovirus genome pLXSH (26), which contains the gene for hygromycin phosphotransferase (a gift from A. D. Miller, Fred Hutchinson Cancer Research Center). The inserts for L(90Z)SH and L(90ZRBD)SH were generated by digestion of envelope subclones pcDNA3.1-90Zenv and pcDNA3.1-90ZRBDenv (43a) with XhoI and BamHI, which cut 163 bases before the envelope translation start site and at the end of the envelope TM subunit, respectively. The insert for L(GALV)SH was generated by digestion of envelope expression construct CIGASenv (46) with XhoI and BamHI, which cut 34 bases before the envelope start site and 2.4 kb downstream of the envelope termination codon, respectively. These fragments were ligated into pLXSH using these same restriction sites. For L(GALV)SH, a 2.3-kb 3′ noncoding BamHI/NotI fragment was subsequently removed to reduce the size of the retrovirus genome. The insert for L(A-MLV)SH was amplified by PCR from pPAM3 (25), a gift from A. D. Miller, using primers containing SalI and BglII sites at their 5′ termini. These primers span the amphotropic murine leukemia virus (A-MLV) envelope start site and termination codon, respectively. The SalI- and BglII-digested PCR product was ligated into pLXSH using the XhoI and BamHI restriction enzyme sites.
Constructs with FeLIX and SUs containing HA epitope tags.
Constructs encoding retrovirus SUs but not the TM domain were generated in a vector (CS2-HA [10]; a gift from S. Tapscott, Fred Hutchinson Cancer Research Center) that contains two copies of the hemagglutinin (HA) epitope tag. Fragments encoding envelope amino acids 1 to 435 of FeLV-A-61E, 1 to 455 of FeLV-B-90Z, 1 to 444 of FeLV-B-90ZRBD, and 1 to 448 of A-MLV (4070A) were amplified by PCR from either full-length proviral clones or envelope subclones using primers containing SacI sites at their 5′ termini. The SacI-digested PCR product was ligated into the SacI site of CS2-HA, and clones were screened to select for those in the correct orientation. The resulting CS2-SU-HA plasmids code for the envelope signal peptide, the entire SU except for the last 10 amino acids (to avoid inclusion of the SU/TM cleavage site), and two copies of the HA epitope in frame at the C terminus. CS2-GALV-SU1–262-HA was made using a similar SacI-based PCR cloning strategy but encodes only amino acids 1 to 262 of the gibbon ape leukemia virus (GALV) SU followed by an identical HA tag. The complete FeLIX open reading frame was amplified by PCR from pCR3.1-FeLIX (2) using primers containing SacI sites at their 5′ termini and inserted into CS2-HA as described above. All constructs generated by PCR were verified by nucleotide sequence analyses.
Preparation of viral supernatants and SU conditioned media.
Viral pseudotypes containing MLV genomes with drug resistance (see below) or reporter genes were generated by transient transfection of 293T cells using a calcium phosphate protocol. Cells were plated 24 h prior to transfection at a density of 1 × 106 to 2.0 × 106 cells per 10-cm-diameter dish. For FeLV-T-61C pseudotypes, cells were transfected with 5 μg of Δpsi-EECC (29) and 5 μg of the retrovirus genome. For all other pseudotypes, cells were transfected with 3.3 μg each of an FeLV 61E-LTR-Δpsi-gag-pol construct (43a), a retrovirus genome, and an envelope expression construct (e.g., CIGASenv, pcDNA3.1-90Zenv, pcDNA3.1-90ZRBDenv, or pSV-AmphoEnv). For pseudotypes used in single-cycle infection assays, we used a murine retrovirus genome containing the gene for β-galactosidase (pRT43.2Tnlsβgal1), provided by M. Eiden (National Institutes of Health). Supernatants were harvested 48 h posttransfection and purified through 0.22-μm-pore-size filters.
Conditioned media containing soluble retrovirus SUs and FeLIX-HA were also generated by transient transfection of 293T cells. For each SU, 10 μg of CS2-SU-HA, CS2-GALV-SU1–262-HA, or CS2-FeLIX-HA plasmid were used. Supernatants were harvested 48 h posttransfection as described above.
Generation of stable cell lines expressing retrovirus envelope proteins.
MDTF-HuPit1, -HuPit2, -FePit1, and -FePit2 (13; Anderson et al., unpublished data) were transduced with A-MLV pseudotypes that packaged murine retrovirus genomes L(90Z)SH, L(90ZRBD)SH, L(GALV)SH, and L(A-MLV)SH. For each Pit-envelope cell line, polyclonal pools were selected and maintained in complete DMEM containing 0.6 mg of G418 and 500 U of hygromycin B/ml.
Infection assays.
Target cells were plated at 1 × 104 to 2.0 × 104 cells per well in 24-well dishes approximately 24 h prior to infection. On the day of infection, the culture medium was replaced with new medium containing 4 μg of Polybrene/ml. In cases in which FeLIX- or SU-conditioned media were used, these supernatants were diluted 1:1 in new medium (i.e., 500 μl conditioned medium in a 1,000-μl total volume) unless otherwise indicated. For the infections in Table 1, FeLIX-conditioned media were harvested from D17 cells expressing FeLIX cDNA (D17-FeLIX) as described in reference 2. Cells were infected with viral pseudotypes that packaged murine retrovirus genome pRT43.2Tnlsβgal1 at a range of dilutions and stained for β-galactosidase expression 48 h postinfection, as described previously (20).
TABLE 1.
Infection of MDTF cells stably expressing Pit receptors in the presence or absence of FeLIX
| Receptor | FeLV-T-61C titer (FFU/ml)a
|
|
|---|---|---|
| No FeLIX | With FeLIX | |
| HuPit1 | <10b | 9.5 × 105 |
| HuPit2 | <10 | <10 |
| FePit1 | <10 | 2.9 × 106 |
| FePit2 | <10 | <10 |
Titers based on challenge with vectors packaging a genome containing the gene for β-galactosidase. FFU, Focus-forming units.
<10 FFU/ml, no blue foci were observed in cells infected with up to 100 μl of cell-free viral supernatant.
Immunoprecipitation and Western blot analysis.
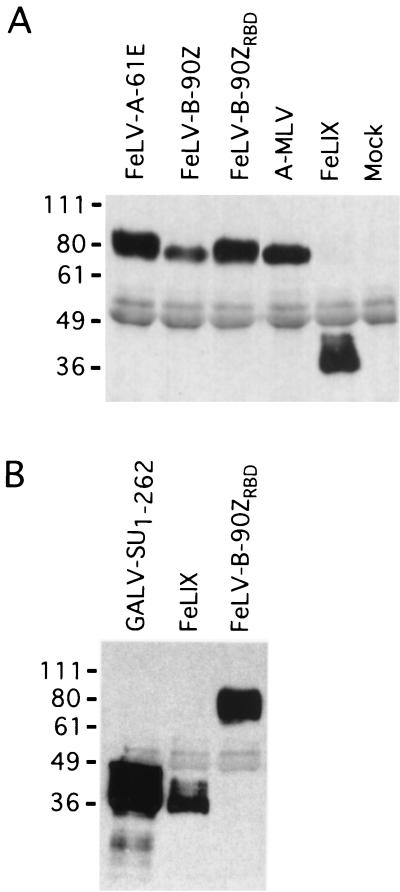
An ascites fluid concentrate of monoclonal antibody HA.11 (Covance, Berkeley, Calif.) was prebound to either protein G-Sepharose (Fig. 3A) or protein A-Sepharose (Fig. 3B) for 2 h at 4°C. One milliliter of conditioned medium containing each HA-tagged SU was precleared with 300 μl of 50% protein A-Sepharose suspension for 3 h at 4°C. Supernatants were then immunoprecipitated with prebound antibody-bead complexes (approximately 7 μl of ascites fluid per reaction) for 2.5 h at 4°C. Immunoprecipitates were washed three times in phosphate-buffered saline (PBS)–0.1% Triton X-100–0.1% NP-40, and bound proteins were eluted by boiling them for 5 min in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer. Proteins were resolved on an SDS-10% PAGE gel and transferred to Immobilion polyvinylidene difluoride membranes (Millipore, Bedford, Mass.). Western blot analysis was performed using a rabbit polyclonal HA.11 antibody (Covance) and horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (Bio-Rad, Hercules, Calif.). Bound antibodies were detected by chemiluminescence (Amersham, Piscataway, N.J.).
FIG. 3.
Detection of HA-tagged retrovirus SUs in conditioned media. (A) Human embryonic kidney 293T cells were transfected with constructs expressing the indicated SU proteins, and cell-free supernatants were harvested 48 h posttransfection. SUs were immunoprecipitated from 1 ml of each supernatant using a monoclonal antibody directed against the HA epitope. One-half of each immunoprecipitate was resolved by SDS-PAGE and analyzed by Western blotting using a polyclonal antibody directed against the same epitope. Mock, sample of cell-free supernatant from cells transfected with an untagged version of FeLIX. Molecular mass markers (in kilodaltons) are indicated to the left. (B) GALV-SU1–262 encodes amino acids 1 to 262 of the GALV SU with a C-terminal HA tag. Immunoprecipitation and Western blot analysis were performed as for panel A except that only one-fifth of each immunoprecipitate was loaded on the gel.
Flow cytometry and SU binding assay.
Cells were washed in PBS, detached from the dish by incubation in PBS–5 mM EDTA, pelleted, and resuspended in complete DMEM. For analysis of surface expression of FeLV envelope proteins, 106 cells were washed once in WB (Hanks buffered saline solution with magnesium and calcium–2% fetal bovine serum). For each wash, cells were pelleted by centrifugation for 5 min in either a swinging-bucket clinical centrifuge at 450 × g or a microcentrifuge at 850 × g and then resuspended in 1 ml of WB and pelleted again. Washed cells were resuspended in 200 μl of WB containing 4 μg of anti-FeLV gp70 monoclonal antibody C11D8 (Custom Monoclonal Antibodies, Sacramento, Calif.). Cells were stained for 1.5 h at 4°C. The cells were then washed twice, resuspended in 150 μl of a 1:100 dilution of r-phycoerythrin-conjugated goat anti-mouse antibody (DAKO, Carpinteria, Calif.), and incubated at 4°C for 45 min. The stained cells were washed again as described before, resuspended in 300 to 500 μl WB, and analyzed using a fluorescence-activated cell sorter (Becton Dickinson, San Diego, Calif.). Dead cells, clumps, and debris were excluded based on forward and side scatter.
For the SU binding assay, cells were detached and washed as described above. One million cells were then incubated with 1 to 500 μl of SU-containing supernatant in a 1-ml total volume at 37°C for 45 min on a rocking platform. The cells were then pelleted and washed with 1 ml of WB. Washed cells were resuspended in 200 μl of a 1:1,000 dilution of an ascites fluid concentrate of monoclonal antibody HA.11 (Covance) and incubated at 4°C for 1 to 1.5 h. Washing, secondary antibody staining, and analysis were performed as described above.
RNA isolation and Northern blot analysis.
Total cellular RNA was harvested from tissues of two FeLV-negative cats. T cells and monocytes were prepared from one of these animals. T cells were prepared by culturing peripheral blood mononuclear cells in the presence of 5 μg of concanavalin A/ml for 3 days followed by culturing with recombinant human interleukin 2 (100 U/ml) for 5 days (1). Monocytes were prepared by culturing marrow mononuclear cells in the presence of recombinant human macrophage colony-stimulating factor (1.5 ng/ml) and recombinant human Flt-3L (100 ng/ml) for 6 to 7 days (19). Feline tissues and cells were kindly provided by J. Abkowitz (University of Washington). RNEasy kits (Qiagen, Valencia, Calif.) were used for RNA isolation from brain, small intestine, monocytes, and T cells. Trizol reagent (Gibco BRL, Grand Island, N.Y.) was used for muscle, kidney, liver, spleen, and lymph node preparations. Ten micrograms of each sample was mixed with 3 volumes of formaldehyde-MOPS (morpholinepropanesulfonic acid) gel loading solution (Ambion, Austin, Tex.) and electrophoresed through a 1% agarose-formaldehyde-MOPS gel. Gels were equilibrated in 20× SSC (1×SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and transferred to nylon membranes by capillary action. 32P-labeled probes were generated by a random-oligomer technique (Amersham) and purified using spin columns. The FeLIX probe was made using an EcoRI fragment from pCR3.1-FeLIX and corresponds to the full-length FeLIX cDNA. The FePit1 probe was made using a 1.4-kb HindIII/BglII fragment from the coding region of the cDNA. Hybridizations were carried out overnight at 65°C in a mixture containing 7% SDS, 0.25 M Na2HPO4, 1 mM EDTA, 1% (wt/vol) bovine serum albumin, 100 μg of salmon sperm DNA/ml, and 106 cpm of probe per ml of buffer. Membranes were washed twice at room temperature with 2× SSC–0.1% SDS, followed by two washes at 58°C with 0.1× SSC–0.1% SDS, and exposed to film with intensifying screens. Filters were stripped of labeled probe according to standard protocols (3).
RESULTS
FeLIX requires Pit1 to facilitate FeLV-T infection.
Although host range and interference studies initially identified Pit1 as an FeLV-B receptor (45), we have shown that certain FeLV-Bs can also utilize the HuPit2 protein (8, 43a). Moreover, all FeLV-Bs examined to date can infect cells using the FePit2 protein, suggesting that FeLV-B is more like the dual-tropic 10A1 group of MLVs in its natural host (Fig. 1) (8, 28; Anderson et al., unpublished data). Because FeLIX is nearly identical to these FeLV-B SUs within the presumed receptor recognition domain, we asked whether Pit2 could also function as a receptor for FeLV-T. We analyzed receptor usage using an assay that detects a single cycle of infection in which we exposed cells to FeLV-T pseudotypes that had packaged a murine retrovirus genome containing the lacZ reporter gene. MDTF cells expressing HuPit1 are susceptible to FeLV-B but resistant to FeLV-T infection. We previously reported that these cells are highly susceptible to infection by FeLV-T when FeLIX is supplied at the time of infection (2). Here we show that MDTF-FePit1 cells are also susceptible to FeLV-T infection in the presence of FeLIX (Table 1). However, we found that MDTFs expressing HuPit2 or FePit2 remained resistant to FeLV-T infection. Therefore, FeLIX-mediated FeLV-T infection is specific to cells expressing Pit1.
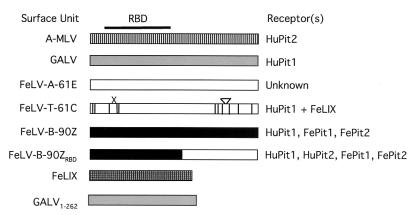
FIG. 1.
Summary of the receptor specificities of feline, murine, and primate type C retrovirus envelopes used in this study. The SU for each virus is shown in schematic form. A-MLV, SU from A-MLV; GALV, SU from GALV. White boxes, FeLV-A-derived sequences; vertical lines, appropriate positions where FeLV-A-61E and FeLV-T-61C differ. The six-amino-acid deletion (X) and six-amino-acid insertion (inverted triangle) in FeLV-T-61C are also shown. FeLV-B-90Z, SU of the 90Z molecular clone. 90ZRBD is a chimeric envelope containing amino acids 1 to 244 from 90Z and the rest from 61E. The codons for FeLIX are 95% identical to those for 90Z within the portion of the envelope gene coding for the mature SU (2). The approximate location of the RBD, as defined for MLVs, is indicated at the top (6). The receptor specificity of each of the viral envelopes is indicated to the right (27, 28, 31, 45, 48, 50). Receptor usage by FeLV-Bs is as described previously (8; Anderson et al., unpublished data).
FeLV-T can only efficiently infect cells expressing Pit1 and an FeLV-B envelope protein.
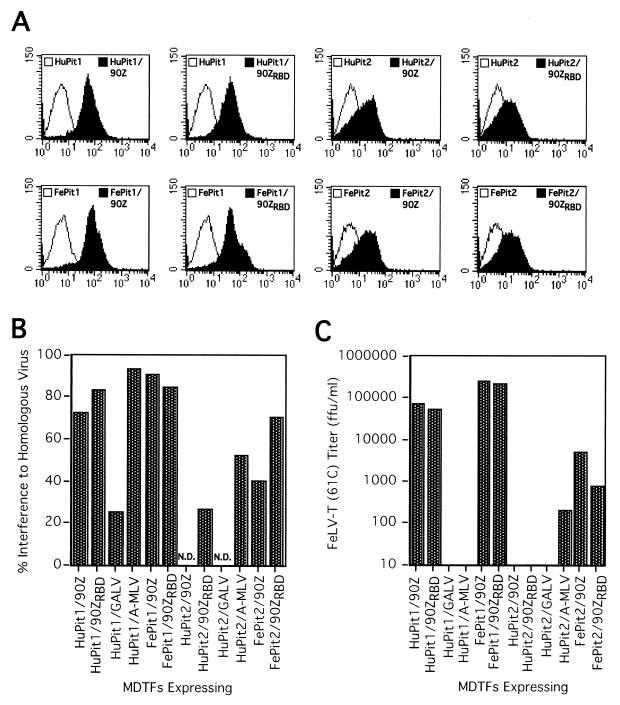
Cells expressing Pit1 and the FeLV-B envelope are susceptible to infection by FeLV-T in the absence of FeLIX, indicating that the FeLV-B SU can functionally substitute for FeLIX (2). Because FeLV-B can also infect cells using Pit2 as a receptor, we asked whether substitution of the FeLV-B envelope for FeLIX could expand the Pit receptor specificity of FeLV-T. Specifically, the 90Z envelope can recognize FePit2, and a chimeric envelope containing a smaller portion of the enFeLV-related sequences (90ZRBD) can utilize both FePit2 and HuPit2 as receptors (8; Anderson et al., unpublished data). We generated MDTF cell lines that express various combinations of Pit receptors (HuPit1, HuPit2, FePit1, and FePit2) and FeLV-B envelopes (90Z and 90ZRBD). The SU could be detected on the surface of each of these cell lines by flow cytometry, although we observed higher surface staining of SU in the HuPit1 and FePit1 cell lines (threefold higher; Fig. 2A). There was partial interference (25 to 90%) to homologous viral challenge in these cell lines, suggesting that, while some surface receptors were bound by SU, others remained available for infection by FeLV-T (Fig. 2B). MDTFs expressing HuPit1 or FePit1 and the FeLV-B envelope were susceptible to infection by FeLV-T when either the 90Z or 90ZRBD envelope was expressed in the target cells (Fig. 2C). However, we did not observe infection of cells expressing HuPit2 with any FeLV-B envelope. This was true even with coexpression of the 90ZRBD envelope, which recognizes the HuPit2 receptor for infection (8). We did detect FeLV-T infection of cells expressing FePit2 and the FeLV-B envelopes, although the infectivity was 50- to 250-fold lower than on corresponding FePit1–FeLV-B envelope cells. We therefore conclude that there is a strong preference for Pit1 as a receptor by these T-tropic variants.
FIG. 2.
Expression and infection analyses of cell lines stably expressing retrovirus envelopes and Pit proteins. (A) Histograms from flow-cytometric analyses of cells stained with monoclonal antibody C11D8, which recognizes an epitope in the SU common to FeLV subgroups A, B, and T (15). In all cases, the x axis is fluorescence intensity (log scale) and the y axis is cell number. Open profiles, parental MDTF-Pit cell lines stained with C11D8 and the secondary antibody; filled profiles, MDTFs expressing envelopes and Pit proteins (upper right corner) stained with C11D8 and the secondary antibody. (B) Interference in stable cell lines expressing Pit-envelope combinations to challenge with homologous virus. Titers were measured as β-galactosidase focus-forming units per milliliter as in Table 1. Percent interference was calculated as (1 − [titer of virus on Pit cell line/titer of virus on Pit-envelope cell line]) × 100. For example, the percent interference by the 90Z envelope in HuPit1–90Zenv cells would be (1 − [FeLV-B-90Z titer on HuPit1 cells/FeLV-B-90Z titer on HuPit1–90Zenv cells]) × 100. N.D., no data, because HuPit2 cells and derivatives are not susceptible to infection by FeLV-B-90Z and GALV. (C) FeLV-T-61C titer in focus-forming units (ffu) per milliliter on stable cell lines expressing Pit-envelope combinations. The data in both panels are representative of at least two independent experiments.
Other retrovirus envelope proteins that use Pit receptors do not facilitate FeLV-T infection.
We next asked whether coexpression of other retrovirus envelopes and their cognate receptors could also facilitate FeLV-T infection. We focused on the GALV and A-MLV envelopes, which utilize HuPit1 and HuPit2 as receptors, respectively (27, 31, 48). Because specific antibodies to these envelope proteins were not available, we could only indirectly measure surface envelope expression in HuPit1-GALV envelope and HuPit2–A-MLV envelope cell lines by using interference. In these cells, we observed partial interference (25 to 50%), as was seen with cells expressing the FeLV envelopes (Fig. 2B). We were also able to rescue a low level of infectious virus from both the GALV envelope and A-MLV envelope cell lines by cotransfecting them with a retrovirus genome and a construct expressing Gag and Pol. This suggests that the envelope is localized to the cell surface (data not shown). In contrast to cells expressing Pit1 and the FeLV-B envelope, cells expressing HuPit1 and the GALV envelope remained resistant to FeLV-T infection (Fig. 2C). Cells expressing HuPit1 and the A-MLV envelope were also resistant to infection, whereas those expressing HuPit2 and the A-MLV envelope permitted a very low level of FeLV-T infection, slightly above background and up to 1,000-fold lower than that permitted by cells expressing FeLV-B envelopes. From these experiments, in which the oligomeric, membrane-associated form of retrovirus envelopes were coexpressed with Pit receptors, we conclude that only the FeLV-B envelope and Pit1 can efficiently mediate FeLV-T infection.
Analysis of FeLV-T cofactor activity by soluble retrovirus SUs.
While FeLIX is similar to the membrane-bound FeLV-B envelope protein, it is secreted from cells and facilitates FeLV-T infection as a soluble cofactor (2). We therefore tested whether the SUs of the retrovirus envelope proteins used above could facilitate FeLV-T infection when supplied, like FeLIX, as soluble proteins in conditioned media. We generated constructs that express nearly the complete SU of each envelope with a C-terminal HA epitope tag. For comparison, we also created an HA-tagged version of FeLIX. Conditioned media containing FeLIX-HA could mediate FeLV-T infection of Pit1 cells, suggesting that the HA tag did not disrupt its function as a cofactor (data not shown). Conditioned media from cells expressing the tagged retrovirus SUs were harvested from transiently transfected cells. Western blot analysis of immunoprecipitates from these conditioned media showed that there were similar levels of the various SUs in the conditioned media, although 90Z SU levels were slightly lower (Fig. 3A).
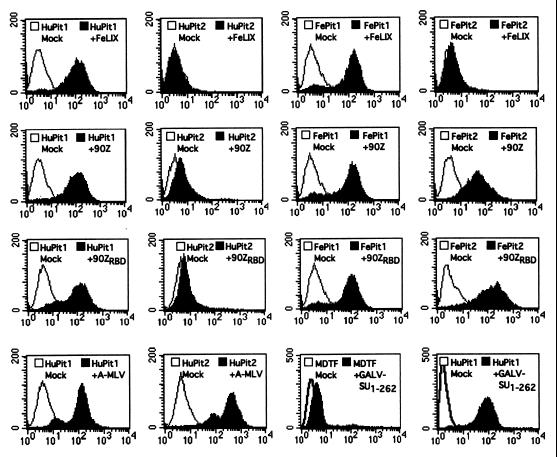
Binding studies were performed with these HA-tagged SUs to confirm that the C-terminal epitope did not alter the ability of the proteins to bind their cognate receptor(s). By flow-cytometric analyses, we could detect FeLIX-HA binding to both HuPit1 and FePit1 but not to either Pit2 homologue (Fig. 4). These binding data may provide insight into the results of the infection studies, which showed that FeLIX can only mediate FeLV-T infection of cells expressing Pit1, not Pit2. The FeLV-B-90Z and -90ZRBD SUs specifically bound to HuPit1 and both feline Pit proteins, consistent with analyses of virus binding and receptor usage (Anderson et al., unpublished data). The A-MLV SU bound MDTF-HuPit1 cells, which most likely reflects binding to the endogenous murine Pit2 protein (49). Consistent with this observation, a greater shift in fluorescence intensity in cells expressing HuPit2 in addition to the endogenous Pit2 protein was observed.
FIG. 4.
Receptor-binding properties of HA-tagged SUs. MDTF or MDTF-Pit cells were incubated with 500 μl of conditioned medium containing the SUs indicated (upper right corner) as described in Materials and Methods. Bound SUs were detected by staining with a monoclonal antibody directed against the HA epitope. In all cases the x axis is fluorescence intensity (log scale) and the y axis is cell number. Mock samples (open profiles), cells incubated in standard media and stained with the same antibody. Pit receptor and SU abbreviations (upper right corner) are as described in the legends to Fig. 1 and 3.
Both of the FeLV-B SUs (90Z and 90ZRBD) could mediate infection of MDTFs expressing either HuPit1 or FePit1 (Table 2). In contrast, neither of the FeLV-B SUs could facilitate FeLV-T infection of FePit2 cells even though both the 90Z and 90ZRBD envelopes can efficiently bind FePit2 (Fig. 4). The 90ZRBD SU, which can recognize HuPit2 (8), could not mediate FeLV-T infection of HuPit2 cells. In a similar manner, addition of the A-MLV SU did not allow FeLV-T infection of MDTFs expressing any of the Pit proteins even though the A-MLV SU can bind to all of the cell lines used.
TABLE 2.
Infection of cells in the presence of conditioned media containing SUs
| Receptor | SU-HAa | FeLV-T-61C titer (FFU/ml)b |
|---|---|---|
| HuPit1 | FeLV-B-90Z | 3.7 × 105 |
| HuPit1 | FeLV-B-90ZRBD | 2.3 × 105 |
| HuPit1 | A-MLV | <10c |
| HuPit1 | GALV-SU1–262 | <10 |
| HuPit1 | FeLIX | 3.1 × 105 |
| HuPit2 | FeLV-B-90Z | <10 |
| HuPit2 | FeLV-B-90ZRBD | <10 |
| HuPit2 | A-MLV | <10 |
| HuPit2 | GALV-SU1–262 | <10 |
| HuPit2 | FeLIX | <10 |
| FePit1 | FeLV-B-90Z | 8.0 × 105 |
| FePit1 | FeLV-B-90ZRBD | 1.0 × 106 |
| FePit1 | A-MLV | <10 |
| FePit1 | FeLIX | 9.2 × 105 |
| FePit2 | FeLV-B-90Z | 20 |
| FePit2 | FeLV-B-90ZRBD | <10 |
| FePit2 | A-MLV | 20 |
| FePit2 | FeLIX | <10 |
Conditioned media containing the indicated SUs were harvested from transiently transfected 293T cells, diluted 1:1, and added at the time of infection.
Titers based on challenge with vectors packaging a genome containing the gene for β-galactosidase. FFU, focus-forming units.
<10 FFU/ml, no blue foci were observed in cells infected with up to 100 μl of cell-free viral supernatant.
Because we were unable to express an HA-tagged version of the full-length GALV SU and because specific antibodies to the GALV envelope were not available, we cannot unequivocally state from the above experiments that the GALV SU does not function with Pit1 as an FeLV-T entry cofactor. To further assess cofactor activity of the GALV SU, we expressed a truncated version of the GALV SU encoding amino acids 1 to 262 with a C-terminal HA epitope tag. This SU fragment, GALV-SU1–262, is similar in size to FeLIX and encompasses the GALV RBD (Fig. 3B). We detected a low level of binding by GALV-SU1–262 to MDTF cells, which likely represents a low-affinity interaction between this SU fragment and the endogenously expressed murine Pit1 (Fig. 4). When the same supernatant was applied to MDTF-HuPit1 cells, we observed a large shift in fluorescence intensity, representing specific binding of the GALV SU fragment to HuPit1. While GALV-SU1–262 could bind Pit1, it was not able to mediate FeLV-T infection of MDTF-HuPit1 cells (Table 2). Taken together with studies of MDTF-Pit1 cells expressing oligomeric GALV Env, these data suggest that, among SUs that bind to Pit1, only those containing sequences derived from endogenous FeLV are able to facilitate FeLV-T infection
The affinity of the soluble cofactor for the transmembrane receptor is not the determinant for FeLV-T receptor specificity.
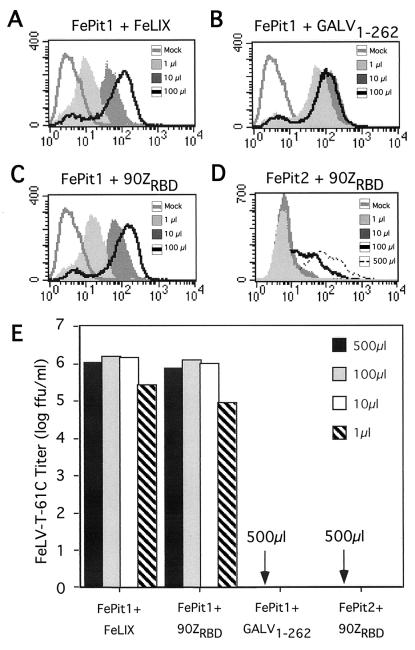
We considered the possibility that the observed specificity in FeLV infection could simply reflect differences in affinities among the soluble cofactors and their respective receptors. To test this hypothesis, we performed cofactor binding and FeLV-T infection assays using different concentrations of various soluble cofactors. FeLIX binding to FePit1 served as a positive control because this is a functional receptor complex for FeLV-T and is presumably the one used by FeLV-T for replication in the cat. We observed that the binding of FeLIX to FePit1 was dose dependent; we could still detect a significant shift in fluorescence intensity with as little as 1 μl of conditioned medium (Fig. 5A). Importantly, the measured levels of binding using 100 and 500 μl of FeLIX supernatant were similar, suggesting that the binding reaction reached saturation at these levels (data not shown). We found that the FeLV-B-90ZRBD SU also bound FePit1 in a dose-dependent and saturable manner, with binding detected over a 500-fold range of SU concentrations (1 to 500 μl; Fig. 5C). Both FeLIX and the 90ZRBD SUs could efficiently mediate FeLV-T infection on MDTF-FePit1 cells in this concentration range, with titers of approximately 105 when as little as 1 μl of conditioned medium containing either cofactor was used (Fig. 5E). We were also able to detect significant binding of the FeLV-B-90ZRBD SU to FePit2 with 100 μl of conditioned medium, although we could not detect binding with 1 to 10 μl (Fig. 5D). When 500 μl of supernatant containing FeLV-B 90ZRBD was used for the binding experiment with FePit2, the shift in fluorescence was similar to that observed for FePit1 with 10 to 100 μl of the same SU. Yet as little as 1 μl of FeLV-B 90ZRBD SU permits entry of FeLV-T with FePit1, while 500 μl of the same supernatant does not permit entry via FePit2 (Fig. 5E). This suggests that even when similar amounts of this FeLV-B SU are bound to the Pit receptors, only FePit1 can permit FeLV-T entry.
FIG. 5.
Receptor binding and FeLV-T cofactor activity of SU fragments at different concentrations. (A to D) MDTF-FePit1 and MDTF-FePit2 cells were incubated with 1 to 500 μl of SU-conditioned media in 1-ml total volumes. Fluorescence-activated cell sorter profiles obtained using the indicated amounts of supernatant are shown. Flow-cytometric analyses of SU fragment binding were performed as described in the legend to Fig. 4 and Materials and Methods. Mock, samples of cells incubated in standard media and stained with the same antibody. In all cases the x axis is fluorescence intensity (log scale) and the y axis is cell number. Abbreviations are as described in the legends to Fig. 1 and 3. (E) Data for a single-cycle infection assay using FeLV-T particles that packaged the gene for β-galactosidase. The total volume of medium in each infection was 1 ml. Variable amounts (1 to 500 μl) of SU-conditioned media were added to the infection for each cofactor. x axis, receptors and cofactor pairs tested. A negative result (arrows) indicates that no blue foci were observed with as much as 100 μl of the FeLV-T virus pseudotype, which corresponds to about 105 particles that can infect cells using FeLIX-Pit1.
To examine whether there is a similar specificity for the cofactor that is independent of its affinity for FePit1, we compared the binding of GALV and that of FeLIX or the FeLV-B SU to Pit1. Using the truncated GALV-SU1–262 we were able to detect binding to FePit1 with 1 μl of conditioned medium (Fig. 5B). The binding of the GALV-SU1–262 fragment to FePit1 reached saturation at 1 to 10 μl, perhaps due in part to higher levels of protein expression in the conditioned media (Fig. 3B). Nonetheless GALV-SU1–262 could not mediate FeLV-T infection with FePit1 even when 500 μl of conditioned medium was used (Fig. 5E). Therefore, even at levels 50- to 100-fold higher than those necessary to saturate the surface receptor, GALV-SU1–262 could not mediate FeLV-T infection. These data indicate that the inability of cofactors other than FeLIX and FeLV-B SUs to mediate FeLV-T infection is not due to a low-affinity interaction between the cofactor and the receptor.
The FeLV-A SU and its receptor do not mediate FeLV-T infection.
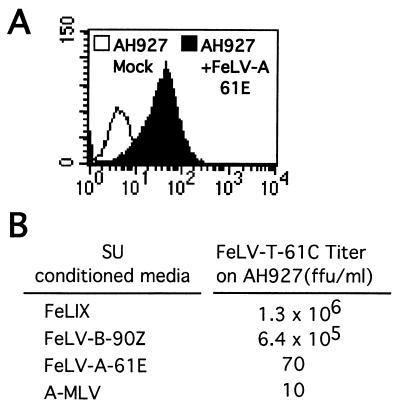
To examine whether the FeLV-A SU could also function with its receptor to mediate FeLV-T infection, we performed infections with AH927 feline fibroblasts. These cells express both FePit1 and the FeLV-A receptor but are very poorly susceptible to infection by FeLV-T because they do not express significant levels of FeLIX (2, 29). As seen with MDTF-HuPit1 and -FePit1 cells, both FeLIX and the FeLV-B-90Z SU could efficiently mediate FeLV-T infection of AH927s, while only background levels of infection were observed when the A-MLV SU was used as the FeLV-T cofactor (Fig. 6B). Consistent with the fact that FeLV-A can readily infect AH927 cells, we could detect binding of the FeLV-A-61E SU to these cells by flow cytometry (Fig. 6A). However, addition of the FeLV-A-61E SU did not render them permissive to FeLV-T infection. Together, our data indicate that the FeLV-T infection pathway is specific in its requirement for Pit1 and either FeLIX or an FeLV-B envelope.
FIG. 6.
Infection of feline fibroblasts in the presence of conditioned media containing retrovirus SUs. (A) Analysis of FeLV-A-61E SU binding to AH927 feline fibroblasts as described in the legend to Fig. 4. (B) FeLV-T-61C titer on AH927 cells in focus-forming units (ffu) per milliliter. Titers are based on challenge with vectors packaging a genome containing the gene for β-galactosidase. HA-tagged SUs were added at the time of infection as a 1:1 dilution of conditioned media harvested from transiently transfected 293T cells.
FeLIX expression is more restricted than FePit1 expression.
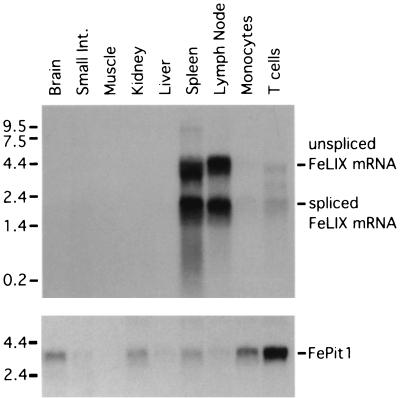
Having shown that FeLIX and Pit1 are the critical cellular proteins required for FeLV-T infection, we analyzed expression of these two proteins in feline tissues. We observed FePit1 expression in a diverse range of tissues by Northern blotting, including kidney, liver, spleen, and lymph node (Fig. 7). We detected higher levels of FePit1 in brain, kidney, spleen, monocytes, and T cells, similar to what has been reported for Pit1 expression in mice (18). In contrast, when we probed the same blot with the FeLIX cDNA, we observed expression of FeLIX and/or related enFeLV envelope genes only in lymphoid tissues, including spleen, lymph node, monocytes, and T cells. These data are consistent with a previous study which showed lymphoid expression of enFeLV envelopes related to FeLIX (23).
FIG. 7.
Northern blot analysis of FeLIX and FePit1 expression in feline tissues. Total cellular RNA was isolated from the indicated tissues. (Top) RNA (10 μg) was loaded in each lane. The filter was probed with a portion of the FeLIX cDNA. Bands corresponding to the predicted unspliced and spliced FeLIX mRNAs are indicated on the right. (Bottom) The same filter was stripped and reprobed with a portion of the FePit1 cDNA. Molecular mass markers (in kilobases) are indicated at the left.
DISCUSSION
Entry by the type C retroviruses is typically directed by interactions between the viral envelope protein and a single host cell receptor. In contrast, FeLV-T requires two cellular proteins to productively infect target cells and to replicate in the infected host. The first is a classic multiple TM receptor protein, while the second, FeLIX, is closely related to the SU of endogenous FeLVs. The unusual nature of this FeLV-T receptor complex suggests that these variants infect cells through a novel mechanism. In this study, we asked whether other related SU-receptor combinations could also facilitate FeLV-T infection. Our results show that only FeLIX and the FeLV-B envelope can efficiently mediate FeLV-T infection and that there is a strong preference for Pit1 as the cell surface receptor, even among FeLV-B envelopes that can efficiently bind to both Pit1 and Pit2 receptors. This specificity is not determined simply by the affinity of the cofactor and the receptor. Rather, it appears to reflect a requirement for a specific interaction between FeLV-T, Pit1, and FeLIX.
Of the six different envelope fragments tested in this study, only those containing sequences derived from endogenous FeLV (FeLIX and FeLV-B SU) were able to facilitate FeLV-T infection. In addition to FeLIX, we found that two different FeLV-B SUs, representative of two distinct classes of FeLV-B found in vivo (34), could mediate FeLV-T infection of cells expressing the Pit1 receptor. In addition, the SU from a related FeLV-B envelope, Gardner-Arnstein (14), could also facilitate FeLV-T infection specifically through Pit1 (data not shown). Although envelopes of the FeLV-B-90Z class can recognize FePit2 (Anderson et al., unpublished data), and in some cases HuPit2 (8), they were not able to facilitate infection of cells expressing these closely related receptors when supplied as soluble cofactors. This was true even when the amount of FeLV-B SU tested was ∼500 times greater than the amount needed to permit FeLV-T entry using FePit1. However, we did observe a low level of infection of cells expressing FePit2 when the 90Z and 90ZRBD SUs were expressed in the target cell membrane. This discrepancy in FeLV-T receptor tropism with soluble SU versus membrane-bound envelope cofactors could reflect differences in local concentrations of SUs or structural issues brought about by expression of the receptor and envelope in the same membrane. These data could also suggest that the trimeric envelope glycoprotein, which is the form expressed when both the SU and TM are included, has increased cofactor activity relative to that for the soluble SU, which is probably a monomer. Thus, in the infected cat, cells expressing Pit2 and infected with FeLV-B may be susceptible to infection by FeLV-T to a limited extent. For FeLIX, the absolute requirement for Pit1 as the transmembrane receptor correlated with the binding properties of the soluble cofactor. By contrast, both the 90Z and 90ZRBD SUs bound FePit2, and the inability of these soluble FeLV-B cofactors to mediate FeLV-T infection on cells expressing these Pit proteins indicates that receptor binding by these cofactors is not sufficient for FeLV-T infection of target cells. These data therefore suggest that Pit1 is necessary for aspects of FeLV-T entry other than cofactor binding.
There is as yet no example other than FeLV-T of a naturally replicating retrovirus that requires a soluble factor for infection. However, Lavillette and coworkers described a mechanism by which MLVs that were engineered to be defective for entry could be rescued by wild-type retrovirus SUs (22). Mutations introduced near the N terminus of the envelope resulted in a postbinding defect that could be rescued in trans by three retrovirus SUs when these soluble envelope fragments were supplied at the time of infection. Rescue was receptor mediated because only target cells that expressed receptors for both the virus and the soluble SU fragment were susceptible to infection. Although FeLV-T envelopes contain similar N-terminal changes and require SU-derived cofactors to infect cells (16, 33, 39), the mechanism of replication-competent FeLV-T infection appears distinct from that of these defective MLVs. First, unlike what is found for the relatively permissive MLV system, only certain cofactors could mediate FeLV-T infection through their respective receptors. Specifically, only Pit1 and either FeLIX or the FeLV-B SU could mediate FeLV-T infection. Coexpression of the GALV and A-MLV envelopes with their respective receptors, Pit1 and Pit2, did not render target cells susceptible to infection by FeLV-T, and the GALV and A-MLV SU fragments did not facilitate infection of cells expressing Pit1 or Pit2 when provided as soluble factors. Second, data from the MLV system suggest that a critical requirement for infection is coexpression of both viral and cofactor receptors on the same cell. For example, the soluble A-MLV SU could rescue a defective ecotropic MLV provided that target cells expressed both Pit2 and the ecotropic MLV receptor, mCAT-1 (22). For FeLV-T, the A-MLV SU was not able to mediate infection of either MDTF-Pit1 or AH927 cells even though these cell lines express both Pit1 and Pit2 proteins. Therefore, the binding of the A-MLV SU to Pit2 was not sufficient to facilitate FeLV-T infection through Pit1 in these cells. In a similar manner, the FeLV-A-61E SU did not render feline fibroblasts susceptible to infection by FeLV-T even though they express both FePit1 and the FeLV-A receptor (29). Therefore, FeLV-T requires a specific combination of cofactor (FeLIX or the FeLV-B SU) and receptor (Pit1) and cannot be “rescued” by the binding of a SU to a different receptor on the target cell. This is in contrast to what has been observed for viruses bearing the mutant MLV SU, which can be rescued by interactions that occur in trans between a variety of soluble SUs and their receptors. It is perhaps not surprising that FeLV-T and the defective MLVs do not have completely analogous modes of entry, given that FeLV-T is a naturally selected variant that is competent for replication in the infected host, whereas the MLVs have specifically been engineered to encode deletions that render them replication defective.
Subsequent studies suggested that soluble MLV SU fragments may mediate trans infection of defective MLVs by interacting with their C-terminal domains (5, 21). Thus, it is possible that rescue of fusion-defective MLVs by soluble SU depends on the concentration of receptors for both the viral envelope and cofactor on target cells, the concentration of the soluble SU fragment, and the affinity of this fragment for both its receptor and the viral envelope. Here we show that the receptor specificity of FeLV-T for Pit1 and FeLIX or the FeLV-B SU is not simply driven by the fact that there is a high-affinity interaction between these two molecules. We found that FeLV-B and GALV SUs bind with similar efficiencies to Pit1, yet the GALV SU and Pit1 cannot act as an FeLV-T receptor complex. This was true even when the concentration of the GALV SU fragment was ∼500-fold higher than that required for FeLIX cofactor activity. This indicates a specific requirement for FeLIX or the FeLV-B SU as a cofactor in the FeLV-T receptor complex. We used a similar approach to show that there is also a specific requirement for Pit1 as part of the FeLV-T receptor complex. We found that an FeLV-B SU that binds to both Pit2 and Pit1 could act as cofactor for FeLV-T infection with Pit1 but not Pit2. One confounding aspect of this comparison was that the FeLV-B SU did not bind as well to cells expressing Pit2 as it did to cells expressing Pit1. However, when we compared the abilities of this cofactor to permit entry at concentrations where levels of binding to the two receptors were equivalent, only Pit1 could permit FeLV-T infection. This suggests that the amount of cofactor bound to the receptor does not determine whether the complex acts as receptor for FeLV-T. These data suggest that FeLV-T may specifically interact with both Pit1 and either the FeLIX or FeLV-B SU cofactor. Specific protein-protein interactions between FeLV-T and both components of the complex would explain why FeLV-B SU–Pit2 and GALV SU-Pit1 cannot function as receptor complexes for FeLV-T. Alternatively, it is possible that the binding of the cofactor to the receptor induces a conformational change in one of these molecules that permits the binding of FeLV-T. For example, the binding of FeLIX may change the conformation of Pit1 in a way that facilitates FeLV-T binding. It is also possible that the binding of FeLV-T SU to either FeLIX or Pit1 could lead to a change in SU that permits a subsequent interaction that is required for fusion. The latter model has some similarities to human immunodeficiency virus (HIV) entry, where binding of CD4 to the HIV SU induces a conformational change in the SU that allows a subsequent interaction with the presumed fusion receptor (7). Studies of the protein-protein interactions between FeLV-T, FeLIX, and Pit1 will be required to discriminate between these potential models.
Retroviruses may evolve in vivo to utilize new receptor proteins, thereby changing their cell tropism. Like 10A1 MLV, A-MLV, GALV, and FeLV-B, FeLV-T uses a phosphate transport protein as a receptor (27, 28, 31, 45, 48, 50). The ability of five different classes of retroviruses to use related receptors suggests that the Pit proteins may possess structural motifs that make them particularly attractive candidates as retrovirus receptors. This may in part reflect the fact that Pit proteins may be somewhat unique among cellular receptors in being able to execute postbinding events in entry (11). Alternatively, these type C retroviruses may have evolved to use the Pit receptors because their widespread expression and functional redundancy in phosphate transport facilitate the establishment of a persistent infection. It is noteworthy that FeLV-B and FeLV-T, both of which evolve from FeLV-A during the course of an infection, utilize a common cell surface receptor to enter target cells. This convergent evolution is particularly intriguing in view of the fact that proteins like FeLIX have been shown to inhibit FeLV-B infection through receptor blockade (23). The ability of FeLV-T in turn to utilize FeLIX as a cofactor for infection may therefore reflect an elegant adaptation to this host defense in vivo.
FeLV-A is found in nearly all natural FeLV infections and is generally considered to be the ecotropic, transmissible form of the virus (40). We hypothesize that FeLV-A is important primarily in the initial stages of FeLV infection, and this may be due to expression of the FeLV-A receptor on cells that are important targets during transmission. It is therefore interesting that the FeLV-A-61E SU was not able to mediate FeLV-T infection of cells permissive for FeLV-A infection. These data suggest that FeLV-A does not play a direct role in FeLV-T replication in the infected cat. These data also suggest that the cells that specifically express the FeLV-A receptor may be less-important target cells for virus replication during the persistent stages of infection and/or in cats that develop immunodeficiency disease as a result of FeLV infection.
The ability of the FeLV-B envelope protein to functionally substitute for FeLIX suggests that there may be a more complex interrelationship between FeLV-T and FeLV-B variants during an in vivo infection. Our analysis of FeLIX expression indicates that lymphoid cells are likely a major susceptible target cell for FeLV-T variants. This finding is consistent with the ability of FeLV-T to infect B cells, T cells, and myeloid cells and to cause deficits in T-cell immunity in infected animals (35–37). Because FeLIX is secreted from cells, we speculate that nearby cells that do not express FeLIX may also be susceptible to infection by FeLV-T if they express Pit1 but not Pit2 (2). Pit1 is more widely expressed, and this would suggest that the natural host range of FeLV-B is much broader than that of FeLV-T. The data presented here, however, imply that cells infected with FeLV-B may also be susceptible to infection by FeLV-T if Pit1 is present, but less so if Pit2 is the primary receptor. Although FeLV-Bs are found in roughly one-third of infected animals, their prevalence approaches 50 to 60% in cats with leukemia and lymphoma, suggesting that FeLV-B may contribute to FeLV-induced neoplasia (30, 47). Based on our data, we hypothesize that coinfection with FeLV-B or coevolution of FeLV-B in vivo could expand the tropism of FeLV-T variants and accelerate the development of FeLV immunodeficiency. The potential role of FeLV-B in FeLV-T infection may provide a mechanism to increase immunosuppression in cats that develop FeLV-associated neoplasia.
ACKNOWLEDGMENTS
We thank Maribeth Eiden and A. Dusty Miller for constructs and helpful discussion, Jan Abkowitz and Kathleen Sabo for the gift of feline tissues and preparation of T cells and monocytes, and Jim Sugai, Heather Cheng, and F. Claire Hankenson for technical assistance and helpful comments.
This work was supported by NIH grant CA 51080. A.S.L. was supported by NIH training grant 2 T32 CA09229.
REFERENCES
- 1.Abkowitz J L, Taboada M, Shelton G H, Catlin S N, Guttorp P, Kiklevich J V. An X chromosome gene regulates hematopoietic stem cell kinetics. Proc Natl Acad Sci USA. 1998;95:3862–3866. doi: 10.1073/pnas.95.7.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson M M, Lauring A S, Burns C C, Overbaugh J. Identification of a cellular cofactor required for infection by feline leukemia virus. Science. 2000;287:1828–1830. doi: 10.1126/science.287.5459.1828. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing and Associates and Wiley-Interscience; 1987. [Google Scholar]
- 4.Bae Y, Kingsman S M, Kingsman A J. Functional dissection of the Moloney murine leukemia virus envelope protein gp70. J Virol. 1997;71:2092–2099. doi: 10.1128/jvi.71.3.2092-2099.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnett A L, Davey R A, Cunningham J M. Modular organization of the Friend murine leukemia virus envelope protein underlies the mechanism of infection. Proc Natl Acad Sci USA. 2001;98:4113–4118. doi: 10.1073/pnas.071432398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Battini J L, Heard J M, Danos O. Receptor choice determinants in the envelope glycoproteins of amphotropic, xenotropic, and polytropic murine leukemia viruses. J Virol. 1992;66:1468–1475. doi: 10.1128/jvi.66.3.1468-1475.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger E A. HIV entry and tropism: the chemokine receptor connection. AIDS. 1997;11(Suppl. A):S3–S16. [PubMed] [Google Scholar]
- 8.Boomer S, Eiden M, Burns C C, Overbaugh J. Three distinct envelope domains, variably present in subgroup B feline leukemia virus recombinants, mediate Pit1 and Pit2 receptor recognition. J Virol. 1997;71:8116–8123. doi: 10.1128/jvi.71.11.8116-8123.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boomer S, Gasper P, Whalen L R, Overbaugh J. Isolation of a novel subgroup B feline leukemia virus from a cat infected with FeLV-A. Virology. 1994;204:805–810. doi: 10.1006/viro.1994.1597. [DOI] [PubMed] [Google Scholar]
- 10.Chen C M, Kraut N, Groudine M, Weintraub H. I-mf, a novel myogenic repressor, interacts with members of the MyoD family. Cell. 1996;86:731–741. doi: 10.1016/s0092-8674(00)80148-8. [DOI] [PubMed] [Google Scholar]
- 11.Cosset F L, Morling F J, Takeuchi Y, Weiss R A, Collins M K, Russell S J. Retroviral retargeting by envelopes expressing an N-terminal binding domain. J Virol. 1995;69:6314–6322. doi: 10.1128/jvi.69.10.6314-6322.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donahue P R, Quackenbush S L, Gallo M V, deNoronha C M C, Overbaugh J, Hoover E A, Mullins J I. Viral genetic determinants of T-cell killing and immunodeficiency disease induction by the feline leukemia virus FeLV-FAIDS. J Virol. 1991;65:4461–4469. doi: 10.1128/jvi.65.8.4461-4469.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eiden M V, Farrell K B, Wilson C A. Substitution of a single amino acid residue is sufficient to allow the human amphotropic murine leukemia virus receptor to also function as a gibbon ape leukemia virus receptor. J Virol. 1996;70:1080–1085. doi: 10.1128/jvi.70.2.1080-1085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elder J H, Mullins J I. Nucleotide sequence of the envelope gene of Gardner-Arnstein feline leukemia virus B reveals unique sequence homologies with a murine mink cell focus-forming virus. J Virol. 1983;46:871–880. doi: 10.1128/jvi.46.3.871-880.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grant C K, Ernisse B J, Jarrett O, Jones F R. Feline leukemia virus envelope gp70 of subgroups B and C defined by monoclonal antibodies with cytotoxic and neutralizing functions. J Immunol. 1983;131:3042–3048. [PubMed] [Google Scholar]
- 16.Gwynn S R, Hankenson F C, Lauring A S, Rohn J L, Overbaugh J. Feline leukemia virus envelope sequences that affect T-cell tropism and syncytium formation are not part of known receptor-binding domains. J Virol. 2000;74:5754–5761. doi: 10.1128/jvi.74.13.5754-5761.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hughson F M. Enveloped viruses: a common mode of membrane fusion? Curr Biol. 1997;7:R565–R569. doi: 10.1016/s0960-9822(06)00283-1. [DOI] [PubMed] [Google Scholar]
- 18.Johann S V, Gibbons J J, O'Hara B. GLVR1, a receptor for gibbon ape leukemia virus, is homologous to a phosphate permease of Neurospora crassa and is expressed at high levels in the brain and thymus. J Virol. 1992;66:1635–1640. doi: 10.1128/jvi.66.3.1635-1640.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kennedy D W, Abkowitz J L. Mature monocytic cells enter tissues and engraft. Proc Natl Acad Sci USA. 1998;95:14944–14949. doi: 10.1073/pnas.95.25.14944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kimpton J, Emerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated β-galactosidase gene. J Virol. 1992;66:2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lavillette D, Boson B, Russell S J, Cosset F L. Activation of membrane fusion by murine leukemia viruses is controlled in cis or trans by interactions between the receptor-binding domain and a conserved disulfide loop of the carboxy terminus of the surface glycoprotein. J Virol. 2001;75:3685–3695. doi: 10.1128/JVI.75.8.3685-3695.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lavillette D, Ruggieri A, Russell S J, Cosset F L. Activation of a cell entry pathway common to type C mammalian retroviruses by soluble envelope fragments. J Virol. 2000;74:295–304. doi: 10.1128/jvi.74.1.295-304.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDougall A S, Terry A, Tzavaras T, Cheney C, Rojko J, Neil J. Defective endogenous proviruses are expressed in feline lymphoid cells: evidence for a role in natural resistance to subgroup B feline leukemia virus. J Virol. 1994;68:2151–2160. doi: 10.1128/jvi.68.4.2151-2160.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miedema F, Meyaard L, Koot M, Klein M R, Roos M L, Groenink M, Fouchier R A M, Van't-Wout A B, Tersmette M, Sciellekens P T A, Schuitemaker H. Changing virus-host interactions in the course of HIV-1 infection. Immunol Rev. 1994;140:35–72. doi: 10.1111/j.1600-065x.1994.tb00864.x. [DOI] [PubMed] [Google Scholar]
- 25.Miller A D, Buttimore C. Redesign of retrovirus packaging cell lines to avoid recombination leading to helper virus production. Mol Cell Biol. 1986;6:2895–2902. doi: 10.1128/mcb.6.8.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller A D, Miller D G, Garcia J V, Lynch C M. Use of retroviral vectors for gene transfer and expression. Methods Enzymol. 1993;217:581–599. doi: 10.1016/0076-6879(93)17090-r. [DOI] [PubMed] [Google Scholar]
- 27.Miller D G, Edwards R H, Miller A D. Cloning of the cellular receptor for amphotropic murine retroviruses reveals homology to that for gibbon ape leukemia virus. Proc Natl Acad Sci USA. 1994;91:78–82. doi: 10.1073/pnas.91.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller D G, Miller A D. A family of retroviruses that utilize related phosphate transporters for cell entry. J Virol. 1994;68:8270–8276. doi: 10.1128/jvi.68.12.8270-8276.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moser M, Burns C, Boomer S, Overbaugh J. The host range and interference properties of two closely related feline leukemia virus variants suggest that they use distinct receptors. Virology. 1998;242:366–377. doi: 10.1006/viro.1997.9008. [DOI] [PubMed] [Google Scholar]
- 30.Neil J C, Fulton R, Rigby M, Stewart M. Feline leukemia virus: generation of pathogenic and oncogenic variants. Curr Top Microbiol Immunol. 1991;171:67–93. doi: 10.1007/978-3-642-76524-7_4. [DOI] [PubMed] [Google Scholar]
- 31.O'Hara B, Johann S V, Klinger H P, Blair D G, Rubinson H, Dunn K J, Saas P, Vitek S M, Robins T. Characterization of a human gene conferring sensitivity to infection by gibbon ape leukemia virus. Cell Growth Differ. 1990;1:119–127. [PubMed] [Google Scholar]
- 32.Overbaugh J, Donahue P R, Quackenbush S L, Hoover E A, Mullins J I. Molecular cloning of a feline leukemia virus that induces fatal immunodeficiency disease in cats. Science. 1988;239:906–910. doi: 10.1126/science.2893454. [DOI] [PubMed] [Google Scholar]
- 33.Overbaugh J, Hoover E A, Mullins J I, Burns D P W, Rudensey L, Quackenbush S L, Stallard V, Donahue P R. Structure and pathogenicity of individual variants within an immunodeficiency disease-inducing isolate of FeLV. Virology. 1992;188:558–569. doi: 10.1016/0042-6822(92)90510-v. [DOI] [PubMed] [Google Scholar]
- 34.Overbaugh J, Riedel N, Hoover E A, Mullins J I. Transduction of endogenous envelope genes by feline leukaemia virus in vitro. Nature. 1988;332:731–734. doi: 10.1038/332731a0. [DOI] [PubMed] [Google Scholar]
- 35.Quackenbush S L, Dean G A, Mullins J I, Hoover E A. Analysis of FeLV-FAIDS provirus burden and productive infection in lymphocyte subsets in vivo. Virology. 1996;223:1–9. doi: 10.1006/viro.1996.0449. [DOI] [PubMed] [Google Scholar]
- 36.Quackenbush S L, Donahue P R, Dean G A, Myles M H, Ackley C D, Cooper M D, Mullins J I, Hoover E A. Lymphocyte subset alterations and viral determinants of immunodeficiency disease induction by the feline leukemia virus FeLV-FAIDS. J Virol. 1990;64:5465–5474. doi: 10.1128/jvi.64.11.5465-5474.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quackenbush S L, Mullins J I, Hoover E A. Colony forming T lymphocyte deficit in the development of feline retrovirus induced immunodeficiency syndrome. Blood. 1989;73:509–516. [PubMed] [Google Scholar]
- 38.Rohn J L, Linenberger M L, Hoover E A, Overbaugh J. Evolution of feline leukemia virus variant genomes with insertions, deletions, and defective envelope genes in infected cats with tumors. J Virol. 1994;68:2458–2467. doi: 10.1128/jvi.68.4.2458-2467.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rohn J L, Moser M S, Gwynn S R, Baldwin D N, Overbaugh J. In vivo evolution of a novel, syncytium-inducing and cytopathic feline leukemia virus variant. J Virol. 1998;72:2686–2696. doi: 10.1128/jvi.72.4.2686-2696.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rohn J L, Overbaugh J. Pathogenic feline retroviruses: feline leukemia virus and feline immunodeficiency virus. In: Chen I S Y, Ahmed R, editors. Persistent viral infections. New York, N.Y: John Wiley and Sons; 1999. pp. 379–408. [Google Scholar]
- 41.Rudensey L M, Kimata J T, Benveniste R E, Overbaugh J. Progression to AIDS in macaques is associated with changes in the replication, tropism, and cytopathic properties of the simian immunodeficiency virus variant population. Virology. 1995;207:528–542. doi: 10.1006/viro.1995.1113. [DOI] [PubMed] [Google Scholar]
- 42.Sarma P S, Log T. Subgroup classification of feline leukemia and sarcoma viruses by viral interference and neutralization tests. Virology. 1973;54:160–169. doi: 10.1016/0042-6822(73)90125-6. [DOI] [PubMed] [Google Scholar]
- 43.Stewart M A, Warnock M, Wheeler A, Wilkie N, Mullins J I, Onions D E, Neil J C. Nucleotide sequences of a feline leukemia virus subgroup A envelope gene and long terminal repeat and evidence for the recombinational origin of subgroup B viruses. J Virol. 1986;58:825–834. doi: 10.1128/jvi.58.3.825-834.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43a.Sugai J, Eiden M, Anderson M M, Van Hoeven N, Meiering C D, Overbaugh J. Identification of envelope determinants of feline leukemia virus subgroup B that permit infection and gene transfer to cells expressing human Pit1 and Pit2. J Virol. 2001;75:6841–6849. doi: 10.1128/JVI.75.15.6841-6849.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tailor C S, Kabat D. Variable regions A and B in the envelope glycoproteins of feline leukemia virus subgroup B and amphotropic murine leukemia virus interact with discrete receptor domains. J Virol. 1997;71:9383–9391. doi: 10.1128/jvi.71.12.9383-9391.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takeuchi Y, Vile R G, Simpson G, O'Hara B, Collins M K, Weiss R A. Feline leukemia virus subgroup B uses the same cell surface receptor as gibbon ape leukemia virus. J Virol. 1992;66:1219–1222. doi: 10.1128/jvi.66.2.1219-1222.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ting Y T, Wilson C A, Farrell K B, Chaudry G J, Eiden M V. Simian sarcoma-associated virus fails to infect Chinese hamster cells despite the presence of functional gibbon ape leukemia virus receptors. J Virol. 1998;72:9453–9458. doi: 10.1128/jvi.72.12.9453-9458.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsatsanis C, Fulton R, Nishigaki K, Tsujimoto H, Levy L, Terry A, Spandidos D, Onions D, Neil J C. Genetic determinants of feline leukemia virus-induced lymphoid tumors: patterns of proviral insertion and gene rearrangement. J Virol. 1994;68:8296–8303. doi: 10.1128/jvi.68.12.8296-8303.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Zeijl M, Johann S V, Closs E, Cunningham J, Eddy R, Shows T B, O'Hara B. A human amphotropic retrovirus receptor is a second member of the gibbon ape leukemia virus receptor family. Proc Natl Acad Sci USA. 1994;91:1168–1172. doi: 10.1073/pnas.91.3.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wilson C A, Farrell K B, Eiden M V. Comparison of cDNAs encoding the gibbon ape leukaemia virus receptor from susceptible and non-susceptible murine cells. J Gen Virol. 1994;75:1901–1908. doi: 10.1099/0022-1317-75-8-1901. [DOI] [PubMed] [Google Scholar]
- 50.Wilson C A, Farrell K B, Eiden M V. Properties of a unique form of the murine amphotropic leukemia virus receptor expressed on hamster cells. J Virol. 1994;68:7697–7703. doi: 10.1128/jvi.68.12.7697-7703.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zavorotinskaya T, Albritton L M. Suppression of a fusion defect by second site mutations in the ecotropic murine leukemia virus surface protein. J Virol. 1999;73:5034–5042. doi: 10.1128/jvi.73.6.5034-5042.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]