Abstract
We investigated the T-cell receptor (TCR) repertoire of CD8+ T cells that recognize the Tax11-19 immunodominant epitope of Tax protein expressed by human T-cell leukemia virus (HTLV-1) that is implicated in the disease HTLV-1-associated myelopathy (HAM/TSP). A panel of Tax11-19-reactive CD8+ T-cell clones was generated by single-cell cloning of Tax11-19/HLA-A*0201 tetramer-positive peripheral blood lymphocytes from an HTLV-1-infected individual. The analyses of TCR usage revealed that the combination of diverse TCR alpha and beta chains could be used for the recognition of Tax11-19 but the major population of T-cell clones (15 of 24 clones) expressed the TCR V beta 13S1 and V alpha 17 chain. We found striking similarities in CDR3 regions of TCR alpha and beta chains between our major group of CD8+ T-cell clones and those originating from different subjects as previously reported, including TCRs with resolved crystal structures. A 3-amino-acid sequence (PG-G) in the CDR3 region of the V beta chain was conserved among all the Tax11-19-reactive T-cell clones expressing V beta 13S1 and V alpha 17 chains. Conserved amino acids in the CDR3 region do not directly contact the Tax11-19 peptide, as corroborated by the crystal structure of B7-TCR, a TCR that is almost identical to VB13S1 clones isolated in this study. Analysis of fine peptide specificity using altered peptide ligands (APL) of Tax11-19 revealed a similar recognition pattern among this panel of T-cell clones. These data suggest that the PG-G amino acids in the CDR3 beta loop provide a structural framework necessary for the maintenance of the tertiary TCR structure.
A precise understanding of the structural basis for T-cell receptor (TCR) recognition of viral antigens among outbred humans remains a critical issue in both the development of vaccination strategies and understanding of the pathogenesis of inflammatory human diseases. While it has long been known that the cytotoxic T-cell response elicited during viral infections is generally focused toward only a few viral epitopes (27), the degree to which common TCR sequences recognize immunodominant class I epitopes among T-cell clones generated either from a single individual or among different subjects sharing major histocompatibility complex (MHC) types has not been well defined.
In the past, T-cell clones have been generated by primary bulk culture, which has led to repertoire skewing by a few, dominant T cells. The synthesis of multimeric MHC molecules loaded with the cognate peptide antigen provides a powerful method that enables the visualization of whole population of T cells recognizing specific viral epitopes (1). The use of multimeric MHC molecules in humans infected with Epstein-Barr virus (EBV), human immunodeficiency virus, or human T-cell leukemia virus type 1 (HTLV-1) revealed unexpectedly high frequencies of CD8+ T cells that respond to viral antigens (3, 5, 21). The identification of antigen-reactive T cells ex vivo by class I MHC multimers followed by single cell T-cell cloning has provided a novel method for generating panels of antigen-reactive T-cell clones that are more representative of the original population than are clones generated by bulk T-cell cloning. The use of HLA-A*0201 tetramers loaded with the EBV epitope GLC identified several recurrent V beta subsets with highly conserved TCR beta CDR3 regions among different subjects. Moreover, their TCR alpha chains comprised the same TCR alpha chain V region, suggesting a hierarchical contribution of TCR alpha chain versus TCR beta chain CDR to the recognition of this particular MHC/peptide complex (18). These surprising data indicate that common TCR sequences may be frequently used among different individuals in the response to EBV.
A second issue related to common TCR recognition sequences involves understanding how a single TCR recognizes peptides that are highly distinct in their primary sequences (2, 13, 26). Understanding the physicochemical interactions between the TCR, MHC, and antigenic peptides has important implications for better prediction of cross-reactivity in autoimmunity and thymic T-cell selection. Analysis of the recent crystal structures of the TCR/MHC/peptide complex has provided a great insight into understanding the degeneracy of TCR recognition of the peptide/MHC complex. Specifically, the comparison of two human TCRs specific for the HLA-A*0201/Tax11-19 complex was recently determined. A prominent feature of the TCR contact surface was a deep pocket that accommodated a tyrosine at position 5 of the Tax peptide. In the two TCRs compared, the B7 TCR was highly specific for aromatic residues while the other A6 TCR P5 pocket was larger, allowing many different residues to be accommodated in the pocket, leading to greater TCR degeneracy (12).
In this study we used HLA-A*0201/Tax11-19 multimers to generate a panel of CD8+ T-cell clones that recognize Tax peptide (epitope 11–19) in an HTLV-1-infected patient. Patients with HTLV-1-associated myelopathy/tropic spastic paraparesis (HAM/TSP) mount a vigorous cytotoxic T-lymphocyte (CTL) response to HTLV-1, and large numbers of virus-specific CD8+ T cells are found in peripheral blood and cerebrospinal fluid (9). Analyses of the TCR repertoire of HTLV-1-specific CD8+ T cells were undertaken to reveal the degree of heterogeneity of the T-cell response to this viral antigen.
As observed after EBV infection, highly restricted TCR sequences by CD8+ T-cell clones with marked homologies in the CDR3 region of TCR V alpha and beta chains were observed across different subjects (8, 24). Specifically, TCRs expressing the V beta 13S1 chain revealed striking sequence similarities in their CDR3 regions while expressing identical TCR alpha chains. These regions showed a high degree of homology to Tax11-19 TCRs that had been previously isolated from other HLA-A*0201-expressing HAM/TSP patients (24) and whose structure had been solved by crystallographic analysis.
These naturally occurring point mutations in the CDR3 beta loop, which can profoundly alter the pattern of antigen recognition (11), allowed a further structure-function analysis of the TCR CDR3 region recognition of the antigen/MHC complex. Three amino acids in the CDR3 region were found to be identical in every T-cell clone examined among these different subjects. Recognition of Tax11-19 peptide analogues was similar among this panel of T-cell clones, suggesting that these common amino acids in the CDR3 region might be involved as framework determinants. This was corroborated by the crystal structure of Tax11-19-specific TCRs, allowing us to conclude that conserved amino acids in the CDR3 beta loop provide a structural framework necessary for the maintenance of tertiary TCR structure.
MATERIALS AND METHODS
CD8+ T-cell clones.
Tax11-19-reactive CD8+ T-cell clones were generated as described by Lim et al. (19). Briefly, blood was obtained from an HLA-A*0201-positive subject with typical HAM/TSP. Peripheral blood mononuclear cells were isolated by Ficoll/Hypaque (Pharmacia, Uppsala, Sweden) density gradient centrifugation followed by washing in RPMI 1640 medium. The cells were stained with fluorescein isothiocyanate-conjugated anti-CD8 monoclonal antibody and phycoerythrin-labeled tetramer HLA-A*0201/Tax11-19 as previously described (3, 19, 20). Tetramer-positive CD8+ T cells were single-cell sorted (Vantage; Becton Dickinson) into 96-U-bottom-well plates and stimulated with 2 μg of phytohemagglutinin PHA (Murex, Dartford, England) per ml in the presence of irradiated (5,000 rads) allogeneic feeder cells in RPMI 1640 medium supplemented with 10% human serum (BioWhittaker, Walkersville, Md.), 4 nM l-glutamine, 100 U of penicillin per ml, 100 mg of streptomycin per ml, and 10 mM HEPES. Two days after PHA stimulation, the culture medium was supplemented with 10% T-stim (Collaborative Biomedicine Products, Bedford, Mass.). The T-cell cultures were maintained for 2 weeks, with medium changes every 3 days.
Peptides.
Tax11-19 peptide (LLFGYPVYV) was synthesized by Quality Controlled Biochemicals, Hopkinton, Mass. Tax11-19 peptide analogues with a single amino acid substitution at position 5 or 8 were synthesized by Chiron Mimotopes (San Diego, Calif.) at a 1-mg scale on pins.
Cytotoxicity assay.
The cytotoxicity of HLA-A*0201/Tax11-19 positive clones was measured by a standard 51Cr release assay. EBV-transformed B cells from an HLA-A*0201 individual were used as target cells. The target cells were labeled by a 1-h incubation with 200 μCi of 51Cr salt, washed in RPMI 1640 medium, and then pulsed for 2 h with Tax11-19 peptide at a concentration of 107 cells/ml and finally with 10 μM Tax11-19 peptide. CD8+ T cells (105) were incubated with 104 target cells for 4 h. Supernatants were assayed for 51Cr release in a 1205 Beta Plate liquid scintillation counter (LKB-Wallac, Gaithersburg, Md.). The percent specific lysis was calculated by the following formula: [(cpm of sample − cpm of spontaneous release)/(cpm of maximum release − cpm of spontaneous release)] × 100.
PCR amplification of TCR chains.
RNA extractions were performed using the RNAzol B method (Teltest Inc., Friendswood, Tex.). RNA was precipitated in the presence of 5 μg of tRNA (Sigma, St. Louis, Mo.) in isopropanol overnight at −20°C. After being washed with 70% ethanol, the pellets were air dried and resuspended in double-distilled H2O. First-strand cDNA synthesis was performed with oligo(dT) in a 11-μl reaction mixture, and the samples were heated to 70°C for 10 min. Then 4 μl of 5× Taq buffer (Perkin-Elmer), 2 μl of 0.1 M dithiothreitol (DTT), and 1 μl each of 10 mM deoxynucleoside triphosphates, 33 U of RNasin, and 200 U of Moloney murine leukemia virus reverse transcriptase (all from Promega, Madison, Wis.) were added. cDNA synthesis was carried out at 42°C for 60 min, and double-distilled H2O was added to a final volume of 200 μl. A 10-μl volume was used for each PCR. The reaction mixtures contained 0.25 μg each of forward and reverse primers, 1 U of Taq polymerase, and 20 μl of a mixture containing deoxynucleoside triphosphates and Taq buffer (Perkin-Elmer, Branchburg, N.J.). Amplifications were done for 35 cycles of 94°C denaturation for 1 min, 60°C annealing for 2 min, and 72°C extension for 3 min, with a final extension step at 72°C for 10 min. Sequences of the primers were previously published (25). PCR products were sequenced by the dideoxy method. The forward and reverse primers were used for sequencing.
RESULTS
TCR repertoire of CD8+ T cells that recognize Tax11-19 peptide.
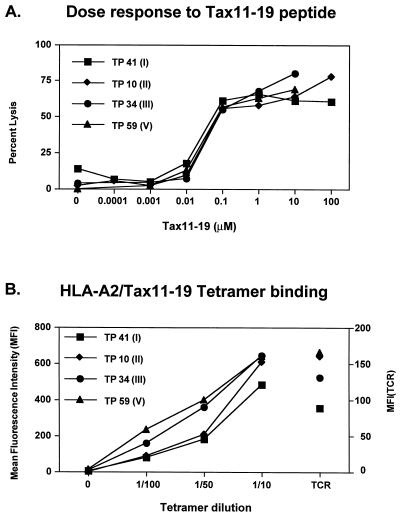
A panel of T-cell clones from a subject with HAM/TSP and recognizing the Tax11-19 peptide were generated by direct single-cell sorting of HLA-A*0201/Tax11-19 multimer-binding T cells ex vivo followed by expansion with PHA and interleukin-2 as recently described (19, 20). All of the CD8+ T-cell clones generated by this technique bound the HLA-A*0201/Tax11-19 multimer and lysed Tax11-19-pulsed HLA-A*0201 target cells (19) (Fig. 1A). The cloning efficiency was 38%, and 25 individual T-cell clones were selected for further analysis.
FIG. 1.
Similar pattern of antigen recognition among T-cell clones expressing TCR BV13S1. (A) Antigen dose-response cytotoxic activity of Tax11-19-reactive CD8+ T-cell clones. HLA-A*0201-expressing EBV-transformed B-cell lines were pulsed with the indicated concentrations of Tax11-19 peptide and used as target cells in a 4-h 51Cr-release assay. The effector-to-target ratio was 10:1. (B) Specific binding of HLA-A*0201/Tax11-19 multimer by CD8+ T-cell clones expressing TCRBV13S1. T-cell clones were incubated with the indicated dilutions of tetramer at 4°C for 1 h. The left y axis indicates the mean fluorescence intensity of multimer binding, while the right y axis shows the mean fluorescence intensity of TCR expression.
The T-cell repertoire was determined by sequencing alpha and beta chains. Sequence analysis of the TCR V beta chain revealed a dominant population expressing TCRBV13S1, but other V beta chains, including TCRBV7, TCRBV8, TCRBV16, TCRBV17, and TCRBV21S1 families, were also used for the recognition of Tax11-19 in the context of HLA-A*0201 (Table 1). Several TCR alpha chains, such as TCRAV1S1, TCRAV2, TCRAV3S1, TCRAV12S2, TCRAV14S1, TCRAV15S1, and TCRAV17S1, were identified in the T-cell clones. The CDR3 lengths ranged from 2 to 8 amino acids for the beta chain and 2 to 4 amino acids for the alpha chain.
TABLE 1.
TCR alpha and beta amino acid sequences of Tax11-19-specific T cells isolated and cloned by the use of HLA-A*0201/Tax11-19 tetramers
| Clonea | TCR | Sequenceb
|
J |
|---|---|---|---|
| VN/Junctional region | |||
| Alpha region | |||
| TP 35 | TCRAV1S1 | Y F C A T V R D G N N T D K L I F G T G T R Q V F P | 34 |
| TP 33, 80 | TCRAV2 | Y Y C A V A H N Y G Q N F V F G P G T R L S V L | 26 |
| TP 21 | TCRAV3S1 | Y F C A V E Q R M V A D K L I F G T G T R L Q V F P | 34 |
| TP 20, 32, 41, 58, 63 | TCRAV4S1 | Y Y Y C I R S G S A R Q L T F G S G T Q L T V L P | 22 |
| TP 43 | TCRAV12S2 | Y L C A V D S G G Y Q K V T F G I G T K L Q V I P | 13 |
| TP 79 | TCRAV12S2 | Y L C A V N I G F G N V L H C G S G T Q V I V L P | 35 |
| TP 27 | TCRAV12S2 | Y L C A V N P P F G N E K L T F G T G T R L T I I P | 48 |
| TP 7 | TCRAV14S1 | Y F C A M M G G G S E K L V F G K G T K L T V N P Y | 57 |
| TP 40 | TCRAV14S1 | Y F C A W D N A G N M L T F G G G T R L M V K P H | 39 |
| TP 26, 45 | TCRAV14S1 | Y F C A Y L N T G N Q F Y F G T G T S L T V I P | 49 |
| TP 60 | TCRAV14S1 | Y F C A Y R T L Y D N F G N E K L T F G T G T R L T I I P | 48 |
| TP 61 | TCRAV15S1 | Y I C A V K E S L V E M R K L T F G T G T R L L T I I P | 48 |
| TP 34 | TCRAV17S1 | Y F C A C F Q G A Q K L V F G Q G T R L T I N P | 54 |
| TP 20, 32, 41, 58, 63, 61, 71, 72, 73, 10, 59 | TCRAV17S1 | Y F C A A F Q G A Q K L V F G Q G T R L T I N P | 54 |
| Beta region | |||
| TP 45 | TCRBV7 | Y L C A S S Q E R L S F G T K N I Q Y F G A G T R L S V L | 2.4 |
| TP 79 | TCRBV8 | Y F C A S S S E Y R S G A N V L T F G A G S R L T V L | 2.6 |
| TP 26, 60 | TCRBV8 | Y F C A S S K P S V M N T E A F F G Q G T R L T V V | 1.1 |
| TP 20, 32, 41, 58, 63 (I) | TCRBV13S1 | Y F C A S S V P G A G E E T Q Y F G P G T R L L V L | 2.5 |
| TP10, 73 (II) | TCRBV13S1 | Y F C A S S S P G Q G N Y E Q Y F G P G T R L T V T | 2.7 |
| TP 34 (III) | TCRBV13S1 | Y F C A S S T P G Q G A Y E Q Y F G P G T R L T V T | 2.7 |
| TP 61 (IV) | TCRBV13S1 | Y F C A S S Y P G Q G E H E Q Y F G P G T R L T V T | 2.7 |
| TP 59, 71, 72 (V) | TCRBV13S1 | Y F C A S S S P G T G V N E Q F F G P G T R L T V L | 2.1 |
| TP 43 | TCRBV13S1 | Y F C A S S K P G L G G T G E L F F G E G S R L T V L | 2.1 |
| TP 48 | TCRBV13S1 | Y F C A S S R P G L A G R N E Q F F G P G T R L T V L | 2.1 |
| TP 27 | TCRBV16S1 | Y F C A S Q L G Q G V H G Y G Y T F G S G T R L T I V | 1.2 |
| TP 33, 80 | TCRBV16S1 | Y F C A S Q D F L S S G A Y N E Q F F G P G T R L T V L | 2.1 |
| TP 7 | TCRBV21S1 | Y L C A S S L T I G G A Q F F G Q G T R L T V V | 1.1 |
| TP 21 | TCRBV21S1 | Y L C A S S F M T D T Q Y F G P G T R L T V L | 2.3 |
| TP 35 | TCRBV21S1 | Y L C A S S L P D T Q Y F G P G T R L T V L | 2.3 |
| TP 40 | TCRAV17S1 | Y L C A S S A R L G Q P Q H F G D G T R L S I L | 1.5 |
T-cell clones expressing TCRBV13S1 were divided into groups (I to Y, indicated in parentheses) depending on the CDR3 region sequence.
N or N(D)J additions for both alpha and beta chains are indicated in bold.
Interestingly, we found that the majority of Tax11-19-specific CD8+ T-cell clones (14 of 25) expressed the BV13S1 TCR and that these T cells exhibited striking similarities in their CDR3 regions (Table 1). In addition, analysis of the TCR alpha chains revealed that 11 of these 14 clones used an identical TCR alpha chain expressing the 17S1 variable chain for the recognition of Tax11-19 peptide. They did not represent the progeny of one clone since there were differences in the CDR3 region of the TCR V beta chain.
The 14 CD8+ T cell clones expressing TCR BV13S1 were divided into five groups based on their CDR3 region sequences (Table 1). Group I (TP 20, TP 32, TP 41, TP 58, and TP 63), with the TCR beta chain CDR3 region sequence YPGAGE, expressed a second TCRAV4S1 chain. The TP 61 T-cell clone with the TCRBV13S1 CDR3 sequence YPGQGEH, expressed TCRAV15S1 alpha chain in addition to TCRAV17S1 (group IV). Clones TP 10, TP 73, TP 59 (group II) and TP 34 (group III) expressed only one TCR V alpha 17S1 chain, as identified by the use of primers that amplify TCR V alpha chains 1 to 18. The T-cell clones in groups II and III expressed the same JB 2.7 and CDR3 regions SPGQGN and TPGQGA, respectively. Clones in group V expressed the SPGTGV sequence in the CDR3 region, the JB 2.1 segment, and the same TCRAV17 chain as the clones from groups I to IV. Two TCR alpha chains observed in groups I and IV of CD8+ T-cell clones (Table 1) were detected at the mRNA level. Since that there are no TCR V alpha chain-specific antibodies available, we were unable to ascertain whether both chains were expressed on the cell surface, thus contributing to two different TCR complexes with different specificities. As shown in Fig. 1A, there was no difference in the dose-response curve of Tax11-19 peptide-pulsed target cells between clones expressing mRNA for two or for one TCR alpha chain. Moreover, there was no difference in the degree of binding of the Tax11-19/HLA-A*0201 tetramer among the different clones (Fig. 1B), thus indicating that even if the second TCR alpha chain is expressed, it does not alter the recognition of the Tax11-19 peptide. In all five groups of clones expressing TCRBV13S1 (total of 12 clones), the first amino acid in the CDR3 regions was uncharged with polar side chains (Tyr, Ser, or Thr).
Potential framework determinants in the TCR CDR3 region used for Tax11-19 recognition.
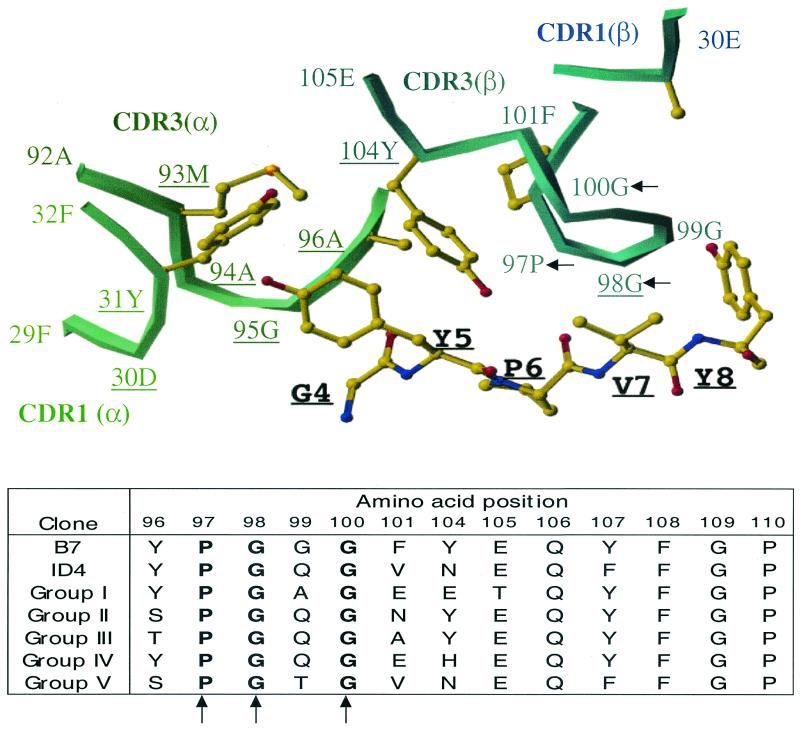
The major population of Tax11-19-reactive CD8+ T-cell clones expressing TCRBV13S1 (groups I to V) maintained conserved Pro at position 97 and Gly at positions 98 and 100 of the CDR3 beta loop. To understand the significance of these conserved amino acids for the recognition of Tax peptide, we took advantage of the TCR/HLA-A*0201/Tax11-19 crystallographic data previously defined using T-cell clone B7 (20). The same usage of the TCR V alpha/beta chain family (TCRBV13S1/TCRAV17S1) and the same amino acid length of CDR3 region between groups I to V and the B7 TCR allowed an indirect analysis of the roles of three conserved amino acids positions on the physicochemical interaction with HLA-A*0201 and Tax11-19 complex. It has been revealed that in the B7-TCR structure, the main TCR contacts are made to residues 5Y and 8Y of the Tax11-19 peptide. In the CDR3 beta region, Tyr at position 104 and Gly at position 98 contribute directly to the peptide binding (Fig. 2, top). Interestingly, the clones belonging to group II and III maintained a Tyr at position 104. In the remaining groups, Tyr was changed to either an uncharged Asn/His or an acidic Glu. Tyr at position 104 makes a hydrophobic contact with Tyr (5Y) of the Tax11-19 peptide. Thus, changes at this position might influence the fine specificity of peptide recognition. In the B7 TCR, Gly at position 98 is within the range of Van der Waals interactions with Pro at position 6, Val at position 7, and Tyr at position 8 of the Tax peptide. In all clones from groups I to V, Gly 98 was maintained. However, Tyr at position 96 and Gly at position 100 do not form a strong contact with Tyr at position 8 of the Tax11-19 peptide, even though the amino acids at these positions are conserved in all of the T-cell clones. It is thus possible that conserved amino acids at positions 96 and 100 in the CDR3 regions contribute to the structural framework of the CDR3 loop, allowing for recognition of the Tax11-19 peptide.
FIG. 2.
Requirement of conserved amino acid positions for recognition of the Tax11-19 peptide. (Top) Crystal structure representation of interactions between TCR-B7 CDR3 regions and HLA-A*0201 complexed with Tax11-19 peptide. Only amino acids 4 to 8 of the Tax11-19 peptide are presented. Amino acids that are conserved in the CDR3 beta chain among T-cell clones are indicated by arrows. Conservation of Pro at position 97 and Gly at positions 98 and 100 in all clones expressing TCRBV13S1 suggest that these residues provide a framework for the tertiary structure of the CDR3 loop. Residues that contact MHC peptide are underlined. Amino acid numbering of TCR alpha and beta chains is as in reference 10. (Bottom) Comparison of CDR3 V beta amino acid sequences of Tax11-19-reactive CD8+ T-cell clones isolated in this study (groups I to V) and previously published (B7 and ID4) (13). Positions that are maintained in all clones are indicated by arrows.
Similar fine specificity of natural CDR3 substitutions to Tax11-19 peptide analogues.
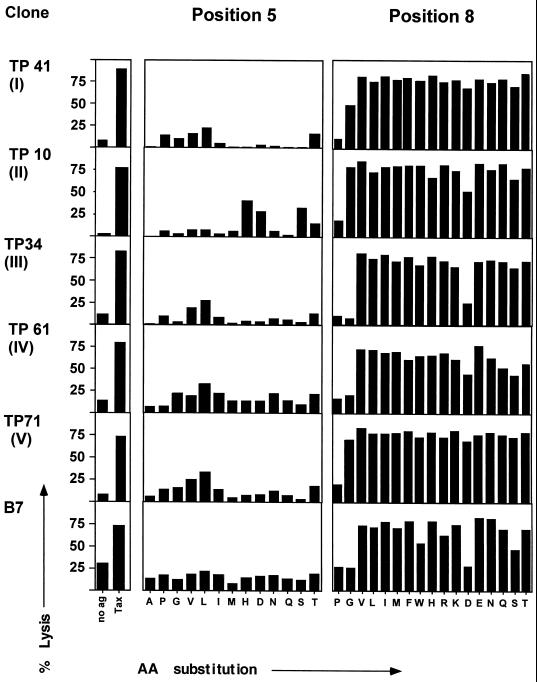
To examine how similar the peptide-binding grooves of TCR are among naturally occurring CDR3 substitutions in TCRBV13S1-expressing CD8+ T-cell clones, we determined the responses of T-cell clones to a series of Tax11-19 peptide analogues. Single amino acid substitutions were made at positions 5 and 8 of the Tax11-19 peptide, since these positions were known to give direct contact with B7 TCR (6). T-cell reactivity was measured as a function of cytotoxicity.
The functional comparison of clones from group I to V and previously isolated clone B7 showed remarkable similarities (Fig. 3). All clones from groups I to V and clone B7 revealed a strong requirement for Tyr at position 5. This is consistent with the results of Hausmann et al. (12), who showed that the B7 clone is exquisitely specific for aromatic residues at this position. The crystal structure of the B7 TCR revealed that Tyr at position 104 of the TCR beta chain makes direct contact with Tyr at position 5 of the Tax11-19 peptide. Clones from groups II and III maintain Tyr at position 104. Even though clones from groups I and IV use Glu or His at position 104, the fine specificity toward recognition of Tax11-19 analogs at position 5 is maintained (Fig. 3).
FIG. 3.
Fine specificity of CD8+ T-cell clones to singly substituted Tax11-19 analogues. T-cell recognition of Tax11-19 peptide analogues at TCR P5 and P8 pockets in Tax11-19-reactive CD8+ T-cell clones expressing strong structural similarities is shown. The peptide recognition was determined by a standard 4-h 51Cr release assay. HLA-A*0201-expressing EBV-transformed B-cell lines were pulsed with peptides at 10 μM and used as target cells. The effector-to-target ratio was 10:1. AA, amino acid.
Interestingly, all of the T-cell clones showed a very degenerate recognition of Tax11-19 peptides with substitutions at position 8 (Fig. 3). As shown in the crystal structure of B7, Tyr at position 8 in the Tax11-19 peptide contacts Gly98 of the TCR V beta chain in the CDR3 loop. All clones from groups I to IV preserve Gly at this position. In all clones, replacement of Tyr8 with Pro8 abolished CTL activity. Likewise, a change from Tyr8 to Gly, Asp, or Ser lessened the CTL activity of B7 and clones from groups III and IV.
These results support the fact that there is a similar structural architecture of the peptide-binding groove among natural CDR3 substitutions.
Different cross-reactivity of natural CDR3 substitutions with the natural peptides.
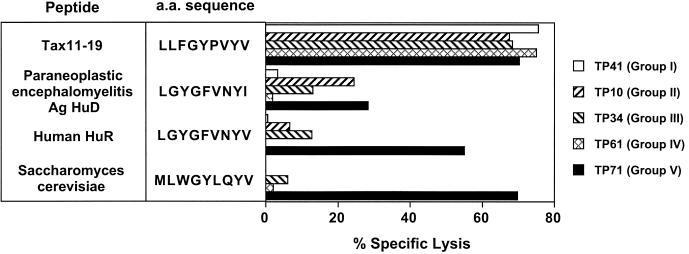
The degenerate recognition is a well-known feature of TCR and a basis of the cross-reactivity of T cells (26). A series of naturally occurring peptides cross-reactive with the Tax11-19 peptide were previously identified (12). We examined the reactivity of our new panel of Tax11-19-reactive T-cell clones to further probe their fine specificity. Of 16 natural peptides with multiple substitutions, 4 could be recognized by the T-cell clones expressing TCRBV13S1 (Fig. 4 and data not shown). However, the recognition pattern of these four peptides was clearly different depending on the different groups of T-cell clones. A T-cell clone in group V showed strong cross-reactivity with human HuR and Saccharomyces cerevisiae, whereas T-cell clones in other groups did not recognize or weakly recognized these peptides. Interestingly, three groups of T-cell clones (groups II, III, and V) recognized the self-central nervous system protein Ag HuD (paraneoplastic encephalomyelitis antigen) to different variable degrees. In summary, while all of the different groups of T-cell clones showed similarities in the recognition of Tax11-19 analogues with a single amino acid substitution, clear differences become apparent when natural peptides with multiple substitutions were analyzed (Fig. 4).
FIG. 4.
Different recognition patterns of CD8+ T-cell clones toward naturally occurring peptides. Differential recognition of natural peptides by CD8+ Tax11-19-reactive T-cell clones expressing BV13S1 is shown. Human and microbial peptides expressing the recognition motif for B7 TCR were previously published (12) and used in this study. Recognition patterns of CD8+ T-cell clones were determined by a 51Cr release assay at a peptide concentration of 10 μM. Of 16 peptides tested, 3 were recognized by CD8+ T-cell clones, and reactivity toward these 3 peptides is shown. As previously published (12), the B7 clone recognized the above peptides with a range of 18 to 20% specific lysis. a.a., amino acid.
DISCUSSION
To study the TCR repertoire of antigen-specific T cells, isolation of antigen-specific T cells is performed as a first step. For this purpose, limiting-dilution culture followed by confirmation of antigen specificity has been commonly used in previous studies. However, some T cells could be deleted from the test pool of the TCR repertoire due to activation-induced cell death. To study a more extensive pool of antigen-specific T cells, we used HLA-A*0201/Tax11-19 tetramer staining followed by direct single-cell sorting and expansion with PHA. This approach generated a diverse panel of CD8+ Tax11-19-specific T-cell clones that were used in the TCR repertoire study. Sequence analysis of TCR revealed that various combinations of TCR alpha and beta chains could be used for the recognition of Tax11-19 (Table 1). That observation is in contrast to previous reports suggesting a more restricted TCR repertoire of Tax11-19-specific CD8+ T cells (24). In these studies, the authors used the limiting-dilution method for the generation of Tax11-19-specific clones. Interestingly, direct TCR sequencing of a bulk population of Tax11-19-specific CD8+ T cells also revealed a more diverse TCR repertoire (22). However, in the aforementioned report, the authors provided the information for V beta chain usage only since they did not study the TCR repertoire at the single-cell level.
Interestingly, the majority of CD8+ T-cell clones generated in our study (14 of 25) expressed TCRBV13S1 TCR, and these cells exhibited striking similarities in their CDR3 regions to previously published TCR sequences (24). A summary of the homologies is presented in Fig. 2 (bottom). TCR sequences of B7 and ID4 clones derived from two different patients with HAM/TSP were previously published by Utz et al. (24). Clone B7 expressed TCRBV13S1 and TCRAV17S1 and exhibited remarkable similarities in the sequences of both alpha and beta chains of clones belonging to groups I to V isolated in our study (Fig. 2, bottom). Specifically, the TCR alpha sequence differed only in two or three amino acids in the CDR3 region: Ala-Met-Glu (AME) in the B7 clone was changed to Ala-Phe-Gln (AFQ) in group I, II, IV, and V T-cell clones and to Cys-Phe-Gln (CFQ) in group III T-cell clones. Perhaps as expected, all these amino acid changes were conservative. The narrowing of the TCR alpha chain repertoire might be explained by data from the resolved crystal structures of TCR-MHC complexes, which show that in some TCRs, all CDR regions of the alpha chains make strong interactions with MHC or peptide. In the crystal structure of B7-TCR, 13 amino acids of the TCR alpha chain make a direct contact with MHC/peptide while only 4 amino acids of the beta chain contribute to MHC/peptide contacts (6). Thus, the “public” TCR sequences observed may be explained by the structural basis of the antigen recognition by TCR.
The comparison of TCR V beta CDR3 region sequences revealed the absolute conservation of Pro at position 97 and Gly at positions 98 and 100 in all T-cell clones expressing TVRVB13S1 (groups I to V), suggesting that these amino acids are critical for the binding to the MHC/Tax11-19 complex. However, the crystallographic data for B7 TCR/HLA-*0201-Tax11-19 showed that Gly at position 98 is within the range of Van der Waals interactions with Tax11-19 peptide contributing specific binding but that the other two amino acids at positions 97 and 100 do not form close contacts with Tax11-19 peptide. It is thus possible that conserved amino acids at positions 97 and 100 in the CDR3 regions contribute to the stability and flexibility of the CDR3 loop, allowing the recognition of Tax11-19 peptide. This observation might indicate that the similarities in the CDR3 regions of antigen-specific T cells might be related to the requirement for the proper conformation of the CDR3 loop of a particular TCR variable chain. Interestingly, the study by Eiraku et al. (7) revealed the presence of related sequences within expanded CD8+ T cells from HLA-A*0201-positive patients infected with HTLV-1. Expanded V beta 13S1 CD8+ T cells also expressed related P-G-X-G sequence in the CDR3 region. The fact that all these clones were isolated from subjects expressing the HLA-A*0201 haplotype suggested the presence of similar selective forces that act on the lymphocyte repertoire selection and may be a result of an imprint of intrathymic peptides expressed in HLA-A*0201 individuals (23).
It has been previously shown that point mutations in the beta chain of CDR3 region can alter the TCR recognition pattern of the antigenic peptide (4, 11, 14–17). Systematic mutagenesis of TCR CDR3 region residues revealed that introduced mutations mostly result in the lost of ability to recognize peptide/MHC or change an overall recognition pattern of mutated TCR. However, our study shows that naturally selected substitutions in the CDR3 beta loops of Tax11-19-reactive CD8+ T-cell clones do not dramatically change the overall antigen specificity but result in subtle changes in antigen fine specificity. This conservation of overall antigen specificity might come from the compensatory mutations in more than two amino acid positions that allow the proper conformation of CDR3 loop for the recognition of specific peptide. Since none of these TCR residues directly contact the peptide in the structure of B7 TCR, we propose that these amino acids are critical for providing a structural framework necessary for the maintenance of the tertiary conformation of the CDR3 loop of the beta chain.
ACKNOWLEDGMENTS
K. D. Bourcier and D. G. Lim contributed equally to this work.
We thank William Biddison for providing the B7 CD8+ T-cell clone.
This work was supported by grants from the National Institute of Health (RO1NS2424710, PO1AI39671, PO1NS38037) and National Multiple Sclerosis Society (RG2172B6 and RG2949A).
REFERENCES
- 1.Altman J D, Moss P A H, Goulder P J R, Barouch D H, McHeyzer-Williams M G, Bell J I, McMichael A J, Davis M M. Phenotypic analysis of antigen-specific T lymphocytes. Science. 1996;274:94–96. [PubMed] [Google Scholar]
- 2.Ausubel L J, Kwan C K, Sette A, Kuchroo J, Hafler D A. Complementary mutations in an antigen peptide allow for crossreactivity of autoreactive T-cell clones. Proc Natl Acad Sci USA. 1996;93:15317–15322. doi: 10.1073/pnas.93.26.15317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bieganowska K, Hollsberg P, Buckle G J, Lim D G, Greten T F, Schneck J, Altman J D, Jacobson S, Ledis S L, Hanchard B, Chin J, Morgan O, Roth P A, Hafles D A. Direct analysis of viral-specific CD8+ T cells with soluble HLA-A2/Tax11-19 tetramer complexes in patients with human T cell lymphotropic virus-associated myelopathy. J Immunol. 1999;162:1765–1771. [PubMed] [Google Scholar]
- 4.Brawley J V, Concannon P. Systematic mutagenesis of TCR complementarity-determining region 3 residues: a single conservative substitution dramatically improves response to both multiple HLA-DR alleles and peptide variants. J Immunol. 1999;163:4946–4952. [PubMed] [Google Scholar]
- 5.Callan M F C, Tan L, Annels N, Ogg G S, Wilson J D K, O'Callaghan C A, Steven N, McMichael A J, Rickinson A B. Direct visualization of antigen-specific CD8+ T cells during primary immune response to Epstein-Barr virus in vivo. J Exp Med. 1998;187:1395–1402. doi: 10.1084/jem.187.9.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ding Y H, Smith K J, Garboczi D N, Utz U, Biddison W E, Wiley D C. Two human T cell receptors bind in a similar diagonal mode to the HLA-A2/Tax peptide complex using different TCR amino acids. Immunity. 1998;8:403–411. doi: 10.1016/s1074-7613(00)80546-4. [DOI] [PubMed] [Google Scholar]
- 7.Eiraku N, Hingorani R, Ijichi S, Machigashira K, Gregersen P K, Monteiro J, Usuku K, Yashiki S, Sonoda S, Osame M, Hall W W. Clonal expansion within CD4+ and CD8+ T cell subsets in human T lymphotropic virus type I-infected individuals. J Immunol. 1998;161:6674–6680. [PubMed] [Google Scholar]
- 8.Elovaara I, Utz U, Smith S, Jacobson S. Limited T cell receptor usage by HTLV-I tax-specific, HLA class I restricted cytotoxic T lymphocytes from patients with HTLV-I associated neurological disease. J Neuroimmunol. 1995;63:47–57. doi: 10.1016/0165-5728(95)00129-8. [DOI] [PubMed] [Google Scholar]
- 9.Elovaara I, Koenig S, Brewah A Y, Woods R M, Lehky T, Jacobson S. High human T cell lymphotropic virus type I (HTLV-I)-specific precursor cytotoxic T lymphocyte frequencies in patients with HTLV-I-associated neurological disease. J Exp Med. 1993;177:1567–1573. doi: 10.1084/jem.177.6.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garboczi D N, Ghosh P, Utz U, Fan Q R, Biddison W E, Wiley D C. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature. 1996;384:134–141. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]
- 11.Goyarts E C, Vegh Z, Kalergis A M, Horig H, Papadopoulos N J, Young A C, Thompson C T, Chang H C, Joyce S, Nathenson S G. Point mutations in the beta chain CDR3 can alter the T cell receptor recognition pattern on an MHC class I/peptide complex over a broad interface area. Mol Immunol. 1988;35:593–607. doi: 10.1016/s0161-5890(98)00056-x. [DOI] [PubMed] [Google Scholar]
- 12.Hausmann S, Biddison W E, Smith K J, Ding Y H, Garboczi D N, Utz U, Wiley D C, Wucherpfennig K W. Peptide recognition by two HLA-A2/Tax11-19-specific T cell clones in relationship to their MHC/peptide/TCR crystal structures. J Immunol. 1999;162:5389–5397. [PubMed] [Google Scholar]
- 13.Hemmer B, Vergelli M, Gran B, Ling N, Conlon P, Pinilla C, Houghten R, McFarland H F, Martin R. Predictable TCR antigen recognition based on peptide scans leads to the identification of agonist ligands with no sequence homology. J Immunol. 1998;160:3631–3636. [PubMed] [Google Scholar]
- 14.Hsu B L, Donermeyer D L, Allen P M. TCR recognition of the Hb(64-76)/I-Ek determinant: single conservative amino acid changes in the complementarity determining region 3 dramatically alter antigen fine specificity. J Immunol. 1996;157:2291–2298. [PubMed] [Google Scholar]
- 15.Kalergis A M, Nathenson S G. Altered peptide ligand-mediated TCR antagonism can be modulated by a change in a single amino acid residue within the CDR3 beta of an MHC class-I restricted TCR. J Immunol. 2000;165:280–285. doi: 10.4049/jimmunol.165.1.280. [DOI] [PubMed] [Google Scholar]
- 16.Kalergis A M, Ono T, Wang F, DiLorenzo T, Honda S, Nathenson S G. Single amino acid replacements in an antigenic peptide are sufficient to alter the V beta repertoire of the responding CD8+ cytotoxic lymphocyte population. J Immunol. 1999;162:7263–7270. [PubMed] [Google Scholar]
- 17.Kasibhatala S, Nalefski E A, Rao A. Simultaneous involvement of all six predicted antigen binding loops of the T cell receptor in recognition of the MHC/antigenic peptide complex. J Immunol. 1993;151:3140–3151. [PubMed] [Google Scholar]
- 18.Lim A, Trautmann L, Peyrat M-A, Couedel C, Davodeau F, Romagne F, Kourilsky P, Bonneville M. Frequent contribution of T cell clonotypes with public TCR features to the chronic response against a dominant EBV-derived epitope: application to direct detection of their molecular imprint on the human peripheral T cell repertoire. J Immunol. 2000;165:2001–2011. doi: 10.4049/jimmunol.165.4.2001. [DOI] [PubMed] [Google Scholar]
- 19.Lim D-G, Bieganowska-Bourcier K, Freeman G, Hafler D A. Examination of CD8+ T cell function in humans using MHC class I tetramers: similar cytotoxicity but variable proliferation and cytokine production among different clonal CD8+ T cells specific to a single viral epitope. J Immunol. 2000;165:6214–6220. doi: 10.4049/jimmunol.165.11.6214. [DOI] [PubMed] [Google Scholar]
- 20.Meyer A L, Trollmo C, Crawford F, Marrack P, Steere A C, Huber B T, Kappler J, Hafler D A. Direct enumeration of Borrelia-reactive CD4 T cells ex vivo by using MHC class II tetramers. Proc Natl Acad Sci USA. 2000;97:11433–11438. doi: 10.1073/pnas.190335897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ogg G S, Jin X, Bonhoeffer S, Dunbar P R, Nowak M A, Monard S, Segal J P, Cao Y, Rowland-Jones S L, Cerundolo V, Hurley A, Markovitz M, Ho D D, Nixon D F, McMichael A J. Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science. 1998;279:2103–2106. doi: 10.1126/science.279.5359.2103. [DOI] [PubMed] [Google Scholar]
- 22.Saito M, Taylor G P, Saito A, Furukawa Y, Usuku K, Weber J N, Osame M, Bangham C R. In vivo selection of T-cell receptor junctional region sequences by HLA-A2 human T-cell lymphotropic virus type 1 Tax11-19 peptide complexes. J Virol. 2001;75:1065–1071. doi: 10.1128/JVI.75.2.1065-1071.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sant'Angelo D B, Waterbury P G, Cohen B E, Martin W D, Van Kaer L, Hayday A C, Janeway C A., Jr The imprint of intrathymic self-peptides on the mature T cell receptor repertoire. Immunity. 1997;7:517–524. doi: 10.1016/s1074-7613(00)80373-8. [DOI] [PubMed] [Google Scholar]
- 24.Utz U, Banks D, Jacobson S, Biddison W E. Analysis of the T-cell receptor repertoire of human T-cell leukemia virus type 1 (HTLV-1) Tax-specific CD8+ cytotoxic T lymphocytes from patients with HTLV-1-associated disease: evidence for oligoclonal expansion. J Virol. 1996;70:843–851. doi: 10.1128/jvi.70.2.843-851.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wucherpfennig K, Zhang J, Witek C, Modabber Y, Hafler D. Clonal expansion and persistance of human T cells specific for an immunodominant myelin basic protein peptide. J Immunol. 1994;152:5581–5592. [PubMed] [Google Scholar]
- 26.Wucherpfennig K W, Strominger J L. Molecular mimicry in T cell-mediated autoimmunity: viral peptides activate human T cell clones specific for myelin basic protein. Cell. 1995;80:695–705. doi: 10.1016/0092-8674(95)90348-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yewdell J W, Bennink J R. Immunodominance in major histocompatibility complex class I restricted T lymphocyte responses. Annu Rev Immunol. 1999;17:51–88. doi: 10.1146/annurev.immunol.17.1.51. [DOI] [PubMed] [Google Scholar]