Abstract
Some studies have reported that the gut microbiota can influence adrenal-related hormone levels. However, the causal effects of the gut microbiota on adrenal function remain unknown. Therefore, we employed a two-sample Mendelian randomization (MR) study to systematically investigate the impact of gut microbiota on the function of different regions of the adrenal gland. The summary statistics for gut microbiota and adrenal-related hormones used in the two-sample MR analysis were derived from publicly available genome-wide association studies (GWAS). In the MR analysis, inverse variance weighting (IVW) was used as the primary method, with MR-Egger, weighted median, and cML-MA serving as supplementary methods for causal inference. Sensitivity analyses such as the MR-Egger intercept test, Cochran’s Q test, and leave-one-out analysis were used to assess pleiotropy and heterogeneity. We identified 27 causal relationships between 23 gut microbiota and adrenal function using the IVW method. Among these, Sellimonas enhanced the function of the adrenal cortex reticularis zone (beta = 0.008, 95% CI: 0.002–0.013, P = 0.0057). The cML-MA method supported our estimate (beta = 0.009, 95% CI: 0.004–0.013, P = 2 × 10− 4). Parasutterella, Sutterella, and Anaerofilum affect the functioning of different regions of the adrenal gland. Notably, pleiotropy was not observed. Our findings revealed that the gut microbiota is causally associated with adrenal function. This enhances our understanding of the gut-microbiota-brain axis and provides assistance in the early diagnosis and treatment of adrenal-related diseases in clinical practice.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-73420-w.
Keywords: Adrenal function, Gut microbiota, Gut-microbiota-brain axis, Causal inference, Mendelian randomization
Subject terms: Microbiology, Endocrinology, Urology
Introduction
Adrenal dysfunction is a common clinical manifestation of adrenal-related diseases. Disorders of adrenal-related hormones (including adrenomedullary hormones, steroid hormones, and catecholamines) originating from the adrenal cortex, such as adrenal adenomas and adrenal cortical carcinomas, or from the adrenal medulla, are frequently encountered in clinical practice1–3. Complications arising from adrenal hormone imbalance pose significant challenges for both patients and healthcare professionals. For instance, high concentrations of catecholamines produced by pheochromocytomas can lead to severe complications during surgery, such as significant fluctuations in blood pressure and arrhythmias, often endangering the patient’s life4. In addition, the overproduction of glucocorticoids by patients with adrenocortical carcinoma not only causes clinical symptoms such as central obesity, moon face, and buffalo hump but is also closely related to postoperative recurrence and survival prognosis5–7. Furthermore, the evaluation of adrenal-related hormone levels not only guides assessing the nature of adrenal masses but also aids in the diagnosis of benign and malignant tumours8. In summary, adrenal hormone levels, which reflect adrenal function, play a crucial role in clinical practice. Therefore, research related to the factors affecting adrenal function is of particular importance in clinical practice.
The gut-microbiota-brain axis is a bidirectional connection between the gut microbiota, gut, and brain, formed through multiple systemic networks. This axis is a major bidirectional pathway that functions through neurological, immune, and endocrine pathways9. The gut-microbiota-brain axis is crucial for maintaining dynamic balance, immune health, and hormone levels in the human body. Signals originating from the brain can influence gastrointestinal activity through both the sympathetic-adrenal-medullary pathway and the hypothalamic-pituitary-adrenal (HPA) axis, which includes gastrointestinal motility, secretion of digestive fluids, and the distribution and gene expression of gut microbiota10,11. In these processes, adrenal-related hormones, such as norepinephrine, epinephrine, and glucocorticoids, play important intermediary roles. It is evident that both the HPA axis and sympathetic-adrenal-medullary pathway are closely associated with adrenal function. In a clinical observational study, Quénéhervé et al. found that more than 30% of patients with chronic adrenal insufficiency experienced gastrointestinal symptoms such as abdominal pain, vomiting, and nausea12. Similar reports have been documented in cases of adrenal crises, with researchers generally attributing these gastrointestinal symptoms to glucocorticoids and mineralocorticoids12–14. Therefore, the adrenal glands not only play an important role in the gut-microbiota-brain axis but also play a significant role in regulating gastrointestinal function and the composition and abundance of the gut microbiota. However, these previous studies have several limitations. Most studies have been conducted on animals, and small sample sizes can lead to biased results. Additionally, clinical observational studies investigating the relationship between gut microbiota and adrenal function are challenging to avoid confounding factors, such as age, environment, dietary habits, and sociocultural characteristics, which are difficult to control effectively in traditional studies. Furthermore, there is a lack of research on the impact of the gut microbiota on adrenal function, leaving the causal relationship between the gut microbiota and adrenal function largely unknown.
MR is a data analysis technique used for the causal inference of aetiology. It utilises genetic variations strongly associated with the exposure factor as instrumental variables (IVs) to assess the causal relationship between the exposure and the outcome15,16. Because genetic variations are assumed to be randomly inherited and unaffected by diseases or other factors, MR can overcome confounding biases that are difficult to avoid in traditional observational studies17. Two-sample Mendelian randomization analysis, as an extension of MR, does not directly analyse individual-level data but instead utilises summary statistics from genome-wide association studies (GWASs). With the increasing number of GWASs related to the gut microbiota and adrenal glands, there is a greater abundance of large-scale summary statistics, which significantly improves the statistical stability and power for conducting relevant two-sample MR analysis18,19. To the best of our knowledge, MR has been extensively applied to investigate the causal relationships between the gut microbiota and several diseases, including Crohn’s disease20, rheumatoid arthritis21, and cancers such as breast, prostate, and endometrial cancer22. However, no research has analysed the causal relationship between the microbiota and adrenal function using MR.
In this study, we systematically investigated the potential causal relationship between these two factors using two-sample Mendelian randomization. We used gut microbiota-related GWAS data available with the MiBioGen consortium as the exposure dataset and utilised six different GWAS datasets from the FinnGen consortium, IEU Open GWAS project, and GWAS Catalog as the outcomes. The six GWAS datasets included adrenal-related hormone levels (adrenomedullin, aldosterone, cortisol, and sex hormones) that reflect adrenal function. Based on these findings, we aimed to provide new insights into the gut-microbiota-brain axis as well as the causal relationship between gut microbiota and adrenal function, with the ultimate goal of aiding in the treatment of adrenal-related diseases.
Methods
Data resource
GWAS summary statistics for the gut microbiota were obtained from the International MiBiogen Consortium, which has conducted the largest genome-wide meta-analysis of gut microbiota composition to date. This study included 18,340 participants from 24 cohorts, with over 70% of them being Europeans. The remaining participants were from other regions, including the Middle East, East Asia, Africa, and other backgrounds. The gut microbiome composition was classified into five categories. In order to investigate the causal relationship between each bacterial population and adrenal function in as much detail as possible, we selected the ‘genus’ category, which represents the lowest level of classification, as the focus of our investigation. In the MiBiogen consortium study, 131 genera had an average abundance of greater than 1%. However, 12 genera were classified as unknown, resulting in the inclusion of 119 gut microbiota genera in our analysis.
GWAS summary statistics for all phenotypes in the outcome were obtained from published GWAS. The ‘Adrenomedullin levels’, representing the level of adrenal medulla function, were sourced from the IEU Open GWAS project. For traits representing adrenal cortex function, GWAS data for ‘hyperaldosteronism’ were obtained from the FinnGen consortium R9 release data. In addition, the GWAS data for ‘Cortisol (plasma) Measurement’, ‘Cortisol (urine) Measurement’, ‘Estradiol measurement’, and Free androgen index’ were obtained from the GWAS Catalog. Most of these GWAS samples consisted of individuals of European descent and were largely independent of each other. For more detailed information, refer to Additional File 1: Table S1.
Instrumental variable selection
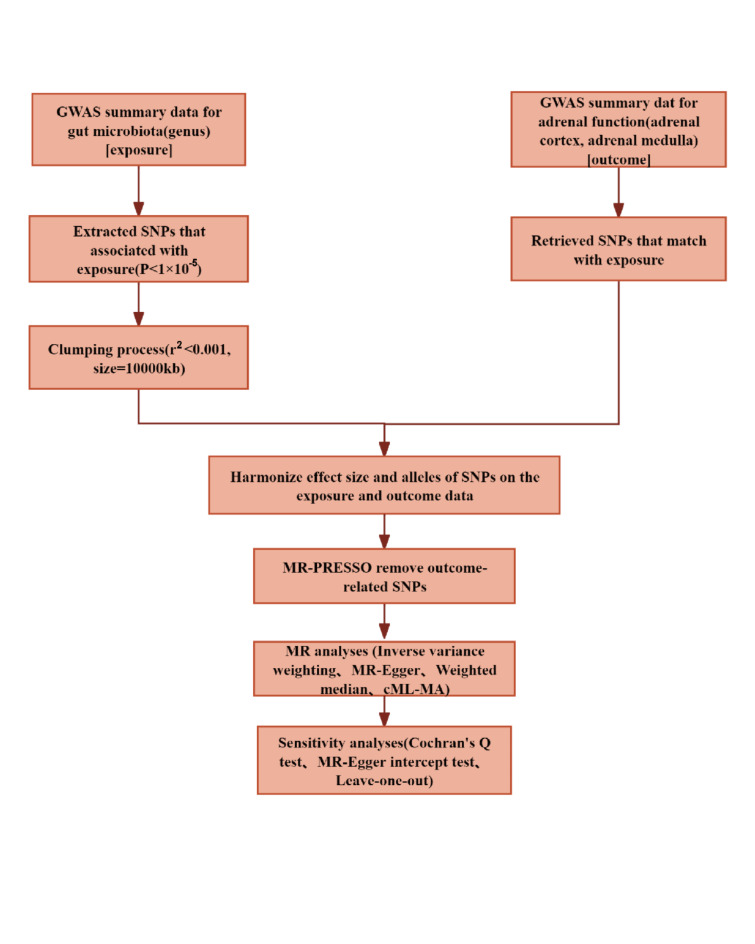
The overall flowchart of this study is shown in Fig. 1. To ensure the authenticity and reliability of the causal relationship between the gut microbiota and adrenal function, we used multiple sets of genetic IVs to reflect the levels of adrenal function. Adrenomedullin levels reflect the function of the adrenal medulla, whereas aldosterone, cortisol, and sex hormones (estradiol and androgen) were used to respectively represent the function of the adrenal zona glomerulosa, fasciculata, and reticularis. Accurate measurement of adrenal function is extremely difficult; therefore, we used these methods.
Fig. 1.
Mendelian randomization study design and workflow. GWAS: genome-wide association study; SNPs: single nucleotide polymorphisms; MR-PRESSO: MR pleiotropy residual sum and outlier; cML-MA: constrained maximum likelihood, and model averaging-based.
The specific steps for selecting the IVs are as follows: Firstly, single nucleotide polymorphisms (SNPs) closely related to the gut microbiota were selected as IVs. Because the number of samples included in the MiBiogen consortium study is limited, we selected SNPs with p-values smaller than the genome-wide significance threshold (1 × 10− 5) as potential IVs17. Secondly, utilising the European-based 1000 Genome Projects reference panel, we employed thresholds of r2 < 0.001 and clumping distance = 10,000 kb to mitigate the impact of linkage disequilibrium effects among SNPs17, which can lead to biased results in Mendelian randomization analysis. Thirdly, we used the R package ‘TwoSampleMR’ to harmonise the data sources, ensuring that the exposure-related SNPs and the SNPs extracted from the outcomes have consistent directions of alleles and removing palindromic SNPs as they cannot be aligned based on the allele frequency. Fourthly, before each MR analysis, the MR pleiotropy residual sum and outlier (MR-PRESSO) tests were used to detect potential horizontal pleiotropy and eliminate outliers23,24. Finally, instrumental variables with F-statistic less than 10 were considered weak instruments and were excluded. We calculated the F-statistic using the following formula: F = beta2/se225,26. The remaining SNPs were used for further MR analysis.
Mendelian randomization analyses
In this two-sample Mendelian randomization analysis, four analytical approaches were employed to assess the causal relationship between gut microbiota and adrenal function: IVW, MR-Egger, weighted median, constrained maximum likelihood, and model averaging-based (cML-MA)17,27. The IVW method combines two or more independent SNPs to minimise the overall variance, with the weight of each independent SNP being inversely proportional to its variance. The estimated value can be interpreted as the weighted slope of the IVs in the weighted linear regression of the exposure factor, with the intercept assumed to be zero. When the selected SNPs are valid IVs, IVW can provide accurate estimates. cML-MA is a novel MR analysis approach that achieves higher statistical power and better control of type I error rates than other MR methods. The MR-Egger regression is based on the assumption that instrument strength is independent of the direct effect (InSIDE), allowing the assessment of the presence of pleiotropy using the intercept term. MR-Egger allows for the presence of pleiotropy in all genetic variations but assumes that pleiotropy is independent of the variant-exposure association28. When certain invalid IVs are present, the weighted median method can still be used to accurately predict the causal relationships29. If the assumption of InSIDE is violated, the weighted median assumption has less bias and lower type-I error rates than the MR-Egger regression, resulting in more accurate predictions of causal relationships29.
According to relevant reports, IVW is superior to other detection methods under certain conditions; therefore, we used IVW as the main method30. cML-MA, which is increasingly recognised and used by researchers, also served as the main MR method in this study to complement and validate the IVW method. Although MR-Egger and weighted medians have lower efficiency (wider CIs)23, they provide reliable predictions of causal relationships across a broader range, thus serving as supplementary checks.
To further evaluate the stability of our initial MR analysis results, we conducted several sensitivity analyses. A funnel plot was used to assess the presence of directional pleiotropy, similar to the approach used in the mate analysis to evaluate publication bias. The MR-Egger intercept test was employed to evaluate horizontal pleiotropy, and a PMR−Egger intercept value less than 0.05 considered the presence of horizontal pleiotropy. Cochran’s Q test was used to detect heterogeneity among the SNPs included in this study. Furthermore, a leave-one-out analysis was performed to identify potentially heterogeneous SNPs among the IVs included in the study and to assess whether our analysis results were driven by individual SNPs31. We also conducted a check using PhenoScanner (www.phenoscanner.medschl.cam.ac.uk), a comprehensive information platform that integrates the associations between genotypes and phenotypes. This was performed to determine whether the SNPs ultimately included in our key MR analysis were associated with potential risk factors, such as inflammatory bowel disease (IBD), ovarian and testicular diseases affecting sex hormone secretion, pituitary-related diseases affecting adrenal cortex function, and depression. We removed SNPs related to these risk factors at the genome-wide significance level to ensure the accuracy and reliability of the final MR analysis.
Statistics
All statistical analyses were performed using the R software (version 4.2.1). Mendelian randomization analyses were conducted using the TwoSampleMR (version 0.5.7) and MRcML (version 0.0.0.9) R packages27. Considering the multiple hypothesis testing conducted, we employed the Bonferroni correction method to establish multiple significance thresholds, defined as 0.05/n, where n represents the number of valid gut microbiota genera included in this study22. In other words, the significance threshold for the p-values was set at 0.0004 (119 genera). Additionally, it should be noted that IVW-derived p-values < 0.05 are still considered indicative of potential causal relationships17,23,32.
Results
Based on the GWAS-correlated p-value threshold (1 × 10− 5) and linkage disequilibrium threshold (R2 < 0.001, clumping window size = 10,000 kb), we initially selected 1531 SNPs as IVs for 119 gut bacterial genera. Details regarding the selected SNPs are provided in Additional file 2: Table S2. The SNPs ultimately used for the MR analysis were all considered strong instruments, as evidenced by their F-statistic values exceeding 10 (Additional file 2: Table S2), indicating the absence of weak instrument bias.
We explored the possible causal relationship between the gut microbiota and adrenal function using a two-sample Mendelian randomization system. Based on the anatomy and functional characteristics of the adrenal glands, we divided them into the adrenal cortex and adrenal medulla regions. Based on the different biological functions in different regions, the adrenal cortex can be further divided into the zona glomerulosa, zona fasciculata, and zona reticularis. We used summary data from GWASs of different adrenal-related hormones to index adrenal function, ensuring a more comprehensive exploration of the causal relationship between gut microbiota and adrenal function. The IVW analysis was used as the primary analytical approach, and cML-MA as a primary supplementary method. The MR results showed that there are potential causal relationships between 23 genera of gut microbiota and adrenal function (Fig. 2, Additional file 2: Tabla S3).
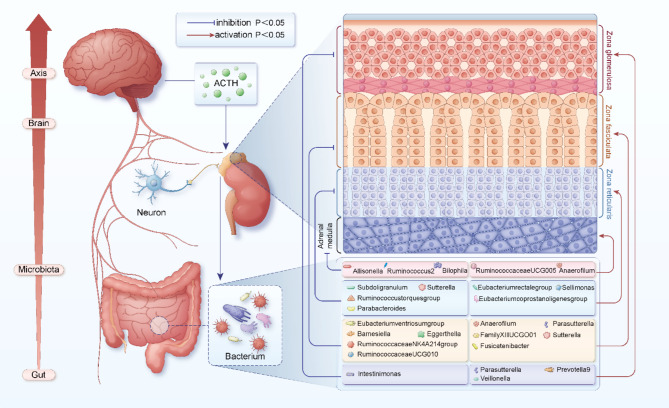
Fig. 2.
The potential relationship among gut microbiota, adrenal function, and the gut-microbiota-brain axis. p-values are derived from the IVW method. Lines in red indicate where the gut microbiota promotes the function of corresponding regions of the adrenal gland (p<0.05); lines in blue indicate where the gut microbiota inhibits the function of corresponding regions of the adrenal gland (p<0.05). ACTH: Adrenocorticotropic Hormone.
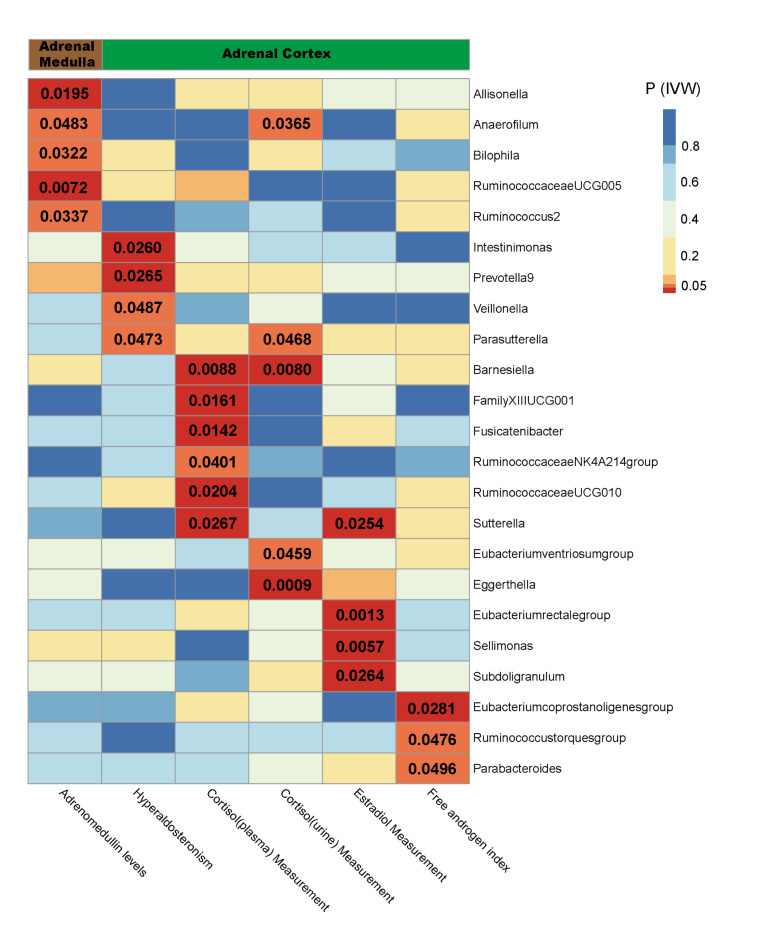
As shown in Table 1, there were potential causal relationships between the five intestinal microbiota and function in the medullary region of the adrenal glands. These include Allisonella (beta= -0.077, 95% CI: -0.141 to -0.012, P = 0.0195), Anaerofilum (beta = 0.070, 95% CI: 0.001 to 0.139, P = 0.0483), Bilophila (beta= -0.115, 95% CI: -0.220 to -0.010, P = 0.0322), Ruminococcus2 (beta= -0.108, 95% CI: -0.208 to -0.008, P = 0.0337) and RuminococcaceaeUCG005 (beta = 0.154, 95% CI: 0.042 to 0.266, P = 0.0072). Among them, Allisonella, Ruminococcus2 and Bilophila inhibited the level of adrenomedullin hormone, whereas Anaerofilum and RuminococcaceaeUCG005 promoted the level of adrenal medulla hormone (Additional file 3: Fig S1). In the zona glomerulosa region of the adrenal cortex, there are suggestive causal relationships between the four gut microbiota genera and adrenal function. These include Intestinimonas (odds ratio = 0.617, 95% CI: 0.403 to 0.944, P = 0.0260), Prevotella9 (odds ratio = 1.538, 95% CI: 1.052 to 2.251, P = 0.0265), Veillonella (odds ratio = 2.002, 95% CI: 1.004 to 3.992, P = 0.0487), and Parasutterella (odds ratio = 1.510, 95% CI: 1.005 to 2.268, P = 0.0473). Except for Intestinimonas, the other three intestinal bacterial genera promoted the function of the adrenal zona glomerulosa region (Additional file 3: Fig S2). In the zona fasciculata region of the adrenal cortex, several gut microbiota genera, including Barnesiella, FamilyXIIIUCG001, Fusicatenibacter, RuminococcaceaeNK4A214group, Sutterella, Anaerofilum, Parasutterella, RuminococcaceaeUCG010, Eubacteriumventriosumgroup, and Eggerthella, showed potential causal relationships with adrenal function (Table 1). Among them, Anaerofilum and Parasutterella are of particular interest because they show potential causal relationships with different regions of adrenal function in the comprehensive MR analysis (Fig. 3). Barnesiella was also of interest because it had a causal relationship with plasma cortisol (beta=-0.201, 95% CI: -0.352 to -0.051, P = 0.0088) and urinary cortisol (beta=-0.420, 95% CI: -0.730 to -0.110, P = 0.0080) levels. Among these, the genera FamilyXIIUCG001, Fusicatenibacter, Sutterella, Anaerofilum, and Parasutterella promoted the function of the adrenal zona fasciculata (Additional file 3: Fig S3).
Table 1.
The mendelian randomization estimates for genetically predicted adrenal function based on the gut microbiome.
| Exposure | Outcome | Number of SNPs | P IVW | P cML_MA | Beta/Or (95% CI) | P MR-PRESSO | P Cochran’s Q | P MR-Egger intercept |
|---|---|---|---|---|---|---|---|---|
| Allisonella | Adrenomedullin levels | 8 | 0.0195 | 0.0137 | -0.077 (-0.141 to -0.012) | 0.846 | 0.839 | 0.692 |
| Anaerofilum | 11 | 0.0483 | 0.1828 | 0.070 (0.001 to 0.139) | 0.654 | 0.629 | 0.889 | |
| Bilophila | 13 | 0.0322 | 0.0212 | -0.115 (-0.220 to -0.010) | 0.492 | 0.475 | 0.651 | |
| Ruminococcus2 | 15 | 0.0337 | 0.0333 | -0.108 (-0.208 to -0.008) | 0.970 | 0.960 | 0.901 | |
| RuminococcaceaeUCG005 | 14 | 0.0072 | 0.0037 | 0.154 (0.042 to 0.266) | 0.308 | 0.270 | 0.346 | |
| Intestinimonas | Hyperaldosteronism | 16 | 0.0260 | 0.0630 | 0.617 (0.403 to 0.944) | 0.926 | 0.914 | 0.615 |
| Prevotella9 | 15 | 0.0265 | 0.1424 | 1.538 (1.052 to 2.251) | 0.944 | 0.935 | 0.491 | |
| Veillonella | 5 | 0.0487 | 0.0098 | 2.002 (1.004 to 3.992) | 0.796 | 0.783 | 0.420 | |
| Parasutterella | 14 | 0.0473 | 0.0203 | 1.510 (1.005 to 2.268) | 0.708 | 0.771 | 0.509 | |
| Barnesiella | Cortisol (plasma) Measurement | 14 | 0.0088 | 0.0077 | -0.201 (-0.352 to -0.051) | 0.559 | 0.524 | 0.488 |
| FamilyXIIIUCG001 | 8 | 0.0161 | 0.0494 | 0.222 (0.041 to 0.402) | 0.570 | 0.533 | 0.736 | |
| Fusicatenibacter | 18 | 0.0142 | 0.0449 | 0.185 (0.037 to 0.333) | 0.567 | 0.532 | 0.969 | |
| RuminococcaceaeNK4A214group | 13 | 0.0401 | 0.0396 | -0.184 (-0.359 to -0.008) | 0.370 | 0.353 | 0.400 | |
| RuminococcaceaeUCG010 | 6 | 0.0204 | 0.0410 | -0.247 (-0.456 to -0.038) | 0.496 | 0.484 | 0.280 | |
| Sutterella | 12 | 0.0267 | 0.0338 | 0.181 (0.021 to 0.340) | 0.470 | 0.447 | 0.932 | |
| Eubacteriumventriosumgroup | Cortisol (urine) Measurement | 15 | 0.0459 | 0.0230 | -0.300 (-0.595 to -0.005) | 0.527 | 0.519 | 0.713 |
| Anaerofilum | 11 | 0.0365 | 0.1189 | 0.218 (0.014 to 0.423) | 0.454 | 0.430 | 0.700 | |
| Barnesiella | 14 | 0.0080 | 0.0069 | -0.420 (-0.730 to -0.110) | 0.350 | 0.353 | 0.073 | |
| Eggerthella | 11 | 0.0009 | 0.0012 | -0.356 (-0.566 to -0.145) | 0.745 | 0.713 | 0.179 | |
| Parasutterella | 15 | 0.0468 | 0.0809 | 0.251 (0.004 to 0.497) | 0.808 | 0.790 | 0.400 | |
| Eubacteriumrectalegroup | Estradiol measurement | 8 | 0.0013 | 0.0542 | 0.019 (0.008 to 0.031) | 0.450 | 0.380 | 0.338 |
| Sellimonas *** | 9 | 0.0057 | 0.0002 | 0.008 (0.002 to 0.013) | 0.283 | 0.234 | 0.431 | |
| Subdoligranulum | 11 | 0.0264 | 0.0056 | -0.011 (-0.020 to -0.001) | 0.534 | 0.521 | 0.987 | |
| Sutterella | 12 | 0.0254 | 0.0256 | -0.009 (-0.018 to -0.001) | 0.808 | 0.801 | 0.361 | |
| Eubacteriumcoprostanoligenesgroup | Free androgen index | 13 | 0.0281 | 0.0582 | 0.041 (0.004 to 0.077) | 0.671 | 0.661 | 0.952 |
| Ruminococcustorquesgroup | 9 | 0.0476 | 0.0016 | -0.041 (-0.082 to 0.000) | 0.582 | 0.584 | 0.768 | |
| Parabacteroides | 6 | 0.0496 | 0.0037 | -0.075 (-0.150 to 0.000) | 0.073 | 0.032 | 0.578 |
***indicates that the p-value remains significant after Bonferroni correction (p < 0.0004). IVW-derived p-values less than 0.05 are considered indicative of potential causal relationships. PIVW, p-value obtained from inverse variance-weighted (IVW) method; PcML_MA, the p-value obtained from constrained maximum likelihood and model averaging-based (cML-MA) method; PMR−PRESSO, the p-value from the MR pleiotropy residual sum and outlier (MR-PRESSO) test; PCochran’s Q, the p-value from Cochran’s Q test; and PMR−Egger intercept, p-value from the MR-Egger intercept test.
Fig. 3.
IVW estimates of the impact of gut microbiota on adrenal function. The heat map illustrates the complexity of the causal relationships between certain gut microbiota and the functions of different regions of the adrenal gland. The colour of each block represents the p-value derived from each MR analysis using the IVW method. Blue and yellow denote p-values greater than 0.05, whereas orange and red denote p-values less than 0.05. PIVW, the p-values obtained through the IVW method.
In the zona reticularis region of the adrenal cortex, there were causal relationships between several gut microbiota and estrogen levels. These include Sutterella (beta=-0.009, 95% CI: -0.018 to -0.001, P = 0.0254), Eubacteriumrectalegroup (beta = 0.019, 95% CI: 0.008 to 0.031, P = 0.0013), Sellimonas (beta = 0.008, 95% CI: 0.002 to 0.013, P = 0.0057), and Subdoligranulum (beta=-0.011, 95% CI: -0.020 to -0.001, P = 0.0264). In the MR analysis between gut microbiota and androgen levels, Eubacteriumcoprostanoligenesgroup (beta = 0.041, 95% CI: 0.004–0.077, P = 0.0281), Ruminococcustorquesgroup (beta=-0.041, 95% CI: -0.082–0.000, P = 0.0476), and Parabacteroides (beta=-0.075, 95% CI: -0.150–0.000, P = 0.0496) also showed suggestive causal relationships. Among them, Eubacteriumrectalegroup, Sellimonas, and Eubacteriumcoprostanoligenesgroup promoted the function of the adrenal zona reticularis, while the others had opposite effects (Additional file 3: Fig S4). Moreover, when conducting MR analysis using the cML-MA method, we found a persistent causal relationship between Sellimonas (beta = 0.009, 95% CI: 0.004–0.013, P = 0.0002) and the function of the adrenal zona reticularis, which remained significant even after Bonferroni correction. It should be noted that even though most of the significant p-values became non-significant after Bonferroni correction, the estimates with IVW-derived p values < 0.05 should also be treated cautiously23,32.
We conducted a sensitivity analysis to assess the stability and credibility of the MR analysis results. Among the 27 causal relationships identified, the MR-Egger intercept test did not indicate horizontal pleiotropy, as all MR-Egger intercept-derived p-values were greater than 0.05. Except for the causal relationship between Parabacteroides and the adrenal cortical zona reticularis, all p-values derived from Cochran’s Q were greater than 0.05, indicating no evidence of heterogeneity. Additionally, it is important to note that since we employed IVW as our primary analysis method, some degree of heterogeneity may be deemed acceptable23,33. We did not detect any outliers among the SNPs included in the MR analysis as all MR-PRESSO p-values were greater than 0.05. The results of the funnel plot and leave-one-out analyses are illustrated in Additional file 3: Fig S5-16, providing evidence that individual SNPs did not influence our MR analysis. Furthermore, these findings suggest that our assessments were credible and not violated.
Discussion
To the best of our knowledge, this is the first study that has implemented a large-scale MR analysis to systematically investigate the causal relationship between gut microbiota and adrenal function. In the present MR study, we utilised GWAS summary statistics from the MiBiogen consortium, IEU Open GWAS project, FinnGen consortium, and GWAS Catalog to explore the causal relationship between gut microbiota and the adrenal medulla as well as the adrenal cortex (zona glomerulosa, zona fasciculata, and zona reticularis). Our results demonstrate that Sellimonas promotes the function of the adrenal cortex zona reticularis and that there are suggestive causal relationships between 22 other gut microbiota genera and adrenal function. Our research findings not only reveal the causal relationship between gut microbiota and adrenal function in the gut-microbiota-brain axis but also contribute to the study of factors influencing adrenal function and adrenal-related diseases.
In recent years, research on the relationship between gut microbiota and human health has grown exponentially. Disruption of the gut microbiome balance has been linked to the development of various diseases, such as neurological disorders, autoimmune diseases, and cancers (including breast and prostate cancers)34–37. The gut-microbiota-brain axis serves as a critical pathway through which the gut microbiota exerts its influence. The adrenal gland plays a significant role in the normal functioning of the gut-microbiota-brain axis. However, the causal relationship between adrenal function and the gut microbiota remains unknown.
Previous studies using animal models have shown that adrenal-related hormone levels, which are indicative of adrenal function, can influence the composition and abundance of the gut microbiota. For instance, Martínez et al. discovered that in mice lacking adrenomedullin expression, there was an increased proportion of the Proteobacteria class and the Coriobacteriales order in their faeces, along with a decreased presence of beneficial human bacteria, such as Lactobacillus gasseri and Bifidobacterium choerinum38. Similarly, Wu et al. found that in mice orally administered dexamethasone sodium phosphate, there were significant changes in the gut microbiome composition, with reductions in the abundances of Actinobacteria, Bacteroidetes, Firmicutes, and α-proteobacteria39. These findings suggest that hormones secreted by the adrenal glands in animal models can regulate gut microbiota. However, whether gut microbiota can affect adrenal function remains unknown. Regarding the influence of the gut microbiota on adrenal function, some studies have shown that the gut microbiota can affect the levels of adrenal-related hormones. For example, researchers have found that a high prevalence of Prevotella can increase the plasma concentrations of ghrelin, which in turn can elevate dopamine levels40. This result has been corroborated in rodent models, where ghrelin injection was found to stimulate motor activity and increase dopamine levels41. Other studies have indicated that the gut microbiota, such as Clostridium, Enterococcus, and Bacteroides, can influence dopamine regulation40,42,43. However, due to the unclear mechanisms by which these gut microbes affect dopamine secretion and considering that dopamine can originate from both the nervous system and the adrenal medulla, it is challenging to determine whether these gut microbes directly affect adrenal function.
Therefore, to systematically and comprehensively explore the potential causal relationship between the gut microbiota and adrenal function, we conducted an extensive MR analysis. Accurate assessment of adrenal function is challenging. Therefore, we used the levels of adrenal-related hormones as proxies for the function of different regions of the adrenal gland, including the adrenal medulla and cortex (glomerulosa, fasciculata, and reticularis zones). Since the production of adrenal-related hormones is not confined to the adrenal glands but can also occur in other locations, such as the ovaries, testes, and nervous system, we conducted checks on the PhenoScanner platform to ensure that the SNPs included in our key MR analysis were not associated with these potential risk factors. Furthermore, we used the levels of various adrenal hormones to determine the overall function of the adrenal glands. This series of steps was performed to minimise bias and increase the reliability of the results.
In our MR study, we identified 27 potential causal relationships between 23 gut microbes and the adrenal function. Some of these gut microbes attracted our attention because of their significant and multiple causal relationships with different regions of the adrenal gland. For instance, results from the IVW analysis indicated that Sellimonas might enhance the function of the adrenal reticularis zone, leading to increased estrogen levels. Furthermore, the results from the cML-MA method supported a causal relationship between Sellimonas and adrenal reticularis zone function. These findings remained significant even after applying the Bonferroni correction. Owing to the susceptibility of estrogen levels in the human body to various confounding factors, such as ovarian influences, we conducted a preliminary query on the PhenoScanner platform before performing our MR analysis. This was to ensure that the SNPs included in our study were not associated with these confounding factors, thereby minimising the impact of variables such as ovarian influences on our estimates. Our findings are consistent with those of related studies on the role of Sellimonas in enhancing estrogen levels. Wei et al. discovered that a high abundance of Sellimonas was associated with the occurrence of ER-positive breast cancer37. Studies indicate that the growth of cancer cells in ER-positive breast cancer depends on estrogen, which plays a crucial role in the initiation and progression of the tumour44. In other words, the causal relationship between the high abundance of Sellimonas and the increased incidence of estrogen receptor-positive breast cancer cannot be separated from the role of estrogen, corroborating our findings. The specific mechanisms underlying the causal relationship between Sellimonas and adrenal function remain unclear. However, related studies have suggested that short-chain fatty acids produced by Sellimonas, especially butyrate, can regulate the expression of corticotropin-releasing hormone receptor 2 (CRHR2) by modulating histone acetylation at the CRHR2 promoter, thereby influencing adrenal function45,46.
Our MR results indicate a causal relationship between Parasutterella and the function of the adrenal zona glomerulosa and zona fasciculata, which can influence aldosterone and cortisol levels. Previous studies have linked changes in the abundance of Parasutterella to diseases such as inflammatory bowel disease (IBD), diabetes, and fatty liver disease47–49. To date, no studies have reported a relationship between Parasutterella and adrenal function. However, a study of the biological characteristics of Parasutterella found that it significantly regulates bile acid and cholesterol49. Cholesterol is the primary substrate in the synthesis of steroid hormones, which are crucial for the synthesis of adrenocortical hormones. Bile acids also facilitate the absorption of dietary cholesterol in the intestine, and cholesterol synthesis in the liver is closely linked to the enterohepatic circulation of bile acids50. The primary pathway for endogenous cholesterol metabolism is the conversion into bile acids51. Thus, the regulatory effect of Parasutterella on bile acid and cholesterol metabolism may be an important pathway through which it influences the levels of adrenocortical hormones (aldosterone and cortisol).
In previous studies, the presence of Parasutterella, Sellimonas, Sutterella, and Barnesiella has been found to be associated with the occurrence of IBD49,52–54. IBD is an idiopathic inflammatory condition that occurs in the gastrointestinal tract and primarily involves Crohn’s Disease (CD) and ulcerative colitis. IBD can lead to symptoms such as abdominal pain, diarrhoea, nutritional disorders, and disruptions in amino acid and electrolyte metabolism55. Particularly, Crohn’s disease often affects the lower part of the small intestine and the right side of the colon, interfering with the absorption of cholesterol and tyrosine from food, subsequently impacting the synthesis of adrenal-related hormones55. Furthermore, studies have shown that inflammatory diseases are related to dysregulation of the corticotropin-releasing hormone (CRH) system56. CRH stimulates the pituitary gland to secrete and release adrenocorticotropic hormones, thereby regulating adrenal function. These factors may be the potential mechanisms by which the aforementioned gut microbes induce changes in adrenal function. Our MR results also demonstrate a causal relationship between Barnesiella and adrenal function, primarily manifested in its ability to reduce cortisol levels in the human body by affecting the function of the adrenal cortex zona fasciculata. Through studies of human and mouse gut microbiota, Roy et al. found that only a few bacteria in the gut microbiome could influence cholesterol metabolism and plasma cholesterol levels57. Barnesiella is among these bacteria, suggesting a potential mechanism by which it affects the zona fasciculata function in the adrenal cortex57. In addition, our MR analysis results suggest a correlation between Sutterella and the function of the adrenal cortex zona fasciculata and zona reticularis. Studies have indicated a positive correlation between Sutterella, obesity, and insulin resistance in Chinese children58. Researchers hypothesised that this might be related to the regulation of aromatic amino acid synthesis (tyrosine, tryptophan, and phenylalanine)58. Tyrosine is a precursor of catecholamine hormone synthesis. Furthermore, cholesterol levels are typically higher in obese populations than in non-obese individuals. Thus, the causal relationship between Sutterella and the adrenal function may be mediated by tyrosine and cholesterol. However, our MR analysis did not show a relationship between Sutterella and adrenal medullary function, which may be due to the incomprehensive representation of adrenal medullary function by the selected adrenal-related hormones. Additionally, our MR results indicate a causal relationship between Anearofilum and both adrenal cortex and medulla functions. Researchers have reported that Anearofilum can affect the function of endocrine organs and the thyroid and is causally related to Graves’ disease59. Research on its relationship with adrenal function is relatively scarce, and its specific mechanisms require further investigation.
Our MR analysis results indicated that in addition to the previously mentioned gut microbes, 17 other gut microbes potentially have a causal relationship with adrenal function. These include RuminococcaceaeUCG005, Prevotella9, Eggerthella, RuminococcaceaeUCG010, and Subdoligranulum. Interestingly, we found that a significant proportion of the gut microbes identified in our MR analysis were associated with depression and other neurological disorders. For example, Sellimonas, Eggerthella, RuminococcaceaeUCG005, Parasutterella, Anearofilum, Eubacteriumventriosumgroup, and Prevotella9 are all related to depression45,60. Studies have shown that butyrate, serotonin, glutamate, and gamma-aminobutyric acid produced by gut microbes are key neurotransmitters involved in the onset of depression45. Severe depression can lead to elevated cortisol levels in the body and increased adrenal gland size and activity61,62. Therefore, these gut microbes could potentially modulate adrenal function by inducing neurological disorders, such as depression.
Consistent with our MR analysis, other studies have indicated that certain gut microbes control adrenal function by regulating the HPA axis. For example, Wu et al. discovered through animal experiments that Enterococcus faecalis can regulate social behaviour in mice by downregulating corticosterone production and suppressing HPA axis activity63. Desbonnet et al. and Eutamene et al. found that Lactobacillus and Bifidobacterium can also modulate HPA axis activity, thereby improving the symptoms of depression and anxiety64,65. Moreover, studies have shown that short-chain fatty acids (SCFAs) such as acetic acid, propionic acid, and butyric acid produced by bacteria during food digestion can reduce the gene activity of HPA axis proteins, thus modulating adrenal function66. There are also indications that SCFAs may activate the HPA axis by regulating intestinal barrier permeability and microbiome-driven inflammatory states, thereby influencing adrenal function67. Some of these gut microbial relationships with adrenal function were not evident in our MR analysis, which might be attributed to the limited number of GWASs on the adrenal gland and insufficient sample sizes in the existing research. Nevertheless, these findings support a causal relationship between gut microbes and adrenal function, enhancing the credibility of our research results.
To the best of our knowledge, this is a large-scale MR analysis to explore the causal relationship between gut microbiota and adrenal function. Our results affirm the existence of a causal relationship between the gut microbiota and adrenal function. Considering that adrenal dysfunction is a common clinical issue, and that adrenal function is crucial for the gut-microbiota-brain axis, this research holds significant implications for further studies on the gut-microbiota-brain axis and related clinical issues.
However, this study has some limitations. Firstly, the participants included in our MR analysis were predominantly European, which limited the generalisability of our findings. The causal relationship between the gut microbiota and adrenal function in other populations remains uncertain. Secondly, to comprehensively explore the impact of the 119 bacterial genera on adrenal function, the SNPs used in our MR analysis did not meet the traditional GWAS significance threshold (p < 5 × 10− 8). Thirdly, the taxonomic classification of bacteria was only at the genus level and not at the more detailed species or strain levels. The results would be more accurate and reliable if gut microbiome GWASs employ advanced shotgun metagenomic sequencing analysis. Fourthly, although some adrenal-related hormones may be associated with sex, we did not stratify our analysis by sex due to limitations in the available data. Fifthly, although we used adrenal-related hormones to reflect the overall adrenal function, we did not analyse all adrenal-related hormones. This is due to the limited number of GWAS on adrenal function, necessitating further research and development. In addition, despite efforts to minimise potential confounders, SNP-related studies may not completely avoid them. Therefore, we used the PhenoScanner platform to ensure that the SNPs included in our final MR study were not associated with known confounders, thereby reducing the bias. Finally, owing to multiple-hypothesis testing, there is an increased risk of Type I errors. Therefore, we employed the Bonferroni correction method to adjust the p-values. However, the stringent nature of the Bonferroni correction might lead to false negatives. To address this, we utilised a novel MR approach, cML-MA, which efficiently controls the risk of Type I errors while maintaining statistical power. This approach serves as the primary supplementary method.
Conclusion
In summary, this is a comprehensive MR analysis to reveal the causal relationship between the gut microbiota and adrenal function. This deepens our understanding of the gut-microbiota-brain axis and provides a direction for research into the relationship between gut microbiota and adrenal function. Furthermore, we hope that in clinical settings, healthcare professionals will become more aware of the influence of gut microbiota on adrenal function, especially during the perioperative management of adrenal-related hormone levels or in the long-term maintenance of oral medication post-surgery for patients with adrenal-related diseases. This could lead to the identification of additional predictors of adrenal function and potentially beneficial microbial communities, which represents the most significant clinical value of our study.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to express our gratitude to the MiBioGen consortium, FinnGen consortium, IEU Open GWAS project, and the GWAS Catalog for providing the GWAS data.
Abbreviations
- CRHR2
Corticotropin-releasing hormone receptor 2
- CRH
Corticotropin-releasing hormone
- cML-MA
Constrained maximum likelihood, and model averaging-based
- GWAS
Genome-wide association study
- HPA
Hypothalamic-pituitary-adrenal
- IBD
Inflammatory bowel disease
- CD
Crohn’s disease
- IVW
Inverse variance weighting
- InSIDE
Instrument strength independent of the direct effect
- IVs
Instrumental variables
- MR-PRESSO
MR pleiotropy residual sum and outlier
- MR
Mendelian randomization
- SNPs
Single nucleotide polymorphisms
- SCFAs
Short-chain fatty acids
Author contributions
Conceptualization: T.L. and H.J.; Investigation and resources: T.L., Z.L., and Y.L.; Methodology and data analysis: T.L. and C.Z.; Writing—original draft: T.L., C.Z.; Critical revision: Z.Y., Y.G., and D.L.; Funding acquisition: Z.Y. All authors have reviewed, revised, and approved the final manuscript.
Funding
This study was funded by the Young and Middle-Aged Discipline Leaders of the Henan Health Commission (2020-17), the Young Elite Scientists Sponsorship Program by the Henan Association for Science and Technology (HAST; 2022HYTP043), and the Young Backbone Teachers Program of Zhengzhou University (2023ZDGGJS083).
Data availability
The GWAS summary data utilized in this article are all sourced from public databases such as the MiBiogen Consortium, IEU Open GWAS project, FinnGen consortium R9 release data, and GWAS Catalog, all of which are publicly accessible. Specific access links can be found in the ‘Methods’ section of the article.
Declarations
Competing interests
The authors declare no competing interests.
Ethics approval
This study used publicly available participant data. Therefore, no separate ethical approval was required for this study.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributing authors: Tonghu Liu and Hongfei Ji contributed equally and fully to this study.
Contributor Information
Dongxiao Li, Email: li_dongxiao@sina.com.
Yukui Gao, Email: gaoyukui@wnmc.edu.cn.
Zechen Yan, Email: yanzechen@foxmail.com.
References
- 1.Else, T. et al. Adrenocortical carcinoma. Endocr. Rev.35, 282–326. 10.1210/er.2013-1029 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Noelting, S. et al. Personalized Management of Pheochromocytoma and Paraganglioma. Endocr. Rev.43, 199–239. 10.1210/endrev/bnab019 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mete, O. et al. Overview of the 2022 WHO classification of adrenal cortical tumors. Endocr. Pathol.33, 155–196. 10.1007/s12022-022-09710-8 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lenders, J. W. M. et al. Pheochromocytoma and paraganglioma: An endocrine society clinical practice guideline. J. Clin. Endocrinol. Metab.99, 1915–1942. 10.1210/jc.2014-1498 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Vanbrabant, T., Fassnacht, M., Assie, G. & Dekkers, O. M. Influence of hormonal functional status on survival in adrenocortical carcinoma: Systematic review and meta-analysis. Eur. J. Endocrinol.179, 429–436. 10.1530/eje-18-0450 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Berruti, A. et al. Prognostic role of overt hypercortisolism in completely operated patients with adrenocortical cancer. Eur. Urol.65, 832–838. 10.1016/j.eururo.2013.11.006 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Fassnacht, M. et al. European society of endocrinology clinical practice guidelines on the management of adrenocortical carcinoma in adults, in collaboration with the European Network for the Study of Adrenal Tumors. Eur. J. Endocrinol.179, G1–G46. 10.1530/eje-18-0608 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Fassnacht, M. et al. Adrenocortical carcinomas and malignant phaeochromocytomas: ESMO-EURACAN clinical practice guidelines for diagnosis, treatment and follow-up. Ann. Oncol.31, 1476–1490. 10.1016/j.annonc.2020.08.2099 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Asadi, A. et al. Obesity and gut-microbiota-brain axis: A narrative review. J. Clin. Lab. Anal.36, e24420. 10.1002/jcla.24420 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moreira, C. G. et al. Bacterial adrenergic sensors regulate virulence of enteric pathogens in the gut. mBio7, e00826-16. 10.1128/mBio.00826-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark, A. & Mach, N. Exercise-induced stress behavior, gut-microbiota-brain axis and diet: A systematic review for athletes. J. Int. Soc. Sports Nutr.13, 43. 10.1186/s12970-016-0155-6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quénéhervé, L. et al. Digestive symptoms in daily life of chronic adrenal insufficiency patients are similar to irritable bowel syndrome symptoms. Sci. Rep.11, 8077. 10.1038/s41598-021-87158-2 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hahner, S. et al. High incidence of adrenal crisis in educated patients with chronic adrenal insufficiency: A prospective study. J. Clin. Endocrinol. Metab.100, 407–416. 10.1210/jc.2014-3191 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Rushworth, R. L., Torpy, D. J. & Falhammar, H. Adrenal crisis. N. Engl. J. Med.381, 852–861. 10.1056/NEJMra1807486 (2019). [DOI] [PubMed] [Google Scholar]
- 15.Davey Smith, G. & Hemani, G. Mendelian randomization: Genetic anchors for causal inference in epidemiological studies. Hum. Mol. Genet.23, R89-98. 10.1093/hmg/ddu328 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sekula, P., Del Greco, M. F., Pattaro, C. & Köttgen, A. Mendelian randomization as an approach to assess causality using observational data. J. Am. Soc. Nephrol.27, 3253–3265. 10.1681/asn.2016010098 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, P. et al. Association between gut microbiota and preeclampsia-eclampsia: A two-sample Mendelian randomization study. BMC Med.20, 443. 10.1186/s12916-022-02657-x (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang, J. et al. Meta-analysis of human genome-microbiome association studies: The MiBioGen consortium initiative. Microbiome6, 101. 10.1186/s40168-018-0479-3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurilshikov, A. et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat. Genet.53, 156–165. 10.1038/s41588-020-00763-1 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li, Z., Chen, Y. & Ke, H. Investigating the causal relationship between gut microbiota and Crohn’s disease: A mendelian randomization study. Gastroenterology166, 354–355. 10.1053/j.gastro.2023.08.047 (2023). [DOI] [PubMed] [Google Scholar]
- 21.Inamo, J. Non-causal association of gut microbiome on the risk of rheumatoid arthritis: A Mendelian randomisation study. Ann. Rheum. Dis.80, e103. 10.1136/annrheumdis-2019-216565 (2021). [DOI] [PubMed] [Google Scholar]
- 22.Long, Y., Tang, L., Zhou, Y., Zhao, S. & Zhu, H. Causal relationship between gut microbiota and cancers: A two-sample Mendelian randomisation study. BMC Med.21, 66. 10.1186/s12916-023-02761-6 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen, X. et al. Kidney damage causally affects the brain cortical structure: A Mendelian randomization study. eBioMedicine72, 103592. 10.1016/j.ebiom.2021.103592 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verbanck, M., Chen, C. Y., Neale, B. & Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet.50, 693–698. 10.1038/s41588-018-0099-7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu, K. et al. Causal relationship between gut microbiota and gastrointestinal diseases: A mendelian randomization study. J. Transl. Med.22, 92. 10.1186/s12967-024-04894-5 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rao, S. et al. A positive causal effect of shrimp allergy on major depressive disorder mediated by allergy- and immune-related pathways in the East Asian population. Nutrients16, 79. 10.3390/nu16010079 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xue, H., Shen, X. & Pan, W. Constrained maximum likelihood-based Mendelian randomization robust to both correlated and uncorrelated pleiotropic effects. Am. J. Hum. Genet.108, 1251–1269. 10.1016/j.ajhg.2021.05.014 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li, P. et al. Association between gut microbiota and preeclampsia-eclampsia: A two-sample Mendelian randomization study. BMC Med.20, 443. 10.1186/s12916-022-02657-x (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bowden, J., Davey Smith, G. & Burgess, S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int. J. Epidemiol.44, 512–525. 10.1093/ije/dyv080 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hartwig, F. P., Davey Smith, G. & Bowden, J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int. J. Epidemiol.46, 1985–1998. 10.1093/ije/dyx102 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bowden, J., Davey Smith, G., Haycock, P. C. & Burgess, S. Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet. Epidemiol.40, 304–314. 10.1002/gepi.21965 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hemani, G., Tilling, K. & Davey Smith, G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet.13, e1007081. 10.1371/journal.pgen.1007081 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xia, X. et al. The causality between gut microbiome and anorexia nervosa: A Mendelian randomization analysis. Front. Microbiol.14, 1290246. 10.3389/fmicb.2023.1290246 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burgess, S. et al. Guidelines for performing Mendelian randomization investigations. Wellcome Open Res.4, 186–186. 10.12688/wellcomeopenres.15555.1 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeng, C., Zhang, C., He, C. & Song, H. Investigating the causal impact of gut microbiota on glioblastoma: A bidirectional Mendelian randomization study. BMC Genom.24, 784. 10.1186/s12864-023-09885-2 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu, Q. et al. Causal relationship between gut microbiota and autoimmune diseases: A two-sample mendelian randomization study. Front. Immunol.12, 746998. 10.3389/fimmu.2021.746998 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hong, W. et al. Gut microbiome causal impacts on the prognosis of breast cancer: A Mendelian randomization study. BMC Genom.24, 497. 10.1186/s12864-023-09608-7 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei, Z. et al. Gut microbiota and risk of five common cancers: A univariable and multivariable Mendelian randomization study. Cancer Med.12, 10393–10405. 10.1002/cam4.5772 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martínez-Herrero, S. et al. Lack of adrenomedullin results in microbiota changes and aggravates azoxymethane and dextran sulfate sodium-induced colitis in mice. Front. Physiol.7, 595. 10.3389/fphys.2016.00595 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu, T. et al. Chronic glucocorticoid treatment induced circadian clock disorder leads to lipid metabolism and gut microbiota alterations in rats. Life Sci.192, 173–182. 10.1016/j.lfs.2017.11.049 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Hamamah, S., Aghazarian, A., Nazaryan, A., Hajnal, A. & Covasa, M. Role of microbiota-gut-brain axis in regulating dopaminergic signaling. Biomedicines10, 436. 10.3390/biomedicines10020436 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cornejo, M. P. et al. Ghrelin recruits specific subsets of dopamine and GABA neurons of different ventral tegmental area sub-nuclei. Neuroscience392, 107–120. 10.1016/j.neuroscience.2018.09.027 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Dürre, P. Physiology and sporulation in clostridium. Microbiol. Spectr.2, Tbs-0010-2012. 10.1128/microbiolspec.TBS-0010-2012 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Hartstra, A. V. et al. Infusion of donor feces affects the gut-brain axis in humans with metabolic syndrome. Mol. Metab.42, 101076. 10.1016/j.molmet.2020.101076 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kwa, M., Plottel, C. S., Blaser, M. J. & Adams, S. The intestinal microbiome and estrogen receptor-positive female breast cancer. J. Natl. Cancer Inst.108, djw029. 10.1093/jnci/djw029 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Radjabzadeh, D. et al. Gut microbiome-wide association study of depressive symptoms. Nat. Commun.13, 7128. 10.1038/s41467-022-34502-3 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang, X. et al. Sodium butyrate facilitates CRHR2 expression to alleviate HPA axis hyperactivity in autism-like rats induced by prenatal lipopolysaccharides through histone deacetylase inhibition. mSystems8, e0041523. 10.1128/msystems.00415-23 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shin, N. R., Whon, T. W. & Bae, J. W. Proteobacteria: Microbial signature of dysbiosis in gut microbiota. Trends Biotechnol.33, 496–503. 10.1016/j.tibtech.2015.06.011 (2015). [DOI] [PubMed] [Google Scholar]
- 49.Blasco-Baque, V. et al. Associations between hepatic miRNA expression, liver triacylglycerols and gut microbiota during metabolic adaptation to high-fat diet in mice. Diabetologia60, 690–700. 10.1007/s00125-017-4209-3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ju, T., Kong, J. Y., Stothard, P. & Willing, B. P. Defining the role of Parasutterella, a previously uncharacterized member of the core gut microbiota. ISME J.13, 1520–1534. 10.1038/s41396-019-0364-5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.di Gregorio, M. C., Cautela, J. & Galantini, L. Physiology and physical chemistry of bile acids. Int. J. Mol. Sci.22, 1780. 10.3390/ijms22041780 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, Y., Yutuc, E. & Griffiths, W. J. Cholesterol metabolism pathways - Are the intermediates more important than the products?. FEBS J.288, 3727–3745. 10.1111/febs.15727 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yuan, X. et al. Depression and anxiety in patients with active ulcerative colitis: crosstalk of gut microbiota, metabolomics and proteomics. Gut Microbes13, 1987779. 10.1080/19490976.2021.1987779 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hiippala, K., Kainulainen, V., Kalliomäki, M., Arkkila, P. & Satokari, R. Mucosal prevalence and interactions with the epithelium indicate commensalism of Sutterella spp.. Front. Microbiol.7, 1706. 10.3389/fmicb.2016.01706 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Arnone, D. et al. Long-term overconsumption of fat and sugar causes a partially reversible pre-inflammatory bowel disease state. Front. Nutr.8, 758518. 10.3389/fnut.2021.758518 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rosen, M. J., Dhawan, A. & Saeed, S. A. Inflammatory bowel disease in children and adolescents. JAMA Pediatr.169, 1053–1060. 10.1001/jamapediatrics.2015.1982 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sukhareva, E. V. The role of the corticotropin-releasing hormone and its receptors in the regulation of stress response. Vavilovskii Zhurnal Genet Selektsii25, 216–223. 10.18699/vj21.025 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Le Roy, T. et al. The intestinal microbiota regulates host cholesterol homeostasis. BMC Biol.17, 94. 10.1186/s12915-019-0715-8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Squillario, M. et al. Gut-microbiota in children and adolescents with obesity: Inferred functional analysis and machine-learning algorithms to classify microorganisms. Sci. Rep.13, 11294. 10.1038/s41598-023-36533-2 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cao, J. et al. A cause-effect relationship between Graves’ disease and the gut microbiome contributes to the thyroid-gut axis: A bidirectional two-sample Mendelian randomization study. Front. Immunol.14, 977587. 10.3389/fimmu.2023.977587 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barandouzi, Z. A., Starkweather, A. R., Henderson, W. A., Gyamfi, A. & Cong, X. S. Altered composition of gut microbiota in depression: A systematic review. Front. Psychiatry11, 541. 10.3389/fpsyt.2020.00541 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zunszain, P. A., Anacker, C., Cattaneo, A., Carvalho, L. A. & Pariante, C. M. Glucocorticoids, cytokines and brain abnormalities in depression. Prog. Neuropsychopharmacol. Biol. Psychiatry35, 722–729. 10.1016/j.pnpbp.2010.04.011 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Misiak, B. et al. The HPA axis dysregulation in severe mental illness: Can we shift the blame to gut microbiota?. Prog. Neuropsychopharmacol. Biol. Psychiatry102, 109951. 10.1016/j.pnpbp.2020.109951 (2020). [DOI] [PubMed] [Google Scholar]
- 64.Wu, W. L. et al. Microbiota regulate social behaviour via stress response neurons in the brain. Nature595, 409–414. 10.1038/s41586-021-03669-y (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Desbonnet, L. et al. Effects of the probiotic Bifidobacterium infantis in the maternal separation model of depression. Neuroscience170, 1179–1188. 10.1016/j.neuroscience.2010.08.005 (2010). [DOI] [PubMed] [Google Scholar]
- 66.Eutamene, H. et al. Synergy between Lactobacillus paracasei and its bacterial products to counteract stress-induced gut permeability and sensitivity increase in rats. J. Nutr.137, 1901–1907. 10.1093/jn/137.8.1901 (2007). [DOI] [PubMed] [Google Scholar]
- 67.van de Wouw, M. et al. Short-chain fatty acids: Microbial metabolites that alleviate stress-induced brain-gut axis alterations. J. Physiol.596, 4923–4944. 10.1113/jp276431 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yousefi, B. et al. Gastrointestinal tract, microbiota and multiple sclerosis (MS) and the link between gut microbiota and CNS. Curr. Microbiol.80, 38. 10.1007/s00284-022-03150-7 (2022). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The GWAS summary data utilized in this article are all sourced from public databases such as the MiBiogen Consortium, IEU Open GWAS project, FinnGen consortium R9 release data, and GWAS Catalog, all of which are publicly accessible. Specific access links can be found in the ‘Methods’ section of the article.