Abstract
Retroviral integration results in the stable and coordinated insertion of the two termini of the linear viral DNA into the host genome. An in vitro concerted two-end integration reaction catalyzed by the Moloney murine leukemia virus (M-MuLV) integrase (IN) was used to investigate the binding and coordination of the two viral DNA ends. Comparison of the two-end integration and strand transfer assays indicates that zinc is required for efficient concerted integration utilizing plasmid DNA as target. Complementation assays using a pair of nonoverlapping integrase domains, consisting of the HHCC domain and the core/C-terminal region, yielded products containing the correct 4-base target site duplication. The efficiency of the coordinated two-end integration varied depending on the order of addition of the individual protein and DNA components in the complementation assay. Two-end integration was most efficient when the long terminal repeat (LTR) was premixed with either the target DNA or the HHCC domain. The preference for two-end integration through preincubation of the HHCC finger with the viral DNA supports the role of this domain in the recognition and/or positioning of the LTR.
During the life cycle of retroviruses, the reverse-transcribed viral DNA is integrated into the host genome in a process catalyzed by integrase (IN). This process can be divided into two steps. First, two terminal nucleotides are removed from the 3′ end of the viral long terminal repeat (LTR), leaving the CA nucleotides exposed (3′ processing) (4, 29, 44, 58). Second, the 3′ OH from the CA attacks the host DNA in a transesterification reaction (strand transfer) (13, 24, 43). The integration of the viral DNA is carried out in a concerted fashion, such that duplication of a short stretch of host DNA results at the site of integration in vivo. In Moloney murine leukemia virus (M-MuLV), the target duplication is 4 bp (for a review, see references 5 and 36). Minimally, IN is sufficient to catalyze the in vitro integration reactions. In vivo, IN is associated within a preintegration complex (2, 3). Additional host factors associated with the integration process, including Ini (42), BAF (50), and HMG I(Y) (26), have been identified.
The retroviral integrase (IN) protein can be divided into three domains through sequence comparison (39). The N-terminal domain contains conserved histidine and cysteine residues, which can coordinate a zinc ion and form an HHCC zinc finger motif (7, 8, 69, 71). Biophysical and biochemical analysis shows that the HHCC domain is involved in protein multimerization (52, 69, 71). Indirect evidence indicates that the HHCC domain is required for the formation of stable IN-LTR complexes (15, 21, 61, 63). The active site, consisting of D-D(35)-E residues, is within the central core domain. All three active-site residues are conserved among retroviruses. Mutation of either one of the aspartic acid residues or the glutamic acid residue abolishes all catalytic activities (17, 22, 48, 62). The C-terminal domain is the least conserved among the three domains and has nonspecific DNA binding activity (23, 47, 55, 64, 68).
The recognition of the viral LTR by IN and the assembly of the integration complex are not yet fully understood. Analysis of chimeric human immunodeficiency virus type 1 (HIV-1)/visna virus or HIV-1/spuma virus IN indicated a role for the catalytic core in the recognition of viral DNA ends and positioning of the host DNA (45, 46, 57). Biochemical and cross-linking studies mapped the CA dinucleotide in close proximity to the catalytic core (30, 33, 34, 38, 49). In contrast, a role for the HHCC region in LTR recognition has been reported for feline immunodeficiency virus/HIV complementation pairs (61). The M-MuLV HHCC region is also required for catalysis of MuLV LTR substrates which lack the 5′ tail (15, 16, 69). These results are consistent with a model in which multiple domains interact with the viral termini. This model is exemplified in the structure of the related Tn5 synaptic complex. In this complex, the N-terminal, C-terminal, and catalytic regions all have contacts with a 20-bp oligonucleotide encoding the Tn5 recognition sequence (14).
To study the integration event, in vitro assays such as 3′ processing and strand transfer reactions have been developed, which utilize purified IN protein and short oligonucleotides containing the sequences of the viral LTR (13, 44, 59). One limitation of these assays is that integration of only one LTR end is examined (one-end integration). In vivo, two LTR ends from the same DNA molecule are integrated into the host DNA in a coordinated fashion (two-end integration). With the M-MuLV, a mutation at one LTR blocks cleavage of both ends by the viral IN in vivo (56). Assays with different efficiencies for concerted two-end integrations have been established (for a review, see reference 35). These assays utilize either mini-viral DNA donors or LTR oligonucleotides. The sources of IN include either virus-infected cell extracts (28), purified viral IN (10, 27, 31, 43, 65, 67), or recombinant IN (1, 10, 32, 35, 37, 54, 60). The addition of proteins including members of the HMG families or nucleocapsid (NC) was found to stimulate these reactions (10, 35). To investigate the function of the M-MuLV HHCC domain, we have modified an in vitro concerted two-end integration assay (60). This assay utilized recombinant M-MuLV IN, LTR oligonucleotide as the donor, and plasmid DNA as the target. Our study shows that zinc was required for efficient two-end integration. The results demonstrate that two mutants, IN 1–105 (containing the HHCC zinc finger domain) and IN 106–404 (containing the central core and the C terminus) could complement in a nonoverlapping fashion, yielding two-end integrants. By varying the order of addition of the IN or DNA components in the complementation assay, different ratios of integration products were obtained. This order-of-addition experiment supports the role of the HHCC domain in LTR recognition and/or positioning.
MATERIALS AND METHODS
Materials.
Crude [γ-32P]ATP (7,000 Ci/mmol) was purchased from ICN. T4 polynucleotide kinase, T4 DNA ligase, and restriction enzymes were obtained from New England Biolabs. Exonuclease-free Klenow fragment of DNA polymerase I was obtained from United States Biochemical. Ni2+-nitrilotriacetic acid agarose was purchased from Qiagen.
Oligonucleotides.
DNA oligonucleotides were prepared by the University of Medicine and Dentistry of New Jersey Biochemistry Department DNA Synthesis Facility and purified by electrophoresis on 20% denaturing polyacrylamide gels. Oligonucleotides used in this study are referred to by their synthesis numbers and were labeled with [γ-32P]ATP by a kinase reaction as previously described (41). Oligonucleotide 9076 (5′-GACTACCCGTCAGCGGGGGTCTTTCATT) and its complementary strand 9075 (5′-AATGAAAGACCCCCGCTGACGGGTAGTC) were used as the blunt-end LTR substrate for two-end integration reactions. Oligonucleotide 9268 (5′-GACTACCCGTCAGCGGGGGTCTTTCA) and its complementary strand 9075, mimicking a precleaved LTR end, were used as the substrate for strand transfer and two-end integration reactions. Oligonucleotide 9904 (5′-CGCTCGAGACTACCCGTCAGCGGGGGTCTTTCA), containing a XhoI site (underlined), was used for PCR to amplify linear two-end integration products. Oligonucleotide 9966 (5′-GACTACCCGTCAGCGGGGGTC) was used to sequence the junction between the LTR and the target DNA.
Purification of M-MuLV integrase.
Recombinant M-MuLV IN (wild type [WT], IN 106–404, and IN 1–105) containing a hexahistidine tag were expressed in Escherichia coli BL21(DE3) (Novagen) and purified by Ni2+-nitrilotriacetate agarose chromatography as previously described (41). WT/Zinc and IN 1–105/Zinc were renatured in the presence of 10 μM ZnCl2 (69). Zinc was omitted in the last step of renaturation. WT/EDTA was renatured in the presence of 1 mM EDTA. The zinc content of WT IN proteins was measured by atomic absorption spectroscopy as previously described (69). The molar ratio of zinc ion to IN was 1.1:1 for WT/Zinc protein and 0.4:1 for WT/EDTA protein. To remove the His tag from IN 1–105, IN 1–105 was digested with thrombin as described previously (69) and tested in two-end integration reactions.
In vitro assays.
Strand transfer and 3′-processing reactions were performed as previously described (41). The reaction buffer contained 20 mM morpholineethanesulfonic acid (MES: pH 6.2), 10 mM dithiothreitol, 10 mM MnCl2, 10 mM KCl, and 10% glycerol. The conditions for two-end integration reaction were the same as those for the strand transfer reaction except that 200 mM KCl and 10% dimethyl sulfoxide (DMSO) were added. The concerted two-end integration assay is a modification of that previously described (60). The LTR oligonucleotide was labeled at the 5′ end by T4 polynucleotide kinase and mixed with a complementary strand at a ratio of 1:2 (labeled oligonucleotide versus complementary strand). The oligonucleotides were annealed by heating for 3 min at 95°C and then cooling to room temperature. Typically, one reaction mixture (30 μl) contained 1 pmol of labeled LTR, 1.2 μg of target plasmid DNA, and 20 pmol of IN protein. Precleaved LTR substrate was used as the donor unless indicated otherwise. Complementation assays were performed by mixing 160 pmol of HHCC finger domain protein (IN 1–105) with 20 pmol of IN 106–404. Unless indicated otherwise, the IN protein and the LTR were mixed and incubated on ice for 5 min and then at 37°C for 5 min, and the target DNA and salt were then added. The reaction mixtures were incubated at 37°C for 2 h and stopped by addition of 10 mM EDTA (pH 8.0)–0.5% sodium dodecyl sulfate and 100 μg of proteinase K per ml at 37°C for 1 h. A 10-μl volume of the reaction mixture was subjected to electrophoresis on a 1% agarose gel. After gel electrophoresis, the gel was dried and exposed to Kodak X-Omat Blue XB-1 film. To examine the smaller LTR-LTR integration products, 2 μl of the reaction mixture was run on a 20% denaturing polyacrylamide gel. For the order-of-addition experiment, the yield of one-end and two-end integration products was quantified by phosphorimaging.
Isolation of two-end integrants and sequence analysis.
A 1.7-kb pGEM-3Zf(+)′ target plasmid was constructed to facilitate the isolation of linear two-end integrants. The 3.2-kb pGEM-3Zf(+) (Promega) plasmid was digested with SspI and AflIII, and the 1.7-kb fragment was isolated and treated with Klenow to fill in the sticky ends. After ligation and transformation into E. coli HB101, the new 1.7-kb plasmid was isolated for use as the target DNA in the two-end integration assay. The two-end integration reaction products were subject to electrophoresis on a 1% agarose gel. The linear 1.7-kb DNA product was excised and isolated using the Freeze-Squeeze kit (Bio-Rad). Linear two-end integrants were PCR amplified using 0.5 U of Taq polymerase (Gibco-BRL) and 20 pmol of primer 9904, carrying a XhoI site at its 5′ terminus. Primer 9904 hybridized to the LTR sequence at each end of the linear two-end integrant and served as both the upstream and downstream PCR primers. The conditions for PCR amplification were 20 mM Tris-HCl (pH 8.4), 50 mM KCl, 1.5 mM MgCl2, 2% DMSO, 2.5% glycerol, 1 mg of bovine serum albumin per ml, and 0.2 mM each deoxynucleoside triphosphate. An initial cycle at 72°C for 10 min was performed to repair the gap resulting from integration and was followed by a 2-min cycle of 95°C and 30 cycles at 95°C for 1 min and 74°C for 3 min (including the annealing step). The linear 1.7-kb PCR product was isolated from a 1% agarose gel, digested with XhoI, extracted with phenol, and precipitated with ethanol. The DNA was ligated to itself and transformed into E. coli HB101 cells. To confirm that the individual clone is from a two-end integration, plasmid DNA was digested with XhoI, introduced with the LTR PCR primer. The parental 1.7-kb plasmid does not encode a XhoI site and has one XmnI site. Only those clones cut by XhoI were scored as two-end integrants. To sequence the junction between the LTR and target DNA, the plasmid DNA was digested with XhoI and XmnI, releasing two fragments which each contained one viral LTR. The individual XhoI-XmnI fragments were then isolated and sequenced with primer 9966, using an AmpliCycle kit from Perkin-Elmer.
RESULTS
Recombinant M-MuLV IN can catalyze concerted two-end integration in vitro.
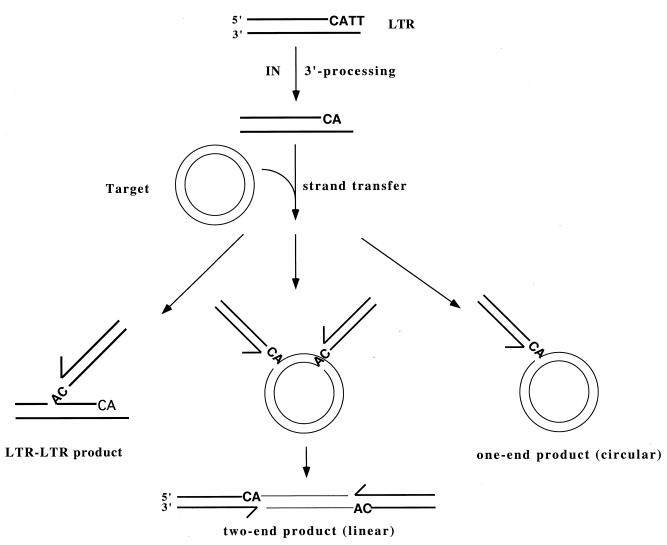
An in vitro concerted two-end integration assay was used to study the mechanism of the retroviral integration. This assay utilized recombinant M-MuLV IN protein, short duplex LTR oligonucleotides as the donor, and circular plasmid DNA as the target (outlined in Fig. 1). Three major products were expected from the assay: integration of the LTR into the LTR (LTR-LTR integration); integration of one LTR end into the plasmid DNA, which remains circular (one-end integration); and integration of two LTR ends into the plasmid DNA in coordination, yielding a linear product (two-end integration). The two larger integration products were visualized after agarose gel electrophoresis, whereas the small LTR-LTR integration products were separated on a 20% denaturing polyacrylamide gel.
FIG. 1.
Schematic illustration of the concerted two-end integration assay. The donor LTR oligonucleotides duplex (thick line) is first 3′ processed by IN, generating the 5′ two-nucleotide tail. In the presence of circular plasmid target DNA, strand transfer proceeds. Three major types of products are formed in this assay: circular one-end integration product, linear two-end integration product, and LTR-LTR integration product. Minor integration products, including cases where two or more LTRs are inserted independently into the same plasmid, are not included in the figure. The size of the DNA is not drawn according to scale.
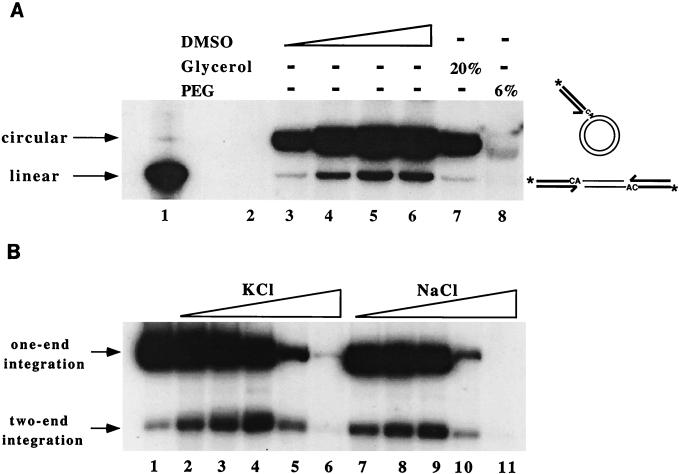
The results of the assay indicate that under reaction conditions defined for the in vitro strand transfer assay (41), the full-length M-MuLV IN can catalyze the concerted two-end integration reactions, indicated by the formation of linear plasmid DNA, but at low efficiency (Fig. 2A, lane 3). Titration of the reaction components improved the efficiency of the two-end integration. DMSO stimulated the formation of both one-end and two-end integration products (lanes 4 to 6), with maximal activity achieved with 10% DMSO. Neither 20% glycerol nor 6% polyethylene glycol (PEG) stimulated the integration activity (lanes 7 and 8). Salt concentrations from 50 to 200 mM specifically stimulated the two-end integration, as evidenced by increase of the two-end integration products and decrease of the one-end integration products (Fig. 2B). Maximal two-end integration was obtained in 200 mM KCl and therefore was used in all the assays in the presence of 10% DMSO.
FIG. 2.
M-MuLV IN catalyzes the concerted two-end integration reaction. Products shown are the result of integration of the 32P-labeled LTR donor into unlabeled plasmid target DNA by WT IN. The positions of the one-end circular lariat-like products and the two-end linear products are indicated. (A) Effect of solvent conditions on the two-end integration reaction. Lanes: 1, 32P-labeled linear pGEM target plasmid DNA; 2, no-IN control; 3 to 6, 0, 5, 10, and 20% DMSO, respectively; 7, 20% glycerol; 8, 6% PEG 8000. (B) Effect of salt concentrations on the two-end integration reaction. Lanes: 1, no salt; 2 to 6, 50, 100, 200, 400, and 600 mM KCl, respectively; 7 to 11, 50, 100, 200, 400, and 600 mM NaCl, respectively.
To confirm that the linear DNA products resulted from the concerted integration events, the linear DNA was isolated, amplified by PCR, and subcloned into E. coli, and the sequence at the site of integration was examined (Table 1). With WT M-MuLV IN, the majority (53%) of the products contained the correct 4-bp duplication, indicating that the linear products represent true concerted integration events. Additional isolates contained duplications of either 3 bp (9%) or 5 bp (22%). This distribution of host target duplication is in agreement with those obtained in vitro for HIV and avian sarcoma virus integrase (1, 31). The remaining 5 of the 32 integrants sequenced contained large duplications or deletions, which can result from two independent one-end integration events into one target DNA. The majority of the integrants were distributed within the nonessential regions between the replication origin and the ampicillin resistance gene of the plasmid (data not shown).
TABLE 1.
Sequencing results of LTR-target junctionsa
| Duplication | No. (%) of products obtained with:
|
||
|---|---|---|---|
| WT | IN 106–404 + IN 1–105/Zinc | IN 106–404 + IN 1–105/EDTA | |
| 4 bp | 17 (53) | 21 (66) | 25 (73) |
| 3 bp | 3 (9) | 1 (3) | 3 (9) |
| 5 bp | 7 (22) | 2 (6) | 4 (12) |
| Others | 5 (16) | 8 (25) | 2 (6) |
| Total | 32 | 32 | 34 |
The DNA containing two LTRs from concerted two-end integration was isolated and sequenced as described in Materials and Methods. The total of 98 clones were from two independent experiments.
Zinc is required for two-end integration catalyzed by the M-MuLV IN.
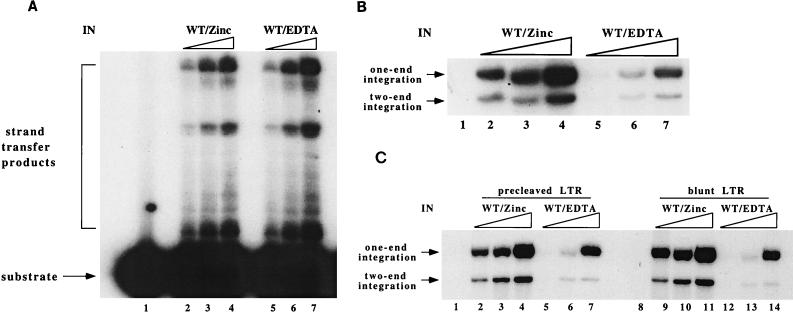
Early study of the M-MuLV IN demonstrated that full-length IN purified in the presence of EDTA was fully active, indicating that zinc was not required for the strand transfer reaction, which utilizes the LTR oligonucleotides as both donor and target DNA (41). To investigate the possible requirement of zinc for the two-end integration assay, recombinant IN renatured in the presence of zinc (WT/Zinc) and recombinant IN renatured in the presence of EDTA (WT/EDTA) were used in both strand transfer and two-end integration reactions. In the strand transfer assay with precleaved LTR oligonucleotide substrates, WT/Zinc and WT/EDTA displayed equivalent levels of activity (Fig. 3A, compare lanes 2 to 4 with lanes 5 to 7). However, in the two-end integration assay, WT/EDTA was much less active than WT/Zinc for the yield of both one-end and two-end integration products (Fig. 3B, compare lanes 2 to 4 with lanes 5 to 7). A two-end integration assay using blunt LTR oligonucleotides produced similar results (Fig. 3C). The LTR-LTR integration and LTR-plasmid integration assays differ in the size and nature of the target DNA; the two-end integration assay utilizes long circular plasmid DNA rather than short oligonucleotides as the target DNA. The difference in activity in the two assays may reflect a defect in utilization of longer DNA target by the WT/EDTA. These results suggest that the zinc influences the formation of a functional IN-LTR-target complex, most probably through the HHCC zinc finger domain.
FIG. 3.
Zinc is required for the efficient two-end integration reaction catalyzed by the M-MuLV IN. (A) Comparison of WT/Zinc with WT/EDTA for the strand transfer reaction (LTR-LTR integration). Lane 1, no-IN control; lanes 2 to 4, 4, 10, and 20 pmol of WT/Zinc; lanes 5 to 7, 4, 10, and 20 pmol of WT/EDTA. (B) Comparison of WT/Zinc and WT/EDTA for the concerted two-end integration reaction. The lanes are the same as in panel A. Equivalent amounts of IN proteins were used in the experiments in panels A and B. (C) Comparison of two-end integration reactions using either precleaved or blunt LTR. Lanes 1 and 8, no-protein control; lanes 2 to 4 and 9 to 11, 4, 10, and 20 pmol of WT/Zinc; lanes 5 to 7 and 12 to 14, 4, 10, and 20 pmol of WT/EDTA. One picomole of LTR substrate was used in all panels.
Complementation of IN domains in the two-end integration reaction.
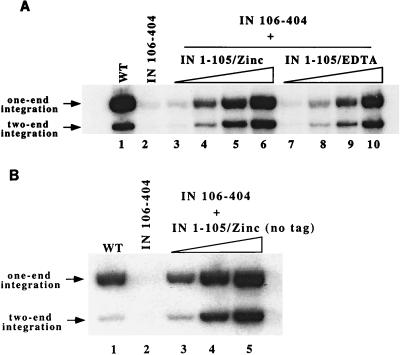
Assembly of a coordinated two-end integration complex requires the precise coordination of the IN multimer with the two viral LTRs and the target DNA. The HHCC domain was previously shown to stimulate the assembly of IN dimers and tetramers (52, 69, 71) and can function in trans as a nonoverlapping domain in a complementation assay with the core and C terminus (61, 69). The ability of two nonoverlapping M-MuLV IN domains, IN 1–105 encoding the HHCC domain and IN 106–404 encoding the core and the C terminus, to function in the concerted two-end integration assay was therefore examined. In this experiment, IN 1–105 and IN 106–404 were mixed first, the LTR was added afterward, and the target DNA and salt were added last. As shown in Fig. 4A, IN 106–404 alone has no two-end integration activity and very low level of one-end integration activity (Fig. 4A, lane 2). Complementation of IN 106–404 with increasing amounts of IN 1–105 yielded both two-end and one-end integrations. Maximal amounts of products were detected at a molar ratio of 8:1 (HHCC: core/C-terminus) (Fig. 4A and data not shown). Either IN 1–105/Zinc (Fig. 4A, lanes 3 to 6) or IN 1–105/EDTA (lanes 7 to 10) could complement IN 106–404. Complementation with excess IN 1–105/EDTA was much more efficient in integration into target plasmid than was the WT IN/EDTA (Fig. 3B). This could be due to the excess IN 1–105, which may compensate for the lack of zinc through protein-protein interactions.
FIG. 4.
Complementation of IN domains for two-end integration reaction. (A) Complementation of IN 106–404 by IN 1–105 for the two-end integration assay. Lane 1, 20 pmol of WT/Zinc; lane 2, 20 pmol of IN 106–404 alone; lanes 3 to 6, titration of IN 1–105/Zinc against 20 pmol of IN 106–404; lanes 7 to 10, titration of IN 1–105/EDTA against 20 pmol of IN 106–404 (the amounts of IN 1–105 used were 2, 10, 40, and 160 pmol, respectively). The positions of one-end and two-end integration products are indicated. (B) Complementation of IN 106–404 by detagged IN 1–105/Zinc for the two-end integration assay. Lane 1, 20 pmol of WT/Zinc; lane 2, 20 pmol of IN 106–404 alone; lanes 3 to 6, titration of 10, 40, and 160 pmol of IN 1–105/Zinc without the His tag against 20 pmol of IN 106–404.
IN 1–105/Zinc without the His tag was also tested for complementation in the concerted integration assay. As shown in Fig. 4B, IN 1–105/Zinc with the tag removed could complement IN 106–404 efficiently to catalyze both one-end and two-end integration events (Fig. 4B, lanes 3 to 5).
The two-end integration products were isolated as linear DNA from the complementation assay and then subcloned and sequenced as described in Materials and Methods. As found with WT IN, more than half the clones had 4-bp duplications at the site of integration (66% for complementation with IN 1–105/Zinc and 73% for complementation with IN 1–105/EDTA [Table 1]), characteristic of the M-MuLV concerted integration in vivo. Similar results were obtained when linear products from complementation with detagged IN 1–105/Zinc were sequenced (data not shown). These results indicate that the assembly process of the nonoverlapping complementing halves of IN is sufficient to catalyze concerted two-end integration reactions.
Efficiency of two-end integration depends on the order of addition of components in the complementation assay.
To investigate the involvement of the HHCC zinc finger domain in the assembly of the IN-DNA complex, the order of addition of the protein and DNA components was varied in the two-end integration assay. There were four major components in this experiment: IN 1–105, IN 106–404, the donor LTR, and the circular target plasmid DNA. Two of the four components were mixed first and incubated at 37°C for 5 min. Then the third component was added, followed by another 5-min incubation at 37°C. The fourth component was added together with the salt (200 mM KCl). After a 5-min incubation on ice, the integration assay was started by incubation at 37°C. Experiments were analyzed for the overall yield as well as the distribution of integration products. Reaction products were analyzed both on agarose gels for integrations into the plasmid DNA (Fig. 5A) and on denaturing polyacrylamide gels for integration into the LTR oligonucleotide target (Fig. 5B). In the assay presented, the precleaved LTR substrates were used as the donor substrate. Similar results were observed using blunt-end LTR oligonucleotides (data not shown).
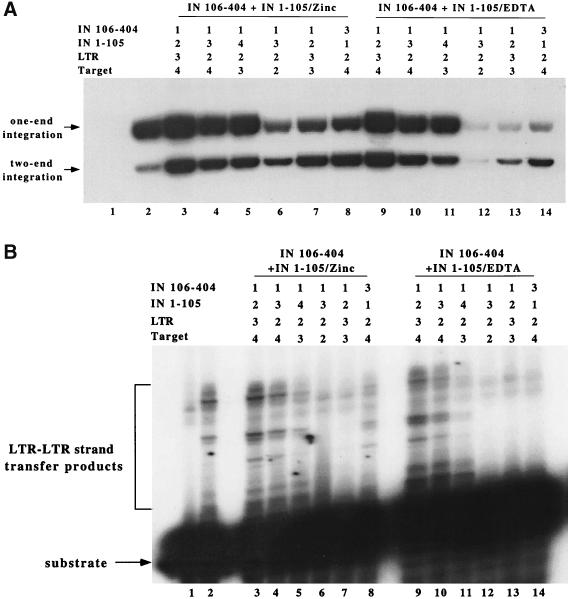
FIG. 5.
Order of addition experiment using precleaved LTR. The four major components used in the assay include IN 1–105, IN 106–404, LTR, and plasmid target DNA. The order by which each component was added is noted as 1 through 4 in the grid at the top of the figure. The same numbers in the reaction indicate that the two components were added together. (A) Integration products separated on an agarose gel. Lane 1, no-protein control; lane 2, 20 pmol of WT/Zinc control; lanes 3 to 8, complementation of IN 106–404 with IN 1–105/Zinc; lanes 9 to 14, complementation of IN 106–404 with IN 1–105/EDTA. The assay used 20 pmol of IN 106–404 and 160 pmol of IN 1–105. (B) Integration products separated on a 20% denaturing polyacrylamide gel. These products arose from LTR-LTR integration. The lanes are the same as in panel A.
The yield of integration products varied dramatically depending on the order of addition of the protein and DNA components. The control WT IN (zinc) produced only a low level of concerted two-end integrants, with the majority of products being single-end integrations: both into the plasmid target DNA (Fig. 5A, lane 2) and the oligonucleotide substrate (Fig. 5B, lane 2). The yield of integration products was greatly enhanced using the reconstituted IN. Complementation assays differ from the WT IN in that the molar ratio of HHCC construct IN 1–105 to IN 106–404 is 8:1. Preincubation of IN 106–404 with IN 1–105 resulted in an overall increase of integration efficiency (Fig. 5, lanes 3 and 9). Reactions which first introduced the LTR to IN 106–404 or the reconstituted IN complex resulted in LTR serving as both the viral donor DNA and the target DNA and yielded a high level of LTR-LTR integration products (lanes 3 and 4 in Fig 5). Interestingly, the incubation of the LTR DNA with either the reconstituted IN protein or IN 106–404 resulted in highly efficient integration into the exogenous plasmid target DNA, which was added in a subsequent step (Fig. 5A, lanes 3 to 5). However, under these conditions, the reactions favored the single-end integrants. Discrimination of the LTR from the target binding site and the preferential assembly of concerted two-end integration complex were obtained by premixing the LTR with the target DNA (Fig. 5A, lanes 6 and 7). Under conditions where the LTR and target DNA were added simultaneously, the ratio of two-end to one-end integration was greatly improved (lanes 6 and 7). This discrimination of viral and target DNA extends to one-end integration into the LTR oligonucleotide, where LTR-LTR integration products were barely detected (Fig. 5B, lanes 6 and 7). Under conditions of premixing of the target and donor DNA, the reconstituted IN again resulted in the higher yield of plasmid integrants (Fig. 5A, compare lane 6 with lane 7). Similar results were obtained when IN 1–105/EDTA was used instead of IN 1–105/Zinc in the order-of-addition experiment (Fig. 5, compare lanes 3 to 8 with lanes 9 to 14). However, the overall integration efficiency was lower with IN 1–105/EDTA, under conditions that favor two-end integration (compare lanes 6 to 8 with lanes 12 to 14).
The order of addition of the LTR substrate with respect to IN 1–105 or IN 106–404, surprisingly, altered the preference for the two-end concerted products. The majority of the products formed after preincubation of the LTR with the HHCC IN 1–105 construct followed by IN 106–404 and the target DNA were concerted two-end products (Fig. 5A, lanes 8 and 14). The DNA sequences of these products were determined, and more than 85% of the isolated integrants had the correct 4-bp duplication. In contrast, single-end integration predominated under conditions where the LTR was incubated first with IN 106–404 and then with IN 1–105 and the target (Fig. 5A, lanes 4 and 10). Although the retroviral HHCC constructs are not reported to have DNA binding activity, preincubation of the viral termini with the HHCC favors the production of concerted two-end products. This effect was observed with IN 1–105 renatured in zinc or in EDTA (Fig. 5A, lanes 8 and 14). The yield of products with IN 1–105/EDTA was lower than the zinc preparation; however, the relative ratio of the products was maintained. The lowest overall yield of integration was observed when IN 106–404 was mixed with the LTR and the target plasmid together in the absence of the HHCC domain (Fig. 5A, lanes 6 and 12). These results strongly support the role of the HHCC domain in the recognition and/or positioning of the LTR. Cumulatively, the results indicate two mechanisms that favor concerted two-end integrations: preincubation of the LTR with the HHCC domain or the presence of the large target DNA at the time of assembly of the LTR complex.
DISCUSSION
Retroviral integration in vivo requires the two LTR ends from the same viral DNA to be joined to the host DNA in a coordinated and staggered fashion (see reference 5 for a review). In vivo cleavage of the M-MuLV LTRs is concerted; one mutation in the U3 LTR blocked the processing of both LTR ends (56). Multiple assays using short oligonucleotides that mimic the LTR termini have been developed, which have greatly assisted in deciphering the mechanism of integration (13, 44, 59). These assays differ from integration in vivo in that the products represent integration of a single viral terminus and the LTR sequence serves as both the donor and the target DNA. Thus, the mechanism developed for recognition of the viral termini must be adjusted to position the LTR sequence into the target site. To overcome these problems, we modified an in vitro concerted two-end integration assay (60) in this study to define the assembly and recognition of the integration complex.
Previously, our laboratory has shown that two nonoverlapping M-MuLV IN domains (IN 1–105 and IN 106–404) could complement each other for strand transfer and 3′-processing reactions (69). This pair of mutants can also catalyze the concerted two-end integration. Six variations in the order of addition of the protein and DNA components were tested and found to profoundly affect the yield and distribution of the integration products. Figure 6 schematically outlines the assembly of functional IN-DNA complex that could take place in the order-of-addition experiment. The IN is diagrammed to contain two binding sites, one for the LTR (the L site) and the other for the target (the T site) DNA. The criteria for binding to either site are not completely understood. Binding determinants for the CA within the LTR termini have been localized to the catalytic core (30, 33, 34, 38, 49). Stable recognition of the LTR requires either the 5′ overhang tail or the HHCC domain (15, 16, 69). The stable association of the LTR with the L site through the HHCC domain is schematically shown by altered positioning of the LTR in Fig. 6. The LTR substrates used in these experiments contain the 5′ tail.
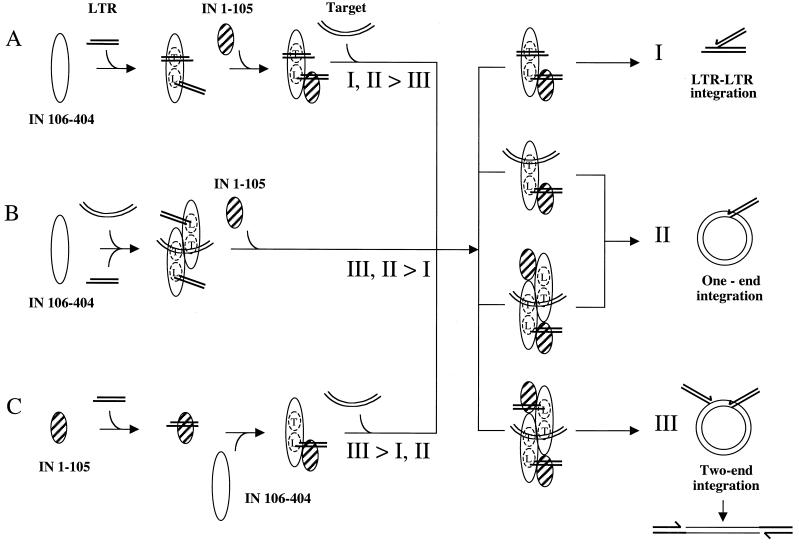
FIG. 6.
Model for the assembly of the IN-LTR-target complex. Three possible pathways of IN-DNA complex assembly are listed as A through C. Three types of integration products are listed as I through III. Although other pathways for assembly may exist, the results of this study support the model presented here. The major products of each pathway are indicated in the figure. The ratio of integration products depends on the order in which the protein and DNA components were added. Pathways A, B, and C correspond to lanes 3 to 5 and 9 to 11, 6 to 7 and 12 to 13, and 8 and 14 in Fig. 5A, respectively. The striped oval represents IN 1–105, either renatured with zinc or EDTA. The open oval represents IN 106–404. Straight double lines represent the LTR. Curved double lines represent the plasmid DNA target. IN is proposed to have distinct binding sites for the LTR and the target, as indicated by L and T, respectively. The IN domains are drawn as monomers and dimers but could represent dimers, tetramers, or higher-order multimers.
Results presented in this paper indicate that the presence of zinc within the HHCC domain influences the target binding as well. IN renatured in EDTA was capable of efficient one-end integration into an oligonucleotide substrate but was much less efficient in integration into the plasmid DNA target. In contrast, IN renatured in the presence of zinc utilized large plasmid DNA with high efficiency. The M-MuLV HHCC domains containing zinc are known to form stable dimers (69). In addition, the HIV-1 IN protein in the presence of zinc forms tetramers (52, 71). Therefore, it is possible that higher-order multimerization induced by the HHCC domain in the presence of zinc may assemble a full-target binding site consisting of two half-target sites from individual core–C-terminus constructs. In vitro strand transfer and target site selection studies support this model. In the absence of the HHCC domain, the M-MuLV IN 106–404 construct (NΔ105) yielded limited integration into one preferential site in the LTR oligonucleotide (40). This indicates that stable association with the short oligonucleotide occurs at only limited positions. The addition of the HHCC domain expanded the target site selection. Another possibility is that the HHCC domain chelated with zinc is directly involved in the target positioning. Indeed, the HHCC domain has been found to be in close contact with the target DNA by UV cross-linking studies (33). However, we do not see these two models, as well as the role of the HHCC domain in positioning the LTR, to be mutually exclusive. The synaptic complex of Tn5 presents a model of how an N-terminal domain could be involved in multiple inter- and intradomain contacts as well as association with DNA (14).
In the first three variations in the order-of-addition experiment, the LTR substrate was initially incubated with either IN 106–404 or the reconstituted IN protein. In the absence of an alternative target, the LTR oligonucleotide will bind to both the LTR and target sites, as depicted in pathway A in Fig. 6. With either IN 106–404 or the reconstituted IN protein, preincubation with the LTR greatly stimulated the yield of one-end and two-end integration products. This stimulation through the ordered addition of substrates follows the biochemical analysis of integration inhibitors, where the target site is proposed to be formed after the assembly of the viral ends (25). However, integration complexes assembled by this pathway are less efficient in two-end integration than in one-end integration. It is interesting that a host protein, barrier to autointegration factor (BAF) (50), has been identified which blocks the ability of MuLV to autointegrate in vitro. This mechanism could allow for the assembly in the cytoplasm of the activated complex containing the viral termini to be temporally and spatially distinct from the binding of the host target DNA in the nucleus.
Two pathways were identified which resulted in the efficient catalysis of two-end integration. The first pathway required the simultaneous presentation of both the viral LTR and the target DNA, schematically diagrammed in Fig. 6, Pathway B. The target DNA may be directed to the target binding site due to the large size of the plasmid DNA. It is possible that the binding of the plasmid DNA facilitates the assembly of the full target binding site by bridging the individual half-sites in the absence of the HHCC domain. Stabilization of the large target DNA to the target binding site prevents the LTR from being used as target DNA, thus decreasing the level of LTR-LTR integrations. The second pathway involved the preincubation of the LTR with the HHCC domain (Fig. 6, pathway C). The positioning of this complex within the catalytic core-C terminus would block nonspecific binding of the LTR to the target binding site. This recognition of the LTR by the HHCC could be to regions upstream of the CA, within the conserved inverted repeat of the M-MuLV. The stable placement of the LTR into the donor binding site would be the result of multiple determinants including the CA, the 5′ tail, and the HHCC domain. This scenario is in good agreement with the observed structure of the Tn5 synaptic complex (14). In the Tn5 synaptic complex, the N terminus of the Tn5 transposase binds to the internal region of the transposon DNA, which is critical for the assembly of the synaptic complex and the coordination of the two DNA ends. In addition, the HHCC domain may stabilize the LTR binding through protein-protein interactions with other IN domains. Further studies defining the DNA binding potential of the M-MuLV HHCC domain are in progress.
Titration of various components in the concerted two-end integration assays revealed differential effects. The highest yield of both one-end and two-end integration was obtained in the presence of 10% DMSO. DMSO is able to significantly increase integration activities of various integrases through the proposed increase in protein-protein interactions (31, 51, 66). In contrast to DMSO, high salt concentrations specifically stimulated concerted two-end integration while suppressing one-end integration, similar to avian myeloblastosis virus IN (66). High salt could affect protein association or protein-DNA interactions. The results of this study indicate that zinc stimulates both one-LTR and two-LTR integration into the plasmid target. Additional studies incorporating zinc into assays using WT IN/EDTA yielded similar results (data not shown). Zinc is thus not discriminatory between one-end and two-end integration; however, it facilitates the use of large target DNA.
The IN HHCC construct used in these studies consists of the first 105 amino acids of the M-MuLV IN. This includes the first 50-amino-acid domain not conserved in other human or avian retroviruses. Circular dichroism analysis of IN 1–105 renatured in EDTA indicated that the domain was structured (69), thereby accounting for the complementation activity of the domain. Atomic absorption analysis of IN 1–105 (EDTA) indicated that 10% of the molecules still contained zinc (69) and may account, in part, for the activity observed. Sequence analysis of concerted two-end integration products catalyzed with IN 1–105/EDTA indicates that the fidelity of the target duplication was equal to or better than that obtained with the WT or IN 1–105/Zinc preparations. It is possible that the increase in structure observed in the presence of zinc may be induced in the IN 1–105/EDTA through alternative protein-protein or protein-DNA contacts. The excess of HHCC domain in the complementation assays may allow for the selection and assembly of active molecules, which produced more efficient two-end integration. This may explain why the complementing pair (IN 1–105 plus IN 106–404) was more efficient than WT IN at two-end integration.
Sequencing of the integrants proved that the linear plasmid DNA seen in the assay was from authentic concerted two-end integration events. Sequence analysis indicated that the majority of the two-end integration products produced with either the full-length WT IN or the reconstituted IN had the 4-bp duplication found in vivo. Secondary products included less stringent duplication of either 3 or 5 bp. This distribution of target site duplications is consistent with those obtained by using other in vitro systems (1, 31). Among the WT IN integrants recovered, four have large duplications of target sequences ranging from 27 to 77 bp and one has a deletion of 96 bp (data not shown). Integration products with large duplications or deletions could arise from two independent one-end integration events.
NMR and X-ray crystallographic structural data for IN subdomains have been obtained (6, 9, 11, 12, 18–20, 53, 70), but no data for a synaptic complex have been obtained. The use of the complementation system provides an alternative method to study the assembly of a reconstituted IN-DNA complex, which is highly active for the catalysis of two-end integration events.
ACKNOWLEDGMENTS
This work is supported by NIH grant R01 CA76545 to M.J.R.
We thank Abram Gabriel, Keith Bupp, and Jennifer Seamon for critically reading the manuscript.
REFERENCES
- 1.Aiyar A, Hindmarsh P, Skalka A M, Leis J. Concerted integration of linear retroviral DNA by the avian sarcoma virus integrase in vitro: dependence on both long terminal repeat termini. J Virol. 1996;70:3571–3580. doi: 10.1128/jvi.70.6.3571-3580.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowerman B, Brown P O, Bishop J M, Varmus H E. A nucleoprotein complex mediates the integration of retroviral DNA. Genes Dev. 1989;3:469–478. doi: 10.1101/gad.3.4.469. [DOI] [PubMed] [Google Scholar]
- 3.Brown P O, Bowerman B, Varmus H E, Bishop J M. Correct integration of retroviral DNA in vitro. Cell. 1987;49:347–356. doi: 10.1016/0092-8674(87)90287-x. [DOI] [PubMed] [Google Scholar]
- 4.Brown P O, Bowerman B, Varmus H E, Bishop J M. Retroviral integration: structure of the initial covalent product and its precursor, and a role for the viral IN protein. Proc Natl Acad Sci USA. 1989;86:2525–9. doi: 10.1073/pnas.86.8.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown P O. Integration. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 161–203. [PubMed] [Google Scholar]
- 6.Bujacz G, Jaskolski M, Alexandratos J, Wlodawer A, Merkel G, Katz R A, Skalka A M. High-resolution structure of the catalytic domain of avian sarcoma virus integrase. J Mol Biol. 1995;253:333–346. doi: 10.1006/jmbi.1995.0556. [DOI] [PubMed] [Google Scholar]
- 7.Burke C J, Sanyal G, Bruner M W, Ryan J A, LaFemina R L, Robbins H L, Zeft A S, Middaugh C R, Cordingley M G. Structural implications of spectroscopic characterization of a putative zinc finger peptide from HIV-1 integrase. J Biol Chem. 1992;267:9639–9644. [PubMed] [Google Scholar]
- 8.Bushman F D, Engelman A, Palmer I, Wingfield P, Craigie R. Domains of the integrase protein of human immunodeficiency virus type 1 responsible for polynucleotidyl transfer and zinc binding. Proc Natl Acad Sci USA. 1993;90:3428–3432. doi: 10.1073/pnas.90.8.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai M, Zheng R, Caffrey M, Craigie R, Clore G M, Gronenborn A M. Solution structure of the N-terminal zinc binding domain of HIV-1 integrase. Nat Struct Biol. 1997;4:567–577. doi: 10.1038/nsb0797-567. [DOI] [PubMed] [Google Scholar]
- 10.Carteau S, Gorelick R J, Bushman F D. Coupled integration of human immunodeficiency virus type 1 cDNA ends by purified integrase in vitro: stimulation by the viral nucleocapsid protein. J Virol. 1999;73:6670–6679. doi: 10.1128/jvi.73.8.6670-6679.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen J C, Krucinski J, Miercke L J, Finer-Moore J S, Tang A H, Leavitt A D, Stroud R M. Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding. Proc Natl Acad Sci USA. 2000;97:8233–8238. doi: 10.1073/pnas.150220297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Z, Yan Y, Munshi S, Li Y, Zugay-Murphy J, Xu B, Witmer M, Felock P, Wolfe A, Sardana V, Emini E A, Hazuda D, Kuo L C. X-ray structure of simian immunodeficiency virus integrase containing the core and C-terminal domain (residues 50–293)—an initial glance of the viral DNA binding platform. J Mol Biol. 2000;296:521–533. doi: 10.1006/jmbi.1999.3451. [DOI] [PubMed] [Google Scholar]
- 13.Craigie R, Fujiwara T, Bushman F. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell. 1990;62:829–837. doi: 10.1016/0092-8674(90)90126-y. [DOI] [PubMed] [Google Scholar]
- 14.Davies D R, Goryshin I Y, Reznikoff W S, Rayment I. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science. 2000;289:77–85. doi: 10.1126/science.289.5476.77. [DOI] [PubMed] [Google Scholar]
- 15.Donzella G A, Jonsson C B, Roth M J. Coordinated disintegration reactions mediated by Moloney murine leukemia virus integrase. J Virol. 1996;70:3909–3921. doi: 10.1128/jvi.70.6.3909-3921.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donzella G A, Leon O, Roth M J. Implication of a central cysteine residue and the HHCC domain of Moloney murine leukemia virus integrase protein in functional multimerization. J Virol. 1998;72:1691–1698. doi: 10.1128/jvi.72.2.1691-1698.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drelich M, Wilhelm R, Mous J. Identification of amino acid residues critical for endonuclease and integration activities of HIV-1 IN protein in vitro. Virology. 1992;188:459–468. doi: 10.1016/0042-6822(92)90499-f. [DOI] [PubMed] [Google Scholar]
- 18.Dyda F, Hickman A B, Jenkins T M, Engelman A, Craigie R, Davies D R. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science. 1994;266:1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- 19.Eijkelenboom A P, Lutzke R A, Boelens R, Plasterk R H, Kaptein R, Hard K. The DNA-binding domain of HIV-1 integrase has an SH3-like fold. Nat Struct Biol. 1995;2:807–810. doi: 10.1038/nsb0995-807. [DOI] [PubMed] [Google Scholar]
- 20.Eijkelenboom A P, van den Ent F M, Vos A, Doreleijers J F, Hard K, Tullius T D, Plasterk R H, Kaptein R, Boelens R. The solution structure of the amino-terminal HHCC domain of HIV-2 integrase: a three-helix bundle stabilized by zinc. Curr Biol. 1997;7:739–746. doi: 10.1016/s0960-9822(06)00332-0. [DOI] [PubMed] [Google Scholar]
- 21.Ellison V, Brown P O. A stable complex between integrase and viral DNA ends mediates human immunodeficiency virus integration in vitro. Proc Natl Acad Sci USA. 1994;91:7316–7320. doi: 10.1073/pnas.91.15.7316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Engelman A, Craigie R. Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J Virol. 1992;66:6361–6369. doi: 10.1128/jvi.66.11.6361-6369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Engelman A, Hickman A B, Craigie R. The core and carboxyl-terminal domains of the integrase protein of human immunodeficiency virus type 1 each contribute to nonspecific DNA binding. J Virol. 1994;68:5911–5917. doi: 10.1128/jvi.68.9.5911-5917.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Engelman A, Mizuuchi K, Craigie R. HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell. 1991;67:1211–1221. doi: 10.1016/0092-8674(91)90297-c. [DOI] [PubMed] [Google Scholar]
- 25.Espeseth A S, Felock P, Wolfe A, Witmer M, Grobler J, Anthony N, Egbertson M, Melamed J Y, Young S, Hamill T, Cole J L, Hazuda D J. HIV-1 integrase inhibitors that compete with the target DNA substrate define a unique strand transfer conformation for integrase. Proc Natl Acad Sci USA. 2000;97:11244–11249. doi: 10.1073/pnas.200139397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Farnet C M, Bushman F D. HIV-1 cDNA integration: requirement of HMG I(Y) protein for function of preintegration complexes in vitro. Cell. 1997;88:483–492. doi: 10.1016/s0092-8674(00)81888-7. [DOI] [PubMed] [Google Scholar]
- 27.Fitzgerald M L, Vora A C, Zeh W G, Grandgenett D P. Concerted integration of viral DNA termini by purified avian myeloblastosis virus integrase. J Virol. 1992;66:6257–6263. doi: 10.1128/jvi.66.11.6257-6263.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujiwara T, Craigie R. Integration of mini-retroviral DNA: a cell-free reaction for biochemical analysis of retroviral integration. Proc Natl Acad Sci USA. 1989;86:3065–3069. doi: 10.1073/pnas.86.9.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujiwara T, Mizuuchi K. Retroviral DNA integration: structure of an integration intermediate. Cell. 1988;54:497–504. doi: 10.1016/0092-8674(88)90071-2. [DOI] [PubMed] [Google Scholar]
- 30.Gerton J L, Brown P O. The core domain of HIV-1 integrase recognizes key features of its DNA substrates. J Biol Chem. 1997;272:25809–25815. doi: 10.1074/jbc.272.41.25809. [DOI] [PubMed] [Google Scholar]
- 31.Goodarzi G, Im G J, Brackmann K, Grandgenett D. Concerted integration of retrovirus-like DNA by human immunodeficiency virus type 1 integrase. J Virol. 1995;69:6090–6097. doi: 10.1128/jvi.69.10.6090-6097.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goodarzi G, Pursley M, Felock P, Witmer M, Hazuda D, Brackmann K, Grandgenett D. Efficiency and fidelity of full-site integration reactions using recombinant simian immunodeficiency virus integrase. J Virol. 1999;73:8104–8111. doi: 10.1128/jvi.73.10.8104-8111.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heuer T S, Brown P O. Mapping features of HIV-1 integrase near selected sites on viral and target DNA molecules in an active enzyme-DNA complex by photo-cross-linking. Biochemistry. 1997;36:10655–10665. doi: 10.1021/bi970782h. [DOI] [PubMed] [Google Scholar]
- 34.Heuer T S, Brown P O. Photo-cross-linking studies suggest a model for the architecture of an active human immunodeficiency virus type 1 integrase-DNA complex. Biochemistry. 1998;37:6667–6678. doi: 10.1021/bi972949c. [DOI] [PubMed] [Google Scholar]
- 35.Hindmarsh P, Leis J. Reconstitution of concerted DNA integration with purified components. Adv Virus Res. 1999;52:397–410. doi: 10.1016/s0065-3527(08)60308-5. [DOI] [PubMed] [Google Scholar]
- 36.Hindmarsh P, Leis J. Retroviral DNA integration. Microbiol Mol Biol Rev. 1999;63:836–843. doi: 10.1128/mmbr.63.4.836-843.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hindmarsh P, Ridky T, Reeves R, Andrake M, Skalka A M, Leis J. HMG protein family members stimulate human immunodeficiency virus type 1 and avian sarcoma virus concerted DNA integration in vitro. J Virol. 1999;73:2994–3003. doi: 10.1128/jvi.73.4.2994-3003.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jenkins T M, Esposito D, Engelman A, Craigie R. Critical contacts between HIV-1 integrase and viral DNA identified by structure-based analysis and photo-crosslinking. EMBO J. 1997;16:6849–6859. doi: 10.1093/emboj/16.22.6849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson M S, McClure M A, Feng D F, Gray J, Doolittle R F. Computer analysis of retroviral pol genes: assignment of enzymatic functions to specific sequences and homologies with nonviral enzymes. Proc Natl Acad Sci USA. 1986;83:7648–7652. doi: 10.1073/pnas.83.20.7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jonsson C B, Donzella G A, Gaucan E, Smith C M, Roth M J. Functional domains of Moloney murine leukemia virus integrase defined by mutation and complementation analysis. J Virol. 1996;70:4585–4597. doi: 10.1128/jvi.70.7.4585-4597.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jonsson C B, Donzella G A, Roth M J. Characterization of the forward and reverse integration reactions of the Moloney murine leukemia virus integrase protein purified from Escherichia coli. J Biol Chem. 1993;268:1462–1469. [PubMed] [Google Scholar]
- 42.Kalpana G V, Marmon S, Wang W, Crabtree G R, Goff S P. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science. 1994;266:2002–2006. doi: 10.1126/science.7801128. [DOI] [PubMed] [Google Scholar]
- 43.Katz R A, Merkel G, Kulkosky J, Leis J, Skalka A M. The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell. 1990;63:87–95. doi: 10.1016/0092-8674(90)90290-u. [DOI] [PubMed] [Google Scholar]
- 44.Katzman M, Katz R A, Skalka A M, Leis J. The avian retroviral integration protein cleaves the terminal sequences of linear viral DNA at the in vivo sites of integration. J Virol. 1989;63:5319–5327. doi: 10.1128/jvi.63.12.5319-5327.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Katzman M, Sudol M. Mapping domains of retroviral integrase responsible for viral DNA specificity and target site selection by analysis of chimeras between human immunodeficiency virus type 1 and visna virus integrases. J Virol. 1995;69:5687–5696. doi: 10.1128/jvi.69.9.5687-5696.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Katzman M, Sudol M. Mapping viral DNA specificity to the central region of integrase by using functional human immunodeficiency virus type 1/visna virus chimeric proteins. J Virol. 1998;72:1744–1753. doi: 10.1128/jvi.72.3.1744-1753.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khan E, Mack J P, Katz R A, Kulkosky J, Skalka A M. Retroviral integrase domains: DNA binding and the recognition of LTR sequences. Nucleic Acids Res. 1991;19:851–860. doi: 10.1093/nar/19.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kulkosky J, Jones K S, Katz R A, Mack J P, Skalka A M. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol Cell Biol. 1992;12:2331–2338. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kulkosky J, Katz R A, Merkel G, Skalka A M. Activities and substrate specificity of the evolutionarily conserved central domain of retroviral integrase. Virology. 1995;206:448–456. doi: 10.1016/s0042-6822(95)80060-3. [DOI] [PubMed] [Google Scholar]
- 50.Lee M S, Craigie R. A previously unidentified host protein protects retroviral DNA from autointegration. Proc Natl Acad Sci USA. 1998;95:1528–1533. doi: 10.1073/pnas.95.4.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee S P, Han M K. Zinc stimulates Mg2+-dependent 3′-processing activity of human immunodeficiency virus type 1 integrase in vitro. Biochemistry. 1996;35:3837–3844. doi: 10.1021/bi952056p. [DOI] [PubMed] [Google Scholar]
- 52.Lee S P, Xiao J, Knutson J R, Lewis M S, Han M K. Zn2+ promotes the self-association of human immunodeficiency virus type-1 integrase in vitro. Biochemistry. 1997;36:173–180. doi: 10.1021/bi961849o. [DOI] [PubMed] [Google Scholar]
- 53.Lodi P J, Ernst J A, Kuszewski J, Hickman A B, Engelman A, Craigie R, Clore G M, Gronenborn A M. Solution structure of the DNA binding domain of HIV-1 integrase. Biochemistry. 1995;34:9826–9833. doi: 10.1021/bi00031a002. [DOI] [PubMed] [Google Scholar]
- 54.McCord M, Stahl S J, Mueser T C, Hyde C C, Vora A C, Grandgenett D P. Purification of recombinant Rous sarcoma virus integrase possessing physical and catalytic properties similar to virion-derived integrase. Protein Expression Purif. 1998;14:167–177. doi: 10.1006/prep.1998.0954. [DOI] [PubMed] [Google Scholar]
- 55.Mumm S R, Grandgenett D P. Defining nucleic acid-binding properties of avian retrovirus integrase by deletion analysis. J Virol. 1991;65:1160–1167. doi: 10.1128/jvi.65.3.1160-1167.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murphy J E, Goff S P. A mutation at one end of Moloney murine leukemia virus DNA blocks cleavage of both ends by the viral integrase in vivo. J Virol. 1992;66:5092–5095. doi: 10.1128/jvi.66.8.5092-5095.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pahl A, Flugel R M. Characterization of the human spuma retrovirus integrase by site-directed mutagenesis, by complementation analysis, and by swapping the zinc finger domain of HIV-1. J Biol Chem. 1995;270:2957–2966. doi: 10.1074/jbc.270.7.2957. [DOI] [PubMed] [Google Scholar]
- 58.Roth M J, Schwartzberg P L, Goff S P. Structure of the termini of DNA intermediates in the integration of retroviral DNA: dependence on IN function and terminal DNA sequence. Cell. 1989;58:47–54. doi: 10.1016/0092-8674(89)90401-7. [DOI] [PubMed] [Google Scholar]
- 59.Sherman P A, Fyfe J A. Human immunodeficiency virus integration protein expressed in Escherichia coli possesses selective DNA cleaving activity. Proc Natl Acad Sci USA. 1990;87:5119–5123. doi: 10.1073/pnas.87.13.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Singh I R, Crowley R A, Brown P O. High-resolution functional mapping of a cloned gene by genetic footprinting. Proc Natl Acad Sci USA. 1997;94:1304–1309. doi: 10.1073/pnas.94.4.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van den Ent F M, Vos A, Plasterk R H. Dissecting the role of the N-terminal domain of human immunodeficiency virus integrase by trans-complementation analysis. J Virol. 1999;73:3176–3183. doi: 10.1128/jvi.73.4.3176-3183.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Gent D C, Groeneger A A, Plasterk R H. Mutational analysis of the integrase protein of human immunodeficiency virus type 2. Proc Natl Acad Sci USA. 1992;89:9598–9602. doi: 10.1073/pnas.89.20.9598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vink C, Lutzke R A, Plasterk R H. Formation of a stable complex between the human immunodeficiency virus integrase protein and viral DNA. Nucleic Acids Res. 1994;22:4103–4110. doi: 10.1093/nar/22.20.4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vink C, Oude Groeneger A M, Plasterk R H. Identification of the catalytic and DNA-binding region of the human immunodeficiency virus type I integrase protein. Nucleic Acids Res. 1993;21:1419–1425. doi: 10.1093/nar/21.6.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vora A C, Chiu R, McCord M, Goodarzi G, Stahl S J, Mueser T C, Hyde C C, Grandgenett D P. Avian retrovirus U3 and U5 DNA inverted repeats. Role of nonsymmetrical nucleotides in promoting full-site integration by purified virion and bacterial recombinant integrases. J Biol Chem. 1997;272:23938–23945. doi: 10.1074/jbc.272.38.23938. [DOI] [PubMed] [Google Scholar]
- 66.Vora A C, Grandgenett D P. Assembly and catalytic properties of retrovirus integrase-DNA complexes capable of efficiently performing concerted integration. J Virol. 1995;69:7483–7488. doi: 10.1128/jvi.69.12.7483-7488.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vora A C, McCord M, Fitzgerald M L, Inman R B, Grandgenett D P. Efficient concerted integration of retrovirus-like DNA in vitro by avian myeloblastosis virus integrase. Nucleic Acids Res. 1994;22:4454–4461. doi: 10.1093/nar/22.21.4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Woerner A M, Klutch M, Levin J G, Marcus-Sekura C J. Localization of DNA binding activity of HIV-1 integrase to the C-terminal half of the protein. AIDS Res Hum Retroviruses. 1992;8:297–304. doi: 10.1089/aid.1992.8.297. [DOI] [PubMed] [Google Scholar]
- 69.Yang F, Leon O, Greenfield N J, Roth M J. Functional interactions of the HHCC domain of Moloney murine leukemia virus integrase revealed by nonoverlapping complementation and zinc-dependent dimerization. J Virol. 1999;73:1809–1817. doi: 10.1128/jvi.73.3.1809-1817.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang Z N, Mueser T C, Bushman F D, Hyde C C. Crystal structure of an active two-domain derivative of Rous sarcoma virus integrase. J Mol Biol. 2000;296:535–548. doi: 10.1006/jmbi.1999.3463. [DOI] [PubMed] [Google Scholar]
- 71.Zheng R, Jenkins T M, Craigie R. Zinc folds the N-terminal domain of HIV-1 integrase, promotes multimerization, and enhances catalytic activity. Proc Natl Acad Sci USA. 1996;93:13659–13664. doi: 10.1073/pnas.93.24.13659. [DOI] [PMC free article] [PubMed] [Google Scholar]