Abstract
Lyssaviruses, the causative agents of rabies encephalitis, are distributed in seven genotypes. The phylogenetically distant rabies virus (PV strain, genotype 1) and Mokola virus (genotype 3) were used to develop a strategy to identify functional homologous interactive domains from two proteins (P and N) which participate in the viral ribonucleoprotein (RNP) transcription-replication complex. This strategy combined two-hybrid and green fluorescent protein–reverse two-hybrid assays in Saccharomyces cerevisiae to analyze protein-protein interactions and a reverse genetic assay in mammalian cells to study the transcriptional activity of the reconstituted RNP complex. Lyssavirus P proteins contain two N-binding domains (N-BDs), a strong one encompassing amino acid (aa) 176 to the C terminus and a weak one in the 189 N-terminal aa. The N-terminal portion of P (aa 52 to 189) also contains a homomultimerization site. Here we demonstrate that N-P interactions, although weaker, are maintained between proteins of the different genotypes. A minimal transcriptional module of the P protein was obtained by fusing the first 60 N-terminal aa containing the L protein binding site to the C-terminal strong N-BD. Random mutation of the strong N-BD on P protein identified three highly conserved K residues crucial for N-P interaction. Their mutagenesis in full-length P induced a transcriptionally defective RNP. The analysis of homologous interactive domains presented here and previously reported dissections of the P protein allowed us to propose a model of the functional interaction network of the lyssavirus P protein. This model underscores the central role of P at the interface between L protein and N-RNA template.
The etiological agents of rabies encephalitis constitute the Lyssavirus genus in the Rhabdoviridae family (38). Phylogenetic analysis distinguished seven genotypes which in turn segregated into two major phylogroups with distinct pathogenic and immunogenic characteristics (1). Phylogroup 1 encompasses genotype 1 (GT1) (classical rabies virus), GT4 (Duvenhage virus), GT5 (European bat lyssavirus type 1), GT6 (European bat lyssavirus type 2), and GT7 (Australian bat lyssavirus). Phylogroup 2 comprises GT2 (Lagos bat virus) and GT3 (Mokola virus). The PV strain rabies virus (GT1) and Mokola virus (GT3) can be considered prototypes for phylogroups 1 and 2, respectively.
Lyssaviruses have a 12-kb nonsegmented negative-strand genomic RNA tightly wrapped by the nucleoprotein (N) (34). This N-RNA complex constitutes the template for the RNA-dependent RNA polymerase (L) and its cofactor, the phosphoprotein (P). All four elements form the ribonucleoprotein (RNP) complex which successively operates in transcriptional then replicative modes during cell infection. The transcription process, characterized by the recognition of gene start and stop signals, produces a short positive-strand leader RNA and five capped and polyadenylated mRNAs encoding, successively, N, P, M (matrix protein), G (glycoprotein), and L. Replication produces genome-length encapsidated positive-strand RNA, which serves in turn as a template for amplification of the negative-strand genomic RNA. The switch from transcription to replication is still debated, but studies on vesicular stomatitis virus (VSV), the working model for rhabdoviruses, implicate the amount of available N protein for genome encapsidation. N alone tends to aggregate nonspecifically in the cytoplasm, and one role of P, via N-P complexes, is to prevent this aggregation and to keep N in a suitable form for specific encapsidation (9, 14, 17, 22, 24, 26, 27). P also participates in the genome expression strategy by mediating attachment of the L polymerase core to the N-RNA template (32). It is thus clear that all four elements of the RNP complex establish close interactions. Understanding these interactions and localizing the protein domains involved would not only elucidate the functional strategy of genome expression but also offer potential targets of pharmacological interest in diseases where vaccination still remains the only efficient prophylactic tool. To date, the lyssavirus N protein, because it is highly refractory to deletion analysis, and L protein, because of its large size, have been poorly studied. The last 566 C-terminal residues of the rabies L protein have been shown to contain the only P protein binding site (6). In addition, the rabies N protein was shown to contain a large NH2 core (amino acids [aa] 1 to 376) encompassing the specific RNA binding site (aa 298 to 352) followed by a trypsin-sensitive site separating a C tail (aa 377 to 450) (18, 20). Structural analysis by electron microscopy showed that one N protein interacts with nine nucleotides (10, 33), that the NH2 core is sufficient to form RNP (18), and that P interacts with the C tail (32). A second P protein binding site has been observed on N (5). The rabies P protein has been, by far, the most extensively studied. Coimmunoprecipitation experiments have identified two independent N-binding sites which were slightly differently mapped in two studies: aa 69 to 177 and aa 173 to 297 (mainly aa 268 to 297) for the CVS strain (5) and aa 1 to 131 (mainly aa 1 to 20) and aa 69 to 273 (mainly aa 250 to 273) for the ERA strain (11). The first 19 N-terminal residues of P also contain the major L-binding site (6). Two cellular kinases, rabies virus protein kinase and protein kinase C, phosphorylate specific serine residues at positions 63 to 64 and 162 to 210 or 271, respectively (15), but this phosphorylation is not required for P oligomerization, which involves a large C-terminal domain mapped between aa 52 and 297 (13). Finally, it was recently demonstrated that the region 139 to 172 of lyssavirus P protein including the DXKSXQ motif interacts strongly with the cytoplasmic dynein light chain (LC8), an element of the microtubule-associated motors involved in minus-end directed axonal transport (19, 28). This could explain how the RNP is transported along the neuron axons from the peripheral site of inoculation to the central nervous system. Despite this substantial amount of data, our understanding of the dynamics of interactions between the four partners of the RNP complex and of their functional roles in transcription and replication remains limited. The present work addresses these questions by combining two-hybrid and green fluorescent protein (GFP)–reverse two-hybrid assays in Saccharomyces cerevisiae to analyze protein-protein interactions and a reverse genetic assay in mammalian cells to assess the functionality of the RNP complex. The functional identification of the interactive domains was performed on proteins from the two divergent lyssaviruses, rabies virus and Mokola virus, in order to delineate an organizational network shared by all members of the Lyssavirus genus.
MATERIALS AND METHODS
Plasmid construction.
Fusion proteins used as bait and prey in two-hybrid assays contained the complete open reading frame (ORF) of the relevant polypeptide, respectively, fused to the Gal4p DNA binding domain (BD) cloned in pAS2ΔΔ (Laboratoire du Métabolisme des ARNs, Institut Pasteur, Paris, France) and to the Gal4p activation domain (AD) cloned in pACTII (Clontech). The constructs used in the reverse genetic assay were inserted behind the T7 promoter in pBluescript SK+/− (Stratagene). All constructs were obtained by PCR of cDNA clones or reverse transcription (RT)-PCR of viral RNA from purified virus using primers with convenient flanking restriction sites. Standard manufacturer's protocols were followed for RNA extraction using the TRIzol reagent (Life Technologies), for RT using Expand Reverse Transcriptase (Roche) and for PCR using Expand high-fidelity PCR system (Roche). Fusion joints and the complete ORF of the PCR-generated fusions were sequenced on an ABI 377 automatic sequencer (Perkin-Elmer).
The full-length PV strain rabies virus P gene from aa 1 through 297 (P-Rab), as well as deletion mutants lacking aa 1 to 57 (P-Rab?58–297), aa 1 to 189 (P-Rab?190–297), aa 176 to 297 (P-Rab?1–175), and aa 1 to 60 fused to aa 176 to 297 (P-Rab?61–175) were inserted into the NcoI restriction site of the vectors pACTII, pAS2ΔΔ, and/or pBluescript SK+/−. The full-length (aa 1 to 450) PV strain rabies virus (N-Rab) and Mokola virus (N-Mok) N genes, the full-length Mokola virus P gene from aa 1 through 303 (P-Mok), as well as its deletion mutants lacking aa 1 to 56 (P-Mok?57–303), aa 1 to 185 (P-Mok?186–303), and aa 177 to 303 (P-Mok?1–176) were inserted into the BamHI restriction site of the vectors pACTII, pAS2ΔΔ, and/or pBluescript SK+/−. The L gene of rabies virus (L-Rab32) was cloned by RT-PCR using specific primers adding NcoI (5′) and EcoRI (3′) restriction sites. The PCR product was inserted into NcoI/EcoRI-digested pBluescript SK+/− containing an NcoI/EcoRI adapter between the BamHI and HindIII sites. All plasmids were amplified in Escherichia coli strain DH5α and purified by chromatography on Qiagen columns.
Yeast two-hybrid analysis.
The two-hybrid system is based on the ability of the yeast transcription factor GAL4 to be divided into two separable and functional domains: an N-terminal domain which binds to specific DNA sequences upstream of activation sequence G (BD) and a C-terminal acidic domain necessary to activate transcription (AD). In order to analyze the interaction between two proteins X and Y these two GAL4 domains are cloned separately in 2μm plasmids fused in frame with proteins X and Y. Plasmids encoding GAL4BD-X (pAS2ΔΔ-X) and GAL4AD-Y (pACTII-Y) were introduced into the S. cerevisiae strain SFY 526 with deletions of trp1, his3, gal4, and gal80, auxotrophic for Leu and Trp, and containing a GAL1-lacZ reporter gene (16). If X and Y interact, GAL4BD and GAL4AD are brought close together, reconstituting a functional GAL4 transcription factor which is able to drive expression of the GAL1-lacZ reporter gene. Thus, the interaction of the tested proteins is assayed by measuring β-galactosidase activity.
Yeast SFY526 was transformed with the two plasmids (pAS2ΔΔ-X and pACTII-Y) using the LiCl procedure (12). Transformed cells were plated on Sabouraud dextrose (SD)-Trp-Leu minimal medium plates, and colonies were streaked 4 to 5 days later on plates of SD minimal medium lacking Trp and Leu (SD-−Trp-−Leu) for β-galactosidase activity assay.
β-Galactosidase assays.
Activation of the LacZ reporter gene has been measured by quantitative liquid assays on permeabilized cells with the LacZ chromogenic substrate ONPG (o-nitrophenyl-β-d-galactoside) (Sigma catalog no. N1127) as described (25). β-Galactosidase units are expressed as the optical density at 405 nm × 1,000/optical density at 600 nm of assayed culture × volume assayed (in milliliters × time in minutes). Assays were done at least in triplicate for each independent transformant. Values represent the mean, and the standard errors were between 10 to 20% of the mean.
P-MokΔ1–176 random mutant library.
The P-MokΔ1–176 random mutant library was fused in frame to a positive selectable marker, the GFP gene (4). This was performed in two steps. First the EGFP gene (Clontech) was amplified with primers FUS-GFP 5′ (5′ CGGGATCCATGGGTAAAGGAGAAGAAC 3′) and FUS-GFP 3′ (5′ GAAGATCTTATTTGTATAGTTCATCCATGCC 3′) and the BamHI-BglII fragment was cloned into the BamHI site of pACTII (Clontech). The resulting pACT-GFP vector contained a polylinker with an SfiI and a BamHI site 5′ of the GFP gene.
In a second step, random mutations of the P-MokΔ1–176 gene were generated using PCR under suboptimal conditions to reduce the fidelity of DNA synthesis by Taq DNA polymerase (Pharmacia). The following primer set was used: primer Sfi (5′ CATATGGCCATGGAGGCC 3′), complementary to the SfiI site of P-MokΔ1–176 in pACTII, and primer FUS-P-MokΔ1–176 (5′ TATGAAGATCTCTCTGCCTCCTCGAGCCGGGCCAT 3′), which introduces a BglII site at the 3′ end of P-MokΔ1–176 gene. The PCR conditions were similar to those in the standard protocol (23) except for a nucleotide bias (1 mM dGTP, dCTP, and dTTP and 0.2 mM dATP, giving a dGTP/dATP ratio of 5) and 0.5 mM Mn2+ (29). P-MokΔ1–176 DNA (2 μg) was used as matrix, and only 20 cycles of amplification were performed. The amplification product was cloned into the pACT-GFP vector using SfiI and BamHI sites. This produced an in-frame fusion of P-Mok?1–176 mutants with EGFP. An overall mutation frequency of 4% was determined by sequencing 10 randomly picked clones. The base substitution matrix is characterized essentially by transitions (T→C, 43%; A→G, 36%) and to a lesser level by transversions (A→T, 11%; T→A, 5%).
Reverse two-hybrid analysis.
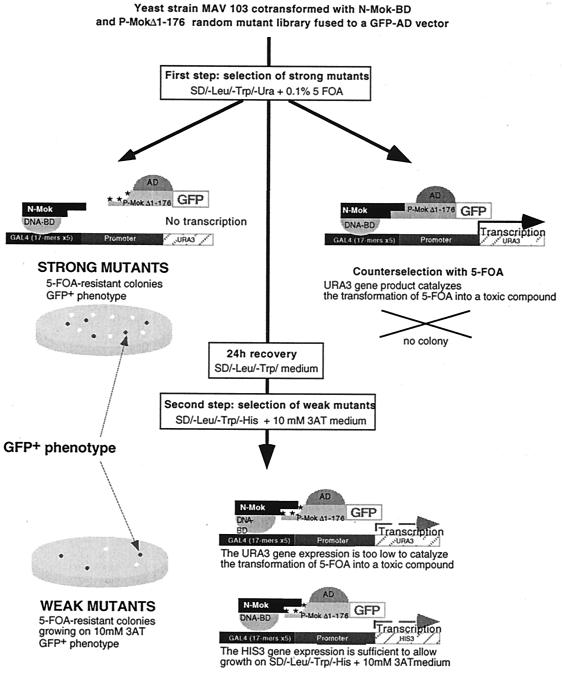
Reverse two-hybrid analyses were performed as previously described in reference 36. Briefly, engineered yeast strain MAV 103 (generous gift of Gibco-BRL), in which URA3 expression is tightly regulated by a promoter containing GAL4 binding sites (SPALn) was cotransformed with N-Mok-GAL4BD and a library of P-MokΔ1–176 mutants. Since GAL4-inducible URA3 alleles confer a fluoroorotic acid (FOA)-sensitive phenotype, growth on medium containing SD-−Leu-−Trp plus 0.1% 5-FOA (catalog no. 16193; Lancaster Synthesis Ltd.) only selects P-MokΔ1–176 mutants altered in their capacity to interact with N-Mok (Fig. 1). In a second step, weakly interactive mutants were selected using medium containing SD-−Leu-−Trp-−His plus 10 mM 3-aminotriazole (3-AT) (Sigma catalog no. A.8056). We developed in parallel a positive selection using GFP fluorescence in order to eliminate noninformative mutants having nonsense mutations, deletions, or insertions.
FIG. 1.
GFP–reverse two-hybrid screening procedure for the GAL4AD P-MokΔ1–176 random mutation library. The EGFP gene (Clontech) was introduced in frame with mutated P-MokΔ1–176 inserts as a positive selectable marker. A titration experiment for both negative and positive phenotypes was preliminarily done with MAV 103 yeast cells expressing wt P-MokΔ1–176–GFP fusion in pACTII and N-Mok in pAS2ΔΔ. This interaction allowed growth on plates with up to 100 mM 3-AT and a total growth inhibition with 0.1% 5-FOA. Under these conditions, strong and weak mutations affecting P-MokΔ1–176 interaction with N-Mok were selected from the mutant library by using a two-step protocol with sequential growth selection (36) combined with a positive selection for strong EGFP fluorescence. First, MAV 103 yeast cells cotransformed with the P-MokΔ1–176 random mutant library and N-Mok were selected on plates containing SD-−Leu-−Trp-−Ura plus 0.1% 5-FOA. Large EGFP-positive yeast colonies (strong mutants) were patched on plates containing SD-−Leu-−Trp plus 0.1% 5-FOA. Colonies on 5-FOA medium were replica-plated to SD-−Leu-−Trp medium to allow recovery for 24 h. They were subjected to a second selection on plates containing SD-−Leu-−Trp-−His plus 10 mM 3-AT, and strongly expressing EGFP colonies (weak mutants) were patched on plates containing SD-−Leu-−Trp-−His plus 10 mM 3-AT. After plasmid rescue, the phenotypes of mutagenized alleles were verified by patching the colonies on selective media and microscopically checking for EGFP fluorescence.
Reverse genetic assay.
The reverse genetic assay was based on the method in reference 31 with modifications (P. Le Mercier et al., submitted for publication). BSR cells (30) were grown in Dulbecco's modified Eagle's medium, Glutamax I (Gibco) supplemented with 8% fetal calf serum (FCS), and gentamicin (40 mg/liter). The cells were plated at 2.5 × 105 cells per well in a 24-well plate and incubated at 37°C for 24 h. The medium was then removed and the cells were infected with T7 recombinant vaccinia virus vTF7-3 (multiplicity of infection of 10 PFU per cell) in Dulbecco's modified Eagle's medium in order to obtain a cytoplasmic expression of T7-RNA polymerase. After 30 min the medium was removed and the cells were transfected with plasmids in which viral N, P, and L ORFs and a minigenome cDNA are under the control of the T7 promoter (1 μg of pT7-N and pT7-P, 0.2 μg of pT7-L-32, and 0.5 μg of minigenome plasmid pDI.mut) using polyethylenimine (PEI) (Aldrich catalog no. 40,872-7). Briefly, 4.5 mg of pure PEI was diluted in 8 ml of water, neutralized with HCl, adjusted to 10 ml, and filtered through a 0.2-μm-pore-size filter (Millipore). PEI and plasmid DNA solutions were each diluted in 50 μl of 150 mM NaCl (PEI/DNA ratio of 1.5), kept 10 min at room temperature, mixed and vortexed, and then added to the cell supernatants without FCS. After 2 h, the transfection medium was removed and fresh medium with 5% FCS was added, and the cells were incubated at 37°C for 48 h. pDI.mut cDNA cassette (Le Mercier et al., submitted) is transcribed by T7-RNA polymerase to produce a PV strain rabies minigenome (RNA-Rab) composed of the trailer and leader sequences, as well as, respectively, the L stop and N start transcription signals flanking an antisense luciferase RNA. In addition, to perfectly mimic the extremities of the wild-type (wt) viral genome, the minigenome RNA is flanked by a hammerhead ribozyme and a hepatitis-delta virus genomic ribozyme at the 5′ and 3′ ends, respectively. The T7-expressed N, P, and L proteins and the RNA minigenome form a functional RNP template resulting in luciferase gene transcription, and thus the amount of luciferase activity is related to the transcriptional activity of the RNP.
Luciferase assay.
At 48 h after transfection, cells were washed with phosphate-buffered saline and overlaid with 200 μl of lysis buffer (25 mM Tris-phosphate [pH 7.8], 2 mM dithiothreitol, 10% glycerol, 10% Triton X-100) for 10 min at room temperature. The extracts were centrifuged 3 min at 4,000 × g, and luciferase expression was measured using a Berthold luminometer by injecting 100 μl of luciferase assay reagent (Promega catalog no. E1501) into 10 μl of each supernatant and counting for 10 s. Luciferase activity was measured in triplicate; values represent the mean (standard error < 10% of the mean).
RESULTS
Two-hybrid intragenotypic analysis.
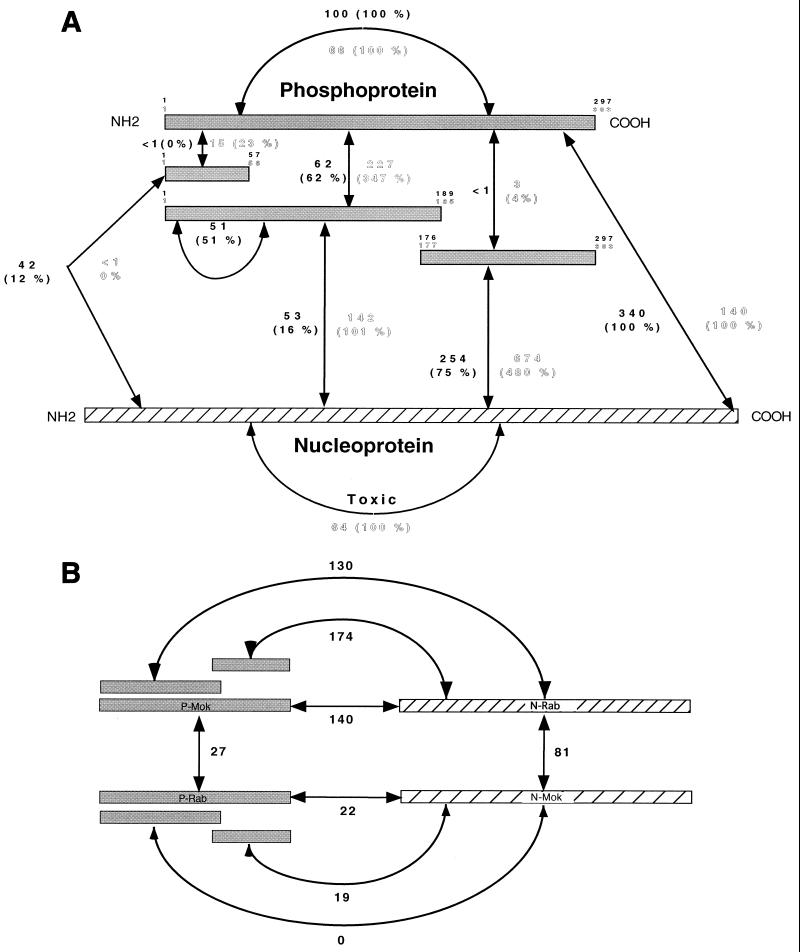
P and N self-association properties were first analyzed for both rabies and Mokola viruses using the two-hybrid method (Fig. 2A). A strong transactivation was obtained when P-Mok-AD and P-Mok-BD were coexpressed (66 U, 100%), indicating P-Mok homo-oligomerization. Similar homo-oligomerization was obtained with P-Rab (100 U, 100%). P protein deletion mutants were tested for their interaction with full-length P (P-full). A dramatic increase of β-galactosidase activity (347%) was observed when the P-Mok N-terminal half (P-MokΔ186–303) was coexpressed with P-Mok-full. This elevated transactivation compared to P-Mok-full is classic in two-hybrid systems (see other examples below) and probably due to better domain folding in the absence of steric hindrance (35). In contrast, the C-terminal half (P-MokΔ1–176) interacted very weakly (4%) with P-Mok-full. A similar interaction pattern was obtained with the P-Rab N-terminal half (P-RabΔ190–297, 62%) and C-terminal half (P-RabΔ1–175, <1%). In addition, the P-RabΔ190–297 was capable of substantial homo-oligomerization (51%). Taken together, these results argue for a lyssavirus P oligomerization domain located between aa 1 and 185 or 189. Since an alignment of the P-Mok and P-Rab amino acid sequences reveals two highly conserved regions around aa 1 to 60 and aa 200 to 280 (Fig. 3), the interaction potential of the highly conserved N-terminal domain was studied. P-MokΔ57–303 interacted poorly with P-Mok-full (23%), and P-RabΔ58–297 failed to interact with P-Rab-full. This suggests that most of the multimerization region has either been deleted in these mutants or that its proper folding requires larger flanking regions.
FIG. 2.
Interactions between lyssavirus N and P proteins measured in the yeast two-hybrid system in S. cerevisiae. Full-length or deletion mutants of the PV strain rabies virus (Rab, GT1, limits in black type) and the Mokola virus (Mok, GT3, limits in outlined type) were fused with either the GAL4AD or the GAL4 DNA BD. Transcription of the reporter gene (LacZ) under the control of the GAL4 DNA-binding regulatory elements indicated interaction between the two fusion proteins of interest. The amount of β-galactosidase indicates the intensity of the interaction. (A) Intragenotypic interactions. β-Galactosidase activities (means of a triplicate) are represented in black letters for Rab and in outlined letters for Mok. The values in parentheses represent the percentage of activity of each interaction compared to interaction between full-length P or N proteins (100%). (B) Intergenotypic N-P interactions expressed in β-galactosidase activity (means of a triplicate).
FIG. 3.
Comparison of lyssavirus P proteins. Sequence alignment of the P proteins of PV strain rabies virus (Rab, GT1) and Mokola virus (Mok, GT3) is shown. Dashes represent gaps introduced to optimize the alignment. Grey boxes outline identical residues (overall identity, 48%).
A similar dissection was impossible to perform with N protein due to its high sensitivity to deletion (20). In addition, lyssavirus N protein is toxic for yeast cells. However, homo-oligomerization was observed for N-Mok-full (64 U). Interactions between full-length P and N proteins were observed with proteins from either virus (P-Rab/N-Rab, 340 U, 100%; P-Mok/N-Mok, 140 U, 100%). When P deletion mutants were assayed, the strongest interaction with homologous N-full was observed with the C-terminal half (P-RabΔ1–175, 75%; P-MokΔ1–176, 480%). Although the N-terminal part of P (P-RabΔ190–297, 16%; P-MokΔ186–303, 101%) also interacted with homologous N-full, the strength of this interaction was fivefold less than that observed with the C-terminal half. These results argue that lyssavirus P proteins have two N-BDs, a stronger one in the C-terminal half (aa 176 or 177 to aa 297 or 303) and a weaker one in the N-terminal half (aa 1 to 185 or 189).
Two-hybrid intergenotypic analysis.
When lyssavirus intergenotypic oligomerizations where analyzed (Fig. 2B), N-Rab-full and N-Mok-full, which are 81% identical (2), showed substantial interaction (81 U) despite their toxicity for yeast. On the other hand, P-Rab-full and P-Mok-full also showed a significant interaction (27 U), suggesting oligomerization between these orthologous proteins that are only 48% identical. Furthermore, P-RabΔ190–297 was sufficient for an interaction with P-Mok-full (data not shown), arguing that the oligomerization motif in the N-terminal half of lyssavirus P proteins is conserved, although it is not part of the highly conserved N-terminal end up to aa 56 or 57, as previously demonstrated (Fig. 2A).
When lyssavirus intergenotypic P-N interaction was analyzed (Fig. 2B) significant β-galactosidase activity (140 U, 100%) was observed with N-Rab-full–P-Mok-full, and as in the intragenotypic context, the C-terminal half, P-MokΔ1–176, provided reproducibly a slightly greater interaction with N-Rab-full (174 U, 124%) than the N-terminal half, P-MokΔ186–303 (130 U, 90%), did. The symmetrical observation was made when N-Mok-full was coexpressed with P-Rab-full (22 U, 100%), P-RabΔ1–175 (19 U, 86%) and P-RabΔ190–297 (0%). Surprisingly, intergenotypic interactions were globally sixfold more intense in the combination P-Mok–N-Rab than P-Rab–N-Mok. The reason for that is unclear but could be related to the observation that nucleoproteins from nonsegmented negative-strand RNA viruses are folded into a large N-terminal globular core and an exposed C-terminal 80 aa tail predicted to interact with the P protein (3, 8, 20, 21, 32). The C tails from N-Rab and N-Mok are only 69% identical compared to 81% for the entire protein, and this variability could explain the differences in P protein binding intensities.
Reverse two-hybrid screening of important residues of the strongest N-BD in the C-terminal half of P.
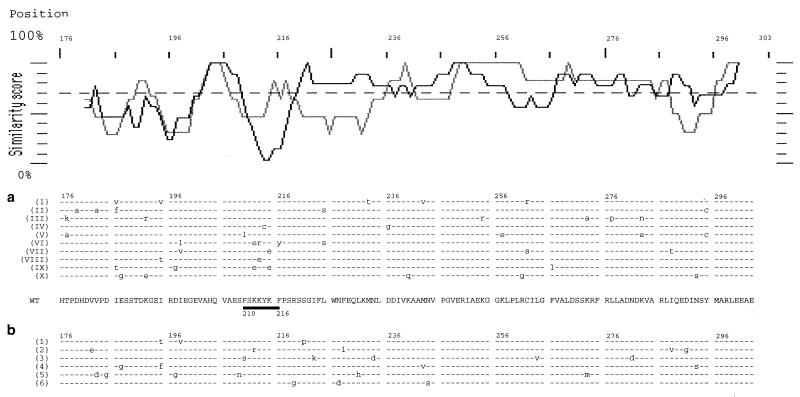
To precisely define which amino acid residues of P are implicated in N-P interaction, a library of mutants was generated by random PCR mutagenesis of the strongest P-Mok N-BD (P-MokΔ1–176). This library was screened using a GFP–reverse two-hybrid system, where dissociation of the interaction is a selective advantage (Fig. 1) (H. Endoh et al., submitted for publication). After having verified that fusion of P-MokΔ1–176 with GFP did not modify its interaction with N-Mok and that the frequency of reversion to 5-FOA resistance was low (0.01%), 1.5 × 106 clones coexpressing P-MokΔ1–176 mutants GFP-AD and N-Mok-BD were plated on medium containing SD-−Leu-−Trp-−URA plus 0.1% 5-FOA. After 24 h of recovery on SD-−Leu-−Trp medium, FOA-resistant colonies (2%) were selected for histidine prototrophy on selection medium lacking His. This selection segregated amino acid changes which partially affect N-P-MokΔ1–176 interaction (weak mutations) (37). Among FOA-resistant colonies, three GFP phenotypes were observed: GFP-negative colonies containing nonsense mutations; GFP faintly positive colonies resulting from read-through of nonsense mutations; and strongly positive GFP colonies. Ten plasmids from the third set of clones were reintroduced into yeast cells containing N-Mok-BD to confirm their 5-FOA resistance and strong GFP positive phenotype, and their inserts were sequenced (Fig. 4a). Six weak binding mutant clone inserts were also sequenced (Fig. 4b). While the distribution of the mutations in weak mutants was essentially random, strong mutants were more frequently mutated in a short lysine-rich stretch (210FSKKYKF216). Of nine mutations affecting this stretch, five corresponded to charge modifications of at least one of the three lysine residues to glutamic acid. We converted all three lysine residues in full-length P-Mok into glutamic acids. In the classical two-hybrid system, this mutant (PS1) protein displayed threefold less interaction with N-Mok (48 U, 34%), strongly supporting the idea that these lysines play a key role in N-P binding but also underlining the coexistence in the N-terminal half of P-Mok of the second independent N-BD, of lower affinity, which remained efficient in PS1. Interestingly, the PS1 mutant interaction with P-Mok was of similar intensity to that of P-Mok self-interaction (70 U, 106%), verifying that the N-terminal half, involved in P oligomerization, is not affected.
FIG. 4.
Comparison of the strong and weak mutants of the strongest N-BD of P-Mok. Similarity profile (upper part) and sequence alignment for the 10 strong (dark line) (a) and 6 weak (grey line) (b) P-Mok Δ1–176 mutants. The corresponding lysine-rich stretch, found to be frequently mutated, is underlined (position 210 to 216).
Functional analysis by reverse genetics.
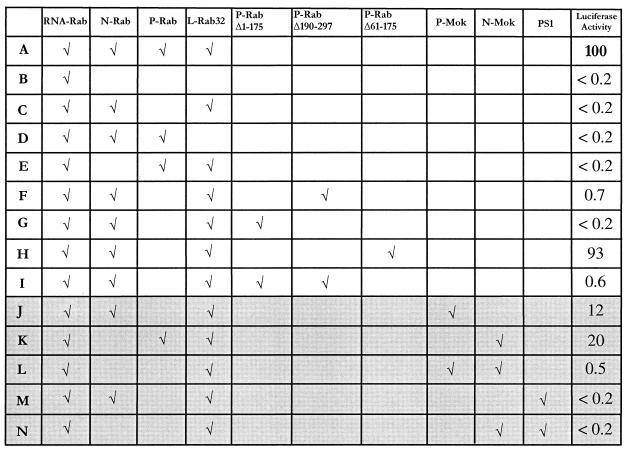
To evaluate the functionality of intra- and intergenotypic RNP complexes, a reverse genetic assay was developed (Le Mercier et al., submitted). Viral N, P, and L cDNAs as well as a viral minigenomic cDNA were cloned in T7 expression vector and cotransfected into T7 recombinant vaccinia virus-infected BSR cells. The minigenomic RNA (RNA-Rab), which is vastly overproduced by T7 RNA polymerase, successively encompasses the following (from the 5′ to the 3′ end): hammerhead ribozyme, rabies virus trailer sequence, antisense RNA from luciferase gene, rabies virus leader sequence, and hepatitis-delta virus genomic ribozyme. This T7 RNA polymerase transcript after ribozyme cleavage perfectly mimics the extremities of the wild type viral genomic RNA. Thus, the minigenomic RNA and the N, P, and L proteins expressed from the respective mRNAs form a functional RNP. This RNP could serve as template for two successive RNA synthetic functions: transcription and then replication. In this assay transcription consists of production of both a small leader RNA and a capped and polyadenylated luciferase RNA whose level is measured by luciferase activity. Only transcription is evaluated in this case, which does not formally exclude a functionality of the RNP complex at the replicative level. In Fig. 5 the reference level corresponds to the luciferase activity obtained with minigenome RNA-Rab and full-length proteins N-Rab, P-Rab, and L-Rab (line A). As shown in lines B to E each component of the RNP complex is required for transcriptional activity. P-RabΔ190–297, which contains the P-oligomerization domain and the weak N-BD, induced a partially active RNP (line F), while P-RabΔ1–175, which contains the strong N-BD, failed (line G). This difference could be explained by the absence in P-RabΔ1–175 of the essential L interacting domain located in the N-terminal 19 aa, as described for the CVS P protein (6). Accordingly, when the N-terminal 60 aa of P-Rab were fused to P-RabΔ1–175 (P-RabΔ61–175) a strong transcriptional activity was restored (line H). Interestingly, simultaneous expression of P-RabΔ1–175 and P-RabΔ190–297 only displays the residual activity observed with P-RabΔ190–297 alone (line I). Since these two fragments did not interact in the two-hybrid assay (data not shown), this underlines the absolute necessity of a physical link between the L-BD and the strong N-binding site to get a functional transcription complex. The substitution of the P-Rab or N-Rab proteins by P-Mok or N-Mok proteins resulted in a heterogenotypic RNP with lower but significant activity (lines J and K) as expected from the sequence divergence (2). Swapping both N-Mok and P-Mok in a minigenome RNA–L-Rab context still decreased the activity (line L), suggesting that the RNA-L complex has stringent requirements for homologous P and/or N. The PS1 P-Mok mutant was unable to reconstitute a functional RNP with either N-Rab or N-Mok (lines M and N), although it maintained a significant interaction with N-Mok, through the weak N-binding site.
FIG. 5.
Functionality of reconstituted RNP measured by reverse genetics. Luciferase activity was measured in triplicate, and values are expressed as a percentage of the reference activity. The reference value (100%) is the activity obtained with minigenomic RNA and full-length N, P, and L proteins of the PV strain rabies virus (Rab, line A). The background luciferase level is noted as <0.2%, and the values of 0.5% or more are significantly above the background level. Lines B to E correspond to control experiments and demonstrate that each component is necessary for transcriptional activity. The results using intergenotypic components (lines J to N) are displayed with a gray background.
DISCUSSION
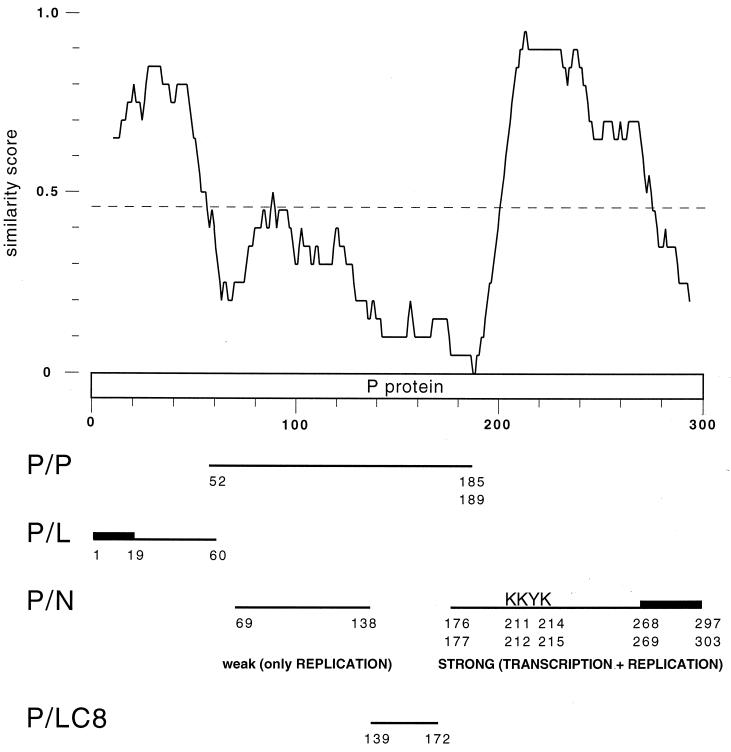
Previous studies have demonstrated interactions between the elements of the rabies virus RNP complex (6, 5, 11, 18, 20, 32). They used cross-linking or coimmunoprecipitation of N, P, and L proteins expressed in bacteria or in eukaryotic cells. These studies focused on different strains of rabies virus from GT1 of the Lyssavirus genus: CVS, ERA, or SADB19. In the present study, we combined an in vivo two-hybrid interactive domain mapping of primarily P but also of N protein, with a reverse genetic assay to analyze the transcriptional functionality of the reconstituted RNP complex. In addition, this combined analysis was applied to two phylogenetically distant lyssaviruses representative of the two principal phylogroups: the PV strain rabies virus (GT1, phylogroup 1) and the Mokola virus (GT3, phylogroup 2) (1). Both intragenotypic and intergenotypic interactions were studied, based on the rationale that N and P proteins coming from the two different viruses should be able to reconstitute genotypically heterogeneous RNPs capable of significant transcriptional activity although lower than that of homogeneous RNP. The observation that P-Mok gave a lower transcriptional activity than N-Mok in an otherwise RNP-Rab context was not surprising since N is the more conserved protein (81% identity), while P proteins from Mokola virus and rabies virus are only 48% identical, with the similarity mostly concentrated in two highly conserved domains restricted to aa 1 to 60 and aa 200 to 280 (Fig. 6).
FIG. 6.
Functional interaction map of lyssavirus phosphoprotein. Similarity profile between the PV strain rabies virus and Mokola virus P proteins (similarity plot program of GCG, window 20 aa). The P protein domains involved in oligomerization or in interaction with L and N lyssavirus proteins or with the dynein LC8 are mapped. The stretches particularly important for the interaction are indicated by thick lines, and the KKYK motif is noted.
Oligomerization properties were demonstrated for both rabies and Mokola virus N and P proteins, in an intragenotypic or an intergenotypic context. Deletion analysis of the P protein suggested that its multimerization domain is located within the N-terminal half (aa 1 to 185 or 189). However, the highly conserved first 57 residues do not appear to be implicated, even though they include a region with coiled-coil forming potential (aa 1 to 30) frequently involved in protein oligomerization (7). This result is in good agreement with that from cross-linking of CVS strain rabies virus P protein mutants produced in bacteria, delineating the oligomerization domain in the region of aa 52 to 297 (13). Taken together, these data suggest that the lyssavirus P protein oligomerization domain is mainly contained in aa 52 to 185 or 189, i.e., in the central highly variable part of the protein (Fig. 6).
Coimmunoprecipitation experiments with P and N proteins have previously shown two independent N-binding sites along the rabies P protein. Using BSR (baby hamster kidney) cell extracts expressing both proteins from the CVS strain (5; D. Blondel, personal communication), these two sites were mapped to aa 69 to 138 and aa 173 to 297, of which the distal part, aa 268 to 297, is absolutely required for N-binding. Using an in vitro coupled transcription-translation system and Sf9 cell extracts expressing both proteins from the ERA strain (11), it was found that the region of aa 1 to 131 was able to bind N when both proteins where synthesized simultaneously, while aa 69 to 273 could bind whether the proteins were produced simultaneously or separately. The distal part of each domain (aa 1 to 20 and aa 250 to 273, respectively) appeared critical for N binding. Our results with both the PV rabies strain and Mokola viruses are consistent with the existence of two independent N-BDs along lyssavirus P protein but clearly modulate their respective functional importance. The carboxy-terminal domain (C-terminal half, aa 176 or 177 to aa 297 or 303) plays the major role in P-N binding since it is about fivefold more intense than that of the amino-terminal domain (N terminal half, aa 1 to 185 or 189). The reverse two-hybrid method demonstrated that the short lysine-rich motif FSKKYKF (position 210 to 216 in P-Mok and 209 to 215 in P-Rab), conserved in the seven different genotypes of lyssaviruses (S. Nadin-Davis, personal communication), was of critical importance for the N-protein binding competence of the P C-terminal domain. However, the triple mutant PS1, corresponding to a full-length P-Mok in which these three K are mutated to E, still binds to N-Mok, although threefold less efficiently than wt P-Mok-full, and is still able to oligomerize with P-Mok-full. This result indicates that the N-terminal domain of P is able to display interactions independently. In summary, our results are in reasonable agreement with the previous results and can be combined with them to predict that the lyssavirus P protein harbors two independent N-binding sites. One site of primary importance residing in the C-terminal half (aa 176 or 177 to aa 297 or 303) and encompassing two very important stretches, the lysine-rich motif (aa 209 or 210 to aa 215 or 216) and the C-terminal tail (aa 268 or 269 to aa 297 or 303). Another site of secondary importance in the N-terminal half probably lies between aa 69 and 138, i.e., possibly partially overlapping with the oligomerization domain (aa 57 to 185 or 189) (Fig. 6). The existence of a third weak N-binding site in the very first 20 aa of P (11) is unclear, but it should be noted that the two-hybrid method revealed a weak interaction between N-Rab and the first 57 N-terminal residues of P-Rab, a result which was not confirmed with the similar domain of P-Mok.
The reverse genetics analysis allowed us to complete the in vivo binding studies by examining transcriptional functionality in a reconstituted RNP complex. Whereas P-Mok gave substantial transcription in an RNP-Rab context, the PS1 mutant did not, suggesting that an efficient N-binding site in the C-terminal half of P is needed to promote transcription. However, the C-terminal domain of P alone, in spite of its strong interaction with N protein, is unable to reconstitute a functional RNP, whereas the N-terminal domain alone had residual transcriptional activity. In addition, the fusion of the first 60 N-terminal aa of P to the carboxy-terminal domain (P-RabΔ61–175) restored full transcriptional functionality, despite the probable lack of the weak N-binding site (aa 69 to 138) as well as the oligomerization domain (aa 52 to 185 or 189) in the mutant. This result supports the idea that the N-terminal L protein binding site, which was mapped to the first 19 residues of P with a stabilizing effect of aa 20 to 52 (6), is essential for P transcriptional function and sufficient when associated with the major N-protein binding site in the C-terminal domain. However, colinearity between these domains is needed since coexpression of P-RabΔ1–175 and P-RabΔ190–297 failed to restore functionality. Thus, deletion of the region of aa 61 to 175, which encompasses the weak N-binding site, is not deleterious for transcription. One can, however, presume that this region is of primary importance for replication, which was not measured in our reverse genetics assay. It was previously shown for VSV that P protein plays a key role during replication by complexing the N protein and keeping it in a convenient form for encapsidation (17). The second weak N-protein binding site could be crucial for this P replicative function. The previous observation that the N-terminal domain (aa 1 to 131) of the ERA rabies virus P protein is only able to bind N when both proteins are synthesized simultaneously (11) supports the argument for this chaperone-like function of the weak N-protein binding site for P on N during replication. A recent model of VSV RNP assembly proposes that an N dimer interacts with one molecule of P and that five such 2:1 N-P complexes form a barrel-like oligomer, corresponding to one turn of the RNP helix (14). The role of the P protein in this model is to promote the correct assembly of the N-oligomer, which in the absence of P forms random aggregates. In contrast, the rabies N protein overexpressed in the baculovirus system naturally forms an N-RNA ring structure without requiring P coexpression and even if the C-terminal tail of N, carrying the P binding site, is deleted. This suggests that the chaperone-like function of lyssavirus P on N would not be crucial for N multimerization in RNP (32).
In summary, the combination of in vivo two-hybrid and reverse genetic approaches associated with an intergenotypic swapping strategy within the Lyssavirus genus has allowed a more precise delineation of interactive domains of the lyssavirus P protein and a definition of their respective functionality in the RNP context. Figure 6 proposes a functional interaction map of this protein, which occupies a nodal role in the network of functional protein-protein interaction by providing a bridge at the interface between N-RNA complex, L, and cellular factors. In this P protein model, the very conserved first 60 aa mainly act for L recruitment, the L binding site being more restricted to the first 19 aa. The consecutive variable domain carries the oligomerization domain (aa 52 to 185 or 189) and a weak N-binding site (aa 69 to 138) which are not required for transcription but are probably implicated in replication. This region also contains the domain of interaction with the cytoplasmic dynein light chain LC8 (aa 139 to 172), a cellular factor implicated in retrograde axonal transport that mediates the RNP transport along the neuron axons (19, 28). Finally, the C-terminal half (aa 176 or 177 to aa 297 or 303), including the region of aa 268 to 297, demonstrated to be important for rabies virus, encompasses the main transcriptionally important N-binding site which contains a very conserved region with the lysine-rich motif FSKKYKF (aa 209 or 210 to aa 215 or 216).
ACKNOWLEDGMENTS
We thank Alain Jacquier and Micheline Fromont-Racine (Unité de Génétique des Interactions Macromoléculaires, Institut Pasteur), Pierre Legrain and Jean-Christophe Rain (Hybrigenics, Paris, France), Marc Vidal (Dana Farber Cancer Institute, Boston, Mass.), and Michael Brasch (Life Technologies) for helpful discussions and the generous gift of yeast strains and plasmid vectors; Philippe Le Mercier for providing the reverse genetic cassette; and Charles Roth for helpful comments and suggestions. We are greatly indebted to Yvette Forteville, Malika Campanaro, and Karine Wiszniowski for technical assistance.
REFERENCES
- 1.Badrane H, Bahloul C, Perrin P, Tordo N. Evidence of two Lyssavirus phylogroups with distinct pathogenicity and immunogenicity. J Virol. 2001;75:3268–3276. doi: 10.1128/JVI.75.7.3268-3276.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bourhy H, Kissi B, Tordo N. Molecular diversity of the Lyssavirus genus. Virology. 1993;194:70–81. doi: 10.1006/viro.1993.1236. [DOI] [PubMed] [Google Scholar]
- 3.Buchholz C J, Retzler C, Homann H E, Neubert W J. The carboxy-terminal domain of Sendai virus nucleocapsid protein involved in complex formation between phosphoprotein and nucleocapsid-like particles. Virology. 1994;204:770–776. doi: 10.1006/viro.1994.1592. [DOI] [PubMed] [Google Scholar]
- 4.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 5.Chenik M, Chebli K, Gaudin Y, Blondel D. In vivo interaction of rabies virus phosphoprotein (P) and nucleoprotein (N): existence of two N-binding sites on P protein. J Gen Virol. 1994;75:2889–2896. doi: 10.1099/0022-1317-75-11-2889. [DOI] [PubMed] [Google Scholar]
- 6.Chenik M, Schnell M, Conzelmann K K, Blondel D. Mapping the interacting domains between the rabies virus polymerase and phosphoprotein. J Virol. 1998;72:1925–1930. doi: 10.1128/jvi.72.3.1925-1930.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curran J, Boeck R, Lin-Marq N, Lupas A, Kolakofsky D. Paramyxovirus phosphoproteins form homotrimers as determined by an epitope dilution assay, via predicted coiled coils. Virology. 1995;214:139–149. doi: 10.1006/viro.1995.9946. [DOI] [PubMed] [Google Scholar]
- 8.Curran J, Homann H, Buchholz C, Rochat S, Neubert W, Kolakofsky D. The hypervariable C-terminal tail of the Sendai paramyxovirus nucleocapsid protein is required for template function but not for RNA encapsidation. J Virol. 1993;67:4358–4364. doi: 10.1128/jvi.67.7.4358-4364.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis N L, Arnheiter H, Wertz G W. Vesicular stomatitis virus N and NS proteins form multiple complexes. J Virol. 1986;59:751–754. doi: 10.1128/jvi.59.3.751-754.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flamand A, Raux H, Gaudin Y, Ruigrok R W H. Mechanisms of rabies neutralization. Virology. 1993;194:302–313. doi: 10.1006/viro.1993.1261. [DOI] [PubMed] [Google Scholar]
- 11.Fu Z F, Zengh Y, Wunner W H, Koprowski H, Dietzschold B. Both the N- and the C-terminal domains of the nominal phosphoprotein of rabies virus are involved in the binding to the N protein. Virology. 1994;200:590–597. doi: 10.1006/viro.1994.1222. [DOI] [PubMed] [Google Scholar]
- 12.Gietz R D, Schiestl R H, Willems A R, Woods R A. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 13.Gigant B, Iseni F, Gaudin Y, Knossow M, Blondel D. Neither phosphorylation nor the amino-terminal part of rabies virus phosphoprotein is required for its oligomerization. J Gen Virol. 2000;81:1757–1761. doi: 10.1099/0022-1317-81-7-1757. [DOI] [PubMed] [Google Scholar]
- 14.Green T J, Macpherson S, Qiu S, Lebowitz J, Wertz G W, Luo M. Study of the assembly of vesicular stomatitis virus N protein: role of the P protein. J Virol. 2000;74:9515–9524. doi: 10.1128/jvi.74.20.9515-9524.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta A K, Blondel D, Choudhary S, Banerjee A K. The phosphoprotein of rabies virus is phosphorylated by a unique cellular protein kinase and specific isomers of protein kinase C. J Virol. 2000;74:91–98. doi: 10.1128/jvi.74.1.91-98.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 17.Howard M, Wertz G. Vesicular stomatitis virus RNA replication: a role for the NS protein. J Gen Virol. 1989;70:2683–2694. doi: 10.1099/0022-1317-70-10-2683. [DOI] [PubMed] [Google Scholar]
- 18.Iseni F, Barge A, Baudin F, Blondel D, Ruigrok R W H. Characterization of rabies virus nucleocapsids and recombinant nucleocapsid-like structures. J Gen Virol. 1998;79:2909–2919. doi: 10.1099/0022-1317-79-12-2909. [DOI] [PubMed] [Google Scholar]
- 19.Jacob Y, Badrane H, Ceccaldi P E, Tordo N. Cytoplasmic dynein LC8 interacts with lyssavirus phosphoprotein. J Virol. 2000;74:10217–10222. doi: 10.1128/jvi.74.21.10217-10222.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kouznetzoff A, Buckle M, Tordo N. Identification of a region of the rabies virus N protein involved in direct binding to the viral RNA. J Gen Virol. 1998;79:1005–1013. doi: 10.1099/0022-1317-79-5-1005. [DOI] [PubMed] [Google Scholar]
- 21.Krishnamurthy S, Samal S K. Identification of regions of bovine respiratory syncytial virus N protein required for binding to P protein and self-assembly. J Gen Virol. 1998;79:1399–1403. doi: 10.1099/0022-1317-79-6-1399. [DOI] [PubMed] [Google Scholar]
- 22.La Ferla F, Peluso R. The 1:1 N-NS protein complex of vesicular stomatitis virus is essential for efficient genome replication. J Virol. 1989;63:3852–3857. doi: 10.1128/jvi.63.9.3852-3857.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leung D W, Chen E, Goeddel D V. A method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction. Technique J Methods Cell Mol Biol. 1989;1:11–15. [Google Scholar]
- 24.Masters P S, Banerjee A K. Resolution of multiple complexes of phosphoprotein NS with nucleocapsid protein N of vesicular stomatitis virus. J Virol. 1988;62:2651–2657. doi: 10.1128/jvi.62.8.2651-2657.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 26.Peluso R. Kinetic, quantitative, and functional analysis of multiple forms of the vesicular stomatitis virus nucleocapsid protein in infected cells. J Virol. 1988;62:2799–2807. doi: 10.1128/jvi.62.8.2799-2807.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peluso R W, Moyer S U. Viral proteins required for in vitro replication of vesicular stomatitis virus defective-interfering particle genome RNA. Virology. 1988;162:369–376. doi: 10.1016/0042-6822(88)90477-1. [DOI] [PubMed] [Google Scholar]
- 28.Raux H, Flamand A, Blondel D. Interaction of the rabies virus P protein with the LC8 dynein light chain. J Virol. 2000;74:10212–10216. doi: 10.1128/jvi.74.21.10212-10216.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rice G C, Goeddel D V, Cachianes G, Woronicz J, Chen E Y, Williams S R, Leung D W. Random PCR mutagenesis screening of secreted proteins by direct expression in mammalian cells. Proc Natl Acad Sci USA. 1992;89:5467–5471. doi: 10.1073/pnas.89.12.5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato M, Maida N, Yoshida H, Urade M, Saito S, Miyazaki T, Shibata T, Watanabe M. Plaque formation of herpes virus hominidis type 2 and rubella virus in variants isolated from the colonies of BHK21/WI-2 cells formed in soft agar. Arch Virol. 1977;53:269–273. doi: 10.1007/BF01314672. [DOI] [PubMed] [Google Scholar]
- 31.Schnell M J, Mebatsion T, Conzelmann K K. Infectious rabies viruses from cloned cDNA. EMBO J. 1994;13:4195–4203. doi: 10.1002/j.1460-2075.1994.tb06739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schoehn G, Iseni F, Mavrakis M, Blondel D, Ruigrok R W H. Structure of recombinant rabies virus nucleoprotein-RNA complex and identification of the phosphoprotein binding site. J Virol. 2001;75:490–498. doi: 10.1128/JVI.75.1.490-498.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas D, Newcomb W W, Brown J C, Hall J S, Hainfeld J F, Trus B L, Steven A C. Mass and molecular composition of vesicular stomatitis virus: a scanning transmission electron microscopy analysis. J Virol. 1985;54:598–607. doi: 10.1128/jvi.54.2.598-607.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tordo N, Charlton K, Wandeler A. Rhabdoviruses: rabies. In: Mahy B W J, Collier L, editors. Microbiology and microbial infections. Vol. 1. London, United Kingdom: Arnold; 1998. pp. 665–692. [Google Scholar]
- 35.Transy C, Legrain P. The two-hybrid: an in vivo protein-protein interaction assay. Mol Biol Rep. 1995;21:119–127. doi: 10.1007/BF00986502. [DOI] [PubMed] [Google Scholar]
- 36.Vidal M, Brachmann R K, Fattaey A, Harlow E, Boeke J D. Reverse two-hybrid and one-hybrid systems to detect dissociation of protein-protein and DNA-protein interactions. Proc Natl Acad Sci USA. 1996;93:10315–10320. doi: 10.1073/pnas.93.19.10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vidal M, Braun P, Chen E, Boeke J D, Harlow E. Genetic characterization of a mammalian protein-protein interaction domain by using a yeast reverse two-hybrid system. Proc Natl Acad Sci USA. 1996;93:10321–10326. doi: 10.1073/pnas.93.19.10321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walker P J, Benmansour A, Dietzgen R, Fang R X, Jackson A O, Kurath G, Leong J C, Nadin-Davies S, Tesh R D, Tordo N, Van Regenmortel M H V, Fauquet C M, Bishop D H L, editors. Rhabdoviridae. Orlando, Fla: Academic Press; 2000. [Google Scholar]