SUMMARY
The nuclear Overhauser effect (NOE) is one of NMR spectroscopy’s most important and versatile parameters. NOE is routinely utilized to determine the structures of medium-to-large size biomolecules and characterize protein-protein, protein-RNA, protein-DNA, and protein-ligand interactions in aqueous solutions. Typical [1H,1H] NOESY pulse sequences incorporate water suppression schemes to reduce the water signal that dominates 1H-detected spectra and minimize NOE intensity losses due to unwanted polarization exchange between water and labile protons. However, at high- and ultra-high magnetic fields, the excitation of the water signal during the execution of the NOESY pulse sequences may cause significant attenuation of NOE cross-peak intensities. Using an evolutionary algorithm coupled with artificial intelligence, we recently designed high-fidelity pulses [Water irrAdiation DEvoid (WADE) pulses] that elude water excitation and irradiate broader bandwidths relative to commonly used pulses. Here, we demonstrate that WADE pulses, implemented into the 2D [1H,1H] NOESY experiments, increase the intensity of the NOE cross-peaks for labile and, to a lesser extent, non-exchangeable protons. We applied the new 2D [1H,1H] WADE-NOESY pulse sequence to two well-folded, medium-size proteins, i.e., the K48C mutant of ubiquitin and the Raf kinase inhibitor protein (RKIP). We observed a net increase of the NOE intensities varying from 30 to 170% compared to the commonly used NOESY experiments. The new WADE pulses can be easily engineered into 2D and 3D homo- and hetero-nuclear NOESY pulse sequences to boost their sensitivity.
Keywords: nuclear Overhauser effect; [1H,1H] NOESY; WADE pulses; GENETICS-AI; pulse design; evolutionary algorithm; artificial intelligence
Introduction
Since its discovery in the early 50s, the nuclear Overhauser effect (NOE) has been an essential parameter for NMR characterization of biomacromolecules1–3. NOE results from the dipolar interactions between two nuclear spins, and its dependence on the internuclear distances has enabled the structure determination of biomacromolecules3–6. The introduction of the transverse relaxation optimized spectroscopy (TROSY) experiment7 has furthered the importance of NOEs, expanding the application of NMR spectroscopy to larger proteins and protein complexes8. More recently, methyl-methyl NOE has emerged as a critical tool for site-specific assignments of side-chain methyl groups in proteins, enabling the structural and dynamic characterization of sizeable supramolecular assemblies9. Finally, a thorough quantitation of NOE (exact NOE or eNOE)6,10 has also led to the identification of multiple conformational states of proteins10 as well as intra-molecular allosteric pathways11.
All biologically relevant NMR studies are performed in aqueous solutions with a concentration of the biomacromolecules less than 1 mM and a proton concentration from the water of 110 M. The latter has represented a long-standing challenge for NMR spectrometer receivers, especially for 1H-detected experiments12,13. Typical homo- and heteronuclear NOE-based pulse sequences, however, are equipped with water suppression schemes, which minimize the water signal and detect [1H,1H] NOE correlations at high sensitivity. Some of these pulse schemes also reduce the unwanted exchange between labile sites (e.g., amide protons) and water, increasing the intensity of NOE cross peaks. In addition, high and ultra-high magnetic fields enhance the longitudinal relaxation of the spin magnetization and augment NOE sensitivity; yet, radiation damping complicates water suppression14,15.
The most common water-suppression schemes used for NOE measurements include selective solvent presaturation, spin-lock pulses, jump-return sequences, selective defocusing of the water signal, and shaped pulses16–20. Other methods are based on the differential relaxation time or diffusion properties between the water and biomolecules12,13. One of the most effective schemes is water presaturation (presat), which consists of a weak continuous radio frequency (RF) field that irradiates the water signal for 1–2 seconds before executing the pulse sequence20. Nonetheless, the weak RF field attenuates the proton resonances in rapid exchange with solvent, e.g., amide protons that constitute proteins’ fingerprint, and the effectiveness of solvent presaturation is significantly reduced by spin diffusion21. The WATERGATE sequence avoids these drawbacks22. WATERGATE is a pulsed gradient spin echo (PGSE)23 sequence with the hard π pulse replaced by the sequence ‘selective π/2 – Hard π – selective π/2’, which inverts all the resonances except the solvent signal. For solute resonances, the effect of the first gradient pulse is reversed by the second gradient with a π pulse applied to the solute resonances. As a result, the dephasing effect of the two gradient pulses is accumulated, and the solvent signal is suppressed. However, the sequence of selective and hard pulses in WATERGATE can cause spectral phase distortions, a problem that was solved using improved versions of WATERGATE17,24 or excitation sculpting (ES) sequence25. Among these sequences, ES is inhomogeneity compensated and its water selectivity and irradiation bandwidth can be easily tailored. Binomial (1–1, 1–3-3–1, etc.)26,27 or binomial-like (3–9-19 or W5)28 sequences have also been used for water suppression in 2D and 3D NOESY experiments29. These symmetric schemes feature a π-shift in the second half of the sequence. When the RF carrier is set on resonance with the water signal, the effective operation of the right half is canceled by the left half of the sequence, and the solute coherences are refocused and detected. Similar to WATERGATE, the solvent resonances in the binomial sequences are suppressed by a pair of dephasing gradients. Notably, the reduced saturation transfer from water to the solute makes binomial sequences favored over water presaturation. Nevertheless, the binomial sequences have limited bandwidth irradiation, representing a problem for biomolecules with broad chemical shift dispersion. Ideally, for biomolecular NMR spectroscopy at high and ultra-high magnetic fields, a water-suppression scheme should have high selectivity for the water signal, broadband irradiation, high fidelity of spin operation, and should be compensated for the different sources of inhomogeneity.
Recently, we developed a new software, GENErator of TrIply Compensated pulSes via Artificial Intelligence or GENETICS-AI, that combines an evolutionary algorithm and artificial intelligence (AI) to create RF pulses with constant amplitude and variable phase shapes30. Using GENETICS-AI, we generated broadband pulses (called WADE for Water irrAdiation DEvoid pulses) that elude water irradiation while exciting a large bandwidth at high-fidelity31. The WADE pulses used as a water-suppression sequence increase the sensitivity of the transverse relaxation optimized (TROSY) HSQC pulse sequence31. Here, we report the implementation of WADE-π/2 (W1F90) and WADE-π (WR4) pulses into a standard 2D [1H,1H] NOESY experiment (i.e., [1H,1H] WADE-NOESY) and show that they significantly enhance the NOE cross peaks intensities for labile, and to a lesser extent, non-labile proton resonances of two well-folded proteins: the K48C mutant of ubiquitin (UbiK48C) and Raf kinase inhibitor protein (RKIP). We show that the intensities of the NOESY cross-peaks follow the typical kinetics of the NOE buildup, with an increase of the cross-peak intensities ranging from 30 to 170%. The WADE pulses can be easily implemented into homo- and hetero-nuclear NOESY-type of experiments to boost their sensitivity.
Material and Methods
Expression and purification of ubiquitin K48C mutant.
Uniformly 15N labeled K48C ubiquitin mutant (UbiK48C) was expressed and purified as reported by Olivier et al.32. Briefly, the recombinant human K48C mutant was generated from the ubiquitin wild-type gene (pRSET) vector using the QuickChange® kit from Stratagene (CA, USA). The protein was expressed in E. coli BL21 (DE3) cells cultured at 30 °C in M9 minimal media, containing 15NH4Cl (Cambridge Isotope Laboratories Inc.) salt as the only nitrogen source. Protein overexpression was induced at an optical density (OD600) above 0.8 by adding 1 mM of isopropyl β-D-1-thiogalactopyranoside (IPTG) and was carried out for 5 hours. The cell suspension was centrifugated at 6370 g for 30 min at 4°C, and the cell pellet was collected, flash-frozen, and stored at −20 °C. For protein purification, the pellet was resuspended in 50 mL of 50 mM sodium acetate (pH 5.0) and 5 mM of β-mercaptoethanol (β-me) buffer, homogenized with a cell grinder, and sonicated using Branson Sonifier 450 (output of 4; duty cycle, 40%) for 10 minutes. Cells debris was pelleted by centrifugation at 45,700g for 40 min at 4°C. The pooled supernatant containing UbiK48C was applied to a Whatman P11 phosphocellulose resin (Sigma-Aldrich) and eluted with a gradient from 0–1 M NaCl. The fractions containing UbiK48C were concentrated and passed through a Sephacryl S-100 resin (GE Healthcare) using 100 mM phosphate buffer at pH 7.0 as a mobile phase. The purified protein was concentrated, lyophilized, and stored at −20 °C. For NMR samples,15N UbiK48C was solubilized in 10 mM sodium acetate buffer (pH 6.0), 100 mM NaN3 and 2 mM dithiothreitol (DTT) to final protein concentration of 0.5 mM.
Expression and purification of RKIP.
Recombinant Raf kinase inhibitor protein (RKIP) was expressed and purified as reported previously by Lee et al.33. Transformed E. coli BL21(DE3) pLysS cells (Invitrogen™) were grown in M9 minimal media containing 15NH4Cl (Cambridge Isotope Laboratories Inc.), at 30 °C. Protein overexpression was initiated by adding 0.4 mM IPTG when the OD600 was above 1.1 and was carried out for 5 hours. The cells were harvested by centrifugation at 6370g for 30 min at 4°C, and the cell pellet was collected and conserved at −20 °C. The cell pellet was then resuspended in 35 mL of 50 mM Tris-HCl pH 8.0, 100 mM NaCl, 20% sucrose, 0.15 mg/ml lysozyme, 1 tablet of protease inhibitor (cOmplete™, Roche Applied Science), 100 U/mL DNAse I (Roche Applied Science), and 5 mM β-me and lysed using French press at 1000 psi. Cell debris was centrifuged at 45,700g for 40 min at 4°C, and the supernatant was batch-bound with Ni2+-NTA agarose affinity resin (Thermofisher) at 4 °C for 3 hours or overnight. The resin/proteins mixture was applied to a gravity column and washed using 50 mM Tris-HCl pH 8.0, 100 mM NaCl, 20% sucrose, 1 mM PMSF, 5 mM β-me. The target protein was eluted in two fractions using the same buffer supplied with 200 mM and 500 mM imidazole, respectively. RKIP gene was cloned into a pET SUMO vector (Invitrogen™), and the His-tagged-SUMO was removed using a stoichiometric amount of recombinant UPL1 protease in 50 mM Tris-HCL pH 8.0, 150 mM NaCl, 5 mM β-me, 0.5 mM PMSF. The cleavage reaction was performed overnight at 4 °C. A reverse Ni2+NTA purification was performed to eliminate contaminants. The fractions containing the un-tagged RKIP (flow-through) were concentrated using a 10 kDa spin concentrator (MilliporeSigma, Life Science) and loaded to a 16/60 Hi Load Superdex 200 (GE Healthcare) size exclusion column (SEC) using 50 mM Tris-HCL pH 8.0, 150 mM NaCl, 2 mM DTT as a mobile phase. The purified protein was concentrated and stored at 4 °C in the SEC buffer supplied with protease inhibitors. 15N labeled RKIP sample was prepared by buffer-exchanging and concentrating the stored protein in 20 mM KH2PO4 (pH 6.5), 10 mM DTT, 10 mM MgCl2, and 1 mM NaN3 buffer supplied with 5% D2O, and 0.5% Pefa block® (Sigma-Aldrich, USA) to a final protein concentration of 0.3 mM.
NMR Spectroscopy.
NMR experiments were performed on an 850 MHz Bruker Avance NEO equipped with a TCI cryoprobe. Ten mixing times were used for the NOESY buildup curves: 25, 50, 75, 100, 150, 200, 300, 400, 500, and 700 ms. The new WADE pulses were implemented into the NOESY experiments available from the Bruker library (noesyfpgpph19/noesyesgpph), where the third π/2 pulse was replaced by a WADE-π/2 (W1F90) pulse, and the WATERGATE 3919 was replaced by the WADE-π (WR4) pulse. Note that the manes of the WADE pulses is reported according to the nomenclature used in our recent paper on GENETICS-AI30. The acquisition parameters consisted of 1024 points in the first dimension and 256 complex points in the indirect dimension. All spectra were processed using NMRPipe34. Both dimensions were processed using a sine bell window function shifted by 90o and zero-filled to double the size for a final matrix of 1024×512.
Results and Discussion
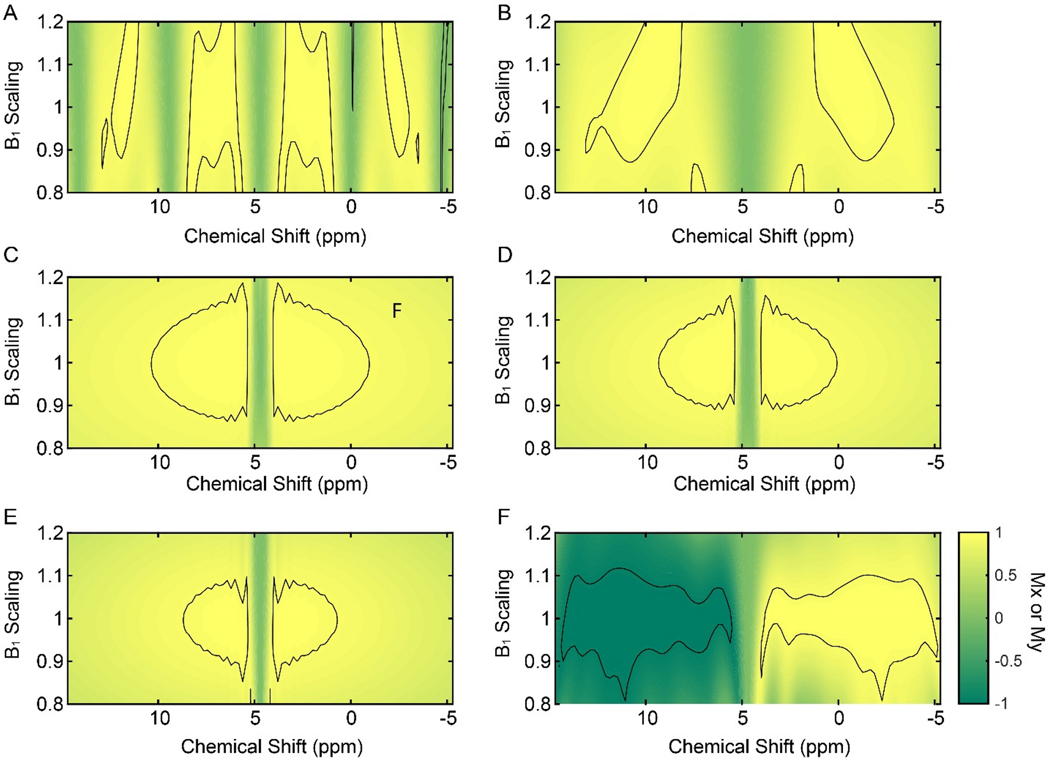
We first evaluated the performance of the most common water suppression schemes implemented in the 2D [1H,1H] NOESY pulse sequences in the Bruker library: Grad-3919-Grad, WATERGATE (Grad-Sel90-Hard180-Sel90-Grad), WATERGATE with flip-back pulses (Sel90Hard90-Grad-Sel90-Hard180-Sel90-Grad)35, and ES pulses (Grad1-Sel180-Hard180-Grad1-Grad2-Sel180-Hard180-Grad2)25. Figure 1 shows the schematics of these pulse sequences and the resulting operators for both water and protein magnetization. We simulated the responses of the transverse magnetization (Mx or My) to the different water-suppression schemes for a bandwidth of 20 kHz varying the RF offset and amplitude (B1±20%, Figure 2). As expected, the Grad-3919-Grad sequence shows a periodic response of Mx within the bandwidth considered. Using an interpulse delay of 200 μs, we obtained an excitation bandwidth relatively narrow (see the areas with fidelity levels of 0.5, Figures 2A). Note that the bandwidth covered by the Grad-3919-Grad sequence can be tailored by changing the interpulse delay, as shown in Figure 2B, though a trade-off is necessary between the bandwidth irradiated and water selectivity. We then analyzed the responses of the transverse components of the magnetization for the WATERGATE and the WATERGATE with water flip-back pulse sequences (Figures 2C and 2D. Unlike the 3919 sequences, the WATERGATE sequences display improved water selectivity and broader irradiation regions with a fidelity level of 0.95. Finally, we tested the ES sequence, which showed a better water selectivity. However, the high-fidelity area is somewhat reduced for the ES sequence (Figure 2E). Overall, the fidelity of the magnetization response for all the water suppression schemes considered has a limited bandwidth, spanning approximately 10 ppm with no scaling of the B1 field.
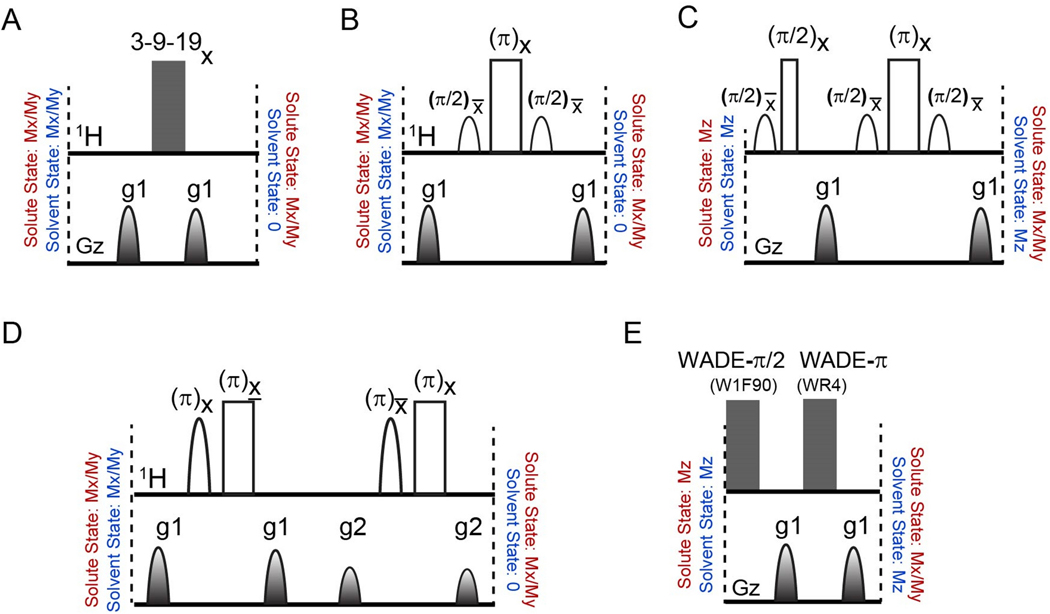
Figure 1.
Water suppression pulse schemes utilized for the 2D [1H, 1H] NOESY experiments. (A) The 3–9-19 sequence with total durations of 3.02 ms and 2.4ms for interpulse delays of 200 μs and 75 μs, respectively. (B) The WATERGATE sequence with a total duration of 6.04 ms using a selective pulse of 2 ms. (C) The WATERGATE sequence with water flip-back pulses for a total duration of 8.03 ms. (D) The excitation sculpting sequence with a total duration of 8.04ms. (E) The WADE sequence with a maximum RF amplitude of 5 kHz and a total duration of 5.01 ms. Unfilled wide and narrow rectangular shapes represent hard π and π /2 pulses, respectively. Unfilled shapes in 1H channel represent water-selective π/2 or π pulses. Filled shapes represent gradient pulses of 1 ms duration.
Figure 2.
Contour plots showing the responses of the transverse magnetization components (Mx or My) to the water suppression schemes showed in Figure 1 as a function of RF amplitude (B1), and for a bandwidth of 25 kHz. (A) Response of the Mx component to the Grad-3919-Grad sequence with an interpulse delay of 200 μs. The total length for the suppression sequence was 3.02 ms. (B) Response of the Mx component to the Grad-3919-Grad sequence with an interpulse delay of 75 μs. The total length for the water suppression sequence was 2.4 ms (C) Response of the Mx component to a WATERGATE sequence (Grad-Sel90-Hard180-Sel90-Grad). The total length for the suppression sequence was 6.04 ms. (D) Response of the My component to the sequence Sel90-Hard90-Grad-Sel90-Hard180-Sel90-Grad. The total length of the suppression sequence was 8.03 ms. (E) Response of the Mx component to the excitation sculpting sequence (Grad1-Sel90-Hard180-Grad1-Grad2-Sel90-Hard180-Grad2). The total length of the water suppression sequence was 8.04 ms. (F) Response of the My component to the W1F90-Grad-WR4-Grad sequence. The total length of the water suppression sequence was 5.01 ms with a RF amplitude of the WADE pulses of 5 kHz. Selective pulses used in the WATERGATE sequences were Sinc pulses of 2 ms and a RF amplitude for the hard pulse of 25 kHz. The contour lines in the plot are shown at a fidelity level of 0.95.
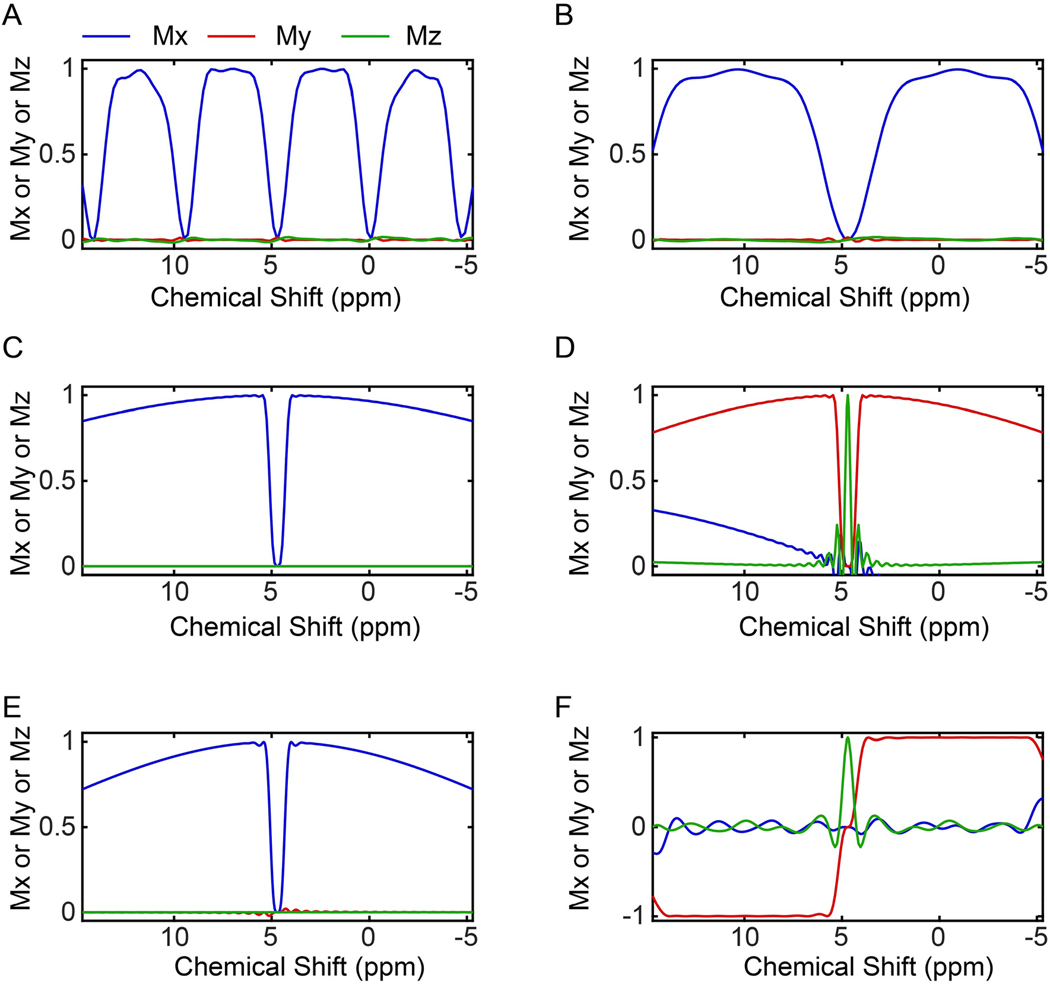
With this in mind, we programmed GENETICS-AI30 to create high-fidelity constant-amplitude pulses that elude water irradiation and achieve broader bandwidths than the classical water-suppression schemes. GENETICS-AI utilizes an evolutionary algorithm to generate a library of optimal phase shapes (OPS) to train an artificial intelligence (AI) module of MatLab® and create optimal solutions to a specific problem30. The WADE pulses designed by GENETICS-AI excite a significantly broader bandwidth with a higher level of fidelity relative to the previous pulse schemes and do not irradiate the water signal31. The response of the transverse magnetization to the WADE pulse scheme is reported in Figure 2F. For clarity, we also present the 1D profiles of the simulated magnetization responses by changing RF offset (Figure 3) and amplitude (Figure 4). Note that the sign of the My response for WR4 pulse is inverted to obtain null irradiation on resonance (water signal), leaving the z-component of the water signal virtually unperturbed (Figures 3F). As a result, the spectrum will display resonances with a positive sign for values of chemical shifts greater than 4.7 ppm and a negative sign for values less than 4.7 ppm. A detailed comparison of the WADE pulse performances with other binomial like sequences is reported in our previous article36.
Figure 3.
Transverse magnetization response as a function of the RF offset to various water suppression schemes used in 2D [1H,1H] NOESY experiments. For simplicity, the offset is reported in ppm. (A) 3–9-19 sequence with an interpulse delay of 200 μs. (B) 3–9-19 sequence with an interpulse delay of 75 μs. (C) WATERGATE sequence. (D) modified WATERGATE with water flip-back pulses. (E). Excitation sculpting sequence. (F) WADE water suppression sequence. The green lines show the Mz magnetization that on-resonance reaches a maximum for the WATERGATE with water flip-back (D) and WADE (F) sequences.
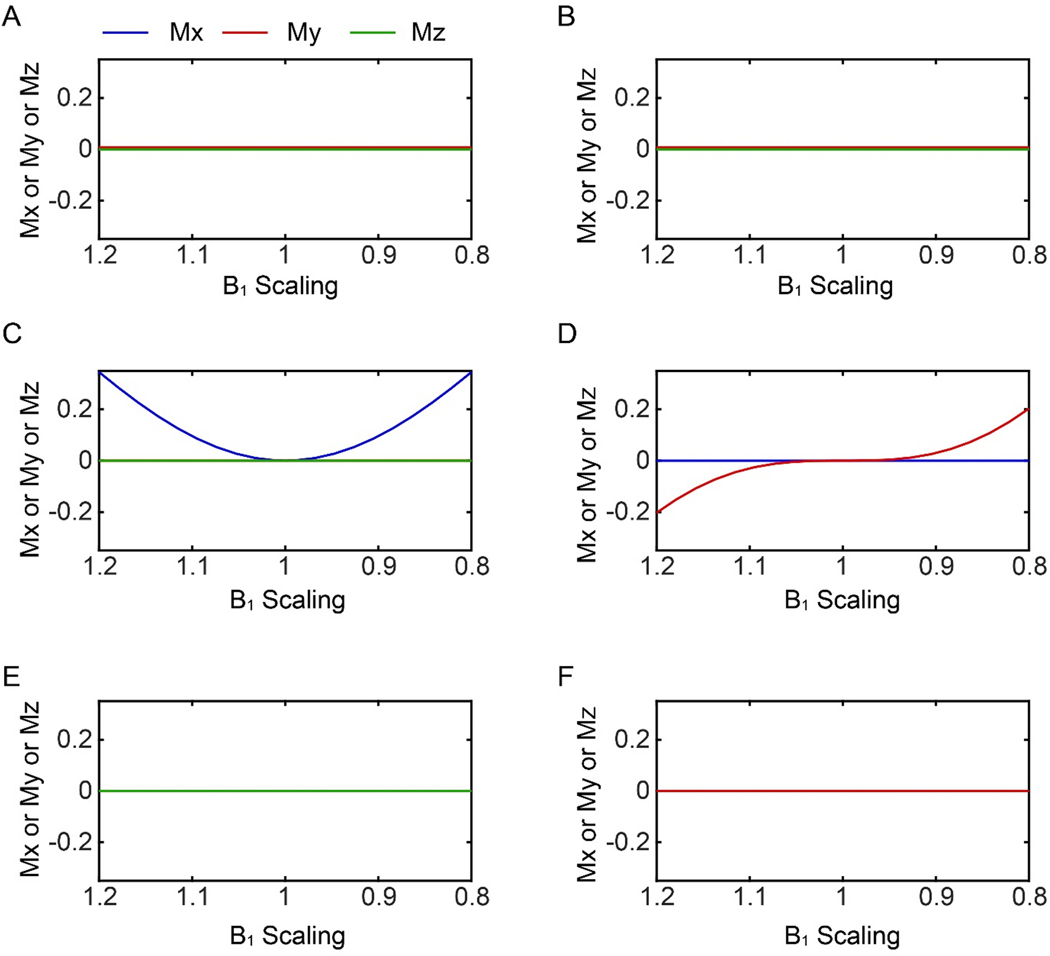
Figure 4.
Responses of the transverse magnetization to the different water suppression schemes used in the [1H,1H] NOESY experiments as a function of the RF amplitude. (A) and (B) 3–9-19 with an interpulse delay of 200 μs, and 75 μs, respectively. (C) WATERGATE sequence, (D) modified WATERGATE sequence (Lippens et al.35). (E) excitation sculpting and (F) WADE-NOESY suppression. Both WATERGATE sequences (C and D) have non-zero transverse water magnetization with RF inhomogeneity.
We then implemented the W1F90 and WR4 pulses into the 2D [1H,1H] NOESY pulse sequence (Figure 5). The general scheme was taken from the noesygpph19 code available in the Bruker sequence library. We did not change the first two hard π/2 pulses that excite the entire 1H bandwidth, including the water signal. We substituted, however, the hard π/2 pulse after the mixing time with a W1F90 pulse (red) and the water-suppression scheme with a WR4 pulse (blue), which is flanked with two gradient pulses along the z-direction to remove spurious magnetization. The W1F90 pulse excites the left and right sides of the bandwidth with high fidelity, leaving a null excitation on-resonance. The My component of the magnetization as a function of the RF offset follows a sigmoidal behavior, with an on-resonance point of inversion. The latter is accomplished by modulating the phase shape, as indicated in Figure 5B. The WR4 pulse for the coherence detection is the same utilized for our previously published [1H,15N] TROSY-HSQC sequence31. The WR4 pulse does not affect the Mx component of the magnetization and operates an inversion operation on both sides of the bandwidth, avoiding water irradiation.
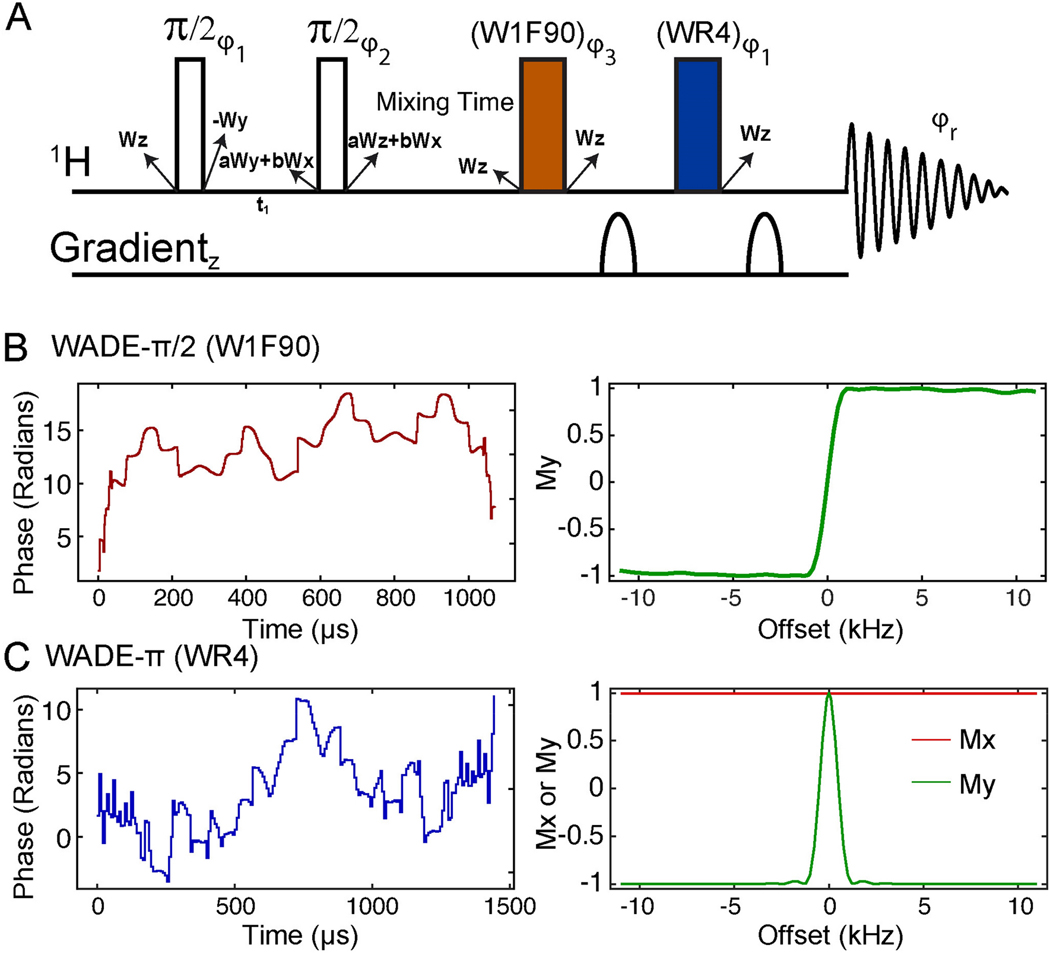
Figure 5.
(A) [1H,1H] WADE-NOESY pulse sequence. The two non-filled rectangles are hard π/2 pulse, and the filled rectangles are WADE pulses. The phases φ1, φ2, φ3, and the receiver phase (φ2r) were {x, -x}, { x, x, x, x, x, x, x, x, -x, -x, -x, -x, -x, -x, -x, -x }, {x, x, -x, -x, -y, -y, y, y}, and { x, -x, -x, x, y, -y, -y, y, -x, x, x, -x, -y, y, y, -y} respectively. The state of water magnetization is given near each pulse. Starting with z magnetization on water (Wz), a mixture of transverse magnetization is created (aWy + bWx) during the t1 evolution. Here we assume no relaxation during the t1 delay. The value of a and b are a function of chemical shift and t1. During the long mixing time the water magnetization returns to the z-direction. Both WADE pulses do not affect the state of water magnetization, which remains along the z-direction during acquisition. (B) Phase shape of the W1F90 pulse and corresponding excitation profile simulated with an initial magnetization Mz. (C) Phase shape of WR4 pulse and its simulated response with initial magnetizations Mx (red) and My (green). The amplitude for both pulses is kept constant at 6.33 kHz.
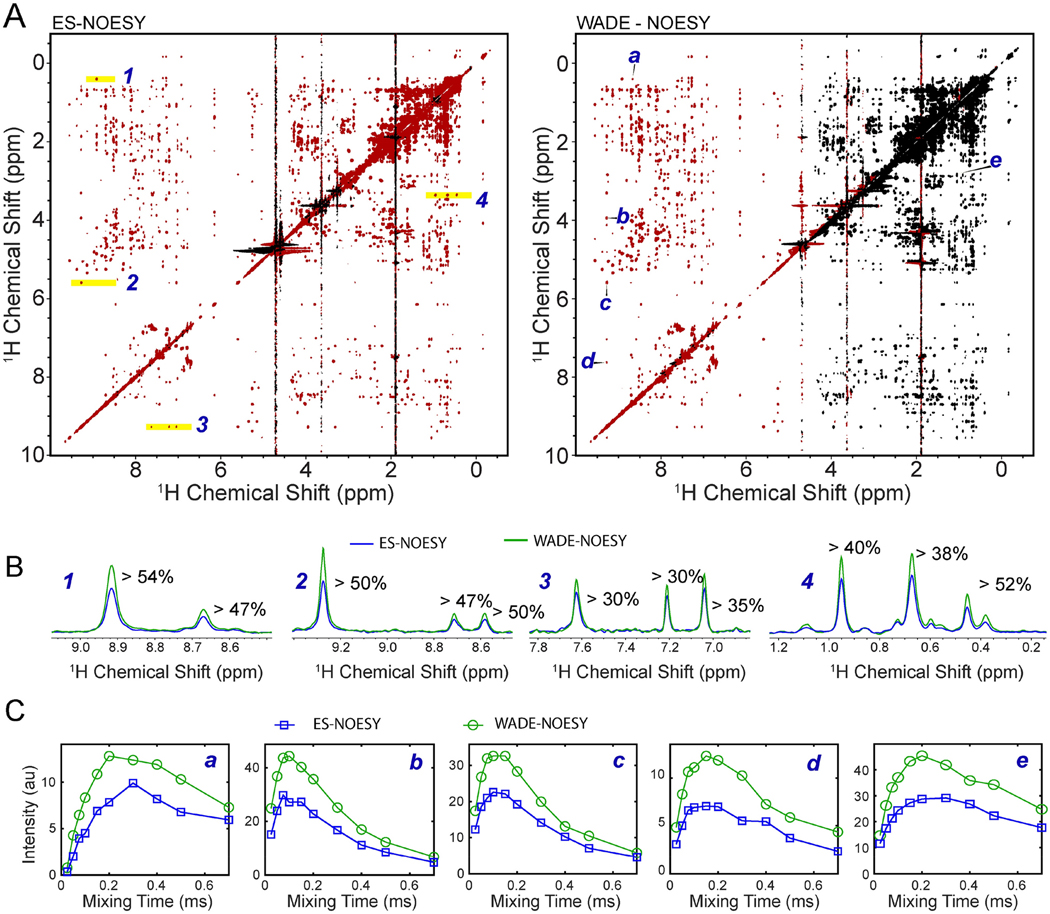
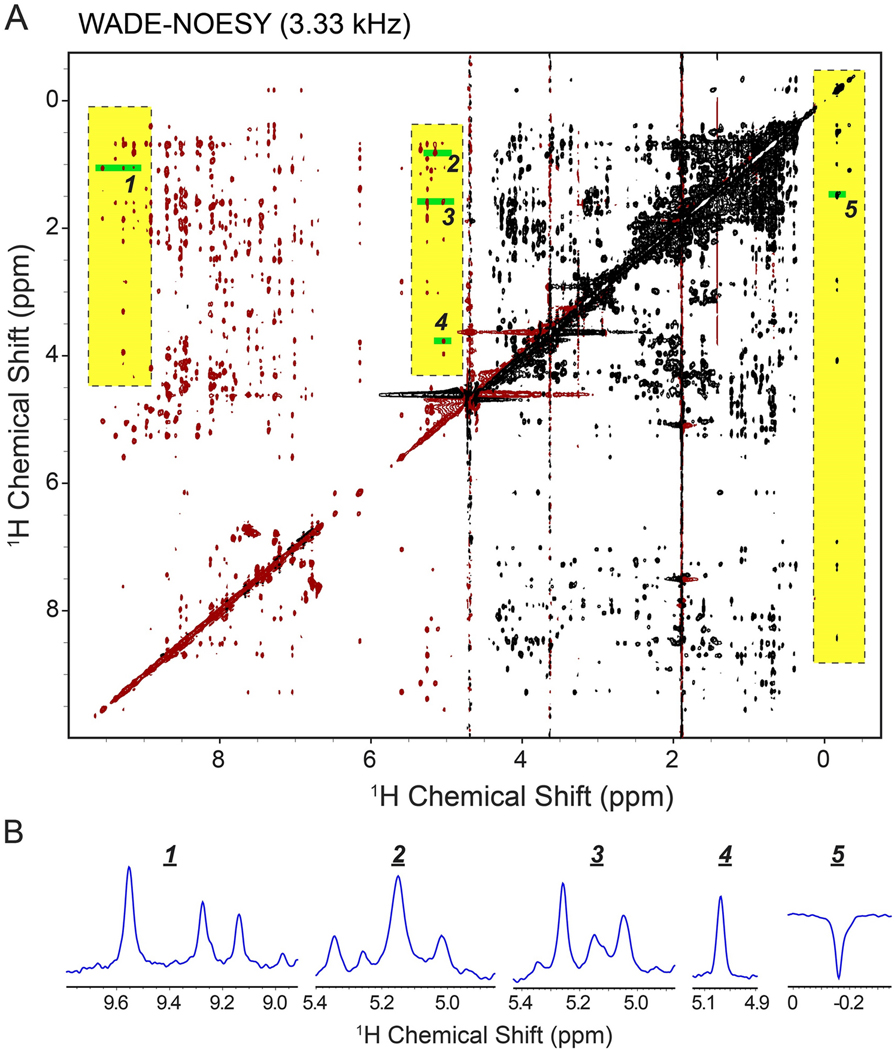
To experimentally assess the performance of the WADE pulses, we compared the 2D [1H,1H] NOESY experiment with ES water suppression (Bruker sequence noesyesgpph) with the [1H,1H] WADE-NOESY pulse sequence UbiK48C. A comparison of the 2D NOESY spectra is shown in Figure 6A. UBIK48C fold comprises a five-stranded β sheet, an α helix, and a short 310 helix; therefore, a significant number of NOEs are expected. The peaks of the [1H,1H] WADE-NOESY spectrum are negative in the range of chemical shifts from −1.0 to 4.7 ppm and positive from 4.7 to 10 ppm, according to the W1F90 pulse excitation profile of Figure 5. Overall, the 2D [1H,1H] WADE-NOESY spectrum contains a higher number of NOEs, particularly in the amide region, featuring several HN-HN contacts absent in the [1H,1H] ES-NOESY experiment. Additional NOE correlations are observed for the resonances at both spectrum edges. The higher number of NOEs in the amide region of UbiK48C can be explained by the W1F90 and WR4 pulses that keep the water magnetization along the z-axis, minimizing the exchange of labile amide protons with water. A closer analysis of the NOE cross-peaks between the amide groups and amide and Hα protons shows that the [1H,1H] WADE-NOESY spectrum has a higher resonance intensity up to 54% relative to the [1H,1H] ES-NOESY spectrum (Figure 6B). We also observed an enhancement of the resonance intensities for the NOE cross-peaks in the aliphatic region of the protein. In this case, the signal enhancement can be attributed to the higher fidelity of the spin operation for the two WADE pulses, especially for peaks at the edges of the spectrum. Overall, we observe an increase in signal intensity of ~50% for these resonances. We then tested the NOE buildup for several resonances as a function of the mixing time (Figure 6C). From the selected residues, it is possible to appreciate the typical kinetics of the NOE buildup and the significant enhancement of the NOE cross peaks intensities relative to the [1H,1H] ES-NOESY spectra. While the peak intensities are enhanced throughout the spectrum, the peak intensities near the water signal are weaker than the corresponding cross peaks in the [1H,1H] ES-NOESY spectrum. However, the latter can be easily overcome by increasing the water selectivity, i.e., reducing the WADE pulses amplitude. This is demonstrated in Figure 7, which shows the [1H,1H] WADE-NOESY spectrum obtained by lowering the WADE pulse amplitude to 3.33 kHz. In this case, the peaks near the water signals are significantly more intense than the ES sequence. Also, the broader irradiation bandwidth of the WADE pulses enhances the peak intensities at both edges of the spectrum relative to the ES sequence (see highlighted regions of Figure 7).
Figure 6.
(A) Comparison of the 2D [1H-1H] NOESY of UBIK48C with a 150 ms mixing time using ES and WADE suppression scheme. The peaks in red are positive and those in black are negative. All experiments were performed on a Bruker 850 MHz spectrometer at 303 K using identical acquisition parameters: 256 number of complex points were acquired in indirect dimension and 8 scans per FID with a relaxation delay of 2 sec. Identical processing parameters were used for both spectra. (B) 1D slices of the 2D [1H-1H] ESNOESY (blue) and [1H-1H] WADE-NOESY (green) for the peaks highlighted in A. (C) NOESY buildup curves [1H-1H] ES-NOESY(blue) and [1H-1H] WADE-NOESY (green) for the resonances marked A.
Figure 7.
(A) [1H-1H] WADE-NOESY of UBIK48C with WADE pulses of 3.33 kHz to improve peak detections near the water signal. The highlighted regions emphasize resonances with a significantly higher intensity than those in the ES-NOESY. The experiment was performed on a Bruker 850 MHz spectrometer at 303 K. 128 number of complex points were acquired in indirect dimension and 4 scans per FID with a relaxation delay of 2 sec. (B) 1D slices from near water and far off resonance regions of the spectrum.
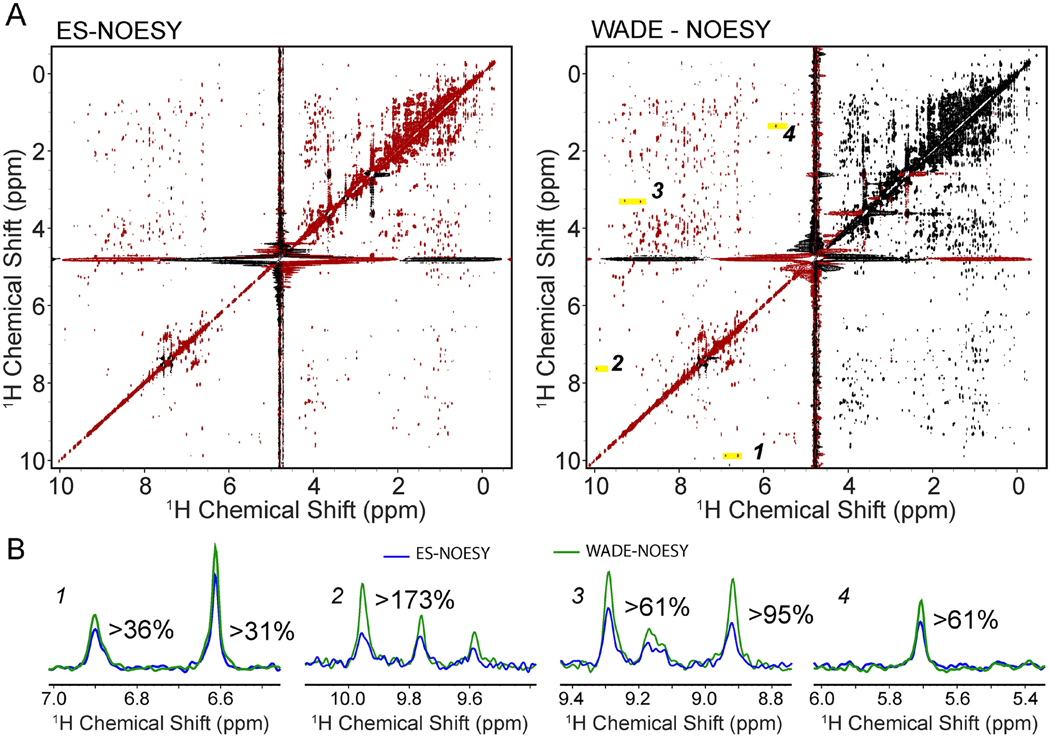
We also compared the performance of the two pulse sequences for RKIP, a well-folded protein involved in kinase signaling33. As for UBIK48C, the number of NOE cross-peaks obtained for RKIP with the [1H,1H] WADE-NOESY experiment is substantially higher than the corresponding [1H,1H] ES-NOESY spectrum (Figure 8A). The latter is even more apparent for peaks at the two edges of the bandwidth, as illustrated in the cross-sections of Figure 8B, where some of the cross-peaks are barely detectable with the [1H,1H] ES-NOESY experiment. In addition, a significant increase in the number and intensity of the cross-peaks is noticeable for the HN-HN region.
Figure 8.
2D [1H,1H] NOESY spectra of RKIP with a 150 ms mixing time using (A) ES as a water suppression sequence and (B) WADE water suppression. Positive peaks are shown in red and negative peaks in black. All experiments were performed on a Bruker 900 MHz spectrometer at 300 K using identical acquisition parameters with 128 complex time points in the indirect dimension and 8 scans per FID. The relaxation delay was set to 2 sec. Both spectra were processed using NMRpipe with identical processing parameters. We have used the sine window function with offsets 0.2 and 0.45 in t2 and t1 dimensions, respectively. NMRpipe solvent filter (SOL) is used with a sine low pass filter. Zero filled the complex FID data to a final size of 1024*512 complex points. A baseline correction using 4th order polynomial is performed in both dimensions. (B) 1D slices were taken from the 2D ES-NOESY (blue) and WADE-NOESY (green) for the peaks highlighted in A.
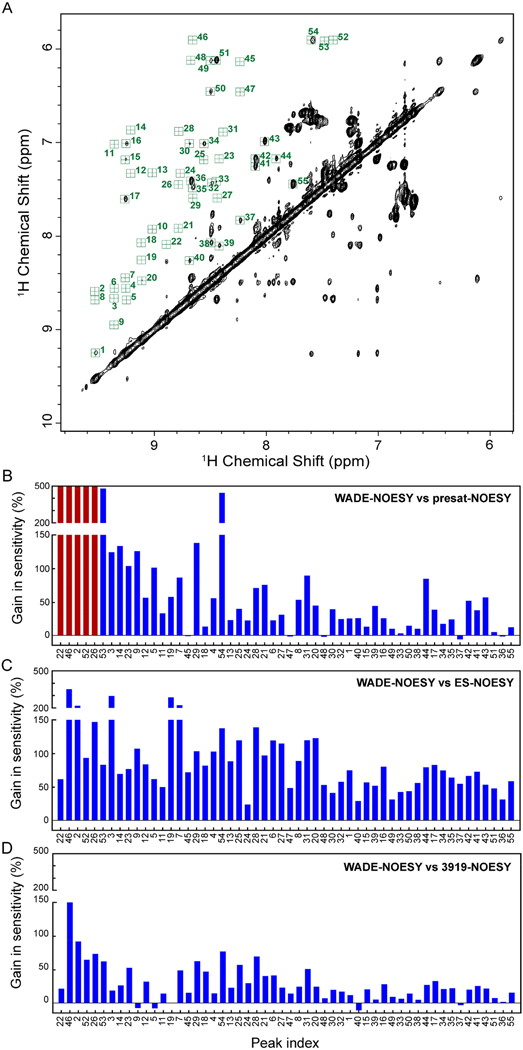
To determine more quantitatively the improvement of the [1H,1H] WADE-NOESY over the other pulse schemes of Figure 1, we analyzed the relative gain in sensitivity. Figure 9A shows the amide region of UbiK48C with the HN-HN cross-peaks indicated by random numbers. The histograms in Figures 9B–D show the differences in the intensities of the cross-peaks relative to the [1H,1H] WADE-NOESY experiment. Compared to the [1H,1H] NOESY with presaturation (presat-NOESY), the [1H,1H] WADE-NOESY shows an average increase of cross-peak intensity of ~60%. The highest gain (>200%) is achieved for labile HN resonances that broaden out partially or entirely due to the exchange with the water signal caused by the weak RF field used in the presaturation sequence. The comparison with ES and 3919 sequences shows an average sensitivity gain of 89% and 30%, respectively. In these latter cases, the higher fidelity of operations for the WADE pulses may explain the signal enhancement.
Figure 9.
Comparison of cross peak intensities for the different pulse sequences. (A) Amide region of UBIK48C. For simplicity, the cross-peaks were indicated with random numbers. Sensitivity gain of the NOE cross peaks of the 2D [1H,1H] WADE-NOESY relative to (B) presat-NOESY (noesyphpr). Red bars represent the peaks that broaden out due to exchange. (C) ES-NOESY (noesyesgpph), and (D) 3919-NOESY (noesygpph19). All experiments were recorded on Bruker 900 MHz spectrometer at 300K with 128 complex time points in indirect dimension and 8 scans per FID. A relaxation delay of 2 sec was used for all the experiments. The spectra were processed with identical processing parameters.
RF pulse design is fundamental for developing NMR spectroscopy at high and ultra-high magnetic fields37. In the past years, many research groups have contributed to designing and developing new pulses or pulse sequences that enhance NMR signal detection for biological macromolecules38–41. However, NMR spectroscopy at ultra-high magnetic fields calls for RF pulses that irradiate larger bandwidths, with high fidelity of spin operations and compensation for instrumental inhomogeneity. We recently showed that a combination of an evolutionary algorithm with AI enables the design of new RF pulses with a constant amplitude to excite broader bandwidth30. These new pulses can be used for a variety of applications, spanning from spin entanglement to solution30,31 and solid-state36 NMR spectroscopy, as well as magnetic resonance imaging30. The design of the WADE pulses is only an example of the new possibilities that the new software GENETICS-AI opened up.
In conclusion, we presented the implementation of W1F90 and WR4 pulses in the 2D [1H,1H] NOESY pulse sequences, demonstrating a significant enhancement in the detection of internuclear NOE. The enhancement is due to the broadband nature of the new pulses and a substantially reduced magnetization exchange between water and labile amide protons. Concomitantly, we observed an increase in sensitivity of NOEs for non-exchangeable protons, which is due to the higher fidelity spin operation of the WADE pulses relative to the commonly used water suppression schemes. We anticipate that the WADE pulses will improve the sensitivity of the recently developed L-PROSY experiments42 for labile protons as well as heteronuclear 3D 15N- or 13C-edited NOESY experiments that are more suitable for larger biomacromolecules.
Acknowledgment
The authors thank Dr. M. Rosner (Ben May Department for Cancer Research, University of Chicago, Chicago, IL, USA) for providing the RKIP plasmid. All the NMR experiments were conducted at the Minnesota NMR Center (University of Minnesota, Minneapolis, MN, USA).
Funding
This work was supported by the National Institute of Health (HL 144130) to G.V. and a subcontract to GV from GM121735 (Marsha Rosner, P.I.).
Footnotes
Disclaimer
The authors declare no conflict of interest concerning the research presented here.
Consent
All the authors have seen and approved the submitted manuscript.
Publisher's Disclaimer: This Accepted Manuscript (AM) is a PDF file of the manuscript accepted for publication after peer review, when applicable, but does not reflect post-acceptance improvements, or any corrections. Use of this AM is subject to the publisher’s embargo period and AM terms of use. Under no circumstances may this AM be shared or distributed under a Creative Commons or other form of open access license, nor may it be reformatted or enhanced, whether by the Author or third parties. By using this AM (for example, by accessing or downloading) you agree to abide by Springer Nature’s terms of use for AM versions of subscription articles: https://www.springernature.com/gp/open-research/policies/accepted-manuscript-terms
Data Availability
The WADE pulses and the [1H,1H] WADE-NOESY pulse sequence are available at the University of Minnesota Repository Site (Link will be available before publication) and GitHub (https://github.com/manuvsub/WADE_NOESY).
References
- 1.Overhauser AW Polarization of nuclei in metals. Physical Review 92, 411 (1953). [Google Scholar]
- 2.Carver TR & Slichter CP Polarization of Nuclear Spins in Metals. Physical Review 92, 212–213 (1953). [Google Scholar]
- 3.Kumar A, Ernst RR & Wüthrich K. A two-dimensional nuclear Overhauser enhancement (2D NOE) experiment for the elucidation of complete proton-proton cross-relaxation networks in biological macromolecules. Biochemical and Biophysical Research Communications 95, 1–6 (1980). [DOI] [PubMed] [Google Scholar]
- 4.Wuthrich K. NMR of Proteins and nucleic acids, (Wiley, 1986). [Google Scholar]
- 5.Kumar A, Wagner G, Ernst RR & Wuthrich K. Buildup Rates of the Nuclear Overhauser Effect Measured by Two-Dimensional Proton Magnetic-Resonance Spectroscopy - Implications for Studies of Protein Conformation. Journal of the American Chemical Society 103, 3654–3658 (1981). [Google Scholar]
- 6.Vogeli B. The nuclear Overhauser effect from a quantitative perspective. Prog Nucl Magn Reson Spectrosc 78, 1–46 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Pervushin K, Riek R, Wider G. & Wüthrich K. Attenuated T2 relaxation by mutual cancellation of dipole–dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proceedings of the National Academy of Sciences 94, 12366–12371 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pervushin KV, Wider G, Riek R. & Wuthrich K. The 3D NOESY-[(1)H,(15)N,(1)H]-ZQ-TROSY NMR experiment with diagonal peak suppression. Proc Natl Acad Sci U S A 96, 9607–12 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zwahlen C. et al. An NMR Experiment for Measuring Methyl−Methyl NOEs in 13C-Labeled Proteins with High Resolution. Journal of the American Chemical Society 120, 7617–7625 (1998). [Google Scholar]
- 10.Vögeli B, Olsson S, Güntert P. & Riek R. The Exact NOE as an Alternative in Ensemble Structure Determination. Biophysical Journal 110, 113–126 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strotz D. et al. Protein Allostery at Atomic Resolution. Angew Chem Int Ed Engl 59, 22132–22139 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Price WS Water Signal Suppression in NMR Spectroscopy. in Annual Reports on NMR Spectroscopy, Vol. 38 (ed. Webb GA) 289–354 (Academic Press, 1999). [Google Scholar]
- 13.Zheng G. & Price WS Solvent signal suppression in NMR. Progress in Nuclear Magnetic Resonance Spectroscopy 56, 267–288 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Krishnan VV & Murali N. Radiation damping in modern NMR experiments: Progress and challenges. Progress in Nuclear Magnetic Resonance Spectroscopy 68, 41–57 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Warren WS & Richter W. Concentrated Solution Effects. in eMagRes. [Google Scholar]
- 16.Nguyen BD, Meng X, Donovan KJ & Shaka AJ SOGGY: solvent-optimized double gradient spectroscopy for water suppression. A comparison with some existing techniques. J Magn Reson 184, 263–74 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Liu M. et al. Improved WATERGATE Pulse Sequences for Solvent Suppression in NMR Spectroscopy. Journal of Magnetic Resonance 132, 125–129 (1998). [Google Scholar]
- 18.Hwang TL & Shaka AJ Water Suppression That Works. Excitation Sculpting Using Arbitrary Wave-Forms and Pulsed-Field Gradients. Journal of Magnetic Resonance, Series A 112, 275–279 (1995). [Google Scholar]
- 19.Chen J, Zheng G. & Price WS A new phase modulated binomial-like selective-inversion sequence for solvent signal suppression in NMR. Magn Reson Chem 55, 115–119 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Hoult DI Solvent peak saturation with single phase and quadrature fourier transformation. Journal of Magnetic Resonance (1969) 21, 337–347 (1976). [Google Scholar]
- 21.Grzesiek S. & Bax A. The Importance of Not Saturating H2o in Protein Nmr - Application to Sensitivity Enhancement and Noe Measurements. Journal of the American Chemical Society 115, 12593–12594 (1993). [Google Scholar]
- 22.Piotto M, Saudek V. & Sklenář V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. Journal of Biomolecular NMR 2, 661–665 (1992). [DOI] [PubMed] [Google Scholar]
- 23.Stejskal EO & Tanner JE Spin Diffusion Measurements: Spin Echoes in the Presence of a Time-Dependent Field Gradient. The Journal of Chemical Physics 42, 288–292 (1965). [Google Scholar]
- 24.Adams RW, Holroyd CM, Aguilar JA, Nilsson M. & Morris GA “Perfecting” WATERGATE: clean proton NMR spectra from aqueous solution. Chem Commun (Camb) 49, 358–60 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Stott K, Stonehouse J, Keeler J, Hwang T-L & Shaka AJ Excitation Sculpting in High-Resolution Nuclear Magnetic Resonance Spectroscopy: Application to Selective NOE Experiments. Journal of the American Chemical Society 117, 4199–4200 (1995). [Google Scholar]
- 26.Hore PJ A new method for water suppression in the proton NMR spectra of aqueous solutions. Journal of Magnetic Resonance (1969) 54, 539–542 (1983). [Google Scholar]
- 27.Hore PJ Solvent suppression in fourier transform nuclear magnetic resonance. Journal of Magnetic Resonance (1969) 55, 283–300 (1983). [Google Scholar]
- 28.Sklenar V, Piotto M, Leppik R. & Saudek V. Gradient-Tailored Water Suppression for 1H-15N HSQC Experiments Optimized to Retain Full Sensitivity. Journal of Magnetic Resonance, Series A 102, 241–245 (1993). [Google Scholar]
- 29.Marion D. et al. Overcoming the overlap problem in the assignment of 1H NMR spectra of larger proteins by use of three-dimensional heteronuclear 1H-15N Hartmann-Hahn-multiple quantum coherence and nuclear Overhauser-multiple quantum coherence spectroscopy: application to interleukin 1 beta. Biochemistry 28, 6150–6 (1989). [DOI] [PubMed] [Google Scholar]
- 30.Manu VS, Pavuluri K, Olivieri C. & Veglia G. High fidelity control of spin ensemble dynamics via artificial intelligence: from quantum computing to NMR spectroscopy and imaging PNAS Nexus In Press(2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Manu VS, Olivieri C, Pavuluri K. & Veglia G. Design and applications of water irradiation devoid RF pulses for ultra-high field biomolecular NMR spectroscopy. Physical Chemistry Chemical Physics (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olivieri C. et al. Simultaneous detection of intra- and inter-molecular paramagnetic relaxation enhancements in protein complexes. J Biomol NMR 70, 133–140 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee J. et al. Raf Kinase Inhibitory Protein regulates the cAMP-dependent protein kinase signaling pathway through a positive feedback loop. Proc Natl Acad Sci U S A 119, e2121867119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delaglio F. et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6, 277–93 (1995). [DOI] [PubMed] [Google Scholar]
- 35.Lippens G, Dhalluin C. & Wieruszeski JM Use of a water flip-back pulse in the homonuclear NOESY experiment. Journal of Biomolecular NMR 5, 327–331 (1995). [DOI] [PubMed] [Google Scholar]
- 36.Gopinath T, Manu VS, Weber DK & Veglia G. PHRONESIS: A One-Shot Approach for Sequential Assignment of Protein Resonances by Ultrafast MAS Solid-State NMR Spectroscopy. Chemphyschem 23, e202200127 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ardenkjaer-Larsen JH et al. Facing and Overcoming Sensitivity Challenges in Biomolecular NMR Spectroscopy. Angew Chem Int Ed Engl 54, 9162–85 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schanda P. Fast-pulsing longitudinal relaxation optimized techniques: Enriching the toolbox of fast biomolecular NMR spectroscopy. Progress in Nuclear Magnetic Resonance Spectroscopy 55, 238–265 (2009). [Google Scholar]
- 39.Guéron M, Plateau P. & Decorps M. Solvent signal suppression in NMR. Progress in Nuclear Magnetic Resonance Spectroscopy 23, 135–209 (1991). [DOI] [PubMed] [Google Scholar]
- 40.Rance M, Loria JP & Palmer A.G.r. Sensitivity improvement of transverse relaxation-optimized spectroscopy. J Magn Reson 136, 92–101 (1999). [DOI] [PubMed] [Google Scholar]
- 41.Zhu G, Kong XM & Sze KH Gradient and sensitivity enhancement of 2D TROSY with water flipback, 3D NOESY-TROSY and TOCSY-TROSY experiments. J Biomol NMR 13, 77–81 (1999). [DOI] [PubMed] [Google Scholar]
- 42.Novakovic M, Cousin SF, Jaroszewicz MJ, Rosenzweig R. & Frydman L. Looped-PROjected SpectroscopY (L-PROSY): A simple approach to enhance backbone/sidechain cross-peaks in (1)H NMR. J Magn Reson 294, 169–180 (2018). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The WADE pulses and the [1H,1H] WADE-NOESY pulse sequence are available at the University of Minnesota Repository Site (Link will be available before publication) and GitHub (https://github.com/manuvsub/WADE_NOESY).