Abstract
The membrane-spanning domain (MSD) of a number of retroviral transmembrane (TM) glycoproteins, including those from the human and simian immunodeficiency viruses (HIV and SIV), have been predicted to contain a charged arginine residue. The wild-type SIV TM glycoprotein is 354 amino acids long. The entire putative cytoplasmic domain of SIV (amino acids 193 to 354) is dispensable for virus replication in vitro, and such truncation-containing viruses are capable of reaching wild-type titers after a short delay. We show here that further truncation of eight additional amino acids to TM185 results in a protein that lacks fusogenicity but is, nevertheless, efficiently incorporated into budding virions. By analyzing a series of nonsense mutations between amino acids 193 and 185 in Env expression vectors and in the SIVmac239 proviral clone, a region of the SIV TM that contains the minimum requirement for glycoprotein-mediated cell-to-cell fusion and that for virus replication was identified. Virus entry and infectivity were evident in truncations to a minimum of 189 amino acids, whereas cell-cell fusion was observed for a protein of only 187 amino acids. Glycoprotein was efficiently incorporated into budding virions in truncations up to 185 amino acids, indicating that such proteins are membrane anchored and are transported to the cell surface. However, truncation of the TM to 180 amino acids resulted in a protein that displays a transport defect and may be retained in the endoplasmic reticulum. Based on our analyses of these mutants, an alternative model for the MSD of SIV is proposed. Our model suggests that membrane-imbedded charged residues can be neutralized by side-chain interactions with lipid polar head groups. As a consequence, the membrane-spanning region can be reduced by more than a helical turn. This new model accounts for the ability of truncations within the predicted MSD to remain membrane anchored and maintain biological activity.
The simian immunodeficiency virus (SIV) was first isolated from captive macaques with immunodeficiency and lymphoma and has since been identified in a number of primate species (7, 17, 18, 36, 52). SIV induces a chronic, wasting immunodeficiency in macaques that is similar to AIDS in humans infected with the human immunodeficiency virus (HIV) (28, 50, 51). For HIV and SIV, as for all retroviruses, the Gag protein is sufficient to produce virus-like particles (31, 42, 54, 68, 69). Incorporation of the envelope glycoprotein is required, however, for those particles to be infectious. The SIV envelope glycoprotein is synthesized as a polyprotein precursor that is cleaved into the surface (SU) and transmembrane (TM) subunits within the medial or trans-Golgi complex by the cellular protease Furin or by a Furin-like convertase (19, 33, 39). Like HIV type 1 (HIV-1), SIV encodes a TM envelope glycoprotein that contains a cytoplasmic domain (CD) that is exceptionally long (>150 amino acids) for a retrovirus. Despite similarities in both sequence and structure of the CD, the two viruses display contrasting phenotypes as a consequence of truncations within this domain. With exceptions, introduction of stop codons into the CD of HIV-1 blocks virus infectivity, whereas the introduction of analogous truncations in SIV results in viruses that manifest enhanced infectivity (23–25, 29, 40, 49, 56, 66, 72, 75, 76). The difference in phenotypes between HIV and SIV truncations appears to be a function of the failure of the HIV truncations to be efficiently incorporated into particles (23, 75) and of enhanced incorporation of truncated SIV glycoproteins (40, 49, 76). Nevertheless, SIV truncations are selected against in macaques, where they rapidly revert to the wild-type sequence (45, 67). Notably, recent reports have demonstrated that glycoprotein incorporation into HIV-1 virions is more permissive in certain cell lines, many of which were previously utilized in the analysis of truncation mutations in the TM (57). It has been suggested that permissiveness, or lack thereof, is linked to some as yet undefined host factor (3, 57). These reports provide some explanation for seemingly paradoxical findings; nevertheless, the mechanism for Env incorporation into HIV and SIV is still not fully understood.
The membrane-spanning domain (MSD) of type I TM proteins, such as a retroviral Env, is defined by a C-terminal, long hydrophobic sequence often bordered on each side by charged residues (10, 30, 32). The long hydrophobic region and the C-terminal charged amino acid(s) are thought to participate in stopping protein translocation into the endoplasmic reticulum (ER). Although charged residues are not always a prerequisite for defining a cellular MSD, the lentiviral glycoproteins encode conserved lysine or arginine residues that have been predicted to define the N- and C-termini of the membrane-spanning region.
The precise boundaries of the MSD and, therefore, the precise length of the CD remain poorly defined. Previous assignments of the borders of MSDs have relied on analyses of the amino acid hydrophobicity and the predicted hydrophobic moment of a helix required to span a membrane of 3- to 4-nm average thickness (11, 12, 21, 22, 46, 48, 59). Mutations in the HIV-1 and SIV TM ectodomain and CD have grossly identified a region that can serve as a membrane anchor. Glycoprotein constructs truncated prior to the putative MSD are often efficiently secreted (40). The requirement for such a membrane anchor is underscored by the finding that glycosylphosphatidylinositol (GPI)-linked glycoproteins can be synthesized and transported in a normal manner and are efficiently incorporated into viral particles, yet such GPI-linked TM proteins are nonfusogenic and do not mediate virus entry (43, 55, 63, 71). However, replacement of the predicted membrane-spanning sequence in HIV-1 Env or vesicular stomatitis virus (VSV) G with heterologous membrane anchors results in chimeras that retain fusogenicity and infectivity in vitro (58, 73). Therefore, a protein domain spanning the plasma membrane is required for fusion, but whether the specific sequence of the domain is critical for biological function remains unclear.
The current prediction for the SIV MSD is 28 amino acids. Given that a functional MSD can be as short as 16 amino acids in VSV G (2) and given the presence of a basic residue (arginine 180) located 17 amino acids within the putative SIV glycoprotein MSD, investigation of the minimal SIV TM CD and MSD sequences is of significant interest. The prediction that the SIV and HIV MSDs contain a centrally located arginine residue (SIV TM amino acid 180; HIV Env amino acid 696) is curious given the phenotype of VSV G glycoproteins with an introduced charge (1). The significance of this seemingly disruptive charged residue in the nonpolar lipid bilayer has been the subject of conflicting reports. Helseth et al. mutated the analogous residue in HIV-1 to serine and described a modest reduction in fusogenicity (35). That finding is consistent with the observation that substitution of a heterologous membrane anchor (CD22), lacking charged residues, for the HIV MSD does not block fusion or infectivity (73). In contrast, Owens et al. reported that any alteration in the HIV-1 TM arginine resulted in significant loss of glycoprotein-mediated fusion (59). Despite its conservation in all HIV and SIV strains, the function of an intramembrane arginine remains unclear.
We and others have shown that C-terminal truncation of the SIV glycoprotein from 354 to 207 or 208 amino acids results in a virus with replication kinetics equivalent to or more rapid than the wild type (40, 49, 76). This truncated glycoprotein is analogous to the mutation in TM spontaneously generated during adaptation of wild-type SIV to human cells (38, 45). Glycoprotein TM207 has been shown to be highly fusogenic and to be incorporated into virions to a higher extent than wild-type Env (40, 49). This mutation expands the in vitro host range of the virus, apparently as a consequence of the increased density of virion-associated glycoprotein (40). Loss of a tyrosine residue (Tyr 196) that is part of a constitutive endocytosis signal in the SIV CD is at least in part responsible for these phenotypes (49, 62, 65). Introduction of a nonsense mutation for amino acid 194 in the SIV TM glycoprotein results in a virus, TM193, that replicates in macaque peripheral blood mononuclear cells (PBMC) with marginally (2 to 4 days) delayed kinetics compared to those of the wild type, despite the loss of 161 amino acids of the cytoplasmic tail. TM193 corresponds to a protein that, under the current assignment of the MSD, has only two cytoplasmic amino acids. Therefore, SIV replication in vitro does not require 161 amino acids of the cytoplasmic tail. Since extensive truncation of the SIV TM to 163 amino acids results in a protein that is efficiently secreted (40), no other TM sequence N-terminal to amino acid 164 can function to span the membrane.
In an effort to define the cytoplasmic border of the SIV MSD, we have introduced sequential truncation mutations into the SIV TM glycoprotein between arginine 180 and arginine 193. We show here that truncations (TM189, TM190, and TM191), which extend into the putative MSD, retain biological activity and are efficiently incorporated into virions. Cell-cell fusion activity is retained by constructs as short as 187 amino acids. Moreover, while glycoproteins truncated to amino acid 185 are membrane anchored and are efficiently incorporated into virions, they are no longer able to induce fusion or mediate virus entry. This study, therefore, defines the minimal C-terminal requirements of the SIV TM glycoprotein required to mediate membrane anchorage, fusion, and infectivity. The results suggest an alternative structure for the SIV TM glycoprotein, with revised borders for the MSD.
MATERIALS AND METHODS
Cell lines and culture.
COS-1 and 293T cells were obtained from the American Type Culture Collection (Manassas, Va.) and maintained in complete medium consisting of Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, 2 mM glutamine, 100 U of penicillin G/ml, and 100 μg of streptomycin sulfate (pen/strep)/ml (all from Life Technologies, Grand Island, N.Y.). Cells were passaged three times weekly and were transfected at 50 to 70% confluency. CCR5/HeLa-CD4-LTR-β-galactosidase (R5 MAGI) cells were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases. This cell line was grown in DMEM complete medium, as above, supplemented with 100 μg of hygromycin B/ml, 200 μg of Geneticin/G418 sulfate/ml, and 1 μg of puromycin/ml (all from Calbiochem, San Diego, Calif.). Cells were maintained in a subconfluent state at all times and were discarded after 20 passages. Primary macaque PBMC were obtained from heparinized venous blood. Mononuclear cells were separated by Ficoll-sodium diatrizoate (LSM; Organon Teknika, Durham, N.C.) density gradient centrifugation. Erythrocytes were lysed in hypotonic buffer, and the leukocytes were stimulated with the lectin concanavalin A (Boehringer-Roche, Indianapolis, Ind.) in RPMI 1640 with 15% fetal calf serum, 2 mM glutamine, pen/strep, and 20 U of interleukin-2 (IL-2)/ml for 72 h. Cells were washed twice in complete medium prior to infection with reverse transcriptase (RT) activity normalized virus. After a 4-h incubation, the cells were washed and placed in 25-cm3 flasks in complete RPMI 1640 supplemented with IL-2 (Boehringer-Roche) at 8 U/ml for the indicated times.
Glycoprotein and proviral expression constructs.
To generate DNA fragments containing the sequential truncation mutations in the SIVmac239 TM glycoprotein, we employed a two-step PCR protocol. Briefly, reverse-complement oligonucleotides were designed to overlap the mutagenesis region and introduce stop codons at each position between amino acids 186 and 190 (see Fig. 1A). The oligonucleotide sequences were TM185, 5′-CCTTAACTTAGCTAGCATTTGTCATATATAGATCACTATTC-3′; TM186, 5′-CCTTAACTTAGCTAGCATTTATACTATATAGATCACTATTC-3′; TM187, 5′-CTGCCTTAACTTAGCTAGCTATTGTACTATATAGATCAC-3′; TM188, 5′-CCCCTGCCTTAACTTAGCTCACATTTGTACTATATAGATC-3′; and TM189, 5′-CCCCTGCCTTAACTTTCATAGCATTTGTACTATATAGATC-3′ (stop codons in reverse complement are underlined). A sense-strand oligonucleotide (5′-CAGAACTGTATCGATTGGAATTGGGAG-3′) was selected to overlap a unique ClaI (underlined) site in SU (nucleotide [nt] 8328). Amplification and simultaneous mutagenesis of this region, using pSRS354, a simian virus 40 promoter-SIV envelope glycoprotein expression vector (23, 40), as a template, produced a fragment of approximately 700 bp, the precise size of which depends on the location of the mutation. This mutant DNA fragment was then used as a primer for PCR along with an oligonucleotide that overlaps a unique SacII site in the Mason-Pfizer monkey virus (MPMV) polyadenylation/constitutive transport element region of the pSRS vector. The resulting DNA fragment was digested with ClaI and SacII and cloned into similarly digested pSRS354. Mutations were verified by DNA sequencing and by expression of the truncated products (see below). The mutations TM190 and TM191 were 3′ of a unique NheI site (nt 8998), allowing the use of a simple amplification/mutagenesis regimen employing the MPMV SacII site described above to create clonable fragments containing these mutations. The mutagenic oligonucleotides were sense orientation: TM190, 5′-GTACAAAT(GCTAGC)TTAGTTAAGGCAGGGG-3′; TM191, 5′-GTACAAAT(GCTAGC)TAAGTAAAGGCAGGG-3′; TM191Ser*, 5′-GTACAAAT(GCTAGC)TTCGTAAAGGCAGGG-3′; and TM192, 5′-CAAAT(GCTAGC)TAAGTTATAGCAGGGGTATAGG-3′, with the stop codons underlined and the NheI site in parentheses. The products of these amplifications were digested with NheI and SacII, gel purified, and cloned into similarly digested pSRS.
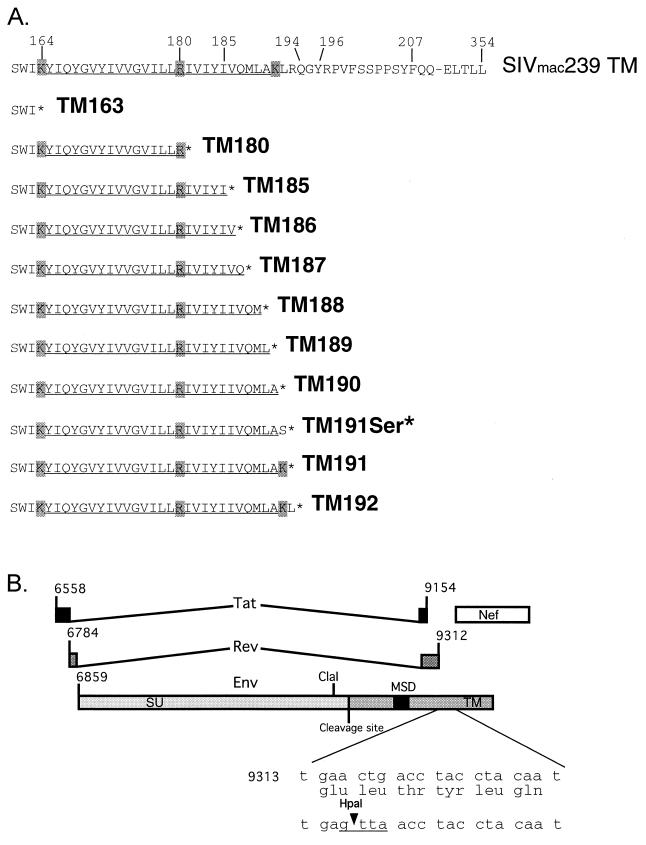
FIG. 1.
Locations of glycoprotein truncation mutants. (A) Truncation mutations were generated by PCR mutagenesis to insert a stop codon (∗) subsequent to the amino acid number (i.e., TM185 has a stop codon in place of amino acid 186). TM191Ser* contains a serine substitution at amino acid 191, followed by a termination signal. Charged amino acids bordering or contained within the MSD are boxed in gray, and the previously defined MSD is underlined. (B) A unique HpaI site was inserted into the C-terminal region of Env in pBR322 SIVmac239 by silent mutagenesis as described in Materials and Methods. The resulting construct is denoted pBR SIV-HpaI. The black box denotes the MSD. The unique ClaI site is located in the C-terminal region of Env at position 8328.
Because the SIVmac239 sequence does not contain useful cloning sites in the region coding for the cytoplasmic tail, we were forced to employ a similar PCR strategy for transfer of the mutant glycoproteins from pSRS to the full-length and infectious proviral vector pBR322 SIVmac239 (kindly provided by Toshi Kodama of the Oregon Regional Primate Center). A unique HpaI site was introduced by silent mutation in a region of the genome 3′ to the truncation mutations that encodes only Env (nt 9314 to 9331). The oligonucleotide used to insert the site was 5′-CCCATATTGTAGGTAGGTTAACTCAGTCCTGAG-3′ (the HpaI site is underlined). This oligonucleotide and the ClaI oligonucleotide (above) were used to prime the amplification of a mutagenesis product to be used as a primer in a subsequent amplification versus reverse-complement oligonucleotide overlapping a unique EcoRI site in pBR322 sequences of the SIV proviral vector. The resulting ClaI-EcoRI fragment containing the introduced unique HpaI cloning site was then cloned into pBR322 SIVmac239 to generate the vector pBR SIV-HpaI (see Fig. 1B). We verified that this vector expressed viral protein normally and equivalent to the level of the parental pBR322 SIVmac239 and that virus derived from the pBR SIV-HpaI construct was equivalently infectious.
After sequencing to verify the truncation mutations in the pSRS background, DNA fragments containing the truncations were amplified by using the ClaI oligonucleotide described above and the oligonucleotide containing the silent mutation that introduces the HpaI site. The resultant fragments, containing the truncation mutations, were digested with ClaI and HpaI, gel purified, and cloned into pBR SIV-HpaI. The entire region encompassed by the oligonucleotides was sequenced to verify the mutation and to eliminate clones that had acquired stochastic mutations as a result of the multiple PCR amplifications necessary to generate the constructs.
All plasmid DNAs were propagated in Escherichia coli strain DH5α and purified by cesium chloride isopycnic gradient centrifugation. Concentrations were calculated by UV spectrometry. All PCR employed the error-correcting polymerase Pfu turbo (Stratagene, La Jolla, Calif.).
Glycoprotein expression and immunoprecipitation.
Mutant pSRS constructs were transfected into COS-1 cells using standard calcium phosphate protocols (6). To verify protein expression, processing, and stability, transfected cells were metabolically labeled. Cells were starved for 30 min with leucine-deficient DMEM (Sigma) and labeled with [3H]leucine (250 μCi/ml) (DuPont, NEN, Boston, Mass.) for 1 h at 37°C. The radioactive protein was chased through the secretory pathway by incubating the cells for 4 h in complete medium. At the completion of the chase, the supernatants were removed and filtered through 0.45-μm-pore-size syringe filters. Lysis buffer was added to a final concentration of 1% Nonidet P-40, 0.1% sodium dodecyl sulfate (SDS), and 0.5% deoxycholate, and the lysate was immunoprecipitated using pooled sera from SIV-infected macaques (1:1,000) (kindly provided by Patricia N. Fultz, University of Alabama at Birmingham). The cells were washed once in ice-cold phosphate-buffered saline (PBS) and then lysed in the buffer described above. Nuclei were pelleted at 14,000 rpm in a Microfuge, and the cleared lysates were transferred to new tubes and immunoprecipitated with pooled macaque sera as described above for the supernatants. Immunoprecipitated proteins from the supernatants and the cell lysates were extracted with formalin-fixed Staphylococcus aureus cells (protein A), and the complexes were washed three times with lysis buffer lacking deoxycholate. A final wash in 20 mM Tris, pH 6.8, was used to remove residual detergent, and the complex was denatured in protein loading buffer (50 mM Tris [pH 6.8], 100 mM dithiothreitol, 2% SDS, 0.1% bromophenol blue, and 10% glycerol). Viral glycoproteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) (8% polyacrylamide). The gels were fixed and impregnated with Enhance (Dupont, NEN). Dried gels were fluorographed at −70°C.
Quantitation of surface expression using biotinylation.
For surface labeling of viral Env proteins, transiently transfected COS-1 cells were metabolically labeled with [35S] Cys/Met protein labeling mix (Dupont, NEN). Cells were pulse-labeled for 30 min and chased for 4 h. The supernatants were removed, and the cells were washed twice with ice-cold PBS containing Mg2+ and Ca2+ (PBS-MC). Cell surface glycoproteins were biotinylated in a modification (64) of the method of Lisanti et al. (53). Surface proteins were labeled for 30 min on ice with 2 ml of Sulfo-NHS-SS-biotin (Pierce, Rockford, Ill.) (100 mg/ml in PBS-MC). The biotin solution was removed, and any excess biotin was quenched in three ice-cold washes with serum-free medium supplemented with 20 mM glycine. The biotinylated cells were then lysed on ice with the lysis buffer described above supplemented with 20 mM glycine. Immunoprecipitations were carried out as described above except that 20 mM glycine was present in all washes. Prior to the final Tris(pH 6.8) wash, 25% of the lysate was removed for total protein analysis. The remainder (75%) was subjected to surface protein analysis as follows. The pellet of antigen, antibody, and protein A was boiled for 5 min in 10% SDS to dissociate the antigen-antibody association. The detergent-protein solution was then diluted to 0.1% SDS in PBS with 20 mM glycine and precipitated a second time with monomeric avidin agarose beads (Pierce). The cell surface protein-avidin complex was washed two times with lysis buffer lacking deoxycholate and supplemented with 20 mM glycine. Surface proteins were electrophoresed on 8% polyacrylamide gels. The gels were fixed, dried, and exposed to phosphor screens for quantification of the steady-state surface to total glycoprotein ratio using a phosphorimager (Molecular Dynamics, Sunnyvale, Calif.) and ImageQuant software. Relative surface protein levels were calculated as follows: ([(MS − BS)/(MT − BT)]/[(WTS−BS)/(WTT−BT)]) × 100%, where M represents the phosphorimaging signal from the mutant glycoprotein and WT represents that from the wild type. Subscripts S or T represent the surface or total glycoprotein measurement, and B is the background for each sample measured within the lane of the gel.
Cell-cell fusion assays.
To evaluate the minimal requirements in the TM glycoprotein for cell-to-cell fusion, COS-1 cells were transiently transfected with each pSRS truncation mutant glycoprotein expression construct that also encodes the SIV Tat protein. Twenty-four hours after transfection, the cells were trypsinized and equivalent numbers of cells for each mutant were added, in triplicate, to 70%-confluent R5 MAGI cells in 12-well plates. Transfected cells were added at a ratio of 1/5 unless noted otherwise. The cells were incubated in DMEM for 24 h to allow glycoprotein-mediated fusion to occur. After the incubation, the cells were fixed in 1% formaldehyde–0.2% glutaraldehyde in PBS, washed two times in PBS, and then stained for β-galactosidase using the method described by Kimpton and Emerman (44). Syncytia were counted by visual microscopy at 100× magnification. Two measures were utilized. The total number of syncytia per field was counted for 24 randomly selected fields, and the data were reported as the mean number of blue foci per field. Alternatively, 18 syncytia were selected at random and the total number of nuclei in each syncytium was counted to determine the number of nuclei/syncytia. The median and quartile spreads of values were determined, and Wilcoxon-rank sum analysis was performed to statistically demonstrate the relationship between mutant and wild-type values for nuclei/syncytia.
RT assays.
RT activity from virus-containing supernatants was assayed as previously described (23). Briefly, 25 μl of 0.45 μm-pore-size-filtered culture supernatant was incubated with 75 μl of reaction cocktail for 90 min at 37°C, at which time the reaction was stopped by the addition of 50 μl of 200 mM sodium pyrophosphate. Reaction mixtures were dot blotted onto NA45 membrane (Schleicher & Schuell, Keene, N.H.), washed two times in 0.5 M sodium phosphate, and quantified using a radioanalytic scanning system (AMBIS Systems, San Diego, Calif.).
Infectivity in R5 MAGI or PBMC.
Proviral constructs were transfected into 293T cells for virus production. Supernatants were filtered and assayed for RT activity at 48 h posttransfection. Supernatants were adjusted for equivalent RT activity and volume prior to addition to the target cells and allowed to adsorb for 4 h. The cells were washed to remove unbound particles and were then placed at 37°C. R5 MAGI cells were infected for 30 h and then stained as described above (44). The R5 MAGI infections were carried out in nine replicates, and five fields at ×100 magnification from each replicate infection were counted for a total of 45 fields per mutant. The average and standard deviation of the number of blue foci per 100× field are reported.
Primary macaque mononuclear cells were isolated as described above. PBMC (3 × 106) were infected with RT-and volume-normalized, 0.45-μm-filtered, virus-containing cell supernatants derived from transfection of 293T cells with SIV proviral constructs. After a 4-h adsorption period, the cells were washed with complete medium containing IL-2 and a 1-ml aliquot of medium was removed (day 1). Aliquots were removed every third day and replaced at each sampling with complete medium (see above) containing IL-2. The aliquots of supernatants were stored at −80°C until the time of assay for RT activity. All infections were carried out in triplicate.
Virus pelleting and analysis of glycoprotein incorporation.
Transfection of 293T cells was carried out as above. As a transfection control, supernatants were assayed for RT activity at 48 h posttransfection. To assay for the incorporation of truncated TM proteins, the transfected cells were then starved for 90 min in leucine-deficient DMEM (Sigma, St. Louis, Mo.), followed by a 16-h labeling with 250 μCi/ml of [3H]leucine (Dupont, NEN) in 3 ml of DMEM with 2% fetal bovine sera (Life Technologies). After being labeled, the virus-containing supernatants were harvested and 0.45-μm filtered. The filtrate was pelleted for 90 min at 100,000 × g through 1 ml of a 28% (wt/wt) sucrose cushion. The supernatant was aspirated, and the pellet was lysed for immunoprecipitation as described above. Viral lysates were immunoprecipitated with sera from SIV-infected macaques, and the proteins were separated on SDS–10% polyacrylamide gels. Proteins were visualized by fluorography.
RESULTS
Synthesis and processing of TM glycoprotein truncation mutants.
Mutations in the SIV TM CD and MSD were generated through oligonucleotide-directed mutagenesis. Stop codons were inserted at positions 186 to 194 in the amino acid sequence of SIV TM. A variation of the truncation to 191 amino acids, TM191Ser*, was also constructed to investigate the requirement for a C-terminal basic amino acid to anchor the SIV MSD (Fig. 1). Mutant glycoproteins expressed in COS-1 cells were metabolically labeled, and the viral proteins were immunoprecipitated with pooled sera from SIV-infected macaques. Immunoprecipitated glycoproteins were analyzed by SDS-PAGE followed by autoradiography (Fig. 2A, left panel). The synthesis and processing of mutant glycoproteins were normal over the course of a 4-h chase. The truncated TM glycoproteins migrated with an apparent molecular mass of 28 kDa on SDS-PAGE. The diffuse signal in the radiograph indicates that, despite extensive truncations, the mutant glycoproteins are efficiently and variably glycosylated. Therefore, C-terminal truncation of Env by as many as six amino acids into the putative MSD (TM185) did not alter the synthesis of the glycoprotein precursor or its transport to the Golgi network for glycosylation and cleavage to the SU and TM subunits. Examination of the culture supernatants revealed that the level of shedding of the gp130 subunit from the glycoprotein complex was not enhanced in the presence of the truncation mutations (not shown). Removal of five additional amino acids (TM180) resulted in a protein that was not processed and appeared to be rapidly degraded, likely due to failure to exit the ER (not shown).
FIG. 2.
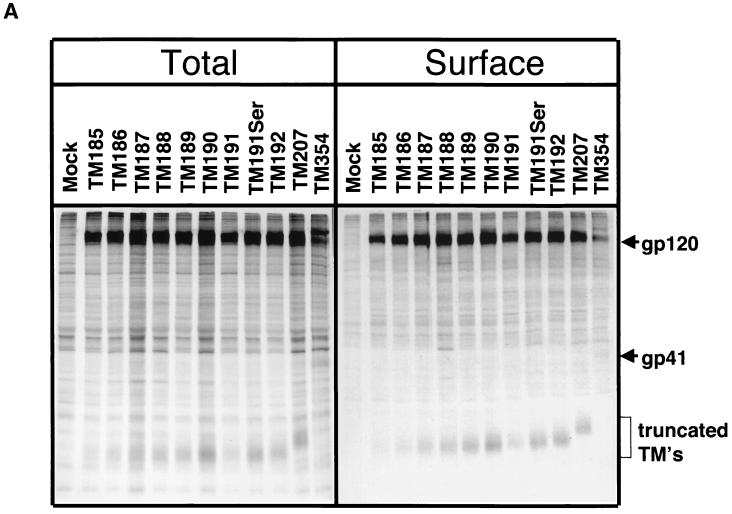
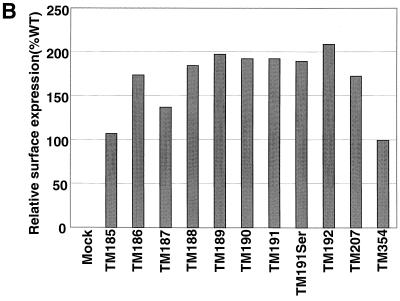
Expression of truncated glycoprotein on the surface of transiently transfected COS-1 cells. Viral glycoproteins were surface labeled with Sulfo-NHS-SS-biotin. (A) Total glycoprotein was immunoprecipitated with SIV-infected macaque sera (left panel). A portion of the immunoprecipitated protein was subjected to streptavidin precipitation to extract the fraction biotinylated and therefore present on the cell surface (right panel). Proteins were separated on 10% polyacrylamide gels. (B) Surface expression relative to the wild type (TM354) for each mutant was calculated by phosphorimager analysis as described in Materials and Methods.
Truncation of the TM glycoprotein into the putative MSD increases surface expression of Env.
Since synthesis and processing of MSD-truncated SIV TM glycoproteins appeared normal, we examined the surface expression of these mutant proteins to determine whether they were transported to, and retained normally in, the plasma membrane. All of the truncation mutations except TM207 removed the SIV TM endocytosis signal centered around Tyr196 (49, 65); therefore, enhanced surface expression of the mutants was anticipated. Indeed, the ratios of surface (biotinylated) to total glycoprotein, as determined by SDS-PAGE and phosphorimaging analysis, indicated that the mutant glycoproteins were expressed on the cell surface at levels ranging from 107 to 209% that of TM354 (Fig. 2B). Therefore, the truncated glycoproteins were present on the cell surface at levels equivalent to or higher than that of the wild-type glycoprotein. Truncation of TM from 207 to 186 amino acids produced little variation in the level of glycoprotein on the cell surface. Thus, it seems likely that, as has been reported for HIV-1, additional motifs C-terminal of this region mediate endocytosis of the wild-type Env protein (74). A mutation in TM that truncates the glycoprotein six amino acids into the putative MSD (TM185) is present on the cell surface at a level equivalent to that of the wild-type. The levels of surface expression are consistent in multiple experiments, and the relative quantitations reported are independent of the protein subunit (SU or TM) used in the calculation of surface-to-total ratios. This finding is further indicative of the maintenance of the SU-TM interaction in the truncated constructs.
Analysis of truncation mutations reveals the minimal sequence necessary to achieve cell-cell fusion.
Because all of the truncation mutants were expressed at nearly equivalent levels on the surface of COS-1 cells, we investigated their ability to mediate cell-cell fusion. Cell-cell fusion was quantified in two ways. In the first method, the number of syncytia in eight fields, from each of three separate transfections and subsequent mixings with R5 MAGI indicator cells, was counted (Fig. 3A). Using this method, truncations of the glycoprotein to 191 amino acids resulted in essentially wild-type levels of fusion; thereafter, fusion decreased progressively with the removal of amino acids. The level of fusion exhibited by TM187 was approximately 50% that of the wild type. Truncation beyond TM 187 resulted in a reduction of fusion by more than 10-fold from the level of the wild type (TM354).
FIG. 3.
Truncated glycoproteins mediate cell-to-cell fusion. Transiently transfected COS-1 cells were mixed with R5 MAGI cells and incubated overnight. The cells were stained for in situ β-galactosidase activity localized in the nucleus. Blue foci were counted for n = 24 fields at ×100. (A) Relative fusion for each of the mutants compared to that mediated by the wild type. The average number of syncytia in mock-transfected wells (0.13/well) was subtracted from each value prior to normalization to the wild-type level. (B) The number of nuclei per syncytia for each mutant was determined by counting the number of nuclei in each multinucleated cell. For TM185, n = 3, since only three syncytia were evident in the entire well in the experiment shown. The box plot shows the maximum and minimum values, denoted by horizontal bars, for each mutant. The first (⧫) and third (□) quartiles (encompassed by the shaded box) and the median value (X) for each mutant are shown. Wilcoxon rank sum analysis was performed to statistically compare each mutant to the wild type (TM354). The probabilities associated with such comparisons are shown along with the sample size in parentheses.
As an alternative quantification of glycoprotein-mediated fusion, the number of nuclei/syncytium for 18 randomly selected syncytia in single wells was determined. For the mock-transfected cells, no multinucleated cells exhibiting blue nuclear staining were identified. As described above, truncation of TM to 191 amino acids resulted in syncytia that were essentially equivalent in size (maximum and average) to those produced by the wild-type glycoprotein (Fig. 3B). The median size of syncytia formed by glycoproteins 191 amino acids or longer was four to six nuclei, with considerable variability inherent in the assay. Truncation of the glycoprotein beyond 191 amino acids resulted in a reduction in the median syncytium size, as well as a dramatic reduction in the upper limit of nuclei/syncytium; no syncytia larger than seven nuclei were observed for TM185 to TM190. The difference between the median number of nuclei/syncytium for each of the mutants TM185 to TM190 and that of the wild type (TM 354) was significant when assessed in a Wilcoxon rank sum analysis (Fig. 3B). Interestingly, the sizes of syncytia generated by TM187 were not significantly different from those produced by TM186 (P = 0.749) (Fig. 3B), despite the fact that the abundance of syncytia per well was approximately fivefold that of TM186.
Truncation mutants within the putative MSD are replication competent.
To facilitate analysis of the impact of the truncations on virus infectivity, each mutation was transferred into the proviral vector pBR SIV-HpaI (Fig. 1B), and virions produced by transfection of 293T cells were used to infect R5 MAGI cells. The time course of the R5 MAGI cell infection was limited to prevent virus from spreading throughout the culture and creating secondary fusion events. This regimen produced largely two- and three-cell foci separated by large numbers of uninfected cells, indicating that each blue syncytium represented an individual infectious event. As in the glycoprotein-mediated fusion assays, levels of infectivity indistinguishable from that of wild-type were evident in truncations to 191 amino acids (Fig. 4A). A three-fold reduction in the level of infectivity was observed when the TM glycoprotein was truncated to 190 amino acids. Despite the reduction in virus infectivity, the TM190 virus displayed infectivity 20-fold that of the background mock-transfected supernatant. Removal of an additional amino acid, TM189, resulted in a reduction in the level of infectivity to 20% that of the wild type and significant variation in the number of infectious events per field. The degree of statistical error associated with the TM189 virus reflects the fact that some of the (n = 45) fields examined did not contain blue foci. The substitution of serine for lysine at position 191 had no effect on the level of infectivity. Viruses containing truncations TM187 and TM188, despite incorporation of a fusogenic glycoprotein, were not capable of mediating infection in this indicator system, clearly demonstrating that the amino acid requirements for fusion and those for infectivity are distinct and separable.
FIG. 4.
Infectivity in R5 MAGI cells and macaque PBMC. (A) Proviral constructs containing the TM truncation mutations were transfected into 293T cells. Virus-containing supernatants were titered by RT analysis. RT-normalized and volume-adjusted supernatants were added to replicate wells of R5 MAGI cells. The cells were incubated for 30 h prior to fixation and staining in situ for β-galactosidase activity. Ten fields at ×100 magnification were counted for each construct, and the results are reported as the mean number of blue foci per field. (B) Concanavalin A-stimulated macaque PBMC were infected with a normalized quantity of each SIV mutant-containing supernatant in triplicate (described in Materials and Methods). Supernatants from the infected macaque cultures were evaluated for RT activity after 15 days in culture.
Mutants containing truncations in the putative MSD replicate in macaque PBMC.
SIV constructs containing C-terminal truncation mutations at residues predicted to lie within the putative MSD were able to infect R5 MAGI cells and also the T-cell line CEMx174 (not shown). To extend this finding, we assessed whether viruses containing these mutations were capable of replication in primary macaque PBMC.
Analysis of supernatant RT activity from infections of macaque PBMC with SIV truncation mutants revealed that the minimum TM amino acid sequence requirement for replication was 189 amino acids (Fig. 4B). This result is consistent with our infectivity studies in R5 MAGI cells (Fig. 4A), where TM189 appeared to be attenuated but capable of virus entry and propagation. The level of TM189 replication in PBMC was equivalent to that of the wild type, whereas in R5 MAGI cells, the level of infectivity was highly variable. Replication kinetics for SIV TM189 in the CEMx174 cell line displayed a delay, as we have noted previously for other truncations (reference 40 and data not shown). As in the R5 MAGI infectivity experiments (Fig. 4A), replication in PBMC also had an absolute requirement for sequences in TM since TM187 and TM188 did not replicate above background levels. The ability of SIV to replicate to wild-type levels in primary PBMC in vitro in the absence of lysine 191 indicated that the glycoprotein function was unaffected by loss of an amino acid that had previously been predicted to be important as a stop-translocation signal. These results suggest that the minimum requirements for membrane anchorage, fusion, and infectivity are distinct within the SIV TM glycoprotein and reside in sequences previously predicted to span the membrane.
Truncated glycoproteins are efficiently incorporated into virions despite the loss of putative MSD sequences.
The infectivity results described above were generated using virus-containing supernatants derived from proviral transfection of 293T cells. Because the inability to infect target cells is often a defect related to failure of mutant glycoproteins to be efficiently incorporated into virus particles, we evaluated the level of glycoprotein incorporation into virions produced from 293T cells. To investigate whether the TM truncation mutants were incorporated into particles, we used 3[H]leucine to metabolically label virus (Fig. 5). The wild-type TM354 glycoprotein contains 49 leucine residues, whereas the truncated TM glycoproteins contain 24 (TM185) to 26 (TM207) leucine residues. The truncated TM glycoprotein, which migrates as a diffuse band of approximately 28 kDa, was clearly visible in immunoprecipitates prepared from pelleted virions. Virus from SIV TM207 contained a similar protein that is 30 to 32 kDa and, thus, provided for identification of the diffuse bands as truncated TM rather than one of the viral accessory glycoproteins. The wild-type TM glycoprotein is approximately 41 kDa and is efficiently incorporated into virus derived from 293T cells. Given that truncations TM185, TM186, TM187, and TM188 appear to be incorporated into virions at levels similar to that of the wild type, it is unlikely that differences in glycoprotein incorporation are the cause of their failure to infect target cells.
FIG. 5.
Incorporation of truncated glycoproteins into SIV virions. Proviral expression constructs containing the SIV MSD truncation mutations were transfected into 293T cells for virus production. The cells were labeled 48 h posttransfection for 16 h with [3H]leucine. Labeled virus particles were pelleted at 100,000 × g through a 28% sucrose cushion. The pellets were lysed and immunoprecipitated as described in Materials and Methods. Immunoprecipitates were analyzed on 10% polyacrylamide gels. The arrows denote the positions of p27 CA, gp120, gp41, and the truncated TM glycoproteins.
DISCUSSION
Defining the precise length and structure of the lentiviral TM glycoprotein MSD is critical to understanding how the protein is synthesized on the ER, folds, and is transported through the secretory pathway. Moreover, this domain is required for anchorage on cellular or viral membranes and is essential for biological activity of the glycoprotein. MSDs have traditionally been assigned on the basis of the hydrophobic moment and amphipathicity of helices and on an ability to neutralize intramembrane charges, if present (27, 34). Unfortunately, definitive atomic structural determinations exist for only a few MSD, chiefly those for the Rhodobacter viridis photosynthetic reaction center and for bacteriorhodopsin (20, 37). Thus, the exact location of and requirements for the MSDs of HIV-1 and SIV have proven difficult to determine.
The predicted MSD of the TM glycoproteins from both SIV and HIV contains a charged arginine residue centrally located in a long hydrophobic stretch of amino acids (Fig. 6). All previous assignments for the MSDs of these TM glycoproteins have placed the arginine residue within the lipid bilayer without explanation of a mechanism by which such a structure would be stabilized (12, 30). It is possible that the lentivirus intramembrane arginine residues may be involved in protein-protein interactions as have been described for assembly and surface expression of the T-cell receptor complex. This complex is stabilized in the secretory pathway through salt bridges between acidic residues in the MSD of CD3 and the basic residues in the MSD helices of T-cell receptor proteins (16). While such an interaction has not been demonstrated for lentivirus Env proteins, we cannot exclude such a possibility at this time, given the promiscuous incorporation of cellular proteins into virus particles. Mutational studies have not resulted in a consensus opinion as to whether the arginine residue is essential to glycoprotein function. Helseth and colleagues suggested that the basic residue is not essential for HIV-1 biological activity since replacement of the arginine at residue 696 with serine did not alter fusogenicity (35). In contrast, Owens et al. (59) described a series of point mutations and small deletions near and encompassing the HIV-1 intramembrane arginine, all of which significantly reduced fusion.
FIG. 6.
Two alternative structures for the SIV MSD. The structure on the right represents an SIV membrane-spanning helix predicted from Kyte-Doolittle analysis of the SIV TM glycoprotein as described by Chakrabarti et al. (12). The membrane-spanning region is from lysine 164 to lysine 191 and contains an arginine in the membrane at position 180. The truncation mutations are indicated and refer to the C-terminal amino acid prior to the stop codon (i.e., TM193 is produced by conversion of amino acid 194 to a stop). An alternative structure supported by the data presented in this study is shown on the left. This model provides a potential mechanism for reduction in the size of the membrane anchor and neutralization of the intramembrane arginine side chain. Interactions of the side chains of arginine 180 and lysine 164 with the phospholipid head groups are indicated by the heavy black lines and the + charge.
Truncation mutations in the SIV cytoplasmic tail, such as those observed following growth in human cells, increase glycoprotein expression on the cell surface, alter replication kinetics (in certain cells), induce higher levels of fusion, and result in higher levels of incorporation into virus particles (40, 49, 61, 70, 76). The effective or actual loss of a constitutive Tyr-based endocytosis signal in the CD at position Tyr196 is apparently responsible for many of these effects (65). We also demonstrated that further truncation of the SIV TM to 193 amino acids resulted in virus that was replication competent in vitro despite a nearly complete lack of CD sequences (40).
Here we have shown that it is possible to truncate the SIV TM glycoprotein into the putative MSD while maintaining different levels of biological function with successive reductions in length. We demonstrated that all of the glycoprotein truncations up to amino acid 185, within the putative MSD, are efficiently synthesized, translocated into the ER, and transported through the Golgi where the polyprotein is cleaved to the SU and TM subunits. This result indicates that the glycoprotein mutants are securely anchored in the membrane throughout the secretory pathway. More importantly, all of the constructs are expressed on the cell surface at levels equivalent to, or greater than, that of the wild-type TM. In analyses of supernatants, we did not observe differences in SU shedding for any of the truncated constructs (data not shown). Further truncation of the SIV TM up to the putative intramembrane arginine residue at position 180 (Fig. 1) resulted in a pronounced transport defect. This mutant, TM180, is not expressed on the cell surface and in preliminary experiments appears to be retained in the ER. The location of the TM180 Env product has proven difficult to determine conclusively, however, since the protein also appears to be rapidly degraded (our unpublished results).
All of the truncated Env proteins, with the exception of TM180, were incorporated into virus particles at levels similar to that of the wild-type Env. This provides further evidence that the mutant constructs are transported normally and are efficient substrates for inclusion into assembling particles. Infectivity, as analyzed in single-round infections in R5 MAGI cells and by replication in macaque PBMC, required a TM glycoprotein of at least 189 amino acids. The level of replication for the TM189-containing virus in PBMC was equivalent to that of the wild type, although in single-cycle R5 MAGI assays, virus containing this truncated Env was significantly attenuated. These data argue that the entire CD and three amino acids of the putative MSD are dispensable for SIV replication.
Further truncation of the glycoprotein abrogated infectivity in both R5 MAGI cells and PBMC. In contrast, cell-cell fusion activity was retained in mutant Env proteins with TM domains of 187 and 188 amino acids. The level of fusion, as defined by the number of syncytia/well, decreased from wild-type levels (TM191) with each successive truncation. When the number of nuclei/syncytium was investigated, high variability was observed in both the median and maximum size of syncytia formed; nevertheless, there was a reduction in both parameters as the truncation extended from TM191 to TM187.
These studies have allowed us to dissect the minimal carboxyl-terminal requirements in the SIV TM glycoprotein for infectivity in vitro, for cell-cell fusion, and for glycoprotein transport. We have shown that each function of the glycoprotein is maintained by a distinct sequence in the TM. Interestingly, the requirement for infectivity (TM189) is more stringent than that for fusion (TM187), which in turn is more stringent than that for membrane anchorage and intracellular glycoprotein transport (TM185). The observation that truncation mutants that are competent for glycoprotein transport and cell surface expression (TM185 and TM186) are highly inefficient in mediating fusion argues that sequences in addition to those necessary to arrest translocation and anchor the protein in the membrane are required for membrane fusion to occur. It is possible that these additional residues are required at some, as yet undefined, early step in fusion, such as the formation or resolution of a hemifusion diaphragm, in order to mediate cytoplasmic mixing. We have not yet resolved why fusogenic Env molecules that are incorporated into virus particles (TM187 and TM188) are defective in mediating virus entry. Env fusogenicity decreases with the length of the TM domain, and, if the probability that sufficient functional Env complexes associate to generate a fusion pore is a stochastic event, it is likely that the efficiency of fusion depends on the effective concentration of functional Env molecules. In cell-to-cell fusion, this constraint may not be so important since additional Env molecules will be constantly transported to the plasma membrane, but in a virus where the number and density of Env molecules are low and fixed, this may be critical.
In an effort to reconcile the data presented here with the results of previous mutagenesis studies of the SIV and HIV MSD and CD as well as with solved membrane-spanning structures, we propose an alternative model for the SIV TM MSD. The assignment of a basic charged residue to the middle of lentivirus predicted MSD sequences is inconsistent with thermodynamic principles of MSDs, and, thus, the border between the C-terminal end of the MSD and the CD has remained unclear. Our model provides an explanation for how truncations, into a region previously thought to lie within the membrane, might maintain biological functionality. Application of the known structures of the photosynthetic reaction center and bacteriorhodopsin together with their amino acid composition within the membrane, provides support for this reorganization of the SIV MSD structure (20, 37). In the photosynthetic reaction center and bacteriorhodopsin a number of basic residues appear to reside in the membrane phase with their side chains positioned to interact with the negatively charged lipid head groups (8, 37). The potential interaction of the basic side chains with the lipid head groups eliminates the requirement for neutralization by an acidic amino acid partner, such as those described in other systems (15, 16). Moreover, the length of arginine/lysine side chains allows for these amino acids to reside as much as 1.5 turns within the MSD and still be neutralized and provide anchorage (8). Indeed, a recent report suggests that basic residues in integrin subunits occupy just such internal positions in the MSD, as determined by a glycosylation mapping technique (5). In our model, the intramembrane arginine 180 residue and perhaps lysine 163 are accommodated within the MSD, providing for a reduction in the length of sequence required to span the bilayer. This hypothesis is supported by the studies of Adams and Rose (2) on the VSV G MSD, which demonstrated that an MSD as short as 16 amino acids was sufficient to anchor the G glycoprotein and allow fusion.
Locating the C-terminal arginine (Arg180) close to the inner boundary of the membrane would effectively extend the CD of gp41. This is consistent with the observations of Sauter et al. (65). These investigators fused the 16 membrane-proximal amino acids (TM residues 191 to 207) of the SIV CD, which contain a well-characterized endocytosis motif (Y196RPV), to the ectodomain and MSD of CD4 and found that the SIV sequences inefficiently mediated endocytosis of CD4. In order for the endocytosis motif to function, it was necessary to precede the SIV sequence with four residues (RCRH) of the CD4 CD. This spacing placed the endocytosis motif seven to eight residues from the MSD, in a context similar to the minimum determined for active endocytosis in other systems (14). The model presented here would also position the tyrosine-based motif at a similar distance from the membrane, consistent with the efficient endocytosis of SIV Env.
If our model is correct, it may be that induction of fusion, such as the conformational changes observed on binding of CD4 and coreceptor (41, 47), requires only those amino acids necessary to form a membrane anchor, but that resolution of a fusion intermediate requires a longer MSD. Recent studies by Armstrong et al. (4), using the influenza virus hemagglutinin (HA) glycoprotein, which demonstrated a stringent length requirement for the hemifusion-to-fusion transition, support this concept. It is also possible that the additional amino acids are required to support an interaction essential to the formation of a fusion pore. Recent data from the study of fusion with influenza virus HA have suggested that the formation of a fusion pore requires a minimal recruitment of glycoprotein oligomers before content mixing is possible (9, 13, 26). It may be that sequences C-terminal to residue 185 provide for the formation of superoligomeric complexes necessary to completely fuse membranes.
Despite the determination of the minimal C-terminal requirements in the SIV TM for the in vitro biological activities described above, it remains unclear whether the specific sequence of the SIV MSD is required for a fully pathogenic virus. The entire MSD of HIV can be replaced with that of the nonfusogenic proteins, CD4 and CD22, which are devoid of charged residues in the MSD (60, 73). Nevertheless, given the conservation of basic charged residues at identical positions in the HIV and SIV MSDs, it seems likely that the MSD sequence or at least the charged residues play an important functional role in the virus life cycle. Thus, while it may be possible to substitute heterologous domains in place of the MSD and generate viruses that replicate in vitro, such chimeras are likely to be attenuated in vivo. The SIV MSD has been selected and conserved to provide functionality to the protein in the processes of membrane fusion and virus entry. The domain is also essential in synthesis and transport of Env during virus assembly. In the absence of atomic structure data for the MSD, our study has allowed the assignment of biological functions to specific sub-domains within this region. Further analyses, particularly with respect to the fusion reaction, are under way to determine how the different truncations of the SIV TM affect the separate steps of membrane fusion.
ACKNOWLEDGMENTS
We are grateful to Patricia Fultz for macaque primary cells and SIV-infected macaque sera. Our sincerest thanks to Toshi Kodama for providing the original pBR322 SIVmac239 clone. We also thank Mike Sakalian for helpful discussions and critical reading of the manuscript. Jeannette Lee of the UAB Center for AIDS Research Biostatistics Core supported by grant P30-AI27767 provided valuable assistance with the statistical analyses.
This work was supported by U.S. Public Health Service grant R37AI33319 to E.H. J.T.W. was supported on NIH Training Grant T32AI07493. Experiments were performed in the Central Virus Culture Core and Molecular Biology Core of the UAB Center for AIDS Research, supported by grant P30-AI27767.
REFERENCES
- 1.Adams G A, Rose J K. Incorporation of a charged amino acid into the membrane-spanning domain blocks cell surface transport but not membrane anchoring of a viral glycoprotein. Mol Cell Biol. 1985;5:1442–1448. doi: 10.1128/mcb.5.6.1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adams G A, Rose J K. Structural requirements of a membrane-spanning domain for protein anchoring and cell surface transport. Cell. 1985;41:1007–1015. doi: 10.1016/s0092-8674(85)80081-7. [DOI] [PubMed] [Google Scholar]
- 3.Akari H, Fukumori T, Adachi A. Cell-dependent requirement of human immunodeficiency virus type 1 cytoplasmic tail for Env incorporation into virions. J Virol. 2000;74:4891–4893. doi: 10.1128/jvi.74.10.4891-4893.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Armstrong R T, Kushnir A S, White J M. The transmembrane domain of influenza hemagglutinin exhibits a stringent length requirement to support the hemifusion to fusion transition. J Cell Biol. 2000;151:425–438. doi: 10.1083/jcb.151.2.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Armulik A, Nilsson I, von Heijne G, Johansson S. Determination of the border between the transmembrane and cytoplasmic domains of human integrin subunits. J Biol Chem. 1999;274:37030–37034. doi: 10.1074/jbc.274.52.37030. [DOI] [PubMed] [Google Scholar]
- 6.Ausubel F M, Brent R, Kingston R E, Moore D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Greene Publishing Associates, Wiley-Interscience; 1987. [Google Scholar]
- 7.Baier M, Werner A, Cichutek K, Garber C, Muller C, Kraus G, Ferdinand F J, Hartung S, Papas T S, Kurth R. Molecularly cloned simian immunodeficiency virus SIVagm3 is highly divergent from other SIVagm isolates and is biologically active in vitro and in vivo. J Virol. 1989;63:5119–5123. doi: 10.1128/jvi.63.12.5119-5123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ballesteros J, Weinstein H. Analysis and refinement of criteria for predicting the structure and relative orientations of transmembranal helical domains. Biophys J. 1992;62:107–109. doi: 10.1016/S0006-3495(92)81794-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bentz J. Minimal aggregate size and minimal fusion unit for the first fusion pore of influenza hemagglutinin-mediated membrane fusion. Biophys J. 2000;78:227–245. doi: 10.1016/S0006-3495(00)76587-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berman P W, Nunes W M, Haffar O K. Expression of membrane-associated and secreted variants of gp160 of human immunodeficiency virus type 1 in vitro and in continuous cell lines. J Virol. 1988;62:3135–3142. doi: 10.1128/jvi.62.9.3135-3142.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chakrabarti L, Emerman M, Tiollais P, Sonigo P. The cytoplasmic domain of simian immunodeficiency virus transmembrane protein modulates infectivity. J Virol. 1989;63:4395–4403. doi: 10.1128/jvi.63.10.4395-4403.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chakrabarti L, Guyader M, Alizon M, Daniel M D, Desrosiers R C, Tiollais P, Sonigo P. Sequence of simian immunodeficiency virus from macaque and its relationship to other human and simian retroviruses. Nature. 1987;328:543–547. doi: 10.1038/328543a0. [DOI] [PubMed] [Google Scholar]
- 13.Chernomordik L, Leikina E, Kozlov M M, Frolov V A, Zimmerberg J. Structural intermediates in influenza haemagglutinin-mediated fusion. Mol Membr Biol. 1999;16:33–42. doi: 10.1080/096876899294733. [DOI] [PubMed] [Google Scholar]
- 14.Collawn J F, Stangel M, Kuhn L A, Esekogwu V, Jing S Q, Trowbridge I S, Tainer J A. Transferrin receptor internalization sequence YXRF implicates a tight turn as the structural recognition motif for endocytosis. Cell. 1990;63:1061–1072. doi: 10.1016/0092-8674(90)90509-d. [DOI] [PubMed] [Google Scholar]
- 15.Cosson P, Bonifacino J. Role of transmembrane domain interactions in the assembly of class II MHC molecules. Science. 1992;258:659–662. doi: 10.1126/science.1329208. [DOI] [PubMed] [Google Scholar]
- 16.Cosson P, Lankford S P, Bonifacino J, Klausner R D. Membrane protein association by potential intramembrane charge pairs. Nature. 1991;351:414–416. doi: 10.1038/351414a0. [DOI] [PubMed] [Google Scholar]
- 17.Daniel M D, Letvin N L, King N W, Kannagi M, Sehgal P K, Hunt R D, Kanki P J, Essex M, Desrosiers R C. Isolation of T-cell tropic HTLV-III-like retrovirus from macaques. Science. 1985;228:1201–1204. doi: 10.1126/science.3159089. [DOI] [PubMed] [Google Scholar]
- 18.Daniel M D, Li Y, Naidu Y M, Durda P J, Schmidt D K, Troup C D, Silva D P, MacKey J J, Kestler H W, Sehgal P K, King N W, Ohta Y, Hayami M, Desrosiers R C. Simian immunodeficiency virus from African green monkeys. J Virol. 1988;62:4123–4128. doi: 10.1128/jvi.62.11.4123-4128.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Decroly E, Vandenbranden M, Ruysschaert J M, Cogniaux J, Jacob G S, Howard S C, Marshall G, Kompelli A, Basak A, Jean F. The convertases furin and PC1 can both cleave the human immunodeficiency virus (HIV)-1 envelope glycoprotein gp160 into gp120 (HIV-1 SU) and gp41 (HIV-1 TM) J Biol Chem. 1994;269:12240–12247. [PubMed] [Google Scholar]
- 20.Deisenhofer J, Michel H. The photosynthetic reaction center from purple bacterium Rhodopseudomonas viridis. Science. 1989;245:1463–1473. doi: 10.1126/science.245.4925.1463. [DOI] [PubMed] [Google Scholar]
- 21.di Marzo Veronese F, DeVico A L, Copeland T D, Oroszlan S, Gallo R C, Sarngadharan M G. Characterization of gp41 as the transmembrane protein coded by the HTLV-III/LAV envelope gene. Science. 1985;229:1402–1405. doi: 10.1126/science.2994223. [DOI] [PubMed] [Google Scholar]
- 22.di Marzo Veronese F, Joseph B, Copeland T D, Oroszlan S, Gallo R C, Sarngadharan M G. Identification of simian immunodeficiency virus SIVmac env gene products. J Virol. 1989;63:1416–1419. doi: 10.1128/jvi.63.3.1416-1419.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dubay J W, Roberts S J, Hahn B H, Hunter E. Truncation of the human immunodeficiency virus type 1 transmembrane glycoprotein cytoplasmic domain blocks virus infectivity. J Virol. 1992;66:6616–6625. doi: 10.1128/jvi.66.11.6616-6625.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Earl P L, Koenig S, Moss B. Biological and immunological properties of human immunodeficiency virus type 1 envelope glycoprotein: analysis of proteins with truncations and deletions expressed by recombinant vaccinia viruses. J Virol. 1991;65:31–41. doi: 10.1128/jvi.65.1.31-41.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Earl P L, Moss B. Mutational analysis of the assembly domain of the HIV-1 envelope glycoprotein. AIDS Res Hum Retrovir. 1993;9:589–594. doi: 10.1089/aid.1993.9.589. [DOI] [PubMed] [Google Scholar]
- 26.Ellens H, Bentz J, Mason D, Zhang F, White J M. Fusion of influenza hemagglutinin-expressing fibroblasts with glycophorin-bearing liposomes: role of hemagglutinin surface density. Biochemistry. 1990;29:9697–9707. doi: 10.1021/bi00493a027. [DOI] [PubMed] [Google Scholar]
- 27.Fasman G D, Gilbert W A. The prediction of transmembrane protein sequences and their conformation: an evaluation. Trends Biol Sci. 1990;15:89–91. doi: 10.1016/0968-0004(90)90187-g. [DOI] [PubMed] [Google Scholar]
- 28.Fauci A S. Multifactorial nature of of human immunodeficiency virus disease: implications for therapy. Science. 1993;262:1011–1018. doi: 10.1126/science.8235617. [DOI] [PubMed] [Google Scholar]
- 29.Freed E O, Martin M A. Domains of the human immunodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J Virol. 1996;70:341–351. doi: 10.1128/jvi.70.1.341-351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gabuzda D, Olshevsky U, Bertani P, Haseltine W A, Sodroski J. Identification of membrane anchorage domains of the HIV-1 gp160 envelope glycoprotein precursor. J Acquired Immune Defic Syndr. 1991;4:34–40. [PubMed] [Google Scholar]
- 31.González S A, Affranchino J L, Gelderblom H R, Burny A. Assembly of the matrix protein of simian immunodeficiency virus into virus-like particles. Virology. 1993;194:548–556. doi: 10.1006/viro.1993.1293. [DOI] [PubMed] [Google Scholar]
- 32.Haffar O K, Dowbenko D J, Berman P W. Topogenic analysis of the human immunodeficiency virus type 1 envelope glycoprotein, gp160, in microsomal membranes. J Cell Biol. 1988;107:1677–1687. doi: 10.1083/jcb.107.5.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hallenberger S, Bosch V, Angliker H, Shaw E, Klenk H-D, Garten W. Inhibition of furin-mediated cleavage activation of HIV-1 glycoprotein gp160. Nature. 1992;360:358–361. doi: 10.1038/360358a0. [DOI] [PubMed] [Google Scholar]
- 34.Hartmann E, Rapoport T A, Lodish H F. Predicting the orientation of eukaryotic membrane-spanning proteins. Proc Natl Acad Sci USA. 1989;86:5786–5790. doi: 10.1073/pnas.86.15.5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Helseth E, Olshevsky U, Gabuzda D, Ardman B, Haseltine W, Sodroski J. Changes in the transmembrane region of the human immunodeficiency virus type 1 gp41 envelope glycoprotein affect membrane fusion. J Virol. 1990;64:6314–6318. doi: 10.1128/jvi.64.12.6314-6318.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henderson L E, Benveniste R E, Sowder R, Copeland T D, Schultz A M, Oroszlan S. Molecular characterization of Gag proteins from simian immunodeficiency virus (SIVMne) J Virol. 1988;62:2587–2595. doi: 10.1128/jvi.62.8.2587-2595.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Henderson R, Baldwin J M, Ceska T A, Zemlin F, Beckman E, Downing K H. Model for the structure of bacteriorhodopsin based on high resolution electron cryo-microscopy. J Mol Biol. 1990;213:899–929. doi: 10.1016/S0022-2836(05)80271-2. [DOI] [PubMed] [Google Scholar]
- 38.Hirsch V M, Edmondson P, Murphey-Corb M, Arbeille B, Johnson P R, Mullins J I. SIV adaptation to human cells. Nature. 1989;341:573–574. doi: 10.1038/341573a0. [DOI] [PubMed] [Google Scholar]
- 39.Hosaka M, Nagahama M, Kim W S, Watanabe T, Hatsuzawa K, Ikemizu J, Murakami K, Nakayama K. Arg-X-Lys/Arg-Arg motif as a signal for precursor cleavage catalyzed by furin within the constitutive secretory pathway. J Biol Chem. 1991;266:12127–12130. [PubMed] [Google Scholar]
- 40.Johnston P B, Dubay J W, Hunter E. Truncations of the simian immunodeficiency virus transmembrane protein confer expanded virus host range by removing a block to virus entry into cells. J Virol. 1993;67:3077–3086. doi: 10.1128/jvi.67.6.3077-3086.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jones P L, Korte T, Blumenthal R. Conformational changes in cell surface HIV-1 envelope glycoproteins are triggered by cooperation between cell surface CD4 and co-receptors. J Biol Chem. 1998;273:404–409. doi: 10.1074/jbc.273.1.404. [DOI] [PubMed] [Google Scholar]
- 42.Karacostas V, Nagashima K, Gonda M A, Moss B. Human immunodeficiency virus-like particles produced by a vaccinia virus expression vector. Proc Natl Acad Sci USA. 1989;86:8964–8967. doi: 10.1073/pnas.86.22.8964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kemble G W, Danieli T, White J M. Lipid-anchored influenza hemagglutinin promotes hemifusion not complete fusion. Cell. 1994;76:383–391. doi: 10.1016/0092-8674(94)90344-1. [DOI] [PubMed] [Google Scholar]
- 44.Kimpton J, Emerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated β-galactosidase gene. J Virol. 1992;66:2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kodama T, Wooley D P, Naidu Y M, Kestler H W I, Daniel M D, Li Y, Desrosiers R C. Significance of premature stop codons in env of simian immunodeficiency virus. J Virol. 1989;63:4709–4714. doi: 10.1128/jvi.63.11.4709-4714.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kowalski M, Potz J, Basiripour L, Dorfman T, Goh W C, Terwilliger E, Dayton A, Rosen C, Haseltine W, Sodroski J. Functional regions of the envelope glycoprotein of human immunodeficiency virus type 1. Science. 1987;237:1351–1355. doi: 10.1126/science.3629244. [DOI] [PubMed] [Google Scholar]
- 47.Kuhmann S E, Platt E J, Kozak S L, Kabat D. Cooperation of multiple CCR5 coreceptors is required for infections by human immunodeficiency virus type 1. J Virol. 2000;74:7005–7015. doi: 10.1128/jvi.74.15.7005-7015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kyte J, Doolittle R. A simple method for displaying the hydrophobic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 49.LaBranche C C, Sauter M M, Haggarty B S, Vance P J, Romano J, Hart T K, Bugelski P J, Marsh M, Hoxie J A. A single amino acid change in the cytoplasmic domain of the simian immunodeficiency virus transmembrane molecule increases envelope glycoprotein expression on infected cells. J Virol. 1995;69:5217–5227. doi: 10.1128/jvi.69.9.5217-5227.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Letvin N L, Daniel M D, Sehgal P K, Desrosiers R C, Hunt R D, Waldron L M, MacKey J J, Schmidt D K, Chalifoux L V, King N W. Induction of AIDS-like disease in macaque monkeys with T-cell tropic retrovirus STLV-III. Science. 1985;230:71–73. doi: 10.1126/science.2412295. [DOI] [PubMed] [Google Scholar]
- 51.Letvin N L, King N W. Immunologic and pathologic manifestations of the infection of rhesus monkeys with simian immunodeficiency virus of macaques. J Acquir Immune Defic Syndr. 1990;3:1023–1040. [PubMed] [Google Scholar]
- 52.Li Y, Naidu Y M, Daniel M D, Desrosiers R C. Extensive genetic variability of simian immunodeficiency virus from African green monkeys. J Virol. 1989;63:1800–1802. doi: 10.1128/jvi.63.4.1800-1802.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lisanti M P, Sargiacomo M, Graeve L, Saltiel A R, Rodriguez-Boulan E. Polarized apical distribution of glycosyl-phosphatidylinositol-anchored proteins in a renal epithelial cell line. Proc Natl Acad Sci USA. 1988;85:9557–9561. doi: 10.1073/pnas.85.24.9557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luo L, Li Y, Kang C Y. Expression of gag precursor protein and secretion of virus-like gag particles of HIV-2 from recombinant baculovirus-infected insect cells. Virology. 1990;179:874–880. doi: 10.1016/0042-6822(90)90159-o. [DOI] [PubMed] [Google Scholar]
- 55.Melikyan G B, White J M, Cohen F S. GPI-anchored influenza hemagglutinin induces hemifusion to both red blood cell and planar bilayer membranes. J Cell Biol. 1995;131:679–691. doi: 10.1083/jcb.131.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mulligan M J, Yamshchikov G V, Ritter G D J, Gao F, Jin M J, Nail C D, Spies C P, Hahn B H, Compans R W. Cytoplasmic domain truncation enhances fusion activity by the exterior glycoprotein complex of human immunodeficiency virus type 2 in selected cell types. J Virol. 1992;66:3971–3975. doi: 10.1128/jvi.66.6.3971-3975.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murakami T, Freed E O. The long cytoplasmic tail of gp41 is required in a cell type-dependent manner for HIV-1 envelope glycoprotein incorporation into virions. Proc Natl Acad Sci USA. 2000;97:343–348. doi: 10.1073/pnas.97.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Odell D, Wanas E, Yan J, Ghosh H P. Influence of membrane anchoring and cytoplasmic domains on the fusogenic activity of vesicular stomatitis virus glycoprotein G. J Virol. 1997;71:7996–8000. doi: 10.1128/jvi.71.10.7996-8000.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Owens R J, Burke C, Rose J K. Mutations in the membrane-spanning domain of the human immunodeficiency virus envelope glycoprotein that affect fusion activity. J Virol. 1994;68:570–574. doi: 10.1128/jvi.68.1.570-574.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Raja N U, Vincent M J, Abdul Jabbar M. Vpu-mediated proteolysis of gp160/CD4 chimeric envelope glycoproteins in the endoplasmic reticulum: requirement of both the anchor and cytoplasmic domains of CD4. Virology. 1994;204:357–366. doi: 10.1006/viro.1994.1540. [DOI] [PubMed] [Google Scholar]
- 61.Ritter G D, Mulligan M J, Lydy S L, Compans R W. Cell fusion activity of the simian immunodeficiency virus is modulated by the intracytoplasmic domain. Virology. 1993;197:255–264. doi: 10.1006/viro.1993.1586. [DOI] [PubMed] [Google Scholar]
- 62.Rowell J F, Stanhope P E, Siliciano R F. Endocytosis of the HIV-1 envelope protein: mechanism and role in processing for association with class II MHC. J Immunol. 1995;155:473–488. [PubMed] [Google Scholar]
- 63.Salzwedel K, Johnston P B, Roberts S J, Dubay J W, Hunter E. Expression and characterization of glycophospholipid-anchored human immunodeficiency virus type 1 envelope glycoproteins. J Virol. 1993;67:5279–5288. doi: 10.1128/jvi.67.9.5279-5288.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Salzwedel K, West J T, Jr, Mulligan M J, Hunter E. Retention of the human immunodeficiency virus type 1 envelope glycoprotein in the endoplasmic reticulum does not redirect virus assembly from the plasma membrane. J Virol. 1998;72:7523–7531. doi: 10.1128/jvi.72.9.7523-7531.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sauter M M, Pelchen-Matthews A, Bron R, Marsh M, LaBranche C C, Vance P J, Romano J, Haggarty B S, Hart T K, Lee W M F, Hoxie J A. An internalization signal in the simian immunodeficiency virus transmembrane protein cytoplasmic domain modulates expression of envelope glycoproteins on the cell surface. J Cell Biol. 1996;132:795–811. doi: 10.1083/jcb.132.5.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shacklett B L, Denesvre C, Boson B, Sonigo P. Features of the SIVmac transmembrane glycoprotein cytoplasmic domain that are important for Env functions. AIDS Res Hum Retrovir. 1998;14:373–383. doi: 10.1089/aid.1998.14.373. [DOI] [PubMed] [Google Scholar]
- 67.Shacklett B L, Weber C J, Shaw K E S, Keddie E M, Gardner M B, Sonigo P, Luciw P A. The intracytoplasmic domain of the Env transmembrane protein is a locus for attenuation of simian immunodeficiency virus SIVmac in rhesus macaques. J Virol. 2000;74:5836–5844. doi: 10.1128/jvi.74.13.5836-5844.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shioda T, Shibuta H. Production of human immunodeficiency virus (HIV)-like particles from cells infected with recombinant vaccinia viruses carrying the gag gene of HIV. Virology. 1990;175:139–148. doi: 10.1016/0042-6822(90)90194-v. [DOI] [PubMed] [Google Scholar]
- 69.Smith A J, Cho M-I, Hammerskjold M-L, Rekosh D. Human immunodeficiency virus type 1 Pr55gag and Pr160gag-pol expressed from a simian virus 40 late replacement vector are efficiently processed and assembled into virus-like particles. J Virol. 1990;64:2743–2750. doi: 10.1128/jvi.64.6.2743-2750.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Spies C P, Ritter G D, Mulligan M J, Compans R W. Truncation of the cytoplasmic domain of the simian immunodeficiency virus envelope glycoprotein alters the conformation of the external domain. J Virol. 1994;68:7840–7845. doi: 10.1128/jvi.68.2.585-591.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weiss C D, White J M. Characterization of stable Chinese hamster ovary cells expressing wild-type, secreted, and glycosylphosphatidylinositol-anchored human immunodeficiency virus type 1 envelope glycoprotein. J Virol. 1993;67:7060–7066. doi: 10.1128/jvi.67.12.7060-7066.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wilk T, Pfeiffer T, Bosch V. Retained in vitro infectivity and cytopathogenicity of HIV-1 despite truncation of the C-terminal tail of the env gene product. Virology. 1992;189:167–177. doi: 10.1016/0042-6822(92)90692-i. [DOI] [PubMed] [Google Scholar]
- 73.Wilk T, Pfeiffer T, Bukovsky A, Moldenhauer G, Bosch V. Glycoprotein incorporation and HIV-1 infectivity despite exchange of the gp160 membrane-spanning domain. Virology. 1996;218:269–274. doi: 10.1006/viro.1996.0190. [DOI] [PubMed] [Google Scholar]
- 74.Wyss S, Berlioz-Torrent C, Boge M, Blot G, Honing S, Benarous R, Thali M. The highly conserved C-terminal dileucine motif in the cytosolic domain of the human immunodeficiency virus type 1 envelope glycoprotein is critical for its association with the AP-1 clathrin adapter. J Virol. 2001;75:2982–2992. doi: 10.1128/JVI.75.6.2982-2992.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu X, Yuan X, McLane M F, Lee T-H, Essex M. Mutations in the cytoplasmic domain of human immunodeficiency virus type 1 transmembrane protein impair the incorporation of Env proteins into mature virions. J Virol. 1993;67:213–221. doi: 10.1128/jvi.67.1.213-221.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zingler K, Littman D R. Truncation of the cytoplasmic domain of the simian immunodeficiency virus envelope glycoprotein increases Env incorporation into particles and fusogenicity and infectivity. J Virol. 1993;66:2824–2831. doi: 10.1128/jvi.67.5.2824-2831.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]