Abstract
A hallmark of addiction is the ability of drugs of abuse to trigger relapse after periods of prolonged abstinence. Here, we describe an epigenetic mechanism whereby chronic cocaine exposure causes lasting chromatin and downstream transcriptional modifications in the nucleus accumbens (NAc), a critical brain region controlling motivation. We link prolonged withdrawal from cocaine to the depletion of the histone variant H2A.Z, coupled with increased genome accessibility and latent priming of gene transcription, in D1 dopamine receptor–expressing medium spiny neurons (D1 MSNs) that relate to aberrant gene expression upon drug relapse. The histone chaperone ANP32E removes H2A.Z from chromatin, and we demonstrate that D1 MSN–selective Anp32e knockdown prevents cocaine-induced H2A.Z depletion and blocks cocaine’s rewarding actions. By contrast, very different effects of cocaine exposure, withdrawal, and relapse were found for D2 MSNs. These findings establish histone variant exchange as an important mechanism and clinical target engaged by drugs of abuse to corrupt brain function and behavior.
ANP32E drives epigenetic priming of D1 MSNs by cocaine.
INTRODUCTION
Chronic cocaine exposure induces lasting alterations in gene regulation in the brain’s motivation and reward circuitry, leading to neuroplastic changes and vulnerability to relapse. Susceptibility to relapse is believed to involve in part stable changes in chromatin in the nucleus accumbens (NAc), a brain region that controls motivated behaviors, that alter transcription during long-term drug withdrawal (1). However, the molecular events that underlie maladaptive gene activity in NAc in cocaine addiction and other substance use disorders remain incompletely understood. Here, we investigated how prolonged withdrawal from chronic exposure to cocaine changes gene regulation and the underlying chromatin landscape in defined cell populations within the NAc and causally linked these changes to cocaine-associated behavioral phenotypes.
The NAc is primarily composed of two generally opposing types of GABAergic projection neurons called medium spiny neurons (MSNs), expressing either D1 dopamine receptors (DRD1s) or D2 dopamine receptors (DRD2s). These distinct cell types exhibit differences in cocaine-related neural activity and effects on behaviors associated with drug exposure: Activation of D1 MSNs promotes reward-associated behaviors, whereas activation of D2 MSNs suppresses them (2–10). Increasing evidence shows that acute cocaine experience induces immediate-early gene expression primarily in D1 MSNs and that the transcription factors cAMP response element–binding protein (CREB) and activator protein-1 (AP-1) drive dopamine-related gene expression programs in the NAc (11–16).
While these data are important, they do not explain the long-lasting nature of cocaine-induced behavioral abnormalities—which have been shown to persist for months after the last cocaine exposure. Here, we show that prolonged drug withdrawal is characterized by latent changes in gene regulation that predominate in D1 MSNs and become apparent upon cocaine reexposure and that this transcriptional “priming” is associated with persistent remodeling of chromatin structure in this cell type. Recent work by our and other laboratories has characterized cocaine-induced changes in histone modifications (17–25), which play critical roles in orchestrating stimulus-related gene activity (17, 25–28) . However, little is known about how epigenetic changes relate to lasting changes in gene regulation in withdrawal and relapse: Prior studies have failed to demonstrate appreciable overlap of any given histone modification to changes in gene expression.
One explanation for such relatively limited overlap is the possibility that the field has been focusing on the wrong histone modifications: studying those histone modifications that are best characterized in other systems (mostly other tissues) as opposed to using an open-ended approach to first identify the histone modifications that are most robustly affected in NAc MSNs in response to cocaine exposure. To overcome this gap in knowledge, we took advantage of unbiased mass spectrometry to characterize in a comprehensive manner withdrawal-associated changes in both histone modifications and the abundance of histone variants, which are emerging as important regulators of gene expression in the adult nervous system (29, 30). Histone variants are non-allelic counterparts of canonical histones that are structurally and functionally distinct, with replication-independent dynamics that are controlled by a network of histone chaperones in postmitotic cells (31).
We demonstrate here that prolonged withdrawal from chronic cocaine causes marked depletion of the histone variant, H2A.Z, in the NAc especially at putative enhancer regions and key neuronal genes that control synaptic plasticity. We show further that genome accessibility is increased prominently at these genes after prolonged withdrawal and linked to aberrant gene expression upon drug relapse, specifically in D1 MSNs. Removal of H2A.Z is dependent on the histone chaperone, acidic leucine-rich nuclear phosphoprotein 32 family member E (ANP32E) (32), and we demonstrate that D1 MSN–selective Anp32e knockdown (KD) prevents cocaine-induced H2A.Z depletion and cocaine-related gene priming and effectively blocks rewarding responses to cocaine. By contrast, chronic cocaine exposure, prolonged withdrawal, and relapse produce very different patterns of transcriptional regulation in D2 MSNs. These findings thus identify circuit-specific epigenetic priming of gene expression in NAc as a critical mechanism of the lasting effects of cocaine on the brain and behavior.
RESULTS
Cocaine challenge following drug withdrawal reveals latent dysregulation of D1 MSN transcription
A principal hallmark of addiction is the ability of drugs of abuse like cocaine to precipitate relapse after periods of prolonged abstinence. An emerging research focus therefore are mechanisms by which neuronal gene regulation in the NAc is primed or desensitized during prolonged withdrawal, leading to aberrant gene expression upon drug relapse. Yet, little is known about how cocaine lastingly affects circuit-specific gene programs in the divergent D1 and D2 MSNs of this brain region. Here, we first compared cell type–specific transcriptional responses to the acute experience of cocaine in drug-naïve mice with the responses elicited by the equivalent dose of cocaine but after 30 days of withdrawal from chronic cocaine exposure (Fig. 1A). To map transcriptional activities in D1 versus D2 MSNs, we used two transgenic mouse lines (Drd1a/Drd2a::EGFP-L10a) that enabled fluorescent-activated nuclei sorting (FANS) of these functionally distinct cell types, combined with RNA sequencing (RNA-seq) (Fig. 1B and fig. S1). Selectivity of this approach was confirmed by signatures of subtype-defining gene transcripts (fig. S2, A to C).
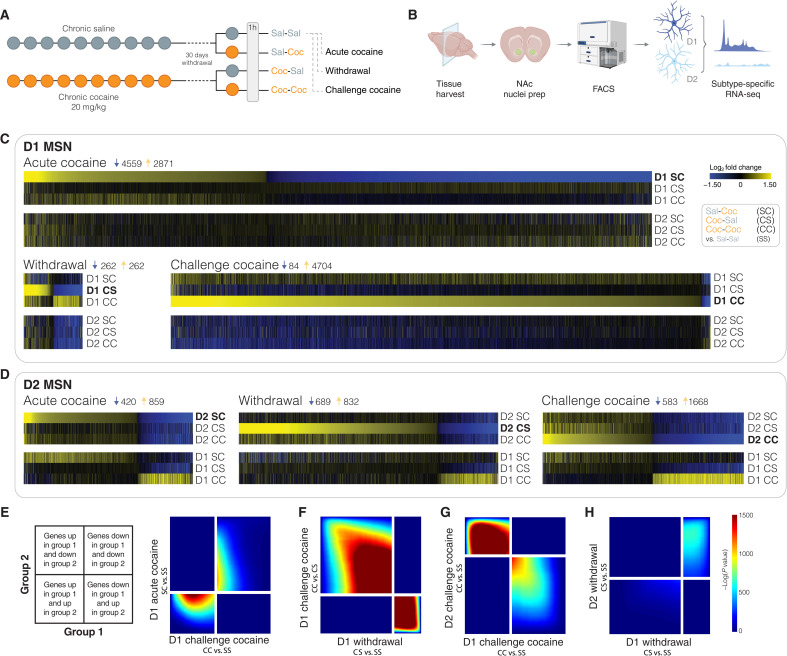
Fig. 1. Latent dysregulation of D1 MSN gene transcription in NAc after prolonged withdrawal from chronic cocaine.
(A) Experimental outline. To characterize cell type–specific changes in gene expression after prolonged cocaine withdrawal, we used two transgenic mouse lines (Drd1a/Drd2a::EGFP-L10a) to purify D1 and D2 MSN nuclei. Following withdrawal (30 days) from chronic saline or cocaine (10 days), animals received either a saline or cocaine (20 mg/kg challenge injection i.p.) to compare the transcriptional response in D1 versus D2 MSNs upon acute cocaine in drug-naïve mice [saline-cocaine (SC)] with that to the same acute cocaine challenge in withdrawal animals [cocaine-cocaine (CC)], and to a saline challenge in withdrawal animals [cocaine-saline (CS)], when compared to control saline animals [saline-saline (SS)]. (B) Nuclei were prepared from NAc of individual mice 1 hour after the last challenge injection, followed by FANS for MSN subtype–specific RNA-seq (six to eight animals per group). FACS, fluorescence-activated cell sorting. (C and D) Heatmaps show genes differentially expressed in (C) D1 MSNs and (D) D2 MSNs after acute cocaine (SC versus SS), withdrawal (CS versus SS), and cocaine challenge (CC versus SS), across all treatment groups, revealing latent changes in gene regulation during withdrawal primarily in D1 MSNs. (E to H) Comparison of transcriptome-wide expression profiles RRHO (key to the left for interpretation), with correction for multiple comparisons using the Benjamini-Hochberg procedure. (E) RRHO comparing transcriptional changes in D1 MSNs after acute cocaine (SC) versus challenge cocaine (CC) confirmed partial directional overlap. (F) Threshold-free comparison of transcriptomes in D1 MSNs indicates that a cocaine challenge (CC) largely reverses changes in gene expression linked to withdrawal (CS). (G) RRHO reveals opposite regulation of gene expression in D2 MSNs compared to D1 MSNs upon a cocaine challenge after withdrawal (CC). (H) RRHO comparing transcriptional changes in D1 and D2 MSNs after cocaine withdrawal (CS) showed no overlap.
We initially evaluated how an acute dose of cocaine in drug-naïve mice (10 daily injections of saline followed by 30 days of withdrawal and then a single cocaine injection, with animals analyzed 1 hour later; termed SC for saline-cocaine) changes the D1 and D2 transcriptomes of the NAc. Compared to D2 MSNs, D1 MSNs showed a pronounced transcriptional response to cocaine involving gene programs related to neural plasticity, such as dendritic spine and postsynaptic density (fig. S3). In animals with a distant history of chronic cocaine, a relapse dose of cocaine after prolonged withdrawal (10 daily cocaine injections with a cocaine challenge injection 30 days later; termed CC or cocaine-cocaine) stimulated gene expression to a substantially greater extent than any other condition, prompting an amplified transcriptional response in D1 MSNs (Fig. 1C). In contrast, cocaine-related gene expression upon relapse remained similar in D2 MSNs when compared to acute cocaine in drug-naïve mice (CC and SC, respectively; Fig. 1D). Notably, withdrawal-related dysregulation of D1 MSN–specific gene expression is latent: The D1 transcriptome remained largely unchanged in withdrawal animals with a priming dose of saline [cocaine-saline (CS)] compared to baseline controls [saline-saline (SS); Fig. 1C]. By contrast, upon a relapse dose of cocaine, many immediate-early genes were primed for rapid induction, and numerous other genes, previously not affected by cocaine in drug-naïve mice, were “primed” during withdrawal and induced by a cocaine challenge (CC; fig. S2, D and E). To evaluate the transcriptome-wide degree of overlap between the differential gene expression profiles upon acute cocaine (SC) versus drug challenge (CC), we performed threshold-free rank-rank hypergeometric overlap (RRHO) analysis. This test showed that the impact of cocaine relapse on the D1 transcriptome, albeit markedly widened and strengthened, corresponded directionally to the effects of its initial acute experience (Fig. 1E). By contrast, withdrawal-related changes in D1 MSN gene expression were largely reversed by cocaine challenge (CS versus CC; Fig. 1F). Compared to D1 MSNs, very different patterns were evident for D2 MSNs, including sets of genes that changed in the opposite direction compared to D1 MSNs (Fig. 1, G and H).
To explore what biological processes and cellular signaling pathways are affected by cocaine relapse (CC), we performed gene ontology (GO) analyses, which revealed that genes stimulated in D1 MSNs not only execute postsynaptic and somatodendritic functions but also have ribosomal and mitochondrial roles (fig. S4A). In contrast, D2 MSNs displayed down-regulation of ribosomal subunit and mitochondrial proteins upon relapse (fig. S4B). Ingenuity Pathway Analysis (IPA) indicates a causal network of CREB1, brain-derived neurotrophic factor, epidermal growth factor, and CAMK1 as upstream regulators, i.e., factors that have been previously implicated in dopamine- and cocaine-related gene transcription within the NAc (table S1) (13, 33–35). Moreover, levodopa and cocaine were predicted as the top upstream chemical activators, highlighting that our subtype-specific transcriptome analyses reflect well-documented regulation of D1 MSNs by cocaine (table S1) (6, 10, 36). Given that these findings reveal latent changes in gene regulation in D1 MSNs after prolonged withdrawal before the relapse dose of cocaine, we set out to explore whether alterations in chromatin structure may underlie this phenomenon.
Chronic cocaine causes lasting changes in D1 MSN chromatin structure
To investigate whether repeated cocaine experience lastingly alters subtype-specific chromatin landscapes, we examined “open” chromatin regions in both D1 and D2 MSN nuclei genome-wide by use of assay for transposase-accessible chromatin with sequencing (ATAC-seq) (Fig. 2A). At baseline (SS), genic chromatin around transcriptional start sites (TSSs) was overall less accessible in D1 MSNs versus D2 MSNs, which is consistent with previous findings of greater D2 transcriptome diversity (37). In contrast, acute cocaine (SC) was linked to a marked genome-wide “opening” of chromatin in D1 MSNs, with a far smaller effect apparent in D2 MSNs. Following chronic cocaine exposure and prolonged withdrawal (CS), D1-specific chromatin opening is remarkably sustained and, upon cocaine relapse (CC), linked to further globally increased accessibility, indicative of transcriptional priming within D1 MSNs (Fig. 2A). By contrast, changes in D2 MSNs were much smaller in magnitude at this genome-wide scale.
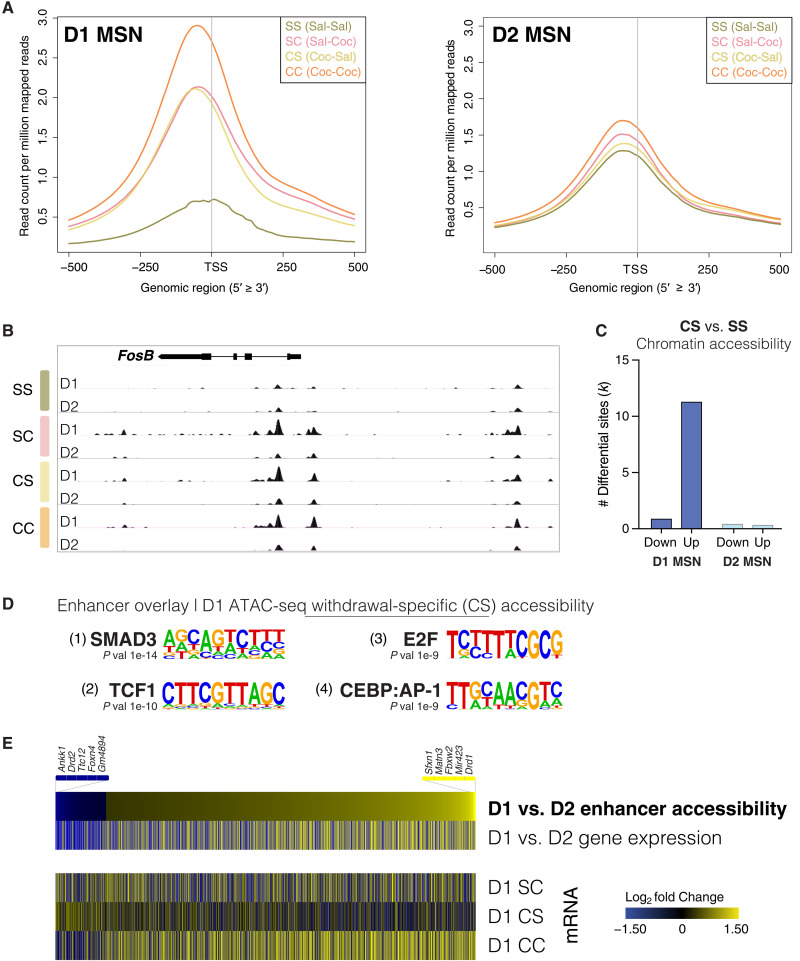
Fig. 2. Withdrawal from chronic cocaine markedly alters chromatin accessibility in D1 MSNs of NAc.
(A) D1 and D2 MSN chromatin accessibility around TSSs was compared across treatment groups using ATAC-seq. SS (control saline), SC (acute cocaine in drug-naïve mice), CS (withdrawal from chronic cocaine), and CC (cocaine challenge following withdrawal from chronic cocaine) (see Fig. 1A). (B) Genome-browser view of the Fosb gene locus showing increased accessibility in D1 MSNs following cocaine exposure, both over its promoter and a putative enhancer region upstream. (C) Differentially accessible genomic loci after prolonged withdrawal from chronic cocaine (CS), showing predominant opening (up) of chromatin in D1 MSNs. (D) Motif analysis on enhancer regions showed increased accessibility specifically in D1 MSNs after withdrawal (Homer de novo motif analysis). (E) Heatmap of differentially accessible enhancers between D1 and D2 MSNs, showing corresponding D1 versus D2 MSN expression of the most proximal genes. The heatmap below shows corresponding gene expression in D1 MSNs with acute cocaine (SC), withdrawal (CS), and cocaine challenge (CC) compared to saline control (SS).
Earlier investigations of drug regulation of gene expression showed that cocaine up-regulates ∆FOSB, encoded by Fosb— a member of the AP-1 family of transcription factors, specifically in D1 MSNs (38, 39). We found that, upon exposure to acute cocaine, both the Fosb core promoter and its upstream enhancer sites displayed increased accessibility in D1 but not D2 MSNs (Fig. 2B). This opening is sustained during prolonged withdrawal and relapse. Fosb is the third-most primed gene in D1 MSNs.
Genome-wide examination of differential gene accessibility following withdrawal confirmed widespread chromatin opening and far fewer sites of decreased accessibility, in D1 MSNs, with a relatively small number of changes observed in D2 MSNs (Fig. 2C and fig. S5). GO analysis revealed that gene cohorts involved in regulating glutamatergic transmission, postsynaptic density assembly, and AMPA receptor activity are more accessible following withdrawal in D1 MSNs compared to drug-naïve mice (table S2). These D1-specific findings corroborate prior studies on whole NAc, which linked changes in AMPA receptors and glutamatergic transmission to cocaine-induced structural plasticity during withdrawal (40–42), along with studies that demonstrate distinct effects of cocaine on glutamatergic function in D1 versus D2 MSNs (8). To uncover more complex regulation and putative transcription factor dynamics under the cocaine treatment paradigms, we performed footprinting analysis using TOBIAS, which implicated the involvement of well-known cocaine-responsive factors that include the AP-1 family members FOS, FOSB, and JUN, as well as previously unknown regulators of cocaine- and withdrawal-responsive gene programs, such as several members of the Kruppel-like factor family of regulatory proteins (table S2). Notably, we found that numerous ionotropic receptors and ion channels, important for neuronal excitability and synaptic plasticity, exhibit transcriptional priming or desensitization following prolonged withdrawal from chronic cocaine, including many glutamate receptor and potassium channel subunits in D1 MSNs (fig. S6).
We next investigated whether the gene priming induced in D1 MSNs involves increased enhancer accessibility following cocaine withdrawal. We developed a machine learning model that was trained on histone mark chromatin immunoprecipitation sequencing (ChIP-seq) data from the ENCODE project. Although enhancers are known to be cell type specific (43), their chromatin signatures are remarkably stable across different cell types, thus enabling their genome-wide discovery from our previously published NAc histone mark ChIP-seq dataset (18). We identified 7796 putative enhancer sites using DeepRegFinder (fig. S7A) (44). In D1 MSNs, these identified enhancers are marked by increased accessibility in withdrawal, potentially priming neuronal gene programs for rapid induction upon cocaine relapse (fig. S7, B and C). To gain insight into the cellular signaling pathways and transcription factors facilitating such latent changes in regulated gene expression, we performed de novo transcription factor motif analysis over enhancer sites that become accessible in withdrawal (Fig. 2D). This approach revealed that the withdrawal-related class of enhancers in D1 MSNs is marked by SMAD3 and early region 2 binding factor (E2F) transcription factor binding elements, as well as by heterodimers of CCAAT enhancer-binding protein (C/EBP) and AP-1, which can recruit coactivators to open chromatin structure (Fig. 2D). Notably, activin-receptor signaling via SMAD3 has been shown to contribute to cocaine-induced transcriptional and morphological plasticity in rat NAc and to be up-regulated following withdrawal from chronic cocaine (24). Further, our previous whole-tissue transcriptome studies across the mouse brain reward circuitry predicted the E2F family of transcription factors as upstream regulators of NAc gene programs whose altered expression upon chronic cocaine or cocaine withdrawal is predictive of addiction-like behavior (18, 45). E2F3a in NAc was directly implicated in controlling transcriptional and behavioral responses to chronic cocaine (46).
To confirm that chromatin remodeling of cocaine-responsive enhancers is linked to gene expression in the NAc, we examined transcriptional activity of nearby genes in both D1 and D2 MSNs. We confirmed that differential enhancer accessibility between these neuronal subtypes is linked to cell type–specific changes in gene expression, with D1-specific enhancer openness linked to increased mRNA expression of the most proximal genes in D1 neurons and vice versa (395 preferentially accessible D2 enhancers and 2878 preferential D1 enhancers in CC; Fig. 2E). Nearby gene expression in D1 neurons was markedly increased following cocaine relapse, compared to respective gene expression induced by acute cocaine or after prolonged withdrawal from chronic cocaine (Fig. 2E). Together, these data indicate that enhancers targeted by SMAD, E2F, and AP-1—all known to interact with the cocaine-responsive chromatin remodeler BRG1 that regulates cocaine-seeking behavior (24)—show increased accessibility in D1 MSNs after prolonged cocaine withdrawal and are linked to priming of neuronal genes for rapid induction upon drug relapse.
Withdrawal from chronic cocaine is associated with marked depletion of H2A.Z
Epigenetic remodeling has emerged as a potent switch for enhancer activity in plasticity-related transcription, with histone protein modifications believed to control enhancer and promoter function in withdrawal and transcription upon drug relapse (1). To systematically characterize global changes in histone modifications across cocaine exposure, withdrawal, and relapse, we took an unbiased approach by using quantitative mass spectrometry on histones purified from NAc nuclei (Fig. 3A and fig. S8A). We calculated the relative abundance of single and combinatorial histone marks on each peptide—as just one example for acetylation and methylation of histone H3 lysine 9 and 14 (H3K9K14), which is reflective of their proportional genomic distribution (fig. S8B). Both acute cocaine and prolonged withdrawal triggered changes in key gene regulatory histone marks, including trimethylation of H3K9, which our laboratory has previously shown to be induced transiently by acute cocaine (17, 23). Beyond histone modifications per se, recent studies implicate dynamic turnover and exchange of histone variants as epigenomic regulators of neural plasticity (47). We observed marked depletion of the replication-independent histone variant, H2A.Z, following withdrawal from chronic cocaine by mass spectrometry (Fig. 3B). H2A.Z, encoded primarily in brain by the H2az1 gene, has multiple roles in transcription during cellular differentiation and development, and stimulus-induced removal of this variant from chromatin was shown to facilitate long-term hippocampal memory, indicating a memory suppressor function in the adult brain (48). Notably, our RNA-seq dataset also revealed significant down-regulation of H2az1 mRNA expression following withdrawal in both D1 and D2 MSNs [D1 CS versus SS log fold change (FC) = −1.3, P = 0.01; D2 CS versus SS log FC = −1.5, P = 0.01; Fig. 3C].
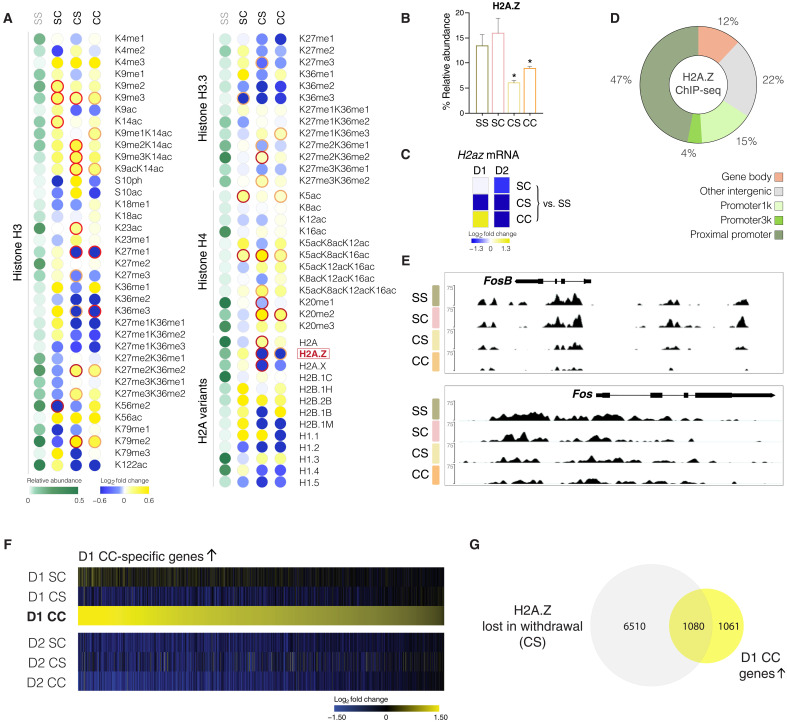
Fig. 3. Mass spectrometry reveals cocaine-induced changes in abundance of posttranslational histone modifications and histone variants in NAc.
(A) Relative abundance of each indicated modification on histone lysine (K) and serine (S) residues, or of each histone variant, is shown in green for saline (SS). Cocaine-induced FCs from SS baseline shown on log2 scale for acute cocaine (SC), withdrawal from chronic cocaine (CS), and cocaine challenge following prolonged withdrawal (CC). Mono/di/trimethylation, me1/2/3; acetylation, ac; ph, phosphorylation; dark red circle, P value < 0.05; light red circle, P value < 0.1. (B) H2A.Z protein abundance in NAc across treatment groups as revealed by histone mass spectrometry. (C) mRNA expression of the H2az1 gene (which encodes H2A.Z) across treatment groups. (D) Genomic distribution of H2A.Z according to ChIP-seq, which indicated primary enrichment of read counts over gene promoters. (E) Genome-browser view of the Fosb (top) and Fos (bottom) gene loci showing H2A.Z ChIP-seq tracks (H2A.Z signal normalized to total H3 ChIP-seq). (F) Heatmap shows MSN subtype–specific mRNA expression of genes that are selectively up-regulated in D1 MSNs with cocaine challenge after prolonged withdrawal from chronic cocaine (CC) and not induced by acute cocaine in drug-naïve mice (SC). (G) Overlap of genes losing H2A.Z following withdrawal from chronic cocaine (CS) and those genes that are sensitized to cocaine challenge in D1 MSNs (shown in Fig. 3F) reveals that roughly half of these relapse-primed genes are marked by H2A.Z loss on withdrawal.
To investigate withdrawal-related changes in H2A.Z abundance genome-wide, we performed ChIP-seq for H2A.Z and, simultaneously, for total histone H3, which was used for chromatin normalization (Fig. 3, D to G). At baseline (SS), genomic H2A.Z binding in the NAc was markedly enriched over promoter elements and flanked TSSs (Fig. 3D), corroborating recent reports for H2A.Z distribution in mouse hippocampus (48). Notably, following prolonged cocaine withdrawal, H2A.Z enrichment was markedly depleted at promoters and NAc-specific enhancer regions, including enhancer elements upstream of Fosb as just one specific example (Fig. 3E, top). In contrast, depletion over Fos was less pronounced (Fig. 3E, bottom).
Loss of H2A.Z was recently shown to increase the accessibility of transcription factor binding sites, predominantly for the AP-1 FOS and JUN families, in cultured human cells (49). These observations suggest that H2A.Z blocks promiscuous transcription factor access and that withdrawal-associated H2A.Z depletion might sensitize genes for induction upon cocaine relapse. Hence, we examined the cohort of genes that are significantly up-regulated in D1 MSNs upon cocaine relapse after withdrawal but that are not induced by acute cocaine in drug-naïve animals (Fig. 3F). We found that these relapse-primed genes, which remain unchanged during withdrawal (in the absence of a cocaine challenge), are characterized by withdrawal-related loss of H2A.Z binding (Fig. 3G) and regulate processes that govern synaptic plasticity, trans-synaptic signaling, and neuron projection morphogenesis (fig. S9A). Computational IPA revealed that these genes are targeted by transcription factors that are well-known to be activated by cocaine in D1 MSNs, including CREB1, C/EBP, and EGR1 (fig. S9B) (12, 22, 50–52).
H2A.Z-specific histone chaperone ANP32E supports gene priming and cocaine reward processing
The histone chaperone ANP32E was recently identified as a highly specific H2A.Z chaperone that removes, but does not deposit, H2A.Z from nucleosomes (32). We found that levels of ANP32E associated with chromatin in NAc are increased after cocaine withdrawal (unpaired t test, P = 0.009; Fig. 4A), suggesting a possible role for this protein in cocaine-induced H2A.Z eviction and downstream transcriptional regulation. To explore whether ANP32E is required for cocaine-induced depletion of H2A.Z, we generated an adeno-associated virus (AAV1) vector containing Cre-dependent small interfering RNAs (siRNAs) to KD Anp32e (siAnp32e) in D1 MSNs of D1-Cre mice. While Anp32e expression is not exclusive to D1 MSNs and is also present in D2 MSNs within the NAc, we focused our investigation of Anp32e function in D1 MSNs considering the pronounced transcriptional priming of cocaine-responsive gene programs revealed by our subtype-specific RNA-seq analyses. Intracranial injection of AAV1-siAnp32e into the NAc leads to effective KD of ANP32E in this region (fig. S10). In addition, we found that the ability of chronic cocaine exposure to deplete H2A.Z from NAc chromatin was abolished completely upon ANP32E KD (fig. S10), thus directly linking cocaine regulation of ANP32E in D1 MSNs to the cocaine-induced depletion of H2A.Z in this brain region.
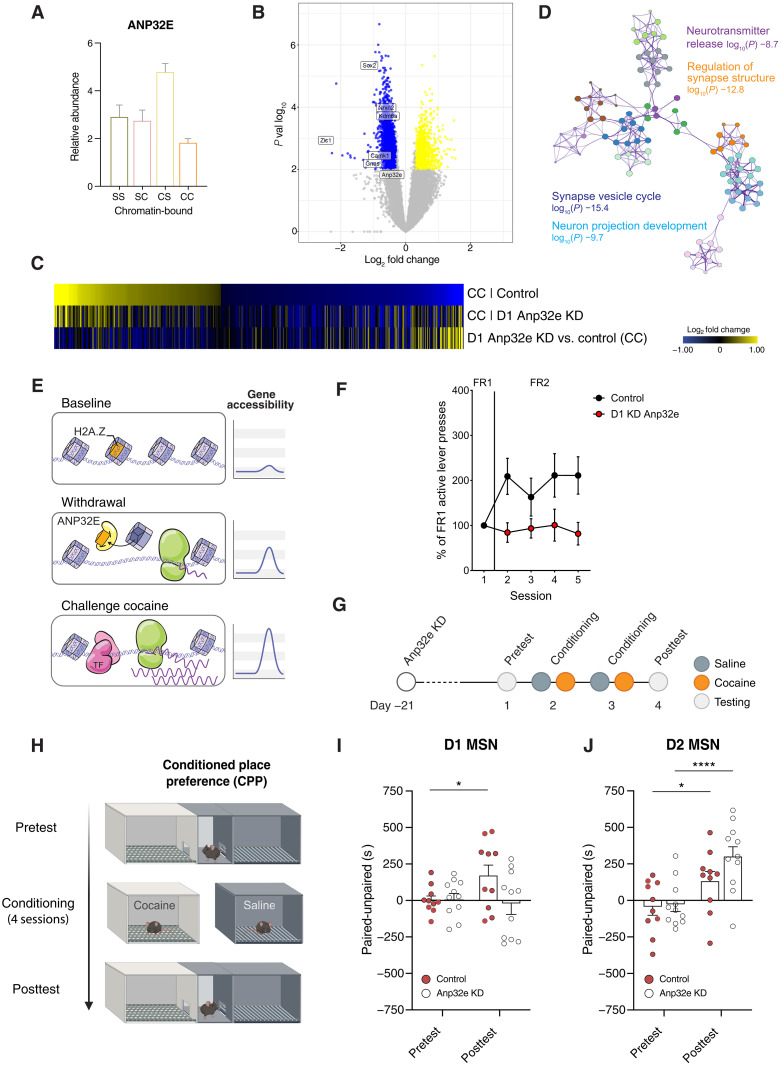
Fig. 4. ANP32E—an H2A.Z-specific histone chaperone—regulates transcriptional and rewarding responses to cocaine.
(A) Mass spectrometry showed increased ANP32E in NAc chromatin with prolonged withdrawal from chronic cocaine (CS) (unpaired t test, P = 0.009, N = 6 per group). (B) Volcano plot shows dysregulated transcripts following cocaine challenge after ANP32E KD in D1 MSNs compared controls, including the plasticity-related genes Sox2, Gnas, Camk1, Nrxn2, and Kdm6a. (C) Heatmap depicts mRNA expression of gene changes in control mice (siScr) after prolonged withdrawal and their loss of transcriptional priming upon ANP32E KD. (D) GO network representing biological processes affected by 1209 transcripts (log2 fold = −0.5, false discovery rate < 0.05) with attenuated regulation postcocaine challenge in D1 MSN–specific ANP32E KD mice. (E) Model: Following repeated cocaine, increased ANP32E mediates H2A.Z eviction, heightening gene responses to cocaine. (F) D1 MSN–specific ANP32E KD suppresses cocaine self-administration. The panel shows the percent of active lever presses during fixed-ratio 2 (FR2) sessions relative to the average response rates during FR1. Control animals increase active lever presses on the FR2 schedule to maintain level of cocaine infusions, but ANP32E KD mice do not modify their lever-pressing behavior upon this contingency change [repeated measures two-way analysis of variance (ANOVA), group × session interaction: F(4,48) = 2.252, not significant; main effect of group: F(1,12) = 5.406, P = 0.0384]. (G) Timeline for cocaine-induced CPP in mice with D1 MSN– or D2 MSN–specific ANP32E KD. (H) Schematic of cocaine-induced CPP. (I) Preference scores for the cocaine-paired chamber in control mice and D1 MSN–specific ANP32E KD mice [F(1,19) = P = 0.04, confirmed by Holm-Šídák’s multiple comparisons test]. Data are mean ± SEM; P values by Wilcoxon matched-pairs signed-rank test. (J) Preference scores for the cocaine-paired chamber in control mice and mice with D2 MSN–specific KD of ANP32E in the NAc [F(19,19) = 2.804, P = 0.015]. Data are mean ± SEM; P values (*P ≤ 0.05; ****P ≤ 0.0001) by Wilcoxon matched-pairs signed-rank test.
To test whether ANP32E is involved in cocaine-related gene priming, we examined how KD of ANP32E in D1 MSNs affects transcriptional responses in the NAc following withdrawal from chronic cocaine and a subsequent cocaine or saline challenge. Our results revealed major transcriptional differences in D1 ANP32E KD animals compared to wild-type mice (siScr control). In the ANP32E KD cohort, numerous genes that we previously found to be primed and greatly up-regulated in response to a cocaine challenge demonstrated less induction than that seen in control animals (Fig. 4, B and C, and fig. S11), suggesting a key role for ANP32E in transcriptional priming linked to prolonged cocaine withdrawal. In contrast to control mice, the transcriptional profile following cocaine challenge of ANP32E KD animals showed suppression of gene programs involved in synaptic transmission, neurotransmitter release, and synapse structure (Fig. 4D), indicating a broad role for ANP32E in cocaine regulation of synaptic function and neural plasticity (Fig. 4E). Our computational analyses identified CREB as a central upstream regulator of cocaine-mediated D1 MSN gene expression. To determine the impact of ANP32E KD on the induction of CREB-regulated genes by cocaine, we analyzed the expression levels of Gria1, Grin2b, Cdk5, CamkIIa, and other CREB targets in D1 MSNs. These genes were found to be mainly trending down or significantly down-regulated compared to siScr control animals, supporting the role of ANP32E in priming these loci (results are summarized in table S1).
Next, we examined the influence of Anp32e expression in D1 MSNs on cocaine self-administration. Mice with D1 ANP32E KD were tested alongside controls on a fixed-ratio 1 (FR1) schedule of reinforcement, followed by an FR2 schedule. Whereas control animals increased their lever pressing behavior with changing contingencies, KD mice did not modify their behavior in response to the escalated effort (FR2) required for cocaine infusion (Fig. 4F). To further investigate how ANP32E affects cocaine-related behaviors, we used an unbiased conditioned place preference (CPP) paradigm, which provides an indirect measure of cocaine reward (Fig. 4, G and H). As predicted, wild-type mice (siScr control) spent significantly more time in the compartment that was associated with cocaine experience in the posttest compared to their pretest [F(1,19) = P = 0.04, confirmed by Holm-Šídák’s multiple comparisons test], indicating an increased preference for the cocaine-paired environment in this group. In contrast, mice with an ANP32E KD in D1 MSNs in NAc did not form a preference for the chamber that was paired with cocaine experience in the posttest (Fig. 4I). These data indicate that ANP32E functions in D1 MSNs of NAc to facilitate cocaine-related reward processing.
We next tested whether ANP32E expression in D2 MSNs might also regulate cocaine CPP, using the Cre-dependent siAnp32e AAV1 in D2-Cre transgenic mice. In contrast to D1 circuit-selective ANP32E KD, which effectively blocked cocaine CPP, we found that the D2-specific KD of ANP32E does not lower cocaine-related reward learning in this behavioral paradigm [F(19,19) = 2.804, P = 0.015; confirmed by Holm-Šídák’s multiple comparisons test; Fig. 4J]. Instead, we observed a trend suggesting that D2-specific KD of ANP32E may enhance cocaine-related reward learning in cocaine CPP, although these results did not reach statistical significance. These findings collectively provide insight into circuit-specific epigenetic and transcriptional priming by ANP32E that is used by cocaine to modify brain function and behavior.
DISCUSSION
We previously showed that prolonged withdrawal from cocaine self-administration results in transcriptional priming of large sets of genes that are up-regulated by cocaine relapse across the brain’s reward circuitry, with especially prominent effects seen in NAc (45). Predicted upstream regulators of this relapse-related gene program included AP-1 and CREB, which have long been implicated in the actions of drugs of abuse (11, 13, 53), along with several transcription factors such as E2F, which has since been validated as well (46). However, these analyses were based on whole-tissue transcriptome data, with still limited information available about the transcriptional effects of cocaine exposure, withdrawal, and relapse selectively in D1 versus D2 MSNs, which display major, in some cases opposite, regulation by the drug (2, 4, 10, 11, 34).
Our cell type–specific data show that acute cocaine and prolonged withdrawal predominantly affect gene regulation in D1 MSNs with more limited responses seen in D2 MSNs. Notably, D1 MSN chromatin accessibility is robustly increased in response to acute cocaine, and this increase is sustained during prolonged withdrawal from chronic cocaine and increased still further upon a relapse dose of the drug. Such increases in chromatin accessibility in D1 MSNs affect prominently both gene regulatory enhancer and promoter elements of genes that regulate neuronal plasticity and metabolism. These findings are akin to a recently reported epigenetic priming mechanism in hippocampus during memory formation and recall, which also involves lasting and widespread opening of chromatin and enhanced transcriptional responses, although no change in respective baseline transcription was observed (54, 55).
In the present study, we found that cocaine-induced changes in D1 MSN chromatin are linked to increased gene expression upon drug relapse. Our bioinformatic analyses predict, again, that CREB and AP-1 transcription factors, among others, are upstream regulators of relapse-related and primed gene expression in D1 MSNs, thus validating prior work on the initial actions of drugs of abuse that implicates these factors in cocaine sensitization and cellular activity (8, 11, 13, 52, 56, 57). These findings support our central hypothesis that cocaine regulation of the epigenetic landscape in NAc neurons determines the inducibility of discrete gene programs that characterize drug relapse. Our data directly establish that a histone variant, H2A.Z, plays a key role in this process.
This study implicates H2A.Z in the actions of cocaine or any other drug of abuse and underscores the value of using unbiased experimental approaches to study the epigenetic basis of transcriptional regulation. Earlier ChIP-seq maps of cocaine regulation of several well-studied histone modifications in NAc revealed relatively limited overlap with changes in gene expression revealed by RNA-seq (18). By contrast, by following our open-ended analyses and identification of H2A.Z depletion as the most prominent histone change in NAc after prolonged withdrawal, we found that well more than 50% of genes in D1 MSNs that are primed for increased expression upon relapse exhibit H2A.Z depletion at their promoter or enhancer regions. Our proteomic analysis identified several additional histone modifications that are prominently regulated in NAc by cocaine withdrawal which now warrant similar characterization in future investigations.
To date, the functional relationship between H2A.Z and transcription remains uncertain, as H2A.Z has been associated with both positive and negative effects on gene expression, with its acetylation having a positive impact (49, 58, 59). Notably, the removal of H2A.Z from chromatin was shown to be critically important for stimulus- and training-induced gene expression in hippocampus (58), and hippocampal H2A.Z depletion resulted in enhanced memory formation. Furthermore, the loss of H2A.Z was recently found to cause a marked increase in the accessibility of AP-1 transcription factor binding sites in cultured cells (49), indicating that H2A.Z may prevent promiscuous transcription factor access and that its removal allows for rapid gene induction. Eviction of this variant is dependent on ANP32E, an H2A.Z-specific histone chaperone (32). Our data show that Anp32e expression in D1 MSNs mediates transcriptional priming observed in the NAc upon cocaine reexposure following prolonged withdrawal. Further, our data show a function for this chaperone in NAc D1 and D2 MSNs in controlling rewarding responses to cocaine, with Anp32e expression (and H2A.Z depletion) in D1 MSNs supporting relapse to cocaine self-administration as well as cocaine CPP but trending to limit cocaine CPP in D2 MSNs.
These data indicate that the KD of ANP32E affects gene expression and behavior even after only a few exposures to cocaine. Recent reports on ANP32E and the importance of the dynamic eviction of H2A.Z from nucleosomes for immediate early gene regulation in the hippocampus and fear memory support this gene regulatory function in the context of acute cocaine (48, 58, 60). Our data reveal that ANP32E KD impairs the transcriptional response to cocaine, attenuating the expression of CREB and AP-1 target genes, which are crucial for neuronal adaptation to cocaine and cause initial behavioral changes. Although these early changes may seem modest, they prime the neurons for altered responses to future drug exposure. In the context of chronic cocaine exposure, such ANP32E-mediated chromatin remodeling may be compounded, leading to more pronounced, lasting epigenome reprogramming and latent transcriptional dysregulation of gene programs controlled by CREB, AP-1, and other factors. These cumulative epigenetic changes are then stabilized during withdrawal, resulting in transcriptional and behavioral changes. This model is supported by studies on chromatin remodeling and immediate early genes. For instance, Maze et al. (61) demonstrated that initial changes in chromatin structure, even from short-term disruptions, alter gene expression and behavior, establishing a foundation for more substantial effects with prolonged exposure.
Overall, histone variants and their chaperones have received limited attention in epigenetic studies of substance use disorders. Our data implicate that ANP32E-mediated H2A.Z histone variant exchange in the NAc in the withdrawal-related epigenome remodeling linked to latent dysregulation of stimulus-induced gene expression upon drug relapse. The success of epigenetic drugs in the treatment of cancer and other conditions suggests the potential feasibility of pharmacologically targeting this ANP32E- and H2A.Z-dependent mechanism. However, small-molecule inhibitors of ANP32E could potentially affect memory functions in the hippocampus, necessitating careful consideration of side effects. Such interventions may require targeting its drug-related functions with short-acting inhibitors during specific windows when ANP32E is engaged, particularly during drug exposure or withdrawal. In addition, exploring downstream gene targets offers alternative strategies in addiction treatment, emphasizing the clinical relevance of our study (62, 63). Together, these findings present epigenetic priming involving ANP32E and H2A.Z depletion as a key mechanism that cocaine engages in modifying brain function and behavior in lasting ways.
MATERIALS AND METHODS
Animals
Adult male C57BL/6 mice (8 to 16 weeks) were housed five per cage on a 12-hour light-dark cycle at a constant temperature (23°C) and had free access to food and water ad libitum. They were assigned randomly to two groups treated for 10 consecutive days with either saline or cocaine (20 mg/kg) intraperitoneal injections and then subjected to a 30-day withdrawal period. Afterward, the two groups were subdivided into another two groups receiving single injections of either saline or cocaine 1 hour before the NAc tissue was harvested to construct a total of four subgroup conditions: control (SS), acute cocaine (SC), withdrawal (CS), and cocaine challenge (CC). For stereotactic surgeries, mice were anesthetized with ketamine (100 mg/kg) and xylazine (10 mg/kg). Syringe needles (33G, Hamilton) were used to bilaterally infuse 1 μl of virus at a flow rate of 0.1 μl/min in head-fixed animals. Coordinates for NAc from bregma were as follows: AP, +1.6 mm; ML, +1.6 mm; DV, −4.5 mm; 10° angle. The viral vectors used in this study were Cre-inducible Anp32e (mouse) siRNA AAV serotype 1 virus (1012 GC/ml) and Cre-inducible scrambled siRNA AAV serotype 1 virus (1012 GC/ml). The Cre-inducible Anp32e (mouse) siRNA is controlled by the U6 TATA-lox1 promoter with enhanced green fluorescent protein expressed from the PGK promoter. To minimize off-target effects, we designed the siRNA against a unique sequence specific to Anp32e and conducted BLAST analysis to confirm its specificity (target sequence: CAGAGTTAGTCCTCGATAATT). The BLAST results indicated that the E value for the siANP32E is >10,000 times lower (higher affinity) than for off-targets, minimizing the risk of off-target effects. All mouse handling was undertaken in accordance with the Institutional Animal Care and Use Committee (IACUC) guidelines at Icahn School of Medicine at Mount Sinai (IACUC protocol number 08-0465).
Western blots
Frozen NAc tissue samples were homogenized and incubated with agitation at 4°C for 30 min in radioimmunoprecipitation assay (RIPA) lysis buffer [10 mM Trizma base, 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 1% Triton X-100, 1% sodium deoxycholate (pH 7.4), and protease inhibitors]. Protein concentrations were determined by Pierce BCA protein assay (Thermo Fisher Scientific, #23225) and were normalized across samples using RIPA lysis buffer. Protein sample volumes were prepared accordingly to contain 25% 4× Laemmli sample buffer (Bio-Rad, #1610747) and 10% 2-mercaptoethanol (Bio-Rad, #1610710) and were heated to 95°C for 5 min before being separated by SDS–polyacrylamide gel electrophoresis with Criterion Precast Gels (Bio-Rad, 4 to 15% tris/glycine) and then transferred onto 0.2-μm nitrocellulose membranes (Bio-Rad, Trans-Blot, midi format). The membranes were blocked in 5% bovine serum albumin (BSA) in tris-buffered saline supplemented with 0.1% Tween 20 (TBST) at room temperature (RT) for 1 hour. Primary antibodies were diluted 1:1000 in 2.5% BSA in TBST and incubated overnight at 4°C. Primary antibodies are listed below. The membranes were washed in TBST thrice for 10 min each, followed by incubation with horseradish peroxidase–conjugated secondary antibodies diluted 1:10000 in 2.5% BSA in TBST at RT for 2 hours. The membranes were washed again three times and imaged with a Fujifilm LAS-4000 imager. Primary antibodies used were anti-H2AZ (GeneTex, GTX108273), anti-ANP32E (OriGene, TA351339), anti-H3 (Abcam, ab1791), and anti-H2AZ (Active Motif, #39013).
Conditioned place preference
An unbiased CPP test was carried out using three chambered CPP Med Associates boxes and software wherein two end chambers have distinct visual (gray versus striped walls) and tactile (small-grid versus large-grid flooring) cues to allow differentiation. All sessions were carried out in a dark room with ambient temperature. On the pretest session, the mice were allowed to explore all three chambers unencumbered for 20 min. Groups were adjusted to counterbalance any preexisting chamber bias. The conditioning was carried out by pairing an injection of saline with one chamber in the morning and a second injection of cocaine (10 mg/kg) with the other chamber in the afternoon for two consecutive days. Each conditioning session lasted 45 min. CPP testing was carried out on the fourth day where each mouse was allowed to explore all three chambers freely for 20 min. Place preference scores were taken as time on the cocaine-paired side–time on the saline-paired side.
Cocaine self-administration procedure
Pretraining
Mice maintained on mild, overnight food restriction were first trained to lever press for a food reward in operant conditioning chambers (Med Associates, Fairfax, VT). Two levers were presented to mice at the beginning of the session, and responding on the active lever led to the delivery of a chocolate pellet (Bio-Serv, Flemington, NJ) according to a FR1 schedule of reinforcement. Responding on the inactive lever had no consequence. Sessions lasted for 1 hour or until 30 rewards had been earned. Mice were trained until acquisition criteria for lever pressing had been met (30 pellets earned in two consecutive sessions with <10 inactive lever responses; four sessions on average required).
Jugular vein catheterization
Following acquisition of responding for food, mice underwent surgical procedures under inhaled isoflurane anesthesia (2%) to implant a preconstructed intravenous catheter (SBD-05C, Strategic Applications Incorporated, Lake Villa, IL) in the right jugular vein. The catheter cannula exited the skin from the animal’s mid-back, and catheter tubing (0.33 mm inner diameter) was threaded subcutaneously over the right shoulder and inserted 1.3 cm into the jugular vein. Ketoprofen (5 mg/kg) was injected subcutaneously following surgery as postoperative analgesia. Mice recovered for 4 days, during which time catheters were flushed once daily with 0.03 ml of heparinized saline (30 U) containing ampicillin antibiotic (5 mg/ml) to forestall infection.
Cocaine self-administration
Following recovery, mice were returned to operant boxes and began cocaine self-administration procedures wherein responding on the active lever, which previously delivered food, now delivered an intravenous infusion of cocaine (0.5 mg/kg per infusion). Mice were first tested on an FR1 schedule of reinforcement for five sessions, followed by an FR2 schedule of reinforcement for four sessions. Infusions were signaled by the activation of a cue light located above the active lever and were followed by a 20-s timeout during which the cue light remained illuminated, and levers remained extended. Responding on the active lever during timeout was recorded but had no consequence. Throughout the session, responding on the inactive lever had no consequence. Catheters were flushed with heparinized saline (30 U) before and after self-administration sessions. Twelve hours after the last self-administration session, catheter patency was tested. Mice were flushed with 0.03 ml of brevital sodium (3 mg/ml) dissolved in saline, followed immediately by 0.03 ml of saline. Patency was confirmed by loss of muscle tone within 3 s following infusion. Mice lacking catheter patency were excluded.
Subcellular protein fractionation
NAc tissue punches were fractionated into cytoplasmic, membrane, soluble nuclear, and chromatin-bound nuclear extracts using the Subcellular Protein Fractionation Kit for Tissues (Thermo Fisher Scientific, #87790). Cytoplasmic extraction buffer, which selectively permeabilizes the cell membrane and releases soluble cytoplasmic contents, was added to the NAc tissues and transferred to Dounce tissue homogenizers (setting B). The tissues were homogenized with 20 strokes and then centrifuged at 500g for 5 min into new tubes with the Pierce Tissue Strainer that removes tissue debris. The supernatants (cytoplasmic extracts) were immediately transferred to clean tubes. The remaining pellets were then mixed and vortexed at maximum setting for 5 s with the membrane extraction buffer, which dissolves plasma, mitochondria, and endoplasmic reticulum/Golgi membranes without solubilizing the nuclear membrane. The mixtures were then centrifuged at 3000g for 5 min. The supernatants (membrane extracts) were again transferred to new tubes. The pellets containing intact nuclei were vortexed at maximum setting for 15 s with the nuclear extraction buffer (NEB) and then incubated at 4°C for 30 min with gentle mixing. They were centrifuged at 5000g for 5 min which yielded the supernatants with soluble nuclear extracts. Last, the chromatin-bound extraction buffer was prepared by adding 5 μl of 100 mM CaCl2 and 3 μl of micrococcal nuclease to 100 μl of RT NEB and then added to the pellet and vortexed again. The tubes were incubated in a 37°C water bath for 15 min followed by vortexing and centrifuging at 16,000g for 15 min. The supernatant (chromatin-bound nuclear extract) was then transferred into new tubes. All extraction buffers contained protease inhibitors (c0mplete mini, EDTA-free, #40774700) and were kept on ice until use.
Chromatin immunoprecipitation sequencing
Chromatin was extracted using the truChIP Chromatin Shearing Kit with formaldehyde and prepared according to the manufacturer’s Low Cell protocol (Covaris PN010179 Rev. O, Dec 2020). A total of 11.1% formaldehyde was made fresh from 16% concentrated formaldehyde. RT 1× fixing buffer A was added to samples, and the 11.1% formaldehyde was then added to make a final concentration of 1% only. Samples were incubated for 5 min on rocker at RT to allow for efficient cross-linking and terminated immediately by adding quenching buffer E and incubating for another 5 min. Supernatants were discarded, and the tissue samples were washed with cold 1× phosphate-buffered saline (PBS) twice. Lysis buffer B was added, and samples were homogenized using the Dounce Tissue homogenizer (~20 strokes) and left to incubate on rocker for 10 min at 4°C. Samples were then centrifuged at 1700g for 5 min at 4°C, and the supernatant was discarded followed by resuspension in wash buffer C and incubated for 10 min at 4°C. Supernatants were discarded after centrifuging at 2000g for 5 min at 4°C, and shearing buffer D3 was added without disturbing the nuclei pellet followed by two rounds of centrifugation at 1700g for 5 min at 4°C while the supernatants were decanted. The nuclei pellets were resuspended in shearing buffer D3 and transferred to Adaptive Focused Acoustics (AFA) microtubes (8 microTUBE-130 AFA Fiber H Slit Strip V2, PN520239) for low cell chromatin shearing using a Covaris ultrasonicator with the following settings: 75.0 peak power; 15.0 duty % factor; 1000 cycles; 11.3 average power for durations of 810.0 and 540.0 s, respectively.
After shearing, samples were transferred into prechilled microcentrifuge tubes and diluted 1:1 with Covaris 2× IP dilution buffer and microcentrifuged at 10,000g for 5 min at 4°C before proceeding to IP with the supernatant. Thirty microliters of beads (Invitrogen Dynabeads Protein G) per IP was aliquoted into 1.5-ml Eppendorf tubes and separated on magnet for ~1 min, and the supernatant was removed by aspiration. The beads were resuspended and mixed in 1 ml of Block solution (1× PBS and 0.5% BSA) by inversion. Supernatants were separated on magnet and aspirated, and the wash process was repeated once more. Last, the beads were resuspended in 250 ml of Block solution and 4 μg/per IP of H2A.Z and H3 antibodies, respectively, and the tubes were rotated at 4°C for 2 hours. The beads were collected on magnet and washed in 1 ml of Block solution for a total of three washes before resuspension in 50 μl/per IP Block solution. Chromatin lysates from each sample were aliquoted, respectively, for H2A.Z and H3 antibody IP into 1.5-ml lo-bind protein Eppendorf tubes, and 50 μl of the beads and 300 μl of block solution were added and mixed by inversion. The IP tubes were rotated at 4°C overnight. The IP/bead mixture was transferred to new prechilled tubes, the beads were collected on magnet, and the supernatant was aspirated. One milliliter of cold RIPA wash buffer [50 mM Hepes-KOH (pH 7.5), 500 mM LiCl, 1 mM EDTA, 1% NP-40, and 0.7% Na-deoxycholate] was added and mixed by inversion. The beads were collected, followed by a total of five RIPA washes before resuspension in 1 ml of final ChIP wash buffer (1× Tris-EDTA buffer and 50 mM NaCl) and subsequent collection on a magnet. The beads were then resuspended in 210 μl of elution buffer [50 mM tris-HCl (pH 8.0), 10 mM EDTA, and 1% SDS] and incubated at 65°C for 30 min with 400 rpm on a thermomixer. The beads were collected again on magnet, and 200 μl of the supernatant was transferred to fresh tubes. The tubes were then boiled at 95°C for 7 min for DNA reverse cross-linking. The DNA were purified using a Qiagen MinElute PCR (polymerase chain reaction) purification kit and eluted with 20 μl of buffer EB [10 mM tris-Cl (pH 8.5)] and transferred to eight-strip PCR tubes.
The library was prepared according to NEBNext Ultra II DNA Library Prep Kit for Illumina (New England Biolabs, #E7645L) protocol. The size distribution of the DNA libraries was validated using the Agilent Bioanalyzer High Sensitivity DNA chip, after which all libraries were sequenced with Genewiz/Azenta on an Illumina NovaSeq S4 machine using a 2 × 150 bp pair-end read configuration to a minimum depth of 30 million reads per sample. Raw reads were processed by trimming the adapter sequences using TrimGalore. Trimmed reads were aligned to the reference genome mm10 using HISAT2 (64). Duplicate fragments were discarded, leaving only high-quality unique reads. Reads were sorted by chromosomal coordinates using samtools (65). MACS (model-based analysis of ChIP-seq) was used with default parameters to obtain sites of H2A.Z (peaks) for each of the four conditions—SC, SS, CS, and CC (66). The distribution of peaks around promoter and gene body regions was visualized using the tool ngs.plot (67). This analysis provides a detailed view of how the peaks are distributed across different genomic features, highlighting areas of chromatin enrichment.
NAc nuclei isolation for FANS
Nuclear samples were obtained from frozen tissue, collected by bilateral NAc punch dissections from 1-mm-thick coronal brain sections using a 14G needle. Virally infected tissue was harvested under fluorescent light. Frozen NAc samples were mechanically dissociated, and cells were lysed using a glass douncer in ice-cold lysis buffer [10.94% (w/v) sucrose, 5 mM CaCl2, 3 mM Mg(CH3COO)2, 0.1 mM EDTA, 10 mM tris-HCl (pH 8), and 1 mM dithiothreitol (DTT), in H2O]. Homogenates were filtered through a 40-μm cell strainer (Pluriselect) into ultracentrifuge tubes (Beckman Coulter), underlaid with 5 ml of high-sucrose solution [60% (w/v) sucrose, 3 mM Mg(CH3COO)2, 10 mM tris-HCl (pH 8), and 1 mM DTT, in H2O]. The sucrose gradient was centrifuged at 24,400 relative centrifugal force for 1 hour at 4°C in a SW41Ti Swinging-Bucket Rotor (Beckman Coulter), and nuclei at the bottom of the gradient were resuspended in PBS. 4′,6-Diamidino-2-phenylindole (DAPI) was added at a concentration of 0.5 μg/ml. Whole cell samples were obtained from fresh tissue, which was rotated at 37°C for 45 min in papain (1 mg/ml) suspended in digestion buffer [5% (w/v) d-trehalose, 0.05 mM d,l-2-amino-5-phosphonovaleric acid (APV), and deoxyribonuclease (DNase) (0.0125 mg/ml), in Hibernate-A (Thermo Fisher Scientific, A1247501)]. Tissue was then placed in FANS buffer [albumin inhibitor (0.58 mg/ml) (Worthington Biochemical, LK003182), 5% (w/v) d-trehalose, 0.05 mM APV, and DNase (0.0125 mg/ml), in Hibernate-A] and triturated using progressively smaller pipette tips. Samples were passed through a 70-μm filter and placed on top of a layer of ovomucoid-albumin (10 mg/ml) in FANS buffer. The pellet was resuspended in FANS buffer, and DAPI was added at a concentration of 0.5 μg/ml.
Assay for transposase-accessible chromatin with sequencing
A published protocol was used for ATAC-seq (68). Nuclei pellets were resuspended gently in transposition reaction mix [25 μl of 2× Tagment DNA (TD) buffer (Illumina, catalog no. FC-121-1030), 2.5 μl of transposase (Illumina, catalog no. FC-121-1030), and 22.5 μl of nuclease-free water] and incubated at 37°C for 30 min followed immediately with purification using a Qiagen MinElute kit whereby transposed DNA was eluted in 10 μl of elution buffer. Transposed DNA fragments were amplified initially using 1× NEBnext PCR master mix (New England Biolabs, catalog no. M0541), 25 μM customized Nextera PCR primers 1 and 2, and 0.6% final concentration of 100× SYBR Green I (Invitrogen, catalog no. S-7563) in a thermocycler with the following parameters: 72°C for 5 min; 98°C for 30 s; 5 cycles of 98°C for 10 s, 63°C for 30 s; and 72°C for 1 min. A side quantitative PCR amplification using a small aliquot of the amplified DNA was used to determine the necessary additional number of cycles to stop amplification before saturation (one-fourth of max fluorescent intensity) to reduce GC and size bias. The remaining DNA samples were amplified to the correct number of cycles and then purified and eluted using a Qiagen PCR Cleanup kit in 20 μl of Elution buffer. Library quality control was performed using an Agilent Bioanalyzer High Sensitivity DNA chip with the samples diluted 1:5. Raw reads from single-end sequencing with a 75-bp read length were processed by trimming the adapter sequences using Cutadapt (69). Trimmed reads were mapped to the reference mm10 genome using HISAT (64). Duplicate fragments were discarded, retaining only unique reads. As no splicing was expected to occur in the ATAC-seq datasets, alignments were performed without the --no-spliced-alignment flag. A comparison of the D1 MSN fastq file alignments with and without the --no-spliced-alignment flag confirmed the minimal difference between the two approaches, with alignment rates of 92.96 and 92.99%, respectively. Reads were sorted by chromosomal coordinates using samtools (65). ngsplot tool was used to generate average profile plots for each sample (67). We used MACS2 to identify sites of chromatin openness (i.e., peaks) in our ATAC-seq samples (66). We used diffReps to identify differentially accessible genomic loci between the CS and SS conditions in D1 and D2 MSNs (70). Sites with a P adjusted cutoff (Padj) of 0.0001 were considered differentially accessible. This stringent cutoff ensures that the identified DARs (differentially accessible regions) are robust and statistically significant. Transcription factor footprinting analysis was performed using TOBIAS (71). For transcription factor binding site enrichment analysis, we used HOMER (hypergeometric optimization of motif enrichment; http://homer.ucsd.edu/homer/). Specifically, we used the findMotifsGenome.pl tool in HOMER to analyze the promoters of the identified differential peaks, which allowed us to identify motifs associated with the DARs.
RNA sequencing
RNA was extracted from frozen TRIzol LS (Ambion) homogenates using the Direct-zol RNA Microprep Kit (Zymo Research) following the manufacturer’s instructions. Ribo-depleted sequencing libraries were prepared with the SMARTer Stranded Total RNA-Seq Kit v2–Pico Input Mammalian (TaKaRa Biotech) following the manufacturer’s instructions and sequenced with Genewiz/Azenta on an Illumina NovaSeq S4 machine using a 2 × 150 bp pair-end read configuration to a minimum depth of 30 million reads per sample. Differential expression was analyzed in R using the DESeq2. For our analysis, we used interaction term models in DESeq2 to account for the combined effects of treatment and genotype or cell type. Specifically, for the ANP32E experiment, an interaction term model was implemented in DESeq2, using the following formula: “~treatment + genotype + treatment:genotype.” For the sorted D1/D2 project, an interaction term model was implemented in DESeq2, using this formula: “~treatment + cell type + treatment:cell type.” These models allowed us to capture the interaction effects and provide a more nuanced understanding of gene expression changes in response to cocaine. Significance cutoffs were of at least 30% expression FC [|log2(FC)| > log2(1.3)] and nominal P < 0.05. By using a nominal P value coupled to FC thresholds, we aimed to balance the detection of significant genes while minimizing false negatives, thereby providing a more comprehensive view of gene regulation. To evaluate the purity of the fluorescence-activated cell sorting–sorted cells from the perspective of the RNA-seq libraries, the cell type components of the RNA-seq samples were estimated using a deconvolution approach, UniCell (72). Briefly, UniCell leverages a deep learning model to deconvolve cell type fractions, with an available database of many cell types. While UniCell does provide a context-free estimation of cell type fractions, cell types of central interest to this study are not provided by UniCell, particularly, individual D1 and D2 cell types. Therefore, UniCell was run in the “Select” mode, in which a user-supplied reference is provided for the purpose of generating contextualized predictions. The user-supplied reference in this case was derived from a study of NAc single-cell RNA (15), which contains several usual brain-related cell types, as well as individual characterization of D1 and D2 cells. To generate the reference, all samples from GSE118020 were downloaded, reprocessed using a standard Seurat pipeline, and subset to those single cells for which the authors provided a cell type assignment. The read counts were combined for all the single cells belonging to a given cell type to create one single pseudo-bulk sample per cell type. To ensure the robustness of our findings, we complemented the differential expression analysis with RRHO plots that were generated using the RRHO2 package implemented in R (github.com/RRHO2/RRHO2) to survey concordant and discordant gene regulation between different conditions (acute cocaine exposure, cocaine challenge, and withdrawal). RRHO2 analysis was performed in R using the “rrho” package, which calculates hypergeometric P values to assess the overlap between the ranked protein lists. The results were visualized as heatmaps using the “heatmap” function in R, where concordant gene expression (both increased or decreased) was represented by activity in the bottom left and top right regions, and discordant gene expression (one increased and the other decreased) was shown in the bottom right and top left regions. The heat intensity in the heatmap reflected the level of overlap in gene regulation. The significance of the overlap was represented by −log10(P value). For the D1/D2-sorted differential analysis and ANP32E KD sequencing, we also calculated the relative enrichment of transcription factor–associated motifs using HOMER (http://homer.ucsd.edu/homer/). We used the “findMotifs” function to examine the promoters of the input list of genes for both known and novel motifs.
Acknowledgments
Funding: Support for this work was provided by grants from the National Institutes of Health, R01DA007359 (E.J.N.), P01DA047233 (E.J.N.), K99AA027839 (P.M.), and R00AA027839 (P.M.) and from the Brain and Behavior Research Foundation (P.M.).
Author contributions: Conceptualization: P.M. and E.J.N. Methodology: P.M., Y.V.d.Z., A.G., M.E., H.K., A.R., B.A.G., C.J.B., and L.S. Investigation: P.M., Y.V.d.Z., A.G., M.E., R.F., H.K., A.R., M.R., A.A.R., C.J.B., and S.S. Visualization: P.M., Y.V.d.Z., C.J.B., M.E., and A.R. Formal analysis: P.M., Y.V.d.Z., C.J.B., M.E., A.R., and S.S. Writing: P.M. and E.J.N. Writing—review and editing: All authors read and commented on the manuscript. Supervision: E.J.N.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The raw FASTQ sequencing data are publicly available in the Gene Expression Omnibus (GEO) with accession nos. GSE272823 (RNA-seq), GSE272369 (ChIP-seq), and GSE272458 (ATAC-seq).
Supplementary Materials
This PDF file includes:
Figs. S1 to S11
Tables S1 and S2
References
REFERENCES AND NOTES
- 1.Walker D. M., Cates H. M., Heller E. A., Nestler E. J., Regulation of chromatin states by drugs of abuse. Curr. Opin. Neurobiol. 30, 112–121 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lobo M. K., Nestler E. J., The striatal balancing act in drug addiction: Distinct roles of direct and indirect pathway medium spiny neurons. Front. Neuroanat. 5, 41 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kravitz A. V., Tye L. D., Kreitzer A. C., Distinct roles for direct and indirect pathway striatal neurons in reinforcement. Nat. Neurosci. 15, 816–818 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.MacAskill A. F., Cassel J. M., Carter A. G., Cocaine exposure reorganizes cell type- and input-specific connectivity in the nucleus accumbens. Nat. Neurosci. 17, 1198–1207 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee K.-W., Kim Y., Kim A. M., Helmin K., Nairn A. C., Greengard P., Cocaine-induced dendritic spine formation in D1 and D2 dopamine receptor-containing medium spiny neurons in nucleus accumbens. Proc. Natl. Acad. Sci. U.S.A. 103, 3399–3404 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luo Z., Volkow N. D., Heintz N., Pan Y., Du C., Acute cocaine induces fast activation of d1 receptor and progressive deactivation of d2 receptor striatal neurons: In vivo optical microprobe [Ca2+]i imaging. J. Neurosci. 31, 13180–13190 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith R. J., Lobo M. K., Spencer S., Kalivas P. W., Cocaine-induced adaptations in D1 and D2 accumbens projection neurons (a dichotomy not necessarily synonymous with direct and indirect pathways). Curr. Opin. Neurobiol. 23, 546–552 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grueter B. A., Robison A. J., Neve R. L., Nestler E. J., Malenka R. C., ∆FosB differentially modulates nucleus accumbens direct and indirect pathway function. Proc. Natl. Acad. Sci. U.S.A. 110, 1923–1928 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dobbs L. K., Kaplan A. R., Lemos J. C., Matsui A., Rubinstein M., Alvarez V. A., Dopamine regulation of lateral inhibition between striatal neurons gates the stimulant actions of cocaine. Neuron 90, 1100–1113 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calipari E. S., Bagot R. C., Purushothaman I., Davidson T. J., Yorgason J. T., Peña C. J., Walker D. M., Pirpinias S. T., Guise K. G., Ramakrishnan C., Deisseroth K., Nestler E. J., In vivo imaging identifies temporal signature of D1 and D2 medium spiny neurons in cocaine reward. Proc. Natl. Acad. Sci. U.S.A. 113, 2726–2731 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Savell K. E., Tuscher J. J., Zipperly M. E., Duke C. G., Phillips R. A., Bauman A. J., Thukral S., Sultan F. A., Goska N. A., Ianov L., Day J. J., A dopamine-induced gene expression signature regulates neuronal function and cocaine response. Sci. Adv. 6, eaba4221 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chandra R., Francis T. C., Konkalmatt P., Amgalan A., Gancarz A. M., Dietz D. M., Lobo M. K., Opposing role for Egr3 in nucleus accumbens cell subtypes in cocaine action. J. Neurosci. 35, 7927–7937 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McClung C. A., Nestler E. J., Regulation of gene expression and cocaine reward by CREB and ΔFosB. Nat. Neurosci. 6, 1208–1215 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Mews P., Calipari E. S., Day J., Lobo M. K., Bredy T., Abel T., From circuits to chromatin: The emerging role of epigenetics in mental health. J. Neurosci. 41, 873–882 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen R., Blosser T. R., Djekidel M. N., Hao J., Bhattacherjee A., Chen W., Tuesta L. M., Zhuang X., Zhang Y., Decoding molecular and cellular heterogeneity of mouse nucleus accumbens. Nat. Neurosci. 24, 1757–1771 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mews P., Sosnick L., Gurung A., Sidoli S., Nestler E. J., Decoding cocaine-induced proteomic adaptations in the mouse nucleus accumbens. Sci. Signal. 17, eadl4738 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maze I., Covingtoni H. E., Dietz D. M., Laplant Q., Renthal W., Russo S. J., Mechanic M., Mouzon E., Neve R. L., Haggarty S. J., Ren Y., Sampath S. C., Hurd Y. L., Greengard P., Tarakhovsky A., Schaefer A., Nestler E. J., Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science 327, 213–216 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng J., Wilkinson M., Liu X., Purushothaman I., Ferguson D., Vialou V., Maze I., Shao N., Kennedy P., Koo J., Dias C., Laitman B., Stockman V., LaPlant Q., Cahill M. E., Nestler E. J., Shen L., Chronic cocaine-regulated epigenomic changes in mouse nucleus accumbens. Genome Biol. 15, 1–18 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.López A. J., Johnson A. R., Euston T. J., Wilson R., Nolan S. O., Brady L. J., Thibeault K. C., Kelly S. J., Kondev V., Melugin P., Kutlu M. G., Chuang E., Lam T. K. T., Kiraly D. D., Calipari E. S., Cocaine self-administration induces sex-dependent protein expression in the nucleus accumbens. Commun. Biol. 4, 883 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rogge G. A., Wood M. A., The role of histone acetylation in cocaine-induced neural plasticity and behavior. Neuropsychopharmacology 38, 94–110 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carpenter M. D., Hu Q., Bond A. M., Lombroso S. I., Czarnecki K. S., Lim C. J., Song H., Wimmer M. E., Pierce R. C., Heller E. A., Nr4a1 suppresses cocaine-induced behavior via epigenetic regulation of homeostatic target genes. Nat. Commun. 11, 504 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wimmer M. E., Fant B., Swinford-Jackson S. E., Testino A., Van Nest D., Abel T., Pierce R. C., H3.3 barcoding of nucleus accumbens transcriptional activity identifies novel molecular cascades associated with cocaine self-administration in mice. J. Neurosci. 39, 5247–5254 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maze I., Feng J., Wilkinson M. B., Sun H., Shen L., Nestler E. J., Cocaine dynamically regulates heterochromatin and repetitive element unsilencing in nucleus accumbens. Proc. Natl. Acad. Sci. U.S.A. 108, 3035–3040 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Z.-J., Martin J. A., Mueller L. E., Caccamise A., Werner C. T., Neve R. L., Gancarz A. M., Li J.-X., Dietz D. M., BRG1 in the nucleus accumbens regulates cocaine-seeking behavior. Biol. Psychiatry 80, 652–660 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Damez-Werno D. M., Sun H., Scobie K. N., Shao N., Rabkin J., Dias C., Calipari E. S., Maze I., Pena C. J., Walker D. M., Cahill M. E., Chandra R., Gancarz A., Mouzon E., Landry J. A., Cates H., Lobo M.-K., Dietz D., Allis C. D., Guccione E., Turecki G., Defilippi P., Neve R. L., Hurd Y. L., Shen L., Nestler E. J., Histone arginine methylation in cocaine action in the nucleus accumbens. Proc. Natl. Acad. Sci. U.S.A. 113, 9623–9628 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bastle R. M., Maze I. S., Chromatin regulation in complex brain disorders. Curr. Opin. Behav. Sci. 25, 57–65 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mews P., Calipari E. S., Cross-talk between the epigenome and neural circuits in drug addiction. Prog. Brain Res. 235, 19–63 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mews P., Donahue G., Drake A. M., Luczak V., Abel T., Berger S. L., Acetyl-CoA synthetase regulates histone acetylation and hippocampal memory. Nature 546, 381–386 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sidoli S., Bhanu N. V., Karch K. R., Wang X., Garcia B. A., Complete workflow for analysis of histone post-translational modifications using bottom-up mass spectrometry: From histone extraction to data analysis. J. Vis. Exp. 17, e54112 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zovkic I. B., Epigenetics and memory: An expanded role for chromatin dynamics. Curr. Opin. Neurobiol. 67, 58–65 (2021). [DOI] [PubMed] [Google Scholar]
- 31.Hammond C. M., Strømme C. B., Huang H., Patel D. J., Groth A., Histone chaperone networks shaping chromatin function. Nat. Rev. Mol. Cell Biol. 18, 141–158 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Obri A., Ouararhni K., Papin C., Diebold M. L., Padmanabhan K., Marek M., Stoll I., Roy L., Reilly P. T., Mak T. W., Dimitrov S., Romier C., Hamiche A., ANP32E is a histone chaperone that removes H2A.Z from chromatin. Nature 505, 648–653 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Graham D. L., Edwards S., Bachtell R. K., DiLeone R. J., Rios M., Self D. W., Dynamic BDNF activity in nucleus accumbens with cocaine use increases self-administration and relapse. Nat. Neurosci. 10, 1029–1037 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Lobo M. K., Covington H. E., Chaudhury D., Friedman A. K., Sun H., Damez-Werno D., Dietz D. M., Zaman S., Koo J. W., Kennedy P. J., Mouzon E., Mogri M., Neve R. L., Deisseroth K., Han M.-H., Nestler E. J., Cell type-specific loss of BDNF signaling mimics optogenetic control of cocaine reward. Science 330, 385–390 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schoenbaum G., Stalnaker T. A., Shaham Y., A role for BDNF in cocaine reward and relapse. Nat. Neurosci. 10, 935–936 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murer M. G., Moratalla R., Striatal signaling in L-DOPA-induced dyskinesia: Common mechanisms with drug abuse and long term memory involving D1 dopamine receptor stimulation. Front. Neuroanat. 5, 51 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kronman H., Richter F., Labonté B., Chandra R., Zhao S., Hoffman G., Lobo M. K., Schadt E. E., Nestler E. J., Biology and bias in cell type-specific RNAseq of nucleus accumbens medium spiny neurons. Sci. Rep. 9, 8350 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lobo M. K., Zaman S., Damez-Werno D. M., Koo J. W., Bagot R. C., DiNieri J. A., Nugent A., Finkel E., Chaudhury D., Chandra R., Riberio E., Rabkin J., Mouzon E., Cachope R., Cheer J. F., Han M.-H., Dietz D. M., Self D. W., Hurd Y. L., Vialou V., Nestler E. J., ∆FosB induction in striatal medium spiny neuron subtypes in response to chronic pharmacological, emotional, and optogenetic stimuli. J. Neurosci. 33, 18381–18395 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nestler E. J., Barrot M., Self D. W., ΔFosB: A sustained molecular switch for addiction. Proc. Natl. Acad. Sci. U.S.A. 98, 11042–11046 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dong Y., Nestler E. J., The neural rejuvenation hypothesis of cocaine addiction. Trends Pharmacol. Sci. 35, 374–383 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Surmeier D. J., Ding J., Day M., Wang Z., Shen W., D1 and D2 dopamine-receptor modulation of striatal glutamatergic signaling in striatal medium spiny neurons. Trends Neurosci. 30, 228–235 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Conrad K. L., Tseng K. Y., Uejima J. L., Reimers J. M., Heng L.-J., Shaham Y., Marinelli M., Wolf M. E., Formation of accumbens GluR2-lacking AMPA receptors mediates incubation of cocaine craving. Nature 454, 118–121 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gallegos D. A., Minto M., Liu F., Hazlett M. F., Aryana Yousefzadeh S., Bartelt L. C., West A. E., Cell-type specific transcriptional adaptations of nucleus accumbens interneurons to amphetamine. Mol. Psychiatry 28, 3414–3428 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramakrishnan A., Wangensteen G., Kim S., Nestler E. J., Shen L., DeepRegFinder: Deep learning-based regulatory elements finder. Bioinform. Adv. 4, vbae007 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Walker D. M., Cates H. M., Loh Y.-H. E., Purushothaman I., Ramakrishnan A., Cahill K. M., Lardner C. K., Godino A., Kronman H. G., Rabkin J., Lorsch Z. S., Mews P., Doyle M. A., Feng J., Labonté B., Koo J. W., Bagot R. C., Logan R. W., Seney M. L., Calipari E. S., Shen L., Nestler E. J., Cocaine self-administration alters transcriptome-wide responses in the brain’s reward circuitry. Biol. Psychiatry 84, 867–880 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cates H. M., Heller E. A., Lardner C. K., Purushothaman I., Peña C. J., Walker D. M., Cahill M. E., Neve R. L., Shen L., Bagot R. C., Nestler E. J., Transcription factor E2F3a in nucleus accumbens affects cocaine action via transcription and alternative splicing. Biol. Psychiatry 84, 167–179 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maze I., Noh K.-M., Soshnev A., Allis C. D., Every amino acid matters: Essential contributions of histone variants to mammalian development and disease. Nat. Rev. Genet. 15, 259–271 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stefanelli G., Azam A. B., Walters B. J., Brimble M. A., Gettens C. P., Bouchard-Cannon P., Cheng H.-Y. M., Davidoff A. M., Narkaj K., Day J. J., Kennedy A. J., Zovkic I. B., Learning and age-related changes in genome-wide H2A.Z binding in the mouse hippocampus. Cell Rep. 22, 1124–1131 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cole L., Kurscheid S., Nekrasov M., Domaschenz R., Vera D. L., Dennis J. H., Tremethick D. J., Multiple roles of H2A.Z in regulating promoter chromatin architecture in human cells. Nat. Commun. 12, 2524 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robison A. J., Nestler E. J., Transcriptional and epigenetic mechanisms of addiction. Nat. Rev. Neurosci. 12, 623–637 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chandra R., Lobo M. K., Beyond neuronal activity markers: Select immediate early genes in striatal neuron subtypes functionally mediate psychostimulant addiction. Front. Behav. Neurosci. 11, 112 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Larson E. B., Graham D. L., Arzaga R. R., Buzin N., Webb J., Green T. A., Bass C. E., Neve R. L., Terwilliger E. F., Nestler E. J., Self D. W., Overexpression of CREB in the nucleus accumbens shell increases cocaine reinforcement in self-administering rats. J. Neurosci. 31, 16447–16457 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Teague C. D., Nestler E. J., Key transcription factors mediating cocaine-induced plasticity in the nucleus accumbens. Mol. Psychiatry 27, 687–709 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gräff J., Tsai L. H., Histone acetylation: Molecular mnemonics on the chromatin. Nat. Rev. Neurosci. 14, 97–111 (2013). [DOI] [PubMed] [Google Scholar]
- 55.Marco A., Meharena H. S., Dileep V., Raju R. M., Davila-Velderrain J., Zhang A. L., Adaikkan C., Young J. Z., Gao F., Kellis M., Tsai L.-H., Mapping the epigenomic and transcriptomic interplay during memory formation and recall in the hippocampal engram ensemble. Nat. Neurosci. 23, 1606–1617 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Muschamp J. W., Nemeth C. L., Robison A. J., Nestler E. J., Carlezon W. A., ΔFosB enhances the rewarding effects of cocaine while reducing the pro-depressive effects of the kappa-opioid receptor agonist U50488. Biol. Psychiatry 71, 44–50 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang J., Zhang L., Jiao H., Zhang Q., Zhang D., Lou D., Katz J. L., Xu M., c-Fos facilitates the acquisition and extinction of cocaine-induced persistent changes. J. Neurosci. 26, 13287–13296 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zovkic I. B., Paulukaitis B. S., Day J. J., Etikala D. M., Sweatt J. D., Histone H2A.Z subunit exchange controls consolidation of recent and remote memory. Nature 515, 582–586 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Giaimo B. D., Ferrante F., Herchenröther A., Hake S. B., Borggrefe T., The histone variant H2A.Z in gene regulation. Epigenetics Chromatin 12, 37 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Narkaj K., Stefanelli G., Wahdan M., Azam A. B., Ramzan F., Steininger C. F. D., Walters B. J., Zovkic I. B., Blocking H2A.Z incorporation via tip60 inhibition promotes systems consolidation of fear memory in mice. eNeuro 5, ENEURO.0378-18.2018 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maze I., Chaudhury D., Dietz D. M., Von Schimmelmann M., Kennedy P. J., Lobo M. K., Sillivan S. E., Miller M. L., Bagot R. C., Sun H., Turecki G., Neve R. L., Hurd Y. L., Shen L., Han M.-H., Schaefer A., Nestler E. J., G9a influences neuronal subtype specification in striatum. Nat. Neurosci. 17, 533–539 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mews P., Cunningham A. M., Scarpa J., Ramakrishnan A., Hicks E. M., Bolnick S., Garamszegi S., Shen L., Mash D. C., Nestler E. J., Convergent abnormalities in striatal gene networks in human cocaine use disorder and mouse cocaine administration models. Sci. Adv. 9, eadd8946 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou Z., Yuan Q., Mash D. C., Goldman D., Substance-specific and shared transcription and epigenetic changes in the human hippocampus chronically exposed to cocaine and alcohol. Proc. Natl. Acad. Sci. U.S.A. 108, 6626–6631 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim D., Paggi J. M., Park C., Bennett C., Salzberg S. L., Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907–915 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nussbaum C., Myers R. M., Brown M., Li W., Shirley X. S., Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shen L., Shao N., Liu X., Nestler E., Ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics 15, 284 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Buenrostro J. D., Giresi P. G., Zaba L. C., Chang H. Y., Greenleaf W. J., Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Martin M., Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12 (2011). [Google Scholar]
- 70.Shen L., Shao N. Y., Liu X., Maze I., Feng J., Nestler E. J., diffReps: Detecting differential chromatin modification sites from chip-seq data with biological replicates. PLOS One 8, e65598 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bentsen M., Goymann P., Schultheis H., Klee K., Petrova A., Wiegandt R., Fust A., Preussner J., Kuenne C., Braun T., Kim J., Looso M., ATAC-seq footprinting unravels kinetics of transcription factor binding during zygotic genome activation. Nat. Commun. 11, 4267 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Charytonowicz D., Brody R., Sebra R., Interpretable and context-free deconvolution of multi-scale whole transcriptomic data with UniCell deconvolve. Nat. Commun. 14, 1350 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S11
Tables S1 and S2
References