Abstract
Cervical cancer (CCa) patients with lymph node (LN) metastasis face poor prognoses and have limited treatment options. Aberrant N6-methyladenosine (m6A) modification of RNAs are known to promote tumor metastasis, but their role in CCa remains unclear. Our study reveals that HNRNPC, an alternative splicing (AS) factor and m6A reader, increases tumor-related variants through m6A-dependent manner, thereby promoting lymphatic metastasis in CCa. We found that HNRNPC overexpression correlates with lymphatic metastasis and poorer prognoses in CCa patients. Functionally, knocking down HNRNPC markedly inhibited the migration and invasion of several CCa cell lines, while supplementing HNRNPC restored the malignant phenotypes of these cells. Mechanistically, HNRNPC regulates exon skipping of FOXM1 by binding to its m6A-modified motif. Mutating the m6A site on FOXM1 weakened the interaction between HNRNPC and FOXM1 pre-RNA, leading to a reduction in the metastasis-related FOXM1-S variant. In conclusion, our findings demonstrate that m6A-dependent alternative splicing mediated by HNRNPC is essential for lymphatic metastasis in CCa, potentially providing novel clinical markers and therapeutic strategies for patients with advanced CCa.
Subject terms: Metastasis, Gynaecological cancer
Introduction
Cervical cancer (CCa) is the fourth most common cancer among women worldwide and the leading cause of death from gynecologic malignancies [1]. Patients with early-stage CCa have a favorable 5-year survival rate exceeding 90% with comprehensive treatment, including surgery and adjuvant therapy [2]. However, the occurrence of lymph node metastasis (LNM) significantly increases recurrence and mortality rates [3], making it a major cause of treatment failure and patient death [4]. Nevertheless, the underlying mechanism of lymphatic metastasis in CCa remains unclear. N6-methyladenosine (m6A) is the most prevalent RNA modification and plays a significant role in post-transcriptional regulation [5]. It has been found to promote tumor metastasis in various cancers [6]. Heterogeneous nuclear ribonucleoprotein C (HNRNPC), a recently identified m6A reader [7], is crucial in regulating precursor RNA (pre-RNA), particularly in the alternative splicing (AS) of pre-mRNA [8]. Accumulating studies have shown that high HNRNPC expression is associated with poor prognosis in several tumors [9–11]. However, its role in gynecological oncology has not been well explored. Our investigation demonstrates that HNRNPC is greatly upregulated in lymph node-positive CCa tissues and is associated with inferior overall survival (OS) and progression-free survival (PFS). Functionally, HNRNPC invigorates lymphatic metastasis of CCa both in vivo and in vitro. Mechanistically, HNRNPC binds to Forkhead box protein M1 (FOXM1) pre-mRNA in an m6A-dependent manner, driving the AS of FOXM1, leading to the high expression of the pro-metastatic variant FOXM1-S. Our study reveals a significant and novel mechanism where HNRNPC, as an AS factor and m6A reader, facilitates lymphatic metastasis in CCa, offering new insights into potential therapeutic targets.
Materials and methods
Human tissue samples
We recruited ten CCa patients with tumors smaller than 2 cm, diagnosed at Sun Yat-sen Memorial Hospital (Table S1). Tumor and adjacent normal tissues were collected and stored in RNA later at −80 °C. Additionally, sixty-one CCa patients diagnosed with FIGO (The International Federation of Gynecology and Obstetrics) IB1-IIIC1 and seventeen patients with benign uterus or ovarian diseases who underwent total hysterectomy were enrolled (Table S1). CCa tumor tissues and normal cervix samples were collected and preserved in RNA later at −80 °C. The formula “” (α = 0.05, P = 48%, δ = 0.1) was used to estimate the sample size of patients for immunohistochemistry (IHC) staining. Ninety-two CCa patients and ninety-six CCa patients, diagnosed via histology and who underwent radical hysterectomy plus pelvic lymphadenectomy at two independent gynecology oncology centers (the First Affiliated Hospital and Sun Yat-sen Memorial Hospital of Sun Yat-sen University), were included for IHC staining. Inclusion criteria included patients with preoperative biopsy pathology confirmed as squamous, adenocarcinoma, or adenosquamous carcinoma, proposed for surgical treatment, free of syphilis, human immunodeficiency virus, and other immune-related diseases, and no history of oral immunosuppressive drugs. The informed consent was obtained, and the ethical approval was granted by the Committees for Ethical Review of Research Involving Human Subjects of Sun Yat-sen University (SYSU-2021-580).
Immunohistochemical (IHC) analysis
Tissue sections were stained with an anti-HNRNPC antibody (1:200, #11760-1, Proteintech) and Horseradish Peroxidase (HRP)-goat-anti-rabbit antibody (Zhong Shan-Golden Bridge Biological Technology). Staining intensity (negative, 0; mild, 1; moderate, 2; severe, 3) and the proportion of positive staining cells (negative, 0%; ≤25%, 1; >25% and ≤50%, 2; >50% and ≤75%, 3; >75% and ≤100%, 4) were multiplied to calculate staining scores.
TCGA and GEO data mining
The clinical profiles and gene expression data of 306 CCa patients were obtained from https://portal.gdc.cancer.gov/. Data from CCa patients in GSE7803, GSE6791, and GSE63514 were sourced from https://www.ncbi.nlm.nih.gov/geo/.
RNA isolation, RT-PCR, and Western Blot
Total RNA and protein were extracted following the protocol of the RNA isolation kit (Esunbio, China) and Protein isolation kit (BestBio, China). Reverse transcription and Polymerase Chain Reaction (PCR) were performed with SYBR qPCR Master Mix kits (Vazyme, China). Primers used in the experiments are listed in Table S2. Western Blot was performed as described in our previous studies.
Cell cultures
Human cervical squamous cancer cell lines Siha, MS751, Caski, and adenocarcinoma cell lines Hela, Hela229 were purchased from the cell bank of Shanghai. Cells were cultured in DMEM or 1640 containing 10% fetal bovine serum (Gibco, America) at 37 °C and were tested for mycoplasma contamination before and during experiments.
RNAi and lentivirus transduction
SiRNA oligonucleotides targeting HNRNPC, Wilms tumor 1 associated protein (WTAP), and negative control siRNA were purchased from GenePharma (Suzhou, China) and are listed in Table S3. SiRNA transfections were conducted according to the manufacturer’s instructions of RNAiMAX (Invitrogen, America). HNRNPC expression was knocked down by stable transduction with pcDNA3.1-puro lentivirus following our previous protocol. FOXM1-L and FOXM1-S variant plasmids, along with FOXM1-m6A-wild-type, and mutation plasmids were packaged into lentivirus to establish stable expression cell lines. Puromycin was used at 2 µg/ml to select stable expression cells.
Cell metastasis assays and proliferative assays
Transwell, wound healing, and proliferative assays were performed as described in our previous studies.
Footpad implantation and popliteal-lymph-node metastasis assay
Four-week-old female BALB/c nude mice were procured from the Experimental Animal Center of Guangdong Province and were randomly allocated to control and experimental groups. The formula “” (α = 0.05, P = 20%, δ = 0.3) was used to estimate the sample size. A total of 1 × 107 cells were suspended in 50 µL of Hanks’ solution and inoculated into the footpads of the mice to build the LNM model. Blinding was used when labeling cell suspension and during inoculation. Fluorescence signals were monitored using an In Vivo Imaging System (IVIS) (PerkinElmer, USA) after 8 weeks. Popliteal lymph nodes were the regional draining lymph nodes and the first site of metastasis from footpad tumor, they were excised and fixed in 4% formalin. Immunohistochemical staining with Pan-CK and hematoxylin and eosin (HE) staining were performed. All animal experimental procedures were approved by the Institutional Animal Care and Use Committee of Sun Yat-sen University (SYSU-IACUC-2021-B1906).
RNA immunoprecipitation (RIP) assay and RIP-qPCR analysis
For the RIP assay and subsequent qPCR analysis, 2 × 107 Hela and Siha cells were lysed using RIP lysis buffer. The RIP assays were performed with the EZ-Magna RIP kit (Millipore, USA). A 5-µg amount of anti-HNRNPC antibody (#11760-1-AP, Proteintech) was used for the RIP incubation.
m6A mutation assay
Hela and Siha cells were plated in six-well plates and transfected with either wild-type or m6A mutant (A-to-C mutation) FOXM1 vectors.
Statistical analysis
Statistical analyses were conducted using SPSS 18.0 software. The F test was applied to estimate homogeneity of variance (P > 0.05 was considered to be homogeneous). The student’s t-test was applied to compare expression differences, and the Chi-Square test assessed the association of clinicopathological features. Kaplan–Meier survival curves and log-rank tests were employed to compare OS and PFS. A significance level of P < 0.05 was considered statistically significant.
Results
HNRNPC correlates with LN metastasis and is an unfavorable prognostic factor in CCa
Analysis of the TCGA database revealed that most m6A-related genes were differentially expressed in CCa compared to normal cervix tissues (Fig. S1A). Among these, HNRNPC expression was higher in CCa tissues than in normal cervix tissues (Fig. 1A, B). Subsequent data mining from GEO and Oncomine databases confirmed the overexpression of HNRNPC in CCa tissues (Figs. S1B, and 1C–E). PCR validation from our cohort also showed higher HNRNPC expression in tumor samples compared to adjacent normal samples (Fig. 1F). Additionally, HNRNPC expression correlated with lymph node (LN) metastatic CCa and was significantly associated with poor prognosis in CCa patients (Fig. 1G, H, Table S4). HNRNPC was overexpressed in LN-positive CCa tissues and metastatic CCa cell lines compared to LN-negative tissues and non-metastatic cell lines (Fig. S1C, D). These expression patterns suggest that HNRNPC may play a potentially pro-LN-metastasis role in CCa.
Fig. 1. HNRNPC overexpression in cervical cancer: clinical and prognostic implications.
A TCGA data reveals elevated HNRNPC expression in tumors across various cancer types compared to normal tissues. B Significantly increased HNRNPC expression observed in TCGA cervical cancers. C–E GEO database analysis confirms high HNRNPC expression in CCa tissues compared to normal tissues (GSE7803, GSE6791, GSE63514). F PCR analysis shows significant upregulation of HNRNPC in CCa tumors compared to adjacent tissues in a local cohort (n = 10). G Local cohort PCR data indicates higher HNRNPC levels in lymphatic metastasis CCa compared to patients without lymph node metastasis and normal cervix. H TCGA data analysis links high HNRNPC RNA expressions to shorter overall survival (n = 292, P value = 0.048). I HNRNPC expression patterns in cervical adenocarcinoma and squamous cell carcinoma from two independent gynecology departments. J Kaplan–Meier curves demonstrate high HNRNPC expression as an unfavorable prognostic factor for overall survival in both local cohorts (cohort1: n = 91, cohort2: n = 96) with P values of 0.007 and 0.008, respectively. *P < 0.05, **P < 0.01, ***P < 0.001. LN lymph node, TCGA The Cancer Genome Atlas, AC adenocarcinoma, SC squamous cell carcinoma.
To further validate HNRNPC’s role in CCa, we conducted IHC staining on paraffin sections of CCa patients from two independent gynecology oncology surgery centers. HNRNPC expression was consistently lower in adjacent normal tissues than in tumors (Fig. S1E). Patients with high HNRNPC expression had higher LN metastasis rates (34.04% and 66.67%) compared to those with low HNRNPC expression (12.25 and 40%) in two cohorts (Tables 1, 2, Fig. S1F). Kaplan–Meier(K-M) curves indicated that HNRNPC expression was an unfavorable risk factor for OS and PFS in CCa patients (Figs. S1G, H, and 1I, J). These findings indicated the potential value of HNRNPC in LN metastasis of CCa.
Table 1.
Clinical-pathologic information of patients in cohort 1.
| Clinical-pathology information | HNRNPC expressions | χ2 value | P value | ||
|---|---|---|---|---|---|
| Low | High | ||||
| Age | <45 | 17 | 14 | 45.125 | 0.049* |
| ≥45 | 32 | 33 | |||
| Pathology | Squamous cell carcinoma | 43 | 42 | 0.081 | 0.960 |
| Adenocarcinoma | 6 | 5 | |||
| Grade | High | 3 | 0 | 4.286 | 0.117 |
| Medium | 12 | 14 | |||
| Low | 34 | 33 | |||
| Stage | I | 40 | 40 | 0.209 | 0.428 |
| II | 9 | 7 | |||
| Lymph node status | Negative | 43 | 31 | 6.629 | 0.01* |
| Positive | 6 | 16 | |||
| LVSI | Negative | 30 | 28 | 0.979 | 0.613 |
| Positive | 8 | 11 | |||
| Myometrial invasion | <1/2 | 15 | 15 | 0.023 | 0.989 |
| ≥1/2 | 34 | 30 | |||
| Parametrial infiltration | Negative | 48 | 42 | 4.537 | 0.103 |
| Positive | 1 | 5 | |||
| Tumor size | ≤4 cm | 45 | 43 | 22.269 | 0.674 |
| >4 cm | 4 | 3 | |||
| Lymph cell infiltration | Negative | 25 | 22 | 0.17 | 0.418 |
| Positive | 24 | 25 | |||
| Vaginal stump involvement | Negative | 48 | 45 | 0.395 | 0.484 |
| Positive | 1 | 2 | |||
LVSI lympho-vascular space involvement.
*P < 0.05.
Table 2.
Clinical-pathologic information of patients in cohort 2.
| Clinical-pathology information | HNRNPC expressions | χ2 value | P value | ||
|---|---|---|---|---|---|
| Low | High | ||||
| Age | <45 | 9 | 10 | 33.093 | 0.159 |
| ≥45 | 26 | 47 | |||
| BMI | ≤24 | 18 | 18 | 107.317 | 0.027* |
| >24 | 17 | 39 | |||
| Pathology | Squamous cell carcinoma | 29 | 49 | 162 | 0.452 |
| Adenocarcinoma | 6 | 8 | |||
| Grade | High | 1 | 4 | 3.741 | 0.189 |
| Medium | 10 | 23 | |||
| Low | 24 | 30 | |||
| Stage | I | 12 | 20 | 3.741 | 0.129 |
| II | 2 | 11 | |||
| III | 21 | 26 | |||
| Lymph node status | Negative | 21 | 26 | 16.438 | 0.021* |
| Positive | 14 | 31 | |||
| LVSI | Negative | 11 | 19 | 0.036 | 0.518 |
| Positive | 24 | 38 | |||
| Endometrial involvement | Negative | 24 | 26 | 4.606 | 0.026* |
| Positive | 11 | 31 | |||
| Myometrial invasion | <1/3 | 5 | 2 | 3.722 | 0.155 |
| ≥1/3 & <2/3 | 5 | 7 | |||
| ≥2/3 | 25 | 48 | |||
| Parametrial infiltration | Negative | 34 | 53 | 0.73 | 0.366 |
| Positive | 1 | 4 | |||
| Tumor size | ≤4 cm | 19 | 38 | 25.458 | 0.264 |
| >4 cm | 16 | 19 | |||
| Lymph cell infiltration | Negative | 17 | 20 | 1.632 | 0.144 |
| Positive | 18 | 37 | |||
| Vaginal stump involvement | Negative | 33 | 55 | 0.247 | 0.492 |
| Positive | 2 | 2 | |||
BMI body mass index, LVSI lympho-vascular space involvement.
*P < 0.05.
HNRNPC enhanced tumor invasion and proliferation in vitro and promoted CCa lymph node metastasis in vivo
Prompted by the above findings, we examined the functions of HNRNPC in CCa cell lines by silencing its expression in Hela229 and MS751 cells (Figs. 2A, and S5). Knockdown of HNRNPC reduced invasion and migration abilities, as shown by Transwell and Wound Healing assays. Additionally, HNRNPC knockdown significantly inhibited the proliferation of CCa cell lines, as detected by clone formation, CCK-8, and EdU assays (Fig. 2B–J).
Fig. 2. Impact of HNRNPC Downregulation on CCa Cell Proliferation, Invasion, and Migration In Vitro.
A Western blot confirms successful downregulation of HNRNPC expression in H229 and MS751 cells using siRNAs; siRNA sequence-1 exhibits higher efficacy. B CCK8 assay demonstrates reduced proliferative capacity of H229 and MS751 cells upon HNRNPC downregulation (OD value = (ODexperiment-ODblank)/((OD0h-ODblank), n = 4)). C, D Clone formation assay reveals decreased cell proliferation following HNRNPC downregulation (n = 3). E, G EdU assay illustrates diminished proliferation of H229 and MS751 cells after HNRNPC silencing (n = 3). F, H Transwell assay shows decreased invasiveness of CCa cells upon HNRNPC downregulation (n = 3). I, J Wound healing assay under serum-free culture conditions indicates reduced migration ability of H229 and MS751 cells 48 h after HNRNPC knocking down. *P < 0.05, **P < 0.01, n.s: P > 0.05. Si1 siRNA sequence-1, si2 siRNA sequence-2.
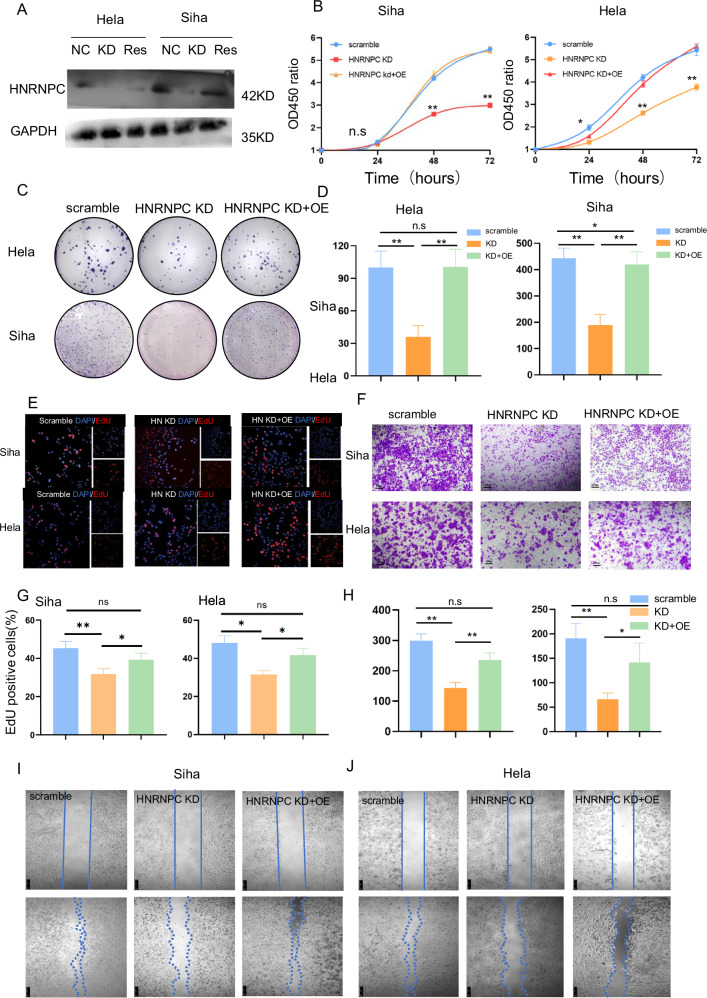
Due to the high basal expression of HNRNPC in CCa cell lines, we were unable to upregulate HNRNPC expression. In order to confirm the function of HNRNPC, we performed rescue assays in Hela and Siha cell lines (Figs. 3A, and S5). The invasion and proliferation abilities decreased in Hela and Siha after HNRNPC knockdown and were restored upon HNRNPC overexpression (Fig. 3B–J).
Fig. 3. HNRNPC drives proliferation, invasion, and migration of CCa cells in vitro.
A Western blot confirms successful rescue of HNRNPC expression in HNRNPC-silenced Hela and Siha cells through HNRNPC cDNA. B CCK8 assay reveals increased proliferation of Hela and Siha cells induced by HNRNPC (OD value = (ODexperiment-ODblank)/((OD0h-ODblank), n = 4)). C, D Clone formation assay demonstrates enhanced cell proliferation upon HNRNPC rescue (n = 3). E, G EdU assay shows increased proliferation of Hela and Siha cells after HNRNPC rescue (n = 3). F, H Transwell assay indicates elevated invasiveness of CCa cells following HNRNPC rescue (n = 3). I, J Wound healing assay under serum-free culture conditions shows heightened migration ability of Hela and Siha cells with HNRNPC rescue. *P < 0.05, **P < 0.01, n.s:P > 0.05. NC negative control, KD knock down of HNRNPC, RES rescue of HNRNPC, OE over-expression of HNRNPC.
To further determine the pro-metastasis function of HNRNPC in vivo, we created stable HNRNPC-knockdown cell lines. A CCa LNM model was constructed by transplanting HNRNPC-knockdown and negative control Hela/Siha cell lines into the footpads of nude mice (Fig. 4A–C). Two months later, we found that downregulation of HNRNPC decreased metastasis in regional draining lymph nodes (Fig. 4D), as confirmed by IHC using Pan-CK antibody (Figs. 4E–L, and S2). Taken together, these results demonstrate that HNRNPC promotes the migration and invasion of CCa cell lines both in vitro and in vivo.
Fig. 4. HNRNPC facilitates CCa metastasis to regional lymph nodes in vivo through the footpad-popliteal lymphatic metastasis model.
A Diagrammatic illustration of footpad injection. B Anatomical atlas displaying footpad tumor and popliteal lymphatic metastasis (green arrow denotes the primary tumor on the footpad of Balb/c nude mice, black arrow indicates metastatic regional lymph nodes on the popliteal). C Fluorescence imaging of footpad tumor and popliteal lymphatic metastasis (white arrow signifies the primary tumor on the footpad, yellow arrow indicates metastatic regional lymph nodes on the popliteal). D Histogram demonstrating reduced in vivo lymph node metastasis rates in HNRNPC knock-down cells (n = 5). E, F Immunohistochemistry (IHC) of pan-CK staining in popliteal lymph nodes of mice injected with Hela scramble cells. G, H Hematoxylin and eosin (HE) staining of popliteal lymph nodes in mice injected with Hela scramble cells. I, J IHC of pan-CK staining in popliteal lymph nodes of mice injected with HNRNPC knocked-down Hela cells. K, L HE staining of popliteal lymph nodes in mice injected with HNRNPC knocked-down Hela cells. NC negative control, KD knock down of HNRNPC, LN negative lymph node, LN+ positive lymph node, HE hematoxylin-eosin staining.
HNRNPC mediated AS of FOXM1 facilitated LN metastasis in CCa
HNRNPC, an AS factor, functions exclusively in the nucleus, as confirmed by IHC staining of tumor tissues, IF staining of CCa cells, and nuclear-cytoplasmic separation experiments (Fig. S3A–C), To understand how HNRNPC promotes LN metastasis through AS regulation, RNA-seq and rMATS analysis were performed in Hela and Siha cells following HNRNPC knockdown (Fig. 5A).
Fig. 5. HNRNPC mediates alternative splicing of multiple molecules, including the key transcriptional factor FOXM1.
A Volcano plots representing differentially expressed genes after HNRNPC silencing in Hela and Siha cells. B GSEA enrichment plots for Hela and Siha cells. C Histograms depicting differentially expressed alternative splicing events in Hela and Siha cells. D Volcano plots illustrating exon skipping events. E Venn diagram depicting shared alternative splicing events in Hela and Siha cells. F Schematic representation of skipped exons (SE) in FOXM1, showing consistent exon retention after HNRNPC knockdown in both cell lines. G PCR validation confirms retention of the FOXM1-L variant after HNRNPC silencing, which is reduced upon HNRNPC overexpression. *P < 0.05, **P < 0.01. KD knock down of HNRNPC, OE over expression of HNRNPC, TF transcription factor, DEG differently expressed gene, SE skipped exon, AS alternative splicing, RI retained intron, MTE mutually exclusive exons, A5SS alternative 5’ splice site, A3SS alternative 3’ splice site.
Kyoto Encyclopedia of Genes and Genomes (KEGG) functional annotation of RNA-seq data indicated significant enrichment of genes in pathways such as steroid biosynthesis, transcriptional misregulation in cancer, and FoxO pathways (Fig. S4A, B). Gene Set Enrichment Analysis (GSEA) analysis indicated that cell adhesion-related-pathways were affected by HNRNPC knockdown (Fig. 5B). Skipped exon (SE) events were the most common AS events in both cell lines (Fig. 5C). HNRNPC acted as either an AS activator or a prohibitor, causing exon inclusion or skipping when HNRNPC was downregulated (Fig. 5D). To uncover how HNRNPC regulates those DEG expressions, we focused on HNRNPC-mediated AS events in transcription factors (TFs) vital to tumor metastasis. After mapping the AS sites and transcriptional levels of TFs in Hela and Siha cells, we found AS of FOXM1 and RPE were regulated in both cell lines while the total RNA levels of these two molecules were stable after HNRNPC silencing (Fig. 5E). Since FOXM1 had a profound impact on tumor progression and development, hence, we chose it for further investigation. AS analysis revealed increased VIIa exon retention of FOXM1 (FOXM1-L), with stable total RNA levels after HNRNPC depletion, validated by qPCR (Figs. 5F-G, and S4C). Consistently, restoration of HNRNPC rescued the VIIa exon skipping of FOXM1 (FOXM1-S).
We detected the expressions of FOXM1-S and FOXM1-L by q-PCR to determine their roles in CCa (Fig. 6A, B). FOXM1-S increased in LN-metastasis CCa tissues but decreased in non-LN-metastasis tissues, while FOXM1-L showed the opposite pattern. FOXM1-S was positively correlated with HNRNPC expression (Figs. 6C, and S3D). Taken together, these results suggest HNRNPC-regulated FOXM1 variants are involved in LN metastasis of CCa. To substantiate the impact of FOXM1 isoforms and delineate the interaction between these two isoforms and HNRNPC in LN metastasis in CCa, we established FOXM1-L and FOXM1-S overexpressing lentivirus and infected them into stable HNRNPC-knockdown cell lines. Transwell assays showed that invasiveness increased with FOXM1-S overexpression, but not with FOXM1-L (Fig. 6D). Western Blot (WB) assay demonstrated that FOXM1-S overexpression rescued the expression of Epithelial-Mesenchymal Transition (EMT) pathway molecules and matrix metalloproteinase (MMP) proteins in HNRNPC-knockdown cell lines (Fig. 6F, and S5). These findings show that FOXM1-S can rescue the pro-metastasis ability of HNRNPC-deficient CCa cells by modulating invasion-associated genes.
Fig. 6. HNRNPC-Mediated Increase in FOXM1-S Variant Plays a Crucial Role in CCa Metastasis.
A RNA levels of FOXM1-L and -S variants in cervical cancer tissues. B RNA levels of FOXM1-L and -S variants in paired samples of cervical cancer and adjacent tissues. C Regression analysis and fitting curve depicting the relationship between HNRNPC and FOXM1-S expressions in cervical cancer samples. D, E Transwell assay results indicating that the FOXM1-S variant rescues cell invasion ability, unlike the FOXM1-L variant. F Western blot results demonstrating elevated MMP proteins and EMT pathway molecules after complementing FOXM1-S rather than FOXM1-L. G RIP-PCR comparing different areas of wild-type and m6A sites mutated FOXM1. *P < 0.05, **P < 0.01, n.s P > 0.05. RIP RNA binding protein immunoprecipitation, wt wild type, mt m6A sites mutation, WTAP KD knocking down WTAP.
HNRNPC promoted the AS of FOXM1 by m6A modification recognition
HNRNPC, an RNA binding protein, recognizes m6A modifications on pre-RNAs. To understand how HNRNPC modulates FOXM1 AS, we first investigated its binding with FOXM1 RNA. Mapping with known HNRNPC binding motifs revealed two potential binding on FOXM1 pre-RNA, one on the VIII exon and another near the VIIa exon. By prediction from websites, we found a cluster of m6A sites on the VIII exon, which was close to the HNRNPC binding motif. RIP-PCR assay demonstrated HNRNPC binding to the VIII exon region of FOXM1 more strongly than to IgG (Fig. 6G). To test the role of m6A modification in HNRNPC-mediated FOXM1 splicing, we examined HNRNPC binding to FOXM1 after m6A site mutation or WTAP si-RNA downregulation in Hela cells. The interaction between HNRNPC and FOXM1 was significantly suppressed after m6A site mutation or WTAP downregulation (Fig. 6G). These results further confirm that HNRNPC promotes LNM through mediating FOXM1 isoform switch in an m6A-dependent way(Fig. 7).
Fig. 7. Hypothesized mechanism diagram: HNRNPC-mediated m6A-dependent binding to FOXM1 promotes lymph node metastasis in CCa through alternative splicing.
In cervical cancer cells, when the VIII exon of FOXM1 undergoes m6A modification, the nuclear-located splicing factor HNRNPC recognizes and binds to the FOXM1 pre-RNA. This interaction promotes the skipping of the VIIa exon of FOXM1, leading to an elevation in the FOXM1-S variant. FOXM1-S variants are then translated into transcriptionally active FOXM1 proteins, promoting the expression of MMP and EMT-related genes, ultimately enhancing lymph node metastasis.
Discussion
CCa is a prevalent and lethal malignancy worldwide [1], with LNM being a significant prognostic factor [5]. In this study, HNRNPC was found to be overexpressed in CCa tissues, especially in LN-metastasis specimens. HNRNPC’s expression level was the independent risk factor of poorer PFS, and OS. Additionally, HNRNPC promoted the invasiveness of CCa cells through in vivo and in vitro assays. Mechanistic studies uncovered the dual role of HNRNPC, acting as both a splicer and an m6A reader. Briefly, HNRNPC recognizes and binds to the polymeric m6A sites on the VIII exon of FOXM1 pre-RNA, modulating the AS of FOXM1 and enhancing the expression of FOXM1-S isoform, thereby promoting LNM of CCa.
HNRNPC is a member of the family of binding proteins for small heterogeneous nuclear RNAs, which perform biological functions in variable splicing complexes with other proteins [8]. In prostate cancer, it promotes the proliferation and migration of tumor cells and correlates with higher tumor stage, Gleason score, and poor prognosis [11]. Bioinformatics analysis of sequencing data from TCGA of glioma [10], oral squamous carcinoma [9], esophageal squamous cell carcinoma [12] shows that the expression of HNRNPC is closely related to patient prognosis, aligning with our findings in CCa. Our study found a discrepancy in LNM rates among patients with high HNRNPC expression in two independent gynecologic oncology centers. This discrepancy may be due to bias in patient sources between the centers. However, the similar findings suggest that HNRNPC has significant predictive value for LNM in CCa.
Existing studies often focus on clinicopathological parameter correlation and prognostic analysis, but further mechanistic studies on phenotypic effects of HNRNPC on tumor cells and its contribution to tumor cell metastasis and poorer prognosis have yet to be reported. A footpad implantation and popliteal lymph nodes metastasis model was used for in vivo experiment. This model aligns with the sequential metastasis pattern observed in CCa. In our future study, a pelvic LNM model will be considered. The findings in this paper suggest that high HNRNPC expression is closely associated with LNM in CCa, which may guide treatment choices. For example, CCa patients with LNM prefer radical radiotherapy over surgery [2], but preoperative pelvic Computed Tomography (CT) has limited ability to discriminate LNM. Therefore, the expression level of HNRNPC in CCa might serve as a potential biomarker for LNM and guide patient treatment. Since the chi-square test used in this study can only confirm a significant correlation between HNRNPC expression and LNM, it is a univariate test and cannot exclude the influence of other factors on this correlation. Therefore, the predictive value of HNRNPC for LNM in CCa needs further validation with larger samples.
HNRNPC has been reported as a classical alternative splicer [8]. In this study, we used sequencing to find that HNRNPC promotes exon skipping of FOXM1 in CCa cell lines, leading to an increase in FOXM1-S production and promoting LNM. Under physiological conditions, FOXM1 RNA is highly folded in three-dimensional structure, hiding HNRNPC binding sites. However, when m6A modification occurs in these regions, RNA accessibility increases, exposing the HNRNPC binding sites. This structural change of RNA due to m6A modification is known as the ‘m6A switch’ [13]. In pancreatic cancer [14], HNRNPC recognizes and binds to the m6A-modified sites on the VII exon in TATA-binding protein-associated factor 8 (TAF8) pre-RNA, promoting exon skipping of TAF8 and liver metastasis. This conclusion is supported by our data that the VIII exon of FOXM1 precursor RNA has a segment of clustered m6A modules, and reducing m6A modification in this region significantly reduces HNRNPC binding to FOXM1, hindering its AS function.
Exon skipping is the most common form of AS. In non-small cell lung cancer, 14th exon skipping in the mesenchymal-epithelial transition factor (MET) gene leads to lower ubiquitination degradation of the MET protein, resulting in sustained activation of the downstream pathway and brain metastasis [15]. In bladder cancer, the splicing factor NONO promotes exon retention of SET domain and mariner transposase fusion gene (SETMAR), increasing the formation of the short transcript SETMAR-S, and inhibiting LNM by increasing the methylation modification of the promoter of EMT-related genes [16].
FOXM1, known as an oncogene [17], has been reported as a transcriptional factor that regulates multiple pathways related to tumor metastasis and epithelial-to-mesenchymal transition (EMT) [17–19]. The 6th and 9th exons of human FOXM1 are alternative exons, resulting in at least four transcripts generated by AS [20]. In our study, FOXM1-L transcript contains the VIIa exon of FOXM1 DNA, and the protein translated from FOXM1-L may lose transcription function due to the fragment insertion [21], while the FOXM1-S transcript does not encompass the VIIa exon [22]. Currently, there are no studies on FOXM1 and LNM, or on the functional differences of FOXM1 isoforms in CCa. In this study, we found that FOXM-L had no obvious biological function in CCa, while FOXM1-S enhanced the metastatic ability of CCa cells by promoting the expression of the EMT pathway and MMPs. Overexpression of FOXM1-S enhanced the invasive ability of HNRNPC-deficient cells. The above results suggest that HNRNPC promotes exon skipping of FOXM1 precursor RNA through the m6A switch, leading to increased expression of FOXM1-S transcripts and promotes LNM of CCa.
Conclusion
Our study indicated that HNRNPC is a potential and promising biomarker for LNM and prognosis in CCa patients. Mechanistically, HNRNPC mediates metastasis and proliferation by regulating AS through the “m6A switch” of FOXM1 pre-RNA. Based on these findings, inhibiting AS of FOXM1 may become a notable treatment decision of CCa LNM.
Supplementary information
Author contributions
All authors have contributed to the manuscript. YL conducted data curation, validation and wrote the original draft. MX performed visualization and formal analysis. ZC provided methodology and validation. YL, CZ, and LY developed methodology. YP and HH did visualization of the data. HL made conceptualization. SY made significant effort in conceptualization, he also reviewed and edit the final version of this manuscript. YL, ZC, and SY played a vital role in acquiring fundings.
Funding information
This work was supported by the National Natural Science Foundation of China (No. 81900428 to Zhi-bo Chen, No. 82273365 to Shu-zhong Yao), the Science and Technology Plan of Guangdong Province (2023A0505050102 to Shu-zhong Yao), Sun Yat-sen University Clinical Research Foundation of 5010 Project (2017006 to Shuzhong Yao), the Medical Scientific Research Foundation of Guangdong Province of China (A2022073 to Yun-yun Liu), Shanwei Science and Technology Plan Project (No. 230523156346866 to Yun-yun Liu).
Data availability
The original data and informations are available to readers promptly upon request to the corresponding author.
Competing interests
The authors declare no competing interests.
Ethical approval and consent to participate
All methods were performed in accordance with the relevant guidelines and regulations. All animal experimental procedures were approved by the Institutional Animal Care and Use Committee of Sun Yat-sen University (SYSU-IACUC-2021-B1906). All experiments involved human tissues were approved by Medical Ethics Committee of Sun Yat-sen University (SYSEC-KY-KS-2021-062), and informed consent was obtained from all participants.
Footnotes
Edited by Gennaro Ciliberto
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Yun-Yun Liu, Meng Xia, Zhi-Bo Chen.
Contributor Information
Huai-Wu Lu, Email: luhuaiwu@mail.sysu.edu.cn.
Shu-Zhong Yao, Email: yaoshuzh@mail.sysu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41419-024-07108-4.
References
- 1.Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73:17–48. [DOI] [PubMed] [Google Scholar]
- 2.Abu-Rustum NR, Yashar CM, Arend R, Barber E, Bradley K, Brooks R, et al. NCCN Guidelines(R) insights: cervical cancer, version 1.2024. J Natl Compr Cancer Netw. 2023;21:1224–33. [DOI] [PubMed] [Google Scholar]
- 3.Matsuo K, Machida H, Mandelbaum RS, Konishi I, Mikami M. Validation of the 2018 FIGO cervical cancer staging system. Gynecol Oncol. 2019;152:87–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown M, Assen FP, Leithner A, Abe J, Schachner H, Asfour G, et al. Lymph node blood vessels provide exit routes for metastatic tumor cell dissemination in mice. Science. 2018;359:1408–11. [DOI] [PubMed] [Google Scholar]
- 5.Zhong S, Guo Q, Chen X, Luo X, Long Y, Chong T, et al. The inhibition of YTHDF3/m(6)A/LRP6 reprograms fatty acid metabolism and suppresses lymph node metastasis in cervical cancer. Int J Biol Sci. 2024;20:916–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang H, Weng H, Chen J. m(6)A modification in coding and non-coding RNAs: roles and therapeutic implications in cancer. Cancer Cell. 2020;37:270–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xing S, Wang J, Wu R, Hefti MM, Crary JF, Lu Y. Identification of HnRNPC as a novel Tau exon 10 splicing factor using RNA antisense purification mass spectrometry. RNA Biol. 2022;19:104–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fischl H, Neve J, Wang Z, Patel R, Louey A, Tian B, et al. hnRNPC regulates cancer-specific alternative cleavage and polyadenylation profiles. Nucleic Acids Res. 2019;47:7580–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang GZ, Wu QQ, Zheng ZN, Shao TR, Chen YC, Zeng WS, et al. M6A-related bioinformatics analysis reveals that HNRNPC facilitates progression of OSCC via EMT. Aging. 2020;12:11667–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang LC, Chen SH, Shen XL, Li DC, Liu HY, Ji YL, et al. M6A RNA methylation regulator HNRNPC contributes to tumorigenesis and predicts prognosis in glioblastoma multiforme. Front Oncol. 2020;10:536875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang S, Xu G, Chao F, Zhang C, Han D, Chen G. HNRNPC promotes proliferation, metastasis and predicts prognosis in prostate cancer. Cancer Manag Res. 2021;13:7263–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Chen W, Pan T, Wang H, Zhang Y, Li C. LBX2-AS1 is activated by ZEB1 and promotes the development of esophageal squamous cell carcinoma by interacting with HNRNPC to enhance the stability of ZEB1 and ZEB2 mRNAs. Biochem Biophys Res Commun. 2019;511:566–72. [DOI] [PubMed] [Google Scholar]
- 13.Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang XT, Li JH, Zhu XX, Huang CS, Gao ZX, Xu QC, et al. HNRNPC impedes m(6)A-dependent anti-metastatic alternative splicing events in pancreatic ductal adenocarcinoma. Cancer Lett. 2021;518:196–206. [DOI] [PubMed] [Google Scholar]
- 15.Sun R, Wang Z, Zhao J, Ren P, Ma J, Guo Y. Optimized detection of unknown MET Exon 14 skipping mutations in routine testing for patients with non-small-cell lung cancer. JCO Precis Oncol. 2023;7:e2200482. [DOI] [PubMed] [Google Scholar]
- 16.Xie R, Chen X, Cheng L, Huang M, Zhou Q, Zhang J, et al. NONO inhibits lymphatic metastasis of bladder cancer via alternative splicing of SETMAR. Mol Ther. 2021;29:291–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liao GB, Li XZ, Zeng S, Liu C, Yang SM, Yang L, et al. Regulation of the master regulator FOXM1 in cancer. Cell Commun Signal. 2018;16:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Balli D, Ustiyan V, Zhang Y, Wang IC, Masino AJ, Ren X, et al. Foxm1 transcription factor is required for lung fibrosis and epithelial-to-mesenchymal transition. EMBO J. 2013;32:231–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dai B, Kang SH, Gong W, Liu M, Aldape KD, Sawaya R, et al. Aberrant FoxM1B expression increases matrix metalloproteinase-2 transcription and enhances the invasion of glioma cells. Oncogene. 2007;26:6212–9. [DOI] [PubMed] [Google Scholar]
- 20.Laoukili J, Stahl M, Medema RH. FoxM1: at the crossroads of ageing and cancer. Biochim Biophys Acta. 2007;1775:92–102. [DOI] [PubMed] [Google Scholar]
- 21.Jia R, Che X, Jia J, Guo J. FOXM1a isoform of oncogene FOXM1 is a tumor suppressor suppressed by hnRNP C in oral squamous cell carcinoma. Biomolecules. 2023;13:1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klinhom-On N, Seubwai W, Sawanyawisuth K, Lert-Itthiporn W, Waraasawapati S, Detarya M, et al. FOXM1c is the predominant FOXM1 isoform expressed in cholangiocarcinoma that associated with metastatic potential and poor prognosis of patients. Heliyon. 2021;7:e06846. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original data and informations are available to readers promptly upon request to the corresponding author.