Abstract
Our progress in understanding mammalian gene function has lagged behind that of gene identification. New methods for mammalian gene functional analysis are needed to accelerate the process. In yeast, the powerful genetic shuffle system allows deletion of any chromosomal gene by homologous recombination and episomal expression of a mutant allele in the same cell. Here, we report a method for mammalian cells, which employs a helper-dependent adenoviral (HD-Ad) vector to synthesize small hairpin (sh) RNAs to knock-down the expression of an endogenous gene by targeting untranslated regions (UTRs). The vector simultaneously expresses an exogenous version of the same gene (wild-type or mutant allele) lacking the UTRs for functional analysis. We demonstrated the utility of the method by using PRPF3, which encodes the human RNA splicing factor Hprp3p. Recently, missense mutations in PRPF3 were found to cause autosomal-dominant Retinitis Pigmentosa, a form of genetic eye diseases affecting the retina. We knocked-down endogenous PRPF3 in multiple cell lines and rescued the phenotype (cell death) with exogenous PRPF3 cDNA, thereby creating a genetic complementation method. Because Ad vectors can efficiently transduce a wide variety of cell types, and many tissues in vivo, this method could have a wide application for gene function studies.
INTRODUCTION
The human genome has been largely sequenced (1,2); however, the identification of human genes will be far more valuable if gene function can be revealed. In general, gene function can be studied with two complementary approaches, genetic and biochemical. For the genetic approach, the effects of genes mutagenized spontaneously or experimentally can be assessed at the molecular, cellular or organ level. For the biochemical approach, gene products can be purified and characterized in vitro using a variety of methods. In addition to these classic approaches, other methods based on the same genetic and biochemical principles have been established. Molecular interactions of gene products can be carried out in yeast using the one-hybrid and two-hybrid systems (3). Methods for the analysis of gene expression, such as the microarray (4), have also been useful for obtaining information about gene function. Recently, RNA interference (RNAi) has developed into an important tool for gene function analysis (5).
The yeast genetic shuffle system (6,7) is a powerful tool for obtaining information about yeast genes through mutational analysis. In this system, any gene on a yeast chromosome can be deleted through homologous recombination and the corresponding gene with a desired mutation can be expressed episomally. Many mammalian genes do not have homologs in yeast, so their functions cannot be inferred from yeast genetic studies. Furthermore, mammalian genes with yeast homologs often encode extra domains and are functionally more complicated than their yeast counterparts. Currently, there are no mammalian methods as convenient as the yeast genetic shuffle system. Mouse models for gene knock-out, knock-in or transgenic expression are used extensively for mammalian gene function analysis (8). More recently, large-scale random mutagenesis of mice has been initiated (9). These approaches, although powerful, are time-consuming and expensive.
A mammalian method similar to the yeast genetic shuffle system would require effective down-regulation of an endogenous mammalian gene and expression of the corresponding mutant gene in the same cell. Here, we report the establishment of such a method, building on advances in RNAi technology (5) and adenoviral vectors (10). Initially, small double-stranded interference RNAs (siRNAs) were synthesized chemically and delivered into cells by transfection (11). Subsequently, DNA-based vectors were developed to express small hairpin RNAs (shRNAs) (12,13). The feasibility of using viral vectors to deliver DNA expressing shRNAs into mammalian cells and animal models has further enhanced the use of this technology in functional genomics, proteomics and gene therapy (14,15).
We chose to use a helper-dependent adenoviral (HD-Ad) vector (10) to deliver shRNAs and genes to cells. The lack of all viral coding sequences in the HD-Ad eliminates the potential for viral gene products to interfere with gene expression in the host cell (the cytopathic effect of cellular entry of the virion is temporary) (16). The large DNA-cloning capacity of the HD-Ad allows several expression cassettes to be incorporated. This is important if a large transgene and more than one shRNA are to be expressed from the same vector. Transgene expression from HD-Ads is relatively stable compared with plasmids, which is critical if experiments are to continue for more than a few days. Finally, adenoviral vectors can transduce a wide variety of cultured cells (including non-dividing cells) with high efficiency, as well as numerous organs of experimental mammals (10). We used the PRPF3 gene (originally called HPRP3) (17,18) to demonstrate the new method. PRPF3 encodes Hprp3p, a key factor involved in RNA splicing (17,18).
MATERIALS AND METHODS
Cell culture
ARPE19 (a gift from R. Hunt, University of South Carolina) and HeLa cells were cultured in DMEM-F12 and α-MEM, respectively, supplemented with 10% fetal bovine serum (FBS). Cells were transduced at 40–60% confluency with virus under serum-free conditions for 2 h, followed by the addition of media to a final concentration of 10% FBS.
Design of shRNA
The target sequences in PRPF3 (GenBank accession no. NM_004698) were selected as follows: 5′-untranslated region (5′-UTR): GGCATGGACAAGAAGAAGGA (shRNA-1); 5′-UTR: GGGGCTGAAGTTTGTGAGGTG (shRNA-2); open reading frame (ORF): GGTGTAGTATTGAGTCCTGTA (shRNA-3); 3′-UTR: GTGTGATCTCAGAACTGTGCCA (shRNA-4); 3′-UTR: GGGAGAATATCTTGCTCCCCT (shRNA-5).
Target sequences were BLAST searched (National Center for Biotechnology Information) against all human sequences in GenBank to verify uniqueness. Five Ts were added to the end of the sequence for efficient RNA polymerase III termination. The oligonucleotides were synthesized, annealed to its inverted repeat (separated by a 6 nt spacer) and cloned into the pBS/U6 vector (Figure 2B). The final constructs were named pBS/U6-shRNA-1, -2, -3, -4 or -5 in reference to their particular shRNA.
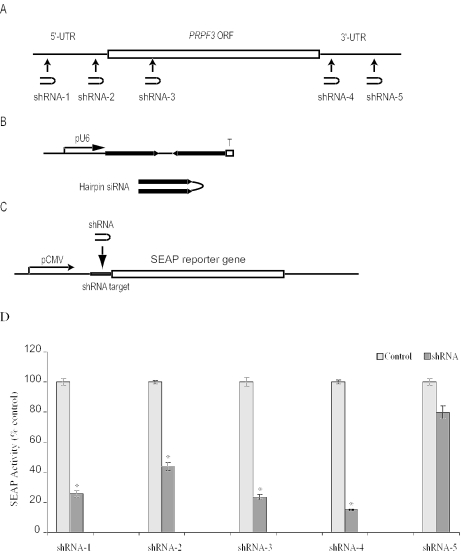
Figure 2.
Design and expression of shRNAs targeting PRPF3. (A) Target sites of shRNAs. (B) Schematic representation of an snRNA expression plasmid. T and pU6 represent termination signal and U6 gene promoter, respectively. (C) Reporter plasmid with an shRNA target site. (D) Inhibition of SEAP reporter expression by shRNAs. Results represent the mean ± SEM of six experiments. Statistical significance was assessed by paired Student's t-test (*P < 0.05).
SEAP reporter system for gene silencing assay
A 21 bp double-stranded oligonucleotide containing the PRPF3 targeted site was inserted into the pCMVSEAP reporter plasmid (19) between the cytomegalovirus promoter and the translation initiation codon. The resulting plasmids, pCMVSEAP-PRPF3-1, -2, -3, -4 or -5, expressed the same level of SEAP activity as the parental plasmid. HeLa cells were cotransfected with the corresponding pCMVSEAP-PRPF3 and pBS/U6-shRNA pair, in a molecular ratio of 1:50, or pCMVSEAP-PRPF3 alone, using PolyFect (Qiagen). SEAP analysis was performed 2 days post-cotransfection as described previously (20).
HD-Ad-F3i preparation
HD-Ad-F3i was made by inserting pBS/U6-shRNA-2 and pBS/U6-shRNA-4 into the pC4HSUvector (21) (Figure 3A). To express PRPF3 exogenously, we used the human ubiquitin C gene promoter (GenBank accession no. D63791). HD-Ad-F3iplus was constructed by introducing the HA-tagged PRPF3 coding sequence, under control of the UbC promoter and followed by the bovine growth hormone poly(A) signal, into HD-Ad-F3i (Figure 4A). Viral vector was produced in 293cre4 cells co-infected with helper virus H14 (21). Virus was amplified through eight serial passages and purified by CsCl gradient centrifugation as described previously (22). The control virus contains the lacZ gene under control of the K18 promoter (23).
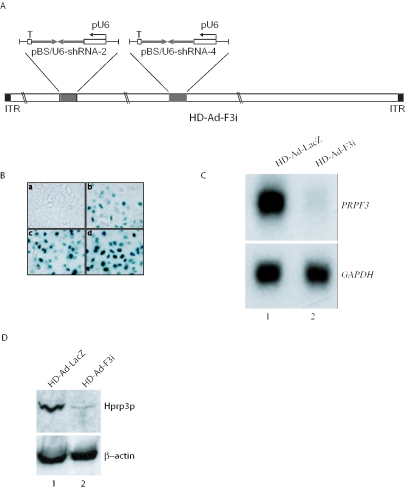
Figure 3.
Reducing PRPF3 expression by HD-Ad-F3i. (A) Schematic representation of the HD-Ad vector expressing shRNAs that target PRPF3. (B) Transduction efficiency of ARPE-19 cells assessed by X-gal staining 2 days after receiving various amounts of HD-Ad-lacZ viral particles per cell (a, 0; b, 2500; c, 5000; d, 10 000). (C) Northern blot of PRPF3 mRNA expression in ARPE-19 cells 2 days after transduction with the control vector (lane 1) or HD-Ad-F3i (lane 2). (D) Western blot of Hprp3p. Antibody against β-actin was used as an internal control.
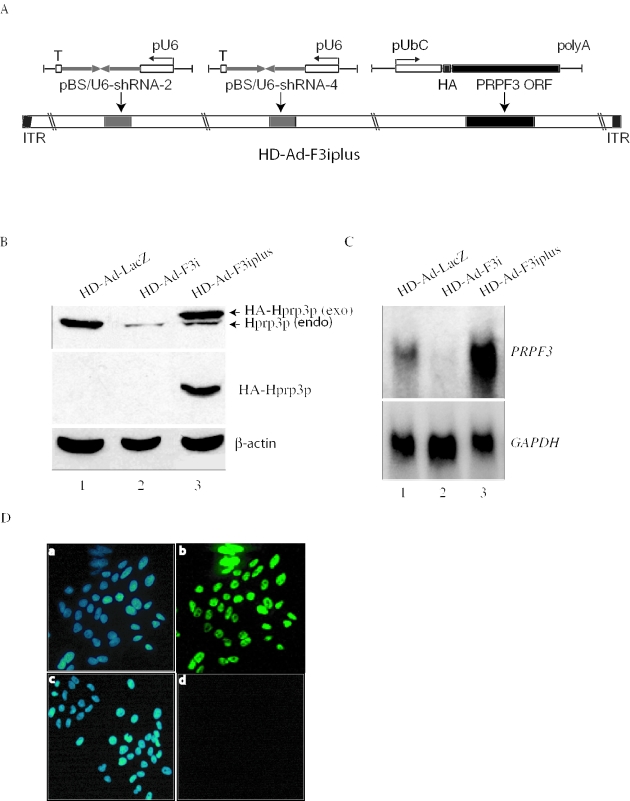
Figure 4.
Complementation of endogenous PRPF3 by exogenous PRPF3. (A) Schematic representation of HD-Ad-F3iplus, which expresses two shRNAs and a copy of HA-tagged PRPF3. ORF, open reading frame; pUbC, human Ubiquitin C promoter; poly(A), bovine growth hormone poly(A) signal. (B) Western blot with antibodies against Hprp3p (upper), HA (middle) and β-actin (lower). Exo, exogenous HA-Hprp3p; endo, endogenous Hprp3p. (C) Northern blot of PRPF3 mRNA expression. Total RNA from ARPE-19 cells transduced with control vector (lane 1), HD-Ad-F3i (lane 2) or HD-Ad-F3iplus (lane 3). (D) Indirect immunostaining micrographs show expression and subcellullar localization of the HA-Hprp3p protein in ARPE-19 cells 48 h post-transduction with Hd-Ad-F3iplus (a and b). As controls, cells were transduced with HD-Ad-lacZ (c and d). Identical fields are shown for DAPI (left panels) and FITC (right panels) channels. The localization of the HA-tagged protein is visualized as green immunofluorescence (b). Cells transduced with HD-Ad-lacZ did not display any immunostaining signal under the FITC channel (d).
Northern blot
Total cellular RNA (30 μg) was separated on a 1% agarose gel containing 0.63 M formaldehyde, transferred to nylon membrane (Roche), and UV cross-linked. For the detection of PRPF3, RPL18 and GAPDH mRNAs, 32P-labeled cDNA probes were prepared by random priming (Amersham Biosciences). RNAs visualized by autoradiography were analyzed using NIH Image software (v 1.62) and normalized to the internal GAPDH control. For the detection of human 18S rRNA, the following oligonucleotide was used: 5′-GGTCCGTCTTGCGCCGGTCCAAGGAATTTCACCTCTAGCGGCGCAATACG-3′ (complementary to RNA sequence 1081–1130, accession no. K03432).
Western blot
Proteins (150 μg) from total cell lysates were resolved by SDS–PAGE and immunoblotted. Protein bands were visualized with the enhanced chemiluminescence system (Amersham Pharmacia Biotech) and quantified using NIH Image software (v 1.62), normalizing to the internal β-actin control. Rabbit antisera against Hprp3p, Hprp4p and U5-116 kD were generated in our laboratory. Mouse anti-HA and anti-β actin monoclonal antibodies were purchased from BABCO (Berkeley, CA) and Abcam (Cambridge, MA), respectively.
Immunostaining
ARPE-19 cells were seeded in six well plates with glass coverslips and transduced with Hd-Ad-F3iplus (5000 particles/cell) or control vector, HD-Ad-lacZ, at 40–60% confluency. Immunostaining was performed at room temperature 48 h post-transduction. The cells grown on coverslips were washed three times with phosphate-buffered saline (PBS) and fixed for 20 min with 4% (w/v) paraformaldehyde in PBS. The plasma membrane was permeablized by 0.2% (v/v) Triton X-100 in PBS for 10 min. The cells were incubated with blocking solution (0.5% BSA in PBS) for 1 h and then with mouse anti-HA monoclonal antibody in blocking solution (0.5% BSA in PBS) for 1 h. The cells were washed three times with PBS and incubated with goat anti-mouse IgG (H + L) fluorescein isothiocyanate (FITC) labeled antibody (GIBCO BRL). Following three washes with PBS, the coverslips were mounted in Vectashield Mounting Medium with DAPI. Fluorescence micrographs were recorded using a CCD camera (DFC 300F, Leica Microsystems, Richmond Hill, Ontario, Canada). Micrographs are representatives of experiments repeated at least three times.
MTT assay
Cell survival was evaluated by a colorimetric 3-4,5-(dimethyl-thyazol-2-yl)-2,5-diphenyltetrazolium (MTT) assay (24) with minor modifications. Briefly, ARPE-19 and HeLa cells transduced with HD-Ad-LacZ, HD-Ad-F3i or HD-Ad-F3iplus were grown in 24 well dishes, in triplicate, for 2, 4, 6 or 8 days. At day 2 post-transduction, cells were trypsinized, counted and similar number of cells re-seeded for each treatment. The procedure was repeated every other day for 8 days. An aliquot of 0.5 ml MTT solution (1 mg/ml in DMEM-F12 or α-MEM; Sigma, St Louis, MO) was then added to each culture well and the plates incubated for 30 min at 37°C. The untransformed MTT was then removed and 0.5 ml 2-propanol added to each well. Absorbance was measured at 570 nm. The data were expressed as a percent of the value obtained from cells transduced with the control vector (HD-Ad-lacZ).
Statistical analysis
Student's t-test or one-way ANOVA followed by Holm's multiple comparison was used to compare data pairs or sets, respectively. Data are presented as mean ± SEM. P < 0.05 was considered significant.
RESULTS
General strategy
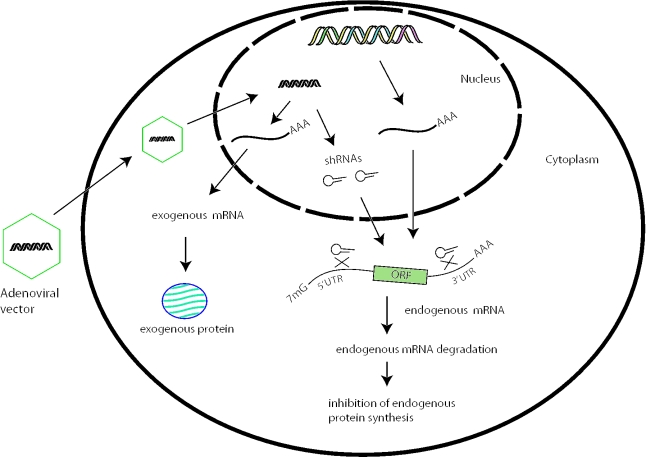
In order to establish a complementation system, it is necessary to knock-down the endogenous gene while simultaneously expressing a copy of the exogenous gene with a desired alteration. As shown in Figure 1, our strategy was to use a viral vector to produce shRNAs that knock-down an endogenous gene by targeting the UTR sequences of the mRNA and to simultaneously express an exogenous version of the gene lacking the UTRs. In order to enhance the silencing power of shRNA, we targeted both 5′- and 3′-UTRs. We used the PRPF3 gene as an example to demonstrate the method. We modified the exogenous PRPF3 cDNA (lacking UTRs) by introducing an HA-epitope tag to the N-terminus coding region. We used the human ubiquitin (UbC) gene promoter to drive exogenous PRPF3 because it is relatively strong and ubiquitously active (25).
Figure 1.
Schematic representation of the mammalian genetic complementation method. An adenoviral vector produces two shRNAs, which reduce the expression of an endogenous gene by targeting both the 5′ and 3′-UTRs of the mRNA. The vector also expresses an exogenous gene lacking the target sequences, complementing the loss of endogenous gene expression.
Testing shRNA efficacy
To identify UTR target sites for knocking down endogenous PRPF3 expression, we examined five selected sequences (Figure 2A). DNA sequences corresponding to each of the shRNAs were cloned into the pBS/U6 vector (12), in which shRNA production is driven by the mouse U6 promoter. Each resulting shRNA is composed of two identical 21 nt sequences in an inverted orientation, separated by a 6 bp loop, with five Ts at the 3′ end for efficient RNA polymerase III termination (Figure 2B).
Next, we designed plasmids to test each shRNA by inserting the target sequence in the 5′ region of the secreted alkaline phosphatase gene (SEAP) (Figure 2C). HeLa cells were transiently cotransfected with each pBS/U6-shRNA expression plasmid, or the control vector, together with the corresponding SEAP reporter plasmid, and SEAP activity was assayed 2 days later. SEAP activity was inhibited 60–80% by shRNAs-1–4, while activity was inhibited less by shRNA-5 (Figure 2D).
Reducing endogenous PRPF3 expression
The reporter assay showed that three of the four shRNAs targeting the UTR sequences of PRPF3 reduced protein expression (Figure 2D). We selected the target sequences for shRNA-2 and shRNA-4 to construct a vector (HD-Ad-F3i) for stable shRNA expression (Figure 3A). To determine whether HD-Ad-F3i effectively and specifically inhibited the expression of endogenous PRPF3, we transduced ARPE-19 cells (a human retinal pigment epithelial cell line) (26) and assayed the levels of PRPF3 mRNA and Hprp3p by northern and western blot. We transduced cells with increasing number of particles of the control vector (HD-Ad-lacZ), followed by X-gal staining. Because 5000 particles/cell transduced >90% of cells (Figure 3B), we selected this value for subsequent transductions. Two days after transduction of ARPE19 cells, the level of PRPF3 mRNA was detected by northern blot. PRPF3 mRNA was dramatically reduced compared with cells transduced with the control vector (Figure 3C). Consistent with the mRNA data, we observed a large decrease in the production of endogenous Hprp3p (Figure 3D). Similar results were observed in D407, A549 and HeLa cell lines (see Supplementary Figure 1 online).
Complementation by exogenous PRPF3 expression
We examined whether an exogenous copy of PRPF3 could be expressed in cells in which endogenous PRPF3 is knocked down. We designed another vector called HD-Ad-F3iplus, which expresses shRNAs that target the endogenous PRPF3 mRNA and also expresses HA-tagged PRPF3 cDNA driven by the human UbC promoter (Figure 4A). The HA-tagged PRPF3 lacks the UTR sequences of the endogenous PRPF3 and is therefore expected to be resistant to the shRNAs. We transduced ARPE-19 cells with HD-Ad-F3iplus and analyzed the levels of protein and mRNA. As shown in Figure 4B, endogenous Hprp3p was greatly decreased 2 days post-transduction with either HD-Ad-F3i or HD-Ad-F3iplus, while the HA-tagged exogenous Hprp3p was expressed in cells transduced with HD-Ad-F3iplus (Figure 4B). The difference in electrophoretic mobility of the endogenous and exogenous Hprp3 proteins, owing to the presence of the HA-tag in the latter, permitted the detection of both proteins using antibodies against Hprp3p. Consistent with the protein data, PRPF3 mRNA was greatly reduced in cells transduced with HD-Ad-F3i (Figure 4C, lane 2). The increase in PRPF3 transcripts in cells transduced with HD-Ad-F3iplus was consistent with the expression of exogenous PRPF3 (Figure 4C, lane 3). This was also evident from the results of immunostaining of ARPE-19 cells transduced with HD-Ad-F3iplus (Figure 4D), which showed 100% cells expressing the HA-tagged Hprp3p.
Effect on other splicing factors
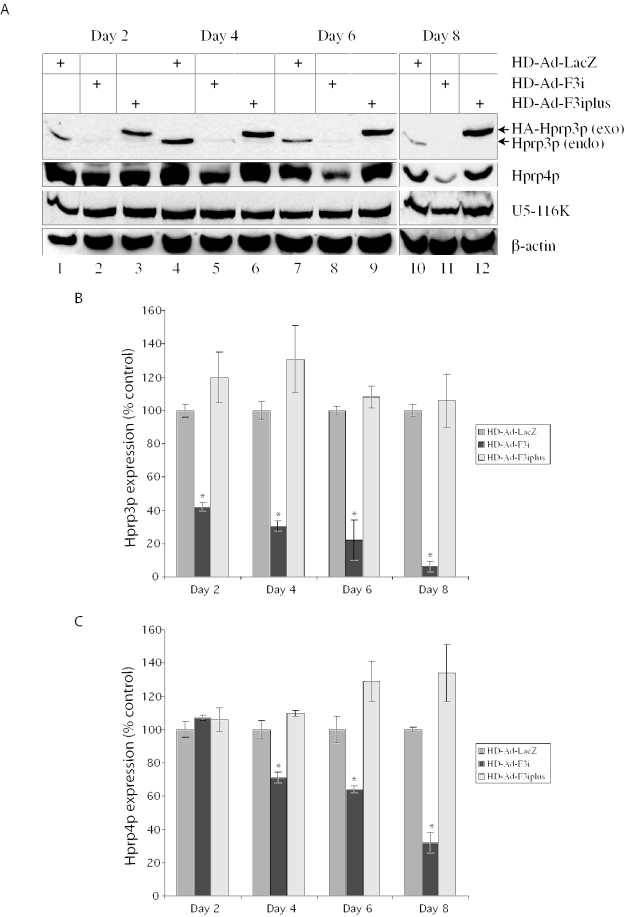
Because Hprp3p is a key component of the U4/U6 snRNP, it is conceivable that its depletion may affect other splicing factor proteins. We addressed this by examining the levels of Hprp4p (a U4/U6 snRNP-associated splicing factor that interacts with Hprp3p) (18,27,28) and the U5-116 kD protein (a splicing factor associated with U5 snRNA) (29). Figure 5A shows the western blot from a representative experiment. Quantitative values were then determined from multiple experiments (Figure 5B and C). There was no noticeable change in the expression of Hprp4p or U5-116 kD in the first 2 days, indicating that the shRNAs did not targeted these genes. However, the steady-state level of Hprp4p gradually decreased from day 4 post-transduction onward, suggesting that Hprp4p is sensitive to depletion of Hprp3p. The level of U5-116 kD protein did not change significantly (Figure 5A). The detrimental effect on Hprp4p due to silencing endogenous PRPF3 was reversed by exogenous Hprp3p expression from HD-Ad-F3iplus (Figure 5A and C).
Figure 5.
Effect of PRPF3 silencing on the steady-state levels of spliceosomal components. (A) Western blot for Hprp3p, Hprp4p and the U5-116 kD protein. Whole-cell extracts were prepared from ARPE-19 cells 2, 4, 6 or 8 days post-transduction with HD-Ad-F3i (lanes 2, 5, 8 and 11), HD-Ad-F3iplus (lanes 3, 6, 9 and 12) or HD-Ad-lacZ (lanes 1, 4, 7 and 10) and immunoblotted. Exo, exogenous HA-Hprp3p; endo, endogenous Hprp3p. (B and C) Hprp3p and Hprp4p levels in cells transduced with HD-Ad-F3i or HD-Ad-F3iplus compared with those in cells transduced with HD-Ad-lacZ, normalized to β-actin. Results represent the mean ± SEM of three experiments. Statistical significance was assessed by one-way ANOVA and Holm's multiple comparison test (*P < 0.05).
Rescuing the survival phenotype of PRPF3 knock-down
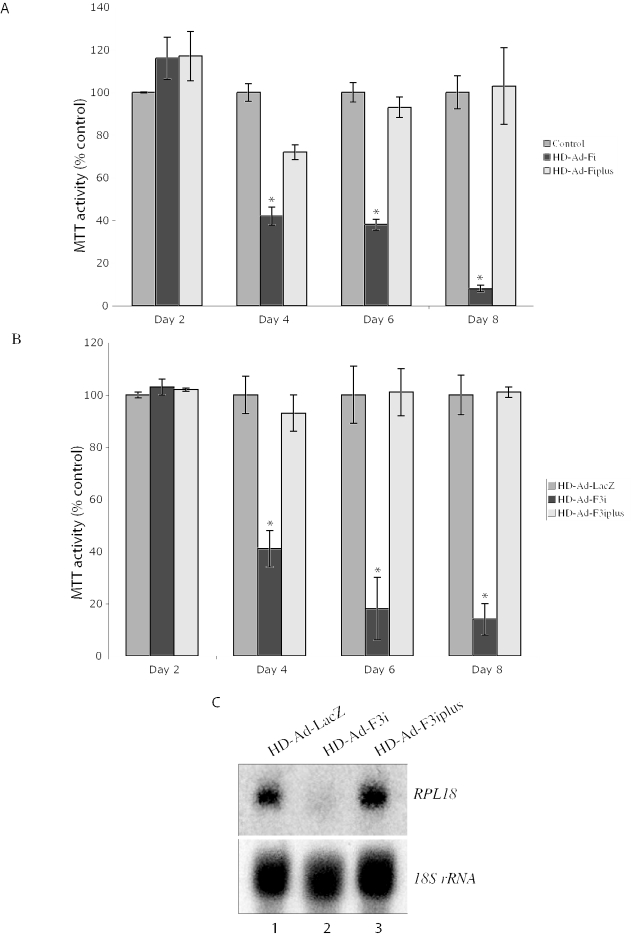
Two different missense mutations in PRPF3 lead to the development of autosomal dominant Retinitis Pigmentosa (adRP). In humans, these mutations appear to only affect photoreceptor cells (30). Although the yeast homolog PRP3 is essential (31), it is possible that human PRPF3 is essential only for the survival of photoreceptor cells; its function may be compensated by other splicing factors in non-photoreceptor cells. To examine whether PRPF3 is essential for the survival of non-photoreceptor cells, we determined the viability of ARPE-19 cells following transduction with HD-Ad-F3i or HD-Ad-F3iplus. As shown in Figure 6A, >90% of cells transduced with HD-Ad-F3i died within 8 days. Cells transduced with HD-Ad-F3iplus survived, indicating that the expression of exogenous PRPF3 can rescue cell death caused by shRNAs targeting endogenous PRPF3 (Figure 6A). Similar results were obtained with HeLa cells (Figure 6B).
Figure 6.
Complementing the phenotype of PRPF3 knock-down by the expression of exogenous PRPF3. Viability of ARPE-19 (A) and HeLa (B) cells was assessed by MTT assay 2, 4, 6 or 8 days post-transduction. Values are relative to cells transduced with the control vector (HD-Ad-lacZ). Results represent the mean ± SEM of three experiments, each in triplicate. Statistical significance was assessed by one-way ANOVA and Holm's multiple comparison test (*P < 0.05). (C) Steady-state levels of RPL18 mRNA and 18S rRNA. Northern blot was performed with total RNA isolated from ARPE-19 cells transduced as indicated.
Because Hprp3p is a splicing factor, we examined whether the cell death in shRNA-targeted cells was related to a deficiency in pre-mRNA splicing. Pre-mRNAs are normally not stable if RNA splicing is blocked, so a reduced steady-state level of mRNA for an intron-containing gene can indicate a splicing deficiency (32,33). We measured mRNA levels of RPL18 (encoding ribosomal protein L18) and 18S rRNA in cells 4 days after transduction with HD-Ad-F3i or HD-Ad-F3iplus. We observed a marked reduction of RPL18 transcripts in cells transduced with HD-Ad-F3i (Figure 6C, lane 2). The RPL18 mRNA level was normal in cells transduced with HD-Ad-F3iplus (Figure 6C, lane 3). The level of 18S rRNA, which is not processed by the typical RNA splicing machinery, was unaffected.
DISCUSSION
In this paper we described the development of a mammalian genetic complementation method, conceptually similar to the yeast genetic shuffle system (6,7), for gene function studies. The strategy described here is to knock-down an endogenous gene and express the exogenous copy in the same cell in a single step. Yeast genetics has made a tremendous contribution to our understanding of eukaryotic genes, but differences in genome complexity make it impossible to fully infer the function of mammalian genes from yeast studies. Many mammalian genes have extra domains for more diverse functions than their yeast counterparts, and some have no yeast homologs. For example, Hprp3p, the gene product of PRPF3 used in this study, has an N-terminus PWI motif (34) that is absent from its yeast homolog (Prp3p). Hprp3p interacts with Hprp4p, which occurs in the yeast homologs (17,18,28), but it also interacts with the splicing factor SPF30 (35), a U2 snRNP-associated protein which has no yeast homolog.
Because our strategy was to target the UTRs of an endogenous gene, the specificity of the knock-down phenotype could be demonstrated by complementation using the wild-type cDNA. Targeting the UTRs also has the advantage that any mutations in the coding region of the targeted gene can be analyzed using the same shRNAs. We chose HD-Ad vectors (36) for delivering gene expression cassettes to mammalian cells because of their high transduction efficiency, high production titer, large cloning capacity, episomal stability and ability to transduce a wide variety of different cell types independent of replicative state. In addition, they can efficiently transduce many organs of experimental mammals (37,38). Our method thus has several unique features for the analysis of gene function in mammalian cells compared with other methods. First, a gene can be analyzed regardless of whether it is essential for cell viability. Second, there is no need to generate knockout cell lines; data can therefore be obtained much more quickly. A viral vector can be produced in 2 weeks once the DNA vector is built (39). Third, the high transduction efficiency (>90%) with sustained transgene expression of our method will allow for the detection of mutant phenotypes more easily by lowering the background arising from non-transduced cells in the culture. Finally, the broad tropism of adenovirus vectors means that this method could be applied to many cell types of various mammals, so long as the target cells are accessible not just those routinely used for genetic manipulation (i.e. mice). However, our method remains to be tested for genes that require only very low levels of expression for their function.
We illustrated the feasibility of the method in multiple cell lines using PRPF3 knock-down and rescue as an example. Mutations in human PRPF3 have been associated with the development of adRP (30), a group of inherited retinal disorders characterized by photoreceptor cell degeneration. Currently, it is not clear how mutations in PRPF3 cause adRP. The autosomal dominant mode of inheritance could be due to a gain of function of the mutant proteins, or haplo-insufficiency of PRPF3 in photoreceptor cells. Mutations in two other splicing genes, PRPF31 (40–42) and PRPC8 (43,44), which code for U4/U6 and U5 snRNP proteins, respectively, also lead to adRP, suggesting that a common splicing mechanism may underlie development of the disease. One hypothesis regarding PRPF3 mutations and adRP is that PRPF3 is essential only in photoreceptor cells. This hypothesis implies that other factor(s) may compensate for reduced PRPF3 function due to the mutation in non-photoreceptor cells. We ruled out this possibility here by demonstrating that PRPF3 is essential in non-photoreceptor cells (ARPE19 and HeLa). Another hypothesis, that photoreceptor cells have low tolerance for a minor reduction in splicing activity, could be examined in mice by subretinal delivery (45) of an HD-Ad to knock-down the endogenous gene while expressing a mutant allele.
In summary, our new method can be used to perform functional analysis of any mammalian genes. In addition, it could be used to create animal models of human diseases by delivering an HD-Ad vector to knock-down an endogenous gene and simultaneously express a mutant version that causes a disease. Finally, our approach may also be used to develop gene therapy for diseases caused by one or more dominant mutations.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Supplementary Material
Acknowledgments
The authors thank Drs James D. Friesen and Roderick McInnes for reviewing the manuscript. This work was supported by Operating Grants from the Canadian Institutes of Health Research, from the Canadian Cystic Fibrosis Foundation and from the Foundation Fighting Blindness-Canada to J.H. J.H. is a CCFF Scholar and holds a Premier's Research Excellence Award of Ontario Canada. Funding to pay the Open Access publication charges for this article was provided by CIHR.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Venter J.C., Adams M.D., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 3.Walhout A.J., Boulton S.J., Vidal M. Yeast two-hybrid systems and protein interaction mapping projects for yeast and worm. Yeast. 2000;17:88–94. doi: 10.1002/1097-0061(20000630)17:2<88::AID-YEA20>3.0.CO;2-Y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shoemaker D.D., Schadt E.E., Armour C.D., He Y.D., Garrett-Engele P., McDonagh P.D., Loerch P.M., Leonardson A., Lum P.Y., Cavet G., et al. Experimental annotation of the human genome using microarray technology. Nature. 2001;409:922–927. doi: 10.1038/35057141. [DOI] [PubMed] [Google Scholar]
- 5.Shi Y. Mammalian RNAi for the masses. Trends Genet. 2003;19:9–12. doi: 10.1016/s0168-9525(02)00005-7. [DOI] [PubMed] [Google Scholar]
- 6.Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- 7.Rothstein R.J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 8.Sato T.N. Gene trap, gene knockout, gene knock-in, and transgenics in vascular development. Thromb. Haemost. 1999;82:865–869. [PubMed] [Google Scholar]
- 9.Rossant J., McKerlie C. Mouse-based phenogenomics for modelling human disease. Trends Mol. Med. 2001;7:502–507. doi: 10.1016/s1471-4914(01)02164-5. [DOI] [PubMed] [Google Scholar]
- 10.Parks R.J. Improvements in adenoviral vector technology: overcoming barriers for gene therapy. Clin. Genet. 2000;58:1–11. doi: 10.1034/j.1399-0004.2000.580101.x. [DOI] [PubMed] [Google Scholar]
- 11.Elbashir S.M., Harborth J., Lendeckel W., Yalcin A., Weber K., Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 12.Sui G., Soohoo C., Affar el B., Gay F., Shi Y., Forrester W.C. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc. Natl Acad. Sci. USA. 2002;99:5515–5520. doi: 10.1073/pnas.082117599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu J.Y., DeRuiter S.L., Turner D.L. RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc. Natl Acad. Sci. USA. 2002;99:6047–6052. doi: 10.1073/pnas.092143499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brummelkamp T.R., Bernards R., Agami R. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell. 2002;2:243–247. doi: 10.1016/s1535-6108(02)00122-8. [DOI] [PubMed] [Google Scholar]
- 15.Davidson B.L., Paulson H.L. Molecular medicine for the brain: silencing of disease genes with RNA interference. Lancet Neurol. 2004;3:145–149. doi: 10.1016/S1474-4422(04)00678-7. [DOI] [PubMed] [Google Scholar]
- 16.Muruve D.A., Cotter M.J., Zaiss A.K., White L.R., Liu Q., Chan T., Clark S.A., Ross P.J., Meulenbroek R.A., Maelandsmo G.M., et al. Helper-dependent adenovirus vectors elicit intact innate but attenuated adaptive host immune responses in vivo. J. Virol. 2004;78:5966–5972. doi: 10.1128/JVI.78.11.5966-5972.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gonzalez-Santos J.M., Wang A., Jones J., Ushida C., Liu J., Hu J. Central region of the human splicing factor hprp3p interacts with hprp4p. J. Biol. Chem. 2002;277:23764–23772. doi: 10.1074/jbc.M111461200. [DOI] [PubMed] [Google Scholar]
- 18.Wang A., Forman-Kay J., Luo Y., Luo M., Chow Y.H., Plumb J., Friesen J.D., Tsui L.C., Heng H.H., Woolford J.L., Jr, Hu J. Identification and characterization of human genes encoding Hprp3p and Hprp4p, interacting components of the spliceosome. Hum. Mol. Genet. 1997;6:2117–2126. doi: 10.1093/hmg/6.12.2117. [DOI] [PubMed] [Google Scholar]
- 19.Koehler D.R., Chow Y.H., Plumb J., Wen Y., Rafii B., Belcastro R., Haardt M., Lukacs G.L., Post M., Tanswell A.K., Hu J. A human epithelium-specific vector optimized in rat pneumocytes for lung gene therapy. Pediatr. Res. 2000;48:184–190. doi: 10.1203/00006450-200008000-00011. [DOI] [PubMed] [Google Scholar]
- 20.Chow Y.H., O'Brodovich H., Plumb J., Wen Y., Sohn K.J., Lu Z., Zhang F., Lukacs G.L., Tanswell A.K., Hui C.C., et al. Development of an epithelium-specific expression cassette with human DNA regulatory elements for transgene expression in lung airways. Proc. Natl Acad. Sci. USA. 1997;94:14695–14700. doi: 10.1073/pnas.94.26.14695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sandig V., Youil R., Bett A.J., Franlin L.L., Oshima M., Maione D., Wang F., Metzker M.L., Savino R., Caskey C.T. Optimization of the helper-dependent adenovirus system for production and potency in vivo. Proc. Natl Acad. Sci. USA. 2000;97:1002–1007. doi: 10.1073/pnas.97.3.1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ng P., Parks R.J., Graham F.L. Preparation of helper-dependent adenoviral vectors. Methods Mol. Med. 2002;69:371–388. doi: 10.1385/1-59259-141-8:371. [DOI] [PubMed] [Google Scholar]
- 23.Toietta G., Koehler D.R., Finegold M., Lee B., Hu J., Beaudet A.L. Reduced inflammation and improved airway expression using helper-dependent adenoviral vectors with a K18 promoter. Mol. Ther. 2003;7:649–658. doi: 10.1016/s1525-0016(03)00059-5. [DOI] [PubMed] [Google Scholar]
- 24.Gerlier D., Thomasset N. Use of MTT colorimetric assay to measure cell activation. J. Immunol. Methods. 1986;94:57–63. doi: 10.1016/0022-1759(86)90215-2. [DOI] [PubMed] [Google Scholar]
- 25.Tsirigotis M., Thurig S., Dube M., Vanderhyden B.C., Zhang M., Gray D.A. Analysis of ubiquitination in vivo using a transgenic mouse model. Biotechniques. 2001;31:120–126. doi: 10.2144/01311rr03. [DOI] [PubMed] [Google Scholar]
- 26.Dunn K.C., Aotaki-Keen A.E., Putkey F.R., Hjelmeland L.M. ARPE-19, a human retinal pigment epithelial cell line with differentiated properties. Exp. Eye Res. 1996;62:155–169. doi: 10.1006/exer.1996.0020. [DOI] [PubMed] [Google Scholar]
- 27.Horowitz D.S., Kobayashi R., Krainer A.R. A new cyclophilin and the human homologues of yeast Prp3 and Prp4 form a complex associated with U4/U6 snRNPs. RNA. 1997;3:1374–1387. [PMC free article] [PubMed] [Google Scholar]
- 28.Lauber J., Plessel G., Prehn S., Will C.L., Fabrizio P., Groning K., Lane W.S., Luhrmann R. The human U4/U6 snRNP contains 60 and 90 kD proteins that are structurally homologous to the yeast splicing factors Prp4p and Prp3p. RNA. 1997;3:926–941. [PMC free article] [PubMed] [Google Scholar]
- 29.Behrens S.E., Luhrmann R. Immunoaffinity purification of a [U4/U6.U5] tri-snRNP from human cells. Genes Dev. 1991;5:1439–1452. doi: 10.1101/gad.5.8.1439. [DOI] [PubMed] [Google Scholar]
- 30.Chakarova C.F., Hims M.M., Bolz H., Abu-Safieh L., Patel R.J., Papaioannou M.G., Inglehearn C.F., Keen T.J., Willis C., Moore A.T., et al. Mutations in HPRP3, a third member of pre-mRNA splicing factor genes, implicated in autosomal dominant retinitis pigmentosa. Hum. Mol. Genet. 2002;11:87–92. doi: 10.1093/hmg/11.1.87. [DOI] [PubMed] [Google Scholar]
- 31.Anthony J.G., Weidenhammer E.M., Woolford J.L., Jr The yeast Prp3 protein is a U4/U6 snRNP protein necessary for integrity of the U4/U6 snRNP and the U4/U6.U5 tri-snRNP. RNA. 1997;3:1143–1152. [PMC free article] [PubMed] [Google Scholar]
- 32.Barrett N.L., Li X., Carmichael G.G. The sequence and context of the 5′ splice site govern the nuclear stability of polyoma virus late RNAs. Nucleic Acids Res. 1995;23:4812–4817. doi: 10.1093/nar/23.23.4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghogawala Z., Choi E., Daly K.R., Blanco L.R., Griffith I.J., Glimcher L.H. An intronic 10-base-pair deletion in a class II A beta gene affects RNA processing. Mol. Cell. Biol. 1989;9:4402–4408. doi: 10.1128/mcb.9.10.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blencowe B.J., Ouzounis C.A. The PWI motif: a new protein domain in splicing factors. Trends Biochem. Sci. 1999;24:179–180. doi: 10.1016/s0968-0004(99)01387-0. [DOI] [PubMed] [Google Scholar]
- 35.Rappsilber J., Ajuh P., Lamond A.I., Mann M. SPF30 is an essential human splicing factor required for assembly of the U4/U5/U6 tri-small nuclear ribonucleoprotein into the spliceosome. J. Biol. Chem. 2001;276:31142–31150. doi: 10.1074/jbc.M103620200. [DOI] [PubMed] [Google Scholar]
- 36.Zhou H., Pastore L., Beaudet A.L. Helper-dependent adenoviral vectors. Methods Enzymol. 2002;346:177–198. doi: 10.1016/s0076-6879(02)46056-9. [DOI] [PubMed] [Google Scholar]
- 37.Cao H., Koehler D.R., Hu J. Adenoviral vectors for gene replacement therapy. Viral Immunol. 2004;17:327–333. doi: 10.1089/vim.2004.17.327. [DOI] [PubMed] [Google Scholar]
- 38.St George J.A. Gene therapy progress and prospects: adenoviral vectors. Gene Ther. 2003;10:1135–1141. doi: 10.1038/sj.gt.3302071. [DOI] [PubMed] [Google Scholar]
- 39.Palmer D., Ng P. Improved System for helper-dependent adenoviral vector production. Mol. Ther. 2003;8:846–852. doi: 10.1016/j.ymthe.2003.08.014. [DOI] [PubMed] [Google Scholar]
- 40.Deery E.C., Vithana E.N., Newbold R.J., Gallon V.A., Bhattacharya S.S., Warren M.J., Hunt D.M., Wilkie S.E. Disease mechanism for retinitis pigmentosa (RP11) caused by mutations in the splicing factor gene PRPF31. Hum. Mol. Genet. 2002;11:3209–3219. doi: 10.1093/hmg/11.25.3209. [DOI] [PubMed] [Google Scholar]
- 41.Vithana E.N., Abu-Safieh L., Allen M.J., Carey A., Papaioannou M., Chakarova C., Al-Maghtheh M., Ebenezer N.D., Willis C., Moore A.T., et al. A human homolog of yeast pre-mRNA splicing gene, PRP31, underlies autosomal dominant retinitis pigmentosa on chromosome 19q13.4 (RP11) Mol. Cell. 2001;8:375–381. doi: 10.1016/s1097-2765(01)00305-7. [DOI] [PubMed] [Google Scholar]
- 42.Wang L., Ribaudo M., Zhao K., Yu N., Chen Q., Sun Q., Wang Q. Novel deletion in the pre-mRNA splicing gene PRPF31 causes autosomal dominant retinitis pigmentosa in a large Chinese family. Am. J. Med. Genet. A. 2003;121:235–239. doi: 10.1002/ajmg.a.20224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Achsel T., Ahrens K., Brahms H., Teigelkamp S., Luhrmann R. The human U5–220kD protein (hPrp8) forms a stable RNA-free complex with several U5-specific proteins, including an RNA unwindase, a homologue of ribosomal elongation factor EF-2, and a novel WD-40 protein. Mol. Cell. Biol. 1998;18:6756–6766. doi: 10.1128/mcb.18.11.6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brown J.D., Beggs J.D. Roles of PRP8 protein in the assembly of splicing complexes. EMBO J. 1992;11:3721–3729. doi: 10.1002/j.1460-2075.1992.tb05457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ali R.R., Reichel M.B., Thrasher A.J., Levinsky R.J., Kinnon C., Kanuga N., Hunt D.M., Bhattacharya S.S. Gene transfer into the mouse retina mediated by an adeno-associated viral vector. Hum. Mol. Genet. 1996;5:591–594. doi: 10.1093/hmg/5.5.591. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.