Abstract
The cis-acting signals required for cleavage and encapsidation of the herpes simplex virus type 1 genome lie within the terminally redundant region or a sequence. The a sequence is flanked by short direct repeats (DR1) containing the site of cleavage, and quasi-unique regions, Uc and Ub, occupy positions adjacent to the genomic L and S termini, respectively, such that a novel fragment, Uc-DR1-Ub, is generated upon ligation of the genomic ends. The Uc-DR1-Ub fragment can function as a minimal packaging signal, and motifs have been identified within Uc and Ub that are conserved near the ends of other herpesvirus genomes (pac2 and pac1, respectively). We have introduced deletion and substitution mutations within the pac regions of the Uc-DR1-Ub fragment and assessed their effects on DNA packaging in an amplicon-based transient transfection assay. Within pac2, mutations affecting the T tract had the greatest inhibitory effect, but deletion of sequences on either side of this element also reduced packaging, suggesting that its position relative to other sequences within the Uc-DR1-Ub fragment is likely to be important. No single region essential for DNA packaging was detected within pac1. However, mutants lacking the G tracts on either side of the pac1 T-rich motif exhibited a reduced efficiency of serial propagation, and alteration of the sequences between DR1 and the pac1 T element also resulted in defective generation of Ub-containing terminal fragments. The data are consistent with a model in which initiation and termination of packaging are specified by sequences within Uc and Ub, respectively.
Herpesviruses have linear double-stranded DNA genomes of 125 to 245 kbp that are circularized upon infection and replicate in an “endless” form, generating branched concatemeric structures. During the assembly of progeny particles, the concatemers are cleaved at specific sites corresponding to the genomic termini and, in a tightly coupled process, the viral DNA is packaged into preformed capsids (reviewed in references 13, 20, and 23).
In the case of herpes simplex virus type 1 (HSV-1), the cis-acting sequences required for cleavage and packaging reside within the terminally redundant region or a sequence (21, 29, 32, 34). The a sequence is 250 to 500 bp long and present as a single copy at the S terminus and one or more tandem copies at the L terminus. In addition, one or more copies are also present in inverted orientation at the junction between the L and S segments (23).
Flanking the a sequences are direct repeats (DR1) of 17 to 20 bp, with single copies of DR1 separating tandem a sequences. The site of genomic cleavage occurs within the DR1 element such that a single copy is regenerated upon ligation of the two genomic ends (see Fig. 1). The central portion of the a sequence comprises multiple tandem reiterations of one or two other short sequence elements (11 to 24 bp) referred to as DR2 and DR4. The quasi-unique sequences, each of ca. 80 bp, which lie between DR1 and either side of the DR2 and DR4 repeats are termed the Ub and Uc elements. In virion DNA the Uc element is adjacent to the L terminus and Ub is adjacent to the S terminus (23). Within the Ub and Uc regions are two domains, defined as pac1 and pac2, respectively, that contain several highly conserved motifs present near the ends of other herpesvirus genomes, including those lacking a terminal redundancy (5, 8, 9, 11). The presence of these conserved sequences near the termini of herpesvirus genomes, the fact that they are brought into close proximity upon circularization, and the fact that in most instances cleavage of concatemeric DNA occurs between the pac1 and pac2 signals led to the suggestion that they have important roles in the cleavage packaging process.
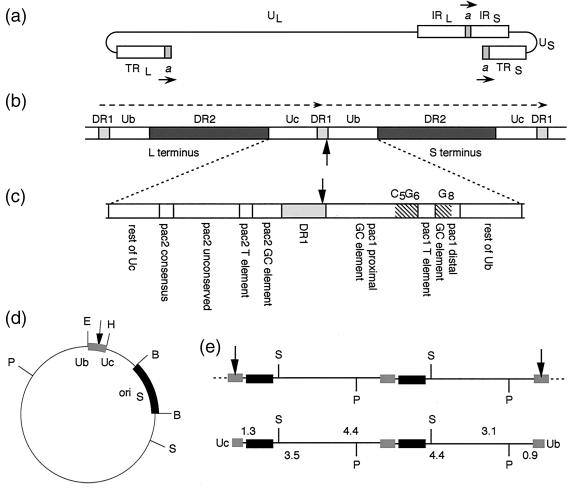
FIG. 1.
Structure and cloning of the HSV-1 Uc-DR1-Ub element. (a) Structure of the HSV-1 genome showing the positions and relative orientations of copies of the a sequence. For simplicity, only single copies are shown at the L terminus and joint. (b) Circularization of linear genomes by direct ligation of the termini brings together two copies of the a sequence (dashed arrows, orientation as in panel a) separated by a single DR1 repeat. The site of ligation, and of cleavage of concatemers, is shown by the arrow. (c) Motifs within the 194-bp Uc-DR1-Ub element spanning tandem a sequences. The poly(G) and poly(C) stretches (within the proximal and distal GC elements) that flank the pac1 T element are indicated by diagonal hatching. (d) Structure of the amplicon, pSA1. oriS and the Uc-DR1-Ub fragment are shown as thickened lines with the site of cleavage indicated by the arrow. E, H, B, S, and P indicate the positions of the EcoRI, HindIII, BamHI, SalI, and PstI restriction endonuclease sites, respectively. (e) The upper line illustrates the structure of a concatemer generated by pSA1 replication (only two complete copies of the monomeric plasmid are depicted). As with full-length HSV-1 genomes, cleavage within DR1 generates linear packaged pSA1 molecules with Uc and Ub at opposite ends (lower line). Digestion of packaged DNA with PstI (P) or SalI (S) yields fragments corresponding to pSA1 monomers plus diagnostic terminal fragments (sizes indicated in kilobase pairs). The larger terminal fragment produced by PstI or SalI cleavage represents the Uc or Ub terminus, respectively.
The analysis of herpesvirus cleavage packaging signals has employed two principal approaches. In the first, plasmids containing a viral origin of DNA replication and putative packaging signal (so-called amplicons) are transfected into tissue culture cells and helper functions are provided either by cotransfection with intact viral DNA or superinfection with virus particles. The amplicon DNA replicates autonomously and, if functional packaging signals are present, it is encapsidated in the form of long concatemeric molecules consisting of head-to-tail repeats of the input plasmid. Encapsidation of the amplicon DNA confers resistance to exogenously added DNase and permits serial propagation as defective genomes in the presence of the helper (10, 22, 29, 32, 34). Alternatively, additional copies of putative cleavage-packaging signals can be inserted at ectopic sites within the viral genome and assessed for functionality by determining whether concatemeric DNA becomes cleaved at novel sites corresponding to the inserted sequences (7, 18, 21, 27, 34). However, particularly in the case of HSV-1, delimiting the sequences required for packaging has frequently been complicated by the repair of mutated sequences through recombination with wild-type copies of the a sequence. There is a strong selection for such repair when amplicons are serially propagated prior to analysis, and sequence homology between ectopically inserted a sequences and the resident genomic copies, possibly involving the DR2-DR4 region, plays an important role (7, 11, 27, 28).
Single copies of the HSV-1 a sequence were initially shown to be sufficient for cleavage and packaging (10, 32, 34). In this instance, the pac1 and pac2 homologies reside at opposite ends of the a sequence separated by up to 300 bp of the DR2 and DR4 repeats and, in order to explain how packaged molecules come to possess an a sequence at each terminus, it is necessary to invoke either specific amplification of a sequences or the “wastage” of DNA lacking a terminal a sequence (10, 11, 34). Circularization of the HSV-1 genome, however, fuses the terminal a sequences and, as shown in Fig. 1b, generates a novel junction (Uc-DR1-Ub) in which the pac2 and pac1 homologies are close together. Indeed, an ∼200-bp fragment corresponding to the Uc-DR1-Ub junction has been shown to represent the minimal functional packaging signal for HSV-1 (22). Moreover, if cleavage of concatemers occurs at the novel junctions between such fused a sequences, genomes with a sequences at each end can readily be generated without the necessity for either a sequence amplification or wastage of DNA. The novel Uc-DR1-Ub junction may therefore represent an important in vivo substrate for the HSV-1 cleavage and packaging machinery.
Conserved pac1 and pac2 motifs were defined by Deiss et al. (11), and their locations within the HSV-1 Uc-DR1-Ub element are illustrated in Fig. 1c. Within Uc a consensus sequence CGCCGCG (pac2 consensus) lies ca. 70 bp from the S terminus, followed in turn by a relatively poorly conserved region (pac2 unconserved), a highly conserved T-rich region (pac2 T element), and a terminal region of high G+C content (pac2 GC element). The 40 to 50 bp of Ub adjacent to the DR1 direct repeat represent a region of the S terminus of high G+C content (pac1 proximal GC element). This is followed by a T-rich element (pac1 T element) and another region of high G+C content (pac1 distal GC element). Characteristically, the pac1 T element is flanked by the sequences CnGn from the proximal GC element and Gn from the distal GC element, and this unit has been proposed to represent the functional pac1 signal (5, 8, 11, 18). Experiments with deletion mutants removing sequences from either end of the HSV-1 a sequence have shown that regions critical for cleavage and packaging reside within both Ub and Uc (11, 22, 27, 34). In addition, strong evidence has been presented that the DR1 element is not essential for cleavage and packaging, with the S and L termini being generated as a result of cleavage at defined distances from the two T elements. This suggests that the T elements or sequences flanking them are of pivotal importance in the process (34). Although several of the analyses were complicated by the occurrence of DNA rearrangements, a 15-bp sequence within pac1 that contributed to the S-terminal cleavage signal was identified (27), and very recently this was shown to lie within a region bound specifically by the HSV-1 DNA packaging protein, UL28 (3).
To date, only in the case of the murine cytomegalovirus (MCMV) packaging signal have the effects of specific mutations within the conserved pac1 and pac2 motifs been analyzed. In that study (18) the wild-type MCMV cleavage signal (generated by fusion of the genomic termini), and mutated forms thereof, were recombined into the viral genome at an ectopic site. Stocks of cell-released virus were derived, and the occurrence of cleavage at the novel signal was determined by analyzing the terminal fragments of virion DNA. Several mutations within both the pac1 and pac2 regions resulted in significant reductions in progeny that had been cleaved at the ectopic signal, while other mutations had little effect. The G tract distal to the pac1 T element, but not the T element itself, was very important for DNA packaging. Within pac2 both the T element and adjacent sequences within the relatively unconserved region were essential for cleavage and packaging, while the pac2 consensus sequence played a contributory role.
In order to analyze the role of individual motifs within the HSV-1 cleavage-packaging signals, we have employed an amplicon-based assay and conditions designed to reduce the opportunity for recombination with helper virus. A minimal cleavage packaging signal (Uc-DR1-Ub), lacking the DR2-DR4 repeats, was incorporated into the amplicon, and encapsidation was examined during a single cycle of infection with helper virus. Using this assay, we have tested the effects of several deletion and substitution mutations within the pac1 and pac2 regions on DNA packaging.
MATERIALS AND METHODS
Cells and viruses.
Baby hamster kidney 21 clone 13 (BHK) cells were grown in Glasgow minimal essential medium (MEM) supplemented with 10% tryptose phosphate broth, 10% newborn calf serum, 100 U of penicillin/ml, and 100 μg of streptomycin/ml (ETC10). Monolayers of BHK cells were set up in ETC10 and, after transfection or infection, were maintained in Glasgow MEM supplemented with 5% newborn calf serum, 100 U of penicillin/ml, and 100 μg of streptomycin/ml (EC5). Stocks of HSV-1 (strain 17 syn+ [16]) and HSV-2 (strain HG52 [12]) were prepared and titrated in BHK cells.
Plasmids.
Plasmid pS1 (30) contains a 540-bp fragment including a functional HSV-1 oriS cloned into the BamHI site of the vector pAT153. Plasmids pY1 and pZ1 contain 1,762- and 2,161-bp HinfI fragments from the junction of the L and S segments of HSV-1 strain 17 syn+, including one and two copies, respectively, of the a sequence (9), cloned between the EcoRI and HindIII sites of pS1 (32). The 194-bp MnlI fragment spanning the junction between the tandem a sequences in pZ1 was isolated, and blunt ends were generated using T4 DNA polymerase in the presence of all four deoxyribonucleoside triphosphates. The resulting fragment (Uc-DR1-Ub element) was inserted, using synthetic linkers, between the EcoRI and HindIII sites of pS1 to generate plasmid pSA1 (Fig. 1d). Site-directed mutagenesis of the Uc-DR1-Ub fragment was performed using the method of Kunkel (14). The EcoRI-to-HindIII fragment of pSA1 was first cloned between the corresponding sites of the vector pTZ18U, and the resulting plasmid, pTZ2, was transferred to Escherichia coli strain CJ236 (Dut− Ung−; New England Biolabs). Uracil-rich single-stranded DNA was prepared after infection with helper phage R408 and annealed to the appropriate mutagenic oligonucleotides. The oligonucleotides were designed to introduce substitution and deletion mutations at various positions within the Uc-DR1-Ub fragment (see Fig. 3) and contained ca. 14 additional bases complementary to the single-stranded DNA on either side of the region being mutated. In the case of the substitution mutations, novel restriction endonuclease sites were incorporated to facilitate screening. After second-strand synthesis, the products were transformed into E. coli XL1-Blue (Stratagene). Colonies containing the desired plasmids were identified by restriction enzyme analysis, and in each case the DNA sequence of the complete Uc-DR1-Ub insert was confirmed. The mutated EcoRI plus HindIII fragments were finally transferred back into pS1, generating the pPH series of plasmids. Plasmid pTZ2 was also digested with SacII or BanII, recircularized, and used to transform E. coli. Colonies containing the expected deletions were identified and the fragments similarly inserted into pS1, generating pPH3 and pPH4, respectively.
FIG. 3.
Sequence of the HSV-1 Uc-DR1-Ub element and of the mutants used in this study. The sequence of the cloned insert in plasmid pSA1 is depicted on three lines (i, ii, and iii) with the characteristic motifs and regions corresponding to those shown in Fig. 1c. The substitutions and deletions in the mutants are indicated below the sequence. The modified sequences of the relevant regions are shown in lowercase for the substitution mutants, and the nucleotides missing in the deletion mutants are indicated by “x.” Plasmid pPH3 contains an 80-bp deletion between the two SacII sites (single underlined) and pPH4 contains a 69-bp deletion between the BanII sites (double underlined).
Transient DNA packaging assay.
Monolayers of BHK cells in 35-mm petri dishes (2 × 106 cells per plate) were transfected by using the calcium phosphate technique followed by treatment with dimethyl sulfoxide (DMSO) at 4 h as previously described (31). Each monolayer received 0.5 ml of precipitate containing 0.5 μg of plasmid and 12 μg of calf thymus carrier DNA. At 2 h after DMSO treatment the cells were infected with 5 PFU of HSV-1 or HSV-2/cell. Incubation was continued for 16 h at 37°C, the medium was removed, the cells were resuspended in TBS (137 mM NaCl, 5 mM KCl, 0.7 mM Na2HPO4, 5.5 mM glucose, 25 mM Tris-HCl [pH 7.4]) and divided into two equal samples which were used to prepare total cellular and DNase-resistant (encapsidated) DNA. The cells from both samples were pelleted and resuspended in 184 μl of RSB (10 mM Tris-HCl [pH 7.5], 10 mM KCl, 1.5 mM MgCl2) containing 0.5% Nonidet P-40 (NP-40). An equal volume of 20 mM Tris-HCl (pH 7.5)–2 mM EDTA–1.2% sodium dodecyl sulfate–1 mg of protease (Sigma grade XIV)/ml was either added immediately (total cell DNA) or after incubation in the presence of 200 μg of DNase I/ml, with occasional mixing, for 20 min at 37°C (encapsidated DNA). After addition of protease, all samples were incubated for 1 h at 37°C, extracted sequentially with phenol and chloroform, and precipitated with ethanol, and the nucleic acids were redissolved in 10 mM Tris-HCl (pH 7.5)–1 mM EDTA containing 5 μg of RNase A and 50 U of RNase T1/ml. DNase-resistant DNA was also similarly prepared from isolated nuclei. In this case the cells were first incubated 10 min on ice in RSB containing 0.5% NP-40 and then the nuclei were pelleted and incubated in 184 μl of RSB containing 0.5% NP-40 and 200 μg of DNase I/ml prior to DNA isolation. Samples of DNA corresponding to the yield from 4 × 105 cells were cleaved with DpnI (which cleaves only unreplicated input plasmid molecules) and a second enzyme (usually EcoRI) that converts the concatemeric products of plasmid replication into monomers. The digested DNA was fractionated by agarose gel electrophoresis and transferred to a Hybond-N membrane (Amersham), and replicated (DpnI-resistant) plasmid DNA was detected by hybridization to a 32P-labeled probe prepared from the vector pAT153. Phosphorimages of Southern blots were acquired using the Personal Molecular Imager and analyzed with Quantity One software (Bio-Rad). In control experiments with DNA from cells that were mock infected after transfection, no DpnI-resistant species were detected, demonstrating that the bands present contain only replicated plasmid molecules.
Serial propagation of amplicons.
Supernatant medium was removed from monolayers of transfected cells at 16 h postinfection, and debris was pelleted by centrifugation for 5 min at 1,300 × g. Fresh monolayers of BHK cells in 35-mm petri dishes were infected with 0.5 ml of the supernatant. At 1 h after virus addition, the inoculum was removed, the residual virus was inactivated with an acid-glycine wash (1), and incubation was continued at 37°C for 18 h in 2 ml of EC5. Total cellular DNA was prepared and analyzed as described above.
RESULTS
Evaluation of the Uc-DR1-Ub element as a packaging signal.
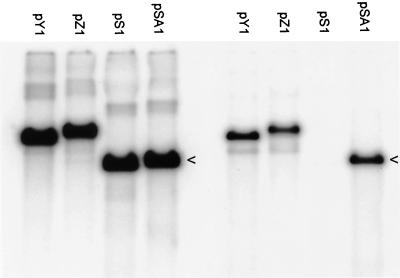
Since Nasseri and Mocarski (22) had previously demonstrated that the Uc-DR1-Ub element could function as an HSV-1 packaging signal, a similar fragment spanning the junction of tandem a sequences of HSV-1 strain 17 syn+ was isolated and cloned between the EcoRI and HindIII sites of the HSV-1 oriS-containing plasmid, pS1. The relative efficiency of this fragment to direct encapsidation of HSV-1 DNA was evaluated in a transient assay employing four plasmids, namely, the parental plasmid pS1 and derivatives containing a single a sequence (pY1), two tandem a sequences (pZ1), or the Uc-DR1-Ub element (pSA1). BHK cells were transfected with each of the plasmids and superinfected with HSV-1 prior to the preparation of total cellular DNA or DNase-resistant DNA. Samples of DNA were digested with EcoRI plus DpnI and analyzed as described in Materials and Methods. Figure 2 shows that, although all four plasmids replicated with similar efficiency (lefthand four lanes), only pY1, PZ1, and pSA1 were readily detectable in the DNase-resistant DNA samples (righthand four lanes). These data confirm the previous observations that single or tandem copies of the a sequence direct packaging with similar efficiency (32) and that the Uc-DR1-Ub element represents a functional packaging signal (22). Moreover, for the first time, they show that in a single cycle of infection the minimal Uc-DR1-Ub fragment functions as efficiently as full-length a sequences containing the DR2 reiterations. Quantification of a large number of experiments indicated that 5 to 10% of replicated pSA1 DNA was generally recovered in the DNase-resistant DNA sample. Although longer exposures allowed the detection of small amounts of packaged pS1 DNA, this represented <1% the amount of packaged pSA1.
FIG. 2.
Replication and packaging of plasmids pS1, pY1, pZ1, and pSA1 in a transient assay. BHK cells were transfected with pS1 (4.2 kbp), pY1 (6.0 kbp), pZ1 (6.2 kbp), or pSA1 (4.4 kbp) as indicated and superinfected with HSV-1, and total (left-hand 4 lanes) and DNase-resistant (right-hand 4 lanes) DNAs were prepared. Aliquots were digested with EcoRI plus DpnI, and the fragments were separated by electrophoresis through an agarose gel. The gel was blotted, the membrane was hybridized to 32P-labeled pAT153 DNA and, after being washed, the membrane was exposed to a phosphorimager screen. The arrowheads indicate replicated (DpnI-resistant) pSA1 monomers.
Analysis of Uc-DR1-Ub mutants.
In order to investigate the role of individual motifs within the Uc and Ub regions, amplicons similar to pSA1 were constructed containing copies of the Uc-DR1-Ub fragment into which substitution and deletion mutations had been introduced. The regions chosen for mutagenesis were essentially those originally defined by Deiss et al. (11), and the DNA sequences of the mutated fragments are shown in Fig. 3. In addition to the plasmids in which individual motifs were altered, two larger deletion mutants removing all of the Uc region (pPH3) and most of Ub (pPH4) were constructed.
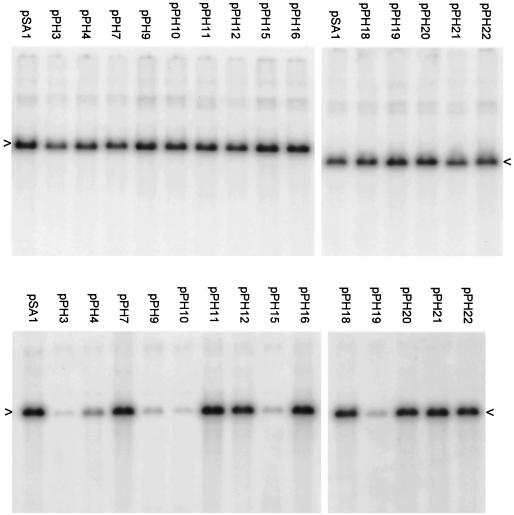
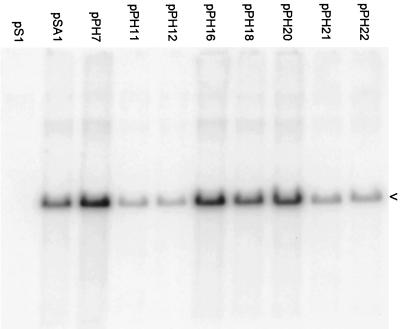
The packaging abilities of the resulting 14 plasmids were compared to those of pSA1 in a transient assay (Fig. 4). The results indicate that each of the mutated plasmids replicated to the same extent as pSA1 (top panels), but there were significant differences in the amount of DNA encapsidated (lower panels). Quantitative data were obtained from three separate experiments in which the full set of mutant plasmids was examined (Table 1). The amount of each plasmid packaged was expressed as a percentage of the pSA1 value and the mean and standard deviation were calculated. The two plasmids most impaired for packaging were pPH3 (Uc deletion) and pPH10 (pac2 T element deletion), which exhibited relative packaging efficiencies of 2.4 and 2.3%, respectively. Plasmids pPH15 (pac2 GC element deletion), pPH9 (pac2 T element substitution), pPH4 (Ub deletion), and pPH19 (pac2 unconserved region deletion) were also significantly impaired (relative packaging efficiencies of 5.0, 6.9, 9.6, and 12.9%, respectively). The remaining eight plasmids all exhibited packaging efficiencies that differed <2-fold from pSA1. The values obtained for pPH21, pPH22 (pac1 proximal GC element deletion and substitution mutants), pPH11, pPH12 (pac1 distal GC element deletion and substitution mutants), pPH20, pPH16, pPH18 (pac2 substitution mutants affecting the unconserved region, the GC element and the consensus sequence), and pPH7 (pac1 T element substitution) were 75, 82, 76, 118, 132, 152, 168, and 177%, respectively.
FIG. 4.
Packaging ability of plasmids containing mutated Uc-DR1-Ub fragments. A transient packaging assay using HSV-1 as helper was performed with the pPH series of plasmids as described in Materials and Methods and in the legend to Fig. 2. The upper and lower panels are for total and DNase-resistant DNA, respectively, and the positions of the replicated amplicon monomers (4.4 kbp) are indicated by arrowheads.
TABLE 1.
Properties of the plasmids used in this study
| Plasmid | Mutation | Mean packaging efficiency ± SD with:
|
Serial propagationc | Generation of Ub terminusd | |
|---|---|---|---|---|---|
| HSV-1a | HSV-2b | ||||
| pSA1 | Wild type | 100 | 100 | ++ | + |
| pPH3 | pac2 (Uc) 80-bp deletion | 2.4 ± 1.6 | 1.2 | ND | ND |
| pPH18 | pac2 consensus substitution | 168 ± 106 | 102 | ++ | + |
| pPH19 | pac2 unconserved deletion | 12.9 ± 5.0 | 26 | ND | ND |
| pPH20 | pac2 unconserved substitution | 132 ± 86 | 58 | ++ | + |
| pPH9 | pac2 T element substitution | 6.9 ± 3.6 | 4.6 | ND | ND |
| pPH10 | pac2 T element deletion | 2.3 ± 0.6 | 1.9 | ND | ND |
| pPH15 | pac2 GC element deletion | 5.0 ± 1.8 | 3.2 | ND | ND |
| pPH16 | pac2 GC element substitution | 152 ± 57 | 105 | ++ | + |
| pPH4 | pac1 (Ub) 69-bp deletion | 9.6 ± 5.8 | 4.3 | ND | ND |
| pPH21 | pac1 proximal GC element deletion | 75 ± 39 | 22 | + | − |
| pPH22 | pac1 proximal GC element substitution | 82 ± 34 | 39 | + | − |
| pPH7 | pac1 T element substitution | 177 ± 55 | 176 | ++ | + |
| pPH11 | pac1 distal GC element deletion | 76 ± 40 | 37 | + | + |
| pPH12 | pac1 distal GC element substitution | 118 ± 65 | 53 | + | + |
Packaging efficiencies were relative to pSA1 and are presented as the mean of three independent determinations ± the standard deviation.
Packaging efficiencies were relative to pSA1 as determined from Fig. 5.
++, serially propagated with similar efficiency to pSA1; +, propagated with reduced efficiency; ND, not done.
+, Ub-containing terminal fragment present in packaged DNA; −, Ub-containing terminal fragment not detected; ND, not done.
These results confirm that the Uc region contains sequences important for DNA packaging and identify the pac2 T element as being of particular importance. Deletions of the elements to either side of pac2 T also significantly impaired encapsidation, but substitution mutations within these regions had relatively little effect, suggesting that the position of the pac2 T element relative to other motifs in the Uc-DR1-Ub fragment may be important for its activity. The removal of most of the Ub region, and part of DR1, impaired packaging ability but, surprisingly, none of the deletion or substitution mutations introduced into the component pac1 elements had a significant effect.
Ability of HSV-2 to function as a helper virus.
As noted above, repair of mutated a sequences may sometimes occur through recombination with wild-type copies (11, 27). Smiley et al. (28) previously demonstrated that HSV-2 was able to recognize the HSV-1 a sequence for DNA cleavage and packaging but that recombinational inversion events did not occur between the two a sequences, presumably as a result of their overall low DNA sequence homology (9).
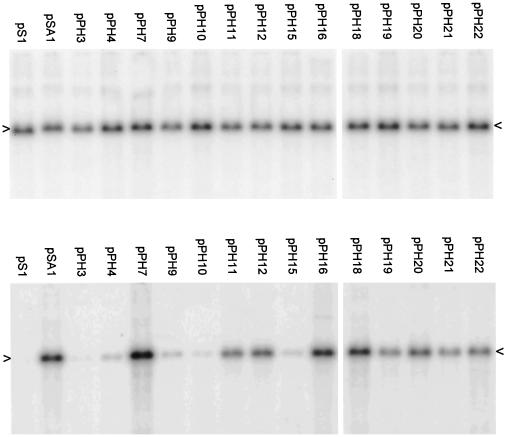
In order to reduce the possibility that recombinational repair might be having a significant influence on the phenotypes of our mutants, we assessed their packaging ability when HSV-1 was replaced by HSV-2 as the superinfecting helper virus. The results (Fig. 5 and Table 1) are very similar to those obtained with HSV-1 helper (Fig. 4 and Table 1). However, the plasmids affecting the pac1 proximal and distal GC elements (pPH21, pPH22, pPH11, and pPH12) all exhibited a small reduction in packaging efficiency relative to pSA1 when HSV-2 was helper, while pPH19 (pac2 unconserved region deletion) showed a small increase. These effects, which were also noted in an independent experiment, are readily illustrated by comparison of the pPH19, pPH21, and pPH22 lanes in Fig. 4 and 5. Thus, although there may be small differences between packaging signal recognition in HSV-1 and HSV-2, these data suggest that it is unlikely that the high-efficiency packaging seen with several mutants is a consequence of recombinational repair.
FIG. 5.
Packaging assay with HSV-2 as superinfecting virus. The assay was performed as in Fig. 4 except that the helper virus was HSV-2. The upper and lower panels are for total and DNase-resistant DNA, respectively, and the positions of the replicated amplicon monomers (4.4 kbp) are indicated by arrowheads.
Further characterization of mutants relatively unimpaired for DNA packaging.
Since the data presented in Fig. 4 showed that eight of the mutants were packaged with a similar efficiency to pSA1, we sought to determine whether the lesions in these plasmids might play a role in amplicon propagation at a stage other than the initial encapsidation process. Supernatant medium from BHK cells that had been transfected with pS1, pSA1, or one of the eight mutants was passaged on fresh cell monolayers, and total DNA was prepared and examined for the presence of replicated plasmid molecules (Fig. 6 and Table 1). In agreement with previous results that showed a requirement for the Uc-DR1-Ub element for serial propagation (22), replicated pSA1 but not pS1 was detected in the total DNA. Plasmids pPH7, pPH16, pPH18, and pPH20 were propagated with the same efficiency as pSA1 but the other four plasmids (pPH11, pPH12, pPH21, and pPH22, carrying alterations to the pac1 distal and proximal GC elements) accumulated to at least fourfold lower levels. When the blot was reprobed to detect fragments of the HSV-1 helper DNA similar levels were detected in each of the lanes. Thus, although alteration of the pac1 distal and proximal GC elements does not appear to affect the amount of DNA encapsidated, the packaged DNA is impaired in its ability to be serially propagated.
FIG. 6.
Serial propagation of amplicons. Samples of cell-released virus from BHK cell monolayers transfected with the indicated plasmids were used to infect fresh cells, and total cellular DNA was prepared and analyzed as described in the legend to Fig. 2. The position of the amplicon monomers (4.4 kbp) is indicated by an arrowhead.
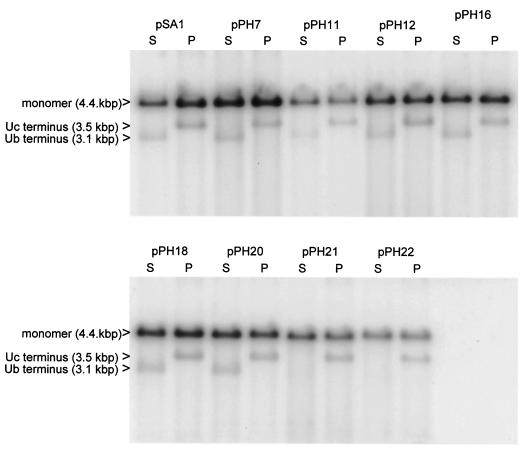
One possible explanation for this above result is that packaged concatemers of pPH11, pPH12, pPH21, or pPH22 may have abnormal termini that could affect their ability to reinitiate infection. To examine the termini of packaged amplicons, the DNase-resistant DNA isolated from the nuclei of transfected cells superinfected with HSV-1 was cleaved with two enzymes, SalI or PstI, that generate distinctive terminal fragments. In addition to amplicon monomers, cleavage of DNase-resistant DNA with SalI would be expected to generate fragments of 3.1 and 1.3 kbp representing the termini bearing the Ub and Uc sequences, respectively (Fig. 1e). The sizes of the corresponding PstI fragments terminating with Ub and Uc are 0.9 and 3.5 kbp, respectively.
The results are shown in Fig. 7 and summarized in Table 1. When DNA from cells that had been transfected with pSA1 was cleaved with SalI or PstI, the larger terminal fragment was readily detected as a band of faster mobility and lower abundance than unit length monomers (upper band). The Uc-containing PstI terminal fragment was detectable in DNA from cells transfected with each of the packaging-competent mutant plasmids. In contrast, although detectable for six of the mutants, the Ub-containing SalI terminal fragment was not apparent in the DNAs of cells transfected with pPH21 or pPH22 (pac1 proximal GC element mutants). Thus, although these two plasmids give rise to DNase-resistant DNA with normal Uc-containing termini, fragments from the other terminus are probably heterogeneous in size and, as a consequence, are not detected as a discrete band. Such heterogeneity might occur through a loss of cleavage specificity or, alternatively, cleavage might occur at the correct position but the terminal sequences may remain susceptible to added DNase because they are not correctly inserted into the capsid.
FIG. 7.
Terminal fragments of packaged amplicon DNA. DNase-resistant DNA, prepared from the nuclei of cells transfected with the indicated plasmids and superinfected with HSV-1, was cleaved with DpnI plus either SalI (S) or PstI (P) and analyzed as described in the legend to Fig. 2. The positions of the larger of the terminal fragments generated by SalI or PstI (3.1 and 3.5 kbp, respectively) and of linear monomeric plasmid are indicated.
In an attempt to estimate the lengths of the packaged monomers, we measured the radioactivity in the bands corresponding to amplicon monomers and terminal fragments (where detectable). Although the presence of a background smear in the lanes made it difficult to determine reliably the amount of terminal fragment, the signal in the amplicon monomers band was generally 10- to 20-fold higher than in the terminal fragment band. If we assume that the signal intensity is proportional to the amount of vector sequence in the DNA fragment, this suggests that the packaged concatemers contain, on average, ca. 12 to 24 monomeric units. None of the mutant plasmids differed significantly from pSA1 in this regard.
DISCUSSION
The transient assay we employed in this study differs in two important aspects from other approaches that have been used to analyze the signals required for herpesvirus DNA packaging. Most significantly, in contrast to previous experiments with amplicons in which mixed virus stocks containing helper and defective genomes were serially propagated prior to analysis (10, 11, 22, 34), we examined DNA packaging during a single cycle of infection. Additionally, the preparations of packaged DNA we analyzed included DNA from the infected cell nucleus.
The approach confers several probable advantages: (i) the opportunity for recombination with the helper virus is minimized; (ii) there is no selection for molecules that may have repaired lethal or debilitating mutations through such a process; (iii) quantitative comparison of the effects of different mutations is facilitated; and (iv) the identification of novel packaging phenotypes is possible. For example, it is conceivable that a class of mutant exists in which significant amounts of DNA are packaged in the nucleus but the resulting nucleocapsids fail to progress to enveloped virions. In an assay dependent on serial propagation, this mutant would exhibit a negative phenotype similar to that of a mutant that was defective in the initiation of packaging. A clear precedent for this type of defect is provided by the studies of defective HSV-1 genomes by Vlazny et al. (35). They reported that capsids in the nuclei contained defective genomes comprising integral numbers of copies of a basic monomeric unit up to the length of a standard HSV-1 genome. However, only the largest defective genomes were detected in the cytoplasm, suggesting the existence of selectivity in the envelopment and translocation of capsids into the cytoplasm. Studies employing ectopic copies of packaging signals to identify cis-acting herpesvirus DNA packaging signals may similarly have failed to differentiate between mutations affecting the initiation and termination of DNA packaging since again DNA from only cytoplasmic or cell-released virus particles was analyzed (18, 27).
We initially confirmed the observation of Nasseri and Mocarski (22) that a 200-bp Uc-DR1-Ub fragment of HSV-1 could act as a packaging signal, and we also showed that it functions with an efficiency similar to that of larger fragments containing one or two copies of the complete a sequence, including the highly reiterated DR2 regions (Fig. 2). Because the small size of the Uc-DR1-Ub fragment and the absence of high-copy-number repeats might be expected to minimize recombination with the helper virus, this fragment was chosen for site-directed mutagenesis experiments. Although we cannot conclusively exclude the possibility that recombination may have occurred between the helper virus and the replicated amplicon DNA to repair the sequences at the termini of packaged molecules, several observations argue that it is unlikely to have had a significant influence on packaging efficiency. First, deletions or substitutions of similar sizes frequently exhibited quite-different phenotypes (e.g., compare pPH7 and pPH9 in Fig. 4). Second, each of the mutants exhibited a similar phenotype when HSV-2, which shares insufficient a sequence homology with HSV-1 to drive recombinational genome isomerization (28), was used as a helper (Fig. 5). Finally, four plasmids with lesions in pac1 (pPH11, pPH12, pPH21, and pPH22), which exhibited wild-type levels of DNA packaging, were impaired in their ability to be serially propagated, and two of these (pPH21 and pPH22) were defective in the generation of a normal Ub-containing terminal fragment (Fig. 6 and 7). If recombinational repair of the lesions had been necessary to achieve high-efficiency packaging, these plasmids would have been expected to be unimpaired in the serial propagation assay and packaged pPH21 and pPH22 DNAs would have been expected to generate normal Ub-containing termini.
In agreement with previous results, both the Ub and Uc regions were shown to play important roles in DNA packaging (11, 22, 27, 34). Both the Ub (pPH4) and Uc (pPH3) deletion mutants nevertheless exhibited residual packaging activity (Fig. 4) at a level greater than that observed with the parental plasmid, pS1, suggesting that there may be some functional redundancy between the regions.
Within the Uc region, the pac2 T element was the single most critical sequence; deletion of this motif had as great an effect as deletion of all of Uc, and the substitution mutant was also significantly impaired. Deletion of the pac2 unconserved region and pac2 GC element resulted in much greater reductions in packaging than substitutions that did not affect the lengths of these regions, suggesting that the position of the pac2 T element relative to other motifs within the Uc-DR1-Ub fragment is likely to be important. These results are broadly similar to those obtained with an ectopic cleavage-packaging signal in MCMV, in which substitution of the pac2 T element or insertions of 6 or 47 bp on either side of it abolished detectable cleavage activity (18). A more significant reduction in activity was noted with an MCMV pac2 consensus substitution mutant than with the corresponding HSV-1 mutant. Since this mutant (pPH18) is also unimpaired in its ability to be serially propagated (Fig. 6), it is possible that any important function of the HSV-1 pac2 consensus may be compensated for by the presence of closely related GC-rich motifs in the adjacent regions of Uc. When deletions were progressively introduced into the Uc side of an a sequence, some reduction in ability to function as a cleavage site was noted, but it was not possible to identify the contribution of individual pac2 elements because of the likely occurrence of recombinational repair (27).
Analysis of the mutants affecting the Ub region yielded several surprising results. Although deletion of the three pac1 elements (and part of DR1) greatly reduced DNA packaging (plasmid pPH4), the substitution and deletion mutants affecting the individual pac1 motifs all exhibited near-wild-type activity (Fig. 4). This paradoxical result is not fully understood. The impairment of pPH4 packaging is unlikely to be a consequence of the removal of the DR1 sequences since it has previously been demonstrated that pac1-directed cleavage can occur in the absence of this element (34). However, it remains possible that the DR1 element plays an important role in the context of a minimal Uc-DR1-Ub packaging signal. Alternatively, there may be functional redundancy among the pac1 motifs, such that the removal of an individual motif has relatively little effect. For example, both the proximal and distal GC elements contain tracts of at least six G residues, and runs of at least three T residues occur in both the proximal GC and T elements.
The results obtained with the mutants affecting the individual pac1 motifs also contrast markedly with data from experiments in which the effects of pac1 mutations on the generation of terminal fragments of HSV-1 or MCMV DNA were examined (18, 27). In the latter case, the poly(G) tracts on either side of the pac1 T element were shown be important for the generation of virions containing genomes cleaved at an ectopically inserted packaging signal. The apparent discrepancy between the sets of data may be explained if these motifs are involved in relatively late stages of the encapsidation process such as the maturation of nuclear DNA-containing capsids into virions. Consistent with this, all four amplicons carrying mutations within the two pac1 GC elements exhibited reduced replication when virus stocks were passaged on fresh cells (Fig. 6), and both mutants with lesions in the proximal GC element were additionally defective in the generation of normal Ub-containing terminal fragments (Fig. 7). However, an alternative possibility that the pac1 GC element mutants may be impaired in the efficiency with which virus particles can initiate new cycles of infection, rather than in virion production per se, cannot be excluded.
Further experiments are clearly required to elucidate the stage(s) at which the pac1 mutants are impaired. This analysis, however, may be more complex with amplicons than ectopic packaging signals. For example, if pac1 is involved in the late stages of DNA packaging, it is probable that the generation of concatemers containing multiple tandem copies of an amplicon may allow packaging to be completed with relatively higher efficiency when the signals retain partial functionality. In addition, packaged amplicons with an abnormal terminus may be able to circularize relatively efficiently by homologous recombination to reinitiate infection even though their termini may be incapable of direct ligation. An interesting approach would be to examine whether the Uc-DR1-Ub mutants allow the rescue of progeny virus when inserted into an HSV-1 bacterial artificial chromosome lacking functional pac signals (24).
It has previously been shown that during herpesvirus DNA packaging only the genomic terminus containing the pac2 signal is found associated with the high-molecular-weight concatemeric DNA (15, 17, 19, 25, 26, 36), and this observation has formed the basis for a proposal that the pac2 sequence controls the initiation and directionality of DNA packaging (19). Our observations that mutations within pac2 have relatively greater effects during the first cycle of infection and that motifs in pac1 may be involved in the late stages of DNA encapsidation are compatible with this model. In the case of HSV-1 genomes, the pac2 signal at the L terminus would promote the initial cleavage of a concatemeric molecule and the initiation of DNA packaging into the preformed capsid. A second cleavage event, driven by pac1, would be necessary for completion of the process and the generation of an authentic S terminus (i.e., the directionality of packaging would be from L to S). It should be noted, however, that until other possible mechanisms can be excluded, this remains a tentative model. Moreover, it remains to be determined whether cleavage reactions are identical at junctions containing only a single a sequence which, unless duplication first occurs, does not contain a Uc-DR1-Ub junction. An attractive feature of the above model, however, is that the independent functioning of the pac1 and pac2 regions may explain how a single a sequence, in which these two signals are separated by several hundred base pairs can efficiently direct cleavage and packaging (7, 10, 11, 27, 32, 34).
The HSV-1 UL28 packaging protein has recently been shown to interact specifically with the pac1 signal and the G tracts at either side of the pac1 T element have been shown to be key elements in the binding (3). Our data are consistent with this interaction being involved at a late stage in packaging, such as the termination cleavage event. Studies with UL28 mutants, however, suggest that no cleavage of concatemers occurs in the absence of UL28, implicating this protein also in the initiation of packaging (2, 6, 33). This suggests that UL28 may also interact with the pac2 signal, as previously reported for the homologous protein of human cytomegalovirus (4), or that binding to the pac1 signal may also be involved in initiation of packaging. In this regard, it is interesting to recall that pac1 mutants that retained at least one of the flanking G tracts were unimpaired for packaging, whereas the mutant lacking the entire Ub region was severely impaired.
ACKNOWLEDGMENTS
We are grateful to Andrew Davison and Frazer Rixon for critical reading of the manuscript, and we thank Leslie Taylor for carrying out DNA sequencing.
P.D.H. was supported by a Medical Research Council Ph.D. Studentship.
REFERENCES
- 1.Abbotts A P, Preston V G, Hughes M, Patel A H, Stow N D. Interaction of the herpes simplex virus type 1 packaging protein UL15 with full-length and deleted forms of the UL28 protein. J Gen Virol. 2000;81:2999–3009. doi: 10.1099/0022-1317-81-12-2999. [DOI] [PubMed] [Google Scholar]
- 2.Addison C, Rixon F J, Preston V G. Herpes simplex virus type 1 UL28 gene product is important for the formation of mature capsids. J Gen Virol. 1990;71:2377–2384. doi: 10.1099/0022-1317-71-10-2377. [DOI] [PubMed] [Google Scholar]
- 3.Adelman K, Salmon B, Baines J D. Herpes simplex virus DNA packaging sequences adopt novel structures that are specifically recognized by a component of the cleavage and packaging machinery. Proc Natl Acad Sci USA. 2001;98:3086–3091. doi: 10.1073/pnas.061555698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogner E, Radsak K, Stinski M F. The gene product of human cytomegalovirus open reading frame UL56 binds the pac motif and has specific nuclease activity. J Virol. 1998;72:2259–2264. doi: 10.1128/jvi.72.3.2259-2264.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Broll H, Buhk H-J, Zimmermann W, Goltz M. Structure and function of the prDNA and genomic termini of the γ2-herpesvirus bovine herpesvirus type 4. J Gen Virol. 1999;80:979–986. doi: 10.1099/0022-1317-80-4-979. [DOI] [PubMed] [Google Scholar]
- 6.Cavalcoli J D, Baghian A, Homa F L, Kousoulas K G. Resolution of genotypic and phenotypic properties of herpes simplex virus type 1 temperature-sensitive mutant KOS tsZ47: evidence for allelic complementation in the UL28 gene. Virology. 1993;197:23–34. doi: 10.1006/viro.1993.1563. [DOI] [PubMed] [Google Scholar]
- 7.Chou J, Roizman B. Isomerization of herpes simplex virus 1 genome: identification of the cis-acting and recombination sites within the domain of the a sequence. Cell. 1985;41:803–811. doi: 10.1016/s0092-8674(85)80061-1. [DOI] [PubMed] [Google Scholar]
- 8.Davison A, Rixon F. Cloning of the DNA of Alphaherpesvirinae. In: Becker Y, editor. Recombinant DNA research and virus. Boston, Mass: Martinus Nijhoff Publishing; 1985. pp. 103–124. [Google Scholar]
- 9.Davison A J, Wilkie N M. Nucleotide sequences of the joint between the L and S segments of herpes simplex virus types 1 and 2. J Gen Virol. 1981;55:315–331. doi: 10.1099/0022-1317-55-2-315. [DOI] [PubMed] [Google Scholar]
- 10.Deiss L P, Frenkel N. Herpes simplex virus amplicon: cleavage of concatemeric DNA is linked to packaging and involves amplification of the terminally reiterated a sequence. J Virol. 1986;57:933–941. doi: 10.1128/jvi.57.3.933-941.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deiss L P, Chou J, Frenkel N. Functional domains within the a sequence involved in the cleavage-packaging of herpes simplex virus DNA. J Virol. 1986;59:605–618. doi: 10.1128/jvi.59.3.605-618.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dolan A, Jamieson F E, Cunningham C, Barnett B C, McGeoch D J. The genome sequence of herpes simplex virus type 2. J Virol. 1998;72:2010–2021. doi: 10.1128/jvi.72.3.2010-2021.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Homa F L, Brown J C. Capsid assembly and DNA packaging in herpes simplex virus. Rev Med Virol. 1997;7:107–122. doi: 10.1002/(sici)1099-1654(199707)7:2<107::aid-rmv191>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 14.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martinez R, Sarisky R T, Weber P C, Weller S K. Herpes simplex virus type 1 alkaline nuclease is required for efficient processing of viral DNA replication intermediates. J Virol. 1996;70:2075–2085. doi: 10.1128/jvi.70.4.2075-2085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGeoch D J, Dalrymple M A, Davison A J, Dolan A, Frame M C, McNab D, Perry L J, Scott J E, Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988;69:1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- 17.McVoy M A, Adler S P. Human cytomegalovirus DNA replicates after early circularization by concatemer formation, and inversion occurs within the concatemer. J Virol. 1994;68:1040–1051. doi: 10.1128/jvi.68.2.1040-1051.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McVoy M A, Nixon D E, Adler S P, Mocarski E S. Sequences within the herpesvirus-conserved pac1 and pac2 motifs are required for cleavage and packaging of the murine cytomegalovirus genome. J Virol. 1998;72:48–56. doi: 10.1128/jvi.72.1.48-56.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McVoy M A, Nixon D E, Hur J K, Adler S P. The ends of herpesvirus DNA replicative concatemers contain pac2 cis cleavage/packaging elements and their formation is controlled by terminal cis sequences. J Virol. 2000;74:1587–1592. doi: 10.1128/jvi.74.3.1587-1592.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mocarski E S. Cytomegaloviruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2447–2492. [Google Scholar]
- 21.Mocarski E S, Roizman B. Structure and role of the herpes simplex virus DNA termini in inversion, circularization and generation of virion DNA. Cell. 1982;31:89–97. doi: 10.1016/0092-8674(82)90408-1. [DOI] [PubMed] [Google Scholar]
- 22.Nasseri M, Mocarski E S. The cleavage recognition signal is contained within sequences surrounding an a-a junction in herpes simplex virus DNA. Virology. 1988;167:25–30. doi: 10.1016/0042-6822(88)90050-5. [DOI] [PubMed] [Google Scholar]
- 23.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2231–2295. [Google Scholar]
- 24.Saeki Y, Ichikawa T, Saeki A, Chiocca E A, Tobler K, Ackermann M, Breakfield X O, Fraefel C. Herpes simplex virus type 1 DNA amplified as bacterial artificial chromosome in Escherichia coli: rescue of replication-competent virus progeny and packaging of amplicon vectors. Hum Gene Ther. 1998;9:2787–2794. doi: 10.1089/hum.1998.9.18-2787. [DOI] [PubMed] [Google Scholar]
- 25.Severini A, Morgan A R, Tovell D R, Tyrrell D L. Study of the structure of replicative intermediates of HSV-1 DNA by pulsed-field gel electrophoresis. Virology. 1994;200:428–435. doi: 10.1006/viro.1994.1206. [DOI] [PubMed] [Google Scholar]
- 26.Slobedman B, Simmons A. Concatemeric intermediates of equine herpesvirus type 1 DNA replication contain frequent inversions of adjacent long segments of the viral genome. Virology. 1997;229:415–420. doi: 10.1006/viro.1997.8447. [DOI] [PubMed] [Google Scholar]
- 27.Smiley J R, Duncan J, Howes M. Sequence requirements for DNA rearrangements induced by the terminal repeat of herpes simplex virus type 1 KOS DNA. J Virol. 1990;64:5036–5050. doi: 10.1128/jvi.64.10.5036-5050.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smiley J R, Lavery C, Howes M. The herpes simplex virus type 1 HSV-1 a sequence serves as a cleavage/packaging signal but does not drive recombinational genome isomerization when it is inserted into the HSV-2 genome. J Virol. 1992;66:7505–7510. doi: 10.1128/jvi.66.12.7505-7510.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spaete R R, Frenkel N. The herpes simplex virus amplicon: a new eukaryotic defective-virus cloning-amplifying vector. Cell. 1982;82:295–304. doi: 10.1016/0092-8674(82)90035-6. [DOI] [PubMed] [Google Scholar]
- 30.Stow N D, McMonagle E C. Characterisation of the TRS/IRS origin of DNA replication of herpes simplex virus type 1. Virology. 1983;130:427–438. doi: 10.1016/0042-6822(83)90097-1. [DOI] [PubMed] [Google Scholar]
- 31.Stow N D, Wilkie N M. An improved technique for obtaining enhanced infectivity with herpes simplex virus type 1 DNA. J Gen Virol. 1976;33:447–458. doi: 10.1099/0022-1317-33-3-447. [DOI] [PubMed] [Google Scholar]
- 32.Stow N D, McMonagle E C, Davison A J. Fragments from both termini of the herpes simplex virus type 1 genome contain signals required for the encapsidation of viral DNA. Nucleic Acids Res. 1983;11:8205–8220. doi: 10.1093/nar/11.23.8205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tengelsen L A, Pederson N E, Shaver P R, Wathen M W, Homa F L. Herpes simplex virus type 1 DNA cleavage and encapsidation require the product of the UL28 gene: isolation and characterization of two UL28 deletion mutants. J Virol. 1993;67:3470–3480. doi: 10.1128/jvi.67.6.3470-3480.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Varmuza S L, Smiley J R. Signals for site-specific cleavage of HSV-1 DNA: maturation involves two separate cleavage events at sites distal to the recognition sequences. Cell. 1985;41:793–802. doi: 10.1016/s0092-8674(85)80060-x. [DOI] [PubMed] [Google Scholar]
- 35.Vlazny D A, Kwong A, Frenkel N. Site-specific cleavage/packaging of herpes simplex virus DNA and the selective maturation of nucleocapsids containing full-length viral DNA. Proc Natl Acad Sci USA. 1982;79:1423–1427. doi: 10.1073/pnas.79.5.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang X, Efstathiou S, Simmons A. Identification of novel herpes simplex virus replicative intermediates by field inversion gel electrophoresis: implications for viral DNA amplification strategies. Virology. 1994;202:530–539. doi: 10.1006/viro.1994.1375. [DOI] [PubMed] [Google Scholar]