Abstract
The human immunodeficiency virus type 1 (HIV-1) Nef protein is an important determinant of AIDS pathogenesis. We have previously reported that HIV-1 Nef is responsible for the induction of a severe AIDS-like disease in CD4C/HIV transgenic (Tg) mice. To understand the molecular mechanisms of this Nef-induced disease, we generated Tg mice expressing a mutated Nef protein in which the SH3 ligand-binding domain (P72XXP75XXP78) was mutated to A72XXA75XXQ78. This mutation completely abolished the pathogenic potential of Nef, although a partial downregulation of the CD4 cell surface expression was still observed in these Tg mice. We also studied whether Hck, one of the effectors previously found to bind to this PXXP motif of Nef, was involved in disease development. Breeding of Tg mice expressing wild-type Nef on an hck−/− (knockout) background did not abolish any of the pathological phenotypes. However, the latency of disease development was prolonged. These data indicate that an intact PXXP domain is essential for inducing an AIDS-like disease in CD4C/HIV Tg mice and suggest that interaction of a cellular effector(s) with this domain is required for the induction of this multiorgan disease. Our findings indicate that Hck is an important, but not an essential, effector of Nef and suggest that another factor(s), yet to be identified, may be more critical for disease development.
The human immunodeficiency virus type 1 (HIV-1) and simian immunodeficiency virus (SIV) Nef proteins are critical for several in vivo and in vitro phenotypes induced by these viruses (for reviews, see references 19, 35, 64, and 82). In vivo, Nef has been shown to be very important for high virus replication and disease progression (36). Young adult rhesus monkeys infected with mutant SIV containing large deletions in nef exhibited low viral load and developed simian AIDS (SAIDS) only rarely (7, 42, 85). Some long-term nonprogressors of HIV-1 infection were also found to harbor a functionally defective Nef protein (5, 21, 44, 58, 70). Similarly, nef-deleted HIV-1 was attenuated, showing lower levels of infection and pathogenicity in SCID-hu mice, although it still induced thymocyte depletion (4, 40, 41). In transgenic (Tg) mice, expression of Nef in T cells through the CD3δ (78), the CD2 (11), and the TcR β chain (49) regulatory regions led to immunodeficiency, loss of T cells, and alteration of T-cell activation. Also, we recently reported that expression of Nef as a transgene in mature and immature CD4+ T cells, and in macrophages and dendritic cells (DC), using the regulatory sequences of the human CD4 gene (CD4C), led to the development of a severe AIDS-like disease in these CD4C/HIV Tg mice (33). This disease is characterized by several pathological changes, such as premature death, wasting, atrophy of lymphoid organs and preferential loss of CD4+ T cells, lymphocytic interstitial pneumonitis, interstitial nephritis (33), and heart disease (Kay et al., submitted for publication). All these features are remarkably similar to those observed in AIDS patients.
Other in vitro studies also indicate that Nef might influence HIV-1 pathogenesis in at least four important ways. (i) Nef has the ability to increase viral replication in primary lymphocytes and macrophages (10, 59, 80) and to enhance virion infectivity in some systems (2, 13, 14, 60, 75; for reviews, see references 19, 31, and 36). (ii) Nef can mediate downregulation of CD4 cell surface expression (reviewed in references 65 and 77), a phenomenon shown to be important for the release of HIV-1 from the cell (45, 67). (iii) Nef can also downmodulate the cell surface expression of major histocompatibility complex class I (MHC-I) molecules (76; for a review, see reference 48), an effect found to protect infected cells from killing by cytotoxic T cells (17). (iv) Finally, Nef can alter T-cell signaling pathways (9, 54, 62, 78). Nef has been found to interact with several signaling molecules: with a serine kinase (8); with a distinct serine/threonine kinase, the Nef-associated kinase (NAK) identified as a member of the p21-activated kinase (PAK) family (52, 53, 63, 71, 74; for reviews, see references 20 and 72); with members of the Src-family of tyrosine kinases, notably Lck (8, 13, 16, 22, 29, 69), Hck (6, 12, 47, 61, 68), Lyn (13, 68), and Fyn (13); and with Vav (23), mitogen-activated protein kinase (29), c-Raf-1 (37), p53 (28), and protein kinase C θ (79). Contrasting results were reported on the interaction of Nef with Lck: some authors could document such binding (13, 16, 22, 29, 69), while others could not (61, 68). Interestingly, Hck was found to bind preferentially and with higher affinity to HIV-1 Nef than did other Src-related kinases (6, 47, 68) and to be activated by such binding (12, 61).
Nef contains several distinct domains, but the proline-rich domain P72XXP75 located at the N-terminal portion of the molecule has attracted much attention. The binding of Nef to Hck occurs through the SH3 domain of Hck and the proline-rich P72XXP75 motif of HIV-1 Nef (12, 13, 15, 30, 47, 68). Similarly, the association of Lck to Nef was found by some workers to require the P72XXP75 motif of Nef and to lead to inhibition of the Lck kinase activity (16, 29), while others found that the Nef P72XXP75 domain was not required for binding to Lck (8, 13). This proline-rich domain of Nef has also been shown to mediate interaction with NAK (43, 57, 84) and Vav (23). Mutations within this proline-rich motif not only abolish activation of and/or binding with NAK (43, 57, 84) and with the Src-related kinases (12, 13, 16, 29, 30, 47, 61, 68) but also prevent several other in vitro effects of Nef, such as altered signaling (25, 27, 39), MHC-I downregulation (27, 56), enhanced viral replication (18, 41), and enhancement of virion infectivity (26), although some studies reported that a mutated proline-rich domain still affected MHC-I downregulation (18) and viral replication (41). There is an apparent consensus that this PXXP motif of Nef is largely dispensable for CD4 downregulation (1, 18, 26, 38, 56, 67, 68), for the ability of Nef to rescue the inhibition of virus release by CD4 (67), and for the producer cell-dependent enhancement of viral entry (81).
In vivo, conflicting results have also been obtained regarding the importance of this proline-rich motif for disease induction. Lang et al. have recently found that this motif in SIV Nef is dispensable for progression to fatal SAIDS (46). In contrast, Khan et al. found that the mutation of the proline-rich motif (P104A and P107A) in SIV Nef was reverted to PXXP, revealing a strong selective pressure for restoration of the SH3 binding domain and suggesting the importance of this motif for Nef function and for the induction of SAIDS (43). In a different model system, the SCID-hu mouse, two other groups concluded that the proline-rich domain of Nef is dispensable for the pathogenicity of HIV-1 (3, 41).
Given this controversy, we constructed Tg mice expressing HIV-1 Nef mutated in this motif and examined one of the cellular effectors, Hck, known to interact with Nef through this proline-rich motif. An HIV-1 Nef allele mutated in the SH3-binding domain was expressed in Tg mice under the same regulatory elements of the human CD4 gene (CD4C), as described before (32, 33). We report here that this domain is critical for inducing the AIDS-like disease observed in CD4C/HIV Tg mice, suggesting that the interaction of one or several cellular factors with this domain is required for the induction of this multiorgan disease. Breeding of wild-type Nef-expressing CD4C/HIV Tg mice on an Hck knockout background prolonged latency but did not abolish disease development, suggesting that Hck is important, but is not one of the cellular effectors which is essential, for disease development in Tg mice.
MATERIALS AND METHODS
Generation of Tg mice.
The CD4C/HIVMuG(AXXA) transgene was constructed by mutating the P72XXP75 residues of the Nef protein to alanines. The mutations of Nef (P72A, P75A) were produced by PCR site-directed mutagenesis, on a SacI-BamHI HIV-1 nef fragment subcloned in a pBS KS vector, using primer 513 (5-TGTCTTAAAGCTACCTGAGCTGTGACTG-3) containing C to G mutations (in boldface type) at nucleotides (nt) 9000 and 9009, to produce P72A and P75A mutations. The P78Q (C to A at nt 9019) mutation spontaneously occurred. Mutations (P72A, P75A, P78Q) were confirmed by sequencing, and the SacI-BamHI fragment was incorporated into the transgene CD4C/HIVMutG DNA backbone used in our previous study (33), to replace the wild-type nef sequences. The DNA transgene was purified and inoculated into 1-day-old (C57BL/6 × C3H)F2 embryos to generate Tg mice as described (33). Three Tg founders were produced, and lines were established from each one by breeding as heterozygotes on the C3H background for three to seven generations.
Mice.
The CD4C/HIVMutG and CD4C/HIVMutA Tg mice (33) and hck−/− knockout mice (51) used in these experiments have been described previously. The hck−/− mice were bred on the C57BL/6 background for several generations and then on the C3H background for 2 generations before being backcrossed with CD4C/HIVMutA Tg mice (C3H for 10 to 12 generations) to generate hck−/− or hck+/− CD4C/HIVMutA Tg and hck−/− and hck+/− non-Tg mice. The Tg mice and their non-Tg littermates were housed under specific-pathogen-free conditions in the same cages. In all experiments the four groups of mice obtained (Tg+ hck−/−, Tg+ hck+/−, non-Tg hck−/−, and non-Tg hck+/−) were littermates.
Antibodies and reagents for fluorescence-activated cell sorting (FACS).
The hybridoma-producing rat anti-mouse B220 (RA36B2) was a kind gift of R. Coffman, DNAX Research Institute of Cellular and Molecular Biology, Palo Alto, Calif. The rat anti-mouse MHC-II (M5-114), rat anti-mouse CD8 (53.6.78), and hamster anti-mouse CD3 (145-2C11) monoclonal antibodies (MAb) were purchased from the American Type Culture Collection (Manassas, Va.). The hybridoma for hamster anti-mouse CD28 (37.51) was a gift of P Hugo. Anti-CD3 MAb was purified using protein G affinity columns. The anti-CD4-phycoerythrin (anti-CD4-PE), anti-CD8-PE, and anti-TcR-fluorescein isothiocyanate (anti-TcR-FITC) were from Cederlane. Irrelevant rat immunoglobulin G1 (IgG1), rat IgG2a, and rat IgG2b MAb were used as isotype controls. Propidium iodide (PI) was from Sigma. The antiphosphotyrosine MAb (4G10) was from Upstate Biotechnology Inc.
Purification of CD4+ T cells.
The peripheral (axillary, inguinal, popliteal, and brachial) lymph nodes were collected to prepare single-cell suspensions. Cells were incubated at 37°C in a humidified 5% CO2 incubator for 1 h to remove adherent macrophages and DC. Nonadherent cells were then harvested and CD4+ T cells were purified by using sheep anti-rat antibody-coated magnetic beads (Dynabeads; Dynal, Oslo, Norway) following a preincubation with an antibody cocktail of hybridoma supernatants. The cocktail contained rat anti-B220 (RA36B2), rat anti MHC-II (M5-114), and rat anti-CD8 (53.6.78). The purity was >92% of TcR+ CD8− T cells as analyzed by flow cytometry (FACS) (Becton Dickinson, San Jose, Calif.).
CFSE fluorescent dye labeling and cell division assay.
CFSE (5-and 6-carboxyfluorescein diacetate succinimidyl ester) was purchased from Molecular Probes Inc. (Eugene, Oreg.). CFSE staining was performed as previously described (83) with slight modifications. Briefly, purified CD4+ T cells were resuspended at 107 cells/ml in phosphate-buffered saline (PBS) without fetal bovine serum (FBS). CFSE was then added at a final concentration of 1 μM, and the cells were incubated at room temperature for 10 min. The reaction was stopped by adding Iscove's modified Dulbecco medium (IMDM; Gibco BRL, Life Technologies, Paisley, Scotland) supplemented with 10% FBS (Gibco). The CD4+ T cells were washed twice and cultured in anti-CD3 (5 μg/ml)-coated flat-bottom 96-well plates at 105 cells/well in 200 μl of the complete medium (IMDM supplemented with 5% FBS, 2 mM l-glutamine, 50 μM β-mercaptoethanol, penicillin [100 U/ml], and streptomycin [100U/ml]) containing anti-CD28 (2 μg/ml) for 3 days.
Flow cytometry.
Flow cytometry was performed using antibodies against various cell surface markers CD4, CD8, TcRαβ, and Thy1.2 for T cells and B220 and Mac-1 for B cells and macrophages, respectively, as described previously (32, 33). Single cell suspension of splenocytes were incubated with hemolytic Gey's solution to remove red blood cells. Splenocytes and harvested CFSE labeled lymph node (LN) cells from cultures were washed twice with PBS containing 2% FBS and 0.05% sodium azide. Nonspecific binding was blocked using a blocking buffer containing 20% FBS, 1× PBS, and 0.05% NaN3. Immunostaining was performed on ice using saturating amounts of MAb. PI was added in a concentration of 1 μg/ml to label dead cells. The FACScan flow cytometer and Cellquest software (Becton Dickinson) were used.
Production of anti-Nef antibodies.
Anti-Nef polyclonal antibodies were raised against a glutathione S-transferase (GST)-Nef fusion protein. To construct the GST-Nef fusion gene, HIV-1 sequences encoding the total Nef (nt 8787 to 9407) were amplified by PCR with MluI and NotI sites at 5′ and 3′ primers, respectively. The PCR product was subcloned in frame into the EcoRI-SalI site of pGEX-4T-1. The fusion gene structure was confirmed by sequencing. The GST fusion protein was expressed and purified on glutathione-Sepharose beads. The purified protein with beads (1 mg) was injected subcutaneously and intramuscularly into female rabbits (2 months old) in four doses. The first three injections were given at 2-week intervals, and the fourth injection was given 2 months later. The animals were killed and serum was collected.
Transgene expression.
The levels of transgene expression were measured first by Northern blot analysis, using 32P-labeled 3.5-kbp SacI (5′-end) and 1.4-kbp HindIII-SacI (nef-specific) NL4-3 HIV-1 probes, as previously described (33). Protein expression was assessed as described previously (33) by Western blot analysis of lymphoid organs extracted in RIPA buffer (10 mM Tris [pH 7.5], 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 1.0% sodium deoxycholate) with protease inhibitors—aprotinin, 2 μg/ml; leupeptin, 2 μg/ml; pepstatin, 1 μg/ml; 1-chloro-3-tosylamido-7amino-l-2-heptanone (TLCK), 50 μg/ml; phenylmethylsulfonyl fluoride (PMSF), 100 μg/ml—and anti-Nef antibodies, prepared as described above (1:2,000 dilution). Antigen-antibody complexes were detected using an ECL detection kit (Amersham). The amount of protein in lysates was quantitated using a Micro BCA assay (Sigma).
Histological and hematological analysis.
A group of 20 Tg animals and an equivalent number of non-Tg littermates were generated from each line and observed for signs of disease (hypoactivity, ruffled hair coat, and respiratory problems). Mice were sacrificed, and lymphoid and nonlymphoid organs were processed for macroscopic and histological assessment and in situ hybridization and compared to CD4C/HIVMutG mice, essentially as previously described (33). Analysis and differential counts were performed on blood from Tg and non-Tg controls as described (32, 33).
Statistics.
Comparison between groups was performed by using one-way analysis of variance (Sigmastat). The data were expressed as means ± standard error (SE) of the mean. P values of ≥0.05 were considered to be not significant.
RESULTS
Construction of CD4C/HIVMutG(AXXA) Tg mice.
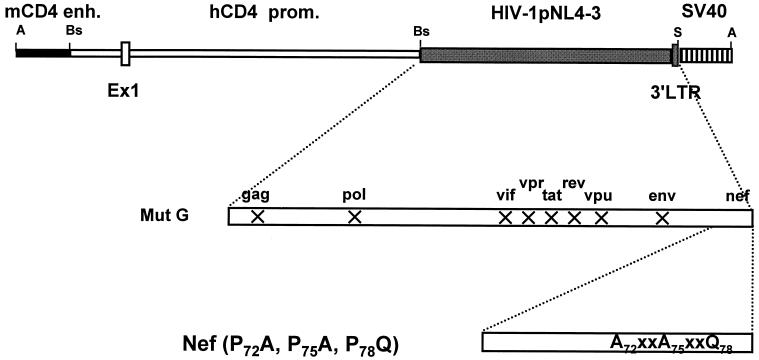
We previously reported that CD4C/HIVMutG Tg mice which express only HIV-1 nef under the control of the chimeric human CD4 promoter-mouse CD4 enhancer regulatory elements (CD4C) develop a severe AIDS-like disease (32, 33). To study the role of the SH3-binding domain of Nef in the development of this disease, three of the four prolines of this motif were mutated in the original HIV-1 NL4-3 (P72A, P75A, P78Q). The mutated nef fragment was then recombined with the CD4C/HIVMutG cassette to replace the wild-type NL4-3 nef sequences (Fig 1). This HIVMutG genome harbors mutations in all the known HIV-1 genes, except in nef (33). Tg mice were constructed, and three founder lines (F42961, F42965, and F42968), designated CD4C/HIVMutG(AXXA), were established and studied. Tg mice were bred on the C3H background with expected Mendelian ratios.
FIG. 1.
Structure of the CD4C/HIVMutG(AXXA) transgene. The CD4C/HIVMutG(AXXA) DNA was constructed as described in Materials and Methods. Abbreviations: mCD4enh., mouse CD4 enhancer; hCD4 prom., human CD4 promoter; SV40, polyadenylation sequences from simian virus 40; Ex1, CD4 gene exon1; X, interruption of the open reading frame of the indicated HIV-1 gene. Abbreviations for restriction sites: A, AatII, Bs BssHII, S, SstI.
Evaluation of transgene RNA expression by Northern analysis on various organs revealed the presence of discrete species of HIV-1 RNA transcripts (8.8 kb full-length, 4.3 kb env-specific and 2.0 kb multiply spliced) and showed that they were more abundant in the thymus than in the spleen or lymph nodes (LNs) and very low (lung and kidney) or negative in several other organs (Fig. 2A and 2B). In addition, in situ hybridization with HIV-1 specific riboprobes showed that expression was present in thymocytes and in peritoneal macrophages (Fig. 3), as expected. Thus, this pattern of expression reflects the cell type specificity of the CD4C regulatory elements as previously documented in CD4C/CD4 (34) and CD4C/HIV (32, 33) Tg mice. As expected, RNA expression was higher in some founders than in others (F42961 > F42968 > F42965). Immunoblot analysis of Tg thymus extracts with anti-Nef antibodies revealed the presence of Nef, at different levels (F42961 > F42968 > F42965), in mice from all three founder lines (Fig. 2C).
FIG. 2.
Northern and Western blot analysis of expression of HIV-1 Nef in CD4C/HIVMutG(AXXA) Tg mice. (A and B) Northern blot analysis of HIV-1 RNA in various tissues of CD4C/HIVMutG(AXXA) Tg mice (2 months old). RNAs from CD4C/HIVMutG Tg mice (F27367) (1.5 months old) serve as a positive control. Total RNAs were hybridized with 32P-labeled HIV-1-specific probes. Abbreviations: T, thymus; S, spleen; H, heart; B, brain; M, muscle; I, intestine; Lu, lung; Te, testis, Lv, liver, L, LN; K, kidney. After hybridization, the blot was washed and rehybridized with an actin probe. (C) Western blot analysis of protein extracts from thymuses from one-month-old Tg and non-Tg littermates from different founders using rabbit anti-Nef antibodies. A thymus from a CD4C/HIVMutG Tg mouse was used as a positive control.
FIG. 3.
In situ detection of HIV-1 expression in CD4C/HIVMutG(AXXA) Tg mice. Thymus from a Tg mouse (A to D) and peritoneal macrophages from Tg (E and F) and non-Tg (G and H) animals were assessed for transgene expression with 35S-labeled antisense (α-sense) and control sense HIV-1 specific riboprobes. (C and D) Bright-field images of the same region shown in dark-field in panels A and B, respectively. Insets to panels C and D are cortex shown at high power. In thymus (B), a more intense hybridization signal is observed over cortical (c) than medullary (m) regions. Tg expression is detected in Tg macrophages (F) but not a non-Tg control macrophages (H) with the α-sense probe. No hybridization signal is seen with the sense probe (A, C, E, and G). Scale bars, 250 μm (A to D) and 50 μm (E to H). Counterstaining was with hematoxylin and eosin.
We have previously documented that both the incidence and the progression of the AIDS-like disease in CD4C/HIVMutG Tg mice correlated very well with the levels of Nef expression (33). Therefore, in the present study, mice from founder lines which expressed mutated Nef at equal (F42968) or higher (F42961) protein levels than the wild-type Nef of CD4C/HIVMutG (F27367) Tg mice (33) were selected for comparative purposes.
Disease fails to develop in CD4C/HIVMutG(AXXA) Tg mice.
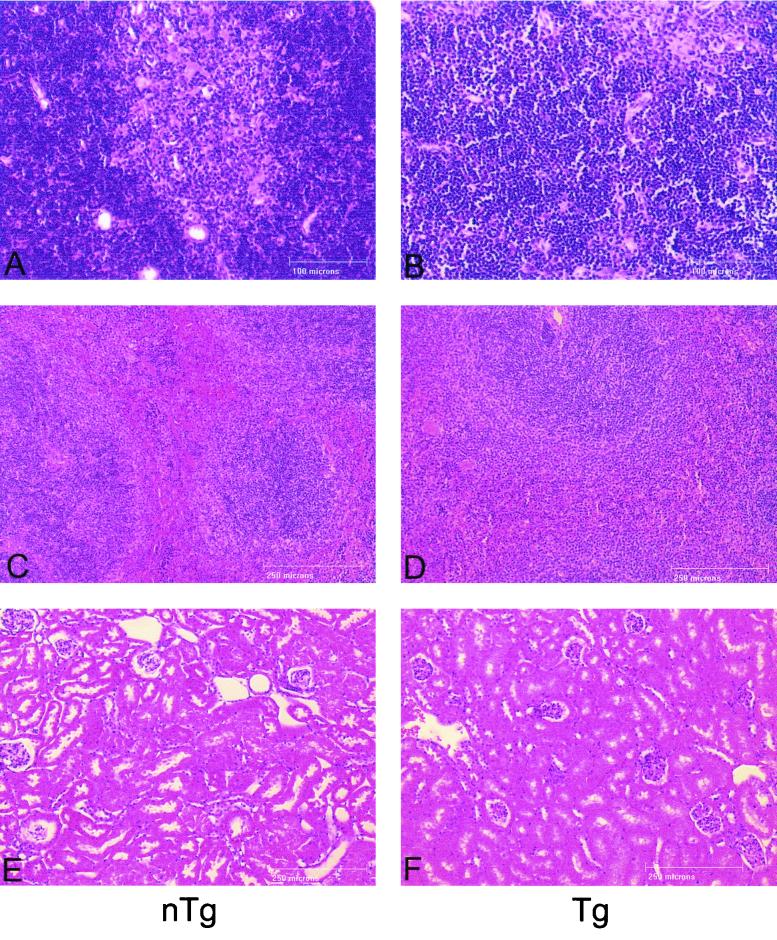
Progeny from the three founder lines of CD4C/HIVMutG(AXXA) Tg mice were routinely monitored for signs of disease such as hypoactivity and weakness, diarrhea, loss of body weight (wasting), edema, and early death. The life span of these Tg mice (n = 30) was comparable to that of their non-Tg littermates (n = 30): they survived in apparent good health until the termination of the experiment (14 months). These Tg mice were further examined at different times of life (at 2, 6, 9, and 14 months old) for evidence of gross and histological phenotypes such as atrophy of lymphoid organs (thymus, spleen, and LN) and kidney and lung diseases, observed previously in Tg mice expressing the wild-type Nef (32, 33). In contrast to the phenotype of CD4C/HIV Tg expressing wild-type Nef, in which the clinical and pathological changes occurred as early as 1 month of age, histopathologic analysis performed on CD4C/HIVMutG(AXXA) Tg mice revealed no differences between Tg and non-Tg animals and specifically no evidence of lymphoid cell depletion with the exception of a small disruption of the architecture of the thymus in a few Tg mice (Fig. 4 and data not shown). Moreover, compared to control non-Tg littermates, there was no change in Tg mice in any of the hematological parameters (hemoglobin, hematocrit, number of red and white blood cells and platelets) measured (data not shown). Also, the differential count of leukocytes (monocytes, eosinophils, basophils, lymphocytes and neutrophils) was comparable in Tg and non-Tg mice (data not shown). These results show that mutation of the proline-rich SH3 binding motif of Nef virtually abolishes the pathogenic potential of this protein in vivo, in Tg mice.
FIG. 4.
Histology of thymus, spleen, and kidney of CD4C/HIVMutG(AXXA) Tg mice. Control non-Tg (A, C, and E) and CD4C/HIVMutG(AXXA) Tg (B, D, and F) tissues were analyzed: thymus (A and B), spleen (C and D), and kidney (E and F). Tg thymus does not exhibit the major pathological changes observed in CD4C/HIVMutG Tg mice but sometimes demonstrates a partial disruption of architecture, with the loss of a clearly defined cortico-medullary junction (B). Tg spleen (D) and kidney (F) demonstrate histology indistinguishable from normal non-Tg controls (C and E, respectively). Scale bars, 100 μm (A and B) and 250 μm (C to F). Counterstaining was with hematoxylin and eosin.
Mutation of the PXXP motif partially attenuated, but did not abolish, the downregulation of the CD4 cell surface expression on lymphoid T cells.
In CD4C/HIVMutG Tg mice expressing wild-type Nef, severe perturbations of the lymphoid cell populations were previously noticed: preferential loss of single-positive CD4+ T cells, increase of CD8+ T cells early in disease and loss of both of these T cell populations later, as well as increase of the B-cell population (33). To assess the effect of transgene expression on cells of the immune system of the CD4C/HIVMutG(AXXA) Tg mice, we first performed quantitation of cell numbers in the lymphoid organs of Tg mice in comparison with Tg mice expressing wild-type Nef and with non-Tg littermates. In our previous studies, we noticed that Tg mice exhibiting moderate levels of Nef became diseased later in life. In these animals, the total cell number in various lymphoid organs was normal at an early stage and became progressively depleted later. Therefore, care was taken to compare Tg lines expressing the same levels of Nef. The cell counts from LN, spleens and thymuses of these CD4C/HIVMutG(AXXA) Tg mice were in the normal range (Table 1).
TABLE 1.
Cell number in lymphoid organs of control and CD4C/HIVMutG(AXXA) Tg mice
| Mouse linea | Total no. of cells (10−6)/organ
|
||
|---|---|---|---|
| Thymus | Spleen | Mesenteric LN | |
| Non-Tgb | 68.9 ± 10 | 73.5 ± 9.7 | 26.5 ± 6.8 |
| CD4C/HIVMutG | 29.6 ± 6.5c | 35.6 ± 9.3c | 6.7 ± 3.1c |
| CD4C/HIVMutG(AXXA) | |||
| F42961 | 72.2 ± 13 | 62.3 ± 12 | 24.3 ± 6.7 |
| F42965 | 69.7 ± 13.8 | 66.3 ± 11.4 | 18.9 ± 5.6 |
| F42968 | 65.8 ± 11.7 | 70.3 ± 13.3 | 20.1 ± 5.4 |
A minimum of nine mice (2 to 6 months old) of each founder line were analyzed.
The non-Tg control values were obtained by pooling the results of all the non-Tg littermates from different lines.
P < 0.05 by using Student's t test.
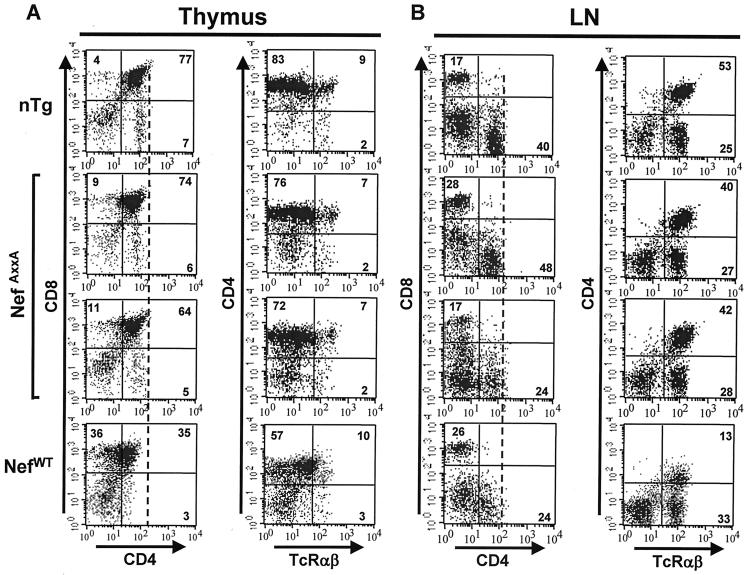
Further cytofluorometric (FACS) analysis was carried out at various time points, on different T cell populations of the thymus and of peripheral lymphoid organs (spleen and LN), with T-cell-specific markers (CD4, CD8, Thy-1, and TCRαβ). This analysis revealed that the only alteration of cell phenotype that was consistently observed in these Tg mice was a partial (30 to 40%) downregulation of the CD4 cell surface protein on single and double positive CD4+ T cells, while no change of the Thy-1 cell surface expression was observed (Fig. 5A and B; Tables 2 and 3). This resulted in a small decrease of the CD4/CD8 cell ratio. This CD4 downregulation phenotype did not increase with time. In addition, two other cell populations studied with different cell markers, B cells (B220) and macrophages (Mac-1), were comparable in CD4C/HIVMutG(AXXA) Tg mice to their non-Tg littermates. Tables 2 and 3 summarize these FACS data.
FIG. 5.
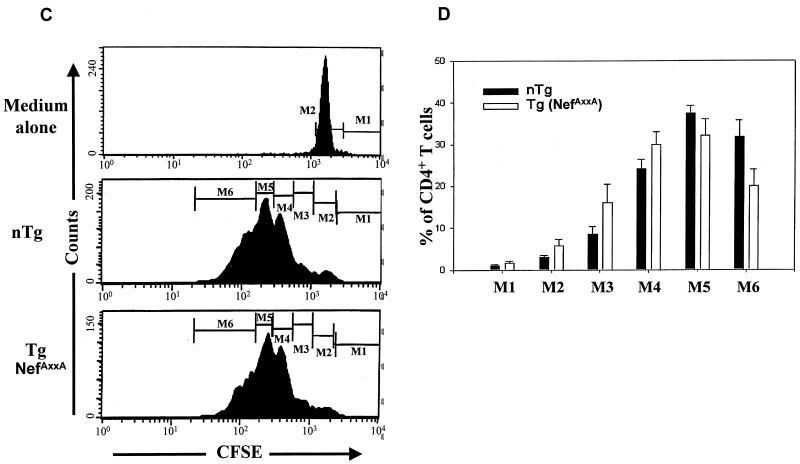
Phenotypic and cell division analysis of T lymphocytes from CD4C/HIVMutG(AXXA) Tg mice. (A and B) FACS analysis of thymic and peripheral T cells. Thymus (A) and mesenteric LN (B) cells from two representative CD4C/HIVMutG(AXXA) Tg mice (F42961) (12 months old) and from CD4C/HIVMutG Tg mice expressing wild-type Nef (positive control) (2 months old) and a non-Tg littermate (12 months old) were analyzed by flow cytometry for the expression of CD4, CD8, and TcRαβ. The percentage of cells found in each quadrant is indicated. A dotted line was drawn across to show the shift of the CD4+ population. Notice that the downregulation of CD4 is more pronounced in CD4C/HIVMutG than in CD4C/HIVMutG(AXXA) Tg mice. (C and D) Cell division kinetics of CD4+ T cells. Fresh single cell suspensions were prepared from peripheral LNs of Tg mice and non-Tg littermates of the F42961 line and labeled by CFSE. After 3 days of culture with or without anti-CD3 plus anti-CD28, the cells were harvested and stained with CD4-PE MAb. Live cells were confirmed by using PI labeling. (C) Histograms show the intensity of CFSE fluorescence of live CD4+ T cells. The top panel represents cell division without the stimulation of antiCD3 plus antiCD28, while the middle and bottom ones show cell division upon the stimulation with antiCD3+ antiCD28. (D) Statistical analysis of CD4+ T-cell division. Comparison was based on the percentage of undivided or divided CD4+ T cells in each peak between Tg mice (n = 6) and non-Tg littermates (n = 6). M1 represents undivided cells and M2 to M6 represents cells that divided 1 to 5 times. No significant difference (P > 0.05) was observed overall when estimated by using analysis of variance.
TABLE 2.
Thymic cell surface marker analysis in CD4C/HIVMutG(AXXA) Tg mice
| Mouse linea | Cell population (%)
|
CD4/CD8 ratio | Mean fluorescence (%)c
|
||||
|---|---|---|---|---|---|---|---|
| Thy 1.2 | CD4+ CD8+ | CD4+ CD8− | CD4− CD8+ | Thy 1.2 | CD4 | ||
| Non-Tgb | 93.4 ± 4.9 | 86.3 ± 3.9 | 8.9 ± 1.5 | 4.3 ± 1.1 | 2.1 ± 0.2 | 100 | 100 |
| CD4C/HIVMutG | 91.6 ± 3.3 | 81.7 ± 2.4d | 5.6 ± 1.5d | 6.1 ± 1.5d | 0.9 ± 0.1d | 99.6 ± 17.6 | 33.3 ± 8.5d |
| CD4C/HIVMutG(AXXA) | |||||||
| F42961 | 96.3 ± 3.4 | 83.7 ± 2.6 | 7.7 ± 1.5d | 5.6 ± 1.5 | 1.4 ± 0.2d | 102.4 ± 14 | 70.2 ± 6.7d |
| F42965 | 97.1 ± 1.7 | 84.6 ± 2.7 | 8.8 ± 1.4 | 6.6 ± 1.6d | 1.3 ± 0.2d | 97.8 ± 10.3 | 70.4 ± 8.2d |
| F42968 | 91.2 ± 3.4 | 87.1 ± 3.1 | 6.4 ± 1.1d | 5.7 ± 1.2d | 1.1 ± 0.1d | 96.5 ± 12.1 | 59.3 ± 8.4d |
FACS analysis was performed on at least nine mice (2 to 12 months old) for each Tg line. The positive control CD4C/HIVMutG Tg mice were 2 to 3 months old.
The non-Tg control values were obtained by pooling the results of all non-Tg littermates from different lines.
The mean fluorescences for Thy 1.2 and CD4 were obtained by calculating the ratio of CD4 or Thy 1.2 staining in Tg thymuses relative to that of non-Tg thymuses (100%). Mean values were then calculated with the values for each line.
P < 0.05 by using Student's t test.
TABLE 3.
Mesenteric LN cell surface marker analysis in CD4C/HIVMutG(AXXA) Tg mice
| Mouse linea | Cell population (%)
|
CD4/CD8 ratio | Mean Fluorescence (%)c
|
||||
|---|---|---|---|---|---|---|---|
| Thy 1.2 | CD4+ | CD8+ | B220 | Thy 1.2 | CD4 | ||
| Non-Tgb | 60.1 ± 8.6 | 46.3 ± 4.8 | 15.8 ± 1.9 | 31.4 ± 4.2 | 2.9 ± 0.2 | 100 | 100 |
| CD4C/HIVMutG | 50.5 ± 4.8d | 20.1 ± 4.1d | 29.9 ± 4.9d | 36.1 ± 4.9 | 0.67 ± 0.1d | 95.6 ± 11.4 | 40.2 ± 9.4d |
| CD4C/HIVMutG(AXXA) | |||||||
| F42961 | 59.7 ± 8.6 | 40.1 ± 4.4d | 26.7 ± 4.4d | 36.1 ± 5.6 | 1.5 ± 0.2d | 107.2 ± 6.1 | 69.3 ± 4.4d |
| F42965 | 65.5 ± 5.6d | 38.1 ± 3.1d | 30.6 ± 4.7d | 35.1 ± 5.1 | 1.2 ± 0.2d | 104.8 ± 14 | 78.5 ± 4.3d |
| F42968 | 60.3 ± 7.7 | 31.5 ± 3.6d | 27.1 ± 4.2d | 37.3 ± 4.7 | 1.2 ± 0.1d | 103.9 ± 10.7 | 85.3 ± 3.5d |
FACS analysis was performed on at least nine mice (2 to 12 months old) for each Tg line. The positive control CD4C/HIVMutG Tg mice were 2 to 3 months old.
The non-Tg control values were obtained by pooling the results of all non-Tg littermates from different lines.
The mean fluorescences for Thy 1.2 and CD4 were obtained by calculating the ratio of CD4 or Thy 1.2 staining in Tg LN relative to that of non-Tg LN (100%). Mean values were then calculated with the values for each line.
P < 0.05 by using Student's t test.
CD4+ T cells from CD4C/HIVMutG(AXXA) Tg mice have a normal capacity to divide after in vitro stimulation.
To examine the function of CD4+ T cells, we assessed their capacity to proliferate using the CFSE-labeling technique (55). After in vitro stimulation with anti-CD3 plus anti-CD28 antibodies, we have recently observed a significant delay in cell division of CD4+ T cells from Tg mice expressing wild-type Nef (CD4C/HIVMutA) compared to control non-Tg mice (see below and Fig. 9). A very similar result was obtained with CD4+ T cells from CD4C/HIVMutG (NefWt) Tg mice (Weng et al., unpublished data). This delayed cell division was absent in cells from CD4C/HIVMutG(AXXA) Tg mice (Fig. 5C and D). As shown, the majority of CD4+ T cells from both non-Tg and Tg mice divided with similar kinetics three (M4) to five (M6) times over a 3-day period. No significant differences were observed in the proportion of CD4+ T cells from non-Tg and Tg mice which remained quiescent or underwent one to five rounds of cell division. This result indicated that the function of CD4+ T cells from CD4C/HIVMutG(AXXA) as scored in this in vitro assay was normal.
FIG. 9.
Cell division kinetics of purified CD4+ T cells from hck−/− CD4C/HIVMutA Tg mice. CD4+ T cells were purified from peripheral LN cells by negative selection and labeled with CFSE, as described in Materials and Methods. After 3 days of culture with anti-CD3 plus anti-CD28, the cells were harvested and stained with PI. Histograms show the intensity of CFSE fluorescence from live (PI negative gate) CD4+ T cells. Quantitative data are shown in percentage of each generation of undivided (M1) and divided (M2 to M6) CD4+ T cells. The results are representative of at least three experiments.
Levels of tyrosine phosphorylated proteins in thymocytes of non-Tg and CD4C/HIVMutG(AXXA) Tg mice are comparable.
We next examined the state of activation of signaling intermediates in thymocytes of CD4C/HIVMutG(AXXA) Tg mice by assessing the phosphotyrosine content of their proteins. We previously reported that Tg mice expressing wild-type Nef exhibit higher levels of tyrosine phosphorylated proteins in their thymocytes, both constitutively and following engagement of the TcR by anti-CD3 (33), indicating a state of constitutive activation and hyperresponsiveness to anti-CD3-TcR engagement. Such a phenotype of total phosphoproteins was not detected in the CD4C/HIVMutG(AXXA) Tg mice, and their thymocytes responded as those from non-Tg control littermates (Fig. 6). For example, no difference in the tyrosine phosphorylation of LAT, an early effector of TcR signaling, could be seen between non-Tg and CD4C/HIVMutG(AXXA) Tg mice, either constitutively or after anti-CD3 stimulation (data not shown).
FIG. 6.
Tyrosine phosphorylation of proteins is not enhanced in thymocytes of CD4C/HIVMutG(AXXA) Tg mice. Thymocytes (5 × 105) from non-Tg and from Tg mice expressing a mutated [CD4C/HIVMutG(AXXA)] or a wild-type (wt) (CD4C/HIVMutG) Nef (2 months old) were stimulated with anti-CD3 for 0 and 5 min. Lysates were then prepared and the immunoblots were probed with antiphosphotyrosine antibodies (4G10). Lanes 1 and 2 represent individual mice from Tg and non-Tg mice (i.e., Tg mice expressing wild-type Nef or Tg mice expressing mutated Nef).
The absence of Hck delays but does not prevent the development of an AIDS-like disease in CD4C/HIV-1 Tg mice.
In vitro studies have shown that the Src-related tyrosine kinase Hck has the ability to interact with Nef, through the P72XXP75 motif (6, 12, 13, 15, 30, 47, 61, 68). However, the physiological relevance of this interaction in vivo remains to be established. In order to directly address whether interaction of Nef with Hck plays a role in vivo in the development of the AIDS-like disease, we took a genetic approach. The CD4C/HIVMutA Tg mice were used for these experiments. These Tg mice harbor an HIV genome with mutations in all genes except env, rev, and nef and develop a severe AIDS-like disease indistinguishable from that of CD4C/HIVMutG (33) and CD4C/HIVWT (32) Tg mice. Therefore, the presence of intact env and rev genes in CD4C/HIVMutA Tg mice does not appear to influence the phenotype induced by Nef. The CD4C/HIVMutA Tg mice were backcrossed with hck knockout (hck−/−) mice (51), to determine whether the ablation of this host gene would prevent partially or totally the appearance of the multiorgan lesions observed in these Tg mice. A group of hck−/− CD4C/HIVMutA Tg mice was generated along with groups of control hck+/− CD4C/HIVMutA Tg mice and hck−/− or hck+/− non-Tg littermates. These mice were observed for up to 12 months. A delay in the mortality rate of hck−/− compared to hck+/− Tg+ mice was observed (Fig. 7A). Interestingly, as many as 23% of hck−/− Tg mice survived up to 12 months, while none of the hck+/− Tg mice survived longer than 9 months. Also, at 6 months of age, only ∼35% of hck−/− Tg were found dead or sacrificed because of the severity of the disease compared to ∼80% of hck+/− Tg mice. However, despite this delayed progression, most of the hck−/− Tg mice still developed the AIDS-like pathological changes that were indistinguishable from those observed in the control hck+/− Tg mice (Fig. 7B) or from those previously reported (32, 33) in the hck+/+ CD4C/HIV Tg mice. No pathological changes were observed in the non-Tg control littermates during these experiments.
FIG. 7.
Development of an AIDS-like disease in CD4C/HIVMutA Tg mice bred on a Hck null background. (A) Cumulative incidence of mortality of hck−/− and hck+/− Tg mice. CD4C/HIVMutA Tg mice were backcrossed to hck gene deficient (hck−/−) mice and observed for a period of up to 12 months. The cumulative incidence of mortality of hck−/− (n = 17) and hck+/− (n = 20) CD4C/HIVMutA Tg mice as well as hck−/− non-Tg mice was plotted. (B) Histopathology in hck−/− Tg mice. Kidney (b), mesenteric LN (d), and thymus (f) from hck−/− Tg mice exhibit typical pathology observed in CD4C/HIV Tg mice (32, 33): interstitial nephritis and glomerulosclerosis, loss of architecture and of lymphoid cells in LN and thymus. (a, c, and e) From corresponding hck−/− non-Tg mice. Scale bars in these panels are valid for their corresponding tissues in panels b, d, and f. Paraffin sections counterstained with hematoxylin and eosin are shown.
FACS analysis of lymphoid populations of these mice revealed a severe loss of CD4+ T cells in hck−/− and hck+/− Tg mice compared to hck−/− or hck+/− non-Tg littermate controls (Fig. 8). Interestingly, in spite of a delay in progression of the disease in hck−/− Tg mice, the extent of the CD4+ T cell loss was similar in Tg mice on hck−/− or hck+/− background. In addition, CD4+ T cells isolated from peripheral LN of hck−/− and hck+/− Tg+ mice revealed a limited capacity to divide in response to in vitro stimulation with anti-CD3 plus anti-CD28 antibodies over a 3-day period (Fig. 9), similar to that observed in wild-type hck+/+ Tg mice (data not shown).
FIG. 8.
FACS analysis of splenocytes from CD4C/HIVMutA Tg mice bred on an hck null background. Total splenocytes were incubated with hemolytic Gey's solution to remove red blood cells. Flow cytometric analyses were based on two-color staining with CD4-PE–TcR-FITC and CD8-PE–TcR-FITC MAb. Quantitation of CD4+ TcR+ or CD8+ TcR+ double positive subpopulations from a live gate are shown in percentage and mean fluorescence intensity (in parentheses). The results are representative of at least three experiments. Note that the CD4 downregulation is not as easily seen in the older Tg animals, which have lost most of their CD4+ T cells, as it is in younger Tg mice expressing wild-type Nef (Fig. 3C).
Together, these results support a partial protection of these Tg mice from the AIDS-like disease in absence of Hck, and emphasize the relative importance of the Nef-Hck interaction in the pathogenesis of AIDS. Importantly, these data also clearly indicate that Hck is not essential for disease development.
DISCUSSION
We have previously reported the development and characterization of a Tg mouse model of AIDS, in which Nef is the principal determinant of AIDS pathogenesis (32, 33). To identify the viral determinants and cellular effectors that contribute to the induction of an AIDS-like disease in these mice, we have started breeding Tg mice on various knockout backgrounds and we have initiated a structure-function study by generating Tg mice expressing a panel of Nef mutant alleles. We report here our in vivo studies on one of these mutants, in the proline-rich P72XXP75 motif of Nef, and on the effect of deleting Hck in these mice.
Tg mice expressing a P72XXP75XXP78 mutated Nef do not develop an AIDS-like disease.
In contrast to the CD4C/HIVMutG Tg mice which express only the wild-type nef gene, and develop a severe AIDS-like disease (33), Tg mice expressing Nef mutated in the SH3-binding domain on an isogenic transgene (CD4C/HIVMutG(AXXA)) fail to develop any disease, except for a partial downregulation of CD4 cell surface expression. Despite a lower level of CD4 molecules at the cell surface, these CD4+ T cells can respond normally to anti-CD3 and anti-CD28 engagements. These results in Tg mice provide strong evidence for a role of this motif, independent of viral replication, in the pathogenesis of AIDS.
It is unlikely that disease development is prevented in these mice as a consequence of a change in cell-specificity of transgene expression for the following reasons. (i) The same regulatory elements (CD4C) were used to express the mutated Nef as the one used to express the wild-type Nef (32, 33), and only three point mutations in the proline-rich motif of Nef along the 24-kbp sequences distinguish CD4C/HIVMutG and CD4C/HIVMutG(AXXA) transgenes; (ii) the same pattern of expression was detected by Northern blot and by in situ hybridization in tissues of CD4C/HIVMutG and CD4C/HIVMutG(AXXA) Tg mice (Fig. 2A and B); (iii) and finally, Tg mice from all the 19 independent lines generated, harboring the CD4C/HIV transgene and expressing the wild-type NL4-3 Nef allele, have developed the typical AIDS-like disease of the CD4C/HIVMutG Tg mice. Taken together, these observations indicate that these CD4C regulatory sequences are able to reproducibly express the reporter HIV-1 genes in the same target cells relevant for disease induction. Therefore, the absence of disease development in CD4C/HIVMutG(AXXA) Tg mice is most likely related to the Nef mutation itself.
Our findings that this mutated Nef allele still induces a partial downregulation of CD4 expression are consistent with previous in vitro studies showing that the proline-rich motif of Nef is largely dispensable for CD4 downregulation (1, 18, 26, 38, 56, 67, 68). Although some studies have shown that mutation of this P72XXP75 has a subtle effect on the CD4 downregulation detectable only at low levels of Nef expression (18), such phenotype was not observed in our CD4C/HIVMutG(AXXA) Tg mice from founders producing different levels of Nef protein: these Tg mice exhibited an equivalent percentage of CD4 downregulation. However, the percentage of CD4 downregulation was lower in Tg mice expressing the mutant Nef than in Tg mice expressing the wild-type Nef. This finding suggests that, in vivo, interaction of Nef with some protein(s) requires an intact proline-rich motif for achieving optimal CD4 downregulation.
The proline-rich P72XXP75 motif of HIV-1 Nef has been found in vitro to be required for interaction with vav (23) and with two important classes of effectors: the Src-related tyrosine kinases (12, 13, 16, 29, 30, 47, 68) and the NNAK, identified as a member of the PAK-family kinases (43, 57, 84). Disruption of the Nef interaction with one or many of these effectors may be responsible for the lack of disease-inducing potential of this mutated Nef protein.
A nef allele mutated in the proline-rich domain has also been studied in another animal model of HIV-1 infection, the SCID-hu mouse (4, 41). In these mice, HIV-1 with the mutated nef gene was found to be as pathogenic as the wild-type HIV-1 strain, in contrast to our results in Tg mice. These contrasting results most likely reflect the fact that each model may score different functions of Nef. It will be important to determine which of these functions reflect best the pathogenesis of Nef in humans. The fact that the CD4C/HIV Tg mice develop a multiorgan disease, with most of the phenotypes associated with human AIDS (32, 33), supports the notion that Nef in these mice is mimicking faithfully its action in human cells.
Since a proline-rich motif similar to the HIV-1 Nef motif exists in SIV Nef, the effect of its mutation has also been studied in vivo and contradictory results have been reported. Lang et al (46) reported that macaques infected with SIV mutated in the PXXP motif progressed to disease before a significant reversion of the mutation occurred in their Nef, strongly suggesting that this motif was dispensable for disease induction. On the other hand, using the same model, Khan et al (43) found that their macaques infected with SIV harboring a Nef mutated in the proline-rich domain did not develop disease, unless revertants of the mutated motif occurred, thus indicating a strong selective pressure to restore this binding site and suggesting an important role for this motif in the development of SAIDS. However, in other in vivo studies with SIV containing a large Nef deletion, the same group has recovered from diseased macaques pathogenic viruses, which contained a novel truncated Nef protein, tNef, deleted among other motifs of PXXP, indicating that this motif is not essential for the pathogenicity of SIV in vivo (73). The reasons for the discrepancies between these studies are not obvious. Nor is it clear to what extent findings with SIV Nef will reflect those with HIV-1 Nef. Although HIV-1 and SIV Nef appear to have the same function in several assays, others reveal differences and these molecules have been found to interact with different effectors (66).
Therefore, the contrasting results obtained with the proline-rich mutant of HIV-1 Nef in Tg mice and of SIV in infected macaques may reflect inherent differences in these two molecules, rather than differences in experimental designs.
A role for Hck, which binds to the HIV-1 P72XXP75 motif, in the progression of an AIDS-like disease in CD4C/HIV Tg mice.
The P72XXP75 motif of HIV-1 Nef has been shown to interact with, among other molecules, the Src-related tyrosine kinases. Among them, Hck and Lyn have the highest binding affinity for Nef (6, 47). As a result of this interaction, Hck kinase activity is enhanced (12, 30, 61). In addition, transformation of fibroblasts in vitro could be demonstrated when Nef and Hck, but not Lyn, were coexpressed (12). These studies have made Hck an attractive candidate as an effector of Nef-induced pathogenesis of AIDS.
However, the significance and relative importance of this Nef-Hck interaction in vivo have remained unclear. Our experiment with Nef expressing Tg mice bred on an hck deficient background addresses this issue. We found that the absence of Hck has no effect on the pathological phenotypes observed in Nef Tg mice, indicating that Hck is dispensable for the development of the AIDS-like disease in Tg mice. Interestingly, however, disease progression was delayed in these hck knockout HIV-1 Tg mice, suggesting that it is the expression of Nef in Hck-expressing cells (monocytes, macrophages and possibly myeloid DC), but not that in T cells (which do not express Hck), that determines the rate of disease progression in these Tg mice, at least to a certain extent. Indeed, the CD4C regulatory elements of the transgene allow expression of HIV-1 in not only CD4+ T cells, but also in cells of the macrophage-dendritic lineage (Fig. 3F) (32–34), thus allowing interaction of Nef with its effectors in these latter cells. The fact that CD4+ T cells were lost to the same extent and at similar rate in hck−/− and hck+/− Tg mice, is consistent with the absence of Hck in T cells. At least two mechanisms may explain the effect of Hck deficiency on disease progression. The metabolism of some of the cell populations normally expressing Hck may be compromised in such a way that Nef signaling will be impaired. Alternatively, a lack of direct interaction between Nef and Hck may be the major molecular defect underlying this phenotype. Our data do not allow us to distinguish between these two possibilities. However, given that Nef-Hck interaction has been well documented, it is tempting to believe that the absence of the Hck partner to bind Nef is an important event in retarding disease progression. The fact that disease progression is only delayed but not totally abrogated in absence of Hck may result from redundancy, a feature inherent to this biological system: Hck may be compensated for by another Src-related kinase. An increase of the specific activity of the Lyn kinase has been detected in hck−/− macrophages (51). A Nef-Lyn interaction in vivo could partially replace the Nef-Hck interaction, since Lyn has also been reported to bind to Nef in vitro (13, 68). Nef may also be able to interact with Fgr in vivo. The hck−/− fgr−/− double mutant mice have been found to be more susceptible to Listeria monocytogenes infection (51) and more resistant to endotoxic shock (50) than hck−/− or fgr−/− single mutant mice, revealing a redundancy of the functions of these molecules in vivo.
Despite these limitations of the model, these studies have enabled us to begin to understand the role of the Nef-Hck interaction in disease progression. Since this interaction is likely to occur in cells of the macrophage-dendritic lineage, this result suggests that one or both of these cell populations are involved in disease progression. The number of cells of these populations may be decreased and/or these cell populations may be abnormally distributed and/or may exhibit altered functions (e.g., secrete altered levels of cytokines or chemokines) which could have a severe impact on CD4+ T-cell function and survival. Further work with hck lyn fgr double or triple mutants (24) should help in determining whether these other Src-related kinases allow to compensate for loss of Hck or whether other non-Src-related kinases are involved in this process.
ACKNOWLEDGMENTS
Z.H. and X.W. contributed equally to this work.
This work was supported by grants from the Medical Research Council of Canada to Z.H. and P.J. We thank Ginette Masse, Karina Lamarre, Benoit Laganiere, Michel Robillard, Chunyan Hu, Viorica Lascau, and Patrick Couture for their excellent technical assistance and Nathalie Tessier for her valuable help in FACS analysis. We thank P. Hugo and R. Coffman for providing reagents. We are grateful to Rita Gingras for typing the manuscript.
REFERENCES
- 1.Aiken C, Krause L, Chen Y L, Trono D. Mutational analysis of HIV-1 Nef: identification of two mutants that are temperature-sensitive for CD4 downregulation. Virology. 1996;217:293–300. doi: 10.1006/viro.1996.0116. [DOI] [PubMed] [Google Scholar]
- 2.Aiken C, Trono D. Nef stimulates human immunodeficiency virus type 1 proviral DNA synthesis. J Virol. 1995;69:5048–5056. doi: 10.1128/jvi.69.8.5048-5056.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aldrovandi G M, Gao L, Bristol G, Zack J A. Regions of human immunodeficiency virus type 1 nef required for function in vivo. J Virol. 1998;72:7032–7039. doi: 10.1128/jvi.72.9.7032-7039.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aldrovandi G M, Zack J A. Replication and pathogenicity of human immunodeficiency virus type 1 accessory gene mutants in scid-hu mice. J Virol. 1996;70:1505–1511. doi: 10.1128/jvi.70.3.1505-1511.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alexander L, Weiskopf E, Greenough T C, Gaddis N C, Auerbach M R, Malim M H, O'Brien S J, Walker B D, Sullivan J L, Desrosiers R C. Unusual polymorphisms in human immunodeficiency virus type 1 associated with nonprogressive infection. J Virol. 2000;74:4361–4376. doi: 10.1128/jvi.74.9.4361-4376.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arold S, O'Brien R, Franken P, Strub M P, Hoh F, Dumas C, Ladbury J E. RT loop flexibility enhances the specificity of Src family SH3 domains for HIV-1 Nef. Biochemistry. 1998;37:14683–14691. doi: 10.1021/bi980989q. [DOI] [PubMed] [Google Scholar]
- 7.Baba T W, Liska V, Khimani A H, Ray N B, Dailey P J, Penninck D, Bronson R, Greene M F, McClure H M, Martin L N, Ruprecht R M. Live attenuated, multiply deleted simian immunodeficiency virus causes AIDS in infant and adult macaques. Nat Med. 1999;5:194–203. doi: 10.1038/5557. [DOI] [PubMed] [Google Scholar]
- 8.Baur A S, Sass G, Laffert B, Willbold D, Cheng-Mayer C, Peterlin B M. The N terminus of Nef from HIV-1/SIV associates with a protein complex containing Lck and a serine kinase. Immunity. 1997;6:283–291. doi: 10.1016/s1074-7613(00)80331-3. [DOI] [PubMed] [Google Scholar]
- 9.Baur A S, Sawai E T, Dazin P, Fantl W J, Cheng-Mayer C, Peterlin B M. HIV-1 Nef leads to inhibition or activation of T cells depending on its intracellular localization. Immunity. 1994;1:373–384. doi: 10.1016/1074-7613(94)90068-x. [DOI] [PubMed] [Google Scholar]
- 10.Blumberg B, Epstein L, de Ronde A, Goudsmit J. HIV1 3′ ORF is open in pathological tissue and accelerates viral replication in primary lymphocytes. Res Virol. 1992;143:63–65. doi: 10.1016/s0923-2516(06)80083-x. [DOI] [PubMed] [Google Scholar]
- 11.Brady H J, Pennington D J, Miles C G, Dzierzak E A. CD4 cell surface downregulation in HIV-1 Nef transgenic mice is a consequence of intracellular sequestration. EMBO J. 1993;12:4923–4932. doi: 10.1002/j.1460-2075.1993.tb06186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Briggs S D, Sharkey M, Stevenson M, Smithgall T E. SH3-mediated Hck tyrosine kinase activation and fibroblast transformation by the Nef protein of HIV-1. J Biol Chem. 1997;272:17899–17902. doi: 10.1074/jbc.272.29.17899. [DOI] [PubMed] [Google Scholar]
- 13.Cheng H, Hoxie J P, Parks W P. The conserved core of human immunodeficiency virus type 1 Nef is essential for association with Lck and for enhanced viral replication in T-lymphocytes. Virology. 1999;264:5–15. doi: 10.1006/viro.1999.9937. [DOI] [PubMed] [Google Scholar]
- 14.Chowers M Y, Spina C A, Kwoh T J, Fitch N J, Richman D, Guatelli J C. Optimal infectivity in vitro of human immunodeficiency virus type 1 requires an intact nef gene. J Virol. 1994;68:2906–2914. doi: 10.1128/jvi.68.5.2906-2914.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collette Y, Arold S, Picard C, Janvier K, Benichou S, Benarous R, Olive D, Dumas C. HIV-2 and SIV nef proteins target different Src family SH3 domains than does HIV-1 Nef because of a triple amino acid substitution. J Biol Chem. 2000;275:4171–4176. doi: 10.1074/jbc.275.6.4171. [DOI] [PubMed] [Google Scholar]
- 16.Collette Y, Dutartre H, Benziane A, Ramos-Morales, Benarous R, Harris M, Olive D. Physical and functional interaction of Nef with Lck. HIV-1 Nef-induced T-cell signaling defects. J Biol Chem. 1996;271:6333–6341. doi: 10.1074/jbc.271.11.6333. [DOI] [PubMed] [Google Scholar]
- 17.Collins K L, Chen B K, Kalams S A, Walker B D, Baltimore D. HIV-1 Nef protein protects infected primary cells against killing by cytotoxic T lymphocytes. Nature. 1998;391:397–401. doi: 10.1038/34929. [DOI] [PubMed] [Google Scholar]
- 18.Craig H M, Pandori M W, Riggs N L, Richman D D, Guatelli J C. Analysis of the SH3-binding region of HIV-1 nef: partial functional defects introduced by mutations in the polyproline helix and the hydrophobic pocket. Virology. 1999;262:55–63. doi: 10.1006/viro.1999.9897. [DOI] [PubMed] [Google Scholar]
- 19.Cullen B R. The role of Nef in the replication cycle of the human and simian immunodeficiency viruses. Virology. 1994;205:1–6. doi: 10.1006/viro.1994.1613. [DOI] [PubMed] [Google Scholar]
- 20.Cullen B R. HIV-1: is Nef a PAK animal? Curr Biol. 1996;6:1557–1559. doi: 10.1016/s0960-9822(02)70770-7. [DOI] [PubMed] [Google Scholar]
- 21.Deacon N J, Tsykin A, Solomon A, Smith K, Ludford-Menting M, Hooker D J, McPhee D A, Greenway A L, Ellett A, Chatfield C, et al. Genomic structure of an attenuated quasi species of hiv-1 from a blood transfusion donor and recipients. Science. 1995;270:988–991. doi: 10.1126/science.270.5238.988. [DOI] [PubMed] [Google Scholar]
- 22.Dutartre H, Harris M, Olive D, Collette Y. The human immunodeficiency virus type 1 Nef protein binds the Src-related tyrosine kinase Lck SH2 domain through a novel phosphotyrosine independent mechanism. Virology. 1998;247:200–211. doi: 10.1006/viro.1998.9244. [DOI] [PubMed] [Google Scholar]
- 23.Fackler O T, Luo W, Geyer M, Alberts A S, Peterlin B M. Activation of Vav by Nef induces cytoskeletal rearrangements and downstream effector functions. Mol Cell. 1999;3:729–739. doi: 10.1016/s1097-2765(01)80005-8. [DOI] [PubMed] [Google Scholar]
- 24.Fitzer-Attas C J, Lowry M, Crowley M T, Finn A J, Meng F, DeFranco A L, Lowell C A. Fcgamma receptor-mediated phagocytosis in macrophages lacking the Src family tyrosine kinases Hck, Fgr, and Lyn. J Exp Med. 2000;191:669–682. doi: 10.1084/jem.191.4.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foti M, Cartier L, Piguet V, Lew D P, Carpentier J L, Trono D, Krause K H. The HIV Nef protein alters Ca(2+) signaling in myelomonocytic cells through SH3-mediated protein-protein interactions. J Biol Chem. 1999;274:34765–34772. doi: 10.1074/jbc.274.49.34765. [DOI] [PubMed] [Google Scholar]
- 26.Goldsmith M A, Warmerdam M T, Atchison R E, Miller M D, Greene W C. Dissociation of the CD4 downregulation and viral infectivity enhancement functions of human immunodeficiency virus type 1 Nef. J Virol. 1995;69:4112–4121. doi: 10.1128/jvi.69.7.4112-4121.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greenberg M E, Iafrate A J, Skowronski J. The SH3 domain-binding surface and an acidic motif in HIV-1 Nef regulate trafficking of class I MHC complexes. EMBO J. 1998;17:2777–2789. doi: 10.1093/emboj/17.10.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greenway A, Azad A, McPhee D. Human immunodeficiency virus type 1 Nef protein inhibits activation pathways in peripheral blood mononuclear cells and T-cell lines. J Virol. 1995;69:1842–1850. doi: 10.1128/jvi.69.3.1842-1850.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greenway A, Azad A, Mills J, McPhee D. Human immunodeficiency virus type 1 Nef binds directly to Lck and mitogen-activated protein kinase, inhibiting kinase activity. J Virol. 1996;70:6701–6708. doi: 10.1128/jvi.70.10.6701-6708.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greenway A L, Dutartre H, Allen K, McPhee D A, Olive D, Collette Y. Simian immunodeficiency virus and human immunodeficiency virus type 1 nef proteins show distinct patterns and mechanisms of Src kinase activation. J Virol. 1999;73:6152–6158. doi: 10.1128/jvi.73.7.6152-6158.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guatelli J C. The positive influence of Nef on viral infectivity. Res Virol. 1997;148:34–37. doi: 10.1016/s0923-2516(97)81910-3. [DOI] [PubMed] [Google Scholar]
- 32.Hanna Z, Kay D G, Cool M, Jothy S, Rebai N, Jolicoeur P. Transgenic mice expressing human immunodeficiency virus type 1 in immune cells develop a severe AIDS-like disease. J Virol. 1998;72:121–132. doi: 10.1128/jvi.72.1.121-132.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hanna Z, Kay D G, Rebai N, Guimond A, Jothy S, Jolicoeur P. Nef harbors a major determinant of pathogenicity for an AIDS-like disease induced by HIV-1 in transgenic mice. Cell. 1998;95:163–175. doi: 10.1016/s0092-8674(00)81748-1. [DOI] [PubMed] [Google Scholar]
- 34.Hanna Z, Simard C, Jolicoeur P. Specific expression of the human CD4 gene in mature CD4+ CD8− and immature CD4+ CD8+ T cells, and in macrophages of transgenic mice. Mol Cell Biol. 1994;14:1084–1094. doi: 10.1128/mcb.14.2.1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harris M. From negative factor to a critical role in virus pathogenesis: the changing fortunes of Nef. J Gen Virol. 1996;77:2379–2392. doi: 10.1099/0022-1317-77-10-2379. [DOI] [PubMed] [Google Scholar]
- 36.Harris M. HIV: a new role for Nef in the spread of HIV. Curr Biol. 1999;9:R459–R461. doi: 10.1016/s0960-9822(99)80282-6. [DOI] [PubMed] [Google Scholar]
- 37.Hodge D R, Dunn K J, Pei G K, Chakrabarty M K, Heidecker G, Lautenberger J A, Samuel K P. Binding of c-Raf1 kinase to a conserved acidic sequence within the carboxyl-terminal region of the HIV-1 Nef protein. J Biol Chem. 1998;273:15727–15733. doi: 10.1074/jbc.273.25.15727. [DOI] [PubMed] [Google Scholar]
- 38.Hua J, Blair W, Truant R, Cullen B R. Identification of regions in HIV-1 Nef required for efficient downregulation of cell surface CD4. Virology. 1997;231:231–238. doi: 10.1006/viro.1997.8517. [DOI] [PubMed] [Google Scholar]
- 39.Iafrate A J, Bronson S, Skowronski J. Separable functions of Nef disrupt two aspects of T cell receptor machinery: CD4 expression and CD3 signaling. EMBO J. 1997;16:673–684. doi: 10.1093/emboj/16.4.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jamieson B D, Aldrovandi G M, Planelles V, Jowett J B, Gao L, Bloch L M C I, Zack J A. Requirement of human immunodeficiency virus type 1 nef for in vivo replication and pathogenicity. J Virol. 1994;68:3478–3485. doi: 10.1128/jvi.68.6.3478-3485.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawano Y, Tanaka Y, Misawa N, Tanaka R, Kira J I, Kimura T, Fukushi M, Sano K, Goto T, Nakai M, Kobayashi T, Yamamoto N, Koyanagi Y. Mutational analysis of human immunodeficiency virus type 1 (HIV-1) accessory genes: requirement of a site in the nef gene for HIV-1 replication in activated CD4+ T cells in vitro and in vivo. J Virol. 1997;71:8456–8466. doi: 10.1128/jvi.71.11.8456-8466.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kestler H W, Ringler D J, Mori K, Panicali D L, Sehgal P K, Daniel M D, Desrosiers R C. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell. 1991;65:651–662. doi: 10.1016/0092-8674(91)90097-i. [DOI] [PubMed] [Google Scholar]
- 43.Khan I H, Sawai E T, Antonio E, Weber C J, Mandell C P, Montbriand P, Luciw P A. Role of the SH3-ligand domain of simian immunodeficiency virus Nef in interaction with Nef-associated kinase and simian AIDS in rhesus macaques. J Virol. 1998;72:5820–5830. doi: 10.1128/jvi.72.7.5820-5830.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kirchhoff F, Greenough T C, Brettler D B, Sullivan J L, Desrosiers R C. Brief report: absence of intact nef sequences in a long-term survivor with nonprogressive HIV-1 infection. N Engl J Med. 1995;332:228–232. doi: 10.1056/NEJM199501263320405. [DOI] [PubMed] [Google Scholar]
- 45.Lama J, Mangasarian A, Trono D. Cell-surface expression of CD4 reduces HIV-1 infectivity by blocking Env incorporation in a Nef- and Vpu-inhibitable manner. Curr Biol. 1999;9:622–631. doi: 10.1016/s0960-9822(99)80284-x. [DOI] [PubMed] [Google Scholar]
- 46.Lang S M, Iafrate A J, Stahl-Hennig C, Kuhn E M, Nisslein T, Kaup F J, Haupt M, Hunsmann G, Skowronski J, Kirchhoff F. Association of simian immunodeficiency virus Nef with cellular serine/threonine kinases is dispensable for the development of AIDS in rhesus macaques. Nat Med. 1997;3:860–865. doi: 10.1038/nm0897-860. [DOI] [PubMed] [Google Scholar]
- 47.Lee C H, Leung B, Lemmon M A, Zheng J, Cowburn D, Kuriyan J, Saksela K. A single amino acid in the sh3 domain of hck determines its high affinity and specificity in binding to hiv-1 nef protein. EMBO J. 1995;14:5006–5015. doi: 10.1002/j.1460-2075.1995.tb00183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Le Gall S, Heard J M, Schwartz O. Analysis of Nef-induced MHC-I endocytosis. Res Virol. 1997;148:43–47. doi: 10.1016/s0923-2516(97)81912-7. [DOI] [PubMed] [Google Scholar]
- 49.Lindemann D, Wilhelm R, Renard P, Althage A, Zinkernagel R, Mous J. Severe immunodeficiency associated with a human immunodeficiency virus 1 NEF/3′-long terminal repeat transgene. J Exp Med. 1994;179:797–807. doi: 10.1084/jem.179.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lowell C A, Berton G. Resistance to endotoxic shock and reduced neutrophil migration in mice deficient for the Src-family kinases Hck and Fgr. Proc Natl Acad Sci USA. 1998;95:7580–7584. doi: 10.1073/pnas.95.13.7580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lowell C A, Soriano P, Varmus H E. Functional overlap in the src gene family: inactivation of hck and fgr impairs natural immunity. Genes Dev. 1994;8:387–398. doi: 10.1101/gad.8.4.387. [DOI] [PubMed] [Google Scholar]
- 52.Lu X, Wu X, Plemenitas A, Yu H, Sawai E T, Abo A, Peterlin B M. CDC42 and Rac1 are implicated in the activation of the Nef-associated kinase and replication of HIV-1. Curr Biol. 1996;6:1677–1684. doi: 10.1016/s0960-9822(02)70792-6. [DOI] [PubMed] [Google Scholar]
- 53.Luo T, Garcia J V. The association of Nef with a cellular serine/threonine kinase and its enhancement of infectivity are viral isolate dependent. J Virol. 1996;70:6493–6496. doi: 10.1128/jvi.70.9.6493-6496.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luria S, Chambers I, Berg P. Expression of the type 1 human immunodeficiency virus Nef protein in T cells prevents antigen receptor-mediated induction of interleukin 2 mRNA. Proc Natl Acad Sci USA. 1991;88:5326–5330. doi: 10.1073/pnas.88.12.5326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lyons A B, Parish C R. Determination of lymphocyte division by flow cytometry. J Immunol Methods. 1994;171:131–137. doi: 10.1016/0022-1759(94)90236-4. [DOI] [PubMed] [Google Scholar]
- 56.Mangasarian A, Piguet V, Wang J K, Chen Y L, Trono D. Nef-induced CD4 and major histocompatibility complex class I (MHC-I) down-regulation are governed by distinct determinants: N-terminal alpha helix and proline repeat of Nef selectively regulate MHC-I trafficking. J Virol. 1999;73:1964–1973. doi: 10.1128/jvi.73.3.1964-1973.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Manninen A, Hiipakka M, Vihinen M, Lu W, Mayer B J, Saksela K. SH3-Domain binding function of HIV-1 Nef is required for association with a PAK-related kinase. Virology. 1998;250:273–282. doi: 10.1006/viro.1998.9381. [DOI] [PubMed] [Google Scholar]
- 58.Mariani R, Kirchhoff F, Greenough T C, Sullivan J L, Desrosiers R C, Skowronski J. High frequency of defective nef alleles in a long-term survivor with nonprogressive human immunodeficiency virus type 1 infection. J Virol. 1996;70:7752–7764. doi: 10.1128/jvi.70.11.7752-7764.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miller M D, Warmerdam M T, Gaston I, Greene W C, Feinberg M B. The human immunodeficiency virus-1 nef gene product: a positive factor for viral infection and replication in primary lymphocytes and macrophages. J Exp Med. 1994;179:101–113. doi: 10.1084/jem.179.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miller M D, Warmerdam M T, Page K A, Feinberg M B, Greene W C. Expression of the human immunodeficiency virus type 1 (HIV-1) nef gene during HIV-1 production increases progeny particle infectivity independently of gp160 or viral entry. J Virol. 1995;69:579–584. doi: 10.1128/jvi.69.1.579-584.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moarefi I, LaFevre-Bernt M, Sicheri F, Huse M, Lee C H, Kuriyan J, Miller W T. Activation of the Src-family tyrosine kinase Hck by SH3 domain displacement. Nature. 1997;385:650–653. doi: 10.1038/385650a0. [DOI] [PubMed] [Google Scholar]
- 62.Niederman T M, Garcia J V, Hastings W R, Luria S, Ratner L. Human immunodeficiency virus type 1 Nef protein inhibits NF-kappa B induction in human T cells. J Virol. 1992;66:6213–6219. doi: 10.1128/jvi.66.10.6213-6219.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nunn M F, Marsh J W. Human immunodeficiency virus type 1 Nef associates with a member of the p21-activated kinase family. J Virol. 1996;70:6157–6161. doi: 10.1128/jvi.70.9.6157-6161.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Peter F. HIV nef: the mother of all evil? Immunity. 1998;9:433–437. doi: 10.1016/s1074-7613(00)80626-3. [DOI] [PubMed] [Google Scholar]
- 65.Piguet V, Schwartz O, Le Gall S, Trono D. The downregulation of CD4 and MHC-I by primate lentiviruses: a paradigm for the modulation of cell surface receptors. Immunol Rev. 1999;168:51–63. doi: 10.1111/j.1600-065x.1999.tb01282.x. [DOI] [PubMed] [Google Scholar]
- 66.Renkema G H, Saksela K. Interactions of HIV-1 nef with cellular signal transducing proteins. Frontiers Biosci. 2000;5:d268–d283. doi: 10.2741/renkema. [DOI] [PubMed] [Google Scholar]
- 67.Ross T M, Oran A E, Cullen B R. Inhibition of HIV-1 progeny virion release by cell-surface CD4 is relieved by expression of the viral Nef protein. Curr Biol. 1999;9:613–621. doi: 10.1016/s0960-9822(99)80283-8. [DOI] [PubMed] [Google Scholar]
- 68.Saksela K, Cheng G, Baltimore D. Proline-rich (PXXP) motifs in HIV-1 Nef bind to SH3 domains of a subset of Src kinases and are required for the enhanced growth of Nef+ viruses but not for down-regulation of CD4. EMBO J. 1995;14:484–491. doi: 10.1002/j.1460-2075.1995.tb07024.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Salghetti S, Mariani R, Skowronski J. Human immunodeficiency virus type 1 Nef and p56lck protein-tyrosine kinase interact with a common element in CD4 cytoplasmic tail. Proc Natl Acad Sci USA. 1995;92:349–353. doi: 10.1073/pnas.92.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Salvi R, Garbuglia A R, Di Caro A, Pulciani S, Montella F, Benedetto A. Grossly defective nef gene sequences in a human immunodeficiency virus type 1-seropositive long-term nonprogressor. J Virol. 1998;72:3646–3657. doi: 10.1128/jvi.72.5.3646-3657.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sawai E T, Baur A, Struble H, Peterlin B M, Levy J A, Cheng-Mayer C. Human immunodeficiency virus type 1 Nef associates with a cellular serine kinase in T lymphocytes. Proc Natl Acad Sci USA. 1994;91:1539–1543. doi: 10.1073/pnas.91.4.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sawai E T, Cheng-Mayer C, Luciw P A. Nef and the Nef-associated kinase. Res Virol. 1997;148:47–52. doi: 10.1016/s0923-2516(97)81913-9. [DOI] [PubMed] [Google Scholar]
- 73.Sawai E T, Hamza M S, Ye M, Shaw K E, Luciw P A. Pathogenic conversion of live attenuated simian immunodeficiency virus vaccines is associated with expression of truncated Nef. J Virol. 2000;74:2038–2045. doi: 10.1128/jvi.74.4.2038-2045.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sawai E T, Khan I H, Montbriand P M, Peterlin B M, Cheng-Mayer C, Luciw P A. Activation of PAK by HIV and SIV Nef: importance for AIDS in rhesus macaques. Curr Biol. 1996;6:1519–1527. doi: 10.1016/s0960-9822(96)00757-9. [DOI] [PubMed] [Google Scholar]
- 75.Schwartz O, Marechal V, Danos O, Heard J M. Human immunodeficiency virus type 1 Nef increases the efficiency of reverse transcription in the infected cell. J Virol. 1995;69:4053–4059. doi: 10.1128/jvi.69.7.4053-4059.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schwartz O, Marechal V, Le Gall S, Lemonnier F, Heard J M. Endocytosis of major histocompatibility complex class i molecules is induced by the hiv-1 nef protein. Nat Med. 1996;2:338–342. doi: 10.1038/nm0396-338. [DOI] [PubMed] [Google Scholar]
- 77.Skowronski J, Greenberg M E, Lock M, Mariani R, Salghetti S, Swigut T, Iafrate A J. HIV and SIV Nef modulate signal transduction and protein sorting in T cells. Cold Spring Harbor Symp Quant Biol. 1999;64:453–463. doi: 10.1101/sqb.1999.64.453. [DOI] [PubMed] [Google Scholar]
- 78.Skowronski J, Parks D, Mariani R. Altered T cell activation and development in transgenic mice expressing the HIV-1 nef gene. EMBO J. 1993;12:703–713. doi: 10.1002/j.1460-2075.1993.tb05704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Smith B L, Krushelnycky B W, Mochly-Rosen D, Berg P. The HIV nef protein associates with protein kinase C theta. J Biol Chem. 1996;271:16753–16757. doi: 10.1074/jbc.271.28.16753. [DOI] [PubMed] [Google Scholar]
- 80.Spina C A, Kwoh T J, Chowers M Y, Guatelli J C, Richman D. The importance of nef in the induction of human immunodeficiency virus type 1 replication from primary quiescent CD4 lymphocytes. J Exp Med. 1994;179:115–123. doi: 10.1084/jem.179.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tokunaga K, Ikuta K, Adachi A, Matsuda M, Kurata T, Kojima A. The cellular kinase binding motifs (PXXP and RR) in human immunodeficiency virus type 1 Nef protein are dispensable for producer-cell-dependent enhancement of viral entry. Virology. 1999;257:285–289. doi: 10.1006/viro.1999.9682. [DOI] [PubMed] [Google Scholar]
- 82.Trono D. HIV accessory proteins: leading roles for the supporting cast. Cell. 1995;82:189–192. doi: 10.1016/0092-8674(95)90306-2. [DOI] [PubMed] [Google Scholar]
- 83.Weston S A, Parish C R. New fluorescent dyes for lymphocyte migration studies. Analysis by flow cytometry and fluorescence microscopy. J Immunol Methods. 1990;133:87–97. doi: 10.1016/0022-1759(90)90322-m. [DOI] [PubMed] [Google Scholar]
- 84.Wiskerchen M, Cheng-Mayer C. HIV-1 Nef association with cellular serine kinase correlates with enhanced virion infectivity and efficient proviral DNA synthesis. Virology. 1996;224:292–301. doi: 10.1006/viro.1996.0531. [DOI] [PubMed] [Google Scholar]
- 85.Wyand M S, Manson K H, Lackner A A, Desrosiers R C. Resistance of neonatal monkeys to live attenuated vaccine strains of simian immunodeficiency virus. Nat Med. 1997;3:32–36. doi: 10.1038/nm0197-32. [DOI] [PubMed] [Google Scholar]