Abstract
Background:
Gut dysbiosis may play a role in immune-mediated diseases, such as psoriasis. There is a growing interest in understanding microbiome influence, with speculations around the importance of an altered gut microbiome linked to the progression to psoriatic arthritis in psoriasis. The objective of this study is to study the gut microbiome in patients with severe psoriatic disease with or without psoriatic arthritis.
Methods:
V3/V4 16S rRNA gene sequencing and bioinformatics analyses were performed with the total DNA extracted from the stool samples of 30 patients with psoriatic disease, 15 of whom had documented psoriatic arthritis.
Results:
We found differences in gut microbiome composition in psoriatic arthritis patients when looking for relative and especially differential abundances. Bacteroidaceae family (P = .02), Bacteroides genus (P = .02), and Bacteroides uniformis (P = .03) were more abundant in psoriatic arthritis patients on differential abundance, adjusted for each taxonomic level. However, the present study did not show significant differences in alpha or beta diversity.
Conclusion:
This study shows different patterns of gut microbiome composition in patients with psoriatic arthritis, with significant overexpression of the Bacteroides genus. This reinforces the microbiome as a field of interest in psoriasis. Nevertheless, it should be noted that some previously described findings related to lower diversity and different clustering between groups could not be demonstrated, probably due to the small number of patients. Additionally, it remains difficult to understand the magnitude of the gut microbiome influence. Is dysbiosis a cause or consequence of the disease? However, the microbiome deserves our attention, especially since it brings different opportunities for intervention through diet, prebiotics and probiotics, pretreatment analysis, prognosis, and even microbiome modulation and transplantation.
Keywords: Psoriasis, psoriatic arthritis, microbiome, gut microbiome, Bacteroides
Introduction
Psoriasis is a chronic, inflammatory, and immune-mediated systemic disease that primarily affects the skin and joints.1-4 It has an estimated prevalence of 2%-3% of the world’s population, varying according to ethnicity and geographic location.5-7 In Brazil, it is estimated to occur in 1.3% of the general population.8,9 Its etiopathogenesis is complex, multifactorial, and not completely understood to the present day. In summary, we can understand it as a disease primarily determined by an aberrant immune response, influenced by a genetic background that is thought to increase predisposition, associated with various environmental stimuli described as possible triggering and/or aggravating factors.10
In approximately 70% of cases, the skin disease precedes joint involvement, and it is estimated that approximately one-third of patients with psoriatic disease will develop psoriatic arthritis over the course of their disease.11 Since psoriatic arthritis is an inflammatory joint disease capable of establishing a progressive and erosive course, early diagnosis is essential to prevent permanent damage to the patient. A short delay of 6 months in the proper diagnosis implies a worse outcome in terms of therapeutic response in the future.12 Researchers often seek to establish good screening tools or even biomarkers that can help the clinical dermatologist in this assessment, improving the window of opportunity for action.1,2,11,13
In 2019, Sher et al14 established a hypothesis of the progressive evolution of psoriatic arthritis, starting from cutaneous disease, in which individuals exposed to genetic and environmental factors would go through a preclinical phase. Soon after, the subclinical stage (imaging exams with early alterations) may progress to a transition phase—prodromal (initial signs and symptoms), culminating in established psoriatic arthritis. The authors posited that in the transition through psoriatic arthritis, changes in the microbiome, such as trauma, certain comorbidities (obesity), and biomechanical stress, could play a role.
We conducted a case-control study seeking a better understanding of the gut microbiome in Brazilian patients with severe psoriatic disease compared to control individuals without psoriatic disease. Based on this, a new analysis was carried out, focused on the group of psoriasis patients, seeking to assess the differences in the gut microbiome in patients with and without psoriatic arthritis, the main objective of the present study.
Methods
This was an observational case-control study developed in a reference outpatient clinic with severe psoriatic disease patients with and without associated psoriatic arthritis. All individuals were aged 18 years or older. The sample was restricted to individuals living in Florianópolis and metropolitan areas (urban area/single geographic unit).
Patients with a clinical and/or histopathological and/or radiological diagnosis of plaque psoriasis and/or psoriatic arthritis were included.1,2 Psoriatic arthritis was diagnosed by a board-certified rheumatologist, respecting the CASPAR criteria.15
We selected only patients with severe disease, classified as 10% or more of affected body surface area and/or a PASI score equal to or greater than 10, and/or a Dermatology Life Quality Index (DLQI) greater than 10.9 Joint disease severity was evaluated using the following scores: Disease Activity in Psoriatic Arthritis Score (DAPSA) and Bath Ankylosing Spondylitis Disease Activity Index (BASDAI).16
We excluded study patients with a history of diarrhea in the last month; those with comorbidities such as inflammatory bowel disease (Crohn’s disease or ulcerative colitis), systemic or autoimmune inflammatory diseases (such as rheumatoid arthritis, systemic lupus, systemic sclerosis, vasculitis, inflammatory myopathies), food allergy, multiple sclerosis, type I diabetes, autism, chronic pancreatitis, and cirrhosis;17-23 active neoplasm; history of surgery with intervention of the intestinal tract; systemic treatment with csDMARDs (methotrexate, sulfasalazine, leflunomide, acitretin, cyclosporine, and JAK inhibitors) or bDMARDs (TNF, IL12, IL17, IL23 inhibitors) in the last 3 months; or use of systemic antibiotics or oral probiotics in the last 3 months. Vegans, vegetarians, and people with a strict raw food diet were also excluded.
Control subjects, without cutaneous and/or articular psoriatic disease, were subject to the same exclusion criteria and were matched by age and sex.
The sample size calculation was based on the methodology described by Zhao et al24 in 2018 and implemented in the R package RnaSeqSampleSize.25 The following assumptions were used: controlling the false discovery rate (FDR) at 5%; a dispersion parameter of the BN distribution of approximately 0.5; an average count per group of at least 50 reads; and similar library sizes between groups. With 15 samples per group, it was possible to detect mean differences (FC = 4) with statistical power above 95%.
Data were collected in medical appointments and electronic medical records only after approval by the ethics committee and agreement and signing of the informed consent by each patient (ethics committee serial number of approval——CAAE: 33498620.6.0000.0115).
Blood samples were collected from patients (not controls) to assess acute-phase inflammatory tests: erythrocyte sedimentation rate (first-hour ESR, in milliliters) and high-sensitivity C-reactive protein (CRP, in milligrams per deciliter). As a complement to the clinical examination performed by the board-certified rheumatologist, ultrasonography of joints, entheses, and nail apparatus was performed, contributing to the definition of psoriatic arthritis at the time of inclusion of the patient in the study. It should be noted that the same rheumatologist evaluated every patient. Additionally, he has a 10-year experience background in rheumatologic ultrasound and was responsible for all ultrasound exams performed in the research.
Examination of the intestinal microbiome was performed by large-scale DNA sequencing using markers from the V3/V4 region of the 16S rRNA gene for taxonomic identification of bacteria extracted and amplified from stool samples.26,27 The results obtained after DNA sequencing were analyzed by the new Neotools v1 bioinformatics pipeline. This pipeline includes 2 advances in the analysis and classification of sequencing data: (1) use of denoise methods, eliminating sequences arising from possible sequencing errors, increasing confidence in the results obtained and (2) new taxonomic classification, based on similarity, using an algorithm called LCA (lowest common ancestor) to determine the lowest possible taxonomic level. The similarity between the sequenced data was analyzed against a database of 16S sequences obtained from complete bacterial genomes (Neoref16Srev6). If there were sequences identical to a single species present in the database, the species taxonomy was assigned. If 2 or more sequences showed similarity of up to 98.7% with different species in the database, the taxonomic assignment was taken to their lowest level of convergence (e.g., genus, family, phylum, and kingdom).
Analyses were performed in R (v. 4.2.0) using the tidyverse (v. 1.3.1) and phyloseq (v. 1.40.0) packages. Alpha- and beta-diversity analyses employed the Shannon index and Bray-Curtis dissimilarity, respectively. Differential abundance analysis used the MicrobiomeStat (v. 1.1), corncob (v. 0.2.0), and DESeq2 (v. 1.36.0) packages, focusing on the taxonomic levels of phylum, family, genus, species, and “lowest” (as detailed as possible for each sequence). P-values were adjusted to control FDR at 5% using the Benjamini-Hochberg procedure. Description of the global microbial profiles was performed using a heatmap with ordered lines, from top to bottom, according to average relative abundance (greatest abundance at the top, top 20 taxonomies). Given the heterogeneity of methods for differential abundance, 3 tools were employed (LinDA, corncob, and DESeq2) and consensus was considered.28-30
Results
From a cohort of 250 patients followed at our psoriasis outpatient clinic, 60 patients were initially selected based on inclusion criteria. Of these, 30 with severe plaque psoriasis were eligible after agreeing to participate and not meeting any of the exclusion criteria. Respecting the sample calculation, the minimum number of 15 patients per group was reached, resulting in 15 psoriasis (Pso) and 15 psoriatic arthritis (PsA) patients. We also included 30 individuals without psoriatic disease as controls. Table 1 displays the demographic characteristics of cases and controls, and Table 2 presents the clinical-epidemiological aspects of our patients.
Table 1.
Clinical-epidemiological profile of our sample (Cases and Controls)
| Cases (n = 30) | Controls (n = 30) | P | |
|---|---|---|---|
| Gender: female (absolute and relative frequencies): |
15 (50%) |
15 (50%) |
|
| Mean age (years) ± SD: |
48.3 (±12.4) |
47.9 (±11.7) |
.172 |
| Minimum-maximum | 28-76 | 24-79 | |
| Comorbidities: yes (absolute and relative frequencies): |
24 (80%) | 22 (73.3%) | .552 |
| Mean BMI (kg/m2) + SD |
29.93 (±5.65) | 27.10 (±4.33) | .226 |
Source: Elaboration of the authors, 2023. BMI, body mass index.
Table 2.
Clinical-Epidemiological Aspects from Our Patients with Severe Psoriatic Disease
|
Pso (n = 15) |
PsA (n = 15) |
P | |
| Gender: female (absolute and relative frequencies) |
9/15 (60%) |
6/15 (40%) |
|
| Mean age (years) ± SD |
47.93 (±13.91) | 48.67 (±10.34) |
.871 |
| Mean disease time progression (years) + SD |
13.60 (±8.29) |
15.87 (±14.29) |
.600 |
| Mean BSA = (minimum and maximum)δ |
22.9% ( 6%-74%) |
24.9% ( 6%-76%) |
.789 |
| Mean PASI δ (minimum and maximum) |
15.88 ( 7.4-23) |
18.28 ( 5.4-38.8) |
.387 |
| DLQI (n = 4):™ | 18 24 |
16 20 |
|
Pso
|
15 (100%) 6/15 (40%) 11/15 (73.3%) |
15 (100%) 7/15 (46.7%) 8/15 (53.3%) |
>.05 |
| Comorbidities: Present (absolute and relative frequencies): |
12/15 (80%) | 12/15 (80%) | 1.0 |
| Mean BMI (kg/m2) + SD: |
32.85 (±4.76) | 27.02 (±5.04) | .003 |
| PsA (absolute and relative frequencies): ∞ Peripheral
|
15 (50%) 7 (46.66%) 12 (80%) 5 (33.33%) |
||
| Mean DAPSA (n = 15): - Remission: Low activity Moderate activity High activity |
23.11 1/15 3/15 6/15 5/15 |
||
| Mean BASDAI (n = 5/15) | 4.98 | ||
| Obesity | 11/15 (73.3%) | 4/15 (26.7%) | <.001 |
| Metabolic syndrome | 7/15 (46.7%) | 7/15 (46.7%) | 1.0 |
| Hypertension | 6/15 (40%) | 6/15 (40%) | 1.0 |
| Diabetes | 2/15 (13.3%) | 2/15 (13.3%) | 1.0 |
| Dyslipidemia | 5/15 (33.3%) | 8/15 (53.3%) | <.001 |
| Hepatic esteatosis | 7/15 (46.7%) | 8/15 (53.3%) | .876 |
| Smoking | 3/15 (20%) | 6/15 (40%) | <.001 |
| Sedentary behavior | 11/15 (73.3%) | 9/15 (60%) | .466 |
| PEST (positive screening for PsA) | 3/15 (20%) | 11/15 (73.3%) | <.001 |
| Mean ESR (mm 1H) | 20.7 | 21.87 | .875 |
| Mean CPR (mg/dL) | 2.77 | 4.67 | .476 |
BASDAI, Bath Ankylosing Spondylitis Disease Activity Index; BMI, body mass index; BSA, body surface area; DAPSA, Disease Activity in Psoriatic Arthritis Score; DLQI, dermatology life quality index; PASI, psoriasis area and severity index; PEST, Psoriasis Epidemiology Screening Tool; PsA, psoriatic arthritis; Pso, plaque psoriasis. δ Those with BSA and PASI <10 had DLQI >10. ∞ PsA: clinical patterns could overlap, and the same patient may present a peripheral pattern, in addition to axial involvement, for example. Source: Elaboration of the authors, 2023.
When comparing PsA and Pso patients, there was no statistically significant difference in terms of mean age and mean time of disease progression. Evolving to clinical characteristics, there was no difference in terms of average PASI and BSA, and also no difference taking into account the presence of scalp, nail, or intertriginous psoriasis. In the present study also, acute phase inflammatory tests were not able to differentiate between patients with and without joint disease (Table 2).
All 30 patients, along with 30 controls included in the study, had their gut microbiome analyzed.
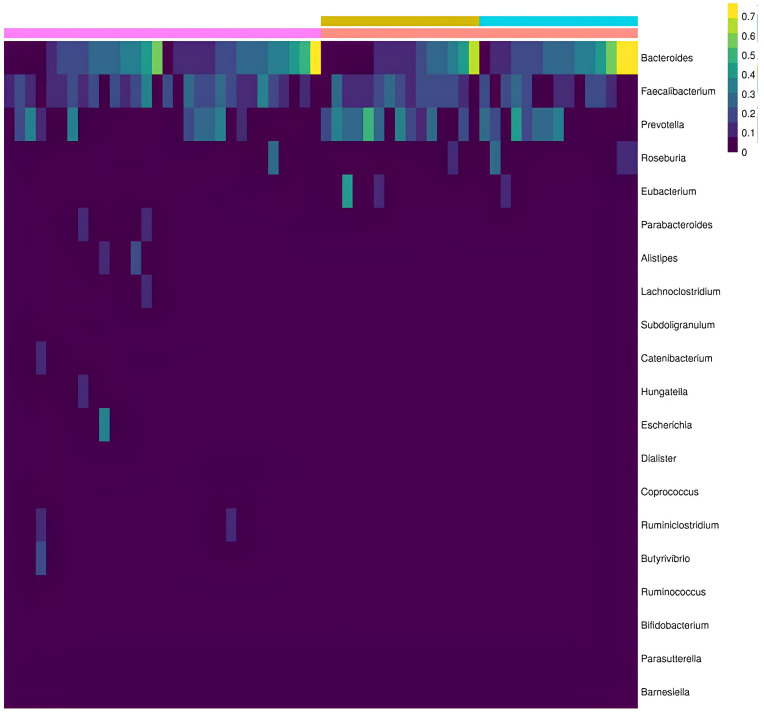
Regarding the relative abundance, according to the taxonomic classification, the following were found in descending hierarchical order: (1) Phyla level: Firmicutes and Bacteroidetes phyla were predominant in both groups, with a greater presence of Proteobacteria and Fusobacteria among patients with psoriatic disease. There was also a slightly greater representation of Bacteroidetes and Proteobacteria among psoriatic arthritis patients. (2) Families: Bactereoidaceae, Ocillospiraceae, and Prevotellaceae families were predominant in both groups. In patients with psoriatic arthritis, there was also a predominance of Lacnospiraceae; among patients without joint involvement, a small predominance of Eubacteriaceae and Bacillaceae was noted; in contrast, there was a higher presence of Enterobacteriaceae in controls without psoriasis. (3) Genus: there was no difference in diversity. A predominance of Bacteroides and Roseburiawas found in PsA patients and Faecalibacterium and Prevotellaamong Pso patients (Figure 1); (4) Species: There was a predominance of Faecalibacterium prausnitzii in both groups. Bacteroides vulgatus, B. stercoris, B. uniformis, and B. plebeius predominated among patients with PsA, while Prevotella copri and Eubacterium rectale predominated among patients with Pso.
Figure 1.
Relative abundance, genus level. Controls (pink bar); psoriatic disease (red bar); as part of psoriatic disease, Pso (yellow bar); Apso (blue bar). The sidebar refers to the magnitude of the reading by color grading (yellow: maximum/purple: minimum). Source: Elaboration of the authors, 2023.
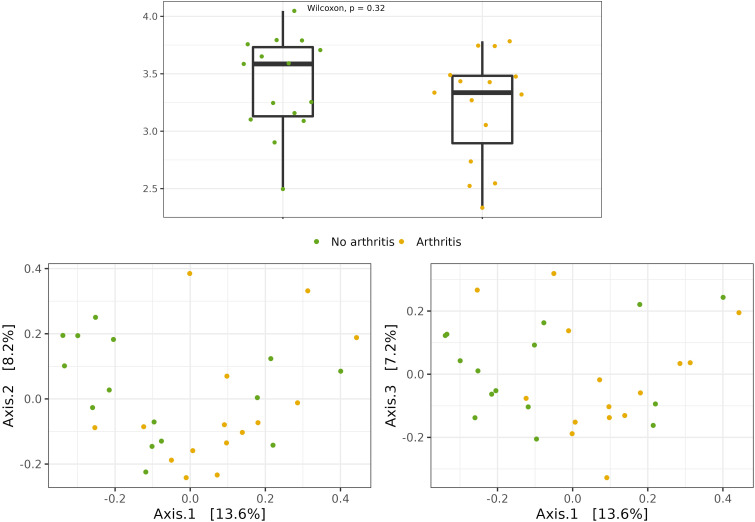
Alpha diversity represents the number of different species in a given sample. It seems that a more diverse microbiome is more resilient and healthy.15,16,33 Although there appears to be a less diverse intestinal microbiome among psoriatic patients, no significant difference was found in both scenarios: psoriatic disease vs. controls (Shannon index; P = .16) and Pso vs. PsA (lower median among those exposed: 3.6 Pso x 3.3 PsA), the difference was not statistically significant (Shannon index; P = .32) (Figure 2A).
Figure 2.
(A) Alpha diversity—Shannon index (P = .32); (B) Beta diversity—Bray‒Curtis dissimilarity index (P = .11); Pso (green); Apso (yellow). Source: Elaboration of the authors, 2023.
On the other hand, a statistically significant difference in the composition of the intestinal microbiome in the psoriasis group as compared to the controls was observed (Bray-Curtis dissimilarity index; P = .031). This different clustering of gut microbiome composition could not be documented when comparing Pro × PsA patients; P = .11 (Figure 2B).
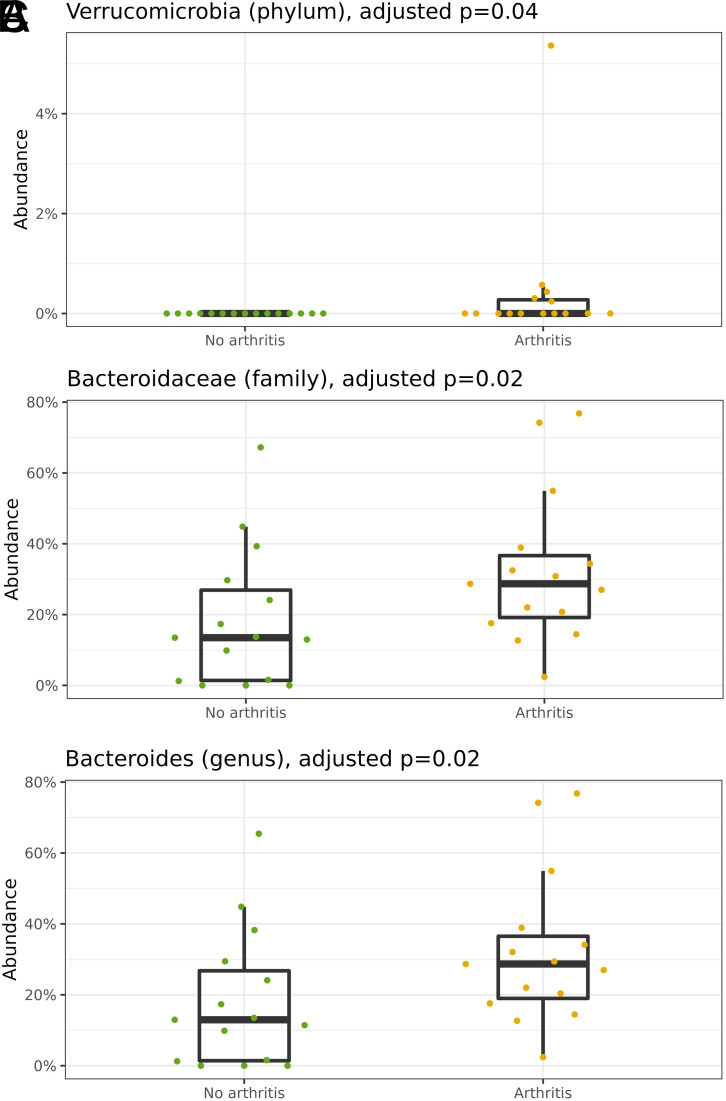
Finally, differential abundance was analyzed and is presented according to taxonomic hierarchical levels in descending order, comparing exposed and unexposed samples. Adjustments were made for each taxonomic level but not necessarily across all taxonomic levels. There was a predominance of the Bacteroidetes phylum among patients with arthritis, which is why the Firmicutes/Bacteroidetes ratio was lower in this group. However, this difference was not statistically significant (P = .11). Nevertheless, at the phylum level, there was a predominance of Verrucomicrobia among PsA patients (P = .04) (Figure 3A).
Figure 3.
(A) Adjusted differential abundance; phylum level: Verrucomicrobia ( P = .04); (B) Adjusted differential abundance; family level: Bacteroidaceae ( P = .02); (C) Adjusted differential abundance; genus level: Bacteroides ( P = .02). Pso (green); Apso (yellow). Source: Elaboration of the authors, 2023.
Moving forward to the family, genus, and species levels, it was verified as a hallmark of the adjusted differential abundance analysis for each level, the greater presence of the Bacteroidaceae family (P = .02) (Figure 3B), Bacteroides genus (P = .02) (Figure 3C), and Bacteroides uniformis species (P = .03) among patients with psoriatic arthritis when compared to patients without joint disease.
Discussion
We carried out a case‒control study with 30 patients with severe psoriatic disease, of whom 15 were diagnosed with psoriatic arthritis, seeking to evaluate the intestinal microbiome based on high-throughput DNA sequencing of the 16S rRNA gene, V3/V4 regions, extracted and amplified from stool samples. We also included 30 individuals without psoriatic disease. All selected individuals, patients, and controls live in urban areas and came from the same geographic unit.
The hypotheses of microorganism participation in the etiopathogenesis of inflammatory arthritis are not recent, especially in rheumatoid arthritis and reactive arthritis but also in spondyloarthritis, including psoriatic arthritis.31 In general, antigenic stimuli are thought to act as a trigger or even a maintenance factor in the autoimmune inflammatory response. Animal models in spondyloarthritis with HLA-B27 and human beta-2 microglobulin-positive transgenic mice, clinically manifested by sacroiliitis, peripheral arthritis, and psoriasiform lesions, did not occur when those animals were raised in sterile environments.32,33 On the other hand, the colonization of the gastrointestinal tract of these same animals by different species of Bacteroides favored the development of joint and cutaneous inflammation.34 Subsequent experimental models documented the development of spondyloarthritis in animals treated with beta-glucans (a prebiotic polysaccharide known to stimulate the growth and proliferation of bacteria and yeast), correlating with changes in the intestinal microbiome.35,36 At the beginning of the twenty-first century, the first evidence linking gastrointestinal inflammation, modifications of the intestinal microbiota, and, consequently, increased permeability of the intestinal barrier with spondyloarthritis was published.37
Advances in high-throughput genetic sequencing techniques have boosted microbiome research along with their influence on human homeostasis, inflammation, and autoimmunity. At the same time, technical improvements allowed for cost reduction and greater accessibility to assessment methodologies, leveraging numerous publications on the subject in the past decade.
Focusing specifically on psoriatic disease, in a pioneering way, Scher et al,38 in 2015, used high-throughput sequencing of the 16S ribosomal gene (same technique as in the present study) to study 16 patients with psoriatic arthritis, 15 with plaque psoriasis, and 17 healthy controls. They demonstrated a decreased diversity of the microbiome among patients with psoriatic arthritis and plaque psoriasis when compared to controls (Shannon index). In parallel, they identified decreased expression of the genera Akkermansia, Ruminococcus, Pseudobutyrivibrio, Parabacteroides, Alistipes, and Coprococcus in patients with psoriatic disease. When comparing patients with and without arthritis, a reduction in Firmicutes, Clostridiales, and Verrucomicrobiales was noted at the phylum level, in addition to an increase in Bacteroidetes in patients with established joint disease.38
In the present study, there was a predominance of the genera Bacteroides and Roseburia in patients with PsA and of Faecalibacterium and Prevotella among patients with PsO, without joint disease. At the species level, it was found that B. vulgatus, B. stercoris, B. uniformis, andB. plebeius predominated among patients with established joint disease. On the other hand, Prevotella copri, and E. rectalepredominated among patients with isolated cutaneous disease. Advancing beyond relative abundance, despite not demonstrating changes in alpha or beta diversity when comparing patients with and without joint disease, our study also documented a predominance of the Bacteroidaceae family (P = .02), genus Bacteroides (P = .02), in addition to species Bacterioides uniform (P = .03) among patients with joint disease. All in adjusted differential abundance.
Scher and colleagues highlighted that a marked decrease in Akkermansia, Ruminococcus, and Pseudobutyvibrio among patients with PsA resembles dysbiosis pattern found in inflammatory bowel disease.38 We did not find these differences, perhaps due to low reading in both studied groups. But, on the other hand, previous studies have linked different species of the genus Bacteroides with inflammatory bowel disease, relating the capacity to produce proteases, especially by the species B. vulgatus, relating to ulcerative colitis, including their expression being proportional to the inflammatory activity of the illness.39
Interestingly, this finding correlates when we return to experimental research, where, in an animal model, it was also demonstrated that colonization of the gastrointestinal tract by different species of Bacteroides favored the development of joint inflammation.34 In the study by Ling et al,40 published in 2022, studying the difference in the intestinal microbiome in patients with psoriatic arthritis (9) and patients with undifferentiated arthritis (10), different groupings of beta-diversity composition were found between the groups. Therefore, the overall microbiome composition was different between groups. Furthermore, there was also greater expression of the Bacteroidetes phylum among patients with psoriatic arthritis, along with Firmicutes, Proteobacteria, and Actinobacteria. Greater expression of the species Megasphaera elsdenii was also identified. In our study, there was no greater expression of Megasphaera spp.
However, Manasso and collaborators,41 in 2020, studying 15 patients with PsA, identified an increase in the orders Clostridiales and Erysipelotrichales, but with a reduction in Bacteroidales when compared to healthy controls. On the other hand, these same authors followed this group after treatment with an anti-TNF-alpha immunobiological, known to be effective in the treatment of psoriatic arthritis, and clinical improvement was accompanied by a reduction of Bacteroidales expression. This, somehow, may support the hypothesis that was formulated in the present study relating the importance of this phylum, order, family, and genus in psoriatic joint disease.
Finally, Xiao and collaborators, in 2024, published the findings of a robust study with 95 individuals, 44 with psoriatic arthritis, 26 with psoriasis, and 25 healthy controls. There was no difference in terms of alpha diversity; however, there were significant grouping differences in beta diversity (Bray–Curtis index) in different scenarios: patients with psoriatic disease vs. controls were grouped together, as well as psoriasis vs. controls and psoriatic arthritis vs. controls. Furthermore, the authors found that the best parameter for differentiating the microbiome of psoriatic disease compared to control individuals was the significant reduction of E. rectale. However, no difference in expression sufficient to differentiate patients with or without joint disease was identified. Conversely, the present study identified greater expression of E. rectale predominated among patients and, when comparing only patients with and without joint disease, those with exclusively cutaneous disease had greater expression of this species in terms of relative abundance.42
Scher and colleagues also showed, at the gender level, a marked decrease in Akkermansia, Ruminococcus, and Pseudobutyvibrio among patients with psoriatic arthritis, with a pattern of intestinal dysbiosis resembling that found in inflammatory bowel disease.38
It is speculated, therefore, that certain patterns of intestinal dysbiosis would determine a decrease in the mucus layer, deregulation of tight junctions, qualitative and quantitative defects in Paneth cells, and increased permeability of the intestinal mucosa. Augmented permeability increases microorganism antigen exposure to the host, especially bacteria (predominantly liposaccharide components of the outer membrane of gram-negative bacteria—LPS). There is a decrease in the population of regulatory T lymphocytes and an increase in the differentiation and expansion of TH1 and TH17 lymphocytes, also increasing the synthesis and secretion of IL17, in addition to a decrease in the production of secretory IgA and short-chain fatty acids. In parallel, this increase in intestinal permeability would allow greater bacterial translocation, with the dissemination of this hypothetical antigenic trigger, inducing inflammation in different sites, including the skin and joints.43,44
Returning to the theory of evolution of psoriatic disease (plaque psoriasis) to joint involvement, in addition to dysbiosis, in the present study, a relationship between dyslipidemia (P < .001) and smoking (P < .001) and joint disease was verified in univariate analysis. Little relevance of acute phase inflammatory tests was seen. A significant association of positive screening with the Psoriasis Epidemiology Screening Tool (PEST) and the presence of psoriatic arthritis was also detected. Another interesting aspect was the demonstration of altered Doppler ultrasound in patients with negative PEST, demonstrating greater sensitivity in the early stages of the disease. There was a tendency towards a higher mean of ultrasensitive quantitative CRP (4.67 × 2.77); however, like the 1-hour ESR, the difference did not reach statistical significance (P = .476). In fact, the literature shows that increased CRP can help in the interpretation of a patient with plaque psoriasis and arthralgia/arthritis; however, when negative, it should not rule out the diagnosis or even be indicative of controlled joint disease or remission.45
We recognize the sample size as an important limitation, even though we have reached the proposed sample calculation.
This study shows different patterns of gut microbiome composition in patients with psoriatic arthritis, with significant overexpression of the Bacteroidacea family Bacteroides genus,and B. uniformes species, similar to previous work on the gut microbiome and psoriatic arthritis. This reinforces the microbiome as a field of interest in psoriasis, especially the gut microbiome, due to its magnitude. Modulating Bacteroides could be a target in the future.
Additionally, we highlight this paper as one of the few conducted in Brazil.
Nevertheless, it should be noted that some previously described findings related to lower diversity and different clustering between groups could not be demonstrated, probably due to the number of patients who could be included in the study. Additionally, it remains difficult to understand the magnitude of the gut microbiome influence on the disease. Is dysbiosis a cause or consequence of the disease? However, the microbiome deserves our attention, especially since it brings different opportunities for intervention through diet, prebiotics and probiotics, pretreatment analysis, prognosis, and even microbiome modulation and transplantation.
Funding Statement
The authors declare that this study received no financial support.
Footnotes
Ethics Committee Approval: This study was approved by the Ethics Committee of Estate Secretary of Health - Santa Catarina/Brazil. July, 2020.
Informed Consent: Written informed consent was obtained from the patients/patient who agreed to take part in the study.
Peer-review: Externally peer reviewed.
Author Contributions: Concept – G.M.A., S.C.; Design – G.M.A., G.R.W.C., S.C.; Supervision – G.R.W.C., S.C.; Resources – G.M.A., G.R.W.C.; Materials – G.M.A., G.R.W.C.; Data Collection and/or Processing – G.M.A., G.R.W.C.; Analysis and/or Interpretation – G.M.A., G.R.W.C., S.C.; Literature Search – G.M.A.; Writing – G.M.A.; Critical Review – G.R.W.C., S.C.
Declaration of Interests: The authors have no conflicts of interest to declare.
References
- 1. Dand N, Mahil SK, Capon F, Smith CH, Simpson MA, Barker JN. Psoriasis and genetics. Acta Derm Venereol. 2020;100(3):adv00030. ( 10.2340/00015555-3384) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Armstrong AW, Read C. Pathophysiology, clinical presentation, and treatment of psoriasis: a review. JAMA. 2020;323(19):1945 1960. ( 10.1001/jama.2020.4006) [DOI] [PubMed] [Google Scholar]
- 3. Nestle FO, Kaplan DH, Baker J. Psoriasis. N Engl J Med. 2009;361(5):469 509. [DOI] [PubMed] [Google Scholar]
- 4. Griffiths CEM, Armstrong AW, Gudjonsson JE, Barker JNWN. Psoriasis. Lancet. 2021;397(10281):1301 1315. ( 10.1016/S0140-6736(20)32549-6) [DOI] [PubMed] [Google Scholar]
- 5. Iskandar IYK, Parisi R, Griffiths CEM, Ashcroft DM, Global Psoriasis Atlas. Systematic review examining changes over time and variation in the incidence and prevalence of psoriasis by age and gender. Br J Dermatol. 2021;184(2):243 258. ( 10.1111/bjd.19169) [DOI] [PubMed] [Google Scholar]
- 6. Rachakonda TD, Schupp CW, Armstrong AW. Psoriasis prevalence among adults in the United States. J Am Acad Dermatol. 2014;70(3):512 516. ( 10.1016/j.jaad.2013.11.013) [DOI] [PubMed] [Google Scholar]
- 7. Michalek IM, Loring B, John SM. A systematic review of worldwide epidemiology of psoriasis. J Eur Acad Dermatol Venereol. 2017;31(2):205 212. ( 10.1111/jdv.13854) [DOI] [PubMed] [Google Scholar]
- 8. Romiti R, Amone M, Menter A, Miot HA. Prevalence of psoriasis in Brazil – a geographical survey. Int J Dermatol. 2017;56(8):e167 e168. ( 10.1111/ijd.13604) [DOI] [PubMed] [Google Scholar]
- 9. Arnone M, Takahashi MDF, Carvalho AVE, et al. Plaque psoriasis diagnostic and treatment guidelines. An Bras Dermatol. 2019;94(2)(suppl 1):S76 S107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hawkes JE, Chan TC, Krueger JG. Psoriasis pathogenesis and the development of novel target immune therapies. J Allergy Clin Immunol. 2017;140(3):645 653. ( 10.1016/j.jaci.2017.07.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gladman DD. Clinical features and diagnostic considerations in psoriatic arthritis. Rheum Dis Clin North Am. 2015;41(4):569 579. ( 10.1016/j.rdc.2015.07.003) [DOI] [PubMed] [Google Scholar]
- 12. Haroon M, Gallagher P, FitzGerald O. Diagnostic delay of more than 6 months contributes to poor radiographic and funcional outcome in psoriatic arthritis. Ann Rheum Dis. 2015;74(6):1045 1050. ( 10.1136/annrheumdis-2013-204858) [DOI] [PubMed] [Google Scholar]
- 13. Christophers E, Barker JNWN, Griffiths CEM, et al. The risk of psoriatic arthritis remains constant following initial diagnosis of psoriasis among patients seen in European dematology clinics. J Eur Acad Dermatol Venereol. 2010;24(5):548 554. ( 10.1111/j.1468-3083.2009.03463.x) [DOI] [PubMed] [Google Scholar]
- 14. Scher JU, Ogdie A, Merola JF, Ritchlin C. Preventing psoriatic arthritis: focusing on patients with psoriasis at increased risk of transition. Nat Rev Rheumatol. 2019;15(3):153 166. ( 10.1038/s41584-019-0175-0) [DOI] [PubMed] [Google Scholar]
- 15. Taylor W, Gladman D, Helliwell P, Marchesoni A, Mease P, Mielants H. Classification criteria for psoriatic arthritis: development of new criteria from a large international study. Arthritis Rheum. 2006;54(8):2665073. [DOI] [PubMed] [Google Scholar]
- 16. Mease PJ. Measures of psoriatic arthritis: tender and Swollen Joint Assessment, Psoriasis Area and Severity Index (PASI), Nail Psoriasis Severity Index (NAPSI), Modified Nail Psoriasis Severity Index (mNAPSI), Mander/Newcastle Enthesitis Index (MEI), Leeds Enthesitis Index (LEI), Spondyloarthritis Research Consortium of Canada (SPARCC), Maastricht Ankylosing Spondylitis Enthesis Score (MASES), Leeds Dactylitis Index (LDI), Patient Global for Psoriatic Arthritis, Dermatology Life Quality Index (DLQI), Psoriatic Arthritis Quality of Life (PsAQOL), Functional Assessment of Chronic Illness Therapy-Fatigue (FACIT-F), Psoriatic Arthritis Response Criteria (PsARC), Psoriatic Arthritis Joint Activity Index (PsAJAI), Disease Activity in Psoriatic Arthritis (DAPSA), and Composite Psoriatic Disease Activity Index (CPDAI). Arthritis Care Res (Hoboken). 2011;63(suppl 11):S64 S85. ( 10.1002/acr.20577) [DOI] [PubMed] [Google Scholar]
- 17. Sartor RB. Microbial influences in inflammatory bowel diseases. Gastroenterology. 2008;134(2):577 594. ( 10.1053/j.gastro.2007.11.059) [DOI] [PubMed] [Google Scholar]
- 18. Yeoh N, Burton JP, Suppiah P, Reid G, Stebbings S. The role of the microbiome in rheumatic diseases. Curr Rheumatol Rep. 2013;15(3):314. ( 10.1007/s11926-012-0314-y) [DOI] [PubMed] [Google Scholar]
- 19. Chen CC, Chen KJ, Kong MS, Chang HJ, Huang JL. Alterations in the gut microbiotas of children with food sensitization in early life. Pediatr Allergy Immunol. 2016;27(3):254 262. ( 10.1111/pai.12522) [DOI] [PubMed] [Google Scholar]
- 20. Chen J, Chia N, Kalari KR, et al. Multiple sclerosis patients have a distinct gut microbiota compared to healthy controls. Sci Rep. 2016;6:28484. ( 10.1038/srep28484) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. De groot PF, Belzer C, Aydin Ö, et al. Distinct fecal and oral microbiota composition in human type I diabetes, an observational study. PLoS One. 2017;12(12):e0188475. ( 10.1371/journal.pone.0188475) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mangiola F, Ianiro G, Franceschi F, Fagiuoli S, Gasbarrini G, Gasbarrini A. Gut microbiota in autismo and mood disorders. World J Gastroenterol. 2016;22(1):361 368. ( 10.3748/wjg.v22.i1.361) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Qin N, Yang F, Li A, et al. Alterations of the human gut microbiome in liver cirrhosis. Nature. 2014;513(7516):59 64. ( 10.1038/nature13568) [DOI] [PubMed] [Google Scholar]
- 24. Zhao S, Li CI, Guo Y, Sheng Q, Shyr Y. RnaSeqSampleSize: real data based sample size estimation for RNA sequencing. BMC Bioinformatics. 2018;19(1):191. ( 10.1186/s12859-018-2191-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McMurdie PJ, Holmes S. Waste not, want not: why rarefying microbiome data is inadmissible. PLOS Comput Biol. 2014;10(4):e1003531. ( 10.1371/journal.pcbi.1003531) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. QIAGEN. [homepage na internet]. QIAamp DNA Stool Mini Kit. [acesso em 30 out 2019]. Available at: https://www.qiagen.com/dk/products/discovery-and-translational-research/dna-rna-purification/dna-purification/genomic-dna/qiaamp-dna-stool-mini-kit/. [Google Scholar]
- 27. Zoetendal EG, Heilig HG, Klaassens ES, et al. Isolation of DNA from bacterial samples of the human gastrointestinal tract. Nat Protoc. 2006;1(2):870 873. [DOI] [PubMed] [Google Scholar]
- 28. Zhou H, He K, Chen J, Zhang X. LinDA: linear models for diferencial abundance analysis of microbiome compositional data. Genome Biol. 2022;23(1):95. ( 10.1186/s13059-022-02655-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Martin BD, Witten D, Willis AD. Modeling microbial abundances and dysbiosis with beta-binomial regression. Ann Appl Stat. 2020;14(1):94 115. ( 10.1214/19-aoas1283) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genom Biol. 2014;15(12):550. ( 10.1186/s13059-014-0550-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stoll ML. Gut microbes, immunity, and spondyloarthritis. Clin Immunol. 2015;159(2):134 142. ( 10.1016/j.clim.2015.05.001) [DOI] [PubMed] [Google Scholar]
- 32. Hammer RE, Maika SD, Richardson JA, Tang JP, Taurog JD. Spontaneous inflammatory disease in transgenic rats expressing HLA-B27 and human beta 2 m: an animal model of HLA-B27-associated human disorders. Cell. 1990;63(5):1099 1112. ( 10.1016/0092-8674(90)90512-d) [DOI] [PubMed] [Google Scholar]
- 33. Taurog JD, Richardson JA, Croft JT, et al. The germfree state prevents development of gut and joint inflammatory disease in HLA-B27 transgenic rats. J Exp Med. 1994;180(6):2359 2364. ( 10.1084/jem.180.6.2359) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rath HC, Herfarth HH, Ikeda JS, et al. Normal luminal bactéria, especially Bacteroides species, mediate chronic colitis, gastrites, and arthritis in HLA-B27/human beta2 microglobulin transgenic rats. J Clin Invest. 1996;98(4):945 953. ( 10.1172/JCI118878) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ruutu M, Thomas G, Steck R, et al. Beta-glucan triggers spondyloarthritis and Crohn’s disease-like ileitis in SKG mouses. Arthritis Rheum. 2012;64(7):2211 2222. ( 10.1002/art.34423) [DOI] [PubMed] [Google Scholar]
- 36. Rehaume LM, Mondot S, Aguirre de Carcer D, et al. ZAP-70 genotype disrupts the relationship between microbiota and host, leading to spondyloarthritis and ileitis in SKG mice. Athritis Rheumatol. 2014;66:2780 2792. [DOI] [PubMed] [Google Scholar]
- 37. Stebbings S, Munro K, Simon MA, et al. Comparison of the fecal microflora of patients with ankylosing spondylitis and controls using molecular methods of analysis. Rhematology (Oxford). 2002;41(12):1395 1401. [DOI] [PubMed] [Google Scholar]
- 38. Scher JU, Ubeda C, Artacho A, et al. Decreased bacterial diversity characterizes the altered gut microbiota in patients with psoriatic arthritis, resembling dysbiosis in inflammatory bowel disease. Arthritis Rheumatol. 2015;67(1):128 139. ( 10.1002/art.38892) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mills RH, Dulai PS, Vázquez-Baeza Y, et al. Multi-omics analyses of the ulcerative colitis gut microbiome link Bacteroides vulgatus proteases with disease severity. Nat Microbiol. 2022;7(2):262 276. ( 10.1038/s41564-021-01050-3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ling CY, Hsu CY, He HR, et al. Gut Microbiota Differences between Psoriatic Arthritis and Other Undifferentiated Arthritis: A Pilot Study; vol 101(28). Baltimore; 2022:e29870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Manasson J, Wallach DS, Guggino G, et al. Interleukin-17 inhibition in spondyloarthritis is associated with subclinical gut microbiome perturbations and a distinctive interleukin-25-driven intestinal inflammation. Arthritis Rheumatol. 2020;72(4):645 657. ( 10.1002/art.41169) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xiao Y, Wang Y, Tong B, et al. Eubacterium rectale is a potential marker of altered gut microbiota in psoriasis and psoriatic arthritis. Microbiol Spectr. 2024;12(4):e0115423. ( 10.1128/spectrum.01154-23) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Van de Wiele T, Van Praet JT, Marzorati M, Drennan MB, Elewaut D. How the microbiota shapes rheumatic diseases. Nat Rev Rheumatol. 2016;12(7):398 411. ( 10.1038/nrrheum.2016.85) [DOI] [PubMed] [Google Scholar]
- 44. Van Praet JT, Donovan E, Vanassche I, et al. Commensal microbiota influence systemic autoimmune reponses. EMBO J. 2015;34(4):466 474. ( 10.15252/embj.201489966) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gialouri CG, Evangelatos G, Pappa M, et al. Normal-C-reactive protein in active psoriatic arthritis: results from real-world clinical practice. Ther Adv Musculoskelet Dis. 2022;14:1759720X221122417. ( 10.1177/1759720X221122417) [DOI] [PMC free article] [PubMed] [Google Scholar]



 Content of this journal is licensed under a
Content of this journal is licensed under a