Abstract
The distal region of the uterine (Fallopian) tube is commonly associated with high-grade serous carcinoma (HGSC), the predominant and most aggressive form of ovarian or extra-uterine cancer. Specific cell states and lineage dynamics of the adult tubal epithelium (TE) remain insufficiently understood, hindering efforts to determine the cell of origin for HGSC. Here, we report a comprehensive census of cell types and states of the mouse uterine tube. We show that distal TE cells expressing the stem/progenitor cell marker Slc1a3 can differentiate into both secretory (Ovgp1+) and ciliated (Fam183b+) cells. Inactivation of Trp53 and Rb1, whose pathways are commonly altered in HGSC, leads to elimination of targeted Slc1a3+ cells by apoptosis, thereby preventing their malignant transformation. In contrast, pre-ciliated cells (Krt5+, Prom1+, Trp73+) remain cancer-prone and give rise to serous tubal intraepithelial carcinomas and overt HGSC. These findings identify transitional pre-ciliated cells as a cancer-prone cell state and point to pre-ciliation mechanisms as diagnostic and therapeutic targets.
Subject terms: Ovarian cancer, Adult stem cells, Cancer models
The cell of origin for high-grade serous carcinoma (HGSC) remains to be determined. Here, the authors characterize the different cell types and states of the mouse uterine tube and identify transitional pre-ciliated cells as a cancer-prone cell state, which could be targeted therapeutically.
Introduction
Ovarian cancer is the sixth leading cause of death for women in the United States1. High-grade serous carcinoma (HGSC) is the most common and aggressive type of ovarian cancer2,3. Over 80% of HGSC are detected at advanced stage and have limited treatment options2,4,5. This is attributed to latent progression of the disease with lack of early symptoms and detection methods2. Detection and treatment of HGSC at earlier stages could be crucial to improving the prognosis of patients with this malignancy. However, identification of new diagnostic markers and therapeutic targets is hindered by our inadequate knowledge about the cells in which HGSC originates and the mechanisms underlying disease initiation.
The location of HGSC initiation has long been debated, but the emerging consensus is that both the ovarian surface epithelium (OSE) and the tubal epithelium (TE) of the uterine tube, also known as the oviduct or Fallopian tube, have potential to progress into HGSC6–11. While cancer-prone stem/progenitor cells have been described for the OSE12,13, the cell of origin of HGSC arising from TE remains unclear. It has been shown that the majority of familial HGSC cases may begin with the appearance of early dysplastic lesions, TP53 signatures, and serous tubal intraepithelial carcinomas (STICs)6,14. These lesions are found exclusively in the distal region of the uterine tube2,14,15. Both TP53 signatures and STICs lack ciliation, express the transcriptional factor PAX8, and harbor mutations in the TP53 gene (also known as Trp53 in the mouse), which encodes for p53. TP53 mutations are the most frequent genetic alterations in HGSC, being present in over 96% of cases16,17. Consistent with these observations, early dysplastic lesions can be induced by inactivation of tumor suppressor genes commonly associated with human HGSC, such as Trp53, Brca1, Brca2, Pten, and Rb1 in Pax8-expressing tubal epithelial cells of the mouse uterine tube18,19.
In both humans and rodents, uterine tubes consist of distal (infundibulum, ampulla, and ampullary-isthmic junction) and proximal regions (isthmus and intramuscular utero-tubal junction). The uterine tube is formed by the simple pseudostratified TE surrounded by a thin stromal layer, two circular smooth muscle layers, and the mesothelium. Two main cellular components of the TE are ciliated cells (also known as multi-ciliated cells), predominantly located in the distal regions of the tube, and secretory cells, which are more abundant in the proximal regions. Additionally, there are basal cells, representing intraepithelial T-lymphocytes and peg cells. Peg cells have been described as either exhausted secretory cells or CD44+ progenitor cells20–22.
Previous mouse lineage-tracing studies have reported that cells expressing Pax8 have the capacity to self-renew and differentiate into ciliated cells in both distal and proximal regions after labeling during embryonic or prepubertal development23. However, recent studies based on immunophenotyping, lineage tracing, and limited single cell RNA-sequencing (scRNA-seq) suggest presence of distinct cell lineages in the distal (Pax8+) and the proximal (Pax2+) regions of the adult mouse uterine tube24.
It is well established that many types of cancer arise from stem cell niches25–27. Previously, using a genetically defined mouse model, we have shown that OSE stem/progenitor cells can be efficiently transformed after inactivation of tumor suppressor genes Trp53 and Rb1 and lead to HGSC formation12,13,28. Notably, tumors arising from non-stem OSE cells were slow-growing and non-metastatic. However, in other cancer types, neoplasms may originate from differentiated or transitional state cells that have acquired some stem cell properties28–30. It has been hypothesized that some ovarian carcinomas may arise from the ciliated cell lineage31. However, no direct experimental data has been offered to support this idea.
Here, we conduct scRNA-seq to establish a comprehensive census of cell types found in the mouse uterine tube. Proximal and distal sections are sequenced separately to investigate characteristics underscoring the distal region’s predisposition towards cancer initiation. By using a combination of computational lineage trajectory projections and genetic cell fate, we identify a TE stem/progenitor cell population and interrogate unique epithelial cell states for their propensity for malignant transformation. These studies reveal that pre-ciliated cells may serve as a specific cell state susceptible to malignant transformation.
Results
Census of cell types of the mouse uterine tube
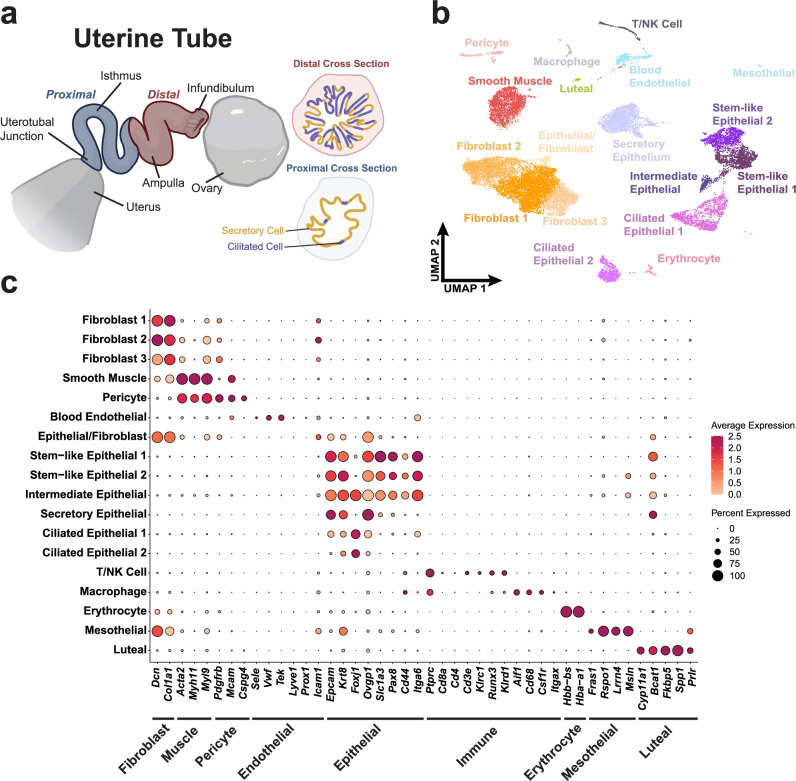
The mouse uterine tube is divided along the distal and proximal axis (Fig. 1a). We collected 62 uterine tubes from 31 adult mice, separated distal and proximal regions, and processed them for scRNA-seq (Supplementary Figs. 1, 2) by region. After sequencing and data preprocessing of distal region samples, 16,583 high-quality cells remained. Following Harmony integration32, we identified 18 clusters by Louvain clustering contributing to epithelial, stromal, and immune compartments, which we visualized using the uniform manifold approximation and projection (UMAP) (Fig. 1b). Features defining each cluster were found using general markers known to define each cell type (Fig. 1c). Among the fibroblast populations were three clusters noted as Fibroblast 1 (Ramp3+, Lsamp+, Htra3+), Fibroblast 2 (Plac9a+, Mgp+, Gpx3+), and Fibroblast 3 (Nbl1+, Cacna1d+, Sfrp2+). Epithelial cells were characterized by epithelial markers (Epcam and Krt8), and further epithelial specificity was achieved with secretory (Ovgp1) and ciliated (Foxj1) markers. Stem-like epithelial clusters were identified separately as Stem-like Epithelial 1 (Slc1a3+, Itga6+, Lamc2+) and Stem-like Epithelial 2 (Cebpd+, Hbegf+, Kctd14+). Additional cell types were noted within our dataset to be mesothelial and luteal cells.
Fig. 1. Census of cell types of the mouse uterine tube.
a Diagram of a partially uncoiled mouse uterine tube. The proximal region contains few ciliated cells and extends from the intratubal junction to the ampulla. The distal region consists of the ampulla and the infundibulum where ciliated cells are abundant. b UMAP visualization of the cell types identified in a pool of 16,583 distal cells from 62 uterine tubes from which high quality sequence data was obtained. c Dot plot representation of genes associated with various tissue types to validate cell type identification. Source data are provided as a Source Data file. a was in part created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs license.
Characterization of distal epithelial cell states
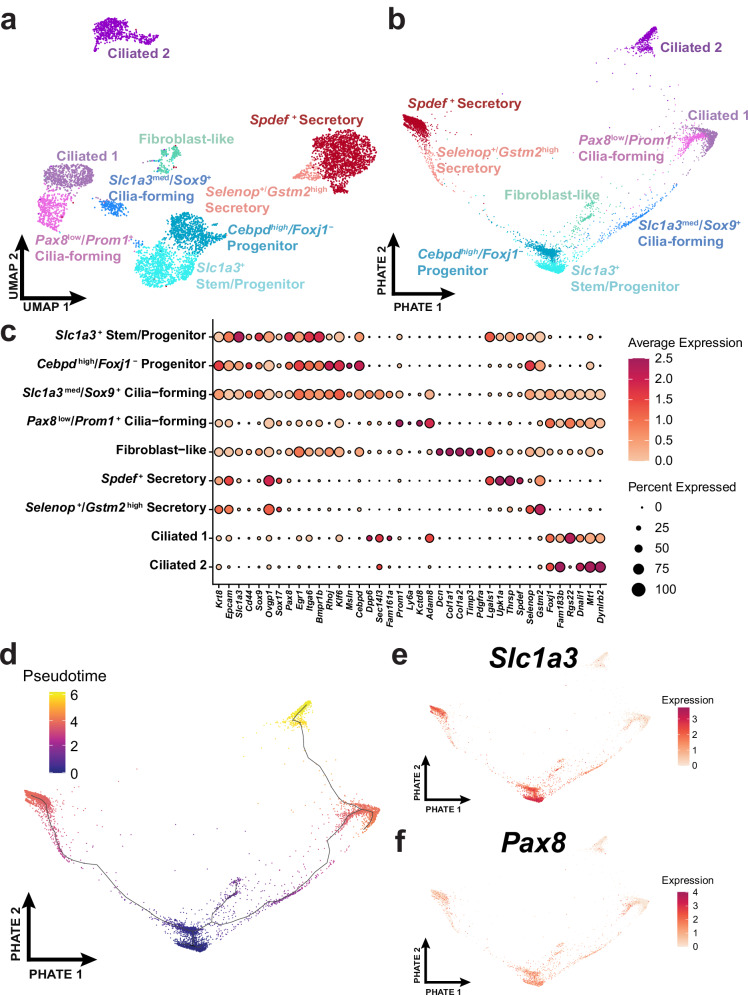
To further investigate the unique cell states of the TE, we subset the epithelial clusters of the distal uterine tube cell types. We specifically subset from the secretory, ciliated, and stem-like epithelial cell clusters (Supplementary Fig. 1). After processing the distal epithelial subset, we identified 9 clusters consisting of ciliated, secretory, stem-like, and transitional cell states (Fig. 2a). We further employed Potential of heat diffusion for affinity-based transition embedding (PHATE)33 to better visualize how tubal epithelial cells progress in a two-dimensional representation similar to UMAP (Fig. 2b). General epithelial markers (Epcam, Krt8), secretory markers (Ovgp1, Sox17), and ciliated markers (Fam183b, Foxj1) were used to classify groups of epithelial cells that are known to reside in the TE. Additional features were found using the Wilcoxon Rank Sum Test to determine cluster-specific markers (Fig. 2c). One cluster within the epithelial subset shared features of fibroblasts, which indicated that this population was most likely a doublet (Supplementary Fig. 3). In agreement with previous studies21,34, ciliated cells and their precursors were mainly located in the distal region (32% distal vs 10% proximal), while secretory cells predominantly populated the proximal region (30% distal vs 65% proximal; Supplementary Table 1). Characterization of the 1,785 high quality proximal cells and the 528 cells within the proximal epithelial subset led to the identification of 6 clusters mainly consisting of secretory cells (Supplementary Fig. 4). Comparison of the clusters identified in distal and proximal regions only showed strong correlation between a subset of secretory cell states (Supplementary Fig. 5).
Fig. 2. Characterization of distal epithelial cell states.
a Distal epithelial cells (n = 6273) from 62 uterine tubes were identified by their Epcam and Krt8 expression and the subset was represented within the UMAP embedding. b A differentiation trajectory among epithelial cells visualized through the PHATE dimensional reduction technique. c Dot plot reflecting highly expressed, specific markers of each identified epithelial cell cluster. d Monocle3 pseudotime analyses calculated over the PHATE embedding. The expression of Slc1a3 (e) and Pax8 (f) visualized over the PHATE embedding. Source data are provided as a Source Data file.
General expression of stem-like markers indicated that Slc1a3 might serve as a potential marker for epithelial stem/progenitor cells that give rise to both secretory and ciliated cells (Fig. 2c and Supplementary Fig. 6). Slc1a3 was previously described as a marker of a cell population containing distinct skin epithelial stem cells35,36. A pseudotime trajectory was calculated with Monocle337 and overlayed on the PHATE embedding (Fig. 2d). In the resulting PHATE representation, Slc1a3 expression is located at the center with secretory and ciliated branches split by the suspected stem/progenitor cell state (Fig. 2e). In this representation, Pax8 expression is present in Slc1a3+ cells and extends towards early cilia-forming cells (Fig. 2f).
In the distal region Pax8 expression was mainly detected in stem/progenitor cell cluster (81% of cells) and transitional pre-ciliogenic state cells (49%). Only 3% of ciliated cells and 28% of secretory cells were Pax8+ (Fig. 2c, and Supplementary Table 1). In the proximal region 67% of putative stem/progenitor cells expressed Pax8, while both secretory and ciliated clusters contained about 29% of Pax8+ cells each (Supplementary Fig. 4 and Supplementary Table 1). Although the capture of Pax8 transcripts may be limited due to dropouts from scRNA-seq, presence of PAX8 expression in SLC1A3+ cells, and its lack in some secretory cells was confirmed by immunostaining (Supplementary Figs. 7 and 8).
Slc1a3+ epithelial cells are stem/progenitor cells for the TE
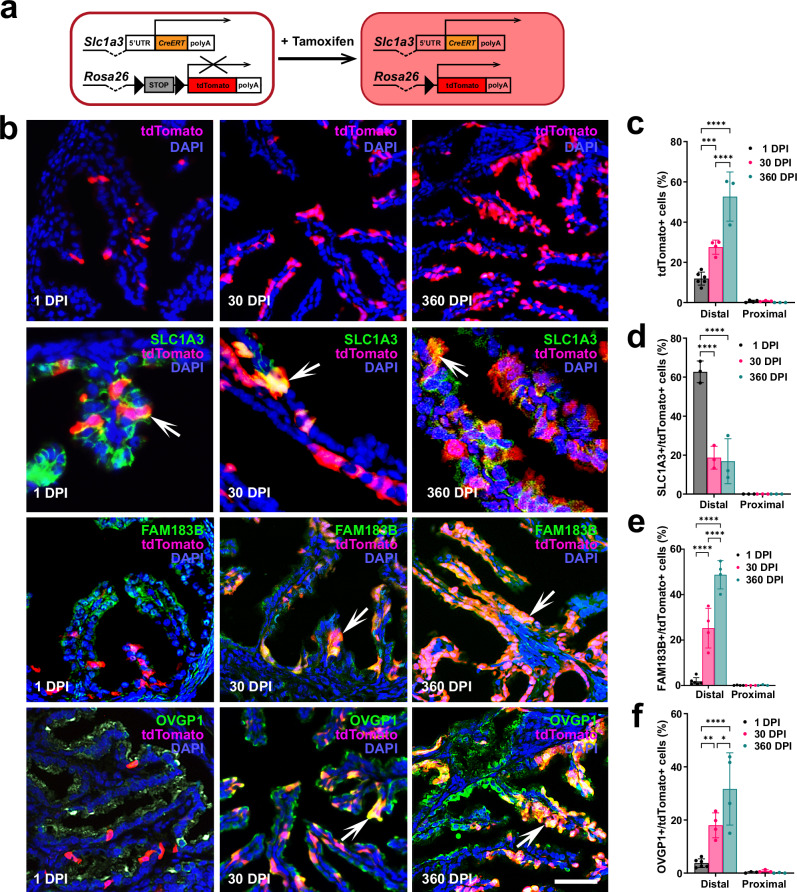
To test if Slc1a3+ epithelial cells are stem/progenitor cells of the TE, cell lineage-tracing and organoid studies were conducted in Slc1a3-CreERT Ai9 mice. In these mice, administration of tamoxifen allows for Cre-loxP mediated induction of tdTomato expression in Slc1a3+ cells (Fig. 3a). One day after tamoxifen induction tdTomato expression was mainly detected in the distal TE (Fig. 3b, c). Within 30 days after tamoxifen injection, tdTomato+ cells expanded to form clusters and persisted for over 1 year (Fig. 3b–f). A fraction of tdTomato cells continued to express SLC1A3 indicative of their long-living potential. Single low dose treatments of tamoxifen yielded corroborating results, in which single cells formed clusters of tdTomato+ cells 30 days after injection (Supplementary Fig. 9). In agreement with recent studies showing no significant impact of estrous cycle on gene expression in the TE38, tamoxifen administration showed no significant impacts on the presence of SLC1A3+ cells 1 and 30 days after injection (Supplementary Table 2).
Fig. 3. Lineage tracing of Slc1a3+ stem/progenitor cells.
a Experimental design. Mice containing Slc1a3-CreERT and Ai9 reporter are injected with tamoxifen to induce expression of modified red fluorescent protein (tdTomato) in cells expressing Slc1a3. b Top row, tdTomato expression (red) 1, 30, and 360 days post induction (DPI) with tamoxifen. Lower rows, tdTomato+ progeny of Slc1a3+ cells express differentiation markers for stem (SLC1A3, orange, arrows), ciliated (FAM183B, orange, arrows), and secretory (OVGP1, orange, arrows) cells. Counterstaining with DAPI (blue). Scale bar represents 50 μm (tdTomato, FAM1A3/tdTomato, OVGP1/tdTomato rows), and 25 μm (SLC1A3/tdTomato row). c Quantification of cells expressing tdTomato in the distal and proximal regions of the uterine tube 1, 30, and 360 DPI. Quantification of cells expressing tdTomato with stem (SLC1A3, d), ciliated (FAM183B, e), or secretory (OVGP1, f) cells 1, 30, and 360 DPI. c–f Two way ANOVA with the Tukey’s multiple comparison post hoc test for all distal regions. c ***P = 0.0003, ****P < 0.0001 (d) ****P < 0.0001 (e) ****P < 0.0001 (f) *P = 0.0146, **P = 0.0058, ****P < 0.0001. Data are presented as mean values ± SD. Biological replicates (mice, n): c Distal 1 DPI n = 6, 30 DPI n = 4, 360 DPI n = 3; Proximal 1 DPI n = 4, 30, and 360 DPI n = 3 each group. d Distal and Proximal n = 3 each group. e Distal 1 DPI n = 6, 30 DPI and 360 DPI n = 4 each group; Proximal 1 DPI n = 4, 30, and 360 DPI n = 3 each group. f Distal 1 DPI n = 6, 30, and 360 DPI n = 4 each group; Proximal n = 3, each group. Source data are provided as a Source Data file.
SLC1A3+ stem/progenitor cells form ciliated organoids with high efficiency
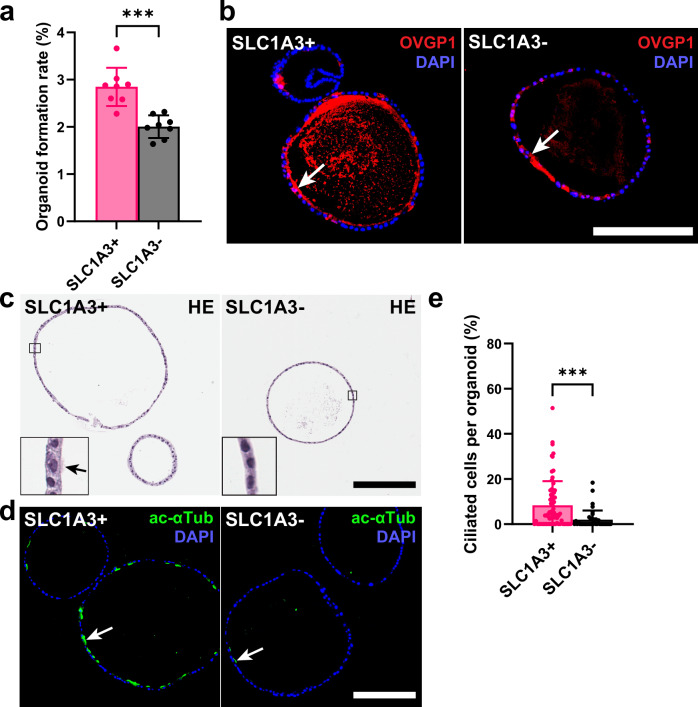
Consistent with previous studies of mouse and human TE20,39,40, organoids were formed by harvesting the mouse distal TE. The organoid-forming ability of tdTomato+ cells from Slc1a3-CreERT Ai9 mice was confirmed with isolation from the distal region after separation by fluorescence-activated cell sorting (FACS) (Supplementary Fig. 10a–d). To further distinguish the SLC1A3+ stem/progenitor cells from others within the TE, we completed magnetic-activated cell sorting (MACS) to split pure epithelial cell pools into SLC1A3+ and SLC1A3- groups (Supplementary Fig. 10e and Supplementary Table 3). Consistent with previous FACS experiments, organoids derived from SLC1A3+ epithelial cells formed organoids at a significantly higher rate as compared to organoids formed by SLC1A3- cells (Fig. 4a). After 14 days of culture, organoids grew to similar sizes (Supplementary Fig. 10f, g) and contained OVGP1+ cells in both groups (Fig. 4b). However, organoids formed from SLC1A3+ cells were marked by a significantly larger population of ciliated cells (Fig. 4c–e), thereby supporting bidirectional differentiation potential of SLC1A3+ cells.
Fig. 4. Organoid formation by SLC1A3+ and SLC1A3- cells.
a Organoid formation rate of distal tubal epithelial cells isolated for SLC1A3 expression by MACS (n = 8). b Organoid sections of day 14 SLC1A3+ and SLC1A3- cell derived organoids stained for secretory marker OVGP1 (red, arrows). Counterstaining with DAPI (blue). c Hematoxylin and Eosin (HE) staining of SLC1A3+ and SLC1A3- cell derived organoids after 14 days of culture. Arrow denotes cilia. d Representative images of ciliation (green, acetylated α-Tubulin) between SLC1A3+ and SLC1A3- cell derived organoids. Counterstaining with DAPI (blue). b–d Scale bar all images 200 μm. e Quantification of ciliated cells between SLC1A3+ (n = 71) and SLC1A3- (n = 40) cell derived organoids. a, e ***P = 0.0002, ****P = 0.0005, two-tailed unpaired t-tests. Data are presented as mean values ± SD. Source data are provided as a Source Data file.
In summary, based on general expression of stem-like markers, computational lineage trajectory projections, cell lineage tracing, and organoid assays, Slc1a3+ epithelial cells represent stem/progenitor cells for the distal TE and contribute to the long-term maintenance of both ciliated and secretory epithelial cells within the uterine tube.
Slc1a3+ epithelial cells are not cancer-prone
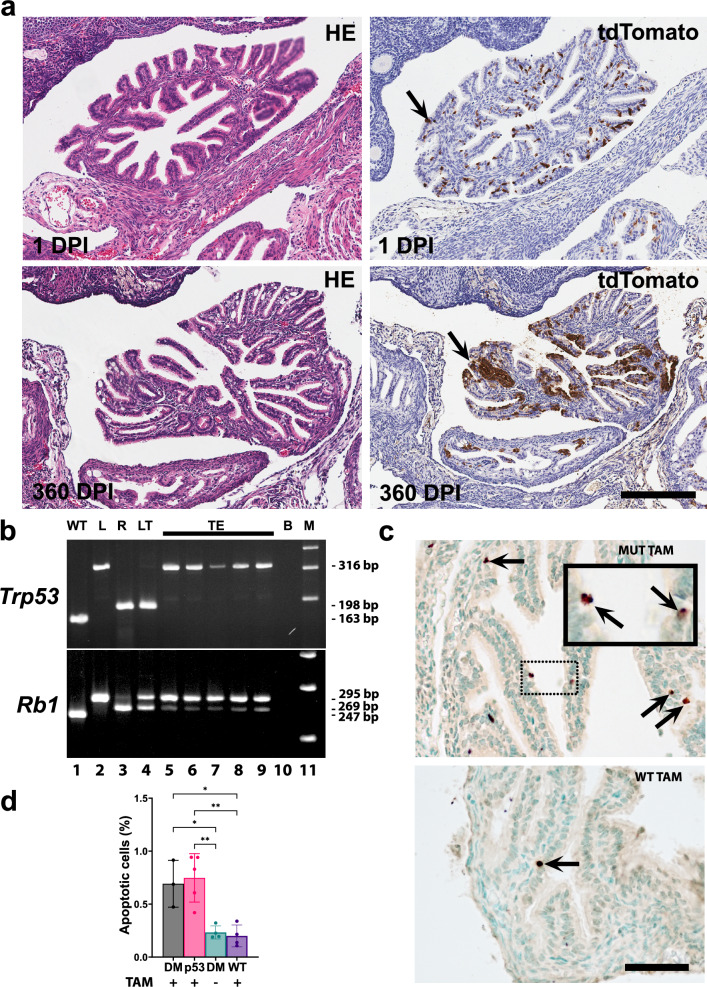
To investigate the role of Slc1a3+ cells in malignant transformation, we have prepared Slc1a3-CreERT Trp53loxP/loxP Rb1loxP/loxP Ai9 mice. However, no TE neoplastic lesions were observed in these mice (n = 19) by 1 year after tamoxifen administration (Fig. 5a). Large clusters of TE cells expressed tdTomato at the time of collection. Cre-mediated excision of Rb1 but not Trp53 was detected by microdissection-PCR assay in all tested samples (Fig. 5b). Evaluation of the TE showed increased apoptotic rate 1 day after tamoxifen administration in mice with either Trp53/Rb1 or Trp53 mutations. (Fig. 5c, d). Thus, Trp53 inactivation leads to elimination of targeted Slc1a3+ cells by apoptosis, thereby explaining their resistance to malignant transformation.
Fig. 5. Slc1a3+ epithelial cells are not cancer-prone.
a tdTomato+ tubal epithelial cells in mice containing Slc1a3-CreERT with floxed Trp53 and Rb1 genes, and an Ai9 reporter 1 and 360 days post induction (DPI) with tamoxifen. Hematoxylin and Eosin (HE) staining (left column) and immunostaining for tdTomato (arrows, brown color), Elite ABC method, hematoxylin counterstaining (right column). Scale bar, all images 200 μm. b PCR analysis of Trp53 and Rb1 gene structure in the same samples of microdissected cells from the tubal epithelium (TE, lanes 5–9) and lung neoplasm (LT, lane 4) of Slc1a3-CreERT Trp53loxP/loxP Rb1loxP/loxP Ai9 mice collected 1 year after tamoxifen induction. Samples with known gene structure (wild-type, WT, lane 1, floxed gene, L, lane 2, and recombinant gene, R, lane 3). 316-, 198-, and 163-bp fragments are diagnostic for floxed, excised, and wild-type alleles of the Trp53 gene, respectively. 295-, 269, and 247-bp fragments are diagnostic for floxed, excised, and wild-type alleles of the Rb1 gene, respectively. B, blank control (lane 10), M, DNA marker (Lane 11). Representative of 5 microdissection-PCR experiments. c Apoptotic cells (arrows) in the tubal epithelium 1 day after tamoxifen induction of Cre-mediated inactivation of Trp53 and Rb1 in Slc1a3-CreERT Trp53loxP/loxP Rb1loxP/loxP Ai9 mice (MUT TAM) and littermates without Slc1a3-CreERT (WT TAM). Dot-line rectangle indicates respective location of cells shown in the inset in the top image. TUNEL assay, methyl green counterstaining. Scale bar, 50 μm and 21 µm, inset. d Quantification of apoptotic cells 1 day after administration of tamoxifen (TAM+) or vehicle (TAM-) in Slc1a3-CreERT Trp53loxP/loxP Rb1loxP/loxP Ai9 mice (DM), Slc1a3-CreERT Trp53loxP/loxP Ai9 mice (p53) or littermates without Slc1a3-CreERT (WT). d One way ANOVA with the Tukey’s multiple comparison post hoc test (DM TAM+ vs. DM TAM-) *P = 0.0192, (DM TAM+ vs. WT TAM+) *P = 0.0123, (p53 TAM+ vs. DM TAM-) **P = 0.0035, (p53 TAM+ vs. WT TAM+) **P = 0.0022. Data are presented as mean values ± SD. Biological replicates (mice, n): DM TAM+ n = 3, p53M TAM+ n = 5, DM TAM- n = 4, WT TAM+ n = 4. Source data are provided as a Source Data file.
Pre-ciliated cells are susceptible to malignant transformation
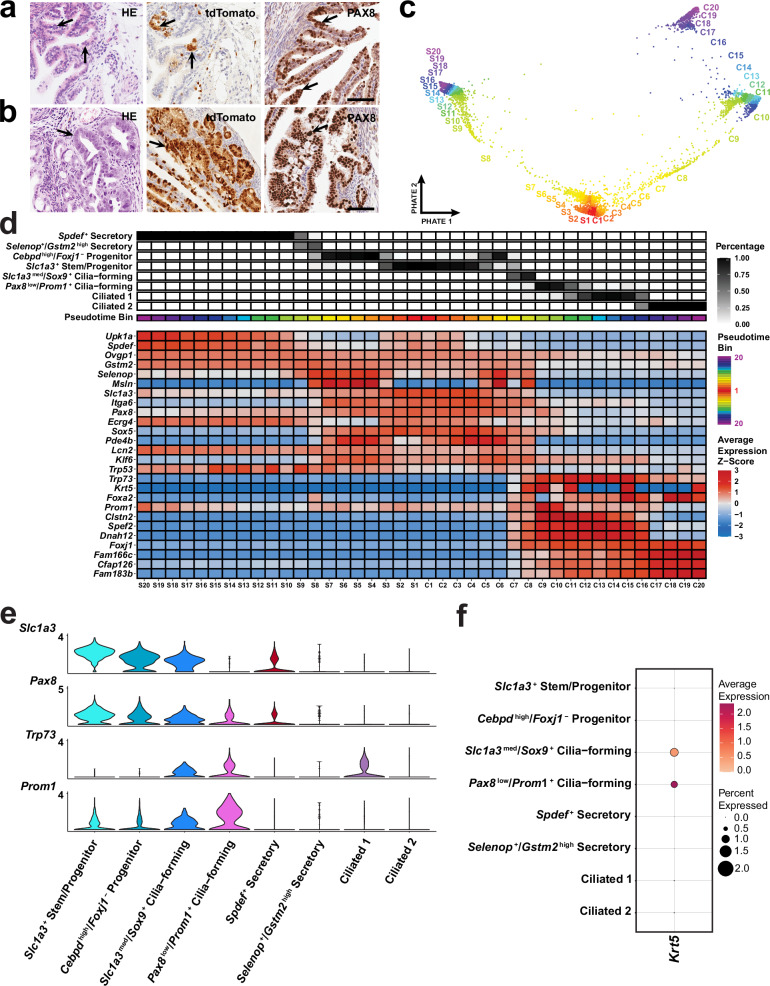
Consistent with previous reports18,19, we have observed that the TE can be transformed by conditional inactivation of Trp53 and Rb1 in cells expressing Pax8 (Fig. 6a, b, Supplementary Table 4). Early dysplastic lesions (6 out of 12 cases), and/or HGSC (3 out of 12 cases) were detected in 58% of mice by 400 days post induction (DPI). These data suggest that a subset of Pax8+ cells is susceptible to malignant transformations, distinct from Slc1a3+ stem/progenitor cells that do not give rise to malignancies.
Fig. 6. Identification of cancer-prone cell states.
Early (a) and advanced (b) neoplastic lesions (arrows) in Pax8-rtTA Tre-Cre Trp53loxP/loxP Rb1loxP/loxP Ai9 mice. Hematoxylin and Eosin (HE) staining (left column) and immunostaining for tdTomato (middle column, brown color), and PAX8 (right column, brown color). Elite ABC method, hematoxylin counterstaining. a, b Scale bar, all images 60 µm. Biological replicates n = 6 (a) and n = 3 (b). c Pseudotime binning along the PHATE embedding to visualize how bins are assigned for 6,273 distal epithelial cells from 62 uterine tubes. d Inferred pseudotime trajectories of secretory and ciliated epithelial cell lineages. The lineages extend from S1 and C1 to S20 and C20 respectively, where S20 and C20 are presumed to be a more differentiated cell state. The percent abundance of each cell type contributing to each pseudotime bin is reflected in black. The average z-scored expression was calculated for each gene to identify genes that best represent smaller transitional states within each lineage. Each pseudotime bin is equally sized and consists of about 150 cells. e Log-normalized expression of Slc1a3, Pax8, Trp53 and Prom1 visualized in a violin plot of epithelial cell clusters. f Dot plot of Krt5 expression among epithelial cell populations. Source data are provided as a Source Data file.
To define a unique population of Pax8-expressing cells implicated in malignant transformation, we leveraged our scRNA-seq analysis to identify a group of cells expressing Pax8 but not Slc1a3 in the distal region of the uterine tube. Strikingly, the only cell state exhibiting Pax8 expression without concomitant Slc1a3 expression displayed features consistent with pre-ciliated cells (Fig. 6c–e).
We subset the branched trajectory of secretory and ciliated cells from Slc1a3+ progenitors and formed 20 pseudotime bins equally divided along the trajectory to total about 150 cells per bin (Fig. 6c, d). We have found that Pax8+, Slc1a3- cells are present as a transitional state along the ciliogenic lineage. Such cells are characterized by expression of Krt5, Prom1, and Trp73 (Fig. 6d–f). Notably, expression of these genes negatively correlates with Trp53 expression, thereby suggesting reduced requirement for Trp53 during ciliogenesis (Fig. 6d and Supplementary Fig. 11). At the same time, the majority of other putative driver genes identified in human HGSC13,41, including Rb1, Nf1, and Pten, are preferentially expressed in stem/progenitor populations along the pre-ciliogenesis trajectory. Expression of Brca1, Brca2 and Csmd3 is mainly present in cilia-forming cells (Supplementary Fig. 11).
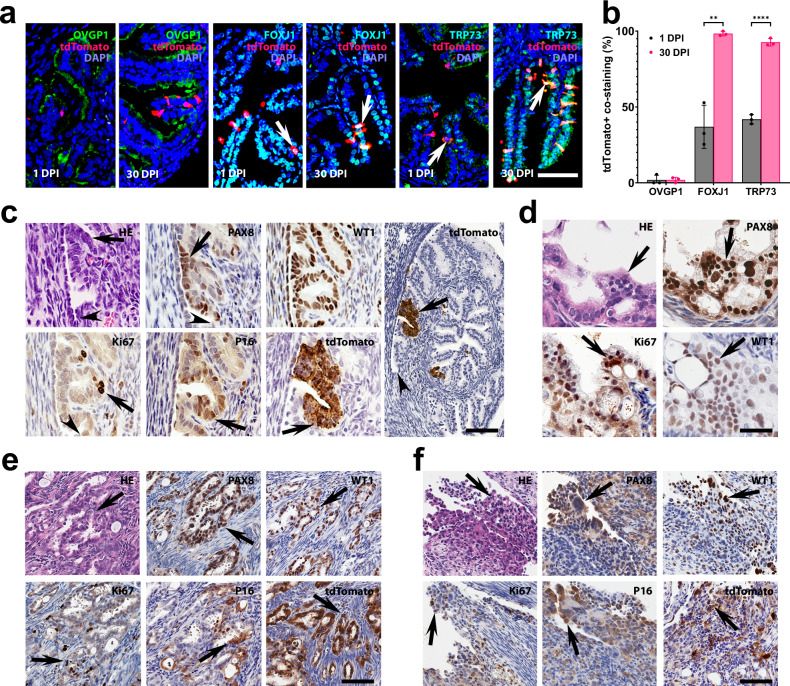
To characterize Krt5+ pre-ciliated cells, we prepared Krt5-CreERT Ai9 mice. In agreement with our single cell transcriptome findings, Krt5+ cells represented a small fraction (0.83%) of pre-ciliated cells (FOXJ1±) of the distal TE (Fig. 7a, b, Supplementary Table 4). Consistent with transitional state of Krt5+ cells, their progeny mainly presented with ciliated marker expression (FOXJ1 and TRP73) 30 days after tamoxifen treatment (Fig. 7a, b). Krt5+ cells of the TE increased in their number as typical for transit-amplifying cells (Supplementary Table 5). Few Krt5+ cells were also detected in the OSE. However, these cells decreased in number by 30 DPI (Supplementary Table 5).
Fig. 7. Transitional pre-ciliated Krt5+ cells are highly susceptible to malignant transformation.
a Detection of ciliation FOXJ1 (green) and TRP73 (green) markers but not secretory marker OVGP1 (green) in Krt5+ (tdTomato) cells and their progeny (arrows) in the distal tubal epithelium 1 and 30 days post-induction (DPI) with tamoxifen in Krt5-CreERT Ai9 mice. b Quantification of Krt5+ cells co-expressing tdTomato with OVGP1, FOXJ1 or TRP73 1 and 30 DPI. **P = 0.0017, ****P < 0.0001, two-tailed unpaired t-tests. Data are presented as mean values ± SD. Biological replicates n = 3 in each group. Source data are provided as a Source Data file. c–f Neoplastic lesions in Krt5-CreERT Trp53loxP/loxP Rb1loxP/loxP Ai9 mice. Early dysplastic lesions (arrows) with mild cellular atypia (c) and more pronounced atypical features, loss of cell polarity, and high proliferative index typical for STICs (d). Arrowheads in (c), TE-mesothelial junctions. Advanced neoplastic lesions (arrows) with stromal invasion (e) and peritoneal spreading (f, arrows). Hematoxylin and Eosin (HE) staining and immunostainings for PAX8, Wilms tumor 1 (WT1), Ki67, p16, and tdTomato (brown color) by Elite ABC method with hematoxylin counterstaining. Scale bar 50 µm (a, c, except for right image, and d) and 100 µm (c), right image, (e, f). Biological replicates n = 5 (c), n = 8 (d), n = 4 (e) and n = 3 (f).
To test if pre-ciliated cells are susceptible to malignant transformation, we prepared Krt5-CreERT Trp53loxP/loxP Rb1loxP/loxP Ai9 mice. By 200 DPI 16 out of 21 mice (76%) developed neoplastic lesions (Fig. 7c–f, Supplementary Fig. 12 and Supplementary Table 4). Early dysplastic lesions were found in 12 out of 21 mice (57%). Such lesions showed a range of features from mild cellular atypia (Fig. 7c and Supplementary Fig. 12a, n = 5) to more pronounced cellular atypia, loss of cell polarity, and high proliferative index typical for STICs (Fig. 7d, n = 8). All dysplastic lesions presented with PAX8 expression and lack of ciliation. They were also marked by elevated expression of HGSC markers, P16, and Wilms tumor 1 (WT1, Fig. 7c). Notably, 9 out of 12 early lesions (75%) were located near TE-mesothelial junctions (Fig. 7c and Supplementary Fig. 12a), similarly to the previously described location of human STICs in close vicinity to the tubal-peritoneal junction14,15.
Four mice (19%) presented with carcinomas similar to human HGSC, according to their slit-like and solid patterns of growth, expression of PAX8, WT1, P16 and high levels of proliferation and stromal invasion (Fig. 7e). Three of such tumors spread peritoneally (Fig. 7f) and invaded ovarian fat pad (Supplementary Fig. 12b). No early dysplastic lesions or overt carcinomas were detected in the OSE.
The earliest atypical lesions were observed at 104 DPI as compared to 154 DPI in experiments with inactivation of Trp53 and Rb1 in Pax8+ cells. According to tdTomato cell labeling at 1 DPI, the pool of targeted Krt5+ cells (0.83%) was significantly smaller than that of Pax8+ cells (19%, P = 0.0007; Supplementary Table 4). These observations support the notion that cells in a transitional state along ciliogenesis are cancer-prone.
Discussion
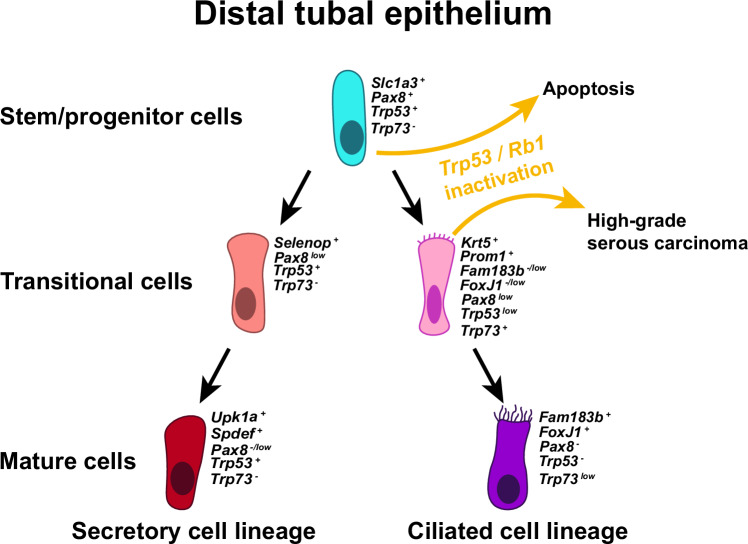
Our single-cell transcriptomics allowed for unique cell states and cell lineage hierarchy to be identified in the context of healthy adult mouse uterine tubes (Fig. 8). Further experimentation has confirmed that Slc1a3+ cells are long living and give rise to both secretory (OVGP1+) and ciliated (FAM183B+) cells in the distal region of the uterine tube. Notably, expression of Pax8 was mainly detected in stem/progenitor cells and transitional cell states. Our findings confirm previous observations that Pax8+ cells can differentiate into ciliated cells23. However, they also indicate that mature secretory cells do not give rise to ciliated cells.
Fig. 8. Cell lineage hierarchy and cancer-prone cell state of the distal tubal epithelium.
The hierarchy begins with Slc1a3+ stem/progenitor cells giving rise to secretory and ciliated cell lineages. Slc1a3+ stem/progenitor cells undergo apoptosis after inactivation of Trp53 alone or together with Rb1, while inactivation of Trp53 and Rb1 in Krt5+ pre-ciliated transitional cells lead to high-grade serous carcinoma. Colors of epithelial cells resemble the colors used to label cells within UMAP embeddings (Fig. 2a).
We could not observe any significant contribution of Slc1a3+ cells to cells of the proximal uterine tube. This can be explained by the existence of developmentally distinct cell lineages forming the proximal region24.
Our current study shows that unlike stem/progenitor cells of the OSE, Slc1a3+ TE stem/progenitor cells do not undergo malignant transformation after Cre-mediated inactivation of Trp53 and Rb1. This was not due to the lack of Cre activity in Slc1a3+ cells. Long-term contribution of tdTomato+ cells to the TE was observed in all uterine tubes by 360 days after induction of Cre expression. Furthermore, all samples contained an excised Rb1 allele. However, we observed only intact floxed Trp53 in tdTomato+ TE cells and increased apoptotic index 1 day after induction of Cre expression. Deletion of Trp53 alone was sufficient to induce apoptosis, thereby supporting the notion that inactivation of Trp53 is incompatible with long-term survival of Slc1a3+ cells.
By comparative evaluation of Pax8+ cells lacking concomitant Slc1a3 expression, we identified a cancer-prone cell population along the ciliogenic lineage. This observation is consistent with prevalence of early dysplastic lesions, TP53 signatures and STICs in the distal, ciliated cell-rich, region of the uterine tube2,14,15. Furthermore, the majority of HGSC putative driver genes13,41, either preferentially expressed in stem/progenitor cells with a bias towards pre-ciliogenic trajectory (Rb1, Nf1 and Pten), or mainly detected in cilia-forming cells (Brca1, Brca2 and Csmd3). Thus, aberrations in these genes may have the most transformation potential in the context of pre-ciliogenic cell state, as opposed to secretory differentiation.
According to our analysis of branched trajectory of secretory and ciliated cells from Slc1a3+ progenitors and cell fate experimental tracing, Krt5+ cells are in a transitional pre-ciliated cell state. Such cells also express Prom1 and Trp73. Consistent with our findings, a previous lineage tracing study suggested that Prom1+ cells represent a state transitional to ciliation42. Our previous studies have shown that ciliated Foxj1+ cells are not transformed by inactivation of Trp53 and Rb119. Hence cancer-susceptibility of TE cells deficient for these genes is limited to a transitional pre-ciliated cell state marked by expression of Krt5, Prom1 and Trp73.
KRT5, PROM1 and TP53 have been linked to human ovarian cancer. Expression of both KRT5 and PROM1 are associated with HGSC progression43,44. TP73, encoding human p73, is upregulated in many cases of epithelial ovarian cancers and modulates the sensitivity of ovarian cancers to chemotherapy (reviewed in ref. 31).
Notably, Trp73 is a critical regulator of ciliogenesis and its inactivation results in ciliopathies45–47. Among downstream targets of Trp73, are micro-RNAs of the miR-34 family48. These micro-RNAs are also an essential component for ciliogenesis49. At the same time, inactivation of mir-34 family of genes is commonly observed in HGSC50, suggesting a link between regulation of ciliogenesis and ovarian cancer.
p73 encoded by TP73/Trp73 is a homolog of the tumor suppressor transcriptional factor p53. Some functions of p53 and p73 are redundant, as evident by the ability of p73 to activate p53-regulated genes in growth suppression and apoptosis induction. However, other functions do remain unique to either p53 or p73, as evidenced by the lack of ciliation abnormalities in mice null for Trp53 and rare mutations of TP73 in cancers51. Consistent with distinct functions of p53 and p73 in TE cells, we have observed that Trp73 expression negatively correlates with Trp53 expression.
Our studies show that Slc1a3+ stem/progenitor cells express Trp53 and its inactivation is incompatible with their survival. Unlike Slc1a3+ cells, transitional pre-ciliated cells express both Trp53 and Trp73. This suggests that Trp73 expression protects cells from apoptosis but not from genomic instability triggered by Trp53, thereby leading to the preferential transformation of pre-ciliated cells. Further studies are required to understand how transition from p53-regulated programs in stem/progenitor cells to p73-controlled ciliogenesis programs may predispose to cancer.
We have observed preferential location of early dysplastic lesions near the junction between TE and mesothelium, in close similarity to their human counterparts14,15. Junction areas (aka epithelial transitional zones) are anatomically defined regions of organs where two different types of epithelial tissue meet. Such junction areas are known to be predisposed to cancer in many locations such as the ovarian hilum region, the gastric squamo-columnar junction, the corneal limbus region, the anal canal, and the uterine cervix. They also contain stem cell niches (reviewed in ref. 26). The existence of a cancer-prone cells in junction sites has been demonstrated definitively in the ovarian hilum region and the gastric squamo-columnar junction12,13,28. Our current observation reinforces the notion that high cancer frequency at the junction areas can be explained by the presence of cancer-prone stem cell niches therein. Further studies should specifically evaluate the mechanisms facilitating preferential transformation of pre-ciliogenic cells at the tubal-mesothelial junction.
There are potential limitations within this study. The use of scRNA-seq may be limited by its low capture efficiency and high level of noise. While scRNA-seq may fail to capture all transcripts in each cell, the technique was sufficient for identifying markers of cell states within the distal TE. Furthermore, organoids were generated from pools of epithelial cells split by their SLC1A3 expression. However, there may be other epithelial cell states that are less likely to form organoids among the SLC1A3+ and SLC1A3- populations, which may impact the organoid formation rate. Finally, we explore cancer-prone cell states by inactivating Trp53 and Rb1. Inactivation of these genes leads to HGSC arising from pre-ciliated TE cells. However, other combinations of driver mutations may transform other cell states and/or produce different types of ovarian carcinoma.
In summary, our study establishes the cell hierarchy, cell lineage dynamics, and identity of stem/progenitor cells in the distal TE. Furthermore, we show resistance of TE stem/progenitor cells to malignant transformation and provide direct experimental evidence for cancer propensity of cells in the transitional pre-ciliated state. These findings explain the preferential appearance of neoplastic lesions in the distal region of the uterine tube and point to the pre-ciliation state as a diagnostic and therapeutic target.
Methods
Experimental animals
The Tg(Slc1a3-cre/ERT)1Nat/J (Slc1a3-CreERT, Stock number 012586), Tg(Pax8-rtTA2S*M2)1Koes/J (Pax8-rtTA, stock number 007176), Tg(tetO-Cre)1Jaw/J (Tre-Cre, stock number 006234), Tg(KRT5-cre/ERT2)2Ipc/Jeldj (K5-Cre-ERT2, Stock number 018394), Gt(ROSA)26Sortm9(CAG-tdTomato)Hze (Rosa-loxp-stop-loxp-tdTomato/Ai9, Stock number 007909), and C57BL6 (Stock number 000664) mice were obtained from The Jackson Laboratory (Bar Harbor, ME, USA). The Trp53loxP/loxP and Rb1loxP/loxP mice, which have Trp53 and Rb1 genes flanked by loxP alleles, respectively, were a gift from Dr. Anton Berns. Mice were bred on a mixed FVB/N and BL6/J background. Their genotype was confirmed by PCR amplification (Supplementary Table 6). Only females were used for our studies of the female reproductive tract. The number of animals used in every experiment is indicated as biological replicates in figure legends and supplementary tables. Animals were euthanized if becoming moribund or developing tumors over 1 cm in diameter. Maximal allowed tumor size/burden was not exceeded. All the experiments and maintenance of the mice followed ethical regulations for animal testing and research. They were approved by the Cornell University Institutional Laboratory Animal Use and Care Committee (protocols numbers 2000-0116 and 2001-0072). Mice were housed within a 10/14 light cycle. The lights came on at 5 a.m. and went off at 7 p.m., the humidity ranged from 30–70 %, and the ambient temperature was kept at 72 °F ± 2 °F.
Doxycycline and tamoxifen induction
For estimation of target cell frequency 6–8 week-old Pax8-rtTA Tre-Cre Ai9 mice received a single dose (12 μl g−1 body weight) of doxycycline (6.7 mg ml−1 in sterile PBS) by intraperitoneal (i.p.) injection. Identical injection schedule was used for tumor induction experiments with 6–10 week-old Pax8-rtTA Tre-Cre Trp53loxP/loxP Rb1loxP/loxP Ai9 mice and control mice. In our cohort, about 10% of TRE-Cre Trp53loxP/loxP Rb1loxP/loxP mice have developed histiocytic sarcomas and have not been included in further analyzes. For tamoxifen induction of Cre expression in cell lineage tracing experiments 6–8 week-old Slc1a3-CreERT Ai9 mice received 1, 3, and 5 daily i.p. injections of tamoxifen (100 μg/g body weight, 8 μl/g body weight, 12.5 mg/ml in corn oil; Sigma-Aldrich, T5648). For low dose lineage tracing experiments, mice were treated with a single i.p. injection of tamoxifen at 10 μg/g body weight). In tumor induction experiments, 6–10 week-old mice Slc1a3-CreERT Trp53loxP/loxP Rb1loxP/loxP Ai9 mice received i.p. injections every 24 h for 3 days. K5-CreERT2 Trp53loxP/loxP Rb1loxP/loxP Ai9 mice of the same age received a single i.p. injection of tamoxifen. For organoid assays after FACS, 5 Slc1a3-CreERT Ai9 mice 6–12 week-old mice per experiment received a single 36 h tamoxifen pulse (100 μg/g body weight as described above). All mice were euthanized by CO2 and further analyses were carried out.
Anatomical nomenclature
“Uterine tube”, “Fallopian tube”, and “oviduct” are terms commonly used by different groups of investigators. The term “uterine tube” precisely describes its location and function as a tube leading from the ovary to the uterus. “Fallopian tube,” named after the 16th-century Italian anatomist Gabriele Falloppio, is a more traditional term, but it’s less descriptive anatomically. “Oviduct” is anatomically descriptive, indicating a duct for the ovum (egg), but it is less specific to human anatomy and is often used in the context of various animals. We use the term “uterine tube” as the most anatomically accurate name for this structure. The “uterine tube” is also the most unifying term allowing to be efficiently used for comparative human/mouse studies common in experimental research.
Pathological evaluation
All mice in carcinogenesis experiments were subjected to gross pathology evaluation during necropsy. Particular attention was paid to potential sites of ovarian carcinoma spreading, such as the omentum, regional lymph nodes, uterus, liver, lung and mesentery. In addition to the ovary, pathologically altered organs, as well as representative specimens of the brain, lung, liver, kidney, spleen, pancreas and intestine, intra-abdominal lymph nodes, omentum and uterus were fixed in 4% PBS buffered paraformaldehyde. They were then evaluated by microscopic analysis of serial paraffin sections stained with hematoxylin and eosin and subjected to necessary immunostainings. All early atypical lesions were diagnosed based on their morphology, staining for PAX8, extent of proliferation and detection of deleted (floxed-out) Trp53 and Rb1 as described earlier52. STICs were identified based on pronounced cellular atypia, the loss of cell polarity, high proliferative index, and Trp53 deletion. The locations of all lesions were determined by three-dimensional reconstruction of 4-mm-thick serial sections as described previously53.
Immunohistochemistry, apoptosis detection, and image analysis
For lineage tracing experiments, Slc1a3-CreERT Ai9 induced mouse tissues were fixed with 4% paraformaldehyde for 1.5 h on ice. After fixation tissues were washed three times 5 min with PBS at room temperature, embedded in Tissue-Tek O.C.T. compound (ThermoFisherScientific), and frozen on dry ice for 10 min. Immunofluorescent detection of FAM183B (IF, dilution 1:150) and OVGP1 (IF, dilution 1:600, 1:800, IHC, 1:800, 1:2000–8000) was performed in 7 μm-thick frozen sections according to standard protocols. For paraffin embedding, tissues were fixed in 4% paraformaldehyde overnight at 4 °C followed by standard tissue processing, paraffin embedding, and preparation of 4 μm-thick tissue sections. For immunohistochemistry, antigen retrieval was performed by incubation of deparaffinized and rehydrated tissue sections in boiling 10 mM sodium citrate buffer (pH 6.0) for 10 min. The primary antibodies against PAX8 (anti-mouse, IF, dilution 1:100, IHC, 1:5000–10,000, anti-rabbit, IF, 1:400, IHC, 1:4000), and tdTomato/RFP (anti-goat, IF, dilution 1:100, anti-rabbit, IF, 1:250, IHC, 1:4000) were incubated at 4 °C for overnight, followed by incubation with secondary biotinylated antibodies (dilution 1:200, 45 min, at room temperature, RT). Modified Elite avidin-biotin peroxidase (ABC) technique (Vector Laboratories, Burlingame, CA, USA; pk-6100) was performed at room temperature for 30 min. Hematoxylin was used as the counterstain. All primary and secondary antibodies used for immunostaining are listed in Supplementary Table 7.
Apoptosis detection was performed on paraffin sections with TUNEL assay following manufacturer instructions (abcam, ab206386). All stained cells were additionally confirmed by morphological recognition of apoptotic bodies, featuring nuclear fragmentation, cytoplasmic shrinkage, and blebbing53,54. More than 1200 cells per sample were scored to estimate the apoptotic index.
For quantitative studies, sections were scanned with a ScanScope CS2 (Leica Biosystems, Vista, CA), 40x objective, a Zeiss LSM 710 Confocal Microscope for tile scans with a pixel dwell of 1.58 μsec and an averaging of 2 using the ZEN (blue edition, Zeiss) software, or a Leica TCS SP5 Confocal Microscope, 20x and 40x objective, followed by the analysis with Fiji software (National Institutes of Health, Bethesda, MD, USA).
Microdissection-PCR
Four μm thick paraffin sections were placed on glass slides, stained with H&E, and parallel sections stained for tdTomato were evaluated under microdissection microscope. Cells from HE stained sections were collected using a 25G1/2 needle, into 0.6 ml Eppendorf tubes filled with lysis buffer, digested in proteinase K, and processed for subsequent PCR amplification12,52,53,55.
TE organoid preparation
Mouse tubal epithelial cells (TE) were isolated from Slc1a3-CreERT Ai9 positive and negative littermates. Single cell suspensions for organoids were adapted from previously described methods7,10,56. Briefly, mouse uterine tubes were collected, washed with wash buffer (Phosphate buffered saline pH 7.4, Thermo Fisher 10010023, 10,000 U/ml Penicillin-Streptomycin, Thermo Fisher 15140122) and digested with digestion buffer (Gibco DMEM/F12, Fisher 11320033; 4 µg/ml Roche Collagenase/Dispase, Sigma 10269638001; 10 µg/ml DNaseI, Sigma 1128459638001) for two rounds (45 min each) at 37 °C. Between rounds, previous digestion buffer was collected and neutralized with 20% FBS containing media in a new tube. New digestion buffer was added to the tissue and mixed vigorously, and digestion allowed to continue. After neutralization, cells were spun down at 600 × g for 5 min at 4 °C and mixed with Matrigel. For every 2 mice (4 uterine tubes), 100 µl of Matrigel would be used and plated along the rim of a 24 well tissue culture plate. Plates were then be incubated in 5% CO2 cell incubator at 37 °C for 60 min, after which Matrigel would be set and organoid media (Gibco Advanced DMEM/F12, Thermo Fisher 12634010; 25% L-WRN Conditioned Medium, ATCC CRL-3276; 12 mM HEPES, Thermo Fisher 15630080; 1% GlutaMax, Thermo Fisher 35050079; 2% B27, Thermo Fisher 17504044; 1% N2, Thermo Fisher 17502048; 10 ng/ml hEGF, Sigma E9644; 100 ng/ml Human FGF-10, 1 mM Nicotinamide, Sigma N0636; 10 µM Y-27632 (ROCKi), Millipore 688000; 2.5 µM TGF-B RI Kinase Inhibitor VI, Millipore 616464) was added.
For passaging, organoids were released from Matrigel with organoid harvesting media (R&D 3700-100-01) by pipetting around the rim of the plate. Released cells were transferred to a 15 ml falcon tube and allowed to chill on ice for 1 h. Cells were then spun down in a refrigerated centrifuge at 600 × g for 5 min at 4 °C. Supernatant was aspirated, and 1 ml of TrypLE (Thermo Fisher 12604013) was added. Cells were then incubated at 37 °C for 10 min and vigorously pipetted. Cells were then recovered with 20% FBS containing media and spun down at 600xg for 5 min at 4 °C. Supernatant was aspirated, organoid media was added, and cells were counted. Typically, cells were plated at 500 cells/µl of Matrigel for expansion. To induce Slc1a3-Cre-ERT mediated recombination, 1 µM 4-Hydroxytamoxifen (Selleck Chem S7827) was supplemented to the media for 48 h.
FACS preparation and analysis
Organoid cells were prepared as described above in organoid preparation. After rescuing cells from digestion, cells were washed in 1x PBS + 1% BSA three times. After the last wash, cells were suspended in 1x PBS + 1% BSA. FACS was performed on a Sony MA900 in two separate experiments with single replicates. Data was acquired using Sony MA-900 complementary software. Analysis was performed using FlowJo software (BD Biosciences). Post FACS, organoids were visualized under a microscope to check for tdTomato expression. Slc1a3-CreERT negative littermates were used to gate negative controls (Supplementary Fig. 13). After gating single cells, Sytox Blue was used to gate for live or dead populations (Supplementary Fig. 13c). From the live cell population, tdTomato+ cells were determined by using Slc1a3-CreERT negative litter mates (Supplementary Fig. 13d). Cells were then plated in Matrigel at a density of 1000 cells per 10 μl. 1000 SLC1A3+ cells and 30,000 SLC1A3- cells were plated. Assessment of organoid forming potential was calculated by counting the number of organoids formed against the number of cells sorted. Each replicate contained uterine tubes pooled from at least 5 mice.
MACS preparation and analysis
Single cell suspensions from the distal mouse uterine tube were prepared as described above from 10 C57BL6 adult female mice aged 6–12 weeks. Magnetic activated cell sorting was performed following the manufacturer protocol (Invitrogen 11533D). Negative selection was first performed to remove stromal cells by incubating cells with biotinylated antibodies against CD45 (Biolegend, 103103, 0.5 µg per million cells), TER-119 (Biolegend, 116203, 0.5 µg per million cells), CD140a (Biolegend, 135910, 0.5 µg per million cells), CD31 (Biolegend, 102503, 0.5 µg per million cells). Positive selection was then performed on the unbound fraction of cells by incubating with a biotinylated antibody against SLC1A3 (Novus biologicals, NB100-1869B, 0.5 µg per million cells). After removal of the unbound fraction, bound cells were released with 200U of DNase I (Sigma, 1128459638001). Cells were counted and equal numbers of cells were plated for both the SLC1A3+ and SLC1A3- populations (100 cells/µl Matrigel). 5000 cells were plated per Matrigel droplet in a 24 well plate. Representative images were taken every few days from plating to day 14. Day 14 organoids were counted for organoid formation rate, organoid size, and sections prepared to assess for expression of organoid markers.
Organoid histological preparation and analysis
Organoids were released from Matrigel with Cultrex Organoid Harvesting Solution and chilled on ice for 1 h as per the passaging protocol. Organoids were then centrifuged in a refrigerated centrifuge at 100 × g for 30 s at 4 °C. Organoids were fixed on ice in 5 ml of 4% PBS-buffered paraformaldehyde while avoiding exposure to light. Organoids were spun down at 100 × g for 30 s and replaced with 1x PBS to rinse. Organoids were spun down once more at 100 × g for 30 s to aspirate the remaining PBS. Organoids were then mixed in 200 µl of HistoGel (Epredia, HG4000012) preheated to 65 °C and dispensed into dome-shaped molds for embedding. HistoGel molds set for 10 min prior to standard tissue processing, paraffin embedding, and preparation of 4-μm-thick tissue sections followed by immunohistochemistry57.
Single-cell RNA-sequencing library preparation
For TE single cell expression and transcriptome analysis we isolated TE from C57BL6 adult female mice at the estrus stage of estrous cycle. In 3 independent experiments a total of 62 uterine tubes were collected. Each uterine tube was placed in sterile PBS containing 100 IU ml−1 of penicillin and 100 µg ml−1 streptomycin (Corning, 30-002-Cl), and separated in distal and proximal regions. Tissues from the same region were combined in a 40 µl drop of the same PBS solution, cut open lengthwise, and minced into 1.5–2.5 mm pieces with 25G needles. Minced tissues were transferred with help of a sterile wide bore 200 µl pipette tip into a 1.8 ml cryo vial containing 1.2 ml A-mTE-D1 (300 IU ml−1 collagenase IV mixed with 100 IU ml−1 hyaluronidase; Stem Cell Technologies, 07912, in DMEM Ham’s F12, Hyclone, SH30023.FS). Tissues were incubated with loose cap for 1 h at 37 °C in a 5% CO2 incubator. During the incubation tubes were taken out 4 times, and tissues suspended with a wide bore 200 µl pipette tip. At the end of incubation, the tissue-cell suspension from each tube was transferred into 1 ml TrypLE (Invitrogen, 12604013) pre-warmed to 37 °C, suspended 70 times with a 1000 µl pipette tip, 5 ml A-SM [DMEM Ham’s F12 containing 2% fetal bovine serum (FBS)] were added to the mix, and TE cells were pelleted by centrifugation 300 × g for 10 min at 25 °C. Pellets were then suspended with 1 ml pre-warmed to 37 °C A-mTE-D2 (7 mg ml−1 Dispase II, Worthington NPRO2, and 10 µg ml−1 Deoxyribonuclease I, Stem Cell Technologies, 07900), and mixed 70 times with a 1000 µl pipette tip. 5 ml A-mTE-D2 was added and samples were passed through a 40 µm cell strainer, and pelleted by centrifugation at 300 × g for 7 min at +4 °C. Pellets were suspended in 100 µl microbeads per 107 total cells, or fewer, and dead cells were removed with the Dead Cell Removal Kit (Miltenyi Biotec, 130-090-101) according to the manufacturer’s protocol. Pelleted live cell fractions were collected in 1.5 ml low binding centrifuge tubes, kept on ice, and suspended in ice cold 50 µl A-Ri-Buffer (5% FBS, 1% GlutaMAX-I, Invitrogen, 35050-079, 9 µM Y-27632, Millipore, 688000, and 100 IU ml−1 penicillin 100 μg ml−1 streptomycin in DMEM Ham’s F12). Cell aliquots were stained with trypan blue for live and dead cell calculation. Live cell preparations with a target cell recovery of 5000–6000 were loaded on Chromium controller (10X Genomics, Single Cell 3’ v2 chemistry) to perform single cell partitioning and barcoding using the microfluidic platform device. After preparation of barcoded, next-generation sequencing cDNA libraries samples were sequenced on Illumina NextSeq500 System.
Download and alignment of single-cell RNA sequencing data
For sequence alignment, a custom reference for mm39 was built using the cellranger (v6.1.2, 10x Genomics) mkref function. The mm39.fa soft-masked assembly sequence and the mm39.ncbiRefSeq.gtf (release 109) genome annotation last updated 2020-10-27 were used to form the custom reference. The raw sequencing reads were aligned to the custom reference and quantified using the cellranger count function.
Preprocessing and batch correction
All preprocessing and data analysis was conducted in R (v.4.1.1 (2021-08-10)). The cellranger count outs were first modified with the autoEstCont and adjustCounts functions from SoupX (v.1.6.1) to output a corrected matrix with the ambient RNA signal (soup) removed (https://github.com/constantAmateur/SoupX). To preprocess the corrected matrices, the Seurat (v.4.1.1) NormalizeData, FindVariableFeatures, ScaleData, RunPCA, FindNeighbors, and RunUMAP functions were used to create a Seurat object for each sample (https://github.com/satijalab/seurat). The number of principal components used to construct a shared nearest-neighbor graph were chosen to account for 95% of the total variance. To detect possible doublets, we used the package DoubletFinder (v.2.0.3) with inputs specific to each Seurat object. DoubletFinder creates artificial doublets and calculates the proportion of artificial k nearest neighbors (pANN) for each cell from a merged dataset of the artificial and actual data. To maximize DoubletFinder’s predictive power, mean-variance normalized bimodality coefficient (BCMVN) was used to determine the optimal pK value for each dataset. To establish a threshold for pANN values to distinguish between singlets and doublets, the estimated multiplet rates for each sample were calculated by interpolating between the target cell recovery values according to the 10x Chromium user manual. Homotypic doublets were identified using unannotated Seurat clusters in each dataset with the modelHomotypic function. After doublets were identified, all distal and proximal samples were merged separately. Cells with greater than 30% mitochondrial genes, cells with fewer than 750 nCount RNA, and cells with fewer than 200 nFeature RNA were removed from the merged datasets. To correct for any batch defects between sample runs, we used the harmony (v.0.1.0) integration method (https://github.com/immunogenomics/harmony).
Clustering parameters and annotations
After merging the datasets and batch-correction, the dimensions reflecting 95% of the total variance were input into Seurat’s FindNeighbors function with a k.param of 70. Louvain clustering was then conducted using Seurat’s FindClusters with a resolution of 0.7. The resulting 19 clusters were annotated based on the expression of canonical genes and the results of differential gene expression (Wilcoxon Rank Sum test) analysis. One cluster expressing lymphatic and epithelial markers was omitted from later analysis as it only contained 2 cells suspected to be doublets. To better understand the epithelial populations, we reclustered 6 epithelial populations and reapplied harmony batch correction. The clustering parameters from FindNeighbors was a k.param of 50, and a resolution of 0.7 was used for FindClusters. The resulting 9 clusters within the epithelial subset were further annotated using differential expression analysis and canonical markers.
Pseudotime analysis
PHATE is dimensional reduction method to more accurately visualize continual progressions found in biological data33. A modified version of Seurat (v4.1.1) was developed to include the “RunPHATE” function for converting a Seurat Object to a PHATE embedding. This was built on the phateR package (v.1.0.7) (https://github.com/scottgigante/seurat/tree/patch/add-PHATE-again). In addition to PHATE, pseudotime values were calculated with Monocle3 (v.1.2.7), which computes trajectories with an origin set by the user37,58–60. The origin was set to be a progenitor cell state confirmed with lineage tracing experiments.
Statistical analyses
Statistical comparisons were performed using a two-tailed unpaired t test and Analysis of Variance (ANOVA) with Tukey-Kramer Multiple Comparisons Test with InStat 3 and Prism 6 software (GraphPad Software Inc., La Jolla, CA, USA).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
We thank Matalin Pirtz, Nikitin lab for critical reading of the manuscript, Peter A. Schweitzer, Director of the Cornell Genomics Facility for his invaluable assistance with single-cell RNA sequencing, Tudorita Tumbar, Cornell Molecular Biology, and Genetics, for providing Slc1a3-CreERT Ai9 mice from her long-term cell lineage tracing experiments and advising on the initial Slc1a3-related experiments, Temirlan Shilikbay, Nazarbayev University, Astana, Kazakhstan for his early contributions to a pilot Slc1a3 project, and Md Mozammal Hossain, Kurtay Ozuner and Derrick Tran for their excellent technical support. This work has been supported by NIH grants (CA182413, CA260115, and CA248524) to A.Y.N., Ovarian Cancer Research Fund grant (327516) to A.Y.N., Sandra Atlas Bass Endowment for Cancer Research to A.Y.N. and J.C.S., the NSF Graduate Research Fellowship Program (GRFP) awarded to C.Q.R. (DGE-2139899) and NIH 1S10RR025502 grant to the Cornell Institute of Biotechnology Imaging Facility for the Zeiss LSM 710 Confocal Microscope.
Author contributions
A.F.N. and A.Y.N. designed experiments; C.Q.R., D.J.F., D.J.P., B.A.H., C.S.A., A.P.A., and S.G. performed experiments; C.Q.R., A.J.D.M., D.W.M., and B.D.C. carried our bioinformatics analyses; D.J.F., L.H.E., and A.Y.N. performed pathological evaluations; J.C.S., and B.D.C. provided resources; A.F.N., C.Q.R. and A.Y.N. wrote the paper.
Peer review
Peer review information
Nature Communications thanks Matthew Ford, Alistair Forrest, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The raw sequencing data generated in this study have been deposited in the Gene Expression Omnibus (GEO) database under accession code GSE252786. All processed Seurat objects for scRNA-seq analysis are available in the Dryad repository at 10.5061/dryad.4mw6m90hm [10.5061/dryad.4mw6m90hm]. All data generated in this study are provided in the Source Data file. The remaining data are available within the Article, Supplementary or Source Data file. Source data are provided with this paper.
Code availability
All code for data preprocessing and figure generation of scRNA-seq data are available through GitHub (https://github.com/coulterr24/MouseTE_scRNA/) and Zenodo (10.5281/zenodo.13830750).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Andrea Flesken-Nikitin, Coulter Q. Ralston.
Change history
11/20/2024
A Correction to this paper has been published: 10.1038/s41467-024-54465-x
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-52984-1.
References
- 1.Siegel, R. L., Giaquinto, A. N. & Jemal, A. Cancer statistics, 2024. CA Cancer J. Clin.74, 12–49 (2024). [DOI] [PubMed] [Google Scholar]
- 2.Seidman, J. D., Ronnett, B. M., Shih Ie, M., Cho, K. R. & Kurman, R. J. in Blaustein’s Pathology of the Female Genital Tract (eds Kurman, R. J., Ellenson, L. H. & Ronnett, B. M.) 841–966 (Springer, 2019).
- 3.Carlson, J. W., Gilks, C. B. & Soslow, R. A. Tumors of the Ovary and Fallopian Tube. Vol. 16 (American Registry of Pathology, 2023).
- 4.Kurman, R. J. & Shih Ie, M. The origin and pathogenesis of epithelial ovarian cancer: a proposed unifying theory. Am. J. Surg. Pathol.34, 433–443 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Landen, C. N. Jr., Birrer, M. J. & Sood, A. K. Early events in the pathogenesis of epithelial ovarian cancer. J. Clin. Oncol.26, 995–1005 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Kim, J. et al. Cell origins of high-grade serous ovarian cancer. Cancers10, pii: E433 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang, S. et al. Both fallopian tube and ovarian surface epithelium are cells-of-origin for high-grade serous ovarian carcinoma. Nat. Commun.10, 5367 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lawrenson, K. et al. A study of high-grade serous ovarian cancer origins implicates the SOX18 transcription factor in tumor development. Cell Rep.29, 3726–3735 e3724 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Hao, D. et al. Integrated analysis reveals tubal- and ovarian-originated serous ovarian cancer and predicts differential therapeutic responses. Clin. Cancer Res.23, 7400–7411 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Lohmussaar, K. et al. Assessing the origin of high-grade serous ovarian cancer using CRISPR-modification of mouse organoids. Nat. Commun.11, 2660 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maniati, E. et al. Mouse ovarian cancer models recapitulate the human tumor microenvironment and patient response to treatment. Cell Rep.30, 525–540 e527 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flesken-Nikitin, A. et al. Ovarian surface epithelium at the junction area contains a cancer-prone stem cell niche. Nature495, 241–245 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamulla, R. J., Nalubola, S., Flesken-Nikitin, A., Nikitin, A. Y. & Schimenti, J. C. Most commonly mutated genes in high-grade serous ovarian carcinoma are nonessential for ovarian surface epithelial stem cell transformation. Cell Rep.32, 108086 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmoeckel, E. et al. LEF1 is preferentially expressed in the tubal-peritoneal junctions and is a reliable marker of tubal intraepithelial lesions. Mod. Pathol.30, 1241–1250 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seidman, J. D. Serous tubal intraepithelial carcinoma localizes to the tubal-peritoneal junction: a pivotal clue to the site of origin of extrauterine high-grade serous carcinoma (ovarian cancer). Int. J. Gynecol. Pathol.34, 112–120 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Kuhn, E. et al. TP53 mutations in serous tubal intraepithelial carcinoma and concurrent pelvic high-grade serous carcinoma-evidence supporting the clonal relationship of the two lesions. J. Pathol.226, 421–426 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee, Y. et al. A candidate precursor to serous carcinoma that originates in the distal fallopian tube. J. Pathol.211, 26–35 (2007). [DOI] [PubMed] [Google Scholar]
- 18.Perets, R. et al. Transformation of the fallopian tube secretory epithelium leads to high-grade serous ovarian cancer in Brca;Tp53;Pten models. Cancer Cell24, 751–765 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu, D. J. et al. Cells expressing PAX8 are the main source of homeostatic regeneration of adult endometrial epithelium and give rise to serous endometrial carcinoma. Dis. Model. Mech.10.1242/dmm.047035 (2020). [DOI] [PMC free article] [PubMed]
- 20.Paik, D. Y. et al. Stem-like epithelial cells are concentrated in the distal end of the fallopian tube: a site for injury and serous cancer initiation. Stem Cells30, 2487–2497 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stewart, C. A. & Behringer, R. R. Mouse oviduct development. Results Probl. Cell Differ.55, 247–262 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Aviles, M., Coy, P. & Rizos, D. The oviduct: a key organ for the success of early reproductive events. Anim. Front.5, 25–31 (2015). [Google Scholar]
- 23.Ghosh, A., Syed, S. M. & Tanwar, P. S. In vivo genetic cell lineage tracing reveals that oviductal secretory cells self-renew and give rise to ciliated cells. Development144, 3031–3041 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Ford, M. J. et al. Oviduct epithelial cells constitute two developmentally distinct lineages that are spatially separated along the distal-proximal axis. Cell Rep.36, 109677 (2021). [DOI] [PubMed] [Google Scholar]
- 25.Flesken-Nikitin, A., Odai-Afotey, A. A. & Nikitin, A. Y. Role of the stem cell niche in the pathogenesis of epithelial ovarian cancers. Mol. Cell Oncol.1, 963431–963437.e963435 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu, D. J. et al. Stem cell pathology. Annu. Rev. Pathol.13, 71–92 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nassar, D. & Blanpain, C. Cancer stem cells: basic concepts and therapeutic implications. Annu. Rev. Pathol.11, 47–76 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Fu, D. J. et al. Gastric squamous-columnar junction contains a large pool of cancer-prone immature osteopontin responsive Lgr5(-)CD44(+) cells. Nat. Commun.11, 84 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friedmann-Morvinski, D. et al. Dedifferentiation of neurons and astrocytes by oncogenes can induce gliomas in mice. Science338, 1080–1084 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwitalla, S. et al. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell152, 25–38 (2013). [DOI] [PubMed] [Google Scholar]
- 31.Rohozinski, J., Diaz-Arrastia, C. & Edwards, C. L. Do some epithelial ovarian cancers originate from a fallopian tube ciliate cell lineage? Med Hypotheses107, 16–21 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods16, 1289–1296 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moon, K. R. et al. Visualizing structure and transitions in high-dimensional biological data. Nat. Biotechnol.37, 1482–1492 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamanouchi, H., Umezu, T. & Tomooka, Y. Reconstruction of oviduct and demonstration of epithelial fate determination in mice. Biol. Reprod.82, 528–533 (2010). [DOI] [PubMed] [Google Scholar]
- 35.Sada, A. et al. Defining the cellular lineage hierarchy in the interfollicular epidermis of adult skin. Nat. Cell Biol.18, 619–631 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghuwalewala, S. et al. Binary organization of epidermal basal domains highlights robustness to environmental exposure. EMBO J.41, e110488 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cao, J. et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature566, 496–502 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winkler, I. et al. The cycling and aging mouse female reproductive tract at single-cell resolution. Cell187, 981–998 e925 (2024). [DOI] [PubMed] [Google Scholar]
- 39.Xie, Y., Park, E. S., Xiang, D. & Li, Z. Long-term organoid culture reveals enrichment of organoid-forming epithelial cells in the fimbrial portion of mouse fallopian tube. Stem Cell Res.32, 51–60 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Rose, I. M. et al. WNT and inflammatory signaling distinguish human Fallopian tube epithelial cell populations. Sci. Rep.10, 9837 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Network, T. C. G. A. R. Integrated genomic analyses of ovarian carcinoma. Nature474, 609–615 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ford, M. J., Harwalkar, K., Kazemdarvish, H., Yamanaka, N. & Yamanaka, Y. CD133/Prom1 marks proximal mouse oviduct epithelial progenitors and adult epithelial cells with a low generative capacity. Biol. Open12, 10.1242/bio.059963 (2023). [DOI] [PMC free article] [PubMed]
- 43.Kaipio, K. et al. ALDH1A1‐related stemness in high‐grade serous ovarian cancer is a negative prognostic indicator but potentially targetable by EGFR/mTOR‐PI3K/aurora kinase inhibitors. J. Pathol.250, 159–169 (2020). [DOI] [PubMed] [Google Scholar]
- 44.Ricciardelli, C. et al. Keratin 5 overexpression is associated with serous ovarian cancer recurrence and chemotherapy resistance. Oncotarget8, 17819–17832 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Logotheti, S. et al. Mechanisms of functional pleiotropy of p73 in cancer and beyond. Front. Cell Dev. Biol.9, 737735 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nemajerova, A. et al. Non-oncogenic roles of TAp73: from multiciliogenesis to metabolism. Cell Death Differ.25, 144–153 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marshall, C. B. et al. p73 Is required for multiciliogenesis and regulates the Foxj1-associated gene network. Cell Rep.14, 2289–2300 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nemajerova, A. et al. TAp73 is a central transcriptional regulator of airway multiciliogenesis. Genes Dev.30, 1300–1312 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Song, R. et al. miR-34/449 miRNAs are required for motile ciliogenesis by repressing cp110. Nature510, 115–120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Corney, D. C. et al. Frequent downregulation of miR-34 family in human ovarian cancers. Clin. Cancer Res.16, 1119–1128 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Levrero, M. et al. The p53/p63/p73 family of transcription factors: overlapping and distinct functions. J. Cell Sci.113, 1661–1670 (2000). (Pt 10). [DOI] [PubMed] [Google Scholar]
- 52.Flesken-Nikitin, A., Choi, K. C., Eng, J. P., Shmidt, E. N. & Nikitin, A. Y. Induction of carcinogenesis by concurrent inactivation of p53 and Rb1 in the mouse ovarian surface epithelium. Cancer Res.63, 3459–3463 (2003). [PubMed] [Google Scholar]
- 53.Nikitin, A. Y. & Lee, W. H. Early loss of the retinoblastoma gene is associated with impaired growth inhibitory innervation during melanotroph carcinogenesis in Rb+/- mice. Genes Dev.10, 1870–1879 (1996). [DOI] [PubMed] [Google Scholar]
- 54.Nikitin, A. Y. et al. Cell lineage-specific effects associated with multiple deficiencies of tumor susceptibility genes in Msh2(-/-)Rb(+/-) mice. Cancer Res.62, 5134–5138 (2002). [PubMed] [Google Scholar]
- 55.Matoso, A., Zhou, Z., Hayama, R., Flesken-Nikitin, A. & Nikitin, A. Y. Cell lineage-specific interactions between Men1 and Rb in neuroendocrine neoplasia. Carcinogenesis29, 620–628, (2008). [DOI] [PubMed] [Google Scholar]
- 56.Ford, M. J., Harwalkar, K. & Yamanaka, Y. Protocol to generate mouse oviduct epithelial organoids for viral transduction and whole-mount 3D imaging. STAR Protoc.3, 101164 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Flesken-Nikitin, A., Harlan, B. A. & Nikitin, A. Y. Transplantation into the mouse ovarian fat pad. J. Vis. Exp.10.3791/54444 (2016). [DOI] [PMC free article] [PubMed]
- 58.Trapnell, C. et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol.32, 381–386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qiu, X. et al. Single-cell mRNA quantification and differential analysis with Census. Nat. Methods14, 309–315 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qiu, X. et al. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods14, 979–982 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequencing data generated in this study have been deposited in the Gene Expression Omnibus (GEO) database under accession code GSE252786. All processed Seurat objects for scRNA-seq analysis are available in the Dryad repository at 10.5061/dryad.4mw6m90hm [10.5061/dryad.4mw6m90hm]. All data generated in this study are provided in the Source Data file. The remaining data are available within the Article, Supplementary or Source Data file. Source data are provided with this paper.
All code for data preprocessing and figure generation of scRNA-seq data are available through GitHub (https://github.com/coulterr24/MouseTE_scRNA/) and Zenodo (10.5281/zenodo.13830750).