Abstract
Introduction
Acute lymphoblastic leukemia (ALL) is among the most common cancers in children. With improvements in combination chemotherapy regimens, the overall survival has increased to over 90%. However, the current challenge is to mitigate adverse events resulting from the complex therapy. Several chemotherapies intercept cancer metabolism, but little is known about their collective role in altering host metabolism.
Objectives
We profiled the metabolomic changes in plasma of ALL patients initial- and post- induction therapy.
Methods
We exploited a biorepository of non-fasted plasma samples derived from the Dana Farber Cancer Institute ALL Consortium; these samples were obtained from 50 ALL patients initial- and post-induction therapy. Plasma metabolites and complex lipids were analyzed by high resolution tandem mass spectrometry and differential mobility tandem mass spectrometry. Data were analyzed using a covariate-adjusted regression model with multiplicity adjustment. Pathway enrichment analysis and co-expression network analysis were performed to identify unique clusters of molecules.
Results
More than 1200 metabolites and complex lipids were identified in the total of global metabolomics and lipidomics platforms. Over 20% of those molecules were significantly altered. In the pathway enrichment analysis, lipids, particularly phosphatidylethanolamines (PEs), were identified. Network analysis indicated that the bioactive fatty acids, docosahexaenoic acid (DHA)-containing (22:6) triacylglycerols (TAGs), were decreased in the post-induction therapy.
Conclusion
Metabolomic profiling in ALL patients revealed a large number of alterations following induction chemotherapy. In particular, lipid metabolism was substantially altered. The changes in metabolites and complex lipids following induction therapy could provide insight into the adverse events experienced by ALL patients.
Keywords: Metabolomics, Lipidomics, Acute lymphoblastic leukemia, Asparaginase phosphatidylethanolamine, Triacylglycerol
1. Introduction
Acute lymphoblastic leukemia (ALL) is one of the most common forms of cancer among children (Hunger & Mullighan, 2015). With the advent of improved combination chemotherapeutic regimens, the survival rate has been transformed from less than 10% in the 1960s to over 90% in the current era (Mulrooney et al., 2019). However, adverse drug reactions during treatment can lead to treatment interruptions and decrease event-free survival (Silverman et al., 2001; Wolthers et al., 2017). These adverse events include infection, thrombosis, bleeding, mucositis, pancreatitis, hypersensitivity reactions, hyperglycemia, hyperbilirubinemia and hyperlipidemia (Silverman et al., 2001).
There are three major phases of ALL therapy: induction, consolidation, and maintenance. The purpose of induction is to eradicate leukemic cells below current detection levels in order to achieve a complete response (Terwilliger & Abdul-Hay, 2017). The main categories of drugs given during induction include corticosteroids, vincristine, doxorubicin, and asparaginase. Several of these drugs intercept cancer metabolism, but little is known about their collective role in altering host metabolism.
To gain greater insights into the metabolic changes that occur during induction therapy and to determine whether these changes might predispose patients to adverse events, we comprehensively profiled plasma metabolites and complex lipids obtained in ALL patients initial- and post-induction therapy by using broad-coverage mass spectrometry methods.
In this study, more than 20% of all detected metabolites and complex lipids were significantly altered after induction therapy. The main changes were in lipid-based moieties, particularly phosphatidylethanolamines (PEs) and triacylglycerols (TAGs), which could contribute to the adverse events in ALL chemotherapy.
2. Materials and methods
2.1. Blood collection and sample preparation
De-identified plasma samples were obtained from patients enrolled in one of two consecutive clinical trials conducted by the Dana-Farber Cancer Institute (DFCI) ALL Consortium: DFCI 05–001 and DFCI 11–001 (Burns et al., 2020; Place et al., 2015). The protocols were approved by the Institutional Review Boards at each participating institution. Written informed consent from each participant’s parent/legal guardian, as well as patient assent when appropriate, was obtained prior to enrollment and initiation of therapy. Plasma samples were obtained only from participants who had consented to the collection and storage of an extra volume of blood for future research. A total of 100 samples were obtained from 50 patients at two time points: (1) initial- and (2) post-induction therapy (Day 32). The samples were obtained during non-fasting conditions. First, blood was collected in an EDTA tube and then sent to the clinical laboratory for processing and storage. For processing plasma, the blood was spun at 1700 g for 7 min at room temperature. The plasma layer was isolated, aliquoted, and then frozen at −80 °C.
2.2. Metabolites and complex lipid detection
Metabolomics profiling was performed on a global metabolomics platform and a lipidomics platform, both of which were developed by Metabolon, Inc. (Durham, NC, USA) (Bridgewater Br, 2014; Ubhi, 2018). All metabolite annotations met the stringent criteria required for a Tier 1 or 2 identification (Sumner et al., 2007).
2.3. Global metabolomics platform
Samples were prepared using an automated MicroLab STAR® system (Hamilton, Reno, NV, USA). Proteins were precipitated with methanol under vigorous shaking for 2 min, followed by centrifugation for 10 min at 2800 rpm. The resulting extract was divided into five fractions: two for analysis via separate reverse phase (RP)/ultrahigh performance liquid chromatography (UPLC)-tandem mass spectrometry (MS/MS) methods with positive ion mode electrospray ionization (ESI), one for analysis via RP/UPLC-MS/MS with negative ion mode ESI, one for analysis via hydrophilic interaction chromatography (HILIC)/UPLC-MS/MS with negative ion mode ESI, and one sample was reserved as a backup.
One aliquot was analyzed using acidic positive ion conditions. In this method, the extract was gradient eluted using water and methanol, containing 0.05% perfluoropentanoic acid (PFPA) and 0.1% formic acid (FA). Another aliquot was also analyzed using acidic positive ion conditions, however, this aliquot was optimized for more hydro-phobic compounds. In this method, the extract was gradient eluted using methanol, acetonitrile, water, 0.05% PFPA and 0.01% FA. Another aliquot was analyzed using basic negative ion optimized conditions. The basic extracts were gradient eluted from the column using methanol, water, and 6.5 mM ammonium bicarbonate at pH 8. The fourth aliquot was analyzed via negative ionization following elution from a HILIC column using a gradient consisting of water and acetonitrile with 10 mM ammonium formate at pH 10.8.
The MS analysis alternated between MS and data-dependent sequential (multistage) mass spectrometry (MSn) scans using dynamic exclusion. The raw data from the MS scans were analyzed through an automated comparison of the features of the ion peaks of the experimental samples to a reference library of chemical standard entries. These entries included retention time, molecular weight (m/z), preferred adducts, in-source fragments, and associated MS spectra. More than 3300 commercially available purified standard compounds were acquired and registered into the laboratory information management systems used for analysis in all platforms to determine their analytical characteristics.
2.4. Lipidomics platform
Lipids were extracted in the presence of deuterated internal standards using an automated BUME extraction (Lofgren et al., 2012).
Flow injection and mass spectrometry (FIA-MS) analysis was performed on a SCIEX 5500 QTRAP (SCIEX, Redwood City, CA, USA) equipped with a SelexION Differential Mobility Separation (DMS) cell (SCIEX, Redwood City, CA, USA), which was operated in the Multiple Reaction Monitoring (MRM) mode using the electrospray in two modes (positive and negative) with a Turbo V ion source. Two plates, Plates 1 and 2, were subjected to a parallel analysis. In Plate 1, phosphatidylcholines, phosphatidylethanolamines, lysophosphatidylcholines, lysophosphatidylethanolamines, and phosphatidylinositols (negative ion mode), as well as sphingomyelins (positive ion mode) were identified. In Plate 2, free fatty acids (negative ion mode) as well as cholesteryl esters, diacylglycerols, triacylglycerols, ceramides, dihydroceramides, hexosylceramides, and lactosylceramides (positive ion mode) were identified.
Individual lipid species were quantified based on the ratio of the signal intensity for the target compounds to the signal intensity for an assigned deuterated internal standard of known concentration. Lipid class concentrations were calculated by taking the sum of all molecular species within a class.
2.5. Data processing
For each metabolite in the global metabolomics platform, the raw abundance values were scaled to set the across-sample median equal to 1. In the lipidomics platform, actual concentrations that were referenced to a standard were determined. In both platforms, only metabolites and complex lipids detected in more than 50% of the samples were included in the analysis. For each of the remaining metabolites and complex lipids, the missing values were imputed by using the smallest corresponding abundance value observed in the dataset (Bridgewater Br, 2014). To normalize the datasets obtained from the initial-induction and post-induction, 8 random samples from the initial-induction were measured in the same batch as the post-induction samples. A total of 129 xenobiotics or unknown metabolites were excluded, resulting in 422 metabolites and 883 complex lipids included in downstream analysis. We identified the unknown metabolites with the same criteria (retention index, mass, and fragmentation pattern). However, the exact chemical natures were unknown because of the lack of the standards or the metabolites not being in the library.
2.6. Statistical analysis
Principal component analysis (PCA) can transform a multitude of variables into a smaller number of components. We performed PCA on each platform to identify any clusters among the samples. In the lipidomics platform, PCA visualization allowed the detection of outliers, and four outlier subjects were excluded (Supplementary Fig. 1). The initial- and post (Day32)-matched samples from the same subjects were used to identify the significantly altered metabolites and complex lipids after the induction therapy. The ratio between the initial- and post-induction values for each molecule was calculated, and then log2 transformed. Three co-variates, age, BMI (age-adjusted percentile), and initial risk for ALL, were identified with the empirical distribution of p-values as influencers. Multivariable linear regression model was performed, adjusting for age, BMI (age-adjusted percentage value), and initial risk for ALL. Both p-values and false discovery rate (FDR) corrected values from Benjamini–Hochberg correction procedure were obtained.
Pathway enrichment analysis was performed using a one-sided Fisher’s exact test, in which metabolic pathways with FDR < 0.05 were considered significantly changed.
Individual levels of metabolites are shown on graphs as means with standard deviations.
2.7. Correlation network analysis
The method employed in this work was described in a previous study (Contrepois et al., 2020). Pairwise Spearman’s rank correlations were calculated using the R package ‘Hmisc’ (v4.1.1) and weighted, undirected networks were plotted with ‘igraph’ (v1.2.6). Correlations with Bonferroni adjusted p-values < 0.05 were included and displayed via the Fruchterman-Reingold method. Nodes were color-coded by the directionality of the change (increased or decreased) (Fig. 5a) or by the classes of the molecules (Fig. 5b, c and Supplementary Fig. 3). Their size represents |log2(fold change)| (Fig. 5a, b and Supplementary Fig. 3), or betweenness centrality (Fig. 5c). The betweenness centrality was calculated by the betweenness function in ‘igraph’.
Fig. 5.
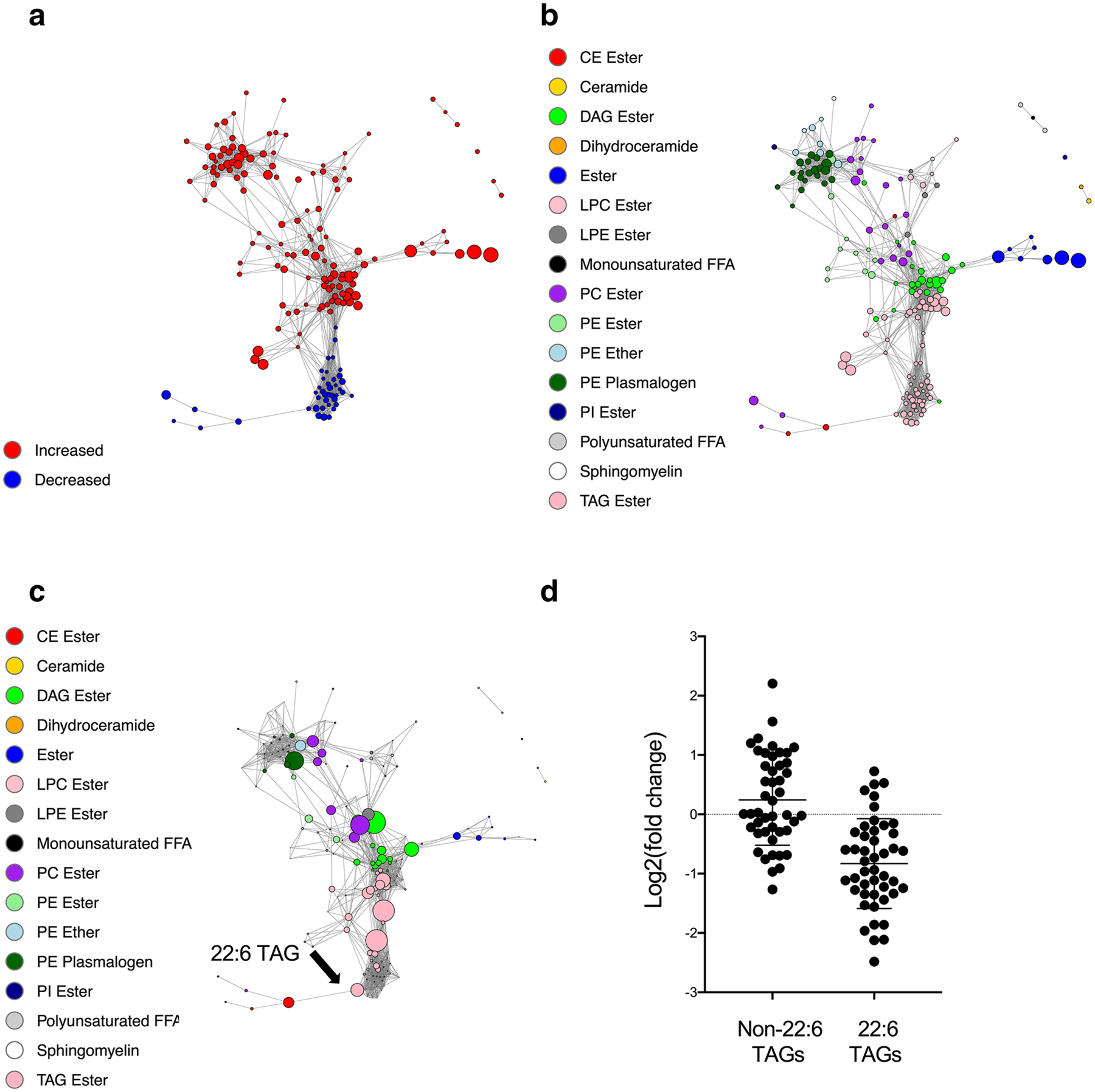
Pairwise Spearman correlation networks of altered complex lipids after induction therapy. a Nodes were color-coded based on the directionality of change (increased: red; decreased: blue), and their size represents |log2(fold change)|. b, c Nodes were color-coded by lipid classes, and their size represents |log2(fold change)| (b), or betweenness centrality (c). d Change in non-22:6 TAGs and 22:6 TAGs after induction therapy
3. Results
3.1. More than 1200 metabolites and complex lipids were detected
An overview of the study with the time points of plasma collection and comprehensive profiling of the metabolome is shown in Fig. 1a.
Fig. 1.
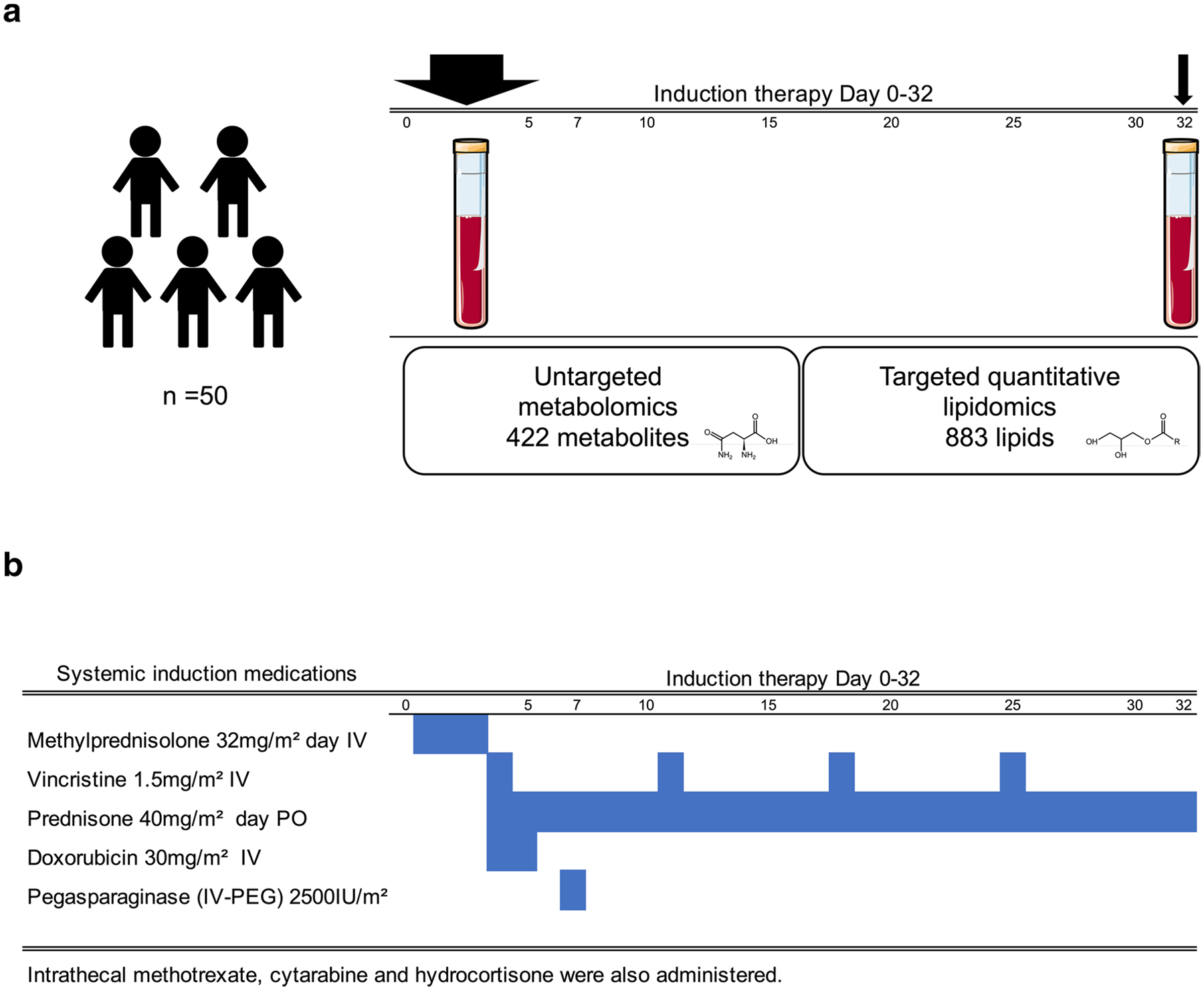
Induction medications and timing of plasma collection. a Overview of the study design, showing the time points of plasma collection and comprehensive profiling of the metabolome using untargeted and targeted metabolomics and lipidomics mass spectrometry-based platforms. Plasma was collected from each patient initial-induction and post- induction. Initial-induction sampling was performed, prior to therapy at Day 1 (protocol 05–001), or at Day 7 (protocol 11–001); and (2) post-induction, at the completion of induction therapy at Day 32. b Schema of the induction regimen
Plasma samples were obtained from 50 newly diagnosed ALL patients who were treated on DFCI protocols 05–001 (n = 33) & 11–001(n = 17) (Burns et al., 2020; Place et al., 2015). The samples were collected at two time points for each subject: (1) prior to therapy (initial-induction) at Day 1 (protocol 05–001), or Day 7 (protocol 11–001); and (2) at the completion of induction therapy (post-induction) at Day 32 (Fig. 1a). Baseline characteristics of the patients are provided in Table 1 and Supplementary Table 1.
Table 1.
Baseline patient characteristics
| Characteristics | |
|---|---|
| Sex (F/M), N | 24/26 |
| Age (years), median (IQR) | 6.5 (8.0) |
| BMI (kg/m2), median (IQR) | 17.1 (4.0) |
| Initial Risk Factor*, N(%) | Standard (29 [58%]) High (21 [42%]) |
| Final Risk Factor**, N(%) | Standard (25 [50%]) High (18 [36%]) Very High (7 [14%]) |
Age rage: 2.4–18.8 years. All the patients received the same induction chemotherapeutic agents
IQR interquartile range
Assigned based on age, presenting leukocyte count, immunopheno-type, central nervous system status
Assigned based on initial risk group, cytogenetics, and end-induction minimal residual disease levels
Comprehensive metabolome profiling was performed using untargeted and targeted metabolomics and lipidomics MS-based platforms. After filtering, a total of 1,305 metabolites and complex lipids were detected across all the samples in the study (Supplementary Table 2a). A list of excluded xenobiotics or unknown metabolites is shown in Supplementary Table 2b. Broad coverage was achieved, with the detection of 422 metabolites across 7 classes and 78 metabolic pathways, and 883 complex lipids across 16 classes.
A timeline of administration and dosing in the induction chemotherapeutic regimen is shown in Fig. 1b.
3.2. A high degree of separation was observed between the initial- and post-induction in global metabolomics and lipidomics platforms
Differential analysis between initial- and post- induction samples revealed a metabolic signature following treatment with 118 metabolites (27.9%) and 172 complex lipids (19.4%) (FDR < 0.05 and |log2(fold change)|> 1). Lists of molecules are shown in Supplementary Tables 3, 4. Differential metabolite and lipid profiles were visualized on PCA plots (Fig. 2 a, b). The significance and impact of metabolite changes are shown in volcano plots to demonstrate the overall metabolic effect of induction therapy (Fig. 2c, d). Out of 118 metabolites, 91 metabolites were increased, and 27 metabolites were decreased (metabolomics platform). Out of 172 complex lipids, 131 lipids were increased, and 41lipids were decreased (lipidomics platform) (Fig. 2e, f). As expected in post-induction samples, asparagine level was decreased upon treatment with asparaginase.
Fig. 2.
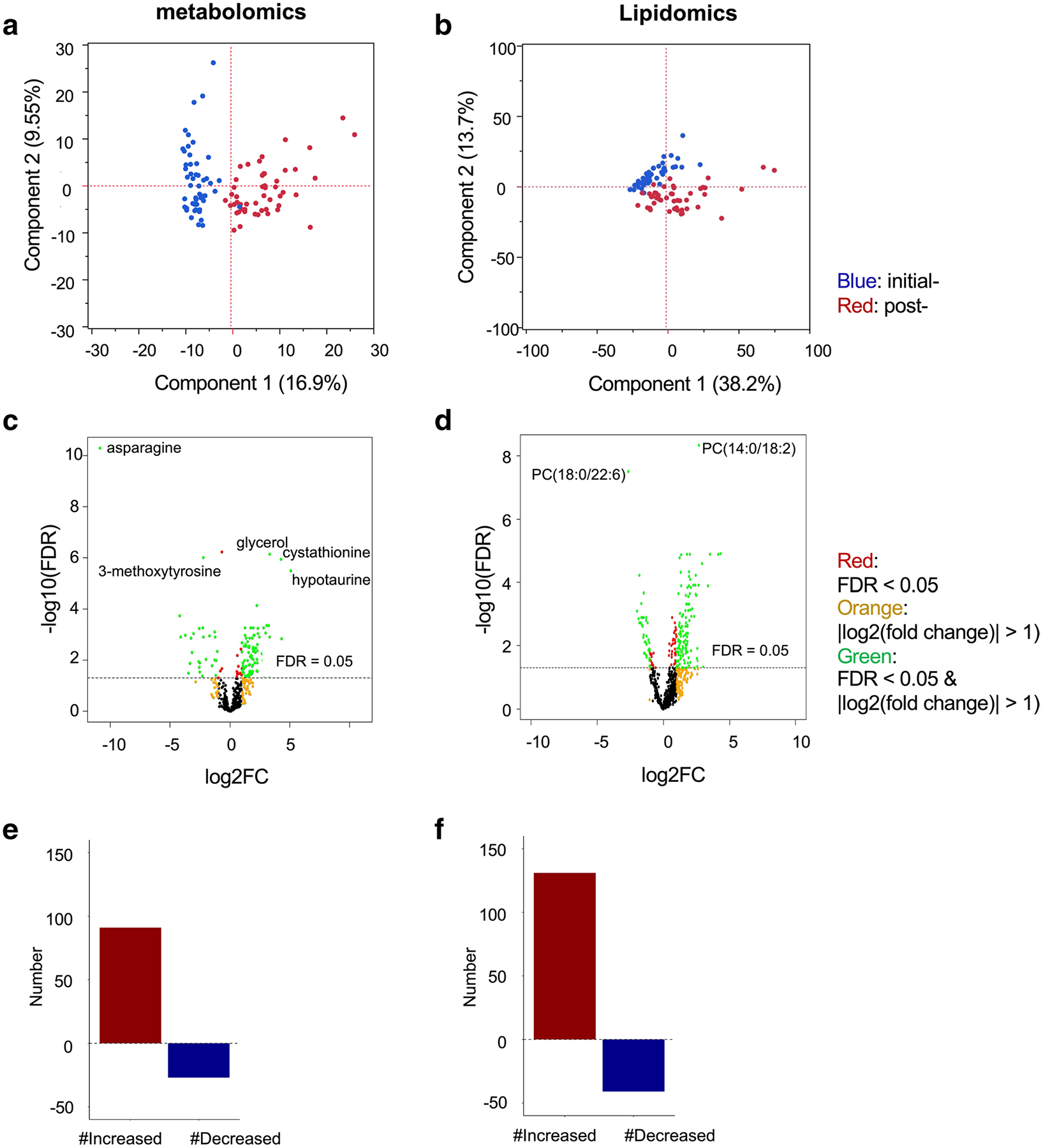
Global metabolomics and lipidomics profiling identifies dramatic differences in plasma metabolites initial- and post- induction therapy. a, c, e Global metabolomics platform. b, d, f Lipidomics platform. a, b Principal component analyses of the paired initial- and post- induction samples. A total of 50 subjects and 46 subjects were analyzed in the global metabolomics and lipidomics platforms, respectively. Blue: initial-induction data; red: post-induction data. c, d Volcano plots. Red: FDR < 0.05; orange: |log2(fold change)|> 1); green: FDR < 0.05 and |log2(fold change)|> 1). e, f The number of altered molecules
Changes in metabolite and lipid classes are shown in Fig. 3a, b. The major classes of identified metabolites were amino acids (n = 151) and lipids (n = 169). In the metabolomics platform, we did not observe a disproportionate change among the various classes of metabolites. However, in the lipidomics platform, there were several trends. We found increases in some classes of membrane lipids, including phosphatidylethanolamine (PE) ester (39.2%), PE plasmalogen (83.3%), PE ether (66.6%), lysophosphatidylcholine (LPC) ester (41.6%), and lysophosphatidylethanolamine (LPE) ester (57.1%).
Fig. 3.
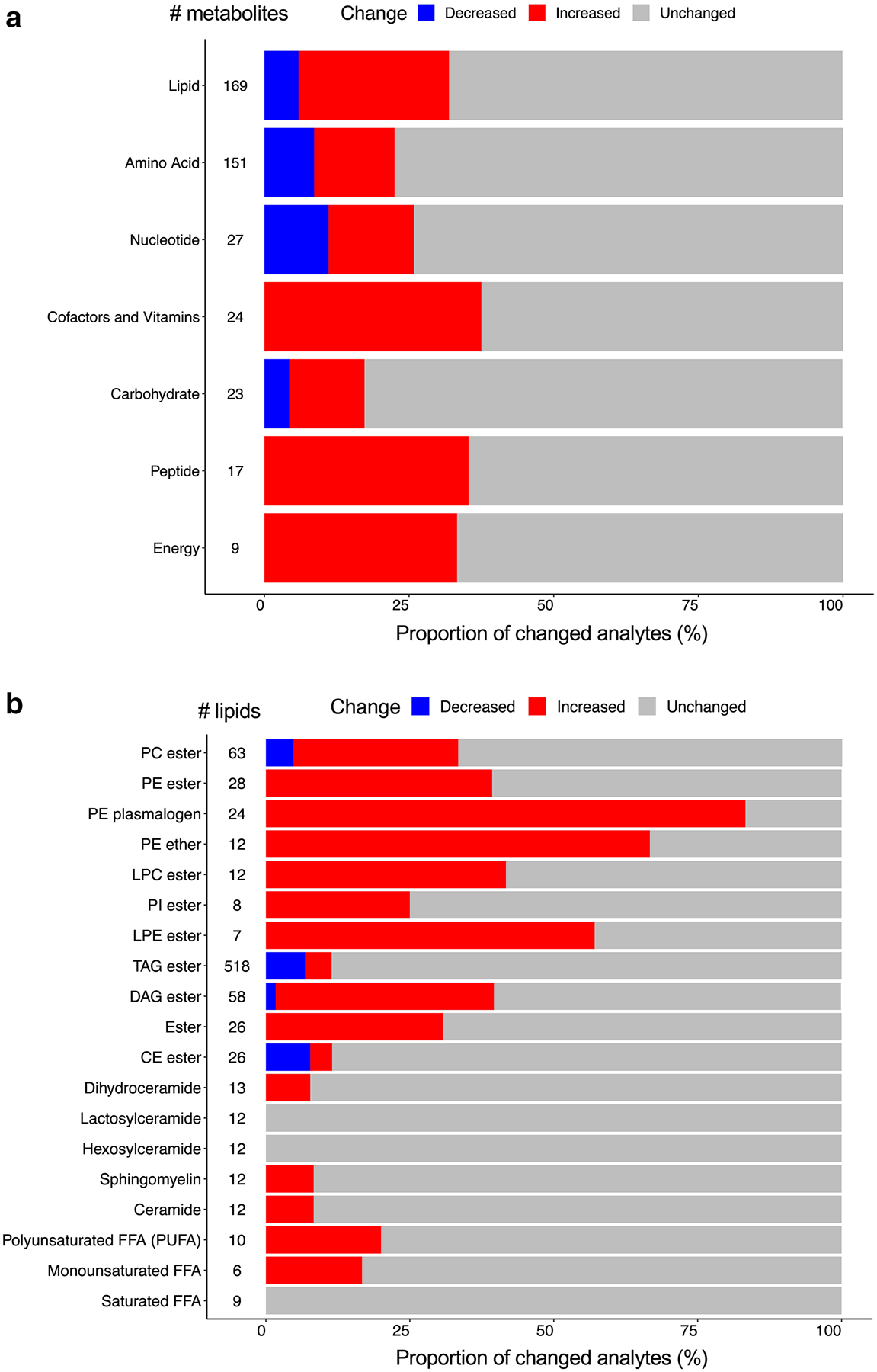
Proportion of classes of altered metabolites and complex lipids. a, b Bar plots showing the percentage of altered metabolites and lipids in each class. a Global metabolomics platform. (b) Lipidomics platform. Blue: decreased; red: increased; gray: unchanged. #: total numbers of metabolites and lipids identified in the class
The effect of induction therapy on the composition of side chain fatty acids in PE plasmalogens is shown in Supplementary Fig. 2. Compared to other fatty acids, 22:6-containing PE plasmalogens were not increased. The same trend was observed in free fatty acids. The fatty acid composition in PE plasmalogens reflects the free fatty acids profile.
3.3. A major change was observed in lipid metabolism
Using metabolic pathways of molecules, we performed pathway enrichment analysis using a one-sided Fisher’s exact test. In the global metabolomics platform, a significant metabolic pathway (FDR < 0.05) was the long chain polyunsaturated fatty acid (omega-3 and omega 6) (Fig. 4a). The same statistical method was applied to the lipidomics platform. The significantly changed lipids were the PE plasmalogen, diacylglycerol (DAG) ester, PE ether, phosphatidylcholine (PC) ester, and PE ester (Fig. 4b).
Fig. 4.
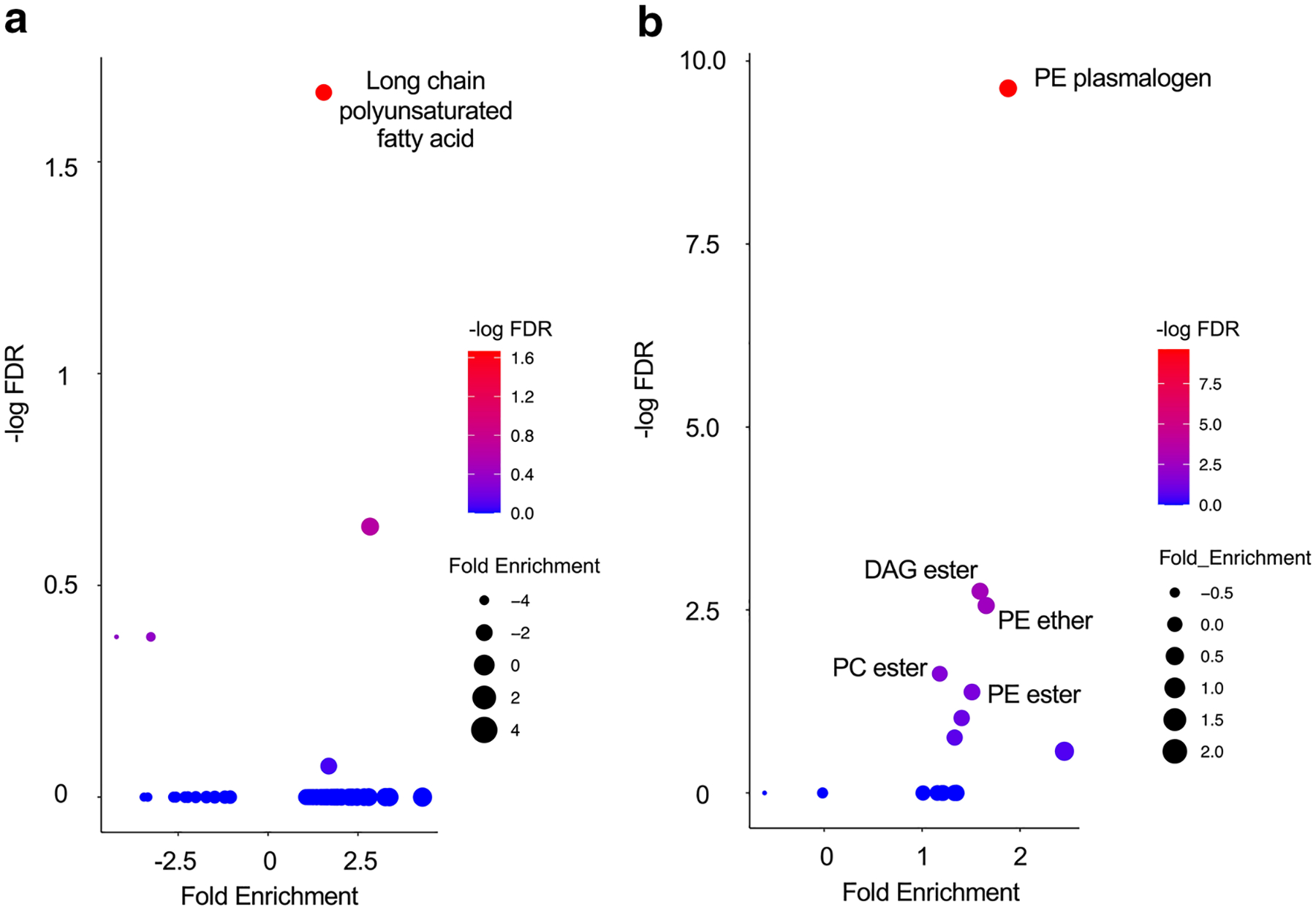
Pathways of altered metabolites and complex lipids. a, b Summary of the pathway enrichment analyses. a Global metabolomics platform. b Lipidomics platform. The y-axis represents −log10 FDR from the pathway enrichment analysis. The x-axis represents the mean of log2 (fold change) of all the significantly changed molecules in the metabolic pathway
These data demonstrate that induction therapy had a strong influence on the lipid profile.
3.4. Network analysis revealed a unique cluster of docosahexaenoic acid (DHA)-containing (22:6) triacylglycerols (TAGs)
We examined which molecules might be candidates for driving the change after induction therapy. All significantly changed metabolites and complex lipids were assessed together. Pairwise Spearman’s rank correlations were calculated, and weighted, undirected networks were plotted. Correlations with Bonferroni adjusted p-values < 0.05 were included and displayed. Among all molecular classes, the lipid molecules correlated with the highest numbers of other molecules (Supplementary Fig. 3). Asparaginase is a key chemotherapeutic agent that converts L-asparagine to aspartic acid (Batool et al., 2016). As expected from the lingering enzymic effect of pegylated (PEG)-asparaginase (Appel et al., 2008; Asselin & Rizzari, 2015), asparagine demonstrated the highest change after induction therapy, as represented by the size of its node, but it did not correlate with other molecules.
Since the major change was observed in lipids, we further focused on the lipidomics platform. Nodes were color-coded by the directionality of the change (increased or decreased) (Fig. 5a) or by the 16 classes of the complex lipids (Fig. 5b, c). The biological classes were preserved as the proximity of the structure of the correlation network. Interestingly, the two centered clusters of TAGs were noted, one of them was increased, and the other was decreased after induction therapy. To identify those molecules that correlated with a higher number of other molecules, we calculated betweenness centrality. The top nodes were determined, including DAG ester (green), TAG ester (light pink), PC ester (purple), and PE plasmalogen (dark green) (Fig. 5c).
Several of 22:6 TAGs were decreased after induction, and one of the 22:6 TAG nodes correlated with a high number of other lipids (Fig. 5c, arrow). To confirm the decreased levels of 22:6 TAGs, all non-22:6 TAGs and 22:6 TAGs from the lipidomics dataset were pooled and analyzed (Fig. 5d). Following induction therapy, 22:6 TAGs were decreased, but non-22:6 TAGs did not change. 22:6 fatty acid is also known as docosahexaenoic acid (DHA), an omega-3 polyunsaturated fatty acid, and a biologically active family of macromolecules (Moloudizargari et al., 2018). This network analysis of lipid nodes indicated that 22:6-TAG was particularly affected during induction therapy.
3.5. Asparaginase exerted a lingering effect on asparagine in the host
Asparaginase is a crucial chemotherapeutic agent used during induction therapy in ALL (Hijiya & van der Sluis, 2016). This enzyme functions as an aminohydrolase, catalyzing the conversion of the L-asparagine to aspartic acid and ammonia (Batool et al., 2016). Moreover, asparaginase has weak glutaminase activity and can contribute to the deamination of glutamine to glutamic acid.
Previous studies on asparaginase pharmacokinetics (Appel et al., 2008; Asselin & Rizzari, 2015) revealed peak activity of PEG-asparaginase at 72–96 h, a half-life of 5.73 ± 3.24 days, and depletion of serum or plasma asparagine for 26–34 days (Asselin & Rizzari, 2015). Those findings are consistent with our data (Supplementary Fig. 4), where post-induction asparagine levels were 7.6% compared to baseline (initial-induction) values. Moreover, after excluding three extreme outlier samples (> 200 times the average of remaining samples), post-induction asparagine levels were less than 1%, while the levels of the three other related-amino acids did not significantly vary (covariate-adjusted, FDR ≥ 0.05). It is possible that asparagine depletion occurred ex vivo in the collection tube after blood collection due to the remaining levels of active asparaginase, but these findings are consistent with the previously reported lingering effect of a single dose of PEG-asparaginase given 25 days prior (Appel et al., 2008; Asselin & Rizzari, 2015). Thus, the low asparagine levels we observed validate the metabolomic panel used in this study.
3.6. Anti-oxidative stress mechanisms were upregulated after induction therapy
Oxidative stress includes reactive oxygen species (ROS), peroxides, and free radicals (Udensi & Tchounwou, 2014). Cancer cells have higher levels of ROS compared to normal cells (Van Loenhout et al., 2020). Several of the induction chemotherapeutic agents also induce oxidative stress (Udensi & Tchounwou, 2014). However, there are anti-oxidative stress mechanisms to counteract oxidative stress, including glutathione (γ-glutamine tripeptide) and other gamma-glutamyl peptides. Indeed, after induction therapy, we observed an increase in three glutathione metabolites and six gamma-glutamyl peptides. (Supplementary Table 3). These data indicate that anti-oxidative stress mechanisms were enhanced after induction therapy.
3.7. Microbial metabolites were changed after induction therapy
The gut microbiota contributes to host physiology through the production of numerous metabolites (Krautkramer et al., 2021). Microbial metabolites have been linked to many diseases, including chronic gastrointestinal diseases, metabolic diseases, and neurological diseases (Martinez et al., 2017). The harsh chemotherapeutic agents used for ALL can disrupt the structural integrity of the gut barrier. Earlier studies on the gut microbiome and pediatric ALL have found significant differences between individuals with ALL and healthy controls at baseline (Nearing et al., 2019). Bile acids are considered microbial metabolites. Primary bile acids are synthesized from cholesterol in the liver. In the distal small intestine and colon, bile acids are subject to deconjugation. Deconjugated bile acids undergo various microbial bio-transformations, leading to a diverse array of secondary bile acids. We observed an increase in two primary bile acid metabolites and one secondary bile acid metabolites. We also identified several microbial amino acids and observed an increase in indoleacetate (Supplementary Table 3). These data indicate that a part of microbial metabolites was changed after induction therapy.
3.8. Chemotherapeutic drug metabolites were not detected after induction therapy
Between initial-induction and post-induction, we did not detect any metabolites of the induction chemotherapeutic drugs that are shown in Fig. 1. This could be due to the timing of the blood sampling. The systemic chemotherapeutic drugs in the regimen have half-lives of a couple of hours to several days: methylprednisolone (2.3 h), prednisone (2–3 h), vincristine (85 h), doxorubicin (20–48 h), and asparaginase (6–7 days) (Wishart et al., 2018). Regarding the pharmacokinetics and the timing of the drug administration, the drug that is most likely to be detected at Day 32 is prednisone. However, both prednisone and the active metabolite of prednisone, prednisolone, were below the limit of detection, which could also be due to the time at which blood was drawn relative to the administration of the drug.
Even though we did not identify the induction chemotherapeutic drugs or their metabolites in our metabolomic panel, we detected the effects of some of these drugs. For example, asparagine levels were decreased due to asparaginase. Endogenous pregnenolone steroid levels were also decreased, likely as a result of adrenal suppression from prednisone.
4. Discussion
Metabolomics measure small molecular chemical entities downstream of genomic, transcriptomic, and proteomic variability, thus providing a highly integrated profile of the biological status of an organism or its condition (Newgard, 2017; Rinschen et al., 2019).
In cancer, its aberrant metabolism has emerged as a major discipline (Andrejeva & Rathmell, 2017). Oncometabolites are endogenous metabolites, and their accumulation initiates or sustains tumor growth and metastasis. The discovery of “oncometabolites” has substantially affected our understanding of cancer (Wishart, 2016). Glutamine is the primary nitrogen source, and most proliferating cells rely on glutamine (Andrejeva & Rathmell, 2017). Asparagine promotes cell proliferation, and asparagine synthetase (ASNS) (Tsai et al., 2020) is frequently overexpressed in cancer. However, ALL cells express low ASNS and rely on exogenous asparagine. This metabolic vulnerability is exploited for ALL therapy with asparaginase which depletes asparagine (Egler et al., 2016). Recent studies suggest that oncometabolites act as signaling molecules or allosteric regulators that control important cell division processes (Fu et al., 2015; Wishart, 2016). In general, if a metabolite has a causal effect on the disease, an enzyme related to its biosynthetic pathway may be a target for treatment. Therefore, the metabolomic insights lead us to discover novel therapeutic drugs (Kang et al., 2018; Wishart, 2016).
In this study, we performed plasma metabolomic profiling of ALL patients at initial- and post-induction therapy timepoints. Utilizing advanced technologies, we identified 1305 metabolites and complex lipids, and more than 20% of them were significantly altered. Pathway enrichment analysis revealed that PE plasmalogen was the most significantly changed lipid metabolism in lipidomic profiling. PEs are the second most abundant glycerophospholipid in eukaryotic cells. They have diverse cellular functions, which include serving as a precursor for phosphatidylcholine (PC) and as a substrate for posttranslational modifications (e.g., Atg8p/LC-3) and influencing membrane topology, and promoting cell and organelle membrane fusion, oxidative phosphorylation, mitochondrial biogenesis, and autophagy (Calzada et al., 2016).
Network analysis demonstrated that the bioactive fatty acids, DHA-containing (22:6) TAGs, were decreased in post-induction therapy. DHA has been linked to the improved efficacy of the chemotherapeutic agents (Siddiqui et al., 2011). The mechanisms involve enhancing drug uptake, lipid peroxidation, and intracellular targets including cyclooxygenase-2, nuclear factor kappa B, and peroxisome proliferator-activated receptor-gamma. Experimental studies have also shown that several steps of the tumorigenic process, including initiation, promotion, latency, growth, and metastasis, can be influenced by polyunsaturated fatty acids (Sam et al., 2017). Moreover, the exposure of ALL cells to fish-oil-derived DHA suppresses the proliferation of leukemia cells (Sam et al., 2017). A small randomized clinical trial of DHA supplementation (total of 22 patients with leukemia or lymphoma) found an increased overall long-term survival (465 days after the start of the chemotherapy) in the DHA-treated group compared to the untreated controls (Chagas et al., 2017).
To our knowledge, the current study is one of the larger studies to date to perform metabolomic profiling initial- and post-induction therapy in children with ALL and the first study to perform both untargeted profiling of metabolites (global metabolomics platform) and targeted quantitative screening of a wide array of lipid classes (lipidomics platform). Four metabolomic studies in leukemia patients have been published in the last decade and provide relevant comparisons with our findings (Bai et al., 2014; Musharraf et al., 2017; Schraw et al., 2019; Tiziani et al., 2013). Tiziani et al. studied peripheral blood and bone marrow metabolomics in 10 children with ALL by using samples obtained at diagnosis and post-induction therapy samples. They identified a total of 102 metabolites. In peripheral blood, 22 were significantly changed after induction therapy. Tiziani et al. also revealed the strongest correlation of lipid metabolism at the diagnosis in comparison of peripheral blood and tumor microenvironment (Tiziani et al., 2013). Bai et al. studied serum in 15 children with ALL prior to therapy, along with another 30 children with ALL who were in remission and 60 healthy donors (Bai et al., 2014). They identified 30 metabolites that were differentially expressed between the ALL patients and healthy donors, and 9 of the metabolites were LPCs. Musharraf et al. studied serum from patients with hematologic dyscrasias (ages not specified), of whom 72 had ALL, 64 had acute myeloid leukemia, 50 had aplastic anemia, and 76 were healthy donors. Only pre-treatment samples were interrogated. Of the 1,425 metabolites identified, 27 were differentially expressed among the four groups, and the ALL patients were the most distinct group (Musharraf et al., 2017). Schraw et al. studied bone marrow in 60 children with ALL who had minimal residual disease (MRD) and 95 without MRD, all at a post-induction therapy time-point. They discovered altered central carbon and amino acid metabolism in MRD-positive patients (Schraw et al., 2019).
The main therapeutic regimen given during induction chemotherapy for ALL includes corticosteroids, vincristine, doxorubicin, and asparaginase. Adverse effects of the chemotherapeutic agents have been described in established databases. Supplementary Table 5 shows a summary of the metabolic adverse events in the evidence-based, multi-database drug search engines, Lexicomp (Lexicomp, Hudson, OH, USA) and Micromedex (IBM, Armonk, NY, USA). Those adverse events include hyperuricemia, hyperglycemia, and hyperlipidemia. We did not observe the altered metabolites related to hyperuricemia or hyperglycemia but did observed an altered lipidomic profile. Pathway enrichment from the lipidomics platform identified PE, DAG and PC as being significantly altered by induction chemotherapy.
Severe lipidemic alterations are a common side effect of treatment with corticosteroids and asparaginase in pediatric patients with ALL (Parsons et al., 1997; Salvador et al., 2018). Salvador et al. found that 34.5% of children and adolescents (119 patients) exhibit hypertriglyceridemia (triglycerides > 500 mg/dL) during induction therapy with corticosteroids and asparaginase. Parsons et al. showed that TAG increases during asparaginase treatment and decreases after the treatment. However, there was no information on the specific TAG altered. In our study, no change in total TAG levels was detected after induction therapy, but this discrepancy may be due to differences in the timing of sample collection.
We also aimed to gain insight into the metabolic changes predisposing ALL patients to adverse events after induction therapy. Asparaginase-associated pancreatitis (AAP) is a severe adverse event caused by a chemotherapeutic agent. AAP is a cause of substantial morbidity with an incidence of between 2 and 10%. Despite low mortality, AAP often results in truncation of asparaginase therapy, which might be associated with an increased risk of leukemia relapse (Wolthers et al., 2017). The direct mechanism behind AAP is unknown, and the available treatments are mainly supportive (Rank et al., 2020). This study identified major changes after induction therapy in lipid-based moieties, particularly in PEs and 22:6 (DHA) TAGs. There are several studies that have linked these lipids to pancreatitis.
In experimental acute pancreatitis models (cerulein-treated rats and acinar cells), DHA treatment inhibits inflammatory mediators. DHA induces the expression of PPAR-γ-target gene, SOD1, and catalase, thereby inhibiting ROS-mediated activation of inflammatory signaling and inflammatory cytokine expression (Song et al., 2017).
Asparaginase is linked to decreased mammalian target of rapamycin complex 1 (mTORC1) signaling (Bunpo et al., 2009). Inhibition of mTORC1 induces a lipid signaling cascade, which decreases PE levels in mitochondrial membranes and promotes proteolysis (MacVicar et al., 2019), and this may cause pancreatic injury. Conversely, activation of mTOR improves autophagic flux and pancreatic injury (Hu et al., 2015).
Patients who develop pancreatitis might have differentially expressed those lipids compared to non- pancreatitis patients due to genetic or environmental factors. Further investigation is needed to determine if these altered lipids are linked to AAP.
The current study has several strengths and some limitations. A major strength is the acquisition of samples from each subject at two time points, initial- and post-induction. This approach allowed for a pair-matched statistical comparisons of the metabolites and complex lipids for each subject, controlling for inter-subject variability. The sample size of 50 subjects is also relatively larger than the sample sizes used in previous metabolomics reports on pediatric leukemia patients. The large-scale sample detection and the quantification of the complex lipids provided a robust platform to analyze data for significant metabolic pathways. Metabolites and complex lipids are known to be influenced by the patients’ prandial state (Smith et al., 2020). Therefore, a limitation is that the subjects were non-fasting. However, to our knowledge, only one pediatric report describes fasting samples (Bai et al., 2014), which attests to the challenges in obtaining fasting samples in this age group. In addition, we obtained plasma, which represents not one system, but a composite of extracellular metabolic changes from several organ systems. Also, further validation with an independent cohort is needed to confirm the results of this study (Davis et al., 2020; Steyerberg & Harrell, 2016).
5. Conclusion
In conclusion, the current plasma metabolomic profiling of ALL patients revealed diverse metabolomic changes after induction therapy. In particular, we identified changes in specific lipids, PEs and (DHA)-containing (22:6) triacylglycerols (TAGs).
Supplementary Material
Acknowledgements
We thank Dr. Kim Ritchey for his guidance in organizing the overall project, and David Mikhail in editing.
Funding
This work was supported by a National Institutes of Health (NIH) award (DK093491 to S.Z.H.) and an Investigator-initiated grant from Servier (to S.Z.H.).
Footnotes
Conflict of interest S.Z.H has equity in Prevcon and serves on the Scientific Advisory Board for Atias. All other authors have no conflict of interest.
Consent to participate: Written informed consent from each participant’s parent/legal guardian, as well as patient assent when appropriate, was obtained prior to enrollment and initiation of therapy. Serum samples were obtained only from participants who had consented to the collection and storage of an extra volume of blood for future research.
Consent for publication: All the authors approved the final edited version of this manuscript.
Ethical approval: De-identified serum samples were obtained from patients enrolled in one of two consecutive clinical trials conducted by the Dana-Farber Cancer Institute (DFCI) ALL Consortium; these trials were the DFCI 05–001 and DFCI 11–001. The protocols were approved by the Institutional Review Boards of each participating institution.
Supplementary Information The online version contains supplementary material available at https://doi.org/10.1007/s11306-021-01814-2.
Data availability
The metabolomics and metadata reported in this paper are available via MetaboLights repository https://www.ebi.ac.uk/metabolights/MTBLS2394, study identifier MTBLS2394.
References
- Andrejeva G, & Rathmell JC (2017). Similarities and distinctions of cancer and immune metabolism in inflammation and tumors. Cell Metabolism, 26, 49–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appel IM, Kazemier KM, Boos J, Lanvers C, Huijmans J, Veerman AJ, van Wering E, den Boer ML, & Pieters R (2008). Pharmacokinetic, pharmacodynamic and intracellular effects of PEG-asparaginase in newly diagnosed childhood acute lymphoblastic leukemia: Results from a single agent window study. Leukemia, 22, 1665–1679. [DOI] [PubMed] [Google Scholar]
- Asselin B, & Rizzari C (2015). Asparaginase pharmacokinetics and implications of therapeutic drug monitoring. Leukaemia & Lymphoma, 56, 2273–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Zhang H, Sun X, Sun C, & Ren L (2014). Biomarker identification and pathway analysis by serum metabolomics of childhood acute lymphoblastic leukemia. Clinica Chimica Acta, 436, 207–216. [DOI] [PubMed] [Google Scholar]
- Batool T, Makky EA, Jalal M, & Yusoff MM (2016). A Comprehensive review on L-Asparaginase and its applications. Applied Biochemistry and Biotechnology, 178, 900–923. [DOI] [PubMed] [Google Scholar]
- Bridgewater Br EAM (2014). High resolution mass spectrometry improves data quantity and quality as compared to unit mass resolution mass spectrometry in high-throughput profiling metabolomics. Journal of Postgenomics Drug & Biomarker Development, 04(2), 1. [Google Scholar]
- Bunpo P, Dudley A, Cundiff JK, Cavener DR, Wek RC, & Anthony TG (2009). GCN2 protein kinase is required to activate amino acid deprivation responses in mice treated with the anti-cancer agent L-asparaginase. Journal of Biological Chemistry, 284, 32742–32749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns MA, Place AE, Stevenson KE, Gutiérrez A, Forrest S, Pikman Y, et al. Identification of prognostic factors in childhood T-cell acute lymphoblastic leukemia: Results from DFCI ALL Consortium Protocols 05–001 and 11–001. Pediatric Blood and Cancer. 10.1002/pbc.28719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzada E, Onguka O, & Claypool SM (2016). Phosphatidylethanolamine metabolism in health and disease. International Review of Cell and Molecular Biology, 321, 29–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chagas TR, Borges DS, de Oliveira PF, Mocellin MC, Barbosa AM, Camargo CQ, Del Moral JAG, Poli A, Calder PC, Trindade E, & Nunes EA (2017). Oral fish oil positively influences nutritional-inflammatory risk in patients with haematological malignancies during chemotherapy with an impact on long-term survival: A randomised clinical trial. Journal of Human Nutrition & Dietetics, 30, 681–692. [DOI] [PubMed] [Google Scholar]
- Contrepois K, Wu S, Moneghetti KJ, Hornburg D, Ahadi S, Tsai MS, Metwally AA, Wei E, Lee-McMullen B, Quijada JV, Chen S, Christle JW, Ellenberger M, Balliu B, Taylor S, Durrant MG, Knowles DA, Choudhry H, Ashland M, … Enslen B (2020). Molecular choreography of acute exercise. Cell, 181, 1112–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis KD, Aghaeepour N, Ahn AH, Angst MS, Borsook D, Brenton A, Burczynski ME, Crean C, Edwards R, Gaudilliere B, Hergenroeder GW, Iadarola MJ, Iyengar S, Jiang Y, Kong JT, Mackey S, Saab CY, Sang CN, Scholz J, … Pelleymounter MA (2020). Discovery and validation of biomarkers to aid the development of safe and effective pain therapeutics: Challenges and opportunities. Nature Reviews. Neurology, 16, 381–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egler RA, Ahuja SP, & Matloub Y (2016). L-asparaginase in the treatment of patients with acute lymphoblastic leukemia. Journal of Pharmacology and Pharmacotherapeutics, 7, 62–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Chin RM, Vergnes L, Hwang H, Deng G, Xing Y, Pai MY, Li S, Ta L, Fazlollahi F, Chen C, Prins RM, Teitell MA, Nathanson DA, Lai A, Faull KF, Jiang M, Clarke SG, Cloughesy TF, … Huang J (2015). 2-Hydroxyglutarate Inhibits ATP Synthase and mTOR Signaling. Cell Metabolism, 22, 508–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijiya N, & van der Sluis IM (2016). Asparaginase-associated toxicity in children with acute lymphoblastic leukemia. Leukaemia & Lymphoma, 57, 748–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu YY, Zhou CH, Dou WH, Tang W, Hu CY, Hu DM, Feng H, Wang JZ, Qian MJ, Cheng GL, & Wang SF (2015). Improved autophagic flux is correlated with mTOR activation in the later recovery stage of experimental acute pancreatitis. Pancreatology, 15, 470–477. [DOI] [PubMed] [Google Scholar]
- Hunger SP, & Mullighan CG (2015). Acute Lymphoblastic Leukemia in Children. New England Journal of Medicine, 373, 1541–1552. [DOI] [PubMed] [Google Scholar]
- Kang YP, Ward NP, & DeNicola GM (2018). Recent advances in cancer metabolism: A technological perspective. Experimental & Molecular Medicine, 50, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krautkramer KA, Fan J, & Backhed F (2021). Gut microbial metabolites as multi-kingdom intermediates. Nature Reviews Microbiology, 19, 77–94. [DOI] [PubMed] [Google Scholar]
- Lofgren L, Stahlman M, Forsberg GB, Saarinen S, Nilsson R, & Hansson GI (2012). The BUME method: A novel automated chloroform-free 96-well total lipid extraction method for blood plasma. Journal of Lipid Research, 53, 1690–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacVicar T, Ohba Y, Nolte H, Mayer FC, Tatsuta T, Sprenger HG, Lindner B, Zhao Y, Li J, Bruns C, Kruger M, Habich M, Riemer J, Schwarzer R, Pasparakis M, Henschke S, Bruning JC, Zamboni N, & Langer T (2019). Lipid signalling drives proteolytic rewiring of mitochondria by YME1L. Nature, 575, 361–365. [DOI] [PubMed] [Google Scholar]
- Martinez KB, Leone V, & Chang EB (2017). Microbial metabolites in health and disease: Navigating the unknown in search of function. Journal of Biological Chemistry, 292, 8553–8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moloudizargari M, Mortaz E, Asghari MH, Adcock IM, Rede-geld FA, & Garssen J (2018). Effects of the polyunsaturated fatty acids, EPA and DHA, on hematological malignancies: A systematic review. Oncotarget, 9, 11858–11875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulrooney DA, Hyun G, Ness KK, Bhakta N, Pui CH, Ehrhardt MJ, Krull KR, Crom DB, Chemaitilly W, Srivastava DK, Relling MV, Jeha S, Green DM, Yasui Y, Robison LL, & Hudson MM (2019). The changing burden of long-term health outcomes in survivors of childhood acute lymphoblastic leukaemia: A retrospective analysis of the St Jude Lifetime Cohort Study. Lancet Haematol, 6, e306–e316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musharraf SG, Siddiqui AJ, Shamsi T, & Naz A (2017). SERUM metabolomics of acute lymphoblastic leukaemia and acute myeloid leukaemia for probing biomarker molecules. Hematological Oncology, 35, 769–777. [DOI] [PubMed] [Google Scholar]
- Nearing JT, Connors J, Whitehouse S, Van Limbergen J, Macdonald T, Kulkarni K, & Langille MGI (2019). Infectious complications are associated with alterations in the gut microbiome in pediatric patients with acute lymphoblastic leukemia. Frontiers in Cellular and Infection Microbiology, 9, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newgard CB (2017). Metabolomics and metabolic diseases: where do we stand? Cell Metabolism, 25, 43–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons SK, Skapek SX, Neufeld EJ, Kuhlman C, Young ML, Donnelly M, Brunzell JD, Otvos JD, Sallan SE, & Rifai N (1997). Asparaginase-associated lipid abnormalities in children with acute lymphoblastic leukemia. Blood, 89, 1886–1895. [PubMed] [Google Scholar]
- Place AE, Stevenson KE, Vrooman LM, Harris MH, Hunt SK, O’Brien JE, Supko JG, Asselin BL, Athale UH, Clavell LA, Cole PD, Kelly KM, Laverdiere C, Leclerc JM, Michon B, Schorin MA, Welch JJ, Lipshultz SE, Kutok JL, … Silverman LB (2015). Intravenous pegylated asparaginase versus intramuscular native Escherichia coli L-asparaginase in newly diagnosed childhood acute lymphoblastic leukaemia (DFCI 05–001): A randomised, open-label phase 3 trial. The Lancet Oncology, 16, 1677–1690. [DOI] [PubMed] [Google Scholar]
- Rank CU, Wolthers BO, Grell K, Albertsen BK, Frandsen TL, Overgaard UM, Toft N, Nielsen OJ, Wehner PS, Harila-Saari A, Heyman MM, Malmros J, Abrahamsson J, Noren-Nystrom U, Tomaszewska-Toporska B, Lund B, Jarvis KB, Quist-Paulsen P, Vaitkeviciene GE, … Schmiegelow K (2020). Asparaginase-associated pancreatitis in acute lymphoblastic leukemia: Results From the NOPHO ALL2008 treatment of patients 1–45 years of age. Journal of Clinical Oncology, 38, 145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinschen MM, Ivanisevic J, Giera M, & Siuzdak G (2019). Identification of bioactive metabolites using activity metabolomics. Nature Reviews Molecular Cell Biology, 20, 353–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador C, Entenmann A, Salvador R, Niederwanger A, Craz-zolara R, & Kropshofer G (2018). Combination therapy of omega-3 fatty acids and acipimox for children with hypertriglyceridemia and acute lymphoblastic leukemia. Journal of Clinical Lipidology, 12, 1260–1266. [DOI] [PubMed] [Google Scholar]
- Sam MR, Esmaeillou M, & Shokrgozar MA (2017). Fish-oil-derived DHA-mediated enhancement of apoptosis in acute lymphoblastic leukemia cells is associated with accumulation of p53, Downregulation of Survivin, and Caspase-3 Activation. Nutrition and Cancer, 69, 64–73. [DOI] [PubMed] [Google Scholar]
- Schraw JM, Junco JJ, Brown AL, Scheurer ME, Rabin KR, & Lupo PJ (2019). Metabolomic profiling identifies pathways associated with minimal residual disease in childhood acute lymphoblastic leukaemia. eBioMedicine, 48, 49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui RA, Harvey KA, Xu Z, Bammerlin EM, Walker C, & Altenburg JD (2011). Docosahexaenoic acid: A natural powerful adjuvant that improves efficacy for anticancer treatment with no adverse effects. BioFactors, 37, 399–412. [DOI] [PubMed] [Google Scholar]
- Silverman LB, Gelber RD, Dalton VK, Asselin BL, Barr RD, Clavell LA, Hurwitz CA, Moghrabi A, Samson Y, Schorin MA, Arkin S, Declerck L, Cohen HJ, & Sallan SE (2001). Improved outcome for children with acute lymphoblastic leukemia: Results of Dana-Farber Consortium Protocol 91–01. Blood, 97, 1211–1218. [DOI] [PubMed] [Google Scholar]
- Smith L, Villaret-Cazadamont J, Claus SP, Canlet C, Guillou H, Cabaton NJ, & Ellero-Simatos S (2020). Important Considerations for Sample Collection in Metabolomics Studies with a Special Focus on Applications to Liver Functions. Metabolites, 10(3), 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song EA, Lim JW, & Kim H (2017). Docosahexaenoic acid inhibits IL-6 expression via PPARgamma-mediated expression of catalase in cerulein-stimulated pancreatic acinar cells. International Journal of Biochemistry & Cell Biology, 88, 60–68. [DOI] [PubMed] [Google Scholar]
- Steyerberg EW, & Harrell FE Jr. (2016). Prediction models need appropriate internal, internal-external, and external validation. Journal of Clinical Epidemiology, 69, 245–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner LW, Amberg A, Barrett D, Beale MH, Beger R, Daykin CA, Fan TW, Fiehn O, Goodacre R, Griffin JL, Hankemeier T, Hardy N, Harnly J, Higashi R, Kopka J, Lane AN, Lindon JC, Marriott P, Nicholls AW, … Viant MR (2007). Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics, 3, 211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T, & Abdul-Hay M (2017). Acute lymphoblastic leukemia: a comprehensive review and 2017 update. Blood Cancer Journal, 7, e577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiziani S, Kang Y, Harjanto R, Axelrod J, Piermarocchi C, Roberts W, & Paternostro G (2013). Metabolomics of the tumor microenvironment in pediatric acute lymphoblastic leukemia. PLoS ONE, 8, e828859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CY, Kilberg MS, & Husain SZ (2020). The role of asparagine synthetase on nutrient metabolism in pancreatic disease. Pancreatology, 20(6), 1029–1034. [DOI] [PubMed] [Google Scholar]
- Ubhi BK (2018). Direct infusion-tandem mass spectrometry (DI-MS/MS) analysis of complex lipids in human plasma and serum using the lipidyzer platform. Methods in Molecular Biology, 1730, 227–236. [DOI] [PubMed] [Google Scholar]
- Udensi UK, & Tchounwou PB (2014). Dual effect of oxidative stress on leukemia cancer induction and treatment. Journal of Experimental & Clinical Cancer Research, 33, 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Loenhout J, Peeters M, Bogaerts A, Smits E, & Deben C (2020). Oxidative stress-inducing anticancer therapies: Taking a closer look at their immunomodulating effects. Antioxidants (Basel), 9(12), 1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart DS (2016). Emerging applications of metabolomics in drug discovery and precision medicine. Nature Reviews. Drug Discovery, 15, 473–484. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, Sajed T, Johnson D, Li C, Sayeeda Z, Assempour N, Iynkkaran I, Liu Y, Maciejewski A, Gale N, Wilson A, Chin L, Cummings R, Le D, … Wilson M (2018). DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Research, 46, D1074–D1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolthers BO, Frandsen TL, Baruchel A, Attarbaschi A, Barzilai S, Colombini A, Escherich G, Grell K, Inaba H, Kovacs G, Liang DC, Mateos M, Mondelaers V, Moricke A, Ociepa T, Samarasinghe S, Silverman LB, van der Sluis IM, Stanulla M, … Toxicity Working G (2017). Asparaginase-associated pancreatitis in childhood acute lymphoblastic leukaemia: An observational Ponte di Legno Toxicity Working Group study. The Lancet Oncology, 18, 1238–1248. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The metabolomics and metadata reported in this paper are available via MetaboLights repository https://www.ebi.ac.uk/metabolights/MTBLS2394, study identifier MTBLS2394.


