ABSTRACT
Escherichia coli O157:H7-adulterated food products are associated with disease outbreaks in humans. Although cattle feces are a source for E. coli O157:H7 contamination, it is unclear if human-associated outbreak isolates differentially colonize and shed in the feces of cattle from that of non-outbreak isolates. It is also unclear if phenotypes, such as biofilm formation, cell attachment, or toxin production, differentiate environmental E. coli O157:H7 isolates from those associated with human illness. The objective of this study was to compare the genotypes and phenotypes of a diverse set of E. coli O157:H7 isolates, with the intent of identifying differences that could inform cattle colonization and fecal shedding, along with virulence potential in humans. Isolates differed in attachment phenotypes on human Caco-2 cells and bovine-derived recto-anal junction squamous epithelial cells, with curli having a strong impact on attachment to the human-derived cell line. The prototypical E. coli O157 isolate EDL933 had the greatest expression of the adhesin gene iha, yet it had decreased expression of the virulence genes stx2, eae, and ehxA compared the lineage I/II isolates RM6067W and/or FRIK1989. Strong or weak biofilm production was not associated with significant differences in cattle colonization or shedding, suggesting biofilms may not play a major role in cattle colonization. No significant differences in cattle colonization and fecal shedding were detected, despite genomic and in vitro phenotypic differences. The outbreak isolate associated with the greatest incidence of hemolytic uremic syndrome, RM6067W, induced the greatest Vero cell cytotoxicity and had the greatest stx2 gene expression.
IMPORTANCE
Foodborne illness has major impacts on global health and imposes financial hardships on food industries. Escherichia coli serotype O157:H7 is associated with foodborne illness. Cattle feces are a source of E. coli O157:H7, and routine surveillance has led to an abundance of E. coli O157:H7 genomic data. The relationship between E. coli O157:H7 genome and phenotype is not clearly discerned for cattle colonization/shedding and improved understanding could lead to additional strategies to limit E. coli O157:H7 in the food chain. The goal of the research was to evaluate genomic and phenotypic attributes of E. coli O157:H7 associated with cattle colonization and shedding, environmental persistence, and human illness. Our results indicate variations in biofilm formation and in vitro cellular adherence was not associated with differences in cattle colonization or shedding. Overall, processes involved in cattle colonization and various phenotypes in relation to genotype are complex and remain not well understood.
KEYWORDS: Escherichia coli, O157:H7, genomics, Shiga toxins, enteric pathogens
INTRODUCTION
Foodborne illnesses in the United States are a significant burden to the welfare of the general populace, besides imposing considerable economic loss to food industry groups. Shiga toxin-producing Escherichia coli (STEC) is a cause of foodborne outbreaks and illnesses associated with STEC are estimated to have an annual cost of 789 million dollars in the U.S, with the O157 serotype responsible for 80% of these costs (1). During the years 2009–2021, 32% of confirmed E. coli O157 serotype outbreaks were attributed to foodborne illness, with beef (31%), vegetable row crops (25%), dairy (10%), and fruit (4%) being the primary food sources (2). Illnesses associated with E. coli O157 serotype have resulted in 76 multistate outbreaks in the US from 2009 to 2021, and in 5,004 illnesses and 30 deaths (2). Although, E. coli O157:H7 is the most prevalent serotype associated with foodborne illness, other commonly implicated STEC serotypes include O26, O103, O111, O121, O145 and O45, which are all considered non-O157 STEC beef adulterants by the USDA-Food Safety and Inspection Service (FSIS) (3). E. coli O157 still contributed to 23% of STEC illnesses during 2015–2017 (4, 5).
Cattle are a primary reservoir of E. coli O157:H7, and unlike in humans, E. coli O157:H7 does not typically cause clinical signs of disease in adult cattle, with infected animals often shedding the organism undetected (6, 7). Although E. coli O157:H7 can be detected in various regions of the bovine intestinal tract, it predominately attaches to bovine epithelial cells at the recto-anal junction (RAJ) (8, 9) and is subsequently shed in feces. It poses a broad contamination risk, including beef products at slaughter and the environment. E. coli can establish itself as a member of many environments, including water, soil, and plants (10–12). Limiting E. coli O157:H7 in cattle to minimize food contamination is important for preventing or reducing disease outbreaks in humans (13).
Currently, E. coli O157:H7 isolates are divided into three main phylogenetic lineages, (lineage I, lineage I/II, and lineage II) based on lineage-specific polymorphism assay 6 (LSPA-6) typing (14). Nucleotide polymorphisms in the lineage I and I/II isolates are associated with pathogenicity in humans, such as a polymorphism in the tir gene at position 255, where a thymine (T) at this position is more associated with clinical human isolates, and an adenine (A) at this position is more associated with bovine isolates (15, 16); however, these broad generalizations are not all encompassing, as E. coli O157:H7 isolates with tir 255 A alleles have been isolated from cases of human illness (15, 17). Current approaches in classifying E. coli O157:H7 recovered from surveillance streams as potential isolates of concern for human illness include the requirement for Shiga toxin genes (stx2a, stx2c, and stx1a) along with the intimin gene eae, with stx2a > stx2c > stx2a + stx1a > stx1a in order of potential illness severity (18). Although consideration for toxin and intimin genes are important, these genes do not fully characterize different E. coli O157:H7 isolates for potential risk to human health or persistence in different environments. Genes other than eae and stx are associated with human clinical E. coli O157:H7 isolates, such as the plasmid-carried ehxA and the O island-associated nle and efa genes (19, 20). The hlyA gene, along with eae and stx2, was present in the majority of E. coli O157:H7 HUS cases in humans (21). Thus, an improved understanding of the relationship between E. coli O157:H7 genome and phenotypes expressed in various settings is important for utilizing data generated from various surveillance efforts.
We present various methods to evaluate E. coli O157:H7 phenotypes with the intent to identify relationships between genome and phenotypes, particularly those associated with cattle colonization and human pathogenicity. Four E. coli O157:H7 isolates from two different LSPA-6 lineages and sources were selected for analysis. The genomes were sequenced, and their phylogenetic relationships to E. coli O157:H7 isolates deposited in the NCBI Pathogen Detection database (https://www.ncbi.nlm.nih.gov/pathogens/) were assessed. Phenotypic assays consisting of cattle colonization and fecal shedding, ex vivo and in vitro cell attachment, biofilm formation, virulence-associated gene expression, and Shiga toxin production were used to evaluate epidemiologically relevant phenotypes, and their relationship to E. coli O157:H7 pathogenic potential. In some cases, differences in phenotypes could be attributed to genomic differences, but much remains to be understood as attachment and biofilm phenotypes did not correlate to cattle colonization, nor did any of the E. coli O157:H7 isolates differentially shed from cattle.
RESULTS
Comparative genomics and isolate characterization
E. coli O157:H7 isolates were selected based upon LSPA-6 typing, isolation source, phylogenetic placement, and association with human illness (Table 1). LSPA-6 lineage I isolates included EDL933 and TW14588, both of which are associated with foodborne outbreaks. LSPA-6 lineage I/II isolates included RM6067W and FRIK1989; RM6067W was associated with a foodborne outbreak, whereas FRIK1989 was isolated from dairy cattle feces and was not associated with human illness.
TABLE 1.
Shiga toxin-producing Escherichia coli O157:H7 isolates used
| Strain | LSPA-6a | Associated environmental source | Outbreak associated | Reference |
|---|---|---|---|---|
| EDL933 | I | Ground beef (1982), USA | Yes | 80 |
| TW14588 | I | Lettuce (2006), USA | Yes | 77 |
| FRIK1989 | I/II | Dairy cattle feces (1991), USA | No | NADCb |
| RM6067W | I/II | Spinach (2006), USA | Yes | 76 |
Lineage-specific polymorphism assay 6.
National Animal Disease Center, USDA.
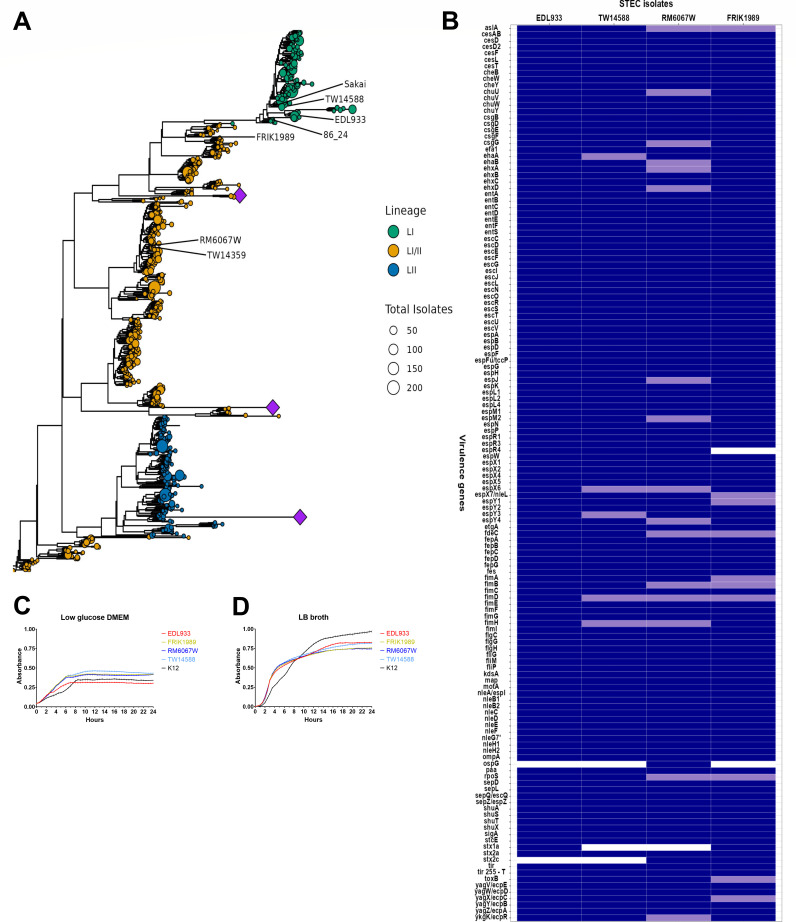
To place these isolates in the context of currently circulating E. coli O157:H7 isolates, we compared genomic sequences from our strains to genomes in the NCBI Pathogen Detection Database https://www.ncbi.nlm.nih.gov/pathogens/. All isolates represented in the analysis possessed the major virulence genes eae and stx, and spanned LSPA-6 lineages I, I/II, and II. LSPA-6 types of isolates were largely congruent with clades present in a core genome-based maximum likelihood phylogenetic tree (Fig. 1A). The foodborne outbreak isolates EDL933, TW14588, and RM6067W separated on the tree in clades with other clinical isolates. The non-clinical isolate FRIK1989 also grouped with clinical isolates on the tree. The currently circulating population of E. coli O157:H7 strains is diverse, and although the isolates represented in this study closely relate to isolates from human infections, they represent a small fraction of the broader phylogenetic diversity present.
Fig 1.
Comparative genomics and isolate characterization. (A) Phylogenetic relationship of E. coli O157:H7 isolates used in this study. A maximum likelihood phylogenetic tree generated from a core genome alignment of 724 representative genomes (one per NCBI SNP cluster) and seven reference genomes that include the four used in the current study (EDL933, TW14588, RM6067W, and FRIK1989). The circles representing the leaves of the tree are sized in proportion to the total number of genomes in that SNP cluster. The leaves are colored according to the LSPA-6 lineage typing scheme. Reference genome positions in the tree are indicated with text. Several clades exhibiting very long branch lengths were collapsed to improve the readability of the tree and are represented by purple diamonds. (B) Heat map depicting major virulence genes/SNPs/INDELs shared by these isolates. Virulence genes were annotated for these isolates’ genomes through PATRIC 3.6.12 (https://www.patricbrc.org) utilizing the virulence factor property and VFBD source within the specialty genes function. Further in silico analysis using geneious prime (https://www.geneious.com/prime/) was employed to ascertain tir 255 T/A allele and subtype stx genes. Snippy (https://github.com/tseemann/snippy) was used to find SNPs/INDELs within genes by using EDL933 as the refence genome. Dark blue indicates the presence of a gene, light blue indicates the presence of a gene with nonsynonymous mutation, and white indicates the absence of a virulence gene. (C&D). Growth profiles of E. coli O157:H7 isolates in low glucose DMEM and LB broth. Absorbance readings were measured on a bioscreen microplate reader for E. coli isolates EDL933, FRIK1989, RM6067W, TW14588, and K12 grown statically at 37°C. Measurements were taken every 30 min using the wideband filter for 24 h. Values shown are background subtracted. (C) Growth curve in low glucose DMEM. (D) Growth curve in LB broth.
Comparison of the four isolate genomes for the presence of known virulence genes revealed few differences, and only the type of stx genes and the Type 3 Secretion System (T3SS) effectors espR4 and ospG (Fig. 1B) were different. Stx1a was identified in EDL933 and FRIK1989, stx2a wass identified in all four isolates, and stx2c was detected in FRIK1989 and RM6067W. All four isolates possessed the T allele at position 255 in the translocated intimin receptor gene (tir), which is associated with virulence in humans (15). The T3SS effector gene espR4 was absent in FRIK1989 but detected in the other three isolates. RM6067W was the only isolate to possess the T3SS effector gene ospG. SNP/INDEL analysis of these isolates using EDL933 as the reference genome indicated the presence of nonsynonymous mutations in several virulence-associated genes (Fig. 1B; Table S1). Of note, TW14588 had a mutation in ehaA, a gene associated with adherence and biofilm formation (22). RM6067W had mutations in csgG, ehaB, and rpoS, which are genes associated with biofilm formation along with the enterohaemolysin genes ehxA and ehxD (23–27). FRIK1989 had a mutation in the rpoS gene distinct from RM6067W, along with a mutation in the toxB gene, which is associated with adherence (28). A detailed list of all polymorphisms is found in Table S1.
It should be noted that this comparison was intended to be between E. coli O157:H7 isolates that possess the full spectrum of virulence genes, so it was not unexpected that FRIK1989 phylogeny was similar to clinical isolates. FRIK1989s inclusion in this comparison was intended to highlight differences or similarities between known human outbreak isolates and an isolate taken from cattle with no known association with clinical illness in humans, but containing the genes attributed to human infection. In addition, FRIK1989 was found to possess a large accessory genome, with FRIK1989 having 419 accessory genes, whereas EDL933, TW14588, and RM6067W have 251, 353, and 364 accessory genes respectively.
The 24-h growth curves measured by changes in optical density in LB broth showed no major deviations between the four tested isolates; however, in low glucose DMEM, EDL933 achieved a lower plateau (Fig. 1C and D).
The four E. coli O157:H7 isolates could be differentiated using polymorphic amplified typing sequences (PATS) (29–31). Using PATS, genomic differences that conferred a distinct DNA fingerprint were identified as shown in Table S2.
Comparing our EDL933 and TW14588 isolate genome assemblies to previously deposited genomes of same named isolates yielded differences in SNPs as well as in the presence and absence of genes (Table S3). Many of these genes are for hypothetical proteins or likely phage related. Our EDL933 has 43 genes that were absent from assemblies GCF_000006665 and GCF_000732965, and our isolate was missing 33 genes that are found in the other assemblies. Using our EDL933 genome as reference, we detected 78 non-synonymous SNPs in GCF_000006665 and 5 non-synonymous SNPs in GCF_000732965. In the comparison of our TW14588 genome to assembly GCF_000155125, there were 175 non-synonymous SNPs when our TW14588 was used as reference, and our TW14588 had 224 genes absent from GCF_000155125, whereas GCF_000155125 has 313 unique genes.
Fecal shedding and recto-anal colonization in cattle
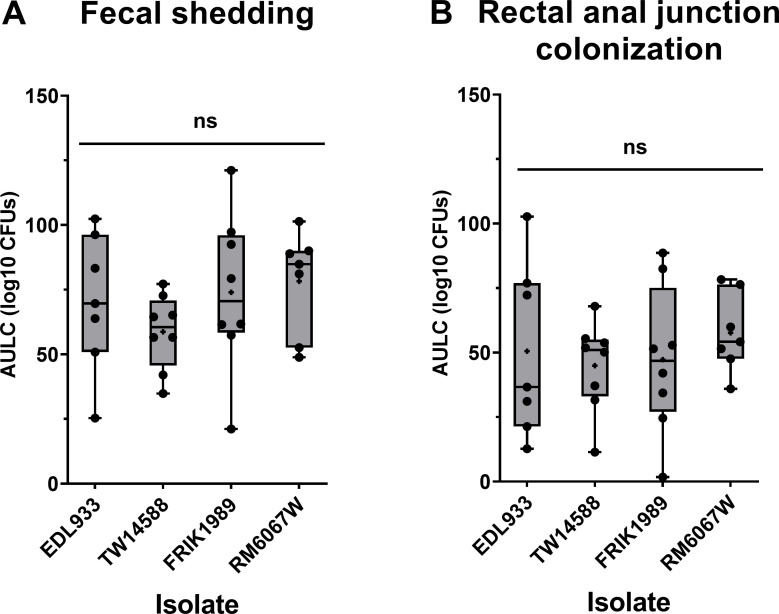
Fecal shedding of the four E. coli O157:H7 isolates from orally inoculated Jersey calves was analyzed using a trapezoid area under the log curve (AULC) for CFUs/g calculated from days 1, 2, 3, 4, 5, 7, 9, 11, and 14 post-inoculation to give an idea of overall shedding. The PATS profile of isolates recovered matched that of the respective inoculum administered to the animals, indicating the lack of pre-existing E. coli O157:H7. There was no significant difference in shedding among isolates over the 14-day period (P = 0.425, Fig. 2A). The time course of the average fecal shedding data for each of the isolates followed the same general pattern with a plateau in CFUs recovered at approximately days 3 to 5 post-inoculation and E. coli O157:H7 recovered gradually dropping over the remaining days of monitoring (Fig. S1A through D). By the end of the 14-day monitoring period, not all calves were negative for E. coli O157:H7, with one of seven calves in EDL933 group (600 CFUs/g feces), two of eight calves in TW14588 group (both by enrichment culture only), and two of eight calves in FRIK1989 group (2400 CFUs/g feces and by enrichment culture only), and three of seven calves in RM6067W group (1150 CFUs/g feces and the other two by enrichment culture only) still shedding.
Fig 2.
E. coli O157:H7 fecal shedding and rectal anal junction mucosa colonization in Jersey calves. Jersey calves were orally inoculated with 6 × 109 CFUs of indicated E. coli O157:H7 isolates (EDL933, TW14588, FRIK1989, or RM6067W), and feces and recto-anal junction mucosal swabs (RAMS) were collected on days 1, 2, 3, 4, 5, 7, 9, 11, and 14 to enumerate O157 levels as indicated in Materials and Methods. Area Under the Log Curve (AULC) was performed (trapezoid area under the curve model; Rstudio version 2021.09.2). (A) Cumulative fecal shedding and (B) cumulative RAMS colonization of respective E. coli O157:H7 isolate. Each point represents an individual animal’s cumulative shedding/colonization in the indicated inoculation group. Statistical analysis was done by one-way ANOVA with Tukey’s comparison of means; no significant (ns) difference in shedding or colonization, (p ≤ 0.05). Mean is indicated with a small “+”
Differences in colonization at the RAJ was assessed through enumeration of E. coli O157:H7 recovered from rectal–anal mucosal swabs (RAMS). Similar to fecal shedding, there were no significant differences in E. coli O157:H7 abundance in RAMS (P = 0.777, Fig. 2B). The time course RAJ colonization trend for all isolates indicated average peak CFU counts from four to seven days post-inoculation, which gradually declined over the remaining monitoring period (Fig. S1E through H). In a comparable manner to fecal counts, by the end of the 14-day monitoring period, not all the calves were negative for E. coli O157:H7, with three of seven calves in EDL933 group (430 CFUs/mL and the other two by enrichment culture only), two of eight claves in TW14588 group (25 CFUs/mL and by enrichment culture only), three of eight calves in FRIK1989 group (230, 20 CFUs/mL, and by enrichment culture only), and six of seven calves in RM6067W (20, 20, and 10 CFUs/mL, and the other three by enrichment culture only) group still colonized. At these latter timepoints several cattle had detectable E. coli O157:H7 at the RAJ but O157 was not detected in the feces.
Assessment of E. coli O157:H7 attachment to epithelial cells
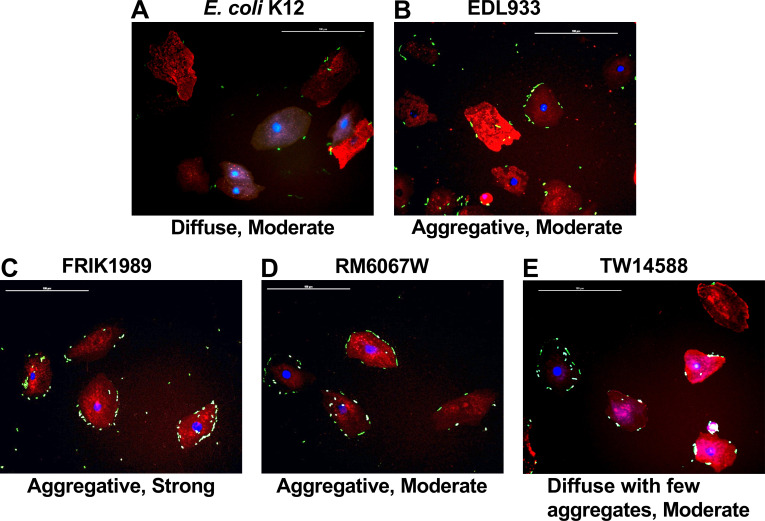
Although the E. coli O157:H7 isolates did not differentially colonize cattle, it was unclear if this phenotype mirrored the ex vivo/in vitro phenotype of attachment to tissue culture cells. All E. coli O157:H7 isolates tested attached to ex vivo bovine RSE (RAJ squamous epithelial) cells (Fig. 3), but the quantity and patterns of adherence were different. As shown in Table 2, the E. coli O157:H7 isolates demonstrated distinct attachment profiles compared with the control, non-pathogenic E. coli K12. All E. coli O157:H7 isolates demonstrated some aggregation on RSE cells compared with E. coli K12, which was attached in a diffuse manner. Of the four E. coli O157:H7 isolates tested only FRIK1989 had a qualitatively strong attachment phenotype to bovine RSE cells (Table 2). The quantitative difference in RSE cell attachment observed between all isolates tested was statistically significant (P < 0.05) for isolates FRIK 1989 (P = 0.0076), RM6067W (P = 0.0121), and EDL933 (P = 0.0139) when compared with E. coli K12. All other comparisons did not yield any significant differences.
Fig 3.
Adherence patterns of E. coli K12 and E. coli O157:H7 isolates on RSE cells. E. coli isolates (K12, EDL933, TW14588, FRIK1989, and TW14588) were incubated with RSE cells at an MOI of 10:1 for 4 h with agitation followed by assessment of cell attachment via immunofluorescence staining and co-localization. Representative immunofluorescent images with RSE cells and E. coli are shown at 400× magnification. Adherence patterns on RSE cells were qualitatively recorded as diffuse or aggregative, with strong or moderate qualifiers as described in the Materials and Methods. E. coli were labeled with FITC (green) conjugated antibodies. RSE cell cytokeratins were labeled with Alexa Flour 594 (red) antibodies. RSE nuclei were stained with DAPI (blue). Scale bar represents 100 µm. Abbreviations: recto-anal junction squamous epithelial (RSE), multiplicity of infection (MOI).
TABLE 2.
Quantitation of bovine recto-anal junction squamous epithelial (RSE) cells with adherent E. coli O157:H7 bacteria
| Bacteria tested | Bacterial adherence pattern | Bovine RSE cells (n = 160)a with adherent bacteria, in the ranges shown, for three different trialsb (MOI(c) = 10:1) | Percent Mean ± standard error of mean, of bovine RSE cells with adherent bacteria in the ranges showne | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Trial I | Trial II | Trial III | |||||||
| >10 | 1–10(d) | >10 | 1–10 | >10 | 1–10 | >10 | 1–10 | ||
| E. coli K12 | Diffuse, moderate | 2 | 130 | 2 | 114 | 0 | 80 | 0.83 ± 0.42 | 67.50 ± 9.21 |
| EDL933 (ATCC 43895) | Aggregative, moderate | 43 | 113 | 57 | 101 | 4 | 145 | 21.67 ± 9.91 | 74.79 ± 8.21 |
| FRIK1989 | Aggregative, strong | 96 | 62 | 94 | 64 | 107 | 53 | 61.88 ± 2.53 | 37.29 ± 2.11 |
| RM6067W | Aggregative, moderate | 75 | 82 | 63 | 94 | 26 | 126 | 34.17 ± 9.22 | 62.92 ± 8.21 |
| TW14588 | Diffuse with few aggregates, moderate | 62 | 93 | 22 | 126 | 2 | 129 | 17.92 ± 11.02 | 72.50 ± 7.21 |
Total number of cells evaluated in each trial.
Each trial had one slide per bacterial group. Each slide in turn had eight technical replicates spotted on it; 20 well-dispersed cells were evaluated per spot.
MOI, multiplicity of infection (106 bacteria:105 cells).
The number of bacteria adhering to each RSE cell is shown as a range of >10, and 1-10. The number of RSE cells without bacteria is not shown.
Percent mean for ranges used to determine “moderate or strong” adherence is shown in bold.
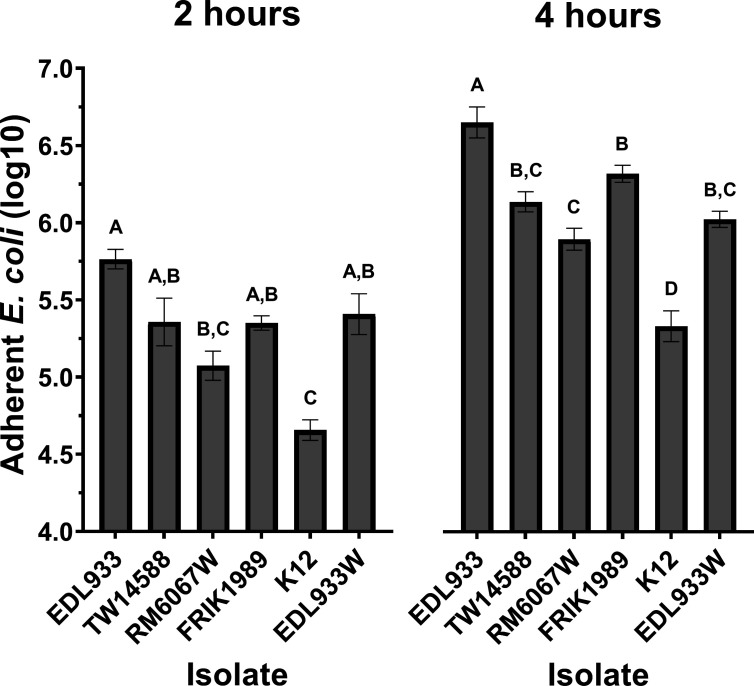
Differences in attachment phenotypes were observed with the E. coli O157:H7 isolates when using the human colon-derived Caco-2 cell line (Fig. 4). All isolates, with the exception of RM6067W, had statistically (P < 0.05) greater adherence than the non-pathogenic control K12 at 2 h. In addition, at 2 h, the only significant difference between the E. coli O157:H7 isolates was between EDL933 (6.3 × 105 CFUs) and RM6067W (1.4 × 105 CFUs) (P = 0.0002). In these experiments a curli-deficient EDL933 isolate named EDL933W was used to test the impact of curli on adherence. At 2 h, there was no significant difference in adherence between EDL933 (6.3 × 105 CFUs) and EDL933W (3.6 × 105 CFUs) (P = 0.1436). Progressing to the 4-h timepoint greater differences in adherence became apparent; all E. coli O157:H7 isolates now had statistically (P < 0.05) greater adherence to the Caco-2 cells than the non-pathogenic control K12. EDL933 had the greatest adherence (5.7 × 106 CFUS) and was statistically different from the other E. coli O157:H7 isolates TW14588 (1.5 × 106 CFUs, P = 0.0003), RM6067W (9.0 × 105 CFUs, P < 0.0001), FRIK1989 (2.2 × 106 CFUs, P = 0.0493), and EDL933W (1.1 × 106 CFUs, P < 0.0001). RM6067W had the lowest adherence of the E. coli O157:H7 isolates, with significantly less adherence than EDL933 (P < 0.0001), and FRIK1989 (P = 0.0053), although not significantly different from TW14588 or EDL933W. At this later timepoint, statistically less adherence of the curli-deficient EDL933W isolate compared with its curli-expressing EDL933 parental isolate (P < 0.0001) was detected, and EDL933W had adherence that did not differ statistically from the other curli-deficient isolates TW14588, RM6067W, and FRIK1989.
Fig 4.
E. coli O157:H7 in vitro adherence to Caco-2 cells. E. coli isolates (EDL933, TW14588, FRIK1989, RM6067W, EDL933W, or non-pathogenic K12) were incubated with Caco-2 cells to assess epithelial cell attachment. EDL933W represents a curli-deficient clone of EDL933. The average number of bacteria initially added to the Caco-2 cultures across all experiments was 1.4 × 105 CFU of each respective isolate. After 2 h and 4 h, non-adherent bacteria were removed via PBS washes, and adherent bacteria were released from cells using 1.0% Triton X-100 with subsequent serial dilution and plating on LB agar for CFU enumeration. Values represent the Log10 mean ± SEM of three independent experiments performed in triplicate wells for each experiment. A one-way ANOVA with Tukey’s comparison of means was performed. Connecting letter report displayed above bars signify differences between E. coli isolate (P ≤ 0.05), with those bars with the same letter having no significant difference between them.
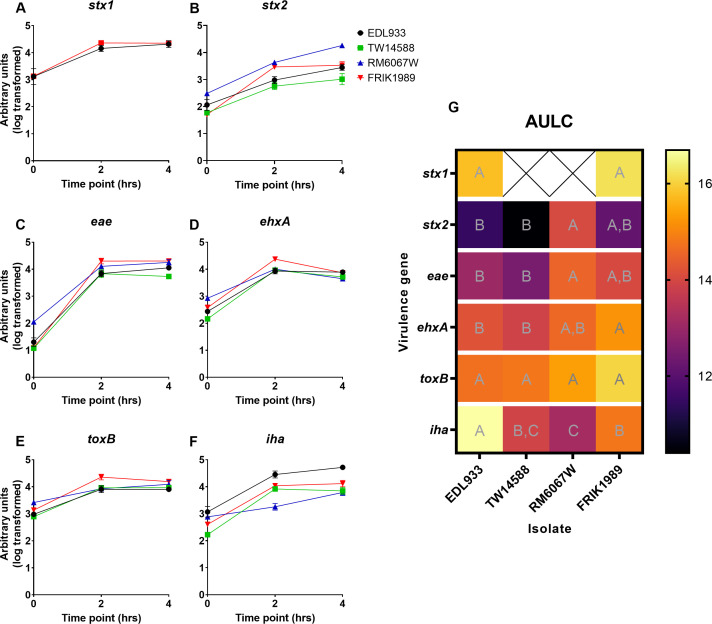
Quantitative RT-PCR
Gene expression of a selection of genes associated with virulence was explored across stationary and exponential growth (Fig. 5). These genes included, stx1, stx2, ehxA, eae, toxB, and iha. Differences in expression were assessed at specific points of growth (2 and 4 h) along with an AULC analysis (Fig. 5G) encompassing the entire time course; the AULC analysis was incorporated to capture overall trends in gene expression as growth phase had a large effect on expression across the isolates. Here, we defined the 2-h timepoint as mid-exponential growth, as it represents the bacteria at 36%–45% of maximal growth as determined in the preceding growth curve, and the 4-h timepoint was defined as being in late-exponential growth as the bacteria would be at 64%–72% maximal growth.
Fig 5.
E. coli O157:H7 quantitative RT-PCR virulence gene expression. Gene expression was assessed during a time course from stationary through late exponential growth with E. coli O157:H7 isolates grown in low glucose DMEM at 37°C. Gene expression was quantified using the relative standard curve method. (A–F) Time course gene expression graphs. (G) Heat map depicting area under the log curve analysis of time course data. A one-way ANOVA with Tukey’s comparison of means was performed. Connecting letter report within each gene row signify statistical differences between E. coli isolates for that gene (P ≤ 0.05), with those with the same letter not being significantly different.
Marked differences in expression of the toxin-encoding genes stx2 and ehxA was noted. Of the stx1-expressing isolates, EDL933 and FRIK1989, there was no significant difference in expression (Fig. 5A). With stx2 (Fig. 5B), which was expressed by all E. coli O157:H7 isolates tested, the isolate with the highest expression was RM6067W which was significantly greater than that of EDL933 (P = 0.0158) and TW14588 (P = 0.0044), but not FRIK1989 (P = 0.0675) with AULC analysis. The greatest expression of ehxA (Fig. 5D) when analyzed by AULC was with FRIK1989, which was significantly greater than EDL933 (P = 0.0090) and TW14588 (P = 0.0034), but not significantly different from RM6067W (P = 0.0839). Interestingly, under stationary growth (time zero), it was RM6067W that has the highest ehxA expression, which was significantly greater than all other isolates (EDL933 P = 0.0011, TW14588 P < 0.0001, FRIK1989 P = 0.0098). A shift in expression occurred under mid-exponential growth phase, as FRIK1989 had the highest ehxA expression [significantly greater than EDL933 (P = 0.0170), TW14588 (P = 0.0413), and RM6067W (P = 0.0382)].
Differences in expression of adherence related genes eae, toxB, and iha were also noted. RM6067W had the greatest expression of eae when compared with the other isolates when analyzed by AULC, which was significantly greater than EDL933 (P = 0.0373) and TW14588 (P = 0.0145) but not FRIK1989 (P = 0.6396) (Fig. 5C). However, the greatest differences were under stationary growth (time zero) as RM6067W eae expression was significantly greater than all other isolates (EDL933 P = 0.0025, TW14588 P = 0.0004, and FRIK1989 P = 0.0005). Under the mid-exponential growth phase, no comparisons yielded significant results. At late exponential growth, TW14588 had significantly less eae expression than EDL933 (P = 0.0242), RM6067W (P = 0.0014), and FRIK1989 (P = 0.0007). With toxB (Fig. 5E), no significant difference in expression was detected with the exception of under stationary growth (time zero). Here, RM6067W has a significantly greater expression than EDL933 (P = 0.0002), TW14588 (P < 0.0001), and FRIK1989 (P = 0.0041); the only other comparison to yield significant differences is TW14588 vs FRIK1989 (P = 0.0101). For iha (Fig. 5F) expression, EDL933 had the highest overall expression in the AULC analysis, which was significantly greater than TW14588 (P = 0.0029), RM6067W (P = 0.0004), and FRIK1989 (P = 0.0137); other comparisons that yielded significant results were RM6067W vs FRIK1989 (P = 0.0297). As all statistical comparisons could not be presented in Fig. 6A, detailed breakdown of all statistical comparisons is presented in Table S4.
Fig 6.
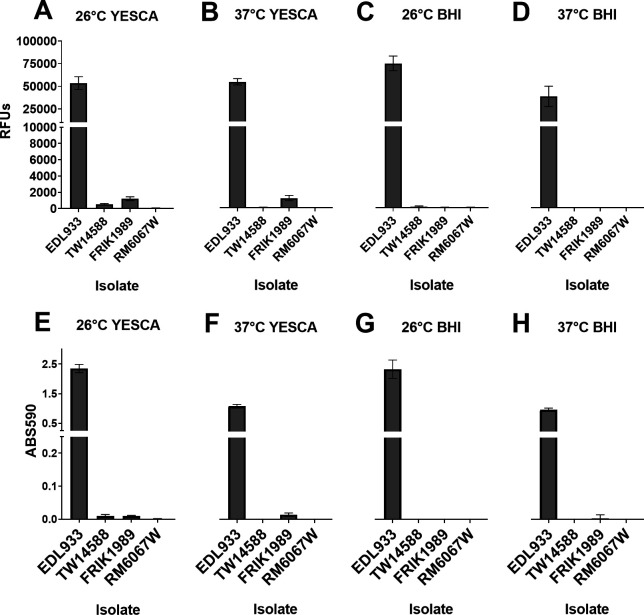
Biofilm production by E. coli O157:H7 isolates under various conditions. E. coli isolates (indicated on x-axis) were cultured statically in 96-well microtiter plates for 5 days at either 26°C or 37°C in YESCA broth or BHI broth. Following removal of non-adherent bacteria, biofilm production was evaluated by either the viability reagent resazurin (A–D) or with crystal violet staining (E and F). Resazurin indicates the viability of cells within the biofilm through the reduction of the non-fluorescent resazurin molecule to the fluorescent molecule resorufin via cellular NADP(H) dehydrogenase. Resazurin (0.03 mg/mL) was incubated with biofilms for 45 min, and relative fluorescence units (RFUs) were measured using 530 excitation and 590 emissions; values shown are background subtracted. Crystal violet binds to both biofilm mass and bacteria indicating the entire biofilm biomass. Crystal violet-stained biofilms were solubilized in 95% ethanol, and absorbance was measured at 590 nm (ABS590); a 1:5 dilution of the solubilized crystal violet was necessary as initial values obtained with the EDL933 isolate maxed out the detector. Values are background subtracted. Values represent the means ± 95% CI of one experiment consisting of three independent cultures per isolate performed in triplicate. Y-axis for B–D are not shown but are identical to A, and for F–H are identical to E Abbreviations: Relative fluorescence units (RFUs), Absorbance (ABS), Brain heart infusion (BHI).
Biofilm production, curli expression, and Shiga toxin production
Biofilm formation is an important consideration for bacterial virulence and persistence in various environments. Biofilm production was assessed for all four E. coli O157:H7 isolates after propagation in both YESCA or BHI broth at both 26°C and 37°C utilizing either crystal violet staining or resazurin reduction (Fig. 6). YESCA broth was chosen as it is a low nutrient and low salt broth that promotes biofilm and curli production (32, 33), whereas BHI represents an enriched media. Resazurin was chosen in addition to crystal violet staining as resazurin reduction provides a way to measure the viability of the cells within the biofilm matrix, whereas crystal violet is an indication of biofilm biomass. EDL933 produced robust biofilms under all test conditions. In YESCA broth, FRIK1989 produced measurable, albeit small, amounts of biofilm at both temperatures, but failed to produce detectable biofilms in BHI at either temperature. TW14588 produced a small amount of biofilm in YESCA broth at 26°C, similar to FRIK1989. No biofilm production was detected under any of the assayed conditions for the RM6067W isolate. In general, measuring these biofilms by either crystal violet staining or resazurin reduction yielded comparable results.
Curli expression by E. coli O157:H7, which can be detected with plating on Congo Red agar, is associated with biofilm production (34). EDL933 produced vibrant red colonies on Congo Red agar, whereas the other three E. coli O157:H7 isolates produced colonies of varying shades of pink, with RM6067W producing the lightest colored colonies (Fig. S2A through E).
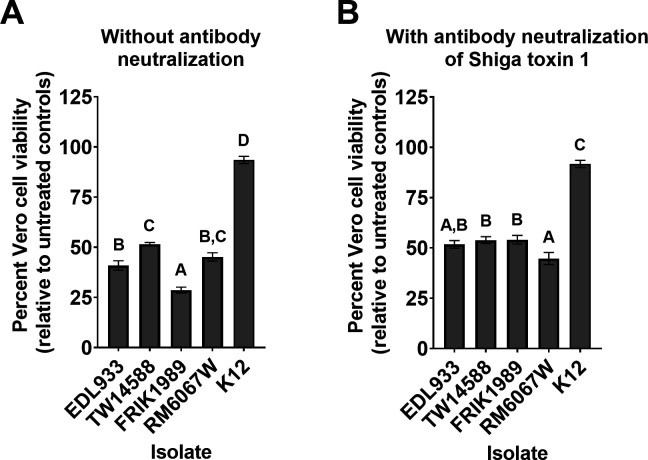
Stx production by E. coli O157:H7 is a major virulence factor for human pathogenesis (35, 36). Stx production by each isolate was assessed using the Vero cell cytotoxicity assay (37, 38) (Fig. 7). Stx cytotoxicity was inferred through the resulting loss in Vero cell viability as measured by the reduction of resazurin for each isolate (37, 38). The greatest Vero cell cytotoxicity resulted from FRIK1989 supernatants with a 71% loss of Vero cell viability, which was significantly different to that of EDL933 (P = 0.0005), TW14588 (P < 0.0001), and RM6067W (P < 0.0001). EDL933 supernatants resulted in a 59% loss of Vero cell viability, which was significantly different compared with TW14588 (P = 0.0027) but not RM6067W (P = 0.4619). TW14588 and RM6067W produced similar levels of Vero cell cytotoxicity at 48% and 55%, respectively (P = 0.1253). No appreciable loss of Vero cell viability was detected with supernatants derived from E. coli K12, an isolate that does not encode stx genes (Fig. 7A). It is important to consider the susceptibility of Vero cells to the different Stx types, as different Stx types are associated with more severe disease in humans (38–40).
Fig 7.
Vero cell cytotoxicity assay for Shiga toxin production by E. coli O157:H7 isolates. Bacterial supernatants were collected from late log phase cultures (6 h) of indicated isolate grown statically at 37°C in low glucose DMEM. Supernatants were added to confluent Vero cells for 48 h. Vero cell viability was determined using resazurin as described in the Materials and Methods. Higher Shiga toxin (Stx) levels are inferred from decreased Vero cell viability, displayed as percent viable relative to non-treated Vero cell controls. (A) Bar graph showing the percent viability of the Vero cells when treated with the 1:40 dilution of the E. coli supernatants with descriptive statistics. (B) Bar graph showing the viability of Vero cells when treated with a 1:40 dilution of the bacterial supernatants pre-incubated with Shiga-like toxin monoclonal antibody (13C4) for 2 h at a 1:50 dilution to neutralize Stx1 from EDL933 and FRIK1989; isolates TW14588 and RM6067W lack stx1. Values represent the mean ± SEM of two experiments performed in triplicate. A one-way ANOVA with Tukey’s comparison of means was performed. Connecting letter report displayed above bars signify differences between E. coli isolate (P ≤ 0.05), with those bars with the same letter having no significant difference between them.
To evaluate the contribution of Shiga toxin 1 vs 2 to Vero cell cytotoxicity, an anti-Stx1 antibody was employed to neutralize Stx1 in supernatants before addition to Vero cells (Fig. 7B). With this approach, RM6067W produced the greatest level of Vero cell cytotoxicity, with a 55% loss in Vero cell viability, which was significantly different to that of TW14588 (P = 0.0437), and FRIK1989 (P = 0.0378), but not EDL933 (P = 0.1763). EDL933, TW14588, and FRIK1989 produced similar levels of Vero cell cytotoxicity at 48%, 46%, and 46%, respectively, which were not significantly different (P ≥ 0.5). The greatest change in Vero cell cytotoxicity with neutralization of Stx1 was for FRIK1989, the isolate that overall had the greatest level of cytotoxicity before Stx1 neutralization.
DISCUSSION
Correlating E. coli O157:H7 genetics to environmental survival and pathogenicity may help predict pathogenic potential of an isolate and assist in limiting widespread human infection (41–43). Although considerable progress in inferring E. coli O157:H7 pathogenesis from genomic data has occurred, the relationship between E. coli O157:H7 genomic diversity and phenotypes expressed in various settings is still poorly understood (15, 44, 45). For example, although Shiga toxin is a known virulence mechanism for E. coli O157:H7 in humans, a collection of non-clinical E. coli O157:H7 isolates demonstrated varying levels of Shiga toxin-induced cytotoxicity, with many isolates producing levels of cytotoxicity comparable to clinical and outbreak isolates (46). Lineage I, I/II, and II E. coli O157:H7 isolates were recovered from cattle considered super-shedders (47). Varied biofilm and curli phenotypes from environmental E. coli O157:H7 isolates are common, with some isolates producing both curli- and curli+ colonies (48, 49). Thus, much remains to be understood in the relationship between genome and phenotype in various settings to assess risk potential of an isolate.
The four E. coli O157:H7 isolates used in this study had the eae and stx2 genes, suggesting all four could cause human illness (50). Although three of the isolates were associated with foodborne illness outbreaks (EDL933, TW14588, and RM6067W), the fourth isolate (FRIK1989) isolated from cow feces was not associated with an outbreak (41, 51, 52). This was despite FRIK1989 belonging to the LSPA-6 lineage I/II (often associated with human illness) and being genetically closely related to clinical E. coli O157:H7 isolates in the NCBI Pathogen Detection database. Regardless of genotype and phenotype, the association with a foodborne outbreak and/or human illness will be dependent on introduction into a human, and many steps in food production and preparation are designed to eliminate that from happening. Another factor to be aware of in studying the relationship between genotype and phenotype is the genome sequence data. In sequencing the isolates used in the current study, several genetic differences were noted when compared with deposited sequences for the same strain. Isolates of EDL933 acquired from different sources are known to display genetic and phenotypic differences (53). Hence, relying solely on public assemblies of the “same strain” may not reveal subtle but key differences when interpolating phenotypic behavior as it relates to genome sequence. Thus, it may be important for each laboratory to sequence their respective isolates under study.
Although the growth curves of the four isolates was unremarkable when grown in LB, when grown in low glucose DMEM, the EDL933 isolates OD measurements indicated a slower rate of growth and lower overall cell density once stationary phase was reached. We attribute these lower OD measurements to EDL933’s unique propensity to flocculate when grown in low glucose DMEM, which was not witnessed in the other three isolates. This flocculating phenotype of EDL933 is likely driven by its propensity to produce curli fimbriae (54) and would have implications in adherence and biofilm formation.
Understanding the relationship between E. coli O157:H7 and its animal reservoir (e.g., cattle) is a crucial step in developing mitigation strategies to reduce E. coli O157:H7 carriage in the animal and potential contamination of food products. There is great variation in reported shedding of E. coli O157:H7 from experimentally inoculated calves across different studies (55, 56). In most studies, a single E. coli O157:H7 isolate is used, which then limits comparisons of different isolates as it relates to shedding and/or colonization. The four E. coli O157:H7 isolates used in the current study were separated both chronologically and by isolation source (41, 51, 52) and tested simultaneously in groups of Jersey calves. Interestingly, no significant differences in colonization or shedding were detected. It was observed that at times during the cattle challenge, E. coli O157:H7 was detected with RAMS, but not in the feces, suggesting colonization will result in sporadic shedding and the absence of E. coli O157:H7 in feces does not necessarily mean the absence of E. coli O157:H7 in the gastrointestinal tract. Sporadic E. coli O157:H7 shedding has been observed in longitudinal cattle herd studies (57). The short plateau and subsequent rapid decline in recovered CFUs from the feces and RAJ, along with the absence of recovered CFUs in the later sampling timepoints, suggest that the E. coli O157:H7 isolates chosen colonized at low levels as many of the positive cattle at later timepoints had E. coli O157:H7 detectable only through enrichment cultivation. Similar colonization trends are observed in other experimentally infected cattle studies (55, 56, 58), suggesting that E. coli O157:H7 colonizes cattle at low CFU burdens or frequent transient colonization is sufficient to maintain E. coli O157:H7 in cattle herds. Studies report the absence of E. coli O157:H7 recovery from feces one day and then the presence of E. coli O157:H7 from feces the next day from the same animal (55, 58), which was observed in several cattle from this study. Given that O157 readily attaches to bovine intestinal cells (59, 60), some amount of colonization is anticipated, and it is unclear how a small amount of shedding relates to contamination at processing.
Although shedding was greater than 104 CFUs/g feces at some time points, which is the amount indicative of a super-shedder (8, 61), shedding at >104 CFUs/g feces was transient and not detected in all cattle within a group. Super-shedding is thought to play a prominent role in environmental and cattle carcass contamination with E. coli O157 (62, 63). RM6067W was the only isolate to have all inoculated cattle shed at >104 CFUs/g feces (i.e., super-shedder levels), but this level of shedding was again transient and for three cattle was only noted at a single sampling timepoint. Given the definition of super-shedder (>104 CFUs/g feces), all four E. coli O157:H7 isolates resulted in the transient presence of super-shedding cattle. The role of biofilm sloughing off in the intestines is suggested to play a role in super-shedding (61), yet here, no significant difference in shedding observed between the poorest biofilm producer RM6067W and the strongest biofilm producer EDL933. The relationship between biofilm formation and sloughing in cattle and the observed in vitro biofilm results is unclear, but our data suggest there is little relationship between the two phenotypes.
Although animal studies are useful to understand the complex interactions between E. coli O157:H7, commensals, and the animal host, they cannot be readily performed. Thus, in vitro attachment to epithelial cells, including primary bovine RAJ squamous epithelial (RSE) cells (59, 60) or Caco-2, could serve as a surrogate to in vivo trials. In this study, the human-derived Caco-2 cell line was used to allow contrast and comparison to the bovine RSE cells along with capturing differences that may be relevant towards cattle versus human virulence. E. coli O157:H7 isolates that persist in cattle herds have strong attachment phenotypes (64). Thus, inferences may be made that E. coli O157:H7 isolates with strong cell culture attachment phenotypes may translate to greater colonization in cattle, although such a relationship has not been clearly determined. Hence, to better approximate RAJ colonization, ex vivo bovine RSE cells were utilized to assess E. coli O157:H7 attachment (59, 60). FRIK1989, RM6067W, and EDL933 had greater quantitative attachment to RSE cells than non-pathogenic K12, but not TW14588. In the case of the Caco-2 cells EDL933 had the strongest attachment phenotype, although for the RSE cells, FRIK1989 had the strongest attachment phenotype. Although the weakest attachment phenotypes across the two models were TW14588 for the RSE cells and RM6067W with the Caco-2 cells. A clear association between the two models in similarity is lacking, but these results may underline how different E. coli O157:H7 genetic backgrounds influence cattle vs human colonization. With the Caco-2 cells, EDL933s aggressive adherence appears to be due in part to curli expression, as its curli deficient clone EDL933W had reduced adherence that was then similar to the other curli deficient isolates. Of interest, RM6067W had the highest overall eae expression but the poorest adherence to the Caco-2 cells. However, RM6067W did have lower expression of the adhesin iha, which has been shown to contribute significantly to Caco-2 adherence (65). Conversely, with the RSE cells curli expression appears unrelated to strong adherence phenotypes as FRIK1989 was the only isolate to have a strong aggregative adherence phenotype. In addition, iha expression may be less relevant to RSE attachment, as EDL933 had the highest expression, and RM6067W had the lowest expression, yet it was TW14588 that had the weakest adherence to RSE cells. It should be noted that TW14588 did have the lowest eae expression; however, eae has not been shown to influence RSE attachment with E. coli O157:H7 (60). The expression results do not point to a possible explanation for FRIK1989s stronger adherence with RSE cells; however, a limited number of genes were examined. The phenotypes derived from these in vitro assays could be useful in probing genetic differences among isolates, but further investigation is needed to address the association with the in vivo results.
Biofilms can facilitate transfer of E. coli O157:H7 from contaminated food to contact surfaces, promote E. coli O157:H7 resistance to disinfectants, enhance environmental survival, and likely play a role in cattle colonization and shedding (61, 66–68). The stark difference in biofilm production between the isolates is expected as biofilm production in E. coli O157:H7 is strain specific (52). EDL933 produced the greatest biofilm mass and showed highest curli expression yet colonized and shed from cattle similar to the other isolates that produced poor biofilms/curli expression. A link between biofilm production and cellular attachment has been proposed (69), and EDL933 also showed the greatest cellular attachment to the Caco-2 cells. FRIK1989 displayed a strong attachment phenotype on the RSE cells yet was a low biofilm producer and low curli producer, indicating that additional elements are involved in cellular attachment versus biofilm formation. Of note, the isolates that produced poor biofilms all had SNPs in genes associated with biofilm production (Fig. 1B; Table S1), however, further empiric evidence would be needed to definitely correlate our results with these specific mutations. Biofilm production may be more relevant in environmental persistence (68), which would be advantageous to the organism in disseminating itself along production chains. EDL933 curli deficient mutants were observed to have poorer persistence on spinach leaves than curli expressing EDL933 (70), and yet we observed the produce-associated isolates (TW14588 and RM6067W) to be poor curli producers. Others have observed that curli expression doesn’t impact internalization into spinach roots using mutants of the 86–24 E. coli O157:H7 isolate (71). This suggests that biofilm and curli expression likely contribute to produce colonization but is dependent on the specific interactions between bacteria and the plant environment. This absence of a strict association between biofilm formation and virulence, as seen here with these E. coli O157:H7 isolates, has also observed with the pathogenic bacteria Listeria monocytogenes (72).
Shiga toxin production is an important consideration for E. coli O157:H7 virulence in humans (50). Although the presence of Stx-encoding genes suggests the potential of E. coli O157:H7 to cause disease in humans, the toxin gene type and expression level play an important role in the severity of the disease (73). For instance, Stx2 subtypes are associated with more severe clinical outcomes (38–40). The four O157 isolates studied had different stx profiles (Fig. 1B). RM6067W that caused the most hemolytic uremic syndrome (HUS) cases in an outbreak (74–76), possessed both stx2a and stx2c, had the highest stx2 gene expression and produced the greatest Vero cell cytotoxicity when Stx1 was neutralized. On the other hand, FRIK1989 also possessed stx2a and stx2c, but stx1a was the major contributor to its Vero cell cytotoxicity. The Vero cell cytotoxicity assay has been traditionally used to infer Stx expression and potential adverse clinical outcome of E. coli O157:H7 isolates. In the absence of these data, only assessing for the presence/absence of various stx genes may not be sufficient to inform virulence potential of various isolates. However, the Vero cell cytotoxicity results may not always correlate with RT-qPCR gene expression results. Although our highest stx2 gene expresser, RM6067W, also caused the highest Vero cell cytotoxicity, EDL933 had significantly reduced stx2 gene expression compared with RM6067W yet was not significantly different in Vero cell cytotoxicity. Additionally, FRIK1989 did not have significantly different stx2 gene expression from RM6067W yet caused significantly less Vero cell cytotoxicity. These differences between RT-qPCR and the Vero cell cytotoxicity results could be the result of the different sampling time points for the two assays, with the RT-qPCR expression sampled from 2- and 4-h exponential phase cultures, whereas the Vero cells were treated with supernatants from 6-h cultures that would be entering the stationary phase; additionally, mRNA levels do not necessarily correlate with protein abundance (77).
Overall, diverse phenotypes within E. coli O157:H7 were appreciated, yet a clear phenotypic relationship to cattle shedding and human disease has not been fully identified. Phenotypic heterogeneity is attributed to improved fitness in virulent bacterial populations (78) and could help explain the variation of observed phenotypes between these E. coli O157:H7 isolates. It was somewhat surprising that differences in shedding were not detected in the animal trial, despite phenotypic differences among the isolates. Of note, TW14588 cattle colonization and fecal shedding counts were on average lower than the other E. coli O157:H7 isolates, and at the same time, TW14588 attachment counts to RSE cells were lower than the other tested E. coli O157:H7 isolates. These data suggest E. coli O157:H7 attachment to RSE cells could be useful in predicting cattle colonization and fecal shedding, although additional testing is warranted, including more cattle per group and more isolates comparable to TW14588, to evaluate the consistency in relationship between in vitro and in vivo phenotypes. Testing additional phenotypic profiles, such as carbon source utilization or acid resistance, may fill in the gaps presented here and help in finding additional phenotypes that can correlate to animal carriage or environmental survival. Determining which isolate has the highest expression of a tested virulence gene is dependent on the growth phase of the isolate. Under stationary growth, an isolate may have higher or lower relative expression to another isolate, but in late exponential growth, this relationship may shift, which emphasizes the importance in defining growth conditions to match the desired environment.
MATERIALS AND METHODS
Isolate selection and culture
Four E. coli O157:H7 isolates (EDL933, TW14588, FRIK1989, and RM6067W) were selected for analysis in this study based on their LSPA-6 lineage and isolation source (Table 1) (51, 75, 76). E. coli K12 was used as an avirulent control in in vitro experiments. E. coli isolates EDL933 (ATCC 43895) and K12 (ATCC 29425) were obtained from the American Type Culture Collection (ATCC, Manassas, VA). E. coli isolates FRIK1989 and TW14588 were obtained from The Thomas S. Whittam STEC Center, Michigan State University, MI. E. coli isolate RM6067W was obtained from Michelle Q. Carter at the Produce Safety and Microbiology Unit, Western Regional Research Center, Agricultural Research Service, USDA. E. coli was grown on LB agar plates at 37°C overnight and then maintained at 4°C; fresh cultures were re-cultured every 2 to 3 weeks. To create the cattle challenge inoculum, the isolates were grown in 10-mL LB broth at 37°C at 190 rpm overnight, these cultures were diluted 1:100 in LB broth and grown at 37°C at 190 rpm for 7.5 h. The cultures were then pelleted, resuspended in 10-mL LB broth supplemented with 10% glycerol, and frozen. The frozen diluted stocks (10 mL) were mixed with 90 mL PBS to create an inoculum for challenge. Vero cells were obtained from the American Type Culture Collection (ATCC CCL-81) and cultured in DMEM (11995–065, Gibco, Thermo Fisher Scientific, Waltham, MA) supplemented with 10% FBS at 37°C and 5% CO2 in T75 tissue culture flasks; flasks were trypsinized and subcultured twice weekly. Caco-2 cells were obtained from the American Type Culture Collection (ATCC HTB-37). Caco-2 cells were cultured at 37°C and 5% CO2 in DMEM (Gibco, Thermo Fisher Scientific, Waltham, MA) supplemented with 10% FBS. T75 flasks of Caco-2 cells were trypsinized and subcultured when confluent.
Growth curves of the E. coli isolates
Growth curves of the isolates were determined using a Bioscreen microplate reader (Growth Curves USA, Piscataway, NJ) to discern differences in growth rate, which could indicate differences in substrate utilization or varied growth characteristics. Cultures were grown overnight in LB broth at 37°C with shaking at 200 rpm from a colony scrape off a LB agar plate. These overnight cultures were standardized to OD600 value of 0.5, and then diluted 1:100 in test media [low glucose DMEM (11054–020, Gibco, Thermo Fisher Scientific, Waltham, MA) or LB broth (L7275-500TAB, Sigma-Aldrich, St. Louis, MO)]. The diluted cultures were pipetted (300 µL) into the wells of a honeycomb plate. The Bioscreen growth conditions were static growth at 37°C with wideband filter readings taken every 30 min for 24 hours.
Polymorphic amplified typing sequence (PATS)
PATS profiles of the test isolates were determined to enable accurate identification of these isolates when recovered from cattle feces and RAJ mucosa swabs. Colony lysates, prepared from each bacterial isolate cultured on LB agar plates, were each tested in triplicate to confirm the PATS profiles as described elsewhere (29–31). Briefly, this bacterial genetic fingerprinting was done using primer pairs targeting the eight polymorphic XbaI-, seven polymorphic AvrII- restriction enzyme sites, and the four virulence genes encoding Shiga toxins 1 and 2 (Stx1 and Stx2), intimin-γ (eae), and hemolysin-A (hlyA); hot start, touchdown PCR was used to generate amplicons from colony lysates (29–31). Amplicons with the putative AvrII- restriction enzyme sites were purified using the QIAquick PCR purification kit (Qiagen, Valencia, CA),and digested with the AvrII restriction enzyme (New England Biolabs, Beverly, MA) to confirm the presence of the restriction site. All reactions were analyzed by electrophoresis on 3% agarose gels stained with ethidium bromide. The presence or absence of amplicons for XbaI and the virulence genes was recorded using “1” and “0”, respectively. The absence of an AvrII amplicon was recorded as “0”, and the presence of restriction site with a SNP as “1”, “2” for an intact restriction site, and “3” for a restriction site duplication (29–31).
DNA extraction & library preparation
The four strains/isolates used in the cattle shedding study and phenotypic assays (RM6067W, TW14588, EDL933, and FRIK1989) were sequenced in-house to provide the most accurate representation of their genotype. Genomic DNA (gDNA) was extracted from the culture used to inoculate calves with the DNeasy blood and tissue Genomic-tip kit (Qiagen, Hilden, Germany). gDNA quality was assessed by Nanodrop spectrophotometry (Thermo Fisher Scientific, Waltham, MA) and a Qubit fluorometer DNA broad range kit (Thermo Fisher Scientific, Waltham, MA). Nanopore libraries were prepared with rapid barcoding kit (SQK-RBK004) according to the manufacturer’s instructions (Oxford Nanopore, Oxford, U.K.). The Nextera Flex kit (Illumina, San Diego, CA) was used to prepare the genomic library for MiSeq sequencing.
Genomic sequencing and assembly
Long-read sequencing was performed on an Oxford Nanopore MinION instrument using a FLO-MIN106 R9.4.1 flow cell for 48 h. Bases were called and data de-multiplexed with Guppy v. 3.1.5 (79). Reads with a quality (Q) score of less than seven were removed from the analysis. Long-read sequencing data were independently assembled with the Flye assembler v2.7 (80, 81). Short-read sequencing was performed on an Illumina MiSeq instrument for 500 cycles (2 × 250) v2 kits with the Nextera Flex protocol. The resultant reads were trimmed and filtered with the BBTools software package (82). Filtered short reads were used to polish the long-read assemblies with Pilon (83), and reads were mapped to confirm the final assemblies with BBMap (82).
In silico analysis of bacterial genomes
The LSPA6 lineages were assigned by a custom script (https://github.com/USDA-FSEPRU/O157LineageAssignment). Virulence genes were screened for each isolate using the Specialty Genes function of PATRIC (https://www.patricbrc.org) utilizing the virulence factor property and VFDB source (84, 85). Polymorphisms in genes were detected using Snippy (86) with EDL933 as the reference genome. Genes of interest were extracted, translated, and aligned using Geneious Prime (https://www.geneious.com/prime/) to visualize polymorphisms in sequences. In silico stx gene subtyping was also performed in Geneious Prime utilizing published subtyping primer sequences (87). Additionally, the genomes were annotated using Bakta v1.8.0, database v5.0, full (88). We compared the assemblies of our isolates to publicly available assemblies with the same strain identifier designations. Two of the NADC isolates had alternative assemblies available, EDL933 and TW14588. We identified SNPs between our assemblies and the public alternatives using Snippy (86) and determined gene presence/absence differences using a pangenome generated by Roary (89). RAxML (90) was used to generate a maximum likelihood phylogenetic tree from core genome alignments.
Phylogenetic tree
We used genomes available in the NCBI pathogen detection project (91) (version PDG000000004.3284) to place our infection genomes in the phylogenetic context of all publicly available E. coli O157:H7 genomes. First, all stx- and eae-positive genomes were identified and downloaded. For a genome to be considered stx positive, it needed to contain both the A and B subunits of either stx1 or stx2, as identified by the AMRFinderPlus (92) tool (part of the NCBI pathogens pipeline). These downloaded genomes were then serotyped with the tool ‘ectyper’ (93). Only genomes with a serotype of “O157:H7” were considered for further analysis. The LSPA-6 lineage typing scheme (14, 94) was used to assign genomes into one of three lineages via a tool available at https://github.com/USDA-FSEPRU/O157LineageAssignment. To assess the large-scale phylogenetic structure of these available genomes, one genome per SNP cluster was randomly selected as a representative. In addition to these representative genomes, the reference genomes E. coli O157:H7 str. Sakai (GCF_000008865.2), E. coli O157:H7 str. 86–24 (GCF_013168095.1) and E. coli O157:H7 str. TW14359 (GCF_000022225.1) and the infection isolates sequenced as part of this publication were added, and a pangenome was constructed using ppanggolin (95). A concatenated multiple sequence alignment was constructed from the core genome, and a maximum likelihood tree was inferred using RAxML (90) using a GTRGAMMA model.
Cattle challenge and shedding enumeration
Ten-month-old Jersey steer and heifer calves were purchased from an Iowa farm and transported to the NADC in Ames Iowa. Upon arrival calves were housed in a BSL2 barn facility. One room was dedicated to each inoculation group, and each room had two pens to house up to two calves per pen. Cross-contamination was reduced by apparel changes between each room by caretakers and researchers. Apparel changes included room specific overalls and boots. New gloves and N95 respirators were also worn in each room. Calves were given ad libitum access to food (alfalfa cubes and hay) and water for the duration of the experiment. All animal protocols were approved by the National Animal Disease Center Animal Care and Use Committee. The calves were pre-screened for the presence of E. coli O157:H7 and were found to be negative based on O157 agglutination testing. The trial was performed as two replicate studies performed in succession. Data were combined after no statistical differences were noted across the replicate trials. For trial 1, four calves per isolate were orally inoculated, and for trial two, four calves were in each group for FRIK1989 and TW14588 , and three calves were in each group for EDL933 and RM6067W. Thus, data from eight calves each were generated for FRIK1989 and TW14588 and seven calves each for EDL933 and RM6067W. Calves were inoculated orally with 10 mL of the prepared inoculum, which contained approximately 6 × 109 total CFUs of each respective isolate. The inoculum was administered using a slip-tip syringe to slowly drip inoculum in the back of the mouth. Rooms were washed down daily to remove fecal material.
Fecal samples were collected 6 or 9 days pre-challenge, on the day of challenge, and on days 1, 2, 3, 4, 5, 7, 9, 11, and 14 post-challenge. Bacterial enumeration of fecal E. coli O157:H7 was performed by direct and enrichment culture on selective media, as described elsewhere (96). Briefly, non-enrichment (direct) E. coli O157:H7 culture enumeration involved placing 10 g of feces in 50 mL of tryptic soy broth supplemented with cefixime (50 µg/L), tellurite (2.5 mg/L), and vancomycin (40 mg/L) (TSB-CTV) with vortexing to generate a homogenous suspension. The fecal suspension was serially log diluted in sterile 0.9% saline and 100 µL spread plated on sorbitol MacConkey agar plates (SMAC) supplemented with 4-methylumbelliferyl-β-D-glucuronide (100 mg/L) (MUG), which were cultured overnight at 37°C. Enrichment culture involved incubating the 10 g of feces in TSB-CTV broth for 18 h at 37°C and 150 rpm, followed by serial log dilution and spread plating as noted for the non-enrichment cultures above with the exception that SMAC-CTMV plates were used. Sorbitol-negative and MUG-negative colonies were subjected to the E. coli O157:H7 latex agglutination test kit (Thermo Fisher Scientific, Waltham, MA) to confirm serotype, with subsequent PATS typing. The limit of detection for fecal direct culture plating was 50 CFUs/g feces; with this in mind, enrichment-only positive cultures were assigned an arbitrary value of 25 CFUs/g feces for subsequent data analysis. Recto-anal junction (RAJ) mucosal colonization by the four test E. coli O157:H7 was evaluated by sampling the RAJ of individual calves with four foam-tipped swabs as described elsewhere (97). The swabs were initially placed in 10 mL of TSB-CTV media for transport back to the laboratory. The tube with swabs was vortexed, and then an additional 40 mL of TSB-CTV media was added and mixed well. Bacterial cultivation and enumeration followed that stated above for fecal E. coli O157:H7 counts. The limit of detection for RAJ direct culture plating was 10 CFUs/mL, with this in mind enrichment only positive cultures were assigned an arbitrary value of 5 CFUs/mL for subsequent data analysis. Cumulative fecal shedding and RAJ colonization were investigated using an area under the log curve analysis, and statistical differences were determined by one-way ANOVA with Tukey’s comparison of means, where P < 0.05 was considered significant (R studio 4.1.2; https://www.rstudio.com).
Ex vivo recto-anal junction squamous epithelial (RSE) cell attachment assay
RSE cell attachment was performed to tests whether it could be a useful surrogate for in vivo colonization experiments and to compare and contrast to attachment phenotypes derived from the human cell line Caco-2, which could underscore different genotypes related to cattle vs human colonization. The RSE cell attachment assays were performed as described elsewhere (33, 60, 98), using bacterial isolates cultured overnight in Dulbecco’s modified Eagle’s medium with low glucose (DMEM-LG; Invitrogen, Carlsbad, CA) at 37°C without aeration, washed, and re-suspended in DMEM with no glucose (DMEM-NG; Invitrogen, Carlsbad, CA) (33, 59, 98). In addition to the test of E. coli O157:H7 isolates, E. coli K12 was included as a comparative control in these assays. Assays were done in triplicates, with eight technical replicates per bacterial isolate per assay. Briefly, RSE cells, collected from bovine rectal–anal junction (RAJ) tissue at necropsies, were suspended in DMEM-NG to a final concentration of 105 cells/mL and mixed with bacteria at a bacteria:cell ratio of 10:1 (33, 59, 98). The mixture was incubated at 37°C with aeration (110 rpm) for 4 h, pelleted, washed, and reconstituted in 100 µL of double-distilled water (dH2O). Eight drops of the suspension (2 µL) were placed on Polysine slides (Thermo Scientific/Pierce, Rockford, IL), dried, fixed, and stained with fluorescent-tagged antibodies specific to the O157 antigen and cytokeratins within the RSE cells (59). The fluorescein isothiocyanate (FITC; green)-labeled goat anti-O157 (KPL, Gaithersburg, MD, USA) antibody targeting the O157 antigen and the mouse anti-(PAN) cytokeratins (AbD Serotec, Raleigh, NC, USA) in combination with Alexa Fluor 594 (red)-labeled goat anti-mouse IgG (H + L; F(ab´)2 fragment) (Invitrogen) targeting the RSE cell cytokeratins were used (59). The primary rabbit anti-E. coli (Thermo Scientific Pierce) antibody and the secondary Alexa Fluor 488 (green)-labeled goat anti-rabbit IgG (H + L; F (ab’)two fragment) (Invitrogen) targeting the anti-E. coli antibody were used to detect E. coli K12 (59).
Attachment patterns on RSE cells were qualitatively recorded as diffuse, aggregative, or nonadherent, and quantitatively as the percentages of RSE cells with or without adhering bacteria (33, 60, 98); attachment was recorded as strongly adherent when more than 50% of RSE cells had 10 adherent bacteria, moderately adherent when 50% or less of the RSE cells had 1 to 10 adherent bacteria, and nonadherent when less than 50% of the RSE cells had only 1 to 5 adherent bacteria. RSE cells with no added bacteria were subjected to the assay procedure and used as negative controls to confirm absence of pre-existing E. coli O157:H7. Quantitative data were compared between isolates for statistical significance using one-way ANOVA with Tukey’s multiple comparisons test; P < 0.05 was considered significant (GraphPad Prism version 8.0.0, GraphPad Software, San Diego, CA).
Congo red binding of the E. coli isolates
Twenty microliter drops of a 1:10 dilution of OD600 0.5 bacterial suspensions in PBS were spotted onto Congo red agar (YESCA agar (0.1% yeast extract, 1% casamino acids, 15 g agar/L), 40 µg/mL Congo red dye, and 6.24 µg/mL Coomassie brilliant blue G-250) and cultivated at 26°C for 48 h (34). Representative photographs were taken; red colonies indicate Congo red binding to curli fimbria, which is associated with adhesion and biofilm formation. A Congo red-negative EDL933 isolate was generated through the serial passage of our Congo red-positive isolate on Congo red agar; the Congo red phenotype of this isolate was stable over successive Congo red passages and produced a PATS profile identical to the parental isolate. This curli-deficient isolate was used in the in vitro cell attachment experiments.
In vitro Caco-2 cell line attachment assay
In vitro cell attachment phenotypes were determined using human-derived Caco-2 cells as means to compare and contrast to the attachment phenotypes derived from the bovine RSE cells. Confluent Caco-2 cells from T75 flask were trypsinized and diluted to 100,000 cells/mL in DMEM (Gibco 11995–065, Thermo Fisher Scientific, Waltham, MA) supplemented with 10% heat-inactivated FBS (Cytiva SH30396.03, HyClone, Logan, UT), with 200 µL of this suspension pipetted into the wells of sterile collagen coated 96-well tissue culture plates (20,000 cells/well) (Biocoat 356407, Corning, Kennebunk, ME). These tissue culture plates were then cultivated at 37°C and 5% CO2 for 15 days to obtain polarized monolayers; media changes were performed every 4 days. To prepare the E. coli, a colony scrape taken from an LB agar plate of E. coli was inoculated into 5 mL of low glucose DMEM (Gibco 11054–020, Thermo Fisher Scientific, Waltham, MA) and grown overnight statically at 37°C. Following overnight growth, a 1:20 dilution of these cultures was made into 5 mL of fresh low glucose DMEM, and these were cultured statically at 37°C for 2 h. The OD600 of the 2-h E. coli cultures was then measured and adjusted to an OD600 0.08, and a 1:3 dilution in low glucose DMEM was made. The media on the Caco-2 cells were removed and replaced with 200 µL of low glucose DMEM, then 10 µL of the diluted E. coli suspensions was added to the Caco-2 cells, the tissue culture plates were centrifuged at 250 rcf for 10 min at room temperature to pellet the bacteria onto the Caco-2 cells. The tissue cultures plates were then placed in an incubator at 37°C and 5% CO2 for either 2 or 4 h to allow for the E. coli to attach to the Caco-2 cells. Following the co-incubation timepoints, the media with non-adherent bacteria were removed, and the wells were washed thrice thrice with PBS to further remove non-adherent bacteria. Then, 200 µL of Triton X-100 (1.0% in PBS) was added for 10 min to lyse the Caco-2 cells and release the adherent bacteria; a serial log dilution was made of these in PBS. The serial dilutions were spotted (3 × 20 µL spots per dilution) onto LB agar plates and grown overnight at 37°C to determine adherent CFU counts. Three independent experiments were performed with triplicate wells for each E. coli isolate. Statistical analysis was done by one-way ANOVA with Tukey’s comparison of means to identify significance in adherent phenotype between E. coli isolates (GraphPad Prism version 9.0.1, GraphPad Software, San Diego, CA).
RNA isolation and quantitative real-time PCR
Gene expression of several shared genes related to adherence and toxin production were assessed to allow association with relevant phenotypes and if differences in expression were associated with gene polymorphisms. E. coli cultures for RNA isolation were prepared as follows. A colony scrape off of an LB agar plate was inoculated into low glucose DMEM (Gibco 11054–020, Thermo Fisher Scientific, Waltham, MA) and cultured overnight statically at 37°C. From these overnight cultures, a 1:20 dilution was made into fresh low glucose DMEM, and these were cultured statically at 37°C for 18 h to achieve stationary growth. At this time, 500-µL aliquots of these cultures were removed and placed in 1-mL RNAProtect Bacteria reagent (QIAGEN, Germantown, MD); these samples represented our stationary phase time zero samples. Immediately after sampling from the 18-h cultures, these cultures were diluted 1:20 in low glucose DMEM and cultured statically at 37°C. At 2 and 4 h, 500-µL aliquots were removed and placed in 1 mL RNAProtect Bacteria reagent; these samples represented mid and late exponential phase cultures. All samples placed in RNAProtect Bacteria reagent were vortexed and incubated at room temperature for 5 min. Following the 5-min incubation, the samples were centrifugated at 5000 rcf for 10 min, then the supernatants were decanted, and the treated cell pellets were stored at −80°C for later RNA isolation. For RNA isolation, the samples were thawed on ice and treated with 200 µL of 1 mg/mL lysozyme (Sigma-Aldrich L-7651, St. Louis, MO) in tris-EDTA buffer pH 8.0 (Sigma-Aldrich, St. Louis, MO), vortexed, and incubated on a shaker for 5 min at room temperature. Following the lysozyme treatment, 700 µL of RLT buffer (QIAGEN, Germantown, MD) with 1% β-mercaptoethanol was added and vortexed. Then, 500 µL ethanol (100%) was added followed by gently mixing. The remaining RNA isolation steps used the RNeasy Mini kit 74106 (QIAGEN, Germantown, MD) following the manufacturer’s protocol with the optional on-column DNase digestion (RNase-Free DNase, Qiagen, Germantown, MD). RNA quality and quantity were measured using a 4200 TapeStation system (Agilent, Santa Clara, CA) following the manufacturer’s protocol for the RNA ScreenTape (Agilent, Santa Clara, CA). Reverse transcription of the RNA used the iScript Reverse Transcription Supermix (Bio-Rad, Hercules, CA) with 250 ng RNA per 20-µL reaction following the manufacturer’s protocol using a Veriti 96-well thermal cycler (Applied Biosystems, Thermo Fisher Scientific, Waltham, MA). Following reverse transcription, the cDNA was diluted 1:2 with PCR grade H2O and stored at −20°C. Quantitative PCR used the iTaq Universal SYBR Green Supermix (Bio-Rad, Hercules, CA) with a Bio-Rad CFX 96 Touch Real-Time PCR system with 2 µL of cDNA and 300 nM primer concentration per 20-µL reaction. Thermocycling conditions were as follows, initial denaturing step 95.0°C for 30 s, then 40 cycles of 95.0°C for 10 s, 55.0°C for 30 s, 72.0°C for 30 s, then plate read. After the 40 cycles, a melting curve from 65.0°C to 95.0°C in 0.5°C increments were included. Primers used are shown in Table 3. Gene expression was determined using a relative standard curve that consisted of a 5 log 5-point standard consisting of pooled cDNA template; PCR efficiencies were also derived from these standards. Initially the ΔΔCt method was chosen but was dropped for the relative standard curve method when it was determined that the reference genes chosen (rpoS and gapA) were not stably expressed across the time course by the isolates. A total of three biological replicates per isolate were included. Statistical analysis was done by one-way ANOVA with Tukey’s multiple comparison test to identify significant difference in gene expression on individual time points and on the AULC for the entire time course (GraphPad Prism version 9.0.1, GraphPad Software, San Diego, CA); for stx1 expression, a t-test was performed as only two isolates expressed this toxin.
TABLE 3.
Primers used for quantitative RT-PCR
| Gene/description | Primer sequence 5’−3’ | PCR efficencya | Reference |
|---|---|---|---|
| stx1/Shiga toxin type I | |||
| GACTGCAAAGACGTATGTAGATTCG | 94% | This publication | |
| TCAATCATCAGTAAAGACGTACCTCC | |||
| stx2/Shiga toxin type II | |||
| ATTAACCACACCCCACCGG | 96% | This publication | |
| TGCCGTATTAACGAACCCG | |||
| iha/Bifunctional siderophore receptor/adhesin | |||
| GCTGGCTATGATCATACTTTC | 102% | This publication | |
| AGCCCCCATTTGTCGCGC | |||
| eae/Intimin | |||
| GCTTACTATTACCGTTCTGT | 93% | This publication | |
| CGTTTTGGCACTATTTGCCC | |||
| toxB/Toxin B | |||
| ATATCGTAAGTAACTCAGGAAAC | 89% | This publication | |
| GTATCCGTCGATGTTATTACTTG | |||
| ehxA/Enterohaemolysin | |||
| AGATGCAGCCCTGACAACAA | 95% | This publication | |
| CCGGTTAATGCACTCACCAG | |||
PCR efficiency represents the average for the primer set across the experiments. PCR efficacy was determined from a 5 log 5-point standard curve using pooled cDNA template.
Biofilm production
Biofilm production was assessed as biofilms have been implicated in environmental persistence and colonization and shedding in cattle. Biofilm production was determined through the commonly used crystal violet stain and viability reagent resazurin on polystyrene microtiter plates (99–101); crystal violet was employed to assess biofilm biomass, whereas resazurin was used to give an indication of cellular viability within the biofilms. Overnight E. coli cultures grown in YESCA broth (0.1% yeast extract and 1% casamino acids) from colony scrapes were diluted to identical OD600 0.5 suspensions and diluted 1:10 in test media [YESCA or Brain Heart Infusion (BHI, CM1135, Oxoid, Basingstoke, UK)]. Two hundred microliters of the suspensions in test media was pipetted into sterile 96-well high-binding flat bottom microtiter plates and maintained at either 26°C or 37°C for 5 days. To quantify biofilm production by crystal violet staining, the media with non-biofilm embedded bacteria were removed, and the wells were washed with PBS twice. The biofilms in the microtiter plates were then heat fixed at 80°C for 30 min. After the plates had cooled, 200 µL of crystal violet (0.1% in H2O) was added for 30 min at room temperature. The stain was removed, and the wells were washed four times with PBS to remove excess stain. After the final PBS wash was removed, the plates were left at room temperature overnight to allow the wells to completely dry. The bound stain was solubilized by adding 200 µL 95% ethanol and absorbance was measured at 590 nm with a BioTek Synergy HTX multimode reader (Agilent, Santa Clara, CA). To quantify biofilm production using resazurin, the growth media were removed, and the wells of the microtiter plate were washed twice with PBS to remove non-embedded bacteria. Two hundred microliters of HBSS was then added to the wells along with 20 µL of 0.3 mg/mL resazurin (R-2127, Sigma, St. Louis, MO). The plates were held at 37°C for 45 min to allow for the reduction of the resazurin, at which time the resazurin reduction was measured at 530 excitation and 590 emission with a Biotek Synergy HTX multimode reader (Agilent, Santa Clara, CA).
Toxin production
Because Shiga toxin is instrumental in the development of disease in humans, we assessed the isolates for toxin production and whether there would be differences among the human outbreak isolates and the cattle isolate. Shiga toxin production by E. coli O157:H7 isolates was determined through the cytotoxic effects of bacterial supernatants on Vero cell (37). Resazurin was used to measure the viability of the treated Vero cells. Vero cells were maintained in DMEM (11995–065, Gibco, Thermo Fisher Scientific, Waltham, MA) supplemented with 10% FBS unless otherwise noted. Two hundred microliters of Vero cells at a cell density of 1.0 × 105 cells/mL was seeded into sterile 96-well tissue culture plates to achieve 20,000 Vero cells per well. These culture plates with Vero cells were cultivated at 37°C and 5% CO2 for two days to allow for confluent monolayers to develop. E. coli was grown overnight at 37°C and 200 rpm in LB broth (L7275-500TAB, Sigma-Aldrich, St. Louis, MO) from a colony scrape taken off an LB agar plate. The overnight cultures were adjusted to the same OD600 0.5 absorbance and diluted 1:100 in low glucose DMEM (11054–020, Gibco, Thermo Fisher Scientific, Waltham, MA); these cultures were incubated statically for 6 h at 37°C. Six hours was chosen for toxin induction as this time period has previously been shown to produce high levels of Shiga toxins (102, 103). These cultures were then centrifuged at 1,600 rcf at room temperature for 10 min. The supernatants were collected and passed through sterile 0.2-µm syringe filters. These filtered supernatants were then tested for cytotoxicity on Vero cells. A dilution series of the supernatants was made in DMEM supplemented with 10% FBS. The media from the confluent Vero cell culture plates were removed and replaced with 100 µL DMEM supplemented with 10% FBS, followed by the addition of 100 µL from the bacterial supernatant dilution series; final dilutions tested were 1:4, 1:40, 1:400, and 1:4000. Controls included Vero cells treated with dilution series of low glucose DMEM sans bacteria cultivation. The Vero cells were then maintained at 37°C and 5% CO2 for 48 h, at which time the media from the Vero cells were removed and replaced with 200-µL phenol red free DMEM supplemented with 5% FBS; reduced serum was used as excessive serum protein has been shown to interfere with resazurin measurements (104). Twenty microliters of 0.3 mg/mL resazurin (R-2127, Sigma, St. Louis, MO) was then added, and the culture plates were incubated for 150 min at 37°C and 5% CO2 to allow for reduction of the resazurin. The reduction of resazurin by viable Vero cells was measured with 530 emission and 590 excitation settings on a BioTek Synergy HTX multimode reader (Agilent, Santa Clara, CA) with Gen5 version 3.10 acquisition software. Using the background subtracted fluorescent readings, the viability of the treatment groups was made relative to the controls [(Test/Control)*100]. Statistical analysis was performed by one-way ANOVA with Tukey’s comparison of means to identify significance in toxin production phenotype between E. coli isolates (GraphPad Prism version 9.0.1, GraphPad Software, San Diego, CA).
As stx2 is associated with worse clinical outcome, anti-stx1 antibody was employed to neutralize the cytotoxic activity of stx1 produced by these O157 isolates in these Vero cell cytotoxicity assays. Bacterial supernatants for testing were produced as stated in the preceding paragraph. Monoclonal anti-stx1 antibody (13C4, Invitrogen, Thermo Fisher Scientific, Waltham, MA) was incubated with 1:20 dilutions of the bacterial supernatants in cell culture media at an antibody ratio of 1:50 for two hours at 37°C with shaking at 100 rpm. The remaining produce followed that outlined in the previous paragraph, with the inclusion of a set of control Vero cells that received the monoclonal antibodies in cell culture media sans bacterial supernatants.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank Bryan Wheeler, Erika N. Biernbaum, Zahra F. Bond, and Lindsey Andersen for their assistance with this research.
This research was supported by appropriated funds from USDA-ARS CRIS 5030–32000-225-00D and an appointment to the Agricultural Research Service (ARS) Research Participation Program administered by the Oak Ridge Institute for Science and Education (ORISE) through an interagency agreement between the United States Department of Energy (DOE) and the United States Department of Agriculture (USDA). ORISE is managed by Oak Ridge Associated Universities (ORAU) under DOE contract number DE-SC0014664. This research used resources provided by the SCINet project of the USDA Agricultural Research Service, ARS project number 0500–00093-001-00-D.
All opinions expressed in this paper are the authors’ and do not necessarily reflect the policies and views of USDA, ARS, DOE, or ORAU/ORISE. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA, ARS, DOE, or ORAU/ORISE. USDA is an equal opportunity provider and employer.
Contributor Information
Indira T. Kudva, Email: indira.kudva@usda.gov.
Crystal L. Loving, Email: crystal.loving@usda.gov.
Luxin Wang, University of California Davis, Davis, California, USA.
Francisco Diez-Gonzalez, University of Georgia Center for Food Safety, Griffin, Georgia, USA.
Kalmia E. Kniel, University of Delaware, Newark, Delaware, USA
DATA AVAILABILITY
Sequencing data associated with the infection isolates’ genome are available through NCBI bioproject accession PRJNA884395.
SUPPLEMENTAL MATERIAL
The following material is available online at https://doi.org/10.1128/spectrum.04140-23.
Fig. S1: Time course of E. coli O157 fecal shedding and RAMS colonization in cattle. Fig S2: E. coli O157 Congo Red agar phenotypes.
SNP differences among tested E. coli O157 isolates.
Table S2: Shows the PATS profiles for tested E coli O157 isolates. Table S4: Shows Quantitative RT-PCR detailed statistics.
Genomic comparison of our E. coli O157 isolates.
An accounting of the reviewer comments and feedback.
ASM does not own the copyrights to Supplemental Material that may be linked to, or accessed through, an article. The authors have granted ASM a non-exclusive, world-wide license to publish the Supplemental Material files. Please contact the corresponding author directly for reuse.
REFERENCES
- 1. Baker CA, Rubinelli PM, Park SH, Carbonero F, Ricke SC. 2016. Shiga toxin-producing Escherichia coli in food: Incidence, ecology, and detection strategies. Food Control 59:407–419. doi: 10.1016/j.foodcont.2015.06.011 [DOI] [Google Scholar]
- 2. Centers for Disease Control and Prevention (CDC) . National outbreak reporting system dashboard. Available from: https://www.cdc.gov/norsdashboard. Retrieved 28 Mar 2024.
- 3. Food Safety and Inspection Service, USDA . 2011. Shiga toxin-producing Escherichia coli in certain raw beef products. Fed Regist 76:2010–0023. [Google Scholar]
- 4. Tack DM, Marder EP, Griffin PM, Cieslak PR, Dunn J, Hurd S, Scallan E, Lathrop S, Muse A, Ryan P, Smith K, Tobin-D’Angelo M, Vugia DJ, Holt KG, Wolpert BJ, Tauxe R, Geissler AL. 2019. Preliminary incidence and trends of infections with pathogens transmitted commonly through food - foodborne diseases active surveillance network, 10 U.S. sites, 2015-2018. MMWR Morb Mortal Wkly Rep 68:369–373. doi: 10.15585/mmwr.mm6816a2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tack DM, Kisselburgh HM, Richardson LC, Geissler A, Griffin PM, Payne DC, Gleason BL. 2021. Shiga toxin-producing Escherichia coli outbreaks in the United States, 2010-2017. Microorganisms 9:1529. doi: 10.3390/microorganisms9071529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lim JY, Li J, Sheng H, Besser TE, Potter K, Hovde CJ. 2007. Escherichia coli O157:H7 colonization at the rectoanal junction of long-duration culture-positive cattle. Appl Environ Microbiol 73:1380–1382. doi: 10.1128/AEM.02242-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cobbold RN, Hancock DD, Rice DH, Berg J, Stilborn R, Hovde CJ, Besser TE. 2007. Rectoanal junction colonization of feedlot cattle by Escherichia coli O157:H7 and its association with supershedders and excretion dynamics. Appl Environ Microbiol 73:1563–1568. doi: 10.1128/AEM.01742-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Naylor SW, Low JC, Besser TE, Mahajan A, Gunn GJ, Pearce MC, McKendrick IJ, Smith DGE, Gally DL. 2003. Lymphoid follicle-dense mucosa at the terminal rectum is the principal site of colonization of enterohemorrhagic Escherichia coli O157:H7 in the bovine host. Infect Immun 71:1505–1512. doi: 10.1128/IAI.71.3.1505-1512.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Low JC, McKendrick IJ, McKechnie C, Fenlon D, Naylor SW, Currie C, Smith DGE, Allison L, Gally DL. 2005. Rectal carriage of enterohemorrhagic Escherichia coli O157 in slaughtered cattle. Appl Environ Microbiol 71:93–97. doi: 10.1128/AEM.71.1.93-97.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Brennan FP, Abram F, Chinalia FA, Richards KG, O’Flaherty V. 2010. Characterization of environmentally persistent Escherichia coli isolates leached from an irish soil. Appl Environ Microbiol 76:2175–2180. doi: 10.1128/AEM.01944-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ishii S, Sadowsky MJ. 2008. Escherichia coli in the environment: implications for water quality and human health. Microbes Environ 23:101–108. doi: 10.1264/jsme2.23.101 [DOI] [PubMed] [Google Scholar]
- 12. Texier S, Prigent-Combaret C, Gourdon MH, Poirier MA, Faivre P, Dorioz JM, Poulenard J, Jocteur-Monrozier L, Moënne-Loccoz Y, Trevisan D. 2008. Persistence of culturable Escherichia coli fecal contaminants in dairy alpine grassland soils. J Environ Qual 37:2299–2310. doi: 10.2134/jeq2008.0028 [DOI] [PubMed] [Google Scholar]
- 13. Duffy G, McCabe E. 2014. Veterinary public health approach to managing pathogenic verocytotoxigenic Escherichia coli in the agri-food chain. Microbiol Spectr 2. doi: 10.1128/microbiolspec.EHEC-0023-2013 [DOI] [PubMed] [Google Scholar]
- 14. Yang Z, Kovar J, Kim J, Nietfeldt J, Smith DR, Moxley RA, Olson ME, Fey PD, Benson AK. 2004. Identification of common subpopulations of non-sorbitol-fermenting, beta-glucuronidase-negative Escherichia coli O157:H7 from bovine production environments and human clinical samples. Appl Environ Microbiol 70:6846–6854. doi: 10.1128/AEM.70.11.6846-6854.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bono JL, Keen JE, Clawson ML, Durso LM, Heaton MP, Laegreid WW. 2007. Association of Escherichia coli O157:H7 tir polymorphisms with human infection. BMC Infect Dis 7:98. doi: 10.1186/1471-2334-7-98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mellor GE, Besser TE, Davis MA, Beavis B, Jung W, Smith HV, Jennison AV, Doyle CJ, Chandry PS, Gobius KS, Fegan N. 2013. Multilocus genotype analysis of Escherichia coli O157 isolates from Australia and the United States provides evidence of geographic divergence. Appl Environ Microbiol 79:5050–5058. doi: 10.1128/AEM.01525-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Haugum K, Brandal LT, Løbersli I, Kapperud G, Lindstedt B-A. 2011. Detection of virulent Escherichia coli O157 strains using multiplex PCR and single base sequencing for SNP characterization. J Appl Microbiol 110:1592–1600. doi: 10.1111/j.1365-2672.2011.05015.x [DOI] [PubMed] [Google Scholar]
- 18. National Advisory Committee on Microbiology Criteria for Foods . 2019. Response to questions posed by the food and drug administration regarding virulence factors and attributes that define foodborne Shiga toxin–producing Escherichia coli (STEC) as severe human pathogens. J Food Prot 82:724–767. doi: 10.4315/0362-028X.JFP-18-479 [DOI] [PubMed] [Google Scholar]
- 19. Buvens G, De Gheldre Y, Dediste A, de Moreau A-I, Mascart G, Simon A, Allemeersch D, Scheutz F, Lauwers S, Piérard D. 2012. Incidence and virulence determinants of verocytotoxin-producing Escherichia coli infections in the Brussels-Capital region, Belgium, in 2008-2010. J Clin Microbiol 50:1336–1345. doi: 10.1128/JCM.05317-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Buvens G, Piérard D. 2012. Virulence profiling and disease association of verocytotoxin-producing Escherichia coli O157 and non-O157 isolates in Belgium. Foodborne Pathog Dis 9:530–535. doi: 10.1089/fpd.2011.1073 [DOI] [PubMed] [Google Scholar]
- 21. Possé B, De Zutter L, Heyndrickx M, Herman L. 2007. Metabolic and genetic profiling of clinical O157 and non-O157 Shiga-toxin-producing Escherichia coli. Res Microbiol 158:591–599. doi: 10.1016/j.resmic.2007.06.001 [DOI] [PubMed] [Google Scholar]
- 22. Wells TJ, Sherlock O, Rivas L, Mahajan A, Beatson SA, Torpdahl M, Webb RI, Allsopp LP, Gobius KS, Gally DL, Schembri MA. 2008. EhaA is a novel autotransporter protein of enterohemorrhagic Escherichia coli O157:H7 that contributes to adhesion and biofilm formation. Environ Microbiol 10:589–604. doi: 10.1111/j.1462-2920.2007.01479.x [DOI] [PubMed] [Google Scholar]
- 23. Barnhart MM, Chapman MR. 2006. Curli biogenesis and function. Annu Rev Microbiol 60:131–147. doi: 10.1146/annurev.micro.60.080805.142106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Uhlich GA, Chen CY, Cottrell BJ, Hofmann CS, Dudley EG, Strobaugh TP, Nguyen LH. 2013. Phage insertion in mlrA and variations in rpoS limit curli expression and biofilm formation in Escherichia coli serotype O157: H7. Microbiol (Reading) 159:1586–1596. doi: 10.1099/mic.0.066118-0 [DOI] [PubMed] [Google Scholar]
- 25. Arnqvist A, Olsén A, Normark S. 1994. σS-dependent growth-phase induction of the csgBA promoter in Escherichia coli can be achieved in vivo by σ70 in the absence of the nucleoid-associated protein H-NS. Mol Microbiol 13:1021–1032. doi: 10.1111/j.1365-2958.1994.tb00493.x [DOI] [PubMed] [Google Scholar]
- 26. Vogeleer P, Tremblay YDN, Jubelin G, Jacques M, Harel J. 2016. Biofilm-forming abilities of Shiga toxin-producing Escherichia coli isolates associated with human infections. Appl Environ Microbiol 82:1448–1458. doi: 10.1128/AEM.02983-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bauer ME, Welch RA. 1996. Characterization of an RTX toxin from enterohemorrhagic Escherichia coli O157:H7. Infect Immun 64:167–175. doi: 10.1128/iai.64.1.167-175.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tatsuno I, Horie M, Abe H, Miki T, Makino K, Shinagawa H, Taguchi H, Kamiya S, Hayashi T, Sasakawa C. 2001. toxB gene on pO157 of enterohemorrhagic Escherichia coli O157:H7 is required for full epithelial cell adherence phenotype. Infect Immun 69:6660–6669. doi: 10.1128/IAI.69.11.6660-6669.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kudva IT, Griffin RW, Murray M, John M, Perna NT, Barrett TJ, Calderwood SB. 2004. Insertions, deletions, and single-nucleotide polymorphisms at rare restriction enzyme sites enhance discriminatory power of polymorphic amplified typing sequences, a novel strain typing system for Escherichia coli O157:H7. J Clin Microbiol 42:2388–2397. doi: 10.1128/JCM.42.6.2388-2397.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kudva IT, Smole S, Griffin RW, Garren J, Kalia N, Murray M, John M, Timperi R, Calderwood SB. 2013. Polymorphic amplified typing sequences (PATS) strain typing system accurately discriminates a set of temporally and spatially disparate Escherichia coli O157 isolates associated with human infection. Open Microbiol J 7:123–129. doi: 10.2174/1874285801307010123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kudva IT, Davis MA, Griffin RW, Garren J, Murray M, John M, Hovde CJ, Calderwood SB. 2012. Polymorphic amplified typing sequences and pulsed-field gel electrophoresis yield comparable results in the strain typing of a diverse set of bovine Escherichia coli O157:H7 isolates. Int J Microbiol 2012:140105. doi: 10.1155/2012/140105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hammar M, Bian Z, Normark S. 1996. Nucleator-dependent intercellular assembly of adhesive curli organelles in Escherichia coli. Proc Natl Acad Sci U S A 93:6562–6566. doi: 10.1073/pnas.93.13.6562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kudva IT, Carter MQ, Sharma VK, Stasko JA, Giron JA. 2017. Curli temper adherence of Escherichia coli O157:H7 to squamous epithelial cells from the bovine recto-anal junction in a strain-dependent manner. Appl Environ Microbiol 83:e02594-16. doi: 10.1128/AEM.02594-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sharma VK, Kudva IT, Bearson BL, Stasko JA. 2016. Contributions of EspA filaments and curli fimbriae in cellular adherence and biofilm formation of enterohemorrhagic Escherichia coli O157:H7. PLoS One 11:e0149745. doi: 10.1371/journal.pone.0149745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. O’Brien AD, Newland JW, Miller SF, Holmes RK, Smith HW, Formal SB. 1984. Shiga-like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea. Science 226:694–696. doi: 10.1126/science.6387911 [DOI] [PubMed] [Google Scholar]
- 36. Tesh VL, O’Brien AD. 1991. The pathogenic mechanisms of Shiga toxin and the Shiga-like toxins. Mol Microbiol 5:1817–1822. doi: 10.1111/j.1365-2958.1991.tb00805.x [DOI] [PubMed] [Google Scholar]
- 37. Gentry MK, Dalrymple JM. 1980. Quantitative microtiter cytotoxicity assay for Shigella toxin. J Clin Microbiol 12:361–366. doi: 10.1128/jcm.12.3.361-366.1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Strockbine NA, Marques LR, Newland JW, Smith HW, Holmes RK, O’Brien AD. 1986. Two toxin-converting phages from Escherichia coli O157:H7 strain 933 encode antigenically distinct toxins with similar biologic activities. Infect Immun 53:135–140. doi: 10.1128/iai.53.1.135-140.1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Orth D, Grif K, Khan AB, Naim A, Dierich MP, Würzner R. 2007. The Shiga toxin genotype rather than the amount of Shiga toxin or the cytotoxicity of Shiga toxin in vitro correlates with the appearance of the hemolytic uremic syndrome. Diagn Microbiol Infect Dis 59:235–242. doi: 10.1016/j.diagmicrobio.2007.04.013 [DOI] [PubMed] [Google Scholar]
- 40. Kulkarni AA, Fuller C, Korman H, Weiss AA, Iyer SS. 2010. Glycan encapsulated gold nanoparticles selectively inhibit shiga toxins 1 and 2. Bioconjug Chem 21:1486–1493. doi: 10.1021/bc100095w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Eppinger M, Mammel MK, Leclerc JE, Ravel J, Cebula TA. 2011. Genomic anatomy of Escherichia coli O157:H7 outbreaks. Proc Natl Acad Sci U S A 108:20142–20147. doi: 10.1073/pnas.1107176108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Eppinger Mark, Mammel MK, Leclerc JE, Ravel J, Cebula TA. 2011. Genome signatures of Escherichia coli O157:H7 isolates from the bovine host reservoir. Appl Environ Microbiol 77:2916–2925. doi: 10.1128/AEM.02554-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rusconi B, Sanjar F, Koenig SSK, Mammel MK, Tarr PI, Eppinger M. 2016. Whole genome sequencing for genomics-guided investigations of Escherichia coli O157:H7 outbreaks. Front Microbiol 7:985. doi: 10.3389/fmicb.2016.00985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Steele M, Ziebell K, Zhang Y, Benson A, Johnson R, Laing C, Taboada E, Gannon V. 2009. Genomic regions conserved in lineage II Escherichia coli O157:H7 strains. Appl Environ Microbiol 75:3271–3280. doi: 10.1128/AEM.02123-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kim J, Nietfeldt J, Benson AK. 1999. Octamer-based genome scanning distinguishes a unique subpopulation of Escherichia coli O157:H7 strains in cattle. Proc Natl Acad Sci USA 96:13288–13293. doi: 10.1073/pnas.96.23.13288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Maldonado Y, Fiser JC, Nakatsu CH, Bhunia AK. 2005. Cytotoxicity potential and genotypic characterization of Escherichia coli isolates from environmental and food sources. Appl Environ Microbiol 71:1890–1898. doi: 10.1128/AEM.71.4.1890-1898.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Arthur TM, Ahmed R, Chase-Topping M, Kalchayanand N, Schmidt JW, Bono JL. 2013. Characterization of Escherichia coli O157:H7 strains isolated from supershedding cattle. Appl Environ Microbiol 79:4294–4303. doi: 10.1128/AEM.00846-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Biscola FT, Abe CM, Guth BEC. 2011. Determination of adhesin gene sequences in, and biofilm formation by, O157 and non-O157 Shiga toxin-producing Escherichia coli strains isolated from different sources. Appl Environ Microbiol 77:2201–2208. doi: 10.1128/AEM.01920-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Parker CT, Kyle JL, Huynh S, Carter MQ, Brandl MT, Mandrell RE. 2012. Distinct transcriptional profiles and phenotypes exhibited by Escherichia coli O157:H7 isolates related to the 2006 spinach-associated outbreak. Appl Environ Microbiol 78:455–463. doi: 10.1128/AEM.06251-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Scheutz F. 2014. Taxonomy meets public health: the case of Shiga toxin-producing Escherichia coli. Microbiol Spectr 2. doi: 10.1128/microbiolspec.EHEC-0019-2013 [DOI] [PubMed] [Google Scholar]
- 51. Wells JG, Davis BR, Wachsmuth IK, Riley LW, Remis RS, Sokolow R, Morris GK. 1983. Laboratory investigation of hemorrhagic colitis outbreaks associated with a rare Escherichia coli serotype. J Clin Microbiol 18:512–520. doi: 10.1128/jcm.18.3.512-520.1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Carter MQ, Brandl MT, Louie JW, Kyle JL, Carychao DK, Cooley MB, Parker CT, Bates AH, Mandrell RE. 2011. Distinct acid resistance and survival fitness displayed by curli variants of enterohemorrhagic Escherichia coli O157:H7. Appl Environ Microbiol 77:3685–3695. doi: 10.1128/AEM.02315-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. S. Hoefler R, Kudva IT. 2021. EDL933 strains of Escherichia coli O157 can demonstrate genetic diversity and differential adherence to bovine recto-anal junction squamous epithelial cells. Open Microbiol J 15:129–138. doi: 10.2174/1874285802115010129 [DOI] [Google Scholar]
- 54. Kay KL, Breidt F, Fratamico PM, Baranzoni GM, Kim G-H, Grunden AM, Oh D-H. 2017. Escherichia coli O157:H7 acid sensitivity correlates with flocculation phenotype during nutrient limitation. Front Microbiol 8:1404. doi: 10.3389/fmicb.2017.01404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cray WC, Moon HW. 1995. Experimental infection of calves and adult cattle with Escherichia coli O157:H7. Appl Environ Microbiol 61:1586–1590. doi: 10.1128/aem.61.4.1586-1590.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sanderson MW, Besser TE, Gay JM, Gay CC, Hancock DD. 1999. Fecal Escherichia coli O157:H7 shedding patterns of orally inoculated calves. Vet Microbiol 69:199–205. doi: 10.1016/s0378-1135(99)00106-6 [DOI] [PubMed] [Google Scholar]
- 57. Shere JA, Bartlett KJ, Kaspar CW. 1998. Longitudinal study of Escherichia coli O157:H7 dissemination on four dairy farms in Wisconsin. Appl Environ Microbiol 64:1390–1399. doi: 10.1128/AEM.64.4.1390-1399.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Brown CA, Harmon BG, Zhao T, Doyle MP. 1997. Experimental Escherichia coli O157:H7 carriage in calves. Appl Environ Microbiol 63:27–32. doi: 10.1128/aem.63.1.27-32.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kudva IT, Dean-Nystrom EA. 2011. Bovine recto-anal junction squamous epithelial (RSE) cell adhesion assay for studying Escherichia coli O157 adherence. J Appl Microbiol 111:1283–1294. doi: 10.1111/j.1365-2672.2011.05139.x [DOI] [PubMed] [Google Scholar]
- 60. Kudva IT, Griffin RW, Krastins B, Sarracino DA, Calderwood SB, John M. 2012. Proteins other than the locus of enterocyte effacement-encoded proteins contribute to Escherichia coli O157:H7 adherence to bovine rectoanal junction stratified squamous epithelial cells. BMC Microbiol 12:103. doi: 10.1186/1471-2180-12-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Munns KD, Selinger LB, Stanford K, Guan L, Callaway TR, McAllister TA. 2015. Perspectives on super-shedding of Escherichia coli O157:H7 by cattle. Foodborne Pathog Dis 12:89–103. doi: 10.1089/fpd.2014.1829 [DOI] [PubMed] [Google Scholar]
- 62. Arthur TM, Brichta-Harhay DM, Bosilevac JM, Kalchayanand N, Shackelford SD, Wheeler TL, Koohmaraie M. 2010. Super shedding of Escherichia coli O157:H7 by cattle and the impact on beef carcass contamination. Meat Sci 86:32–37. doi: 10.1016/j.meatsci.2010.04.019 [DOI] [PubMed] [Google Scholar]
- 63. Chase-Topping M, Gally D, Low C, Matthews L, Woolhouse M. 2008. Super-shedding and the link between human infection and livestock carriage of Escherichia coli O157. Nat Rev Microbiol 6:904–912. doi: 10.1038/nrmicro2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Carlson BA, Nightingale KK, Mason GL, Ruby JR, Choat WT, Loneragan GH, Smith GC, Sofos JN, Belk KE. 2009. Escherichia coli O157:H7 strains that persist in feedlot cattle are genetically related and demonstrate an enhanced ability to adhere to intestinal epithelial cells. Appl Environ Microbiol 75:5927–5937. doi: 10.1128/AEM.00972-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bielaszewska M, Middendorf B, Tarr PI, Zhang W, Prager R, Aldick T, Dobrindt U, Karch H, Mellmann A. 2011. Chromosomal instability in enterohaemorrhagic Escherichia coli O157:H7: impact on adherence, tellurite resistance and colony phenotype. Mol Microbiol 79:1024–1044. doi: 10.1111/j.1365-2958.2010.07499.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ryu J-H, Beuchat LR. 2005. Biofilm formation by Escherichia coli O157:H7 on stainless steel: effect of exopolysaccharide and curli production on its resistance to chlorine. Appl Environ Microbiol 71:247–254. doi: 10.1128/AEM.71.1.247-254.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Silagyi K, Kim S-H, Lo YM, Wei C. 2009. Production of biofilm and quorum sensing by Escherichia coli O157:H7 and its transfer from contact surfaces to meat, poultry, ready-to-eat deli, and produce products. Food Microbiol 26:514–519. doi: 10.1016/j.fm.2009.03.004 [DOI] [PubMed] [Google Scholar]
- 68. Vogeleer P, Tremblay YDN, Mafu AA, Jacques M, Harel J. 2014. Life on the outside: role of biofilms in environmental persistence of Shiga-toxin producing Escherichia coli. Front Microbiol 5:317. doi: 10.3389/fmicb.2014.00317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Puttamreddy S, Minion FC. 2011. Linkage between cellular adherence and biofilm formation in Escherichia coli O157:H7 EDL933. FEMS Microbiol Lett 315:46–53. doi: 10.1111/j.1574-6968.2010.02173.x [DOI] [PubMed] [Google Scholar]
- 70. Macarisin D, Patel J, Bauchan G, Giron JA, Ravishankar S. 2013. Effect of spinach cultivar and bacterial adherence factors on survival of Escherichia coli O157:H7 on spinach leaves. J Food Prot 76:1829–1837. doi: 10.4315/0362-028X.JFP-12-556 [DOI] [PubMed] [Google Scholar]
- 71. Macarisin D, Patel J, Sharma VK. 2014. Role of curli and plant cultivation conditions on Escherichia coli O157:H7 internalization into spinach grown on hydroponics and in soil. Int J Food Microbiol 173:48–53. doi: 10.1016/j.ijfoodmicro.2013.12.004 [DOI] [PubMed] [Google Scholar]
- 72. Borucki MK, Peppin JD, White D, Loge F, Call DR. 2003. Variation in biofilm formation among strains of Listeria monocytogenes. Appl Environ Microbiol 69:7336–7342. doi: 10.1128/AEM.69.12.7336-7342.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Gamage SD, Patton AK, Hanson JF, Weiss AA. 2004. Diversity and host range of Shiga toxin-encoding phage. Infect Immun 72:7131–7139. doi: 10.1128/IAI.72.12.7131-7139.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Rangel JM, Sparling PH, Crowe C, Griffin PM, Swerdlow DL. 2005. Epidemiology of Escherichia coli O157:H7 outbreaks, United States, 1982-2002. Emerg Infect Dis 11:603–609. doi: 10.3201/eid1104.040739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Centers for Disease Control and Prevention (CDC) . 2006. Ongoing multistate outbreak of Escherichia coli serotype O157:H7 infections associated with consumption of fresh spinach--United States, September 2006. MMWR Morb Mortal Wkly Rep 55:1045–1046. [PubMed] [Google Scholar]
- 76. Center for Infectious Disease Research and Policy UoM . 2007. FDA finds Taco John’s E coli strain on California farms. Available from: https://www.cidrap.umn.edu/news-perspective/2007/01/fda-finds-taco-johns-e-coli-strain-california-farms. Retrieved 31 Aug 2022.
- 77. Maier T, Güell M, Serrano L. 2009. Correlation of mRNA and protein in complex biological samples. FEBS Lett 583:3966–3973. doi: 10.1016/j.febslet.2009.10.036 [DOI] [PubMed] [Google Scholar]
- 78. Weigel WA, Dersch P. 2018. Phenotypic heterogeneity: a bacterial virulence strategy. Microbes Infect 20:570–577. doi: 10.1016/j.micinf.2018.01.008 [DOI] [PubMed] [Google Scholar]
- 79. Wick RR, Judd LM, Holt KE. 2019. Performance of neural network basecalling tools for Oxford Nanopore sequencing. Genome Biol 20:129. doi: 10.1186/s13059-019-1727-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kolmogorov M, Yuan J, Lin Y, Pevzner PA. 2019. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol 37:540–546. doi: 10.1038/s41587-019-0072-8 [DOI] [PubMed] [Google Scholar]
- 81. Lin Y, Yuan J, Kolmogorov M, Shen MW, Chaisson M, Pevzner PA. 2016. Assembly of long error-prone reads using de Bruijn graphs. Proc Natl Acad Sci U S A 113:E8396–E8405. doi: 10.1073/pnas.1604560113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Bushnell B. 2014. BBTools software package. http://sourceforge net/projects/bbmap.
- 83. Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wattam AR, Abraham D, Dalay O, Disz TL, Driscoll T, Gabbard JL, Gillespie JJ, Gough R, Hix D, Kenyon R, et al. 2014. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res 42:D581–D591. doi: 10.1093/nar/gkt1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Wattam AR, Davis JJ, Assaf R, Boisvert S, Brettin T, Bun C, Conrad N, Dietrich EM, Disz T, Gabbard JL, et al. 2017. Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res 45:D535–D542. doi: 10.1093/nar/gkw1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Seemann T. Snippy. Available from: https://github.com/tseemann/snippy. Retrieved 20 Dec 2022.
- 87. Scheutz F, Teel LD, Beutin L, Piérard D, Buvens G, Karch H, Mellmann A, Caprioli A, Tozzoli R, Morabito S, Strockbine NA, Melton-Celsa AR, Sanchez M, Persson S, O’Brien AD. 2012. Multicenter evaluation of a sequence-based protocol for subtyping Shiga toxins and standardizing Stx nomenclature. J Clin Microbiol 50:2951–2963. doi: 10.1128/JCM.00860-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Schwengers O, Jelonek L, Dieckmann MA, Beyvers S, Blom J, Goesmann A. 2021. Bakta: rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Microb Genom 7:000685. doi: 10.1099/mgen.0.000685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Page AJ, Cummins CA, Hunt M, Wong VK, Reuter S, Holden MTG, Fookes M, Falush D, Keane JA, Parkhill J. 2015. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31:3691–3693. doi: 10.1093/bioinformatics/btv421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. National Center for Biotechnology Information (NCBI) . 2016. The NCBI pathogen detection project on National Library of Medicine (US) National Center for Biotechnology Information. Available from: https://www.ncbi.nlm.nih.gov/pathogens. Retrieved 18 Jun 2022.
- 92. Feldgarden M, Brover V, Haft DH, Prasad AB, Slotta DJ, Tolstoy I, Tyson GH, Zhao S, Hsu CH, McDermott PF, Tadesse DA, Morales C, Simmons M, Tillman G, Wasilenko J, Folster JP, Klimke W. 2019. Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob Agents Chemother 63:e00483-19. doi: 10.1128/AAC.00483-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Bessonov K, Laing C, Robertson J, Yong I, Ziebell K, Gannon VPJ, Nichani A, Arya G, Nash JHE, Christianson S. 2021. ECTyper: in silico Escherichia coli serotype and species prediction from raw and assembled whole-genome sequence data. Microb Genom 7:000728. doi: 10.1099/mgen.0.000728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Zhang Y, Laing C, Steele M, Ziebell K, Johnson R, Benson AK, Taboada E, Gannon VPJ. 2007. Genome evolution in major Escherichia coli O157:H7 lineages. BMC Genomics 8:121. doi: 10.1186/1471-2164-8-121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Gautreau G, Bazin A, Gachet M, Planel R, Burlot L, Dubois M, Perrin A, Médigue C, Calteau A, Cruveiller S, Matias C, Ambroise C, Rocha EPC, Vallenet D. 2020. PPanGGOLiN: depicting microbial diversity via a partitioned pangenome graph. PLOS Comput Biol 16:e1007732. doi: 10.1371/journal.pcbi.1007732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Kudva IT, Oosthuysen ER, Wheeler B, Loest CA. 2021. Evaluation of cattle for naturally colonized Shiga toxin-producing Escherichia coli requires combinatorial strategies. Int J Microbiol 2021:6673202. doi: 10.1155/2021/6673202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Rice DH, Sheng HQ, Wynia SA, Hovde CJ. 2003. Rectoanal mucosal swab culture is more sensitive than fecal culture and distinguishes Escherichia coli O157:H7-colonized cattle and those transiently shedding the same organism. J Clin Microbiol 41:4924–4929. doi: 10.1128/JCM.41.11.4924-4929.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Mir RA, Brunelle BW, Alt DP, Arthur TM, Kudva IT. 2020. Supershed Escherichia coli O157:H7 has potential for increased persistence on the rectoanal junction squamous epithelial cells and antibiotic resistance. Int J Microbiol 2020:2368154. doi: 10.1155/2020/2368154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Peeters E, Nelis HJ, Coenye T. 2008. Comparison of multiple methods for quantification of microbial biofilms grown in microtiter plates. J Microbiol Methods 72:157–165. doi: 10.1016/j.mimet.2007.11.010 [DOI] [PubMed] [Google Scholar]
- 100. Van den Driessche F, Rigole P, Brackman G, Coenye T. 2014. Optimization of resazurin-based viability staining for quantification of microbial biofilms. J Microbiol Methods 98:31–34. doi: 10.1016/j.mimet.2013.12.011 [DOI] [PubMed] [Google Scholar]
- 101. Crémet L, Corvec S, Batard E, Auger M, Lopez I, Pagniez F, Dauvergne S, Caroff N. 2013. Comparison of three methods to study biofilm formation by clinical strains of Escherichia coli. Diagn Microbiol Infect Dis 75:252–255. doi: 10.1016/j.diagmicrobio.2012.11.019 [DOI] [PubMed] [Google Scholar]
- 102. Zhang X, McDaniel AD, Wolf LE, Keusch GT, Waldor MK, Acheson DW. 2000. Quinolone antibiotics induce Shiga toxin-encoding bacteriophages, toxin production, and death in mice. J Infect Dis 181:664–670. doi: 10.1086/315239 [DOI] [PubMed] [Google Scholar]
- 103. Rocha LB, Piazza RMF. 2007. Production of Shiga toxin by Shiga toxin-expressing Escherichia coli (STEC) in broth media: from divergence to definition. Lett Appl Microbiol 45:411–417. doi: 10.1111/j.1472-765X.2007.02214.x [DOI] [PubMed] [Google Scholar]
- 104. Goegan P, Johnson G, Vincent R. 1995. Effects of serum protein and colloid on the AlamarBlue assay in cell cultures. Toxicol In Vitro 9:257–266. doi: 10.1016/0887-2333(95)00004-r [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1: Time course of E. coli O157 fecal shedding and RAMS colonization in cattle. Fig S2: E. coli O157 Congo Red agar phenotypes.
SNP differences among tested E. coli O157 isolates.
Table S2: Shows the PATS profiles for tested E coli O157 isolates. Table S4: Shows Quantitative RT-PCR detailed statistics.
Genomic comparison of our E. coli O157 isolates.
An accounting of the reviewer comments and feedback.
Data Availability Statement
Sequencing data associated with the infection isolates’ genome are available through NCBI bioproject accession PRJNA884395.