Abstract
Thymic lymphomas induced by Moloney murine leukemia virus (MMLV) have provided many examples of oncogene activation, but the role of tumor suppressor pathways in these tumors is less clear. These tumors display little evidence of loss of heterozygosity, and MMLV is only weakly synergistic with the Trp53 null genotype, suggesting that viral lymphomagenesis involves mechanisms which do not require mutational loss of Trp53 function. To explore this relationship in greater depth, we infected CD2-myc transgenic mice with MMLV and examined the role of Trp53 in the genesis of these tumors. Most (19 of 27) of the tumors from MMLV-infected, CD2-myc Trp53+/− mice retained the wild-type Trp53 allele in vivo while tumors of uninfected CD2-myc Trp53+/− mice invariably showed allele loss from a significant fraction of primary tumor cells. The functional integrity of the Trp53 gene in these tumors was indicated by ongoing allele loss or selection for mutational stabilization during in vitro propagation and by the radiosensitivity of selected Trp53+/− tumor cell lines. An inverse correlation was noted between retention of the wild-type Trp53 allele and expression of p19ARF, providing further evidence of negative-feedback control of the latter by p53. However, expression of p19ARF does not appear to be counterselected in the absence of p53, and its integrity in Trp53+/− tumors was indicated by its transcriptional upregulation on Trp53 wild-type allele loss in vitro in selected tumor cell lines. The role of MMLV was investigated further by analysis of proviral insertion sites in tumors of CD2-myc transgenic mice sorted for Trp53 genotype. A proportion of tumors showed insertions at Runx2, an oncogene which has been shown to collaborate independently with CD2-myc and with the Trp53 null genotype, and at a novel common integration site (ptl-1) on chromosome 8. Genotypic analysis of the panel of tumors suggested that neither of these integrations is functionally redundant with loss of p53, but it appears that the combination of the MMLV oncogenic program with the CD2-myc oncogene relegates p53 loss to a late step in tumor progression or in vitro culture. While the means by which these tumors preempt the p53 tumor suppressor response remains to be established, this study provides further evidence that irreversible inactivation of this pathway is not a prerequisite for tumor development in vivo.
Neonatal infection of mice with Moloney murine leukemia virus (MMLV) is potently oncogenic, leading to the development of thymic lymphoma within 3 to 4 months (9). Much evidence suggests that the virus plays a direct role in the initiation and maintenance of these tumors, which display clonal integrations of proviral sequences adjacent to proto-oncogenes such as c-myc, Pim-1, or Gfi-1. Indeed, analysis of common proviral insertion sites in MMLV-induced lymphomas has led to the identification of a large number of genes which can act as dominant oncogenes in the malignant transformation of lymphocytes (reviewed in reference 16). The importance of insertional mutagenesis and transcriptional activation of host cell genes in the pathogenesis of MMLV-induced tumors is underlined by the fact that the transcriptional enhancer elements within the viral long terminal repeats are key determinants of its oncogenic potency and targeting to the lymphoid compartment (10, 20, 21).
In contrast, the role of tumor suppressor pathways in MMLV tumors is virtually unknown. A genome-wide scan for loss of heterozygosity in MMLV-induced tumors of BRAKJ/F1 mice showed that this was an infrequent event, with no evidence of changes affecting the murine Trp53 locus on chromosome 11 (19). We explored this issue directly by examining the effect of the Trp53−/− genotype on MMLV lymphomagenesis. Although MMLV infection and the Trp53−/− genotype independently led to thymic lymphomas with similar kinetics and phenotypic characteristics, we found that the combination was only weakly additive (1). Furthermore, there was no obvious effect of the Trp53−/− genotype on the spectrum of proto-oncogenes targeted by MMLV in the resultant lymphomas. From these observations we postulated that MMLV lymphomagenesis bypasses the need for genetic inactivation of p53 (1).
Recent studies show that the p53 pathway is coupled to oncogene overexpression. The p19ARF protein, the product of one of the overlapping reading frames at the p16INK4a/p19ARF locus, appears to stabilize p53 through antagonism of its interaction with Mdm2 (36). Moreover, the expression of p19ARF has been reported to be induced in response to overexpression of myc and viral oncogenes such as E1A (6, 37). It appears, therefore, that the p53 pathway acts to protect cells from the consequences of inappropriate proliferative signals as well as genotoxic insult. As p53 itself appears to down-regulate p19ARF as well as Mdm2, the p19ARF-Mdm2-p53 pathway represents a feedback loop which integrates and coordinates the cellular response to a variety of conflicting growth signals (8). The importance of this fail-safe mechanism is indicated by the rapid onset of tumors in mice carrying an overexpressed myc oncogene in combination with defective p53 (4, 13).
To explore the role of MMLV further, we tested the effects of viral infection in mice that carry a CD2-myc oncogene and were either homozygous or heterozygous for an inactivated Trp53 allele. We found that neonatal infection with MMLV could accelerate tumor onset even on the highly tumor-prone CD2-myc Trp53−/− background and that the lack of functional p53 had little or no discernible effect on the range of genes targeted by proviral insertion. The most striking effect, however, was seen in CD2-myc Trp53+/− mice, where viral infection promoted the outgrowth of tumors which retained the wild-type Trp53 allele in apparently functional form. From these results we hypothesize that MMLV leukemogenesis involves uncoupling of the hyperproliferative effects of oncogenes such as myc from the fail-safe mechanisms mediated by p53.
MATERIALS AND METHODS
Transgenic mice
The targeted inactivation of the Trp53 gene and introduction into the mouse germ line have been described previously: the null mice were derived by homologous recombination in 129/Sv murine stem cells, crossed onto a C57BL/6 background (7), and subsequently crossed with NIH mice. The generation of transgenic mice carrying human c-myc under the CD2 locus control region has also been described (35): in short, a linearized CD2-myc plasmid construct was microinjected into (C57BL/6 × CBA/Ca)F2 fertilized eggs. Trp53−/− and CD2-myc mice were crossed to generate progeny heterozygous for both Trp53 and CD2-myc. These F1 animals were crossed to give mice of all six possible genotypes and also backcrossed with Trp53 null mice to produce approximately equal numbers of Trp53−/−/CD2-myc-positive, Trp53+/−/CD2-myc-positive, Trp53−/−, and Trp53+/− mice. Mice were infected within 24 h of birth with 105 infectious units of MMLV (34). Animals were routinely monitored and sacrificed when cachexia was noted.
Establishment of cells in culture.
Tumor tissues were disaggregated in RPMI 1640 medium (Gibco Life Technologies) using scalpel blades. Lymphocytes were separated on a Ficoll-Paque density gradient (Pharmacia) at 3,000 rpm for 10 min, washed in RPMI, and cultured in RPMI containing 10% fetal calf serum, penicillin, streptomycin, 10 mM HEPES, and 50 μM 2-mercaptoethanol.
DNA hybridization analysis.
High-molecular-weight DNA was isolated from mouse tissues and cultured cells using either guanidinium hydrochloride or the Nucleon kit (Scotlab, Coatbridge, Scotland). Digestion of the DNA with restriction enzymes, electrophoresis, hybridization, and autoradiography were carried out as previously described (26). Transfer to Hybond-N membranes was carried out according to the manufacturer's recommendations (Amersham Pharmacia). Probes were radiolabeled by random priming using [α-32P]dCTP (3,000 Ci/mmol; Amersham Pharmacia) to specific activities of 2 × 108/μg or greater. The Trp53 null and wild-type (wt) alleles were detected, along with the pseudogene on BamHI-digested DNA by Southern blotting using a PCR-generated Trp53 exon 4 probe described previously (1). The CD2-myc transgene was detected as a 25-kb band on reprobing the same blots with the CD2-myc probe described previously (31). Tumors, kidney metastases, and cell lines were screened for rearrangements in the Pim-1 locus using a probe previously described (clone A) and in the bmi-1 and pal-1 loci using mouse bmi-1 cDNA clone 13.1 and the pal-1 probe 11A2 (34). Tumors, kidney metastases, and cell lines were also screened for rearrangements at Gfi-1, Ahi-1, and Evi-5 as described previously (10, 22, 27) and at til-1 using the til-1E probe (32). Kidney metastases were screened for rearrangements at the loci mentioned above and at Frat-1 (17). Rearrangements of the T-cell receptor (TCR) β-chain gene and the immunoglobulin heavy-chain (IgH) gene were determined using a 496-bp PCR-derived fragment of the Cβ gene, derived from the 1.2-kb fragment of clone 86T5 (12) and a 1.7-kb EcoRI-BamHI fragment of plasmid J11 (24), respectively. A PCR probe for the U3 region of the MMLV long terminal repeat was derived using primers U3a, CCACCTGTAGGTTTGGCAAGC, and U3b, GGTCATTTCCAGGTCCTTGG, and used for estimation of proviral copy number. Rearrangements at exon 1β of p19ARF were examined by Southern blotting using AflII-digested tumor DNA and an exon 1β-specific probe (8).
Western analysis.
Levels of p53 in cell lines were investigated by Western blotting as described elsewhere (5) using the Pab240 monoclonal antibody (Oncogene Research Products) and a horseradish peroxidase (HRP)-conjugated secondary anti-mouse antibody (Sigma). The antibody complex was visualized by enhanced chemiluminesence (Amersham Pharmacia). The cell line BW5147 was used as a positive control for Trp53 expression (European Collection of Animal Cell Cultures, Salisbury, United Kingdom). p19ARF was detected using a rabbit polyclonal primary antibody raised against an internal peptide (residues 54 to 75) (R562; AbCam Ltd.) and detected using HRP-conjugated goat anti-rabbit secondary (Sigma). p21WAF1 was detected using a goat polyclonal primary antibody (sc397-G; Santa Cruz Biotechnology) and an HRP-conjugated anti-goat secondary antibody (Sigma). p16INK4a was detected using a rabbit polyclonal primary antibody (sc1207; Santa Cruz Biotechnology) and a goat anti-rabbit-HRP conjugate (Sigma). β-actin was also detected as a loading control using a goat polyclonal antibody (sc1616; Santa Cruz Biotechnology).
RT-PCR analysis.
Levels of Mdm2 and p19ARF transcript were investigated in primary tumor cells and tumor cell lines by reverse transcription (RT)-PCR: first- strand cDNA was generated from total RNA isolated using RNAzolB (Tel-Test Inc.) with enhanced avian myeloblastosis virus reverse transcriptase (Sigma) or Superscript-enhanced MMLV reverse transcriptase (Gibco-BRL). For Mdm2, 2 μl of first-strand reaction mixture was placed in a standard 50-μl PCR mixture containing primers Mdm2cn (5′ CAGCTTCGGAACAAGAGACTC 3′) and Mdm2d (5′ CTGTGCTCCTTCACAGAGAAAC 3′) each at 10 μM. PCR was carried out using Taq polymerase (Perkin-Elmer) at 35 cycles of 45 s of denaturation at 94°C, 45 s of annealing at 60°C, and 90 s of elongation at 72°C. p19ARF transcripts were detected using identical conditions except that the primers were the same as those used in a previous study (28). The RT-PCR product from one representative reaction was cloned and sequenced to ensure that the product was as expected.
Flow cytometry.
Flow cytometry for cell phenotype was carried out as described previously using R-phycoerythrin-conjugated anti-mouse CD4 (Serotec), fluorescein isothiocyanate-conjugated anti-mouse CD8 (Serotec/Sigma) and Quantum Red- or fluorescein isothiocyanate-conjugated anti-mouse CD3 (Serotec/Sigma). For the detection of apoptosis, 106 cells were gamma irradiated (5 Gy), cultured as normal for 4 h, harvested by centrifugation, and washed in phosphate-buffered saline. The cells were resuspended and incubated for 15 min in a buffer (10 mM HEPES [pH 7.4], 140 mM NaCl, 5 mM CaCl2) containing fluorescein-conjugated annexin V (Roche Molecular Biochemicals). Control cells established from a CD2-myc-induced thymic lymphoma were treated with 10 μM dexamethasone (Sigma) for 24 h prior to analysis. Annexin V staining increased from 21 to 68% following treatment. All samples were analyzed on a Coulter Epics Elite.
Statistics.
Survival analysis of tumor-prone animals was carried out using the Mann-Whitney rank sum test.
Nucleotide sequence accession number.
The sequence data have been submitted to the DDBJ/EMBL/GenBank databases under accession no. AJ302067.
RESULTS
Collaboration of MMLV and Trp53 loss in T-cell lymphomas of CD2-myc mice: effects on rate of tumor development and tumor cell growth in vitro.
Our previous studies of CD2-myc mice showed that the combination of overexpressed myc and an inactive Trp53 gene is strongly synergistic in promoting lymphoid tumors in mice (4). However, the clonal nature of the tumors which emerged indicated that further selective events are operative in the genesis of these lymphomas. This led us to ask whether neonatal infection of these highly tumor-prone mice with MMLV could reduce the latent period for tumor development still further and whether the virus could be used as a probe for the complementing oncogenic pathways involved.
CD2-myc mice bred onto a Trp53−/− background and littermate controls were infected with MMLV (34) and sacrificed when clinical signs of neoplastic disease were noted. All infected animals developed thymic lymphoma although one Trp53−/−, CD2-myc MMLV-infected mouse also had testicular carcinoma. The lymphomas were characterized by gross thymic enlargement and frequent metastatic spread to other lymphoid organs, especially the spleen and lymph nodes. Metastatic spread to nonlymphoid organs was also evident: 91% of Trp53−/− mice, 78% of Trp53+/− mice, and 62% of Trp53+/+ mice with thymic lymphoma had metastatic cells in the kidney, as demonstrated by Southern blot analysis of kidney DNA using TCR and IgH probes (data not shown).
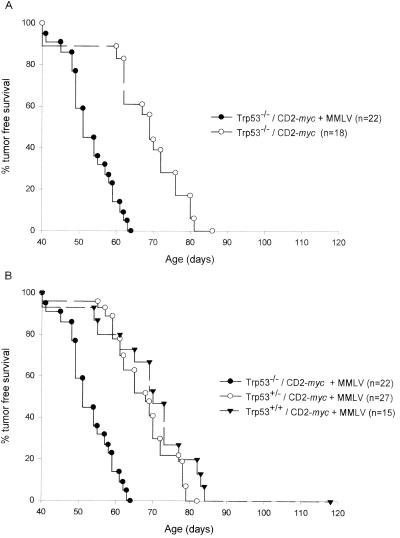
Comparison of the kinetics of tumor development shows that Trp53−/− CD2-myc mice infected with MMLV developed thymic lymphoma significantly faster than uninfected controls (P < 0.001) (Fig. 1A). Figure 1B shows that the status of Trp53 strongly influenced the latency of tumor development in MMLV-infected CD2-myc mice, with Trp53−/− animals showing significantly reduced survival compared with that of heterozygotes for Trp53 (P < 0.001). Trp53+/− mice seemed to develop tumors with reduced latency compared with that of Trp53+/+ littermates, but this difference was not statistically significant, in contrast to the accelerated onset seen in MMLV-infected Trp53+/− mice compared with Trp53+/+ controls in the absence of CD2-myc (1).
FIG. 1.
(A) MMLV infection accelerates the development of thymic lymphoma in Trp53−/− CD2-myc mice. Tumor-free survival was significantly decreased in MMLV-infected Trp53−/− CD2-myc mice compared to that in uninfected mice of the same genotype (P < 0.001). (B) Loss of p53 influences the latency of tumor development in infected CD2-myc mice. Trp53−/− CD2-myc MMLV-infected mice develop tumors significantly faster than Trp53+/− or Trp53+/+ CD2-myc MMLV-infected mice (P < 0.001 for both).
Flow cytometry analysis showed that MMLV-induced tumors from CD2-myc mice of all Trp53 genotypes were of T-cell origin. The tumor phenotypes from both these groups were similar to those observed previously in MMLV-infected, CD2-myc mice, i.e., CD3+ CD4+ CD8+, with a subset of CD3+ CD8+ SP tumors (31). The conclusion from this phenotypic analysis is that germ line inactivation of Trp53 does not grossly alter the tumor cell phenotype in MMLV-infected CD2-myc mice.
As reported previously, the lack of functional p53 in Trp53−/− mice has a significant effect on the propagation of lymphoma cells in vitro. Trp53-deficient, CD2-myc transgenic thymic lymphoma cell lines were easily established in culture, without addition of exogenous growth factors (4). Those with a single functional allele were also predisposed to immortalization. Cultures of MMLV-induced, CD2-myc tumor cells were established from 17 of the 21 Trp53−/− tumors and 18 of the 29 Trp53+/− tumors in this study.
MMLV promotes the development of tumors that retain functional, wild-type Trp53 in CD2-myc Trp53+/− mice.
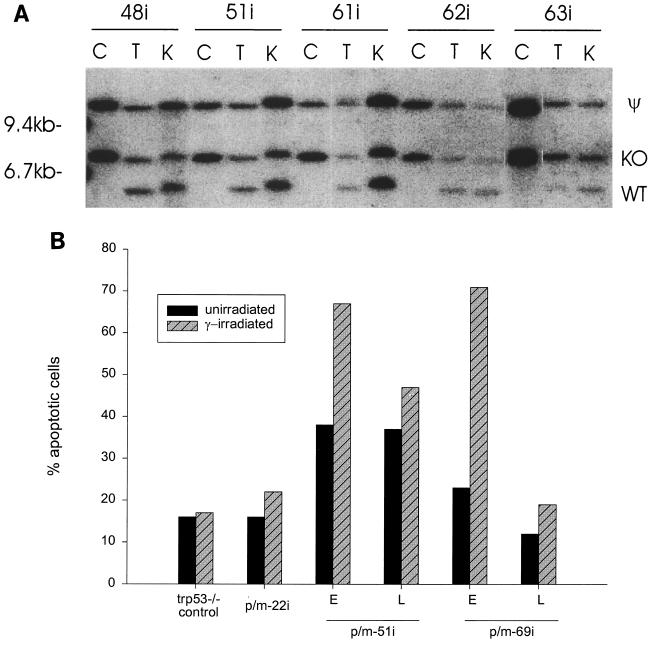
If MMLV infection can bypass the requirement for inactivation of Trp53 in lymphoma development, we postulated that there should be no selective advantage for loss of the Trp53 wt allele in MMLV-induced tumors in Trp53+/− mice. This prediction was borne out by Southern blot analyses of Trp53 in MMLV-infected, CD2-myc lymphomas. Examples of these analyses are shown in Fig. 2A, and the results are summarized in Table 1. Extensive loss of the Trp53 wt allele was observed in 13 of 13 tumors from Trp53+/− CD2-myc mice (4), while only 2 of 27 of the equivalent MMLV-infected lymphomas showed complete loss of the Trp53 wt allele, with a further 6 of 27 showing very slight loss of the wt Trp53 allele. However, cell lines established from Trp53+/− tumors ultimately lost the wild-type allele or showed evidence of mutational stabilization and functional loss. In accord with a previous study, we found no evidence of Trp53 inactivation by proviral insertion or other gross rearrangements.
FIG. 2.
(A) Loss of the Trp53 wild-type allele (WT) is uncommon in MMLV-induced, Trp53+/− CD2-myc primary tumors (T) and in kidney metastases (K) but is common in the resulting established cell lines (C). The pseudogene (ψ) and homologously recombined, knocked-out allele (KO) are indicated. Tumors judged not to have lost the wild-type allele were found to have wt Trp53 alleles with intensity of at least 90% of that of the knockout allele by Southern blot analysis. Loss of the Trp53 wild-type allele is evident in p/m63i cell line, tumor, and kidney DNA but retention of the Trp53 wild-type allele is seen in p/m48i, p/m51i, p/m61i, and p/m62i tumor and kidney DNA. (B) Apoptosis response to gamma irradiation in Trp53+/− cell lines by flow cytometry. Induction of apoptosis, as identified by annexin V staining, was found in early passages (E) of p/m51i and p/m69i cell lines following irradiation (hatched bar). A greatly reduced apoptotic response to gamma irradiation was observed in later passages (L) of these cell lines, coinciding with loss of the Trp53 wt allele as shown by Southern blot analysis. Gamma-irradiated Trp53−/− cells were included as a negative control, and dexamethasone-treated lymphoma cells were used as a positive control.
TABLE 1.
Retention of the Trp53 wild-type allele in spontaneous and MMLV-induced thymic lymphomas in CD2-myc Trp53+/− mice
| MMLV status | No. of tumors with Trp53 allele retentiona/no. tested |
|---|---|
| Infected | 19/27 |
| Uninfected | 0/13 |
Tumors were judged to have retained the Trp53 allele if the intensity of the wild-type band was >90% of that of the Trp53 pseudogene on the same Southern blot (Fig. 2A). Tumors regarded as having LOH showed reductions of 30 to 100%.
Although in vitro culture selects strongly for loss of the p53 pathway, we were able to establish three lines (p/m22i, p/m51i, and p/m69i) which retained an ostensibly wild-type allele for a prolonged period of culture. The fact that the Trp53 wild-type allele was ultimately lost in two of these lines (p/m51i and p/m69i) provides prima facie evidence that it was functional in the primary tumor and early-passage cells. These cell lines were also examined for their apoptotic response to gamma irradiation by annexin V staining. Early-passage cell lines showed a strong response (Fig. 2B), while the other cell line (p/m22i, which expresses high levels of p53 by Western blotting, consistent with a mutant protein [data not shown]) was refractory to this stimulus. The presence of a mutant allele in this cell line is confirmed by the retention of the allele by Southern blotting on long-term culture. The presence of a functional Trp53 gene in early passages of p/m51i and p/m69i cells was further confirmed by the detection of p21WAF1 on Western blots of cell extracts from the gamma-irradiated cells (data not shown).
p19ARF expression in CD2-myc lymphomas is inversely correlated with the presence of an intact Trp53 wt allele and can be reactivated on Trp53 loss in vitro.
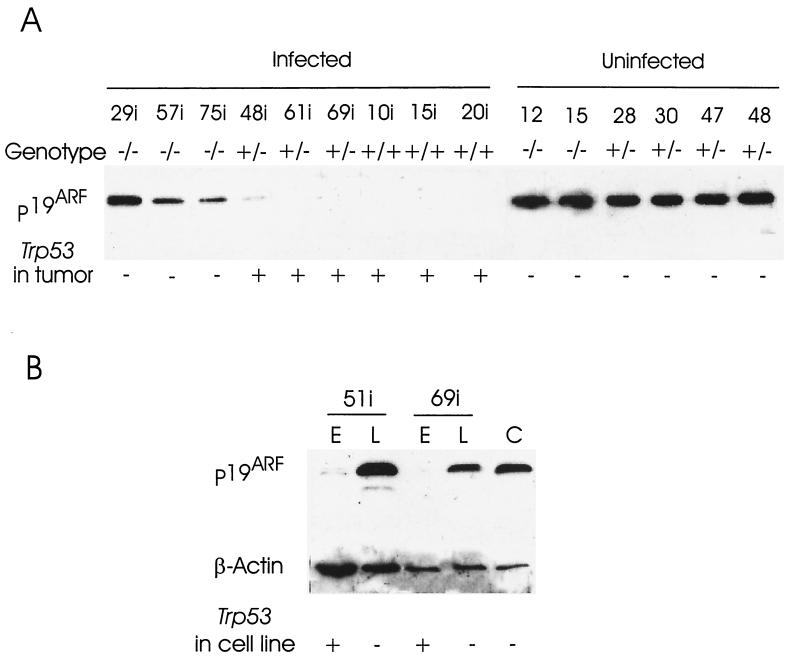
The elucidation of the p19ARF-p53 regulatory loop has revealed another means by which the p53 pathway can be compromised in tumors (8). Moreover, the evidence that this pathway couples overexpression of myc to the p53 response (37) suggested that it may be particularly relevant to this study, as the Trp53−/− genotype has a profound effect on the development of lymphomas in CD2-myc mice (4). Southern blot analysis of the lymphoma series revealed no gross deletions or rearrangements of the p16INK4a/p19ARF locus (not shown). We therefore examined the expression of p19ARF and p16INK4a, the products of alternative and overlapping exons at the p16INK4a/p19ARF locus (28). p19ARF was readily detected by Western blotting and showed a mainly bimodal expression pattern, being either strongly expressed or below detectable levels. Only a few tumors failed to conform to this pattern, showing a low level of p19ARF protein, for example p/m48i (Fig. 3A and Table 2). Regulation of p19ARF expression appeared to be mainly at the transcriptional level, as RT-PCR analysis yielded results essentially identical to those of the Western blots (not shown). Western blot analysis revealed no detectable expression of p16INK4a in cell lines overexpressing p19ARF, suggesting that the two products of the p16INK4a/p19ARF locus are not coordinately expressed (data not shown).
FIG. 3.
Western analysis of p19ARF. (A) Expression of p19ARF is commonly down-regulated in MMLV-induced CD2-myc primary tumors from Trp53+/− and Trp53+/+ mice but not in those from Trp53−/− littermates. In contrast, p19ARF expression is strong in spontaneous primary tumors from Trp53−/− and Trp53+/− mice. It should be noted that the spontaneous tumors (p/m28, 30, 47, 48) from Trp53+/− mice lost the Trp53 wt allele. Gels were routinely run in parallel and stained using Coomassie blue to ensure that there were equal amounts of protein in each lane on the Western blots. The presence or absence of the Trp53 wild-type allele is indicated in each case. (B) Repression of p19ARF expression in the presence of p53. p19ARF expression is repressed in early passages (E) of two cell lines which maintain functional p53, but the repression is not irreversible since on later passage (L), p53 is lost and p19ARF expression is comparable to that of a control cell line (C) that lacks functional p53. Amounts of protein loaded are indicated by β-actin staining.
TABLE 2.
p19ARF expression in thymic lymphomas of MMLV-infected and control mice sorted by Trp53 and CD2-myc genotype
| Genotype | No. of tumors expressing p19ARF/no. tested
|
||
|---|---|---|---|
| CD2-myc carrier, MMLV infected | CD2-myc carrier, noninfected | Lacking CD2-myc, MMLV infected | |
| Trp53−/− | 19/19 | 11/12 | 8/8 |
| Trp53+/− with LOH | 4/8 | 13/13 | 12/13 |
| Trp53+/− without LOH | 4/19 | ||
| Trp+/+ | 0/13 | NDa | 3/9 |
ND, not determined.
The most striking conclusion of this analysis was the strong inverse correlation between the presence of a functional Trp53 gene and expression of p19ARF in primary lymphomas, as noted elsewhere (8). p19ARF was not detected in any of the primary lymphomas arising in MLV-infected Trp53+/+ mice carrying the CD2-myc transgene. Its expression in 4 of 19 Trp53+/− tumors displaying no grossly measurable loss of heterozygosity (LOH) at the Trp53 locus could be explained in one case by loss of function due to a p53 stabilizing mutation (p/m22i), while the other three expressed low levels (p/m32i, p/m47i, and p/m48i), consistent with an emergent minor clone which had lost or mutated the Trp53 wild-type allele. Expression was detected in a higher proportion of Trp53+/− lymphomas which displayed measurable loss of the wild-type allele (4 of 8). The fact that not all scored positive suggests either that there is a time lag between loss of p53 and p19ARF expression or that some of these tumors acquire multiple lesions in the p19ARF-p53 pathway.
This question was explored further in two Trp53+/− lymphoma cell lines, which retained the wild-type allele on early passage (p/m51i and p/m69i) and which were examined for expression of p19ARF before and after they had lost the wild-type Trp53 allele. As shown in Fig. 3B, early-passage (passage 3) cell lines showed little (p/m69i) or no (p/m51i) detectable p19ARF expression, while the late-passage (passage 22) cell lines expressed high levels. Analysis of these lines for rearrangement of TCR and IgH genes established their clonal nature and confirmed that the late-passage cells had not been overgrown by an independently transformed cell clone (not shown). These results are consistent with the view that p19ARF is intact but transcriptionally inactive in the primary lymphoma and early-passage cells. Loss of p53 function appears to derepress p19ARF both in vivo and in vitro.
The fact that p19ARF expression was detected in 3 of 9 lymphomas of MLV-infected wild-type mice was somewhat unexpected and implies that lesions in p53 or its downstream negative regulatory pathway might be more common than previously suspected in these tumors.
Analysis of MMLV target genes in CD2-myc Trp53−/− lymphomas reveals Runx2/til-1 and ptl-1, a novel insertion locus on mouse chromosome 8.
The tumors induced by MMLV in CD2-myc mice were monoclonal or oligoclonal as judged by Southern blotting using TCR and IgH probes, even on the Trp53−/− background. The number of fragments detected with the U3 probe indicated that the tumors were composed of a small number of clones, each of which contained a small number of clonal proviral integrations (Table 3). These data suggested that mutagenic events mediated by proviral insertion were most likely responsible for the acceleration of tumorigenesis by MMLV.
TABLE 3.
Clonality and proviral integrations in MMLV-accelerated CD2-myc tumors sorted according to Trp53 genotype
| Trp53 genotype | Avg clonalitya | Avg proviral copy no.b | No. of tumors/no. tested with insertions at:
|
||
|---|---|---|---|---|---|
| Til-1/Runx2 | Pim-1 | ptl-1 | |||
| −/− | 3.33 | 6.55 | 4/19 | 0/19 | 1/19 |
| +/− | 2.85 | 6.46 | 8/26c | 2/26 | 1/26 |
| +/+ | 2.69 | 6.62 | 4/13 | 0/13 | 0/13 |
Assessed by rearrangement of IgH genes.
Assessed by U3 LTR probe.
Rearrangements at Til-1 were detected in 4 of 8 tumors which had retained the Trp53 wild-type allele and also in 3 of 8 tumors which had lost it. One tumor displayed a rearranged Til-1 allele and a mutant Trp53 allele (p/m22i). Both Pim-1-rearranged tumors showed no LOH at Trp53.
In an attempt to identify the mutagenic targets that might be responsible for the acceleration of tumor onset by MMLV, we screened each tumor using a panel of known insertion sites, including Ahi-1, Bmi-1, Pim-1, Til-1, Evi-5, and Pal-1/Gfi-1. Kidney metastases were also screened for rearrangements at these loci and also for rearrangements at the tumor progression gene Frat-1. The data in Table 3 show that Til-1/Runx2 was rearranged in a number of tumors: three of eight of these Til-1 rearranged tumors showed LOH at Trp53, four of eight showed no LOH, and one had a mutant Trp53 allele (p/m22i). This gene is also a frequent target of MMLV activation in virus-accelerated tumors of CD2-myc mice (32). A small number of Pim-1 rearrangements (2 of 58) were present in these tumors, as noted previously in Trp53+/+ MMLV-induced CD2-myc tumors (31). Neither of the tumors showed LOH at Trp53.
The relatively low hit rate at known integration sites suggested that novel gene targets might be involved. To investigate further, genomic libraries were constructed using DNA from two MMLV-induced CD2-myc Trp53−/− tumor cell lines (p/m6i and p/m29i), which both harbored three proviral integrations and lacked rearrangements at any of the viral insertion loci mentioned above. Junction fragments were cloned from the p/m6i library as described previously (33). Two novel integration sites were identified from this tumor cell line and were termed ptl-1 and ptl-2 (for proviral insertion in T-lymphoma). The ptl-1 locus was identified as a common integration site since it was rearranged in a kidney metastasis of an independently derived tumor (p/m27i). The two novel sites mapped to chromosomes 8 (ptl-1) and 3 (ptl-2) (logarithm of odds scores of 13 and 3, respectively; European Interspecific Collaborative Backcross; HGMP, Hinxton Hall, United Kingdom).
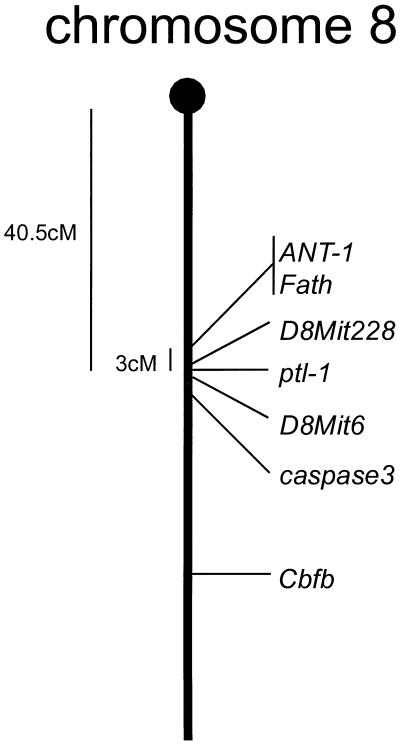
While no gene has yet been identified at ptl-1, the locus is close to markers D8Mit6, D8Mit228, and the two adjacent genes Ant1 and Fath that colocalize on mouse chromosome 8 in a region syntenic with human chromosome 4q34-q35 (11), as shown in Fig. 4. Genomic clones containing the integration site were obtained by subcloning BAC clones positive for a single copy probe for ptl-1, and approximately 7 kb of sequence was determined.
FIG. 4.
Chromosomal localization of ptl-1. The locus was mapped to chromosome 8 with a logarithm of odds score of 13. ptl-1 mapped close to markers D8Mit6 and D8Mit228, 40 centimorgans from the centromere.
Table 3 lists the identified viral insertion targets sorted according to the Trp53 genotype of the tumor-bearing mouse. From inspection of these results it is clear that neither Til-1/Runx2 nor ptl-1 is functionally redundant with Trp53 loss, as the hit rate in Trp53−/− mice was not significantly different from that of other genotypic groups. Insertions at Pim-1 were found in two Trp53+/− tumors but were absent from the Trp53−/− group. Although a similar trend has been noted previously in Trp53 mutant mice lacking the myc transgene (1), the low numbers fail to reach statistical significance. In this respect it is interesting that the average number of proviral integrations was not markedly different in the three Trp53 genotypes, suggesting that the genetic program targeted by MMLV in accelerated CD2-myc lymphomas may be unaffected by Trp53 gene dose (Table 3).
DISCUSSION
In CD2-myc mice, thymic lymphomas arise stochastically with a lifetime incidence of 3 to 18% (31). Neonatal infection of these mice with MMLV leads to 100% tumor incidence with much earlier onset (average time, 71 days). A remarkably similar effect on tumor incidence and onset is seen when CD2-myc mice are crossed onto a Trp53−/− background, with all mice succumbing to T-cell lymphoma at an average age of 72 days (4). In contrast, MMLV infection of Trp53−/− mice shows only a weakly additive effect on the kinetics of tumor development (1). From these observations we suggested that the MMLV lymphomagenic pathway may be at least partially overlapping and functionally redundant with p53 loss. This study reinforces these findings and shows further that MMLV significantly reduces the selective pressure on the p53 pathway in CD2-myc mice, promoting the outgrowth of tumors that retain the p53 gene and preserve at least some of its downstream functions. However, the pressure on p53 is not lost entirely and becomes evident when tumor cells are propagated in vitro.
According to Knudson's two-hit model for inherited tumor suppressor gene function, loss of both copies of the gene is a prerequisite for tumor outgrowth (18). This model applies imperfectly to p53, for which there is evidence that the loss of one allele can predispose to tumorigenesis and some tumors arise without loss of the wild-type allele (35). An extensive study of wild-type Trp53 loss in primary tumors of Trp53 heterozygous mice showed that tumors arising in older mice are more likely to retain the wild-type allele in a functional state, regardless of the type of malignancy, with rates of loss declining from 60 to around 15% in tumors arising at 21 months of age (35). In contrast, we found that 100% of lymphomas in CD2-myc Trp53+/− mice had undergone significant loss of the wild-type allele at the primary tumor stage, despite their relatively late onset, suggesting that in this case p53 inactivation is a rate-limiting event which is required for the growth of tumors to clinically detectable size. However, in CD2-myc Trp53+/− mice infected with MMLV, this trend was reversed, with tumors arising at an early age with a relatively low rate of allele loss (25%).
How might MMLV relieve the selective pressure for loss of p53 in lymphomagenesis? In contrast to Friend MLV-induced erythroleukemias (25), we found no evidence of virus-induced rearrangements of the Trp53 gene itself. Moreover, several lines of evidence indicate that the normal Trp53 allele is functionally intact in most primary tumors of MMLV-infected CD2-myc Trp53+/− mice. First, Southern blot analyses showed that the normal allele was lost during the establishment of cell lines from the primary tumors. The defective targeted Trp53 allele and the pseudogene were unaffected, showing that this loss was not merely the result of random karyotypic variations in aneuploid tumor cells. Secondly, two lymphoma cell lines that retained the wild-type allele for a significant prolonged period of culture showed an intact p53 response as revealed by expression of p21WAF1 and apoptotic markers following exposure to ionizing radiation. It appears that the tumor suppressor function of p53 is either quiescent or neutralized in these tumors, despite the presence of the constitutively expressed CD2-myc gene.
The expression status of the p16INK4a/p19ARF locus was of interest in these tumors since p19ARF, one of the overlapping products of this locus, can stabilize p53 through antagonism of its interaction with Mdm2 (36) and has been reported to be induced in response to overexpression of myc and viral oncogenes such as E1A (6, 37). We found that p19ARF was highly expressed in virtually all Trp53−/− tumors and was commonly observed in Trp53+/− lymphomas, which had demonstrable loss of the wild-type p53 allele, indicating that the postulated negative-feedback loop connecting p53 and p19ARF is operative in T-lymphoma cells (8). Two Trp53+/− cell lines that lost the wild-type allele on prolonged culture showed concomitant up-regulation of p19ARF expression, providing further evidence that p53 was intact prior to allele loss and showing that nonexpression of p19ARF in the corresponding primary tumors was not due to the deletions, nonsense mutations, or DNA methylation which can affect the p16INK4a/p19ARF locus in human tumors of various lineages (reviewed in reference 29).
The lack of apparent pressure to lose p19ARF expression in MMLV-infected Trp53−/− tumors suggests that its p53-independent apoptotic effects noted in primary B cells (8) are either inoperative in T cells or are neutralized by the MMLV lymphomagenic program in T lymphomas. In this respect it is interesting that screening of MMLV insertion sites in our tumor series revealed no hits at the Bmi-1 locus, a gene collaborating with myc that represses transcription of the p16INK4a/p19ARF locus (15). While activation of Bmi-1 by MMLV is a common feature of B-cell lymphomas of Eμ-myc mice (34), it is very rare in MMLV-induced T-cell lymphomas, including virus-accelerated tumors of CD2-myc mice (33). Intriguingly, this lineage preference is echoed in the spectrum of tumors which arise in Ink4a−/− mice, which develop either B-cell lymphomas or sarcomas (30), while Trp53−/− mice develop mainly T-cell lymphomas (7).
It is conceivable, nevertheless, that the combination of MMLV functions with the CD2-myc transgene overcomes the requirement for p53 inactivation during the early stages of neoplastic transformation. A precedent is provided by a prostate cancer model in which the combination of myc and ras appears to render p53 loss redundant (23). Screening of known MMLV insertion sites in this tumor series revealed a significant rate of insertions at Runx2, a gene collaborating with myc that was identified previously in CD2-myc mice (32). However, insertions at this locus were distributed across Trp53 genotype groups, showing that Runx2 activation is not functionally equivalent to p53 loss. In accord with this interpretation, recent evidence shows that a CD2-Runx2 oncogene is strongly synergistic with the Trp53−/− genotype (3). Screening for new insertion sites revealed a novel common insertion locus, ptl-1, which we mapped to mouse chromosome 8. The relevant target gene for these insertions has not yet been identified, but, as noted for Runx2, these insertions were also found in Trp53−/− lymphomas. While insertions at Pim-1 revealed an apparent complementary pattern with Trp53 genotype, the low number of cases recorded indicates that caution should be exercised in drawing any conclusions at this stage.
As MMLV-accelerated tumors of CD2-myc mice appear to be refractory to p19ARF-p53 surveillance, a further possibility which merits consideration is that the MMLV lymphomagenic program uncouples the p19ARF-p53 response to myc overexpression, without necessarily preventing its induction by other stimuli. According to this model, the p19ARF-p53 pathway is in waiting mode in MMLV-induced lymphomas, as it appears to be in normal cells which express only very low levels of p19ARF (29). The later loss of p53 and its functions on in vitro propagation or in metastatic tumors in vivo could then be accounted for by the induction of the p19ARF-p53 pathway in response to growth crisis or suboptimal levels of essential survival factors, and the accumulation of p19ARF in Trp53−/− lymphoma cells could be a consequence of loss of p53-mediated repression rather than its de novo induction by myc. In support of this notion, it has been shown recently that p19ARF can be induced in human tumor cell lines by serum deprivation as well as oncogene-mediated hyperproliferation (14). The intriguing possibility that myc overexpression is uncoupled from p19ARF-p53 in MMLV-accelerated lymphomas may be testable by the use of regulatable myc transgenes (2) in combination with MMLV.
ACKNOWLEDGMENTS
We are grateful to the Leukaemia Research Fund and the Cancer Research Campaign for support of this work.
We also thank the European Collaborative Interspecific Backcross (EUCIB, Cambridge, United Kingdom) for their invaluable assistance. We thank Marie Anne Duffy, Margaret Bell, and Monica Cunningham for technical assistance and Ming Hu and Francois Vaillant for help and advice.
REFERENCES
- 1.Baxter E W, Blyth K, Donehower L A, Cameron E R, Onions D E, Neil J C. Moloney murine leukemia virus-induced lymphomas in p53- deficient mice: overlapping pathways in tumor development? J Virol. 1996;70:2095–2100. doi: 10.1128/jvi.70.4.2095-2100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blyth K, Stewart M, Bell M, James C, Evan G, Neil J C, Cameron E R. Sensitivity to myc-induced apoptosis is retained in spontaneous and transplanted lymphomas of CD2-mycER mice. Oncogene. 2000;19:773–782. doi: 10.1038/sj.onc.1203321. [DOI] [PubMed] [Google Scholar]
- 3.Blyth K, Terry A, Mackay N, Vaillant F, Bell M, Cameron E R, Neil J C, Stewart M. Runx2: a novel oncogenic pathway revealed by in vivo complementation and retroviral tagging. Oncogene. 2001;20:295–302. doi: 10.1038/sj.onc.1204090. [DOI] [PubMed] [Google Scholar]
- 4.Blyth K, Terry A, O'Hara M, Baxter E W, Campbell M, Stewart M, Donehower L A, Onions D E, Neil J C, Cameron E R. Synergy between a human c-myc transgene and p53 null genotype in murine thymic lymphomas–contrasting effects of homozygous and heterozygous p53 loss. Oncogene. 1995;10:1717–1723. [PubMed] [Google Scholar]
- 5.Crouch D H, Lang C, Gillespie D A H. The leucine zipper motif of avian c-myc is required for transformation and autoregulation. Oncogene. 1990;5:683–689. [PubMed] [Google Scholar]
- 6.De Stanchina E, McCurrach M E, Zindy F, Shieh S-Y, Ferbeyre G, Samuelson A V, Prives C, Roussel M F, Sherr C J, Lowe S W. E1A signalling to p53 involves the p19ARF tumour suppressor. Genes Dev. 1998;12:2434–2442. doi: 10.1101/gad.12.15.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donehower L A, Harvey M, Slagle B L, McArthur M J, Montgomery C A, Butel J S, Bradley A. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 8.Eischen C M, Weber J D, Roussel M F, Sherr C J, Cleveland J L. Disruption of the ARF-Mdm2–p53 tumour suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 1999;13:2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fan H. Leukemogenesis by Moloney murine leukemia virus: a multistep process. Trends Microbiol. 1997;5:74–82. doi: 10.1016/S0966-842X(96)10076-7. [DOI] [PubMed] [Google Scholar]
- 10.Gilks C B, Bear S E, Grimes H L, Tsichlis P N. Progression of interleukin-2 (IL-2)-dependent rat T-cell lymphoma lines to IL-2-independent growth following activation of a gene (Gfi-1) encoding a novel zinc finger protein. Mol Cell Biol. 1993;13:1759–1768. doi: 10.1128/mcb.13.3.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grewal P K, Hewitt J E. Fath, the murine homologue of the Drosophila fat tumour suppressor gene, maps to chromosome 8. Mamm Genome. 1997;8:383–384. doi: 10.1007/s003359900450. [DOI] [PubMed] [Google Scholar]
- 12.Hedrick S M, Cohen D I, Nielsen E A, Davis M M. Isolation of cDNA clones encoding T cell specific membrane associated proteins. Nature. 1984;308:149–153. doi: 10.1038/308149a0. [DOI] [PubMed] [Google Scholar]
- 13.Hsu B, Marin M C, el-Nagger A K, Stephens L C, Brisbay S, McDonnell T J. Evidence that c-myc mediated apoptosis does not require wild-type p53 during lymphomagenesis. Oncogene. 1995;11:175–179. [PubMed] [Google Scholar]
- 14.Inoue R, Asker C, Klangby U, Pisa P, Wiman K G. Induction of the human ARF protein by serum starvation. Anticancer Res. 1999;19:2939–2943. [PubMed] [Google Scholar]
- 15.Jacobs J J L, Kieboom K, DePinho R A, van Lohuizen M. The oncogene and Polycomb-group gene Bmi-1 regulates cell proliferation and senescence through the INK4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 16.Jonkers J, Berns A. Retroviral insertional mutagenesis as a strategy to identify cancer genes. Biochim Biophys Acta. 1996;1287:29–57. doi: 10.1016/0304-419x(95)00020-g. [DOI] [PubMed] [Google Scholar]
- 17.Jonkers J, Korswagen H C, Acton D, Breuer M, Berns A. Activation of a novel proto-oncogene, Frat-1, contributes to progression of mouse T-cell lymphomas. EMBO J. 1997;16:441–450. doi: 10.1093/emboj/16.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knudson A G. Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci USA. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lander J K, Fan H. Low-frequency loss of heterozygosity in Moloney murine leukemia virus-induced tumors in BRAKF1/J mice. J Virol. 1997;71:3940–3952. doi: 10.1128/jvi.71.5.3940-3952.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewis A F, Stacy T, Green W R, Taddesse-Heath L, Hartley J W, Speck N A. Core-binding factor influences the disease specificity of Moloney murine leukemia virus. J Virol. 1999;73:5535–5547. doi: 10.1128/jvi.73.7.5535-5547.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Golemis E, Hartley J W, Hopkins N. Disease specificity of nondefective Friend and Moloney murine leukemia viruses is controlled by a small number of nucleotides. J Virol. 1987;61:693–700. doi: 10.1128/jvi.61.3.693-700.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liao X B, Du Y B, Morse H C, Jenkins N A, Copeland N G. Proviral integrations at the Evi-5 locus disrupt a novel 90kDa protein with homology to the Tre2 oncogene and cell cycle regulatory proteins. Oncogene. 1997;14:1023–1029. doi: 10.1038/sj.onc.1200929. [DOI] [PubMed] [Google Scholar]
- 23.Lu X, Park S H, Thompson T C, Lane D P. Ras-induced hyperplasia occurs with mutation of p53, but activated ras and myc together can induce carcinoma without p53 mutation. Cell. 1992;70:153–161. doi: 10.1016/0092-8674(92)90541-j. [DOI] [PubMed] [Google Scholar]
- 24.Marcu K B, Banerji J, Penncavage N A, Lang R, Arnheim N. 5′ flanking region of immunoglobulin heavy chain constant region genes displays length heterogeneity in germlines of inbred mouse strains. Cell. 1980;22:187–196. doi: 10.1016/0092-8674(80)90167-1. [DOI] [PubMed] [Google Scholar]
- 25.Munroe D G, Peacock J W, Benchimol S. Inactivation of the cellular p53 gene is a common feature of Friend virus-induced erythroleukemia: relationship of inactivation to dominant transforming alleles. Mol Cell Biol. 1990;10:3307–3313. doi: 10.1128/mcb.10.7.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neil J C, Hughes D, McFarlane R, Wilkie N M, Onions D E, Lees G, Jarrett O. Transduction and rearrangement of the myc gene by feline leukaemia virus in naturally occurring T-cell leukaemias. Nature. 1984;308:814–820. doi: 10.1038/308814a0. [DOI] [PubMed] [Google Scholar]
- 27.Poirier Y, Kozak C, Jolicoeur P. Identification of a common helper provirus integration site in Abelson murine leukemia virus-induced lymphoma DNA. J Virol. 1988;62:3985–3992. doi: 10.1128/jvi.62.11.3985-3992.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quelle D E, Zindy F, Ashmun R A, Sherr C J. Alternative reading frames of the INK4a tumour suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell. 1995;83:993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 29.Serrano M. The INK4a/ARF locus in murine tumorigenesis. Carcinogenesis. 2000;21:865–869. doi: 10.1093/carcin/21.5.865. [DOI] [PubMed] [Google Scholar]
- 30.Serrano M, Lee H-W, Chin L, Cordon-Cardo C, Beach D, De Pinho R A. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 31.Stewart M, Cameron E R, Toth S, Campbell M, McFarlane R, Onions D E, Neil J C. Conditional expression and oncogenicity of human c-myc transgene linked to a human CD2 dominant control region. Int J Cancer. 1993;53:1023–1030. doi: 10.1002/ijc.2910530628. [DOI] [PubMed] [Google Scholar]
- 32.Stewart M, Terry A, Hu M, O'Hara M, Blyth K, Baxter E W, Cameron E R, Onions D E, Neil J C. Proviral insertions induce the expression of bone-specific isoforms of PEBP2alpha A (CBFA1): evidence for a new myc collaborating oncogene. Proc Natl Acad Sci USA. 1997;94:8646–8651. doi: 10.1073/pnas.94.16.8646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stewart M, Terry A, O'Hara M, Cameron E R, Onions D E, Neil J C. til-1: a novel proviral insertion locus for Moloney murine leukaemia virus in lymphomas of CD2-myc mice. J Gen Vir. 1996;77:443–446. doi: 10.1099/0022-1317-77-3-443. [DOI] [PubMed] [Google Scholar]
- 34.van Lohuizen M, Verbeek S, Scheijen B, Wientjens E, van der Gulden H, Berns A. Identification of cooperating oncogenes in Eμ-myc transgenic mice by provirus tagging. Cell. 1991;65:737–752. doi: 10.1016/0092-8674(91)90382-9. [DOI] [PubMed] [Google Scholar]
- 35.Venkatachalam S, Shi Y-P, Jones S N, Vogel H, Bradley A, Pinkel D, Donehower L A. Retention of wild-type p53 in tumours from p53 heterozygous mice: reduction of p53 dosage can promote cancer formation. EMBO J. 1998;17:4657–4667. doi: 10.1093/emboj/17.16.4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y, Xiong Y, Yarbrough W G. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppressor pathways. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 37.Zindy F, Eischen C M, Randle D H, Kamijo T, Cleveland J L, Sherr C J, Roussel M F. Myc signalling via the ARF tumour suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]