Abstract
A natural hepatitis B virus (HBV) variant associated with seroconversion from HBeAg to anti-HBe antibody contains two nucleotide substitutions (A1764T and G1766A) in the proximal nuclear hormone receptor binding site in the nucleocapsid promoter. These nucleotide substitutions prevent the binding of the retinoid X receptor α (RXRα)–peroxisome proliferator-activated receptor α (PPARα) heterodimer without greatly altering the efficiency of binding of hepatocyte nuclear factor 4 (HNF4) to this recognition sequence. In addition, these nucleotide substitutions create a new binding site for HNF1. Analysis of HBV transcription and replication in nonhepatoma cells indicates that RXRα-PPARα heterodimers support higher levels of pregenomic RNA transcription from the wild-type than from the variant nucleocapsid promoter, producing higher levels of wild-type than of variant replication intermediates. In contrast, HNF4 supports higher levels of pregenomic RNA transcription from the variant than from the wild-type nucleocapsid promoter, producing higher levels of variant than of wild-type replication intermediates. HNF1 can support variant virus replication at a low level but is unable to support replication of the wild-type HBV genome. These observations indicate that the replication of wild-type and variant viruses can be differentially regulated by the liver-specific transcription factors that bind to the proximal nuclear hormone receptor binding site of the nucleocapsid promoter. Differential regulation of viral replication may be important in the selection of specific viral variants as a result of an antiviral immune response.
The hepatitis B virus (HBV) genome is a partially double-stranded 3.2-kb DNA molecule (14, 30). The unusual structure of the viral genome reflects the replication cycle of the hepadnaviruses (30, 54). Nuclear covalently closed circular 3.2-kbp HBV DNA is transcribed to produce a greater-than-genome-length pregenomic 3.5-kb RNA that is reverse transcribed by the HBV polymerase to generate encapsidated viral genomic DNA (14, 30, 54). The core polypeptide and the viral polymerase are both encoded by the HBV 3.5-kb transcripts (36). Therefore, transcriptional regulation of the synthesis of the HBV 3.5-kb RNAs also controls the level of viral DNA synthesis.
The level of 3.5-kb HBV RNA synthesis is determined by the rate of transcription from the nucleocapsid promoter. The regulatory elements controlling transcription from the nucleocapsid promoter have been extensively characterized (17, 23, 26–28, 39, 58–61). Recently it was shown that nuclear hormone receptors are a major determinant in regulating HBV 3.5-kb RNA synthesis and viral replication (52). The most important recognition element involved in controlling viral pregenomic RNA synthesis and viral replication is the proximal nuclear hormone receptor binding site in the nucleocapsid promoter (52).
Viral variants containing two nucleotide substitutions (A1764T and G1766A) in the proximal nuclear hormone receptor binding site in the nucleocapsid promoter are associated with seroconversion from HBeAg to anti-HBe antibody, suggesting the variants have a selective advantage over the wild-type virus during an antiviral immune response (35). It has been suggested that variant viruses synthesize reduced levels of HBeAg and therefore are less susceptible to elimination by an immune response directed against nucleocapsid epitopes (2, 19, 35, 45). This possibility was supported by the observation that variant viruses synthesized a lower level of the precore 3.5-kb RNA that encodes HBeAg (2, 19, 32).
In this study, the properties of the variant viral genome were characterized further using a recently developed viral replication system with which the effects of individual liver-enriched transcription factors can be investigated (52). The nucleotide substitutions in the proximal nuclear hormone receptor binding site of the nucleocapsid promoter dramatically alter the extent to which HBV replicates in response to hepatocyte nuclear factor 4 (HNF4) and retinoid X receptor α (RXRα)–peroxisome proliferator-activated receptor α (PPARα) heterodimers. The wild-type genome replicates to higher levels in response to the ectopic expression of RXRα-PPARα, whereas the variant genome replicates to higher levels in response to the ectopic expression of HNF4. These changes correlate with the loss of RXRα-PPARα binding to the proximal nuclear hormone receptor recognition sequence in the nucleocapsid promoter of the variant genome. These results indicate that the replication of wild-type and variant viruses can be differentially regulated by nuclear hormone receptors and suggest that the presence of an antiviral immune response in the liver may select specific viral variants in which pregenomic RNA synthesis is preferentially responsive to specific nuclear hormone receptors. Therefore, increased HNF4 activity or decreased RXRα-PPARα activity due to the changes in the physiology of hepatocytes during an immune response may contribute to the selection of variants (A1764T and G1766A) with an altered nuclear hormone receptor binding site.
MATERIALS AND METHODS
Plasmid constructions.
The steps in the cloning of the plasmid constructions used in the transfection experiments were performed by standard techniques (43). HBV DNA sequences in these constructions were derived from plasmid pCP10, which contains two copies of the HBV genome (subtype ayw) cloned into the EcoRI site of pBR322 (9). The firefly luciferase (LUC) reporter gene in these constructions was derived from plasmid p19DLUC (40). Plasmid CpLUC contains one complete HBV genome located directly 5′ to the promoterless LUC reporter gene such that the expression of the LUC gene is governed by the HBV nucleocapsid promoter (40). Details of the construction of the nucleocapsid promoter deletion plasmid CpΔ1805-1374LUC have been described previously (60).
Plasmids CpTALUC and CpΔ1805-1374TALUC, containing the A1764T and G1766A nucleotide substitutions in the proximal nuclear hormone receptor binding site of the core promoter, were generated using a Chameleon double-stranded, site-directed mutagenesis kit (Stratagene Cloning Systems, La Jolla, Calif.) according to the manufacturer's instructions. Plasmids CpTALUC and CpΔ1805-1374TALUC are derivatives of the CpLUC and CpΔ1805-1374LUC constructs, respectively. The TA mutation converted the 13-nucleotide proximal nuclear hormone receptor binding site sequence located between −28 and −16 (nucleotide coordinates 1757 and 1769) from AGGTTAAAGGTCT to AGGTTAATGATCT (changes are shown in bold type). The sequences of the mutations introduced into the nucleocapsid promoter constructs were verified by dideoxynucleotide sequencing (44).
Plasmids pHBVTATALUC, CpFL(3)TATALUC, CpFL(4)TATALUC, CpFLTA(3)TATALUC, CpFLTA(4)TATALUC, and PS1pHNF1(1)TATALUC were constructed by inserting synthetic double-stranded oligonucleotides into sites in the polylinker of p19DLUC. pHBVTATALUC was constructed by inserting a double-stranded oligonucleotide containing the large surface antigen promoter TATA-box element, produced by annealing the oligonucleotides CTATATTATATAAGAGAGAAGCT and TCTCTCTTATATAATATAGGTAC (spanning HBV coordinates 2773 to 2791) into the SacI and KpnI sites of p19DLUC in the same orientation as that in which the TATA-box element occurs in the HBV genome (37). CpFL(3)TATALUC, CpFL(4)TATALUC, CpFLTA(3)TATALUC, CpFLTA(4)TATALUC, and PS1pHNF1(1)TATALUC were made by inserting one to four copies (as indicated in the construct designations) of the HBV CpFL, CpFLTA, and PS1pHNF1 (37, 39) double-stranded oligonucleotides into the unique HindIII site of pHBVTATALUC. The oligonucleotide pairs used to generate the CpFL, CpFLTA, and PS1pHNF1 double-stranded oligonucleotides were TCGAGATTAGGTTAAAGGTCTTTGTACTAG and TCGACTAGTACAAAGACCTTTAACCTAATC (oligonucleotide CpFL; HBV coordinates 1751 to 1778), TCGAGATTAGGTTAATGATCTTTGTACTAG and TCGACTAGTACAAAGATCATTAACCTAATC (oligonucleotide CpFLTA; HBV variant coordinates 1751 to 1778), and AGCTAGTTAATCATTACTTC and AGCTGAAGTAATGATTAACT (oligonucleotide PS1pHNF1; HBV coordinates 2719 to 2734). The CpFL and CpFLTA double-stranded oligonucleotides span the nucleocapsid promoter proximal nuclear hormone receptor binding site (39), and the PS1pHNF1 double-stranded oligonucleotide spans the large surface antigen HNF1 binding site (38). The sequence of each construct was verified by dideoxynucleotide sequencing (44).
The HBV DNA 4.1-kbp construct, which contains 1.3 copies of the HBV genome, includes the viral sequence from nucleotide coordinates 1072 to 3182 plus 1 to 1990. This plasmid was constructed by cloning the NsiI/BglII HBV DNA fragment (nucleotide coordinates 1072 to 1990) into pUC13, generating pHBV(1072-1990). Subsequently, a complete copy of the 3.2-kbp viral genome linearized at the NcoI site (nucleotide coordinates 1375 to 3182 plus 1 to 1374) was cloned into the unique NcoI site (HBV nucleotide coordinate 1374) of pHBV(1072-1990), generating the HBV DNA 4.1-kbp construct. The HBV DNA 3.9-kbp construct, which contains 1.2 copies of the HBV genome, includes the viral sequence from nucleotide coordinates 1239 to 3182 plus 1 to 1990. This plasmid was constructed by cloning the SphI/BglII HBV DNA fragment (nucleotide coordinates 1239 to 1990) into pUC13, generating pHBV(1239-1990). Subsequently, a complete copy of the 3.2-kbp viral genome linearized at the NcoI site (nucleotide coordinates 1375 to 3182 plus 1 to 1374) was cloned into the unique NcoI site (HBV nucleotide coordinate 1374) of pHBV(1239-1990), generating the HBV DNA 3.9-kbp construct.
Plasmids 4.1TAmut and 3.9TAmut were derived by introducing the A1764T and G1766A nucleotide substitutions into the proximal nuclear hormone receptor binding site of the core promoter in the HBV DNA 4.1-kbp and 3.9-kbp constructs, respectively, using the Chameleon double-stranded, site-directed mutagenesis kit according to the manufacturer's instructions. The nucleotide substitutions introduced into the nucleocapsid promoter constructs were verified by dideoxynucleotide sequencing (44). Both nucleocapsid promoter regions in these terminally redundant HBV constructs were mutated for this analysis.
The pMTHNF1α, pCMVHNF4, pRS-hRXRα, and pCMVPPARα-G vectors express HNF1α, HNF4, RXRα, and PPARα-G (see below) polypeptides from the rat HNF1α, rat HNF4, human RXRα, and mouse PPARα-G cDNAs, respectively, using the mouse metallothionein I promoter (pMT), the cytomegalovirus immediate-early promoter (pCMV), or the Rous sarcoma virus long terminal repeat (pRS) (4, 29, 33, 38). The PPARα-G polypeptide contains a mutation in the PPARα cDNA changing Glu282 to Gly that may decrease the affinity of the receptor for the endogenous ligand. Consequently, this mutation increases the peroxisome proliferator-dependent (i.e., clofibric acid-dependent) activation of transcription from a peroxisome proliferator response element-containing promoter (33) and was used in this study to demonstrate the peroxisome proliferator-dependent transcriptional transactivation of the nucleocapsid promoter. The pGEXHNF4, pGEXRXRα, and pGEXPPARα vectors express in Escherichia coli DH5α glutathione S-transferase (GST) fusion proteins which include the complete HNF4, RXRα, and PPARα polypeptide sequences (49). Expression and subsequent affinity purification of the GST fusion proteins using glutathione-agarose were performed as described previously (49).
Cells and transfections.
The human hepatoma cell lines Huh7, HepG2, and HepG2.1 and the mouse fibroblast cell line NIH 3T3 were grown in RPMI 1640 medium–10% fetal bovine serum at 37°C in 5% CO2–air. Transfections using LUC reporter gene constructs were performed as previously described (15, 50), except that six-well plates containing approximately 3 × 105 cells per well were used. The transfected DNA mixture comprised 5 μg of a LUC plasmid and 0.25 μg of pCMVβ, which served as an internal control for transfection efficiency. pCMVβ directs the expression of the E. coli β-galactosidase (β-gal) gene using the cytomegalovirus immediate-early promoter (Clontech Laboratories, Palo Alto, Calif.). When appropriate, the DNA mixture also included 0.5 μg of the HNF1α, HNF4, RXRα, and PPARα-G expression vectors, pMTHNF1α, pCMVHNF4, pRS-hRXRα, and pCMVPPARα-G, respectively, or the control expression vectors, pMT, pCMV, and pRS. The DNA was removed 4 to 6 h after transfection, and the cells were washed with 2 ml of medium. Fresh RPMI 1640 medium containing all-trans-retinoic acid and clofibric acid at final concentrations of 1 μM and 1 mM, respectively, was added to the cells as required. Cell extracts were prepared 40 to 48 h after transfection. Cells were lysed in 150 μl of lysis buffer (0.1 M potassium phosphate [pH 7.8], 0.2% [vol/vol] Triton X-100), and the cell debris was pelleted by centrifugation for 2 min at 13,000 rpm in an Eppendorf 5417C microcentrifuge. The supernatant was assayed for LUC activity essentially as previously described (7) and for β-gal activity using a Galacto-Light kit (Tropix, Inc.) as instructed by the manufacturer. The level of β-gal activity observed was not specifically affected by any of the exogenously expressed transcription factors. The LUC activities were normalized to the β-gal activity in each transfection experiment.
Transfections for viral RNA and DNA analysis were performed as previously described (31) using 10-cm plates containing approximately 106 cells. DNA and RNA isolation was performed 3 days posttransfection. The transfected DNA mixture was composed of 10 μg of HBV DNA (4.1 or 3.9 kbp) plus 1.5 μg of the liver-enriched transcription factor expression vectors pMTHNF1α, pCMVHNF4, pRS-hRXRα, and pCMVPPARα-G (39, 42). Controls were derived from cells transfected with HBV DNA and the pCMV expression vector, lacking a liver-enriched transcription factor cDNA insert (39). All-trans-retinoic acid and clofibric acid at 1 μM and 1 mM, respectively, were used to activate the nuclear hormone receptors RXRα and PPARα.
Characterization of HBV transcripts and viral replication intermediates.
Transfected cells from a single plate were divided equally and used for the preparation of total cellular RNA and viral DNA replication intermediates as described previously (51), with minor modifications. For RNA isolation (5), the cells were lysed in 1.8 ml of 25 mM sodium citrate (pH 7.0)–4 M guanidinium isothiocyanate–0.5% (vol/vol) sarcosyl–0.1 M 2-mercaptoethanol. After the addition of 0.18 ml of 2 M sodium acetate (pH 4.0), the lysate was extracted with 1.8 ml of water-saturated phenol plus 0.36 ml of chloroform-isoamyl alcohol (49:1). After centrifugation for 30 min at 3,000 rpm in a Sorval RT6000 apparatus, the aqueous layer was precipitated with 1.8 ml of isopropanol. The precipitate was resuspended in 0.3 ml of 25 mM sodium citrate (pH 7.0)–4 M guanidinium isothiocyanate–0.5% (vol/vol) sarcosyl–0.1 M 2-mercaptoethanol and precipitated with 0.6 ml of ethanol. After centrifugation for 20 min at 14,000 rpm in a Eppendorf 5417C microcentrifuge, the precipitate was resuspended in 0.3 ml of 10 mM Tris HCl (pH 8.0)–5 mM EDTA–0.1% (wt/vol) sodium lauryl sulfate and precipitated with 45 μl of 2 M sodium acetate plus 0.7 ml of ethanol.
For the isolation of viral DNA replication intermediates, the cells were lysed in 0.4 ml of 100 mM Tris HCl (pH 8.0)–0.2% (vol/vol) Nonidet P-40. The lysate was centrifuged for 1 min at 14,000 rpm in an Eppendorf 5417C microcentrifuge to pellet the nuclei. The supernatant was adjusted to 6.75 mM magnesium acetate–200 μg of DNase I/ml and incubated for 1 h at 37°C to remove the transfected plasmid DNA. The supernatant was readjusted to 100 mM NaCl–10 mM EDTA–0.8% (wt/vol) sodium lauryl sulfate–1.6 mg of pronase/ml and incubated for an additional 1 h at 37°C. The supernatant was extracted twice with phenol, precipitated with 2 volumes of ethanol, and resuspended in 100 μl of 10 mM Tris HCl (pH 8.0)–1 mM EDTA. RNA (Northern) and DNA (Southern) filter hybridization analyses were performed using 10 μg of total cellular RNA and 30 μl of viral DNA replication intermediates, respectively, as described previously (43).
RNase protection assays were performed using a Pharmingen Riboquant kit and riboprobes were synthesized using an Ambion Maxiscript kit as described by the manufacturers. Transcription initiation sites for the HBV 3.5-kb transcripts were examined using 20 μg of total cellular RNA and a 333-nucleotide (HBV coordinates 1990 to 1658) 32P-labeled HBV riboprobe. As an internal control for the RNase protection analysis, a 32P-labeled mouse ribosomal protein L32 gene riboprobe spanning 101 nucleotides of exon 3 was used (10). All riboprobes contained additional flanking vector sequences of 40 to 90 nucleotides that were not protected by HBV RNA.
Nuclear extracts and gel retardation analysis.
Nuclear extracts were prepared from Huh7 cells and mouse liver as described previously (8, 41, 48). Gel retardation analysis was performed essentially as described previously (38). One nanogram of 32P-labeled double-stranded oligonucleotide (described in the section on plasmid constructions) was incubated with 9 μg of nuclear extract prior to 4% polyacrylamide gel electrophoresis and autoradiography. When gel retardation competition analysis was performed, the cell extract was preincubated with 750 ng of cold double-stranded competitor oligonucleotide for 15 min prior to the addition of the 32P-labeled double-stranded oligonucleotide. The control double-stranded oligonucleotides derived from the nucleocapsid promoter, CpHNF4 (HNF4 site 1) and CpE (HNF3 site 2), and the enhancer 1/X-gene promoter, XpHNF4, have been described previously (23, 39).
RESULTS
Seroconversion from HBeAg- to anti-HBe antibody-positive status is not always associated with clearance of HBV infection. In the absence of viral clearance, this serological change is often associated with the selection of viral variants that synthesize reduced levels of HBeAg (1, 18, 20, 35). The most common variant contains a nucleotide substitution in codon 28 of the precore polypeptide that converts the tryptophan TGG codon to the amber TAG stop codon. Termination of the synthesis of the precore polypeptide at codon 28 results in a viral variant that cannot synthesize HBeAg (1, 18, 34). Another HBV variant associated with reduced HBeAg synthesis contains two nucleotide substitutions (A1764T and G1766A) in the proximal nuclear hormone receptor binding site in the nucleocapsid promoter (18, 35). This variant produces reduced levels of HBeAg because these nucleotide substitutions are associated with reduced levels of precore RNA synthesis but do not greatly affect pregenomic RNA synthesis and viral replication (2, 19, 32, 35). The mechanism by which the two nucleotide substitutions (A1764T and G1766A) produce the altered levels of the precore and pregenomic RNAs presumably reflects changes in the binding of nuclear hormone receptors to the nucleocapsid promoter.
Identification of the transcription factors binding to the variant proximal nuclear hormone receptor binding site in the nucleocapsid promoter.
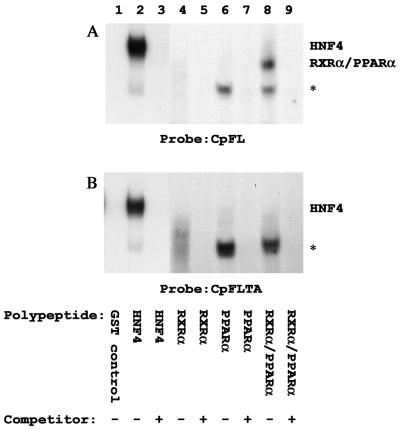
A previous study demonstrated that HNF4, RXRα-PPARα heterodimers, COUPTF1, and ARP1 bound to the proximal nuclear hormone receptor binding site in the nucleocapsid promoter (39). Therefore, gel shift analysis was performed to determine if these nuclear hormone receptors bound to the variant proximal nuclear hormone receptor binding site (Fig. 1, 2, and 3). Initially, the binding of the purified recombinant HNF4, RXRα, and PPARα polypeptides to the wild-type and variant proximal nuclear hormone receptor binding sites in the nucleocapsid promoters was examined by gel shift analysis using the double-stranded oligonucleotides CpFL and CpFLTA, respectively (Fig. 1 and 3). Purified recombinant HNF4 bound to both the wild-type and the variant sites, although the variant site appeared to bind HNF4 to a slightly lesser extent (Fig. 1, lane 2). A complex was observed between the GST-RXRα or GST-PPARα polypeptide preparations and the CpFL or CpFLTA double-stranded oligonucleotides (marked with an asterisk in Fig. 1, lanes 4 to 7). A complex migrating in this position has been observed with several different affinity-purified GST fusion protein preparations. Unlike the RXRα-PPARα heterodimer complex (Fig. 1, lane 8), an antibody to RXRα fails to supershift this complex (A. McLachlan, unpublished data), suggesting that it is a complex formed with a contaminating E. coli protein. However, as previously observed (39), in the presence of both the RXRα and the PPARα polypeptides, the RXRα-PPARα heterodimer complex appears as the predominant complex with CpFL, confirming that this heterodimeric nuclear hormone receptor binds to this nucleocapsid regulatory element (Fig. 1A, lane 8). In contrast, the RXRα-PPARα heterodimer complex was not formed with the CpFLTA double-stranded oligonucleotide, indicating that the two nucleotide substitutions in the variant sequence inhibit RXRα-PPARα binding to the proximal nuclear hormone receptor binding site in the nucleocapsid promoter (Fig. 1B, lane 8).
FIG. 1.
Gel retardation and complex inhibition analyses of the HBV wild-type and variant nucleocapsid promoter CpFL and CpFLTA regulatory elements with purified recombinant nuclear hormone receptors. 32P-labeled, double-stranded oligonucleotides CpFL (A) and CpFLTA (B) and purified recombinant GST (GST control; lane 1), GST-HNF4 (HNF4; lanes 2 and 3), GST-RXRα (RXRα; lanes 4 and 5), GST-PPARα (PPARα; lanes 6 and 7), and GST-RXRα plus GST-PPARα (RXRα/PPARα; lanes 8 and 9) were used for these analyses. Unlabeled, double-stranded oligonucleotides CpFL (A) and CpFLTA (B) were used as competitor DNAs to demonstrate the specificity of the observed complex (lanes 3, 5, 7, and 9). An asterisk indicates the position of a complex observed with several different affinity-purified GST fusion protein preparations, suggesting that it is a complex formed with a contaminating E. coli protein.
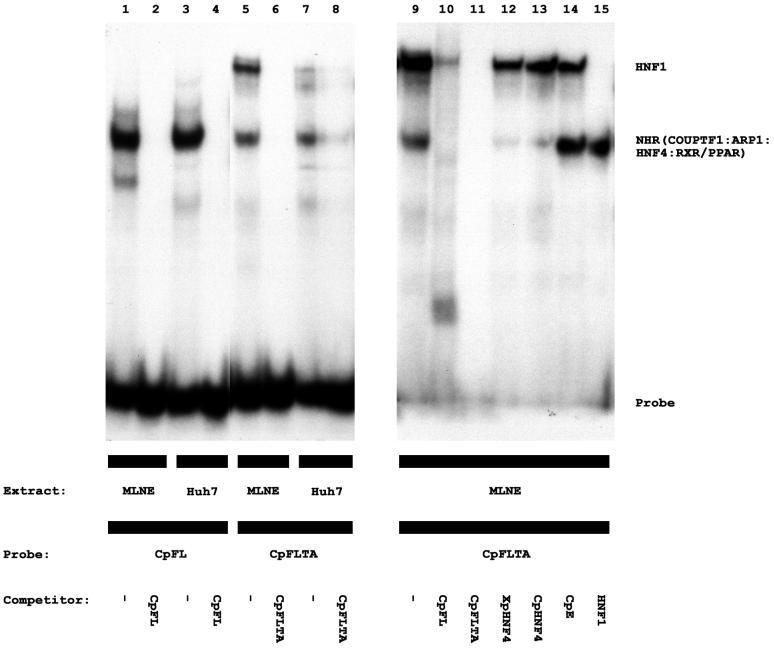
FIG. 2.
Gel retardation and complex inhibition analyses of the HBV wild-type and variant nucleocapsid promoter CpFL and CpFLTA regulatory elements with Huh7 cell and mouse liver nuclear extracts. 32P-labeled, double-stranded oligonucleotides CpFL (lanes 1 to 4) and CpFLTA (lanes 5 to 15) were used for these analyses with a Huh7 cell nuclear extract (lanes 3, 4, 7, and 8) and a mouse liver nuclear extract (MLNE; lanes 1, 2, 5, 6, and 9 to 15). Unlabeled, double-stranded oligonucleotides were used as competitor DNAs to demonstrate the specificity of the observed complexes. CpFL (lanes 2, 4, and 10), wild-type nucleocapsid promoter nuclear hormone receptor binding site (39); CpFLTA (lanes 6, 8, and 11), variant nucleocapsid promoter nuclear hormone receptor binding site; XpHNF4 (lane 12), enhancer 1/X-gene promoter nuclear hormone receptor binding site (39); CpHNF4 (lane 13), nucleocapsid promoter HNF4 binding site 1 (39); CpE (lane 14), nucleocapsid promoter HNF3 binding site 2 (23); HNF1, large surface antigen promoter HNF1 binding site (38). NHR, nuclear hormone receptor.
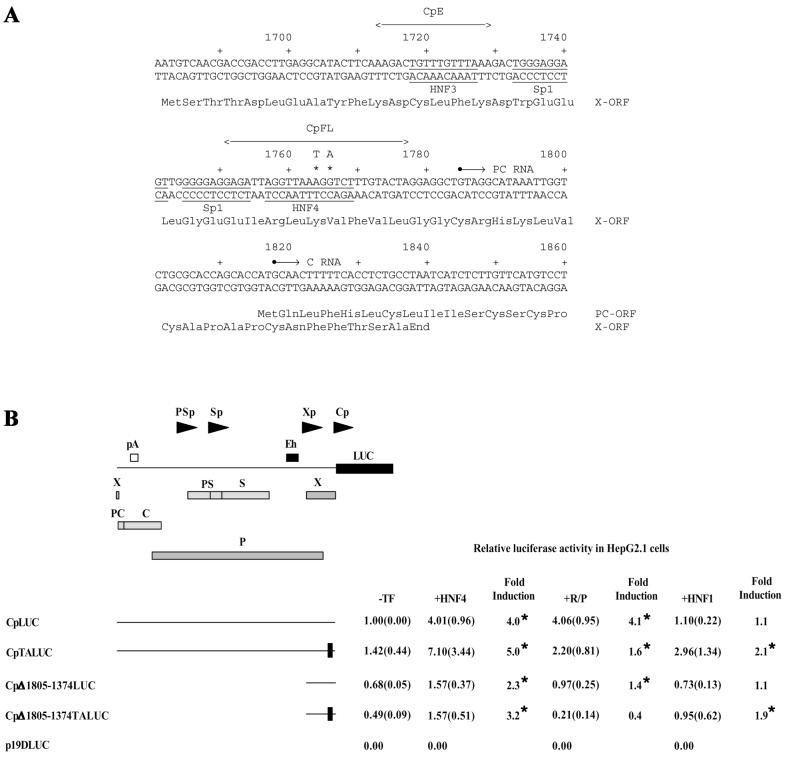
FIG. 3.
Functional analysis of the HBV wild-type and variant nucleocapsid promoters. (A) Nucleotide sequence of the HBV nucleocapsid promoter region (subtype ayw) (13). The nucleotide coordinates are derived from the GenBank database, and their positions relative to a predominant transcription initiation site (+1; nucleotide coordinate 1785) are shown (21, 22, 47, 53, 55). The underlined sequences represent the Sp1 recognition sequences (61), the HNF3 recognition sequence (23), and the HNF4:RXRα-PPARα nuclear hormone receptor recognition sequence (39). The sequences of the HBV double-stranded oligonucleotides CpE and CpFL are indicated. The A1764T and G1766A nucleotide substitutions in the nuclear hormone receptor binding site of the variant HBV genome are indicated above the wild-type HBV sequence. These nucleotide substitutions change lysine at position 130 of the X-gene polypeptide to methionine (Lys130Met) and valine at position 131 to isoleucine (Val131Ile). X-ORF, X-gene open reading frame; PC-ORF, precore open reading frame. (B) Diagrammatic representation of construct CpLUC. Arrowheads indicate the positions and directions of transcription from the HBV large surface antigen (PSp), major surface antigen (Sp), enhancer 1/X-gene (Xp), and nucleocapsid or core (Cp) promoters. Boxes indicate the positions of the HBV enhancer 1 sequence (Eh), HBV polyadenylation sequence (pA), X-gene ORF (X), presurface antigen ORF (PS), surface antigen ORF (S), precore ORF (PC), core ORF (C), polymerase ORF (P), and luciferase ORF (LUC). Below the diagram are shown relative activities of the wild-type and variant nucleocapsid promoters in HepG2.1 cells, in the absence (−TF) or presence of ectopically expressed HNF4 (+HNF4) polypeptide, RXRα-PPARα (+R/P) polypeptides, and HNF1α (+HNF1) polypeptide, using the expression vectors pCMV, pCMVHNF4, pRS-hRXRα, pCMVPPARα-G, and pMTHNF1α. All-trans-retinoic acid and clofibric acid at 1 μM and 1 mM, respectively, were used to activate the nuclear hormone receptors RXRα and PPARα. The activities are reported relative to the activity of the full-length promoter construct, CpLUC, in the absence of ectopically expressed polypeptides. The standard deviation for the mean is shown in parentheses. An asterisk indicates that the fold induction is statistically significant, as determined by Student's t test (P < 0.05). The internal control used to correct for transfection efficiencies was pCMVβ. The horizontal lines indicate the HBV sequences present in the CpLUC plasmids. CpLUC contains the HBV sequences from nucleotide coordinates 1805 to 3182 and 1 to 1804 (nucleotide sequences are designated by using coordinates derived from the GenBank genetic sequence database). The HBV sequences deleted from the various plasmids are designated by nucleotide coordinates. The locations of the A1764T and G1766A nucleotide substitutions in the nuclear hormone receptor binding site of the variant HBV genome are indicated by solid boxes.
The analysis with the recombinant polypeptides does not indicate the nature of all the factors that interact with the wild-type and variant proximal nuclear hormone receptor binding sites in the nucleocapsid promoters. In an attempt to characterize further the members of the nuclear hormone receptor superfamily of transcription factors that can interact with the CpFL and CpFLTA regulatory elements of the nucleocapsid promoter, gel shift and complex inhibition analyses were performed using Huh7 cell and mouse liver nuclear extracts (Fig. 2). Previously, it had been shown that COUPTF1, ARP1, HNF4, and RXRα-PPARα present in these extracts bound to the CpFL double-stranded oligonucleotide (39). A complex is formed with the CpFLTA double-stranded oligonucleotide that migrates in the same manner as the complex formed with the CpFL double-stranded oligonucleotide (Fig. 2). This result indicates that nuclear hormone receptors, most likely COUPTF1 and ARP1, which represent the predominant binding activities in Huh7 cells (39), can bind to the variant binding site, although apparently with a slightly reduced efficiency (Fig. 2). Complex inhibition analysis with nuclear hormone receptor binding sites indicates the specificity of this complex (Fig. 2, lanes 2, 4, 6, 8, and 10 to 13). The CpFLTA double-stranded oligonucleotide forms an additional, slower-migrating complex with the nuclear extracts (Fig. 2, lanes 5, 7, 9, 10, and 12 to 14). The formation of this complex is not inhibited by several double-stranded oligonucleotides that bind nuclear hormone receptors (Fig. 2, lanes 10, 12, and 13) but is inhibited by a double-stranded oligonucleotide that binds HNF1 (Fig. 2, lane 15). This observation supports the previous finding that the variant nuclear hormone receptor binding site can also interact with the HNF1 transcription factor (25).
HNF4 but not RXRα-PPARα transactivates expression from the variant HBV nucleocapsid promoter.
Ectopic expression of HNF4 or RXRα-PPARα activates transcription from the wild-type nucleocapsid promoter approximately fourfold in construct CpLUC, which contains the complete HBV genome located upstream of the LUC reporter gene (Fig. 3B). In contrast, in construct CpTALUC, HNF4 activates transcription from the variant nucleocapsid promoter approximately fivefold, whereas RXRα-PPARα minimally increases transcription from the variant nucleocapsid promoter (Fig. 3B). Similarly, HNF4 activates transcription from the nucleocapsid promoter in the CpΔ1805-1374LUC and CpΔ1805-1374TALUC constructs two- to threefold, indicating that the nucleotide substitutions in the proximal nuclear hormone receptor binding site do not affect promoter activity to a large extent. In contrast, the minimal RXRα-PPARα-mediated activation of transcription from the nucleocapsid promoter observed with the CpΔ1805-1374LUC construct was not apparent when the CpΔ1805-1374TALUC construct was examined (Fig. 3B). These observations support the suggestion that the proximal nuclear hormone receptor binding site in the nucleocapsid promoter of the variant genome cannot bind RXRα-PPARα and therefore is not responsive to transcriptional modulation by these specific transcription factors. In addition, the variant nucleocapsid promoter is very modestly responsive to HNF1α, unlike the wild-type nucleocapsid promoter (Fig. 3B). This effect does not involve the HNF1 site in the large surface antigen promoter, as the nucleocapsid promoter in the wild-type HBV genome is not activated by HNF1α. In addition, deletion of the large surface antigen promoter HNF1 binding site does not prevent the increase in transcription from the nucleocapsid promoter in the CpΔ1805-1374TALUC construct caused by HNF1α. This result indicates that HNF1α binding to the variant nucleocapsid promoter permits this transcription factor to modestly increase transcription from this promoter.
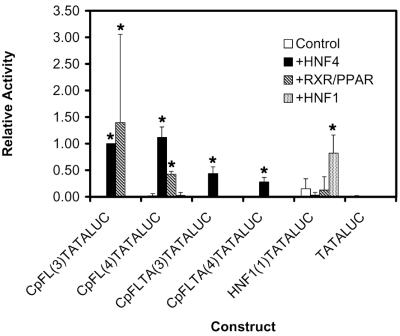
The analysis of the nucleocapsid promoter indicated that the variant nuclear hormone receptor binding site has altered transcription factor binding properties relative to the wild-type site and that these alterations have functional consequences for the modulation of the level of transcription from the nucleocapsid promoter by HNF4, RXRα-PPARα, and HNF1α. To verify further the functional properties of the wild-type and variant nuclear hormone receptor binding sites, they were examined for their capacity to mediate transcriptional activation by HNF4, RXRα-PPARα, and HNF1α in the context of a TATA-box element. The CpFL and CpFLTA double-stranded oligonucleotide synthetic promoter elements were cloned upstream of a TATA-box element and the LUC open reading frame (ORF) and were tested for their transcriptional activities in HepG2.1 cells in the presence or absence of the HNF4, RXRα-PPARα, and HNF1α expression vectors (Fig. 4). In the absence of the expression vectors, the constructs containing the wild-type nuclear hormone receptor site [(constructs CpFL(3)LUC and CpFL(4)LUC)] and the variant nuclear hormone receptor site [constructs CpFLTA(3)LUC and CpFLTA(4)LUC)] had very little transcriptional activity. The wild-type nuclear hormone receptor sites supported transcription from the minimal promoter constructs in response to HNF4 and RXRα-PPARα expression. The variant nuclear hormone receptor sites supported transcription from the minimal promoter constructs in response to HNF4 expression but not in response to RXRα-PPARα expression. These results demonstrate that the nucleotide substitutions in the variant nuclear hormone receptor site prevent RXRα-PPARα binding and therefore prevent this heterodimer from activating transcription from this element. In contrast, the variant nuclear hormone receptor site binds HNF4 to a lesser extent than the wild-type site, and this fact is reflected in the reduction in HNF4-mediated activation of transcription from the minimal promoter constructs (Fig. 4). Transcription from the large surface antigen promoter HNF1 binding site (HNF1TATALUC) was activated by the ectopic expression of the HNF1α transcription factor, whereas the variant nuclear hormone receptor site [constructs CpFLTA(3)LUC and CpFLTA(4)LUC] failed to support HNF1α-dependent transcription from the minimal promoter. These results suggest that the binding of HNF1α to this site is relatively weak and that the importance of HNF1α interacting with this site is apparent only in the context of the nucleocapsid promoter.
FIG. 4.
Functional analysis of the HBV wild-type and variant nucleocapsid promoter nuclear hormone receptor binding sites. The constructs examined contain the CpFL (wild-type nucleocapsid promoter proximal nuclear hormone receptor binding site), CpFLTA (variant nucleocapsid promoter proximal nuclear hormone receptor binding site), and PS1pHNF1 (large surface antigen promoter HNF1 site) double-stranded oligonucleotides cloned into the minimal promoter construct, pHBVTATALUC. The number of copies of the oligonucleotide in the construct is shown in parentheses in the construct name. Relative activities of the constructs in HepG2.1 cells, in the absence (Control) or presence of ectopically expressed HNF4 (+HNF4) polypeptide, RXRα-PPARα (+RXR/PPAR) polypeptides, and HNF1α (+HNF1) polypeptide, using the expression vectors pCMV, pCMVHNF4, pRS-hRXRα, pCMVPPARα-G, and pMTHNF1α are indicated. All-trans-retinoic acid and clofibric acid at 1 μM and 1 mM, respectively, were used to activate the nuclear hormone receptors RXRα and PPARα. The transcriptional activities are reported relative to that of plasmid CpFL(3)LUC in the presence of the HNF4 expression vector; this plasmid is designated as having a relative activity of 1.0. The standard deviation of the mean is indicated by an error bar. An asterisk indicates that the increase in relative activity is statistically significant, as determined by Student's t test (P < 0.05). The internal control used to correct for transfection efficiencies was pCMVβ.
Nuclear hormone receptors differentially regulate wild-type and variant HBV transcription and replication.
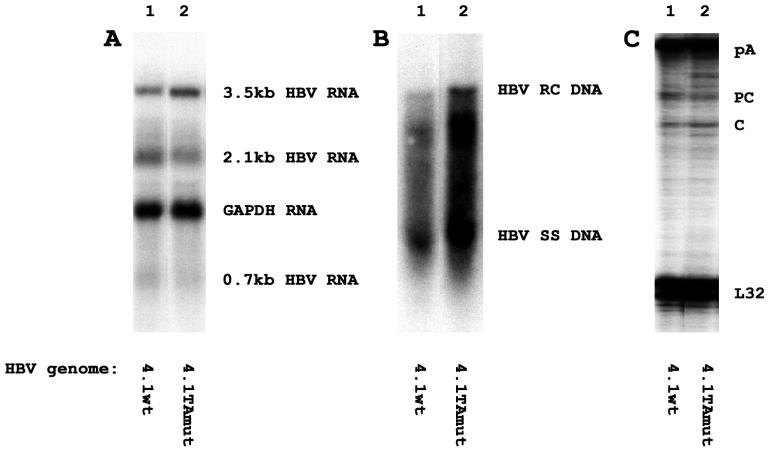
Wild-type and variant HBV DNAs are transcribed and replicated in the differentiated hepatoma cell line, HepG2 (Fig. 5). The levels of HBV 3.5- and 2.1-kb transcripts are very similar for the wild-type and variant HBV DNA 4.1-kbp constructs (Fig. 5A). The level of viral replication intermediates synthesized from the variant HBV DNA 4.1-kbp construct is less than twofold higher than that of the replication intermediates synthesized from the wild-type HBV DNA 4.1-kbp construct (Fig. 5B). This very modest increase in replication may occur as a result of subtle alterations in the levels of the precore and pregenomic RNAs transcribed from these constructs. Consistent with previous observations (2, 19, 32, 35), the variant construct supports a slightly lower level of transcription from the precore RNA initiation site and a slightly higher level of transcription from the pregenomic RNA initiation site than the wild-type construct (Fig. 5C). The modest increase in the level of pregenomic RNA synthesis can account for the slightly higher level of replication observed with the variant construct in HepG2 cells.
FIG. 5.
Transcription and replication of wild-type and variant HBV DNA 4.1-kbp constructs in the HepG2 cell line. Cells were transiently transfected with the wild-type HBV DNA 4.1-kbp construct (4.1wt; lanes 1) and the variant HBV DNA 4.1-kbp construct (4.1TAmut; lanes 2). (A) RNA (Northern) filter hybridization analysis of HBV transcripts. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript was used as an internal control for RNA loading per lane. (B) DNA (Southern) filter hybridization analysis of HBV replication intermediates. HBV RC DNA, HBV relaxed circular DNA; HBV SS DNA, HBV single-stranded DNA. (C) RNase protection analysis performed to map the transcription initiation sites of the HBV precore (PC) and pregenomic or core (C) transcripts. The HBV probe also protected a fragment (pA) derived from the 3′ ends of all the HBV RNAs that terminated at the HBV polyadenylation site. A riboprobe detecting the ribosomal gene L32 transcripts was included as an internal control.
The role of specific liver-enriched transcription factors in regulating the relative level of synthesis of the precore and pregenomic RNAs could not be analyzed with hepatoma cells, which express a variety of endogenous liver-enriched transcription factors. In an attempt to determine the role of specific liver-enriched transcription factors in controlling the level of synthesis of the precore and pregenomic RNAs from the wild-type and variant viral genomes, viral transcription and replication were examined in mouse NIH 3T3 fibroblasts (Fig. 6). In this system, viral replication is dependent on the ectopic expression of nuclear hormone receptors (52). Viral transcription and replication were observed using both wild-type and variant HBV DNA 4.1- and 3.9-kbp constructs (Fig. 6). The level of the 3.5-kb HBV RNA correlated with the level of viral replication when the wild-type HBV DNA constructs were examined (Fig. 6, lanes 1 to 6). However, the relative level of viral replication observed using the variant HBV DNA 4.1-kbp construct and ectopic expression of HNF4 was higher than expected based on the level of the HBV 3.5-kb RNA transcribed from this template (Fig. 6A and B, lanes 8 and 11). In addition, the relative level of viral replication observed using the variant HBV DNA constructs and ectopic expression of RXRα-PPARα was lower than expected based on the level of the HBV 3.5-kb RNA transcribed from this template (Fig. 6, lanes 9 and 12). As a result of these differences between the wild-type and variant HBV genomes, RXRα-PPARα supports a higher level of viral replication from the wild-type HBV DNA constructs than from the variant HBV DNA constructs. In contrast, HNF4 supports a higher level of viral replication from the variant HBV DNA constructs than from the wild-type HBV DNA constructs (Fig. 6B and D). Therefore, alterations in the relative activities of the HNF4 and RXRα-PPARα transcription factors due to changes in the physiological conditions in the liver could result in the preferential amplification of a specific viral genotype.
FIG. 6.
Nuclear hormone receptors activate wild-type and variant HBV replication in the mouse fibroblast cell line NIH 3T3. Cells were transiently transfected with the wild-type HBV DNA 4.1-kbp construct (4.1wt; A and B, lanes 1 to 6), the variant HBV DNA 4.1-kbp construct (4.1TAmut; A and B, lanes 7 to 12), the wild-type HBV DNA 3.9-kbp construct (3.9wt; C and D, lanes 1 to 6), and the variant HBV DNA 3.9-kbp construct (3.9TAmut; C and D, lanes 7 to 12) and liver-enriched transcription factor expression vectors as indicated. (A and C) RNA (Northern) filter hybridization analysis of HBV transcripts. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript was used as an internal control for RNA loading per lane. (B and D) DNA (Southern) filter hybridization analysis of HBV replication intermediates. HBV RC DNA, HBV relaxed circular DNA; HBV SS DNA, HBV single-stranded DNA. All-trans-retinoic acid and clofibric acid at 1 μM and 1 mM, respectively, were used to activate the nuclear hormone receptors RXRα and PPARα (+lig).
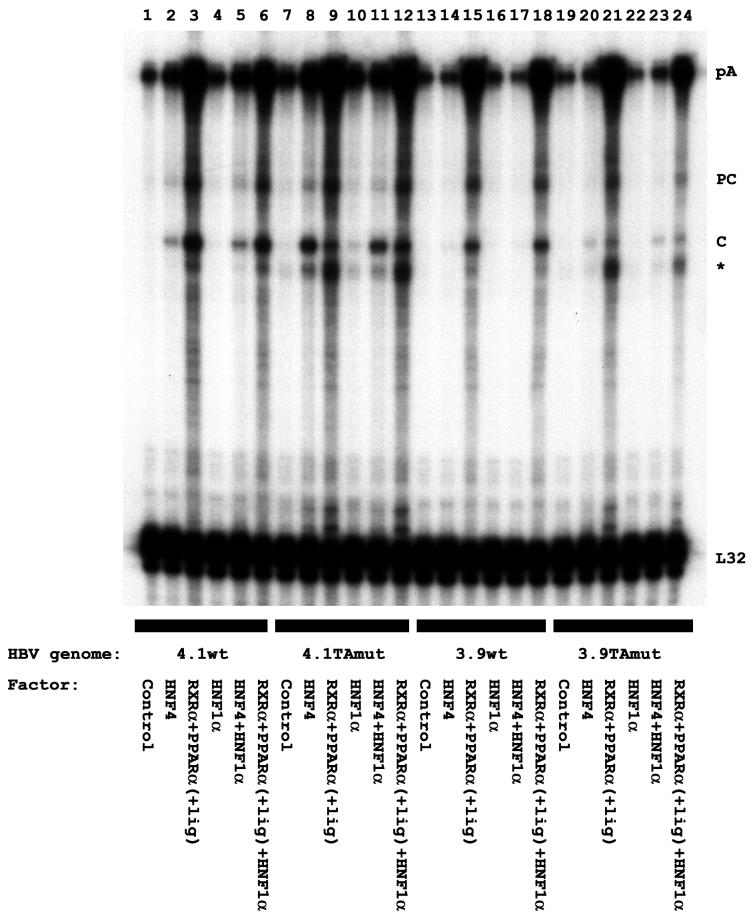
The reason that the variant HBV DNA constructs failed to demonstrate a correlation between the level of the HBV 3.5-kb RNA and viral replication was investigated further by examining the relative levels of the precore and pregenomic RNAs (Fig. 7). The wild-type HBV DNA constructs transcribed the precore and pregenomic RNAs at a ratio of approximately 0.9 when activated by the ectopic expression of either HNF4 or RXRα-PPARα. In contrast, the variant HBV DNA constructs transcribed the precore and pregenomic RNAs at ratios of approximately 0.4 and 1.5 when activated by the ectopic expression of HNF4 and RXRα-PPARα, respectively. Therefore, the variant genome is preferentially transcribed from the pregenomic RNA initiation site by the ectopic expression of HNF4 and preferentially transcribed from the precore RNA initiation site by the ectopic expression of RXRα-PPARα. These observations may explain the higher level of replication relative to the level of HBV 3.5-kb RNA detected when transcription from the variant genome is activated by HNF4. Likewise, the lower level of replication relative to the level of HBV 3.5-kb RNA observed when transcription from the variant genome is activated by RXRα-PPARα reflects the higher proportion of precore RNA present in the 3.5-kb transcripts.
FIG. 7.
Effect of liver-enriched transcription factors on the initiation site of the HBV 3.5-kb RNA transcribed from the wild-type and variant HBV genomes. Mouse NIH 3T3 fibroblasts were transiently transfected with the wild-type HBV DNA 4.1-kbp construct (4.1wt; lanes 1 to 6), the variant HBV DNA 4.1-kbp construct (4.1TAmut; lanes 7 to 12), the wild-type HBV DNA 3.9-kbp construct (3.9wt; lanes 13 to 18), and the variant HBV DNA 3.9-kbp construct (3.9TAmut; lanes 19 to 24) and liver-enriched transcription factor expression vectors as indicated. RNase protection analysis was performed to map the transcription initiation sites of the HBV precore (PC) and pregenomic or core (C) transcripts. The HBV probe also protected a fragment (pA) derived from the 3′ ends of all the HBV RNAs that terminated at the HBV polyadenylation site. The protected fragment indicated with an asterisk was generated as a result of the cleavage of the pA protected fragment at the site of the discontinuity between the wild-type probe and the variant HBV RNA. A riboprobe detecting the ribosomal L32 gene transcripts was included as an internal control. All-trans-retinoic acid and clofibric acid at 1 μM and 1 mM, respectively, were used to activate the nuclear hormone receptors RXRα and PPARα (+lig).
Examination of the ratio of precore to pregenomic RNAs indicates that nuclear hormone receptors can influence the synthesis of these two transcripts from the variant HBV genome and thus alter the level of viral replication. Viral replication is dependent on the level of pregenomic RNAs and also may be modulated by HBeAg encoded by precore transcripts (16, 46). To investigate the relative importance of these two factors in determining the level of viral replication, the level of viral replication relative to the level of pregenomic RNA synthesis was examined (Fig. 6 and 7). The level of viral replication correlated with the level of pregenomic RNA synthesis, except for the variant HBV DNA constructs activated with RXRα-PPARα (Fig. 6B and D, lanes 9 and 12, and Fig. 7, lanes 9, 12, 21, and 24). In these instances, the level of viral replication was lower than expected based on the abundance of the pregenomic RNA. However, the level of the precore RNA synthesized was also high, especially relative to that of the pregenomic RNA, suggesting that the precore RNA or the HBeAg that it encodes may be inhibiting viral replication.
As expected, HNF1 did not affect viral transcription or replication from the wild-type HBV DNA constructs. However, HNF1 can support low levels of synthesis of the HBV 3.5-kb RNA and viral replication from the variant HBV DNA 4.1-kbp construct (Fig. 6A and B, lane 10). These results suggest that binding of HNF1 in the location of the nucleocapsid promoter proximal nuclear hormone receptor binding site can support pregenomic RNA synthesis and viral replication to a modest extent (Fig. 6B, lane 10, and Fig. 7, lane 10). The variant HBV DNA 3.9-kbp construct failed to support detectable HNF1-mediated pregenomic RNA synthesis and viral replication (Fig. 6D, lane 10, and Fig. 7, lane 22), presumably because the enhancer 1 region also contributes to the observed level of transcription from the nucleocapsid promoter in this system. However, it appears that although HNF1 does not alter HNF4-mediated transcription and replication from the variant HBV constructs (Fig. 6, lanes 8 and 11), HNF1 may act in synergy with RXRα-PPARα to activate viral replication (Fig. 6, lanes 9 and 12). The level of the HBV 3.5-kb RNA synthesized is not affected to a large extent by the additional ectopic expression of HNF1 (Fig. 6A and C, lanes 9 and 12). However, HNF1 may subtly alter the ratio of precore to pregenomic RNAs in favor of the pregenomic RNA (Fig. 7, lanes 9, 12, 21, and 24). This increase in the level of the template for viral replication and the reduction in the level of HBeAg from the precore RNA may account for the resulting increase in viral replication (Fig. 6B and D, lanes 9 and 12). These results indicate that the wild-type and variant HBV genomes are differentially transcribed by the liver-enriched transcription factors HNF4, RXRα-PPARα, and HNF1, resulting in different levels of viral replication in response to these factors. These results also suggest that the physiological state of hepatocytes may differentially influence the rates of replication of wild-type and variant viral genomes.
DISCUSSION
The mechanisms determining the emergence of viral variants in response to immune selection may be important in considering possible therapeutic interventions designed to prevent their pathophysiological consequences. Several HBV variants with altered amino acid sequences in immunodominant epitopes of the major surface antigen have been identified and presumably escape elimination because they are not recognized by the normal antibody responses to the viral envelope proteins (18). Similarly, viral variants that fail to express HBeAg may not be efficiently cleared from patients in whom the immune response is primarily directed against nucleocapsid antigens (1, 18, 34, 35). Consistent with this suggestion, various viral variants have been obtained from patients who have seroconverted from HBeAg-positive to anti-HBe antibody-positive status. The most common mutations observed in these viral variants contain in the precore coding region a stop codon that prevents the synthesis of HBeAg (1, 18, 34). A second mutation that is also associated with seroconversion to anti-HBe antibody-positive status is a double nucleotide substitution (A1764T and G1766A) in the proximal nuclear hormone receptor binding site of the nucleocapsid promoter (Fig. 3). This mutation has been reported to reduce HBeAg synthesis by inhibiting the synthesis of the precore RNA without greatly affecting the level of the pregenomic RNA or HBV DNA synthesis (2, 19, 32, 35). Interestingly, this double nucleotide substitution is often found associated with the precore stop codon mutation (1), suggesting that alterations in the nuclear hormone receptor binding site may regulate HBV transcription and replication by mechanisms more complex than simple reduction of precore RNA and HBeAg syntheses.
The effects of the nucleotide substitutions in the proximal nuclear hormone receptor binding site of the nucleocapsid promoter on transcription factor binding were initially characterized by gel shift analysis (Fig. 1 and 2). This analysis demonstrated that HNF4 bound both wild-type and variant recognition elements, whereas the RXRα-PPARα heterodimer bound only the wild-type sequence. Gel shift analysis with Huh7 cell and mouse liver nuclear extracts also indicates that COUPTF1 and ARP1 bind to both wild-type and variant nuclear hormone receptor binding sites. These observations are distinct from previous results suggesting that the variant recognition site could not bind nuclear hormone receptors (3, 25). The reasons for these differences are unclear. However, this analysis did indicate that the variant recognition site binds HNF1 (Fig. 2), as previously described (25).
Functional analysis using reporter gene constructs confirmed that HNF4 and RXRα-PPARα activate transcription by binding to the wild-type proximal nuclear hormone receptor recognition sequence, whereas HNF4 but not RXRα-PPARα activates transcription by binding to the variant recognition sequence (Fig. 3 and 4). HNF1 also activates transcription to a limited extent from the variant nucleocapsid promoter (Fig. 3), although it is not able to activate transcription from the variant recognition sequence in the context of a minimal promoter (Fig. 4). These observations suggest that HNF1 interacts weakly with the variant recognition sequence.
The consequences of the nucleotide substitutions in the proximal nuclear hormone receptor recognition sequence for HBV transcription and replication in human hepatoma cells have been somewhat variable (2, 19, 32, 35). However, the majority of the results suggest that HBV transcription is affected to a limited extent. The level of the precore RNA is somewhat decreased, whereas the level of the pregenomic RNA is relatively unchanged or is slightly increased (Fig. 5). As a consequence of these subtle alterations, viral replication is modestly increased (Fig. 5). A possible explanation for the reported differences is suggested by the effect of specific nuclear hormone receptors on precore and pregenomic RNA syntheses and viral replication (Fig. 6 and 7). Analysis performed under slightly different conditions, even with the same hepatoma cell line, may lead to different levels of activities of the HNF4 and RXRα-PPARα transcription factors that could affect both the levels of precore and pregenomic RNAs and consequently viral replication.
Analysis of the HBV genome in a nonhepatoma cell line permits the effects of individual liver-enriched transcription factors on the regulation of viral transcription and replication to be analyzed (52). Using this approach, it has been possible to establish the relative importance of various transcription factors to the regulation of wild-type and variant viral transcription and replication (Fig. 6 and 7). Most importantly, it is apparent that replication of the wild-type and variant viruses is differentially regulated by HNF4 and RXRα-PPARα (Fig. 6). RXRα-PPARα preferentially activates wild-type viral replication, whereas HNF4 preferentially activates variant viral replication. Therefore, under circumstances in which RXRα-PPARα is more active than HNF4 in regulating HBV transcription, the wild-type virus might be expected to predominate over the variant virus. In contrast, when HNF4 is more active than RXRα-PPARα in regulating HBV transcription, the variant virus might be expected to be predominant relative to the wild-type virus. If the activities of these factors are important in the selection of the variant virus, it might be expected that HNF4 activity would increase relative to RXRα-PPARα activity in the liver during an anti-HBV immune response.
Nuclear hormone receptors regulate viral replication by modulating both the absolute level of transcription from the nucleocapsid promoter and the relative levels of precore and pregenomic RNAs (Fig. 6 and 7). RXRα-PPARα activates HBV 3.5-kb RNA synthesis to a greater extent than does HNF4. For the wild-type HBV genome, these nuclear hormone receptors do not alter the relative levels of the precore and pregenomic RNAs synthesized. In contrast, HNF4 preferentially activates transcription from the pregenomic RNA initiation site and RXRα-PPARα preferentially activates transcription from the precore RNA initiation site of the variant HBV genome (Fig. 7). These data account for the higher level of variant replication supported by the ectopic expression of HNF4 compared with RXRα-PPARα. In addition, these observations support the suggestion that the binding of HNF4 or RXRα-PPARα to the proximal nuclear hormone receptor binding site may inhibit the formation of a preinitiation transcription complex in this region of the nucleocapsid promoter and therefore restrict transcription from the precore RNA initiation site (57). In contrast, the binding of a preinitiation transcription complex to this region may mediate RNA synthesis from the precore RNA initiation site (56). As the proximal nuclear hormone receptor binding site of the nucleocapsid promoter of the variant viral genome cannot bind RXRα-PPARα, the preinitiation transcription complex may have more direct access to this site, resulting in preferential transcription from the precore initiation site. This situation would generate the observed higher relative level of precore RNA than of pregenomic RNA (Fig. 7).
It appears unlikely that the amino acid differences in the X-gene polypeptides (Lys130Met and Val131Ile) encoded by the wild-type and variant HBV genomes contribute to the differences in the levels of replication of these viral genomes in response to the nuclear hormone receptors. The levels of the HBV 3.5-kb RNA synthesized from the wild-type and variant HBV genomes are similar when transcription is activated by HNF4 (Fig. 6). A similar result is observed when transcription is activated by RXRα-PPARα (Fig. 6). These findings suggest that if the wild-type and variant X-gene polypeptides modulate the level of transcription from the nucleocapsid promoter, their effects are similar. In addition, the relative levels of precore and pregenomic RNAs transcribed from the wild-type HBV genome in response to HNF4 and RXRα-PPARα are similar (Fig. 7). This result indicates that the wild-type X-gene polypeptide does not modulate the relative utilization of the transcription initiation sites when HBV 3.5-kb RNA synthesis is activated by different nuclear hormone receptors. Consequently, it appears unlikely that the variant X-gene polypeptide is responsible for transcription mediated by HNF4 initiating preferentially at the pregenomic RNA initiation site and transcription mediated by RXRα-PPARα initiating preferentially at the precore RNA initiation site of the variant HBV genome (Fig. 7).
The observation that wild-type and variant viral replication is differentially regulated by ligand-dependent nuclear hormone receptors suggests that the transcriptional regulation of these viruses may have an important role in the observed emergence of the variant virus during an immune response directed against viral antigens. Activated Kupffer cells secrete a variety of cytokines and lipid mediators of the immune response, including leukotrienes and prostaglandins, putative physiological ligands of PPARα and PPARγ (6, 11, 12, 24). Therefore, it is possible that extracellular stimuli generated during an antiviral immune response alter the physiology of the hepatocytes in a manner modulating the relative activities of transcription factors, including HNF1, HNF4, RXRα, and PPARα. These changes may differentially modulate wild-type and variant HBV replication, contributing to the immune selection of the HBV variant with the double nucleotide substitution (A1764T and G1766A) in the proximal nuclear hormone receptor binding site of the nucleocapsid promoter.
ACKNOWLEDGMENTS
We are grateful to Eric F. Johnson (The Scripps Research Institute, La Jolla, Calif.) for plasmids pGEXHNF4, pGEXRXRα, pGEXPPARα, pCMVHNF4, and pCMVPPARα-G; Ronald M. Evans (The Salk Institute, La Jolla, Calif.) for plasmid pRS-hRXRα; and Riccardo Cortese (Instituto di Ricerche di Biologia Molecolare, Rome, Italy) for plasmid pB1.1 (rat HNF1α cDNA).
This work was supported by a postdoctoral fellowship from the West China University of Medical Sciences, Chengdu, People's Republic of China, to H.T. and Public Health Service grant AI30070 from the National Institutes of Health.
Footnotes
Publication 13968-CB from The Scripps Research Institute.
REFERENCES
- 1.Akahane Y, Yamanaka T, Suzuki H, Sugai Y, Tsuda F, Yotsumoto S, Omi S, Okamoto H, Miyakawa Y, Mayumi M. Chronic active hepatitis with hepatitis B virus DNA and antibody against e antigen in the serum. Disturbed synthesis and secretion of e antigen from hepatocytes due to a point mutation in the precore region. Gastroenterology. 1990;99:1113–1119. doi: 10.1016/0016-5085(90)90632-b. [DOI] [PubMed] [Google Scholar]
- 2.Buckwold V E, Xu Z C, Chen M, Yen T S B, Ou J H. Effects of a naturally occurring mutation in the hepatitis B virus basal core promoter on precore gene expression and viral replication. J Virol. 1996;70:5845–5851. doi: 10.1128/jvi.70.9.5845-5851.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buckwold V E, Xu Z C, Yen T S B, Ou J H. Effects of a frequent double-nucleotide basal core promoter mutation and its putative single-nucleotide precursor mutations on hepatitis B virus gene expression and replication. J Gen Virol. 1997;78:2055–2065. doi: 10.1099/0022-1317-78-8-2055. [DOI] [PubMed] [Google Scholar]
- 4.Chen D, Lepar G, Kemper B. A transcriptional regulatory element common to a large family of hepatic cytochrome P450 genes is a functional binding site of the orphan receptor HNF-4. J Biol Chem. 1994;269:5420–5427. [PubMed] [Google Scholar]
- 5.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 6.Devchand P R, Keller H, Peters J M, Vazquez M, Gonzalez F J, Wahli W. The PPARα-leukotriene B4 pathway to inflammation control. Nature. 1996;384:39–43. doi: 10.1038/384039a0. [DOI] [PubMed] [Google Scholar]
- 7.De Wet J R, Wood K V, DeLuca M, Helinski D R, Subramani S. Firefly luciferase gene: structure and expression in mammalian cells. Mol Cell Biol. 1987;7:725–737. doi: 10.1128/mcb.7.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubois M F, Pourcel C, Rousset S, Chany C, Tiollais P. Excretion of hepatitis B surface antigen particles from mouse cells transformed with cloned viral DNA. Proc Natl Acad Sci USA. 1980;77:4549–4553. doi: 10.1073/pnas.77.8.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dudov K P, Perry R P. The gene family encoding the mouse ribosomal protein L32 contains a uniquely expressed intron-containing gene and an unmutated processed gene. Cell. 1984;37:457–468. doi: 10.1016/0092-8674(84)90376-3. [DOI] [PubMed] [Google Scholar]
- 11.Forman B M, Chen J, Evans R M. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors α and δ. Proc Natl Acad Sci USA. 1997;94:4312–4317. doi: 10.1073/pnas.94.9.4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forman B M, Tontonoz P, Chen J, Brun R P, Spiegelman B M, Evans R M. 15-Deoxy-delta12,14-prostaglandin J2 is a ligand for the adipocyte determination factor PPARgamma. Cell. 1995;83:803–812. doi: 10.1016/0092-8674(95)90193-0. [DOI] [PubMed] [Google Scholar]
- 13.Galibert F, Mandart E, Fitoussi F, Tiollais P, Charnay P. Nucleotide sequence of the hepatitis B virus genome (subtype ayw) cloned in E. coli. Nature. 1979;281:646–650. doi: 10.1038/281646a0. [DOI] [PubMed] [Google Scholar]
- 14.Ganem D, Varmus H E. The molecular biology of the hepatitis B viruses. Annu Rev Biochem. 1987;56:651–693. doi: 10.1146/annurev.bi.56.070187.003251. [DOI] [PubMed] [Google Scholar]
- 15.Graham F L, Van der Eb A J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 16.Guidotti L G, Matzke B, Pasquinelli C, Shoenberger J M, Rogler C E, Chisari F V. Hepatitis B virus (HBV) precore protein inhibits HBV replication in transgenic mice. J Virol. 1996;70:7056–7061. doi: 10.1128/jvi.70.10.7056-7061.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo W, Chen M, Yen T S B, Ou J-H. Hepatocyte-specific expression of the hepatitis B virus core promoter depends on both positive and negative regulation. Mol Cell Biol. 1993;13:443–448. doi: 10.1128/mcb.13.1.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Günther S, Fischer L, Pult I, Sterneck M, Will H. Naturally occurring variants of hepatitis B virus. Adv Virus Res. 1999;52:25–137. doi: 10.1016/s0065-3527(08)60298-5. [DOI] [PubMed] [Google Scholar]
- 19.Günther S, Piwon N, Will H. Wild-type levels of pregenomic RNA and replication but reduced pre-C RNA and e-antigen synthesis of hepatitis B virus with C(1653)→T, A(1762)→T and G(1764)→A mutations in the core promoter. J Gen Virol. 1998;79:375–380. doi: 10.1099/0022-1317-79-2-375. [DOI] [PubMed] [Google Scholar]
- 20.Harrison T J, Zuckerman A J. Variants of hepatitis B virus. Vox Sang. 1992;63:161–167. doi: 10.1111/j.1423-0410.1992.tb05094.x. [DOI] [PubMed] [Google Scholar]
- 21.Honigwachs J, Faktor O, Dikstein R, Shaul Y, Laub O. Liver-specific expression of hepatitis B virus is determined by the combined action of the core gene promoter and the enhancer. J Virol. 1989;63:919–924. doi: 10.1128/jvi.63.2.919-924.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu K-Q, Siddiqui A. Regulation of the hepatitis B virus gene expression by the enhancer element I. Virology. 1991;181:721–726. doi: 10.1016/0042-6822(91)90906-r. [DOI] [PubMed] [Google Scholar]
- 23.Johnson J L, Raney A K, McLachlan A. Characterization of a functional hepatocyte nuclear factor 3 binding site in the hepatitis B virus nucleocapsid promoter. Virology. 1995;208:147–158. doi: 10.1006/viro.1995.1138. [DOI] [PubMed] [Google Scholar]
- 24.Kliewer S A, Lenhard J M, Willson T M, Patel I, Morris D C, Lehmann J M. A prostaglandin J2 metabolite binds peroxisome proliferator-activated receptor τ and promotes adipocyte differentiation. Cell. 1995;83:813–819. doi: 10.1016/0092-8674(95)90194-9. [DOI] [PubMed] [Google Scholar]
- 25.Li J, Buckwold V E, Hon M W, Ou J H. Mechanism of suppression of hepatitis B virus precore RNA transcription by a frequent double mutation. J Virol. 1999;73:1239–1244. doi: 10.1128/jvi.73.2.1239-1244.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li M, Xie Y H, Wu X, Kong Y Y, Wang Y. HNF3 binds and activates the second enhancer, ENII, of hepatitis B virus. Virology. 1995;214:371–378. doi: 10.1006/viro.1995.0046. [DOI] [PubMed] [Google Scholar]
- 27.López-Cabrera M, Letovsky J, Hu K-Q, Siddiqui A. Multiple liver-specific factors bind to the hepatitis B virus core/pregenomic promoter: trans-activation and repression by CCAAT/enhancer binding protein. Proc Natl Acad Sci USA. 1990;87:5069–5073. doi: 10.1073/pnas.87.13.5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.López-Cabrera M, Letovsky J, Hu K-Q, Siddiqui A. Transcriptional factor C/EBP binds to and transactivates the enhancer element II of the hepatitis B virus. Virology. 1991;183:825–829. doi: 10.1016/0042-6822(91)91019-d. [DOI] [PubMed] [Google Scholar]
- 29.Mangelsdorf D J, Ong E S, Dyck J A, Evans R M. Nuclear receptor that identifies a novel retinoic acid response pathway. Nature. 1990;345:224–229. doi: 10.1038/345224a0. [DOI] [PubMed] [Google Scholar]
- 30.McLachlan A. Molecular biology of the hepatitis B virus. Boca Raton, Fla: CRC Press, Inc.; 1991. [Google Scholar]
- 31.McLachlan A, Milich D R, Raney A K, Riggs M G, Hughes J L, Sorge J, Chisari F V. Expression of hepatitis B virus surface and core antigens: influences of pre-s and precore sequences. J Virol. 1987;61:683–692. doi: 10.1128/jvi.61.3.683-692.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moriyama K, Okamoto H, Tsuda F, Mayumi M. Reduced precore transcription and enhanced core-pregenome transcription of hepatitis B virus DNA after replacement of the precore-core promoter with sequences associated with e antigen-seronegative persistent infections. Virology. 1996;226:269–280. doi: 10.1006/viro.1996.0655. [DOI] [PubMed] [Google Scholar]
- 33.Muerhoff A S, Griffin K J, Johnson E F. The peroxisome profilerator-activated receptor mediates the induction of CYP4A6, a cytochrome P450 fatty acid omega-hydroxylase, by clofibric acid. J Biol Chem. 1992;267:19051–19053. [PubMed] [Google Scholar]
- 34.Naoumov N V, Schneider R, Grötzinger T, Jung M C, Miska S, Pape G R, Will H. Precore mutant hepatitis B virus infection and liver disease. Gastroenterology. 1992;102:538–543. doi: 10.1016/0016-5085(92)90101-4. [DOI] [PubMed] [Google Scholar]
- 35.Okamoto H, Tsuda F, Akahane Y, Sugai Y, Yoshiba M, Moriyama K, Tanaka T, Miyakawa Y, Mayumi M. Hepatitis B virus with mutations in the core promoter for an e antigen-negative phenotype in carriers with antibody to e antigen. J Virol. 1994;68:8102–8110. doi: 10.1128/jvi.68.12.8102-8110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ou J-H, Bao H, Shih C, Tahara S M. Preferred translation of human hepatitis B virus polymerase from core protein- but not from precore protein-specific transcript. J Virol. 1990;64:4578–4581. doi: 10.1128/jvi.64.9.4578-4581.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raney A K, Easton A J, McLachlan A. Characterization of the minimal elements of the hepatitis B virus large surface antigen promoter. J Gen Virol. 1994;75:2671–2679. doi: 10.1099/0022-1317-75-10-2671. [DOI] [PubMed] [Google Scholar]
- 38.Raney A K, Easton A J, Milich D R, McLachlan A. Promoter-specific transactivation of hepatitis B virus transcription by a glutamine- and proline-rich domain of hepatocyte nuclear factor 1. J Virol. 1991;65:5774–5781. doi: 10.1128/jvi.65.11.5774-5781.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raney A K, Johnson J L, Palmer C N A, McLachlan A. Members of the nuclear receptor superfamily regulate transcription from the hepatitis B virus nucleocapsid promoter. J Virol. 1997;71:1058–1071. doi: 10.1128/jvi.71.2.1058-1071.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raney A K, Milich D R, Easton A J, McLachlan A. Differentiation specific transcriptional regulation of the hepatitis B virus large surface antigen gene in human hepatoma cell lines. J Virol. 1990;64:2360–2368. doi: 10.1128/jvi.64.5.2360-2368.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raney A K, Milich D R, McLachlan A. Characterization of hepatitis B virus major surface antigen gene transcriptional regulatory elements in differentiated hepatoma cell lines. J Virol. 1989;63:3919–3925. doi: 10.1128/jvi.63.9.3919-3925.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raney A K, Zhang P, McLachlan A. Regulation of transcription from the hepatitits B virus large surface antigen promoter by hepatocyte nuclear factor 3. J Virol. 1995;69:3265–3272. doi: 10.1128/jvi.69.6.3265-3272.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 44.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sato S, Suzuki K, Akahane Y, Akamatsu K, Akiyama K, Yunomura K, Tsuda F, Tanaka T, Okamoto H, Miyakawa Y, Mayumi M. Hepatitis B virus strains with mutations in the core promoter in patients with fulminant hepatitis. Ann Intern Med. 1995;122:241–248. doi: 10.7326/0003-4819-122-4-199502150-00001. [DOI] [PubMed] [Google Scholar]
- 46.Scaglioni P P, Melegari M, Wands J R. Posttranscriptional regulation of hepatitis B virus replication by the precore protein. J Virol. 1997;71:345–353. doi: 10.1128/jvi.71.1.345-353.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sells M A, Zelent A Z, Shvartsman M, Acs G. Replicative intermediates of hepatitis B virus in HepG2 cells that produce infectious virions. J Virol. 1988;62:2836–2844. doi: 10.1128/jvi.62.8.2836-2844.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sladek F M, Zhong W, Lai E, Darnell J E., Jr Liver-enriched transcription factor HNF-4 is a novel member of the steroid hormone receptor superfamily. Genes Dev. 1990;4:2353–2365. doi: 10.1101/gad.4.12b.2353. [DOI] [PubMed] [Google Scholar]
- 49.Smith D B, Johnson K S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 50.Sorge J, Wright D, Erdman V D, Cutting A E. Amphotropic retrovirus vector system for human cell gene transfer. Mol Cell Biol. 1984;4:1730–1737. doi: 10.1128/mcb.4.9.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Summers J, Smith P M, Huang M, Yu M. Morphogenetic and regulatory effects of mutations in the envelope proteins of an avian hepadnavirus. J Virol. 1991;65:1310–1317. doi: 10.1128/jvi.65.3.1310-1317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tang H, McLachlan A. Transcriptional regulation of hepatitis B virus by nuclear hormone receptors is a critical determinant of viral tropism. Proc Natl Acad Sci USA. 2001;98:1841–1846. doi: 10.1073/pnas.041479698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Waisman A, Aloni Y, Laub O. In vitro regulation of human hepatitis B virus core gene transcription. Virology. 1990;177:737–744. doi: 10.1016/0042-6822(90)90540-8. [DOI] [PubMed] [Google Scholar]
- 54.Will H, Reiser W, Weimer T, Pfaff E, Buscher M, Sprengle R, Cattaneo R, Schaller H. Replication strategy of human hepatitis B virus. J Virol. 1987;61:904–911. doi: 10.1128/jvi.61.3.904-911.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yaginuma K, Shirakata Y, Kobayashi M, Koike K. Hepatitis B virus (HBV) particles are produced in a cell culture system by transient expression of transfected HBV DNA. Proc Natl Acad Sci USA. 1987;84:2678–2682. doi: 10.1073/pnas.84.9.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu X M, Mertz J E. Promoters for synthesis of the pre-C and pregenomic mRNAs of human hepatitis B virus are genetically distinct and differentially regulated. J Virol. 1996;70:8719–8726. doi: 10.1128/jvi.70.12.8719-8726.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu X M, Mertz J E. Differential regulation of the pre-C and pregenomic promoters of human hepatitis B virus by members of the nuclear receptor superfamily. J Virol. 1997;71:9366–9374. doi: 10.1128/jvi.71.12.9366-9374.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yuh C-H, Ting L-P. C/EBP-like proteins binding to the functional box-α and box-β of the second enhancer of hepatitis B virus. Mol Cell Biol. 1991;11:5044–5052. doi: 10.1128/mcb.11.10.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang P, McLachlan A. Differentiation-specific transcriptional regulation of the hepatitis B virus nucleocapsid gene in human hepatoma cell lines. Virology. 1994;202:430–440. doi: 10.1006/viro.1994.1359. [DOI] [PubMed] [Google Scholar]
- 60.Zhang P, Raney A K, McLachlan A. Characterization of the hepatitis B virus X- and nucleocapsid gene transcriptional regulatory elements. Virology. 1992;191:31–41. doi: 10.1016/0042-6822(92)90163-j. [DOI] [PubMed] [Google Scholar]
- 61.Zhang P, Raney A K, McLachlan A. Characterization of functional Sp1 transcription factor binding sites in the hepatitis B virus nucleocapsid promoter. J Virol. 1993;67:1472–1481. doi: 10.1128/jvi.67.3.1472-1481.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]