Abstract
Pachytene piRNAs, a Piwi-interacting RNA subclass in mammals, are hypothesized to regulate non-transposon sequences during spermatogenesis. Caenorhabditis elegans piRNAs, the 21URNAs, are implicated in regulating coding sequences; the messenger RNA targets and biological processes they control during spermatogenesis are largely unknown. We demonstrate that loss of 21URNAs compromises homolog pairing and makes it permissive for nonhomologous synapsis resulting in defects in crossover formation and chromosome segregation during spermatogenesis. We identify Polo-like kinase 3 (PLK-3), among others, as a 21URNA target. 21URNA activity restricts PLK-3 protein to proliferative cells, and expansion of PLK-3 in pachytene overlaps with the meiotic defects. Removal of plk-3 results in quantitative genetic suppression of the meiotic defects. One discrete 21URNA inhibits PLK-3 expression in late pachytene cells. Together, these results suggest that the 21URNAs function as pachytene piRNAs during C. elegans spermatogenesis. We identify their targets and meiotic events and highlight the remarkable intricacy of this multi-effector mechanism during spermatogenesis.
C. elegans piRNAs regulate the progression of male meiosis and germ cell fidelity.
INTRODUCTION
Faithful production of gametes, either egg or sperm, from germ cells is essential for fertility. Each germ cell requires careful coordination of several developmental processes from the regulation of germline stem cells and gametogenesis to the protection of the germline genome. Previous work has established that small noncoding RNA (ncRNA) populations, such as microRNAs (miRNAs), endogenous small interfering RNAs (endo-siRNAs), and Piwi-interacting RNAs (piRNAs), all play key roles in regulating the development of germ cells through posttranscriptional gene regulation (1–3).
First studied in Drosophila where they were found to derive primarily from repetitive sequences and transposable elements, a well-defined role of one class of piRNAs is to suppress transposon activity (4, 5). Subsequently, piRNAs were identified in rat (6) and mouse genomes (7–9), and, in these animals, piRNAs were also found to include many sequences mapping to repeated sequences including transposable elements.
Despite diversity in sequence origin between and within organisms, all piRNAs depend on their associated PIWI argonaute proteins for accumulation and function. Studies of PIWI protein mutants in many animals have uncovered an essential role for piRNAs in fertility, particularly during spermatogenesis. For example, in mouse, mutants for PIWI-homologs MIWI (10) and MILI (11) result in spermatogenic arrest followed by apoptosis and the loss of post-meiotic germ cells rendering the males sterile.
Studies of mammalian piRNAs have suggested a somewhat broader functional role than seen in the early fly studies. Mammalian piRNAs are separated into at least two distinct classes based on spatiotemporal expression and their genomic origin. “Pre-pachytene piRNAs” are primarily derived from transposon-based sequences and are expressed in gonocytes and spermatogonia (12). By contrast, “pachytene piRNAs” are depleted of transposon sequences and are expressed during the pachytene stage in spermatocytes as well as round spermatids (12). The distinctions between the two classes reflect a specialization in function, whereby pre-pachytene piRNAs target transposons in pre-meiotic germ cells and pachytene piRNAs may regulate other transcripts such as gene-encoding mRNAs during meiosis. Yet, while a role has been determined for pre-pachytene piRNAs in regulating transposon expression in primordial germ cells, identification of specific function and targets for pachytene piRNAs has remained elusive. Multiple studies in mice have observed that loss of pachytene piRNA regulators results in a distinct spermatid arrest phenotype (10, 13, 14). More recently, Wu et al. (15) supported these findings by demonstrating that loss of the major piRNA cluster pi6 resulted in a spermiogenic defects accompanied by a reciprocal increase in mRNAs associated with spermiogenesis. However, poor sequence conservation among pachytene piRNAs between species has made it difficult to identify individually relevant piRNAs, and even deletion of major piRNA clusters can yield no discernable effect on fertility (16, 17).
Study of pachytene piRNAs has been largely limited to vertebrates (principally mice). Despite this, piRNAs in C elegans, also known as 21URNAs (18, 19), were originally speculated by Wang and Reinke (18) to be analogous to the MIWI-associated pachytene piRNAs based on lack of sequence-matching to transposons. Multiple groups studying mutants of the worm PIWI-homolog piwi-related gene 1 (prg-1) have demonstrated that 21URNAs suppress the expression of only specific classes of transposons (19–21). Further, global regulation of transposons is largely independent of 21URNAs (22). Cornes et al. (23) have recently suggested that Caenorhabditis elegans 21URNAs may initiate transcriptional silencing of spermatogenic genes during larval stages in C. elegans hermaphrodites. In that study, many previously identified piRNA targets showed no changes in transcription, leading to the question of what targets these 21URNAs regulate and what processes they control during spermatogenesis.
Here, we use a combination of developmental and genomic analysis to interrogate the function of spermatogenic 21URNAs and the C. elegans piwi homolog prg-1 in the male C. elegans germline. We show that prg-1 controls male fertility through the piRNA pathway, and loss of this function leads to distinct defects in meiotic progression, pairing of homologous chromosomes, crossover formation, and chromosome segregation. Small-RNA sequencing identified many potential targets of 21URNAs in the male germline, including Polo-like kinase 3 (plk-3), which we studied further to understand the spatiotemporal regulatory mechanisms of piRNAs. Notably, we found that the expression pattern of PLK-3 protein is restricted to the progenitor zone, but loss of prg-1 or a single piRNA target site permits ectopic expression in pachytene cells, coincident with meiotic phenotypes. In addition, we find that deletion of one 21URNA targeting site on PLK-3 results in early onset of PLK-3 protein expression in late pachytene. Defining pachytene piRNAs as Piwi-dependent small RNAs that regulate physiological meiotic events during pachytene, these data lead us to propose that C. elegans male 21URNAs can function as spermatogenic pachytene piRNAs. Last, we show that piRNAs regulate protein translation to restrict PLK-3 protein function in the male germline.
RESULTS
C. elegans male fertility is dependent on the prg-1 PAZ domain
Previous studies have indicated that PRG-1 is essential for fertility in both male and hermaphrodite animals (18, 19). Unlike flies (24, 25) and mice (26, 27), the essential function of the piRNA pathway in worms does not require the endonuclease-mediated slicing function of the PIWI domain (28). However, at least one group has reported that loss of PRG-1 slicing function results in fertility defects (29). Thus, whether the endonuclease function contributes to the regulation of male fertility or whether PRG-1 could have functions independent of canonical piRNA targeting remains to be evaluated. To determine the protein domain through which PRG-1 functions to regulate male fertility, we used CRISPR-Cas9 genome editing to introduce targeted mutations into the prg-1 gene (Fig. 1B; fig. S1, A to D; and Materials and Methods). To generate an endonuclease-dead allele, we introduced a single-nucleotide edit to substitute aspartic acid 583 within the PIWI domain for alanine. This residue is part of a highly conserved catalytic domain D-D-H (Asp-Asp-His) motif found in Argonaute-family proteins and has been previously shown to be essential for endonuclease function in vitro (30, 31); therefore, we refer to this allele as “endonuclease-dead” or prg-1(EnD). To assess a piRNA-dependent role for PRG-1, we deleted a 238-nucleotide (nt) region in the PRG-1 gene that codes for the RNA binding PAZ domain, which is essential to bind to piRNAs (fig. S1 and table S7) (32). We ensured that the deletion was generated in-frame and ascertained that the truncated protein maintained the coding frame. To confirm that the single point mutation for endonuclease-dead allele or the large deletion to remove the PAZ domain did not affect PRG-1 protein expression or localization, these mutants were generated in a backcrossed line where PRG-1 was tagged at its endogenous locus with V5::mCherry (Materials and Methods).
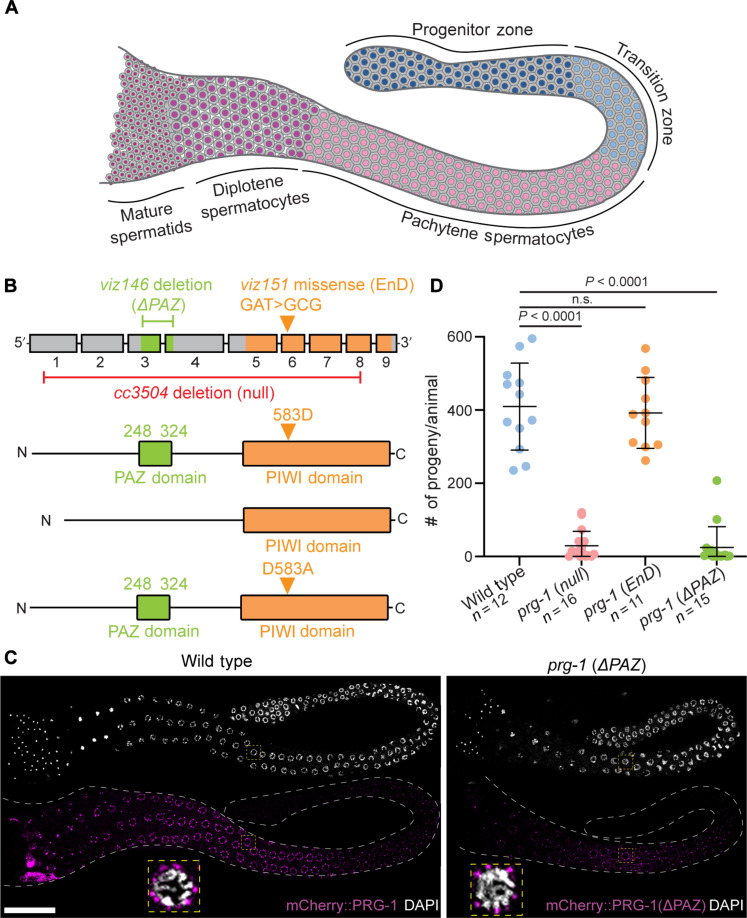
Fig. 1. Loss of the PRG-1 PAZ domain regulates male fertility while retaining germ granule association.
(A) Schematic of a male C. elegans germline displaying the spatiotemporal arrangement of germ cell development. Cells of the progenitor zone (oriented left in the schematic) are located at the most distal end of the germline and enter meiosis in the TZ. Progression through meiosis I continues in the pachytene and diplotene stages before meiotic divisions giving rise to the mature spermatids in the most proximal portion of the germline. (B) prg-1 gene structure with corresponding protein structure. PAZ-domain (green) and PIWI-domain (orange) functions were ablated with targeted CRISPR mutations (Materials and Methods) creating ΔPAZ and RNase-dead (RD) alleles. (C) Representative 4′,6-diamidino-2-phenylindole (DAPI)–stained (white) adult male germlines displaying expression of wild-type PRG-1 and PRG-1(ΔPAZ) (magenta) and subcellular localization around the nucleus in pachytene cells (inset, yellow boxes). (D) Quantitative analysis of male fertility in prg-1 mutant alleles. Statistical significance was calculated by Student’s t test; mutant groups were compared to wild type. Scale bar, 50 μm. Each experiment was conducted at least in triplicate and over at least 25 to 30 germlines analyzed each time.
Analysis of prg-1(EnD) and prg-1(ΔPAZ) alleles showed that both PRG-1 endonuclease-dead and ΔPAZ mutant protein accumulate in meiotic cells in perinuclear granules, similar to wild-type PRG-1 protein (Fig. 1C and fig. S1E), a localization noted previously for PRG-1 in several studies, and consistent with observed association with the germ granules (18, 19). Because the observed PRG-1 pattern was not affected by mutations in the endonuclease domain or the PAZ domain either spatially or at the subcellular level (Fig. 1C and fig. S1E), we conclude that neither domain is critical for the localization of PRG-1 at the germ granules in male germ cells.
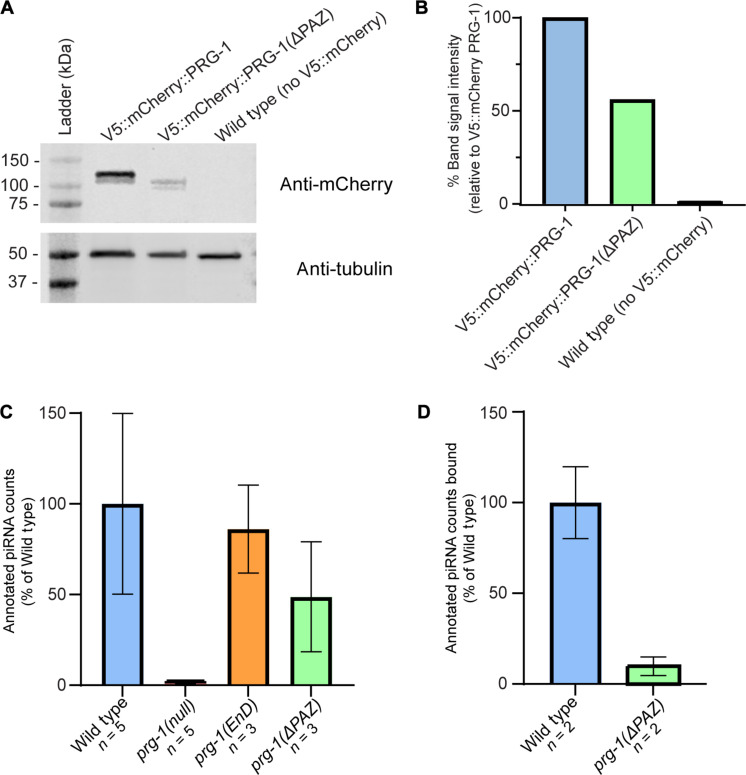
To investigate the contribution of the PAZ domain to PRG-1 stability and piRNA loading, we performed a Western blot analysis as well as an RNA immunoprecipitation (IP) experiment using wild-type PRG-1 and ΔPAZ PRG-1 (Materials and Methods). We observe that deletion of the PAZ domain results in 50% reduction in the total protein level, relative to the wild-type PRG-1 protein levels, and normalized with a tubulin loading control (Fig. 2, A and B). RNA IP (Materials and Methods) analysis of the interacting piRNAs eluted from equal amounts of wild-type and ΔPAZ PRG-1 protein revealed that there was ~90% reduction in the interacting piRNAs in ΔPAZ PRG-1 protein relative to wild-type PRG-1, (Fig. 2D, fig. S2, data S2, and table S2), normalizing each sample reads by total number of reads that mapped to the entire C. elegans genome (data S2). These data suggest that loss of the PAZ domain results in reduced stability of the protein as well as a significant reduction in piRNA interaction despite being localized to P granules. Thus, we consider the ΔPAZ PRG-1 allele as a reduction of function allele.
Fig. 2. Loss of the PRG-1 PAZ domain reduces PRG-1 expression and disrupts piRNA loading.
(A) Western blot data demonstrating reduction of PRG-1 expression after PAZ-domain deletion relative to a tagged control strain. (B) Quantification of band signal intensity from Western blot after normalization to tubulin loading control. Signal intensity is given as a percentage relative to tagged control strain signal. (C) Quantification of piRNA counts from sequenced samples as a percentage of average piRNA reads from wild-type samples, normalized to total mapping reads. (D) Quantification of piRNA counts from RNA IP experiments as a percentage of average piRNA reads from wild-type samples, normalized to total mapping reads.
To determine whether loss of PAZ domain and endonuclease mutations in PRG-1 result in reduced small-RNA levels, we next performed small-RNA sequencing of the wild-type, prg-1(ΔPAZ), and prg-1(EnD) alleles, from both males and hermaphrodites. Hermaphrodites were used in this experiment as internal control against males. Small-RNA sequencing revealed that there was no significant reduction in piRNA levels in the prg-1(EnD) alleles, as opposed to prg-1(null) allele, which was completely depleted in piRNAs (Fig. 2C and fig. S3). In addition, we observed that loss of the PAZ domain led to 60% reduction in piRNA levels in hermaphrodites and 52% reduction in males (figs. S3C and S2C and tables S1 and S3). The reads were normalized for each sample with total number of reads that mapped to the entire C. elegans genome (data S4). We ascribe the difference in piRNA interaction versus change in overall piRNA levels in the ΔPAZ PRG-1 allele to the difference in the assays used and caution against a direct comparison between these assays. Nevertheless, these data allow us to conclude that loss of the PAZ domain generates a strong reduction of function allele and results in reduction in piRNA loading (Fig. 2D) and piRNA levels (Fig. 2C). Thus, we proceeded with the analysis of the phenotypes during spermatogenesis with these alleles.
To determine the fertility of male prg-1 mutants, we mated mutant males from prg-1(cc3504) [Materials and Methods, referred to here on forward as prg-1(null)] (33) as well as the prg-1(EnD) and prg-1(ΔPAZ) alleles (Materials and Methods) to female worms and assayed the number of progeny produced over several days relative to wild-type males in the same experiment (Materials and Methods). We observed that complete loss of prg-1 as well as loss of the PAZ domain from males led to a significant reduction in progeny produced, while the endonuclease-dead allele resulted in no significant change in progeny numbers relative to wild-type males (Fig. 1D). Together, these data lead us to conclude that prg-1 contributes to male fertility in C. elegans primarily through the PAZ domain and thus likely a piRNA-mediated function not requiring the endonuclease activity.
C. elegans prg-1 regulates meiotic processes in pachytene
To determine the underlying cause of the reduced male fertility in the prg-1(null) and prg-1(ΔPAZ) animals, we performed high-definition analysis of meiotic progression on dissected adult male germlines from each of the mutant animals and compared them to wild-type and prg-1(EnD) germlines (Materials and Methods). Consistent with reduced fertility, germlines from prg-1 null and ΔPAZ mutants were significantly smaller relative to wild-type or prg-1(EnD) mutant germlines. This observation led us to ask two questions: (i) Were specific stages of germ cells missing in the prg-1 mutant alleles? (ii) Was there was a defect in meiotic progression itself? In the event of the latter, meiotic germ cells with defects in different processes, such as pairing or recombination, might progress at distinct rates from wild type, overall leading to shrinkage in the size of the germlines (Fig. 3A). To distinguish these possibilities, we used a combination of meiotic markers [pSUN-1 (34), HIM-3 (35), and SYP-1 (36)], to define four specific meiotic stages: transition zone (TZ), early pachytene, mid-late pachytene, and diplotene (fig. S4E). Specifically, pSUN-1 marks TZ (34) and early pachytene, while HIM-3 and SYP-1 denote chromosome synapsis (36), which is a process that begins after pairing of homologous chromosomes and defines the pachytene stage. Thus, we used these markers in combination with traditional staging methods based on chromosomal morphology (37, 38) to define TZ (pSUN-1+ cells with crescent-shaped chromatin morphology), early pachytene (pSUN-1+ cells with incomplete HIM-3/SYP-1 synapsis and paired chromatin morphology), mid-late pachytene (full HIM-3/SYP-1 synapsis and paired chromatin morphology), and diplotene (HIM-3/SYP-1 desynapsis and condensing chromatin morphology).
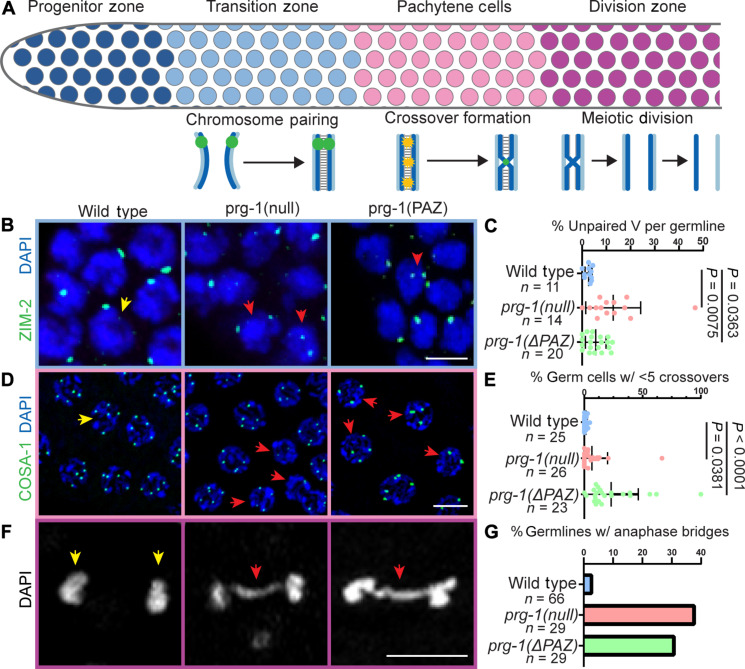
Fig. 3. Multiple meiotic processes are defective in prg-1 mutants.
(A) Schematic of meiotic progression and meiotic events by germ cell stage. Homolog pairing occurs in the TZ, followed by crossover formation in pachytene stage. Meiotic divisions occur in the division zone following diplotene stage. (B) Immunolocalization of ZIM-2 (green) in DAPI-stained nuclei (blue) of TZ cells. Two foci in a nucleus indicates unpaired homologs. One focus in a nucleus indicates paired homologs. Red arrowheads indicate nuclei with unpaired homologs. (C) Quantification of the percentage of germ cells with unpaired homologs per germline analyzed. (D) Localization of GFP::COSA-1 (green) in late pachytene nuclei stained with DAPI (blue). Red arrowheads indicate nuclei with fewer than five crossovers. (E) Quantification of the percentage of germ cells with less than five crossovers per nucleus per germline analyzed. (F) Representative DAPI (DNA, white) images of meiotically dividing germ cells from the division zone. Red arrowheads indicate anaphase bridges. (G) Quantification of percentage of germlines displaying meiotically dividing cells with anaphase bridges. Statistical significance was calculated by Student’s t test, mutant groups were compared to wild type. Scale bars, 5 μm. Each experiment was conducted at least in triplicate and over at least 25 to 30 germlines analyzed each time and at least 500 germ cells each time.
We observed that the prg-1 mutant germlines expressed the meiotic markers assayed and exhibited similar germ cell chromosome morphologies as wild-type germlines (fig. S4, A to D), suggesting that there was no gross loss of any specific stage of meiosis upon loss of prg-1 or PAZ domain. Because all germ cell stages were present, we next considered whether meiotic progression was altered. In the C. elegans gonad, rows of germ cells are sequentially arranged in a spatiotemporal gradient where earlier stages of meiosis are more distally located in the germline and later stages are more proximally located (Fig. 1A). In this manner, the number of cell rows per germ cell stage is constant unless meiotic errors alter progression from one stage to the next producing either fewer or more cell rows. Therefore, we quantified the number of cell rows per each germ cell stage of meiosis to assay meiotic progression (Materials and Methods). We found that the prg-1(null) and prg-1(ΔPAZ) mutants exhibit fewer pachytene germ cell rows, resulting in a shorter pachytene stage (fig. S4E). Because of the spatiotemporal nature of the C. elegans germline, the static observation of fewer pachytene cell rows in the prg-1 mutant male germlines indicate that germ cells are progressing to the diplotene stage of meiosis at a potentially earlier spatial position. Thus, while we find that all stages of meiotic germ cells are present in prg-1 mutant germlines, progression between these stages is altered, suggesting that the underlying meiotic processes that define these stages have been altered or are defective.
To test this hypothesis and determine the meiotic processes that may be affected by the loss of prg-1 function, we assayed homologous chromosome pairing, synapsis, and crossover formation that occur preceding and during pachytene stage of meiosis I (Fig. 3A). In the TZ, germ cells enter meiosis I and homologous chromosomes undergo meiotic pairing, which is essential for events of synapsis and homologous recombination that occur during pachytene in C. elegans (37). In C. elegans, pairing of homologous chromosomes is mediated by cis-acting DNA sequence motifs known as pairing centers (PCs) (37). Associated with these motifs are a family of zinc-finger proteins including ZIM-1, -2, -3, and HIM-8, which control the pairing process (39). ZIM-2 associates with the PC of chromosome V and facilitates its use as marker to assess proper pairing of homologs: Two ZIM-2 foci indicate individual, unpaired homologs, whereas a single focus denotes the completion of pairing. We assayed for ZIM-2 loci to determine and assess pairing of chromosome V in wild-type and prg-1 mutants. We observed that prg-1(null) and prg-1(ΔPAZ) display a reduction in paired chromosomes assayed as an increase in number of germ cells with more than one ZIM-2 focus (Fig. 3, B and C), suggesting that loss of prg-1 function results in reduced homologous pairing.
Despite a reduction in pairing, we observed that the chromosomes synapsed (fig. S4, A to D). One hypothesis is that homolog pairing is delayed, but another possibility is that synapsis is occurring with nonhomologous chromosomes. Normally, these events will be cleared by activation of meiotic checkpoints and apoptosis in the germline. However, C. elegans males lack meiotic checkpoints and apoptosis (40); thus, we hypothesized that if chromosomes were synapsing with nonhomologs, then it should lead to a lower number of crossover designations in pachytene since only homologous pair of chromosomes designate productive crossovers, which are essential for recombination and chromosome segregation (37). To assess for the formation of crossovers, we endogenously tagged the protein COSA-1 (Materials and Methods), which localizes to and mediates the formation of crossovers in C. elegans (41). C. elegans males have five autosomes and an X chromosome, which does not pair since males bear the karyotype XO. Thus, male pachytene germ cells should have five COSA-1 foci per nucleus (42). Analysis of green fluorescent protein (GFP)::COSA-1 foci in the male germ cells from wild-type and prg-1 mutant animals revealed that loss of prg-1 or PAZ function results in an increased number of pachytene stage germ cells with less than five crossovers (Fig. 3, D and E).These data indicate that loss of prg-1 function or PAZ-domain activity results in difficulties in designating crossovers consistent with defects in chromosome pairing and suggest that synapsis may be occurring between nonhomologous chromosomes, which collectively affect meiotic progression.
While several germ cells displayed lower number of crossovers, consistent with lack of appropriate pairing between homologs, there were others that displayed five crossovers per nucleus (Fig. 3). These observations suggest one of two possibilities (i) that some chromosomes did pair appropriately with their homologs and formed productive crossovers with their partners, as expected, or (ii) were forming crossovers with nonhomologous chromosomes, as hypothesized above. If the latter where the crossovers were formed between non-homologous chromosomes, then it would result in chromosome segregation errors during meiotic divisions, or aneuploidy during meiosis. To assess whether the crossover designation was abnormal leading to defects in chromosome segregation during meiotic division, which would have a direct impact on fertility (43), we assayed for anaphase bridges in meiotic divisions in the region of the male germline following diplotene. In wild-type germlines, we observe few anaphase bridges (Fig. 3, F and G), while in prg-1 mutant germlines, we observe an increased frequency of germlines with anaphase bridges. The presence of anaphase bridges indicates aberrant chromosome segregation. Together, these data demonstrate a role for prg-1 PAZ domain–dependent processes in meiotic pairing, crossover designation, meiotic progression, normal chromosome segregation, and fertility of male gametes.
The piRNA pathway regulates genes involved in meiosis and germ cell development during spermatogenesis
In C. elegans, silencing initiated by piRNAs and PRG-1 leads to the biogenesis of complementary endo-siRNA populations belonging to the worm 22G siRNA class (44). The 22G endo-siRNAs bind to the WAGO-family Argonautes resulting in silencing of target RNAs that can be transcriptional and/or posttranscriptional. To determine the nature of the targets and the corresponding molecular pathways through which piRNAs may regulate meiotic events during spermatogenesis, we sequenced small RNAs from males to identify 22G siRNAs that are absent in prg-1 mutants (Materials and Methods, data S4, and table S4).
To identify targets depleted for prg-1–dependent 22G siRNAs, we focused on prg-1(null) and compared these with wild-type animals (Fig. 4A). We first mapped the absolute number of 22G siRNA reads to C. elegans cDNAs, normalizing each sample reads by total number of reads that mapped to the entire C. elegans genome (data S4). Next, we compared reads between wild-type and prg-1(null) animals on a gene-by-gene basis to determine differences in mapped 22G siRNAs. To determine significantly altered 22G siRNA populations, we applied an arbitrary threshold based on two criteria: (i) whether there was at least a fourfold difference [2log (2)] in mapped 22G siRNAs and (ii) whether there were at least 50 reads total mapping to a particular gene. Given our interest in understanding piRNA-mediated silencing of endogenous targets, we focused on the candidates with lower 22G siRNA read levels in prg-1(null) samples (table S4). Overall, with this stringent cutoff, we identified 217 genes with reduced 22G siRNAs that mapped to them (table S4).
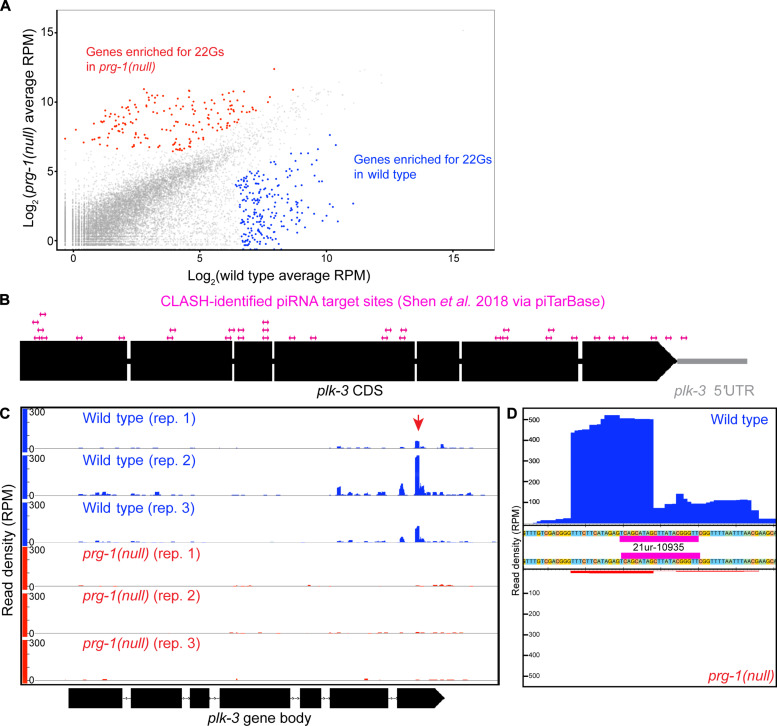
Fig. 4. The piRNA pathway targets genes involved in meiosis and germ cell development, including plk-3, a member of the PLK family.
(A) Scatterplot of mapped 22G reads per C. elegans gene in matched wild-type versus prg-1 mutant samples. Highlighted genes met the 2log (2) cutoff for enrichment of 22G reads in either wild-type (blue) or prg-1 mutant (red). (B) Schematic displaying CLASH-identified piRNA binding sites within the plk-3 coding sequence. (C) Histogram for 22G reads mapped to plk-3 in wild-type (blue) or prg-1 mutant (red) replicates. Red arrowhead denotes cluster of 22G reads that coincided with 21ur-10935 binding site. (D) Magnification of histogram including all wild-type (top, blue) or prg-1 mutant (bottom, red) 22G reads mapping to plk-3 around the location of the 21ur-10935 binding site (magenta).
After identifying the corresponding mRNA targets of down-regulated 22G siRNAs (data S2), we cross-referenced the potential mRNA targets with known germline microarray as well as RNA sequencing–based databases (45, 46) to focus on germline expressed genes. We reasoned that these genes would likely be up-regulated if they were true targets. In this analysis, we identified genes such as glh-1 and pan-1 (fig. S5, A and B), which are germ granule components essential for fertility (47, 48). We also found genes known or predicted to function in meiotic processes such as rec-1 (fig. S5C), demonstrated to be involved in double-strand break formation and crossover distribution in worms (49, 50), mlh-1 (fig. S5D), a worm homolog of the human mismatch repair gene MLH-1, and plk-3 (Fig. 4C), a worm member of the Polo like Kinase (PLK) family of unknown function.
To better understand how these genes are targeted by piRNA-dependent 22G siRNAs, we mapped the location of the small-RNA reads on the target genes and assayed their distribution. As expected, some genes that are targeted by 22G siRNAs have small-RNA reads localized to discrete clusters (Fig. 4C and fig. S5, A to D), indicating sequences within the gene that served as templates for endo-siRNA biogenesis. Comparatively, some of the 22G siRNA clusters are depleted in the prg-1(null) mutant, allowing us to identify which 22G siRNAs are dependent on the activity of prg-1 and, thus, piRNAs. In addition, the location of the 22G siRNAs themselves suggests that those RNA sequences are likely targeting sites for piRNAs and thus ideal for downstream validation. To explore whether the presence of the 22G siRNA reads was coincident with a piRNA target site, we cross-referenced 22G siRNA read clusters on a per-gene basis against results from pirTarBase. pirTarBase features both algorithm-predicted and experimentally validated piRNA binding sites determined through the cross-linking, ligation, and sequencing of hybrids (CLASH) method (51, 52).
Among target mRNAs investigated for piRNA sites, we found that there was a lack of consensus among piTarBase prediction and CLASH-identified sites, so we focused on CLASH-identified sites since we reasoned that these sites likely reflect direct in vivo piRNA-mRNA binding events. We used plk-3 as a case study for our analysis of candidate mRNA target. plk-3 has 55 piRNA target sites listed in piTarBase, 21 of which are algorithm-predicted and 34 are CLASH-identified. These 34 CLASH-identified sites were distributed throughout the plk-3 coding sequence, including a site in the 3′ untranslated region (Fig. 4B). Only a single targeting site was found to coincide with a large cluster of 22G reads: a CLASH-verified site for the piRNA 21ur-10935 in the last exon of the gene (Fig. 4C). Zooming in on this target site, many of the 22G siRNAs mapped to the middle of the 21ur-10935 piRNA target sequence (Fig. 4D), a feature that is thought to define a “real” piRNA-generated 22G endo-siRNAs (52).
In C. elegans, Reed and Barucci et al. (22, 53) found that piRNAs indirectly regulate histone loci through 22G endo-siRNAs in hermaphrodites. To assess whether histone loci were similarly targeted by 22G endo-siRNAs in the male germlines, we assayed the histone locus to determine whether there was depletion of 22G endo-siRNAs (fig. S5E). We observed that unlike the hermaphrodites, in male germlines, 22G endo-siRNAs accumulated normally at the histone loci, suggesting that the histone loci were not targets of the male piRNAs. The observations that (i) plk-3 locus loses 22G endo-siRNAs, (ii) has CLASH-identified piRNA sites and (iii) one CLASH-identified site, in particular, the 21ur-10935 accumulates 22G endo-siRNAs in the middle, a criterion that is considered a critical feature of 22G endo-siRNAs that are produced by piRNAs (52), and (iv) 21ur-10935 is depleted from both prg-1(null) and prg-1(ΔPAZ) alleles, led us to hypothesize that PLK-3 may be a direct target of piRNAs in the male germline. Thus, we proceeded with testing plk-3 as a piRNA target during spermatogenesis.
PLK-3 is regulated by the piRNA pathway to restrict its expression to the distal germline
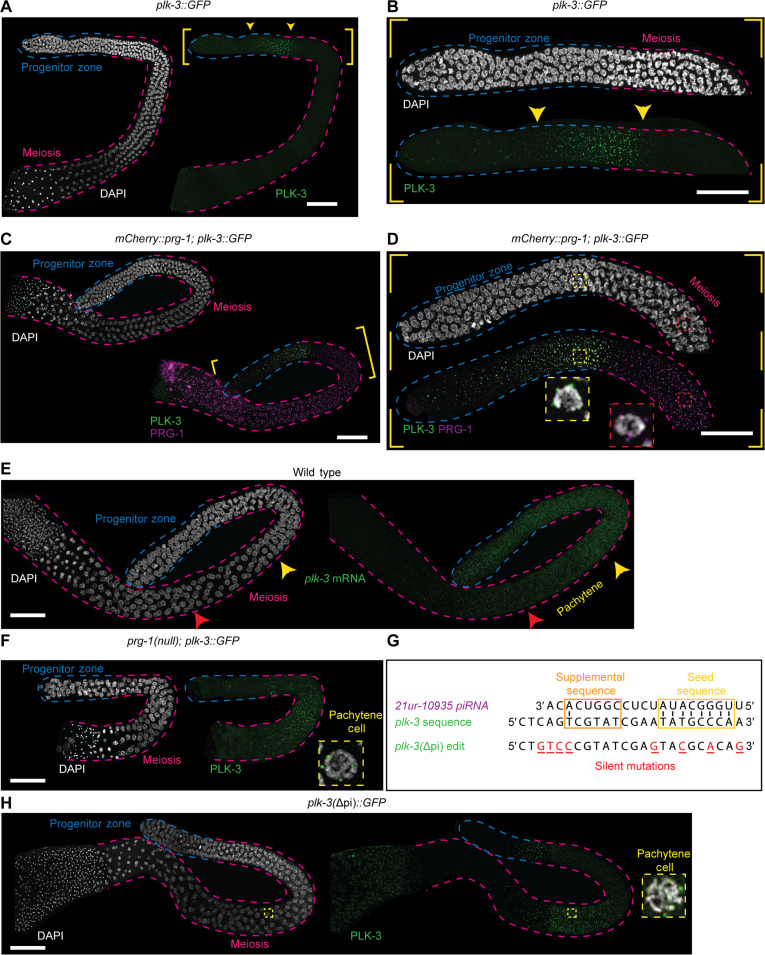
To determine whether plk-3 is regulated by piRNAs in the male germline and infer the contribution of the CLASH-validated piRNA site on plk-3, we tagged plk-3 at its endogenous locus with GFP to observe its protein expression in the germline. The PLK-3::GFP did not present with any overt phenotypes, suggesting that the knock-in did not disrupt function. We found that PLK-3::GFP is expressed in the distal region of the germline, particularly at the end of the progenitor zone where germ cells begin to transition into meiosis (Figs. 1A and 5, A and B). Within these cells, PLK-3 localizes to the perinuclear region in puncta like germ granules (Fig. 5, C and D). As germ cells enter early pachytene, PLK-3 expression tapers and is no longer visible. This expression was notable since PLK-3 protein expression seemed restricted to the proliferative cells, whereas PRG-1 is expressed weakly in the progenitor zone where PLK-3 accumulation is strongest, but markedly increases until the pachytene region where PLK-3 is not observed (Fig. 5D). Given that PRG-1 is required to initiate piRNA silencing the negative correlation between PLK-3 protein expression and PRG-1 accumulation, we hypothesized that PRG-1–dependent 21URNAs spatially restricted PLK-3 to the proliferative region.
Fig. 5. PLK-3 expression is restricted to the distal germline by piRNA function.
(A and B) Wild-type adult male germline expressing PLK-3::GFP (green) stained with DAPI (white). (A) PLK-3::GFP is expressed in the distal regions of the germline in the areas noted by the yellow arrowheads. (B) Zoomed in region of the distal germline from (A), displaying PLK-3::GFP expression between the progenitor zone and meiotic regions of the germline. (C and D) Wild-type adult male germline expressing PLK-3::GFP (green) and V5::mCherry::PRG-1 (magenta) stained with DAPI (white). (D) Zoomed in distal germline from (C), showing that PLK-3 expression is spatially expressed before and almost mutually exclusive with PRG-1 expression at the end of the progenitor zone, but PRG-1 expression is highest in the meiotic region. Inset 1 (yellow, dashed lines) shows a single TZ nucleus with visible PLK-3 perinuclear granules, whereas inset 2 (red, dashed lines) shows a pachytene nucleus with PRG-1 perinuclear granules. (E) Wild-type adult male germline stained with DAPI (white) and plk-3 HCR-FISH, showing expression of plk-3 mRNA throughout the distal regions with the mRNA expression significantly reduced in mid/late pachytene (red arrowhead). (F) A prg-1(null) adult male germline expressing PLK-3::GFP (green) stained with DAPI (white). Inset (yellow, dashed lines) displays single pachytene nucleus in the meiotic region expression PLK-3::GFP. (G) Schematic displaying the plk-3 coding sequence where 21ur-10935’s target sequence maps. Below, plk-3(Δpi) designates CRISPR-edited sequence with specific base changes noted (red, underlined). Predicted base-pairing within seed sequence (yellow box) and supplemental sequence (orange box) is noted. (H) Germline with PLK-3(Δpi)::GFP (green) stained with DAPI (white). Inset (yellow, dashed lines) displays pachytene nucleus in the meiotic region expressing PLK-3(Δpi)::GFP. Scale bars, 50 μm. Each experiment was conducted at least in triplicate and over at least 25 to 30 germlines analyzed each time.
To test the hypothesis that piRNAs restricted PLK-3 expression to the progenitor zone and away from meiotic germ cells, we assayed for plk-3 mRNA using hybridization chain reaction–fluorescence in situ hybridization (HCR-FISH; Materials and Methods). mRNA FISH analysis revealed that plk-3 mRNA accumulates from the progenitor zone continuously throughout pachytene (Fig. 5E, yellow and red arrows, fig. S6). This would suggest that the sharp restriction of PLK-3 protein to the progenitor zone cells was likely due to translational inhibition of PLK-3 protein in TZ of meiosis and pachytene, or a rapid turnover of PLK-3 protein in this region. To assess whether the restriction of PLK-3 protein to the progenitor zone was dependent on piRNAs, we crossed in the PLK-3::GFP into prg-1 mutant backgrounds. We observed that complete loss of PRG-1, as well as loss of the PAZ domain, led to an accumulation of PLK-3 in the meiotic pachytene germ cells into late pachytene, and the PLK-3::GFP localizes to perinuclear structures (Fig. 5F and fig. S7), leading us to conclude that the restriction of PLK-3 to the progenitor zone was because of piRNAs. Because plk-3 mRNA is expressed throughout pachytene in wild-type, prg-1(null), and prg-1(ΔPAZ) domain mutants (Fig. 5E and fig. S6A), we infer that piRNAs are unlikely to regulate the transcription of plk-3 but either inhibit the translation of the plk-3 mRNA directly or control its translation or protein turnover in an indirect manner.
To determine whether PLK-3 protein expression in pachytene was dependent on piRNAs, we focused on the CLASH-identified piRNA site located at the 3′ end of plk-3 that we had correlated with a cluster of 22G siRNAs (Fig. 4D). The presence of an unrelated overlapping piRNA locus within 21ur-10935 precluded the possibility of deleting the piRNA itself. Instead, we introduced mutations into the coding sequence of PLK-3::GFP to disrupt the target site for 21ur-10935. Seven silent mutations were introduced throughout the 21ur-10935 target sequence (Fig. 5G), with three mutations being in the seed and supplemental regions that are thought to be particularly important for targeting (Materials and Methods) (52). We call this allele plk-3(Δpi).
The plk-3(Δpi) allele is fertile, with normal progression of meiosis in males, and does not present with any gross defects in fertility (Fig. 6D). To determine whether the mutation has any consequences for regulation, we assayed for PLK-3::GFP. We observed that loss of 21ur-10935 leads to ectopic PLK-3::GFP protein expression only in late pachytene stage cells (Fig. 5H). As observed for wild-type and prg-1(null) and PAZ domain deletion, there was no change of plk-3 mRNA expression as assayed by FISH (fig. S6B).
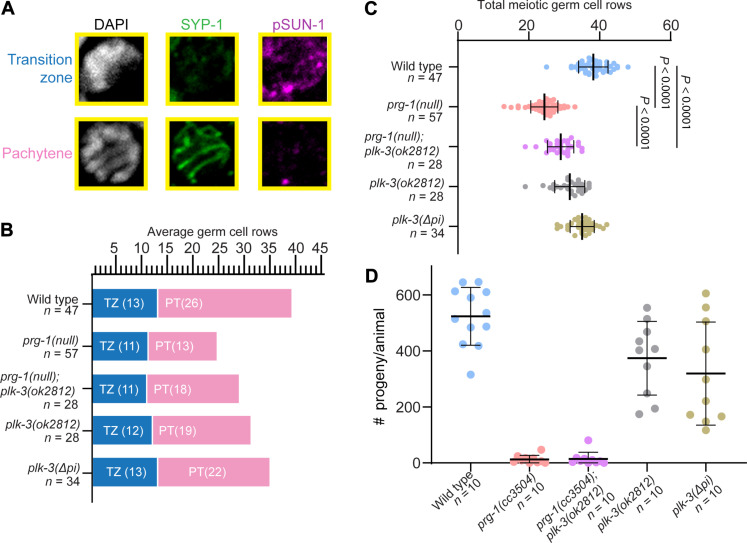
Fig. 6. Loss of plk-3 partially rescues prg-1 meiotic progression defects.
(A) Representative images of TZ and pachytene germ cells stained with DAPI (white), SYP-1 (green), and pSUN-1 (magenta). (B) Graphical representation of average TZ and pachytene cell rows per genotype across germlines analyzed. (C) Quantification of the average total meiotic germ cell rows. Numbers graphed are averages across germlines analyzed. Statistical significance between groups was calculated by one-way analysis of variance (ANOVA) with Bonferroni correction; comparisons not shown on graph: Wild type versus plk-3(ok2812), P < 0.0001; wild type versus plk-3(Δpi), P = 0.0039; prg-1(null);plk-3(ok2812) versus plk-3(Δpi), P < 0.0001; prg-1(null) versus plk-3(ok2812), P < 0.0001; prg-1(null) versus plk-3(Δpi), P < 0.0001; plk-3(ok2812) versus plk-3(Δpi), P = 0.0050. (D) Brood sizes for corresponding genotypes from (B) and (C). Each experiment was conducted at least in triplicate and over at least 25 to 30 germlines analyzed each time.
This result was interesting for two reasons: (i) It suggests that 21ur-10935 piRNA targets plk-3 mRNA only in late pachytene cells and inhibits its protein expression, and (ii) distinct piRNAs likely regulate PLK-3 expression in distinct stages of germ cell development. We speculate that loss of the plk-3 piRNA targeting does not affect meiotic progression or fertility because 21ur-10935 only results in onset of late pachytene expression of PLK-3, while the meiotic defects of pairing and crossover designation would occur only if there was PLK-3 activity in TZ and early and mid-pachytene. Thus, 21ur-10935 seems to repress PLK-3 expression in late pachytene for a function that we currently do not understand, but because we observe an onset of PLK-3 expression upon loss of this targeting site, it does allow us to conclude that plk-3 is directly regulated by piRNAs. It is likely that one, or more, of the other 33 piRNAs that target plk-3 regulate its expression in early and mid-pachytene and the TZ. Together, we conclude that PLK-3 is a target of piRNAs and that its spatiotemporal expression is intricately controlled by the piRNA pathway.
Loss of plk-3 partially suppresses meiotic progression defects in the prg-1 mutant
The observation that plk-3 expression in meiotic germ cells is controlled by the piRNA pathway leads to the hypothesis that the meiotic dysfunction in prg-1 mutants arises, at least in part, from the ectopic expression of piRNA targets. If true, we predict that removing plk-3 function from the meiotic region could restore the meiotic progression defects of the prg-1 mutants. We tested this hypothesis by removing plk-3 function through introducing a plk-3 loss-of-function mutant (Materials and Methods) into prg-1 mutants to observe whether the reduction in PLK-3 would rescue the meiotic progression defects observed in PRG-1 mutants. We found that loss of plk-3 from the prg-1 mutant germlines partially rescues the meiotic progression defects. The prg-1;plk-3(ok2812) mutant germlines are not as reduced in size as prg-1(null) germlines alone (Fig. 6C), and pachytene stage of meiosis is spatially expanded in the double mutant, relative to prg-1 single mutant (Fig. 6B). However, as expected, removal of plk-3 function while partially restoring the meiotic progression defects in the prg-1(null) mutants does not restore their fertility (Fig. 6D and fig. S8) to that of plk-3(ok2812) levels (which is completely fertile). This could be because multiple other targets may need to be restored for onset of complete fertility in the prg-1 mutant animals. These observations suggest that the meiotic progression defects in the prg-1 mutant male germline are, in part, alleviated by the absence of plk-3. We note here that genetic interactions while quantitative (where neither mutation has an absolute effect on the phenotype being measured) present challenges in assignment of upstream-versus-downstream epistasis (54). Nonetheless, the double-mutant phenotype indicates a consequential interaction between the plk-3 and the piRNA regulatory machinery and is supportive of a model in which the piRNA pathway functionally mitigates the potential for meiotic dysregulation that would occur from expression of PLK-3 in meiosis.
DISCUSSION
Spermatogenic 21URNAs are a C. elegans equivalent of mammalian pachytene piRNAs
Although pachytene piRNAs are considered a mammalian phenomenon, we propose that this work, and previous studies, collectively support C. elegans 21URNAs as a model for mammalian pachytene piRNAs based on several criteria. (a) Much like MIWI, which associates with pachytene piRNAs in mice, PRG-1 is temporally present during meiotic pachytene (18, 19, 55). (b) In both mice and C. elegans, phenotypes arise in early spermatids upon loss of pachytene piRNAs and 21URNAs, respectively, reinforcing that their functions are specifically important during the pachytene stage before spermiogenesis (18, 23, 56). In this study, we delineate (i) the distinct meiotic processes that are affected by loss of prg-1 during pachytene stage of meiosis and identify the earliest events misregulated resulting in loss of male fertility. (ii) We find that the meiotic processes that are affected by loss of prg-1 correlate with the temporal expression of PRG-1 and their targets, both of which had been previously undescribed. (iii) Last, while many groups have postulated a role for pachytene piRNAs and 21URNAs in the regulation of protein-coding genes, identifying definitive links between individual pachytene piRNAs and their targets has been challenging. To date, in C. elegans, the piRNA 21UX-1 was identified to regulate the dosage of the sex determination switch gene xol-1 representing at least one known functional target (57).
We found that distinct piRNAs can regulate a single gene in a spatially combinatorial manner. Complete loss of piRNAs led to an expansion of PLK-3 protein into early, mid, and late pachytene. plk-3 mRNA is normally expressed throughout pachytene, suggesting that loss of the piRNAs led to at least two distinct phenomena. In the first case, loss of piRNAs led to either loss of translational inhibition of plk-3 mRNA pachytene and allowed for protein expression directly or loss of piRNAs affected a protein turnover pathway, which led to the accumulation of PLK-3 protein indirectly in pachytene. In the second case, loss of 21-ur-10935 led to specific expression of PLK-3 protein only in this stage, suggesting that 21-ur-10935 likely controls localized plk-3 translation in late pachytene (58). These are intriguing observations since they suggest that the small RNAs regulate localized protein expression of the target, a phenomenon that has not been described before. Together, these data suggest that pachytene piRNAs function to restrict mRNA expression in space and time with plk-3 serving as an example for such targeting that had previously been proposed only indirectly (15). Together, we provide evidence for the term pachytene piRNA, which was coined over a decade ago, in C. elegans, wherein the piRNAs are so defined because they encompass both the functional property of regulating germ cell development during pachytene stages in spermatogenesis (ultimately resulting in loss of fertility and spermatid formation) as well as a functional definition of a piRNA that targets protein-coding genes.
Sexual dimorphism in the role of the piRNA pathway in the regulation of germ cell development
If pachytene piRNAs regulate meiotic pachytene and protein-coding genes in the C. elegans male germline, what do piRNAs control in the female germline? In hermaphrodites, studies have found that piRNAs and prg-1 regulate the maintenance of transgenerational fertility. Reed et al. (53) and Barucci et al. (22) found that in the absence of piRNAs, downstream small-RNA machinery generates 22G siRNAs that target and silence the expression of endogenous genes, particularly histones, which are then inherited by the next generation through the cytoplasm (22). However, in this case, the absence of piRNAs themselves did not directly lead to the regulation of targets but rather affected the homeostasis of small-RNA pathways, which led to an increase in 22G siRNAs targeting histones causing their gradual loss over generations leading to transgenerational sterility. Similarly, Wahba et al. (33) showed that the loss of piRNAs from oocytes leads to an amplification of siRNA populations that suppress ribosomal RNAs and contribute to loss of fertility across generations. Together, these studies identify a role for the C. elegans piRNA pathway in maintaining a balance between small-RNA pathways to prevent erroneous silencing of essential genes in the female germline.
This role is distinct from the function uncovered in this study for male germlines, which is in direct regulation of individual targets (like PLK-3) that affects male fertility. Work from Cornes et al. (23) showed that piRNAs may regulate transcription during larval spermatogenic phase (of a hermaphrodite). In that study, they investigated piRNA targets such as Y80D3A.8 to find that their expression correlates with spermatogenesis. Intriguingly, they observed that prg-1 and hrde-1 mutants had an increase in the number of Y80D3A.8-expressing germ cells. However, the lack of post-meiotic cells in these mutants suggests that this expansion of Y80D3A.8-expressing cells may be a result of a general delay in germline development, as expression levels of Y80D3A.8 do not differ significantly between wild-type spermatogenic cells and prg-1 or hrde-1 mutants. An advantage of the present study being carried out solely in the male germline is the elimination of the hermaphrodite sex switch as a confounding factor, whereby we found that prg-1 male germlines although smaller, contained all spermatogenic cell types. Conversely, we also observed that in larval hermaphrodite germlines, germline development was delayed, leading to fewer cells overall and fewer spermatogenic cells (fig. S9.) Thus, spatial expression of a piRNA target to determine that PLK-3’s expansion in the prg-1 mutant is independent of factors such as differences in developmental timing.
Choi et al. (56), showed that the transcription factor SNPC-1.3 specifically generates male-specific piRNA transcripts, with loss of snpc-1.3 leading to strong fertility defects in the first generation. The piRNAs and the male sterility phenotypes overlap with the piRNAs identified in this study, suggesting that the male and the female piRNAs regulate distinct targets and affect different biological functions. The male-specific piRNAs directly regulate specific germline targets and meiotic events, while the female-specific piRNAs control small-RNA homeostasis and indirectly control targets such as histone loci to result in transgenerational sterility.
The sexually dimorphic differences in regulation of piRNAs and in their regulation of male and female germ cells remain an active field of investigation. The vast range of tested and predicted targets of both male and female piRNAs adds to the challenge of assigning specific cause of loss of fertility in either case. Thus, a bottleneck to understanding the function of piRNAs in either system remains in drawing a link between individual piRNAs and their targets. Identification of specific germ cell target such as PLK-3 during meiosis sets the stage for future studies to identify specific targets and link them to specific biological processes, which will likely bring more clarity and thus resolution to the function of pachytene piRNAs in males and females.
plk-3 as a model for understanding the role of the piRNA pathway in the spatiotemporal regulation of genes in the germline
Given the paucity of piRNA targets that have been directly studied, plk-3 presents an opportunity to further understand how piRNA regulation interacts with genes that function in the germline and meiosis. PLK-3 is member of the PLK family, of which plk-1 and plk-2 have previously been identified to regulate meiotic pairing of homologous chromosomes, respectively. In these studies, RNA interference–mediated depletion of plk-3 yielded no obvious meiotic phenotypes (59, 60). We find that PLK-3 protein accumulates in the progenitor zone and drops sharply as germ cells enter meiosis, suggesting that PLK-3 does not function during meiosis normally, and since PLK-3 does not accumulate in meiotic stages of wild-type male germlines, its depletion should not lead to any obvious meiotic phenotypes. In line with this reasoning, expansion of PLK-3 into meiotic germ cells upon loss of prg-1 suggests that it is the gain of PLK-3 in male meiosis that leads to defects rather than its loss. This model is supported by the partial rescue of meiotic progression in the prg-1;plk-3. Given that PRG-1–dependent 22G siRNAs regulate many different genes (Fig. 4A), there is clearly redundancy in their function supported by the partial rescue of meiotic progression upon loss of plk-3.
In addition to helping uncover a function for PLK-3 in likely “poisoning” events of meiotic progression if expressed in pachytene stage of male meiosis, we also uncovered that piRNAs restrict the expression of their target gene(s) in space and time. Before this study, only one study (57) had uncovered a functional target of piRNAs in worms—xol-1, which acts as a sex switch regulator. In our study, we find that piRNAs restrict the expression of PLK-3 protein and keep it off in meiosis in wild-type animals whereby loss of all the piRNAs (in a prg-1 mutant) results in expansion PLK-3 protein into meiosis (Fig. 7). Further, we find that this restriction of PLK-3 occurs in two distinct manners. (i) plk-3 mRNA is expressed from the progenitor zone until late pachytene (Fig. 4H). Thus, we hypothesize that the piRNAs inhibit PLK-3 protein from expressing in meiosis in early stages of pachytene. (ii) In late pachytene, deletion of piRNA targeting site 21ur-10935 results in expression of PLK-3 protein only in late pachytene, suggesting that 21ur-10935 regulates plk-3 mRNA by posttranscriptional degradation since there is no transcription at this stage of male meiosis (58). In this manner, plk-3 appears to reveal a previously unidentified mechanism for the piRNA pathway in the silencing of genes during early meiotic stages, a mechanism reminiscent of how miRNAs regulate their target genes through posttranscriptional degradation.
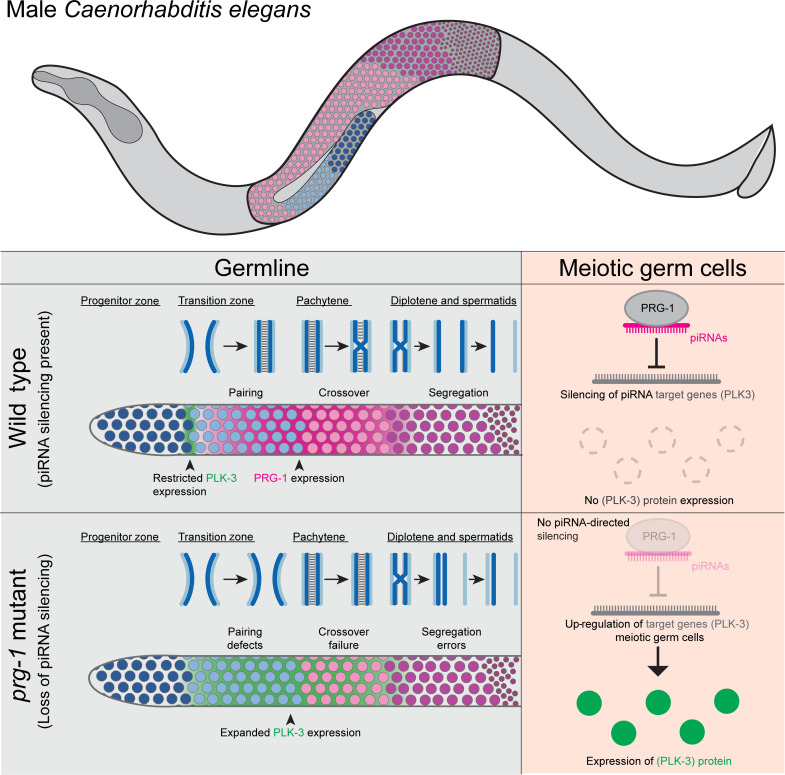
Fig. 7. Pachytene piRNAs control spatiotemporal gene expression in the male germline.
Schematic model for regulation of spatiotemporal gene expression in the male germline by the piRNA pathway. In wild-type animals, PRG-1 is expressed most highly beginning in the TZ through the pachytene stage. This expression corresponds with essential meiotic processes such as pairing of homologous chromosomes and crossover formation. Conversely, PLK-3 is only expressed between the progenitor zone and the early TZ. In PRG-1 mutants, the loss of piRNAs leads to meiotic defects and desilencing of PLK-3 during meiotic stages. Together, our data suggest that loss of piRNA silencing such as PLK-3 during meiosis underlies observed meiotic defects.
Last, many of the predicted piRNA targets identified in this study include either genes ubiquitously expressed in the germline such as glh-1 or genes with known predicted functions in meiotic processes such as rec-1. In the case of the ubiquitously expressed germline genes, it is likely that piRNAs fine tune their expression during pachytene, as shown for xol-1. In all cases, further studies of the function of direct piRNA targets like plk-3 will be essential to more clearly understand how and why their regulation by the piRNA pathway is essential to male fertility.
MATERIALS AND METHODS
C. elegans maintenance
All strains (table S3) in this study were maintained at 20°C on plates made from nematode growth medium [NGM: NaCl (3 g/liter), peptone (2.5 g/liter), agar (20 g/liter), cholesterol (1 ml/liter) (5 mg/liter in ethanol), 1 M CaCl2 (1 ml/liter), 1 M MgSO4 (1 ml/liter), and 1 M KPO4 buffer (25 ml/liter, pH 6.0) in distilled water] and fed OP50 Escherichia coli. Unless otherwise noted, experiments were conducted by placing mid-L4 stage male and hermaphrodite animals at 25°C and then collecting F1 mid-L4 stage male progeny approximately 3 days later. Collected progeny were maintained at 25°C for 24 hours after collection until dissection. We chose 25°C as the temperature to conduct all the fertility assays since argonaute protein family mutants have been shown to exhibit strong fertility phenotypes at 25°C (55).
Male fertility assay
To assess male fertility, males from strains being assayed and fog-2(oz40) female animals were collected at the mid-L4 at 25°C and separately aged 12 hours to early adulthood. Adult animals were then placed onto smaller mating plates (35-mm NGM plates with OP50) at a ratio of three males to one female to ensure successful mating. After 12 hours, the male animals were discarded, and the female was transferred to fresh plate and allowed to lay eggs. Female animals were then transferred every 12 hours onto fresh plates until all females in the assay stopped laying eggs. After every transfer, the plates containing the eggs were maintained at 25°C, and live progeny that hatched from these eggs were counted. Total live progeny laid by an individual mated female was then calculated and used as a readout for male fertility.
Germline dissection and fixation of worms for imaging of tagged proteins or immunofluorescence
Male worms aged 24 hours past mid-L4 stage were dissected as previously described (61). Dissected germlines were fixed in 3% formaldehyde diluted in 100 mM K2HPO4 (pH 7.2) for 10 min at room temperature then washed three times with PBST (0.1% Tween-20 in phosphatebuffered saline).
To maintain fluorescence for experiments in which GFP- or mCherry-tagged protein strains were imaged, washed germlines were immediately transferred to a 6 mm–by–50 mm Kimble tube in with 50 μl of PBST and stained with 4′,6-diamidino-2-phenylindole (DAPI, 2 μg/ml) for 10 min in the dark at room temperature. Germlines were washed three times with PBST then mounted in 5 μl of PBST on an agarose pad made of 2% agarose on a 25 mm–by–75 mm glass microscope slide. Samples were sealed with nail polish and a 22 mm–by–22 mm coverslip and allowed to settle for 10 min in the dark at room temperature before imaging.
For experiments involving immunofluorescence, washed germlines were transferred to a 6 mm–by–50 mm Kimble tube in 5 μl of PBST and permeabilized with 100% methanol overnight at −20°C. Methanol was removed, and samples were washed three times with 1 ml of PBST. Following the three washes, 50 to 100 μl of 30% normal goat serum (NGS) was added to the tubes, and the samples were allowed to block overnight at 4°C. After blocking, 30% NGS was removed, and primary antibodies diluted to their final concentration in 30% NGS were added to the tube and allowed to incubate overnight at 4°C. After the removal of primary antibody, the samples were washed three times with 1 ml of PBST and secondary antibody diluted to their final concentration in 30% NGS was added to the tubes and allowed to incubate for 2 hours at room temperature in the dark. Secondary antibody was removed, and samples were washed three times with 1 ml of PBST. During the final wash, DAPI (at 2 μg/ml) was added to the tubes and the tubes were allowed to incubate for 15 min in the dark at room temperature. DAPI was removed from the samples, and then a single drop (~15 μl) of Vector Laboratories VECTASHIELD Antifade Mounting Medium (SKU: H-1000-10) was added to samples. The stained germlines were then mounted in VECTASHIELD on a 2% agarose pad on a microscope slide and sealed with a glass coverslip and nail polish.
The following primary antibodies with associated concentrations were used in this study: mouse anti-major sperm protein (1:800) (Developmental Studies Hybridoma Bank), guinea pig anti-pSun-1 (1:800) [V. Jantsch (34)], rabbit anti-HIM-3 (1:50 as Alexa Fluor 488 conjugate) (Novus, catalog no.53470002), rabbit anti–SYP-1 (1:200) [A. M. Villeneuve (36)], and guinea pig anti–ZIM-2 (1:1000) [A. Dernburg (39)]. To use anti–HIM-3 and anti–SYP-1 simultaneously for assessment of synapsis, anti–HIM-3 was conjugated directly to Alexa Fluor 488 using an Invitrogen Alexa Fluor 488 Microscale Protein Labeling Kit (catalog no. A30006) and added after post-secondary antibody PBST washes to prevent cross-reaction. The following secondary antibodies were used in this study: goat anti-mouse Alexa Fluor 555 (Invitrogen catalog no. A-21422), goat anti-rabbit Alexa Fluor 555 (Invitrogen catalog no. 11008), and goat anti-guinea pig Cy5 (Invitrogen catalog no. A-11076).
Confocal microscopy and image processing
Images were captured with a Zeiss LSM 900 confocal microscope and acquired as z-stacks (0.19 μm) using a Zeiss Plan-Apochromat 63×/1.4 oil objective and Zeiss Zen micro-imaging software. Z-stacks were acquired with overlapping boundaries to allow for reassembly of montage images from individual focal planes in Adobe Photoshop 2020. Montage images were processed identically before assembly in FIJI (62) where maximum projection images were created and levels were adjusted.
Isolation and sequencing of low-input small-RNA samples
Because of low fertility of prg-1 mutant worms, we performed isolation and sequencing of small-RNA samples derived from 20 to 30 male worms per genotype as previously described (33). In brief, adult male worms were picked into TRIzol. RNA was isolated and purified with reagents from mirVana miRNA isolation kits (Invitrogen, catalog no.AM1560). Samples were then treated with Cap-Clip Acid Pyrophosphatase (CellScript) to enrich for 22G siRNAs during cloning and small-RNA sequencing. Libraries were prepared using a TruSeq Small RNA Library Preparation Kit (Illumina) and then sequenced on a Miseq Genome Analyzer (Illumina).
Processing and mapping of small-RNA reads
Library adapter sequences were trimmed from small-RNA reads using cutadapt (63), and quality of reads was assessed using FastQC. A custom Python script(HQAlign) was used to align trimmed reads to C. elegans protein-coding genes, ncRNAs, and the entire genome itself. A readout was produced of the number of reads that mapped to genes (in the case of protein-coding genes or ncRNAs) or to the genome. For small RNAs that mapped to multiple regions of the genome, such as repetitive regions, a first champion mapping strategy is used to produce the best match for a read. Reads counts mapping to protein-coding genes and ncRNAs for each sample were normalized to total reads from that sample that mapped to the genome.
HCR-FISH for plk-3 mRNA expression
Adult male worms (24 hours past mid L4) were dissected as described above. Fixation was performed with 3% paraformaldehyde for 15 min followed by methanol treatment overnight at −20°C. After methanol, the germlines were processed as previously described (64) with minor alterations (65). The samples were then washed with probe wash buffer as described, and probe hybridization was finally performed for both plk-3 probe (gene of interest) as well as pgl-1 probe, a gene known to be expressed in the germline, which we have previously used as a positive control.
Generation of PCR-product repair template for GFP-tag knockin strains
Repair templates for GFP-knockin strains were generated using DNA purified from the previously described plasmid pJA245 (66) as base. Plasmid DNA was extracted from pJA245 bacteria using a QIAprep Spin Miniprep Kit (Qiagen, catalog no.27106) and then used for both cosa-1 and plk-3 repair templates.
For cosa-1, the following primers were used to amplify, via polymerase chain reaction (PCR) with high-fidelity KOD polymerase (Novagen, catalog no.71316), the GFP-coding sequence, and introduce both a linker sequence and 5′ and 3′ homology arms to the targeted region of the cosa-1 sequence:
GTGAAAAATCGTGAAAACTGAACTGAAGTGTCAATGAGTAAAGGAGAAGAACTTTTCACTGGAGTTG (forward), ATTTTATTTTGAAACGACAACTCACCGAGAACTTGACATCGATGCTCCCGATCCTGCCGATGCTCCCGATGCTCCCGATCCTGCCGATGCTCCCGATGCACCCGATCCTGCCGATGCTCCCGAGGCTCCAGCTCCACCTCCACCTCCTTT (reverse). The resultant product of 1047 base pair (bp) was evaluated for proper size on a gel then purified using a QIAquick Gel Extraction Kit (Qiagen, catalog no.28706).
For plk-3, the following primers were used to amplify, via PCR with high-fidelity KOD polymerase (Novagen, catalog no.71316), the GFP-coding sequence, and introduce both a linker sequence and 5′ and 3′ homology arms to the targeted region of the plk-3 sequence:
GCTCCACTTGCTTCCTCCAGATATCTCGCAGGAGGATCTGGAGGCGGTTCTGGCGGAGGTTCTGGTATGAGTAAAGGAGAAGAACT (forward), ATTATTCACAGATTAGTTGTGTAGCATTTATTTGTATAGTTCGTCCATGC (reverse). The resultant product of 963 bp was evaluated for proper size on a gel and then purified as described above.
CRISPR-Cas9–mediated generation of V5::mCherry-tagged, ΔPAZ, and RNase-dead prg-1 alleles
All alleles were generated at the endogenous prg-1 locus using a co-CRISPR-Cas9 method previously described (67). To tag PRG-1 with V5::mCherry, a CRISPR cut site was identified at the N terminus allowing for insertion of a sequence containing a V5 epitope tag, the mCherry fluorescent protein, and a flexible linker after the first 18 nt of the coding sequence. The corresponding CRISPR RNA (crRNA) was ordered from Dharmacon: TGGCATCTGGAAGTGGTCGC, 1-bp downstream the beginning of the prg-1–coding region. The repair template sequence contained the desired insertion and 35-nt homology arms on the outside of the cut site. The insertion disrupts the crRNA recognition sequence preventing additional recutting by Cas9 after editing. An injection mix was made as previously described (67) using repair template obtained from Updike D and injected into N2 animals at the young adult stage. The F1 progeny of roller animals were screened for the insertion by PCR using the following primers: CGC ACC GAT CGA TTC TGG AT (forward) and TGC TGT AGT CGC TTT GAG TCA (reverse). After PCR, wild-type animals yield a single band of ~200 bp, homozygous insertion animals yield a single band of ~1100 bp, and heterozygous insertion animals yield bands of both ~200 and ~1100 bp. Homozygous worms with the accurate insertion were used to establish founder lines. Founders were backcrossed at least three times with N2 strain before use in experiments or the generation of other alleles.
Domain-specific prg-1 mutants were generated by introducing targeted mutations into the V5::mCherry::PRG-1 strain generated above. To generate the ΔPAZ allele, we identified CRISPR cut sites flanking the 238 nt that encode the PAZ domain to remove this sequence. The corresponding CRISPR crRNAs were ordered from Dharmacon:
AAAATGAATGAGATGTACGG, 824 bp downstream the beginning of the prg-1–coding region.
CCAGTCGGGAAGCACAGTTC, and 1120 bp downstream the beginning of the prg-1–coding region.
We used a repair template sequence that contained a single-nucleotide insertion flanked by 35 nt homology arms outside the sequence deletion. The single-nucleotide insertion allows for the removal of the PAZ-domain sequence without altering the reading frame. PAMs for the crRNAs are located within the deleted region; thus, Cas9 recutting would be prevented after successful deletion. This resulted in the following single-stranded oligo donor (ssODN):
CCACAACGCGTTCAAGAGAAAATGAATGAGATGTACCTGTGCTTCCCGACTGGGCTAACGGATGAGATGCG. After injection into the N2 strain, roller F1 progeny were screened by PCR using the following primers: GGA ACT ACG ATC GTG CCA GA (forward) and CCC ACT CTG CTT GCT TTC CT (reverse). After PCR, wild-type animals yield a single band of ~600 bp, homozygous deletion animals yield a single band of ~300 bp, and heterozygous insertion animals yield bands of both ~600 and ~ 300 bp. Homozygous founders were isolated and backcrossed at least three times with N2 strain before use in experiments.
To generate the ribonuclease (RNAse)–dead allele, we identified CRISPR cut sites near the codon of aspartic acid 583 to introduce a mutation to substitute this amino acid for alanine. The corresponding CRISPR crRNAs were ordered from Dharmacon: TACCACGACTCGACATTGAA, 1986 bp downstream of the beginning of the prg-1–coding region TTCCTTTCAATGTCGAGTCG, and 1991 bp downstream of the beginning of the prg-1–coding region. The repair template contained the desired edit as well as homology arms of 36 and 26 nt outside of the cut sites. A single mutation located in the codon for aspartic acid 583 substitutes the amino acid for alanine. In addition, four silent mutations serve the purpose to mutate the crRNA recognition sites to prevent recutting by Cas9, as well as introducing a DraI restriction digest site to aid in screening. This resulted in the following ssODN with a missense mutation (bold) and silent mutations (lowercase, DraI site underlined):
AAAACACAATGATCGTCGGCTACGCTCTGTACCACGAtTCaACaTTaAAAGGAAAAACTGTCGGTGCTTGCGT.
After injection into the N2 strain, roller F1 progeny were screened by PCR using the following primers: CCATTCCCAACCAATGCGTG (forward) and GTAGAGGATCAAGCGGCTCG (reverse). PCR results amplify a single 481-bp band. After overnight restriction digest with 5 U/rxn of DraI (New England Biolabs) at 37°C, PCR product yields two bands at 246 and 235 bp if homozygously edited or three bands at 481, 246, and 235 bp if heterozygously edited. PCR product from unedited animals remains completely refractory to DraI digest. Homozygous founders were isolated and backcrossed at least three times with N2 strain before use in experiments.
CRISPR-Cas9–mediated generation of GFP-tagged cosa-1 allele
We tagged cosa-1 at its endogenous locus using the same co-CRISPR-Cas9 method used to generate the prg-1 alleles noted in the section above. To tag COSA-1 with GFP, a CRISPR cut site was identified at the C terminus allowing for the insertion of a sequence containing GFP and a flexible linker immediately after the start codon of the cosa-1 gene. The corresponding CRISPR crRNA was ordered from Dharmacon: AAGTGTCAATGTCAAGTTCT, beginning 8 bp before the start codon sequence.
The repair template contained the desired insertion as well as 36- and 39-nt homology arms on the outside of the cut site on the 5′ and 3′ end of the insertion, respectively. The insertion disrupts the crRNA recognition site to prevent recutting by Cas9.
After injection into the N2 strain, roller F1 progeny were screened by PCR using the following primers: AAACTTAGGCTCTGGTCTCGTG (forward), CACCTTCACCCTCTCCACTG (reverse), and GAACCTGATTCGCTGCTGA (reverse). After PCR, wild-type animals yield a single band of 505 bp; homozygous insertion animals yield two bands of 1477 and 824 bp; and heterozygous insertion animals yield three bands of 1477, 824, and 505 bp. Homozygous worms with the accurate insertion were used to establish founder lines. Founders were backcrossed at least three times with N2 strain before use in experiments or the generation of other alleles.
CRISPR-Cas9–mediated generation of GFP-tagged and ΔpiRNA plk-3 alleles
All alleles were generated at the endogenous plk-3 locus using the same co-CRISPR-Cas9 method used to generate the prg-1 alleles noted in the section above. To tag PLK-3 with GFP, a CRISPR cut site was identified at the C terminus allowing for insertion of a sequence containing GFP and a flexible linker just before the terminating codon of the coding sequence. The corresponding CRISPR crRNA was ordered from Dharmacon: CATTTATGCGAGATATCTGG, 17 bp upstream the end of the plk-3–coding region. The repair template sequence contained the desired insertion and 35-nt homology arms on the outside of the cut site. The insertion disrupts the crRNA recognition sequence preventing additional recutting by Cas9 after editing. Injections were performed as described for prg-1 alleles. F1 progeny of roller animals were screened for the insertion by PCR using the following three primers: CAA CCA GGA GCA TGT CGT CT (forward), CAC CTT CAC CCT CTC CAC TG (forward), and TGC GAA AAG GTT GCA AGG TT (reverse). After PCR, wild-type animals yield a single band of 578 bp; homozygous insertion animals yield two bands of ~1482 and 239 bp; and heterozygous insertion animals yield three bands of 1482, 578, and 239 bp. Homozygous worms with the accurate insertion were used to establish founder lines. Founders were backcrossed at least three times with N2 strain before use in experiments or the generation of other alleles.
To generate the ΔpiRNA allele, we identified CRISPR cut sites near the 21ur-10935–binding site to introduce single-nucleotide edits disrupting this sequence. The corresponding CRISPR crRNA were ordered from Dharmacon: TACGACTGAGATACTTCTTT, 137 bp upstream of the end of the plk-3–coding region. The 91-bp repair template contained a 31-bp sequence containing the desired edits as well as 5′ and 3′ homology arms of 30 nt. The edits consist of eight silent mutations that (i) disrupt the piRNA binding site and (ii) disrupt crRNA recognition sequences to prevent additional recutting by Cas9 after editing. This resulted in the following ssODN with silent mutations (red), seven of which were located within the 21ur-10935 target site mutations (underlined):
CGCAAACATATTGACAAACAGCTGCCCAAAGAAGTATCTGTCCCGTATCGAGTACGCACAGGCCAAAATTAAATTGCTTCGTCCTACAAAC.
After injection into the N2 strain, roller F1 progeny were screened by PCR using the following primers: CAC CTT CAC CCT CTC CAC TG (Reverse) and TCT TGT GAG TTG AAG TGT ATC CTT T (Forward), TTG GGC ATA TTC GAT ACG ACT G (Reverse), AGT ATC TGT CCC GTA TCG AGT AC (Forward). After PCR, wild-type animals yield two bands of 465 and 197 bp; heterozygously edited animals yield three bands of 465, 297, and 197; and homozygously edited animals yield two bands of 465 and 297 bp. Homozygous founders were isolated and backcrossed at least three times with N2 strain before use in experiments.
Western blot analysis
Whole worm protein extract and Western blot analysis were performed as described previously (55). To maximize protein, adult hermaphrodites cultured at 25°C were washed from plates of respective genotypes using M-9 buffer then boiled for 2 min with 2× loading dye at 95°C. Western blot analysis was also performed as described (55) with mouse anti-mCherry (1:500) and mouse anti-tubulin (1:500) followed by anti-mouse horseradish peroxidase–conjugated secondary antibody (1:5000). Since both primary antibodies are mouse-derived, membranes were first probed with anti-mCherry, imaged, and then stripped with Abcam mild stripping buffer [recipe: 15 g of glycine, 1 g of SDS, 10 ml of Tween-20 in 1 liter of distilled water, pH adjusted to 2.2] before probing with anti-tubulin. Protein levels for each genotype were normalized using the corresponding tubulin levels. For quantification of band intensity, Western blots were converted to grayscale images in Adobe Photoshop and intensity of the bands was measured using LI-COR mage Studio 1.0 Western blot analysis. The intensity of the tubulin bands was used as the control to calculate the relative amount of mCherry signal and expressed in percentage as arbitrary units.
RNA IP
For all RNA IPs, we used 5 mg of worm lysate prepared identically to lysates used in Western blot analysis. For the IP of v5::mCherry-tagged PRG-1 and PRG-1 mutants, 20 μl of V5-Trap magnetic agarose beads (Chromotek) were equilibrated by washing once in 1 ml of ice-cold IP buffer in 5-ml tubes. Worm lysate from each genotype was then added to tubes of beads, and IP buffer was added to a final volume of 5 ml. Protease and phosphatase inhibitors were added, and then the reactions were allowed to rotate at 4°C for 1 hour. After incubation, the tubes were placed on magnetic racks and supernatant was removed. The beads were then washed by adding 5 ml of ice-cold IP buffer, rotating at 4°C for 5 min. The tubes were then set on a magnetic rack to allow for the removal of the supernatant, and then the entire wash process was repeated for a total of five washes. After the final wash, the beads were stored at −80°C until RNA isolation.
RNA isolation from RNA IP samples for sequencing
Beads were spun down at 4°C before adding 90 μl of 1 M glycine. Tubes were agitated and vortexed before spinning the beads down and transferring the supernatant to a new tube containing 9 μl if 1 M tris (pH 10). The elution process was repeated a total of three times for each bead sample, and then all eluates from each corresponding sample were combined to obtain a single eluate for each genotype. Next, 1000 μl of TRIzol and 200 μl of chloroform was added to each sample, and tubes were shaken by hand before centrifugation at 12,000g for 15 min at 4°C. The resultant aqueous phase from each sample (~900 μl) was transferred to new tubes, and an equal volume of isopropanol was added along with 20 μg of glycogen. After mixing, the samples were incubated at −20°C for 30 min. Following incubation, the samples were centrifuged at 16,000g for 30 min at 4°C. The supernatant from each sample was carefully removed without disturbing the RNA pellet. Each pellet was washed with 1 ml of ice-cold 70% ethanol, vortexed, and then allowed to rotate at 4°C for 10 min. The samples were then centrifuged again at 16,000g for 30 min at 4°C. As much of the supernatant as possible was removed from the samples before letting the RNA pellet air dry for 10 min at room temperature. Last, the pellet was resuspended in 20 μl of RNase-free water and allowed to dissolve for 10 min at 65°C. The RNA concentration of each sample was measured by spectrophotometer before the samples were sent for small-RNA sequencing.
Quantification and statistical analysis
Defining meiotic stages and assessing meiotic progression
Meiotic stages in male C. elegans germlines were defined on the basis of a combination of (i) chromosome morphology as assessed from DAPI staining and (ii) the presence or absence of antibody staining for meiotic markers. On the basis of these indicators, four distinct meiotic stages were defined: TZ, early pachytene, mid/late pachytene, and diplotene.
TZ germ cells have crescent-shaped DAPI morphology and stain positively for pSun-1, a component of the pairing machinery. However, TZ germ cells do not display positive indication of synapsis as measured by presence and perfect colocalization of HIM-3 and SYP-1 on chromosomes. Early pachytene germ cells stain positive for pSUN-1 and display thread-like chromosome morphology characteristic of this stage. The beginning of pachytene staged is defined by completion of synapsis; thus, HIM-3 and SYP-1 are localized to chromosomes and colocalize indicating completed synapsis. Mid-late pachytene stage germ cells have pachytene chromosome morphology and display complete synapsis but do not stain positively for p-SUN-1. Last, diplotene cells display progressive desynapsis, which is visualized as HIM-3 staining present on chromosomes and lack of SYP-1, as well as progressive condensation of DNA. These features are indicative of chromosome remodeling that precedes meiosis I and II.
Because of the linear spatiotemporal nature of the C. elegans germline, disturbances in meiotic progression can be assayed for by counting the number of cell rows per each germ cell stage and comparing to a wild-type control. Meiotic cell row counting was performed on confocal images using Zeiss Zen Blue analysis software. The first meiotic cell row was defined as the first TZ germ cell row where most cells in that row met the criteria to be considered TZ germ cells, whereas counting concluded at the last diplotene germ cell row. Boundaries between germ cell stages were determined by the same criteria where a germ cell row was categorized on the basis of what the identity of most germ cells in that row. Germ cell rows per stage were added and averaged across several germlines and compared among genotypes.
Assay for pairing of homologous chromosomes
Pairing of homologous chromosomes was assessed by analysis of wild-type and mutant anti–ZIM-2 immunofluorescence images. ZIM-2 localizes to the PC region of C. elegans chromosome V and is involved in the pairing of homologous chromosomes during the TZ stage. When chromosome V is unpaired, ZIM-2 is visualized as two individual foci, whereas when paired, chromosome V is indicated by a single ZIM-2 focus. Thus, anti–ZIM-2 staining can be used as a proxy to assay for defects in chromosome pairing. For each germline of each genotype, germ cells were either scored as having unpaired chromosome V (2-foci) or paired chromosome V (1-focus). The percentage of germ cells with unpaired chromosome V was calculated for each germline, and the average percentage of germlines for each genotype were reported.
Assay for crossover formation
Formation of crossovers was assessed by analysis of images from wild-type and mutant strains containing GFP::COSA-1. In C. elegans, COSA-1 localizes to the site of crossovers and mediates their formation. Each pair of homologous chromosomes in late pachytene contains exactly one COSA-1 focus for a total of five foci per nucleus in male germ cells due to the lack of a paired X chromosome. Thus, to assay for the formation of crossovers, we observed germ cells from dissected germlines expressing GFP::COSA-1 using Zeiss Zen Blue software and categorized these germ cells as either those that formed the appropriate number of crossovers (>5 GFP::COSA-1 foci) or those that did not appropriately form crossovers (<5 GFP::COSA-1 foci). The number of germ cells failing to achieve five crossovers per germline was calculated as a percentage of total germ cells per germline, and then the percentages were average across germlines for a given genotype.
Analysis of chromosome segregation defects
Chromosome segregation defects were assessed from images of germlines from wild-type and mutant strains that were stained for DAPI. Meiotic divisions occur spatially proximal in the C. elegans male germline after diplotene germ cell rows but largely distal to the spermatid population. To catch rare division events, germlines were dissected and observed for the presence of dividing cells in anaphase. Germlines that contained dividing cells in anaphase were then assayed for segregation defects, whereas germlines without dividing cells were excluded from analysis. Germlines with anaphase cells were then assessed for the presence of chromosome segregation defects in the form of anaphase bridges. The percentage of germlines containing at least one dividing cell with an anaphase bridge among total germlines analyzed was reported.
Statistical analysis of small-RNA reads
To determine protein-coding genes for which there were significant differences in small-RNA reads, we first applied a cutoff where we considered only protein-coding genes for which there was a 2log (2) fold difference in reads between wild-type and mutant samples. We then assessed the significance of fold differences using a Bayesian model as previously described (68).
Analysis of siRNA mapping to protein-coding genes and piRNA binding sites
To visualize where 22G siRNAs mapped within protein-coding genes, the Integrated Genome Viewer (IGV) (69) was used. The mapping of siRNAs was then cross-referenced with piRTarbase (51). Candidate piRNA target sites were then entered into IGV to evaluate overlap with siRNA reads.
Acknowledgments
We thank D. Updike for reagents and helpful suggestions with the generation of the prg-1 mCherry CRISPR-Cas9 lines, A. Villeneuve for SYP-1 antibody, V. Jantsch for pSUN-1 antibody, and A. Dernburg for the ZIM-2 antibody. J.O. and S.A. would like to thank members of the Arur laboratory, D. Chu, S. Hernault, J. Yanowitz, and the C. elegans small-RNA community for helpful discussions through the course of this study.
Funding: J.O. is a Howard Hughes Medical Institute Gilliam Fellow. The work is funded by the National Institute of General Medicine Sciences (R35 GM140933 to S.A. and R35GM130366 to A.Z.F.). Some strains were provided by the CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440).
Author contributions: J.O. designed the study, conducted the experiments, interpreted data, and wrote the manuscript. L.W. conducted experiments, interpreted data, and edited the manuscript. J.S. conducted experiments and interpreted data. S.-Y.C. conducted experiments and interpreted data. A.Z.F. interpreted data and edited the manuscript. S.A. designed the study, interpreted data, and wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Strains generated in this study will be deposited in the Caenorhabditis elegans Resource Center (CGC). Small-RNA sequencing data have been deposited at GEO and are publicly available as of the date of publication under accession number GEO: GSE245025. Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, S.A. (sarur@mdanderson.org).
Supplementary Materials
The PDF file includes:
Figs. S1 to S9
Tables S5 and S6
Legends for tables S1 to S4, and S7
Legends for data S1 to S4
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S4, and S7
Data S1 to S4
REFERENCES AND NOTES
- 1.Bartel D. P., Metazoan microRNAs. Cell 173, 20–51 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Czech B., Munafo M., Ciabrelli F., Eastwood E. L., Fabry M. H., Kneuss E., Hannon G. J., piRNA-guided genome defense: From biogenesis to silencing. Annu. Rev. Genet. 52, 131–157 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okamura K., Lai E. C., Endogenous small interfering RNAs in animals. Nat. Rev. Mol. Cell Biol. 9, 673–678 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aravin A., Gaidatzis D., Pfeffer S., Lagos-Quintana M., Landgraf P., Iovino N., Morris P., Brownstein M. J., Kuramochi-Miyagawa S., Nakano T., Chien M., Russo J. J., Ju J., Sheridan R., Sander C., Zavolan M., Tuschl T., A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442, 203–207 (2006). [DOI] [PubMed] [Google Scholar]
- 5.Vagin V. V., Sigova A., Li C., Seitz H., Gvozdev V., Zamore P. D., A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313, 320–324 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Lau N. C., Seto A. G., Kim J., Kuramochi-Miyagawa S., Nakano T., Bartel D. P., Kingston R. E., Characterization of the piRNA complex from rat testes. Science 313, 363–367 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Girard A., Sachidanandam R., Hannon G. J., Carmell M. A., A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442, 199–202 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Aravin A. A., Sachidanandam R., Girard A., Fejes-Toth K., Hannon G. J., Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316, 744–747 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Grivna S. T., Beyret E., Wang Z., Lin H., A novel class of small RNAs in mouse spermatogenic cells. Genes Dev. 20, 1709–1714 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deng W., Lin H., Miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev. Cell 2, 819–830 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Kuramochi-Miyagawa S., Kimura T., Ijiri T. W., Isobe T., Asada N., Fujita Y., Ikawa M., Iwai N., Okabe M., Deng W., Lin H., Matsuda Y., Nakano T., Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development 131, 839–849 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Fu Q., Wang P. J., Mammalian piRNAs: Biogenesis, function, and mysteries. Spermatogenesis 4, e27889 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X. Z., Roy C. K., Dong X., Bolcun-Filas E., Wang J., Han B. W., Xu J., Moore M. J., Schimenti J. C., Weng Z., Zamore P. D., An ancient transcription factor initiates the burst of piRNA production during early meiosis in mouse testes. Mol. Cell 50, 67–81 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng K., Wang P. J., Blockade of pachytene piRNA biogenesis reveals a novel requirement for maintaining post-meiotic germline genome integrity. PLOS Genet. 8, e1003038 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu P. H., Fu Y., Cecchini K., Ozata D. M., Arif A., Yu T., Colpan C., Gainetdinov I., Weng Z., Zamore P. D., The evolutionarily conserved piRNA-producing locus pi6 is required for male mouse fertility. Nat. Genet. 52, 728–739 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Homolka D., Pandey R. R., Goriaux C., Brasset E., Vaury C., Sachidanandam R., Fauvarque M. O., Pillai R. S., PIWI slicing and RNA elements in precursors instruct directional primary piRNA biogenesis. Cell Rep. 12, 418–428 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Xu M., You Y., Hunsicker P., Hori T., Small C., Griswold M. D., Hecht N. B., Mice deficient for a small cluster of Piwi-interacting RNAs implicate Piwi-interacting RNAs in transposon control. Biol. Reprod. 79, 51–57 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Wang G., Reinke V., A C. elegans Piwi, PRG-1, regulates 21U-RNAs during spermatogenesis. Curr. Biol. 18, 861–867 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Batista P. J., Ruby J. G., Claycomb J. M., Chiang R., Fahlgren N., Kasschau K. D., Chaves D. A., Gu W., Vasale J. J., Duan S., Conte D. Jr., Luo S., Schroth G. P., Carrington J. C., Bartel D. P., Mello C. C., PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol. Cell 31, 67–78 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wallis D. C., Nguyen D. A. H., Uebel C. J., Phillips C. M., Visualization and quantification of transposon activity in Caenorhabditis elegans RNAi pathway mutants. G3 9, 3825–3832 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Das P. P., Bagijn M. P., Goldstein L. D., Woolford J. R., Lehrbach N. J., Sapetschnig A., Buhecha H. R., Gilchrist M. J., Howe K. L., Stark R., Matthews N., Berezikov E., Ketting R. F., Tavare S., Miska E. A., Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol. Cell 31, 79–90 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barucci G., Cornes E., Singh M., Li B., Ugolini M., Samolygo A., Didier C., Dingli F., Loew D., Quarato P., Cecere G., Small-RNA-mediated transgenerational silencing of histone genes impairs fertility in piRNA mutants. Nat. Cell Biol. 22, 235–245 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cornes E., Bourdon L., Singh M., Mueller F., Quarato P., Wernersson E., Bienko M., Li B., Cecere G., piRNAs initiate transcriptional silencing of spermatogenic genes during C. elegans germline development. Dev. Cell 57, 180–196.e7 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han B. W., Wang W., Li C., Weng Z., Zamore P. D., Noncoding RNA. piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. Science 348, 817–821 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang W., Han B. W., Tipping C., Ge D. T., Zhang Z., Weng Z., Zamore P. D., Slicing and binding by Ago3 or Aub trigger Piwi-bound piRNA production by distinct mechanisms. Mol. Cell 59, 819–830 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reuter M., Berninger P., Chuma S., Shah H., Hosokawa M., Funaya C., Antony C., Sachidanandam R., Pillai R. S., Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature 480, 264–267 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Fazio S., Bartonicek N., Giacomo M., Abreu-Goodger C., Sankar A., Funaya C., Antony C., Moreira P. N., Enright A. J., O’Carroll D., The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature 480, 259–263 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Bagijn M. P., Goldstein L. D., Sapetschnig A., Weick E. M., Bouasker S., Lehrbach N. J., Simard M. J., Miska E. A., Function, targets, and evolution of Caenorhabditis elegans piRNAs. Science 337, 574–578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim K. W., Tang N. H., Andrusiak M. G., Wu Z., Chisholm A. D., Jin Y., A neuronal piRNA pathway inhibits axon regeneration in C. elegans. Neuron 97, 511–519.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song J. J., Smith S. K., Hannon G. J., Joshua-Tor L., Crystal structure of Argonaute and its implications for RISC slicer activity. Science 305, 1434–1437 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Rivas F. V., Tolia N. H., Song J. J., Aragon J. P., Liu J., Hannon G. J., Joshua-Tor L., Purified Argonaute2 and an siRNA form recombinant human RISC. Nat. Struct. Mol. Biol. 12, 340–349 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Lingel A., Simon B., Izaurralde E., Sattler M., Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature 426, 465–469 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Wahba L., Hansen L., Fire A. Z., An essential role for the piRNA pathway in regulating the ribosomal RNA pool in C. elegans. Dev. Cell 56, 2295–2312.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woglar A., Daryabeigi A., Adamo A., Habacher C., Machacek T., La Volpe A., Jantsch V., Matefin/SUN-1 phosphorylation is part of a surveillance mechanism to coordinate chromosome synapsis and recombination with meiotic progression and chromosome movement. PLOS Genet. 9, e1003335 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zetka M. C., Kawasaki I., Strome S., Muller F., Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes Dev. 13, 2258–2270 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.MacQueen A. J., Colaiacovo M. P., McDonald K., Villeneuve A. M., Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes Dev. 16, 2428–2442 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lui D. Y., Colaiacovo M. P., Meiotic development in Caenorhabditis elegans. Adv. Exp. Med. Biol. 757, 133–170 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woglar A., Villeneuve A. M., Dynamic architecture of DNA repair complexes and the synaptonemal complex at sites of meiotic recombination. Cell 173, 1678–1691.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Phillips C. M., Dernburg A. F., A family of zinc-finger proteins is required for chromosome-specific pairing and synapsis during meiosis in C. elegans. Dev. Cell 11, 817–829 (2006). [DOI] [PubMed] [Google Scholar]
- 40.Jaramillo-Lambert A., Harigaya Y., Vitt J., Villeneuve A., Engebrecht J., Meiotic errors activate checkpoints that improve gamete quality without triggering apoptosis in male germ cells. Curr. Biol. 20, 2078–2089 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yokoo R., Zawadzki K. A., Nabeshima K., Drake M., Arur S., Villeneuve A. M., COSA-1 reveals robust homeostasis and separable licensing and reinforcement steps governing meiotic crossovers. Cell 149, 75–87 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Checchi P. M., Engebrecht J., Heteromorphic sex chromosomes: Navigating meiosis without a homologous partner. Mol. Reprod. Dev. 78, 623–632 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.R. S. Hawley, in Genetic Recombination, R. Kucherlapati, G. Smith, Eds. (American Society of Microbiology, 1988), pp. 497–527. [Google Scholar]
- 44.Lee H. C., Gu W., Shirayama M., Youngman E., Conte D. Jr., Mello C. C., C. elegans piRNAs mediate the genome-wide surveillance of germline transcripts. Cell 150, 78–87 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reinke V., Gil I. S., Ward S., Kazmer K., Genome-wide germline-enriched and sex-biased expression profiles in Caenorhabditis elegans. Development 131, 311–323 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Ortiz M. A., Noble D., Sorokin E. P., Kimble J., A new dataset of spermatogenic vs. oogenic transcriptomes in the nematode Caenorhabditis elegans. G3 4, 1765–1772 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gruidl M. E., Smith P. A., Kuznicki K. A., McCrone J. S., Kirchner J., Roussell D. L., Strome S., Bennett K. L., Multiple potential germ-line helicases are components of the germ-line-specific P granules of Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 93, 13837–13842 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao G., Deeb F., Mercurio J. M., Parfenova A., Smith P. A., Bennett K. L., PAN-1, a P-granule component important for C. elegans fertility, has dual roles in the germline and soma. Dev. Biol. 364, 202–213 (2012). [DOI] [PubMed] [Google Scholar]
- 49.Rose A. M., Baillie D. L., A mutation in Caenorhabditis elegans that increases recombination frequency more than threefold. Nature 281, 599–600 (1979). [DOI] [PubMed] [Google Scholar]
- 50.Chung G., Rose A. M., Petalcorin M. I., Martin J. S., Kessler Z., Sanchez-Pulido L., Ponting C. P., Yanowitz J. L., Boulton S. J., REC-1 and HIM-5 distribute meiotic crossovers and function redundantly in meiotic double-strand break formation in Caenorhabditis elegans. Genes Dev. 29, 1969–1979 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu W. S., Brown J. S., Chen T. T., Chu Y. H., Huang W. C., Tu S., Lee H. C., piRTarBase: A database of piRNA targeting sites and their roles in gene regulation. Nucleic Acids Res. 47, D181–D187 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shen E. Z., Chen H., Ozturk A. R., Tu S., Shirayama M., Tang W., Ding Y. H., Dai S. Y., Weng Z., Mello C. C., Identification of piRNA binding sites reveals the argonaute regulatory landscape of the C. elegans germline. Cell 172, 937–951.e18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reed K. J., Svendsen J. M., Brown K. C., Montgomery B. E., Marks T. N., Vijayasarathy T., Parker D. M., Nishimura E. O., Updike D. L., Montgomery T. A., Widespread roles for piRNAs and WAGO-class siRNAs in shaping the germline transcriptome of Caenorhabditis elegans. Nucleic Acids Res. 48, 1811–1827 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Avery L., Wasserman S., Ordering gene function: The interpretation of epistasis in regulatory hierarchies. Trends Genet. 8, 312–316 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seroussi U., Lugowski A., Wadi L., Lao R. X., Willis A. R., Zhao W., Sundby A. E., Charlesworth A. G., Reinke A. W., Claycomb J. M., A comprehensive survey of C. elegans argonaute proteins reveals organism-wide gene regulatory networks and functions. eLife 12, e83853 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Choi C. P., Tay R. J., Starostik M. R., Feng S., Moresco J. J., Montgomery B. E., Xu E., Hammonds M. A., Schatz M. C., Montgomery T. A., Yates J. R. III, Jacobsen S. E., Kim J. K., SNPC-1.3 is a sex-specific transcription factor that drives male piRNA expression in C. elegans. eLife 10, e60681 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tang W., Seth M., Tu S., Shen E. Z., Li Q., Shirayama M., Weng Z., Mello C. C., A sex chromosome piRNA promotes robust dosage compensation and sex determination in C. elegans. Dev. Cell 44, 762–770.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ragle J. M., Morrison K. N., Vo A. A., Johnson Z. E., Hernandez Lopez J., Rechtsteiner A., Shakes D. C., Ward J. D., NHR-23 and SPE-44 regulate distinct sets of genes during Caenorhabditis elegans spermatogenesis. G3 12, jkac256 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chase D., Serafinas C., Ashcroft N., Kosinski M., Longo D., Ferris D. K., Golden A., The polo-like kinase PLK-1 is required for nuclear envelope breakdown and the completion of meiosis in Caenorhabditis elegans. Genesis 26, 26–41 (2000). [DOI] [PubMed] [Google Scholar]
- 60.Harper N. C., Rillo R., Jover-Gil S., Assaf Z. J., Bhalla N., Dernburg A. F., Pairing centers recruit a Polo-like kinase to orchestrate meiotic chromosome dynamics in C. elegans. Dev. Cell 21, 934–947 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gervaise A. L., Arur S., Spatial and temporal analysis of active ERK in the C. elegans germline. J. Vis. Exp. 117, e54901 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Martin M., Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2011). [Google Scholar]
- 64.Choi H. M. T., Schwarzkopf M., Fornace M. E., Acharya A., Artavanis G., Stegmaier J., Cunha A., Pierce N. A., Third-generation in situ hybridization chain reaction: Multiplexed, quantitative, sensitive, versatile, robust. Development 145, dev165753 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Trimmer K. A., Zhao P., Seemann J., Chen S. Y., Mondal S., Ben-Yakar A., Arur S., Spatial single-cell sequencing of meiosis I arrested oocytes indicates acquisition of maternal transcripts from the soma. Cell Rep. 42, 112544 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zeiser E., Frokjaer-Jensen C., Jorgensen E., Ahringer J., MosSCI and gateway compatible plasmid toolkit for constitutive and inducible expression of transgenes in the C. elegans germline. PLOS ONE 6, e20082 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Paix A., Folkmann A., Seydoux G., Precision genome editing using CRISPR-Cas9 and linear repair templates in C. elegans. Methods 121–122, 86–93 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Maniar J. M., Fire A. Z., EGO-1, a C. elegans RdRP, modulates gene expression via production of mRNA-templated short antisense RNAs. Curr. Biol. 21, 449–459 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Robinson J. T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E. S., Getz G., Mesirov J. P., Integrative genomics viewer. Nat. Biotechnol. 29, 24–26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S9
Tables S5 and S6
Legends for tables S1 to S4, and S7
Legends for data S1 to S4
Tables S1 to S4, and S7
Data S1 to S4