Abstract
Single-amino-acid mutations in Sindbis virus proteins can convert clinically silent encephalitis into uniformly lethal disease. However, little is known about the host gene response during avirulent and virulent central nervous system (CNS) infections. To identify candidate host genes that modulate alphavirus neurovirulence, we utilized GeneChip Expression analysis to compare CNS gene expression in mice infected with two strains of Sindbis virus that differ by one amino acid in the E2 envelope glycoprotein. Infection with Sindbis virus, dsTE12H (E2-55 HIS), resulted in 100% mortality in 10-day-old mice, whereas no disease was observed in mice infected with dsTE12Q (E2-55 GLN). dsTE12H, compared with dsTE12Q, replicated to higher titers in mouse brain and induced more CNS apoptosis. Infection with the neurovirulent dsTE12H strain was associated with both a greater number of host genes with increased expression and greater changes in levels of host gene expression than was infection with the nonvirulent dsTE12Q strain. In particular, dsTE12H infection resulted in greater increases in the levels of mRNAs encoding chemokines, proteins involved in antigen presentation and protein degradation, complement proteins, interferon-regulated proteins, and mitochondrial proteins. At least some of these increases may be beneficial for the host, as evidenced by the demonstration that enforced expression of the antiapoptotic mitochondrial protein peripheral benzodiazepine receptor (PBR) protects neonatal mice against lethal Sindbis virus infection. Thus, our findings identify specific host genes that may play a role in the host protective or pathologic response to neurovirulent Sindbis virus infection.
The host response to viral infection represents a complex orchestration of divergent pathways designed to eradicate the virus and benefit the host. However, many pathways that are involved in antiviral defense can also have untoward effects on the host, including cytotoxic T-lymphocyte responses, cytokine responses, and apoptosis, resulting in either dysfunction or death of infected or neighboring uninfected cells. The potential for host pathology due to vigorous antiviral responses is of particular importance for tissues containing vital nonrenewable cell populations, such as the central nervous system (CNS) (reviewed in reference 50). Therefore, an important challenge in understanding the pathogenesis of CNS viral infections is the elucidation of specific host responses that play protective and/or pathologic roles.
With the use of high-density DNA microarrays, it is possible to define changes in gene expression that underlie the host response to viral pathogens and to gain specific insights into the molecular nature of the host pathways that govern viral pathogenesis. To date, however, the use of DNA microarray technology to measure host gene expression during viral infections has been restricted primarily to cells grown in culture. For example, previous studies have described alterations of cellular mRNAs in human foreskin fibroblasts infected with human cytomegalovirus (64), in CD4-positive T cells infected with human immunodeficiency virus type 1 (18), and in human embryonic lung cells infected with herpes simplex virus type 1 (48). However, little is known about the molecular profile of the host transcriptional response to viral infections in the intact host or about the precise physiologic role that alterations in host gene expression may play in viral pathogenesis.
Sindbis virus, a message-sense RNA virus in the alphavirus genus, provides an excellent model system for studying the complex interrelationships between host gene expression and viral pathogenesis. Both host factors and specific viral genetic determinants have been identified that play a critical role in regulating the ability of Sindbis virus to cause neurologic disease in mice. The most important known host factor is age; wild-type Sindbis virus results in a rapidly fatal encephalomyelitis in newborn mice but produces no clinically apparent disease in older mice (30). Interestingly, wild-type Sindbis virus can undergo neuroadaptive mutations which overcome the host age-dependent restriction to virulence (22, 38), and a single-amino-acid mutation at position 55 of the E2 envelope glycoprotein can produce lethal disease in older mice (59, 60). Thus, by using DNA microarray technology to compare the host cellular response in different aged mice infected with Sindbis virus or in mice infected with different strains of Sindbis virus that vary in neurovirulence, it may be possible to identify host genes that are important in modulating viral pathogenesis.
An additional advantage of the Sindbis virus model system is that an efficient strategy has already been developed for studying the effects of specific host cell genes on viral neuropathogenesis (reviewed in reference 24). The strategy involves the use of chimeric recombinant double-subgenomic Sindbis virus constructs that express heterologous genes in virally infected neurons in vivo. Using this approach, it has been shown that enforced expression of several different host cell genes, including bcl-2 (37) beclin 1 (41), bax (39), and SMN (31), protects mice against fatal Sindbis virus encephalitis whereas enforced expression of other host genes increases the likelihood of animal death (H. H. Jiang, W. Jiang, and B. Levine, unpublished data). Therefore, this approach could potentially be useful in determining whether cellular mRNAs that are found to be differentially regulated in virulent and avirulent Sindbis virus infections play a protective or pathologic role in viral pathogenesis.
In this study, we used a Sindbis virus mouse encephalitis model to study how the host cellular gene response differs in a nonlethal and a lethal infection. Our findings identified specific host genes that are differentially regulated during neurovirulent Sindbis virus infection. Furthermore, we used the Sindbis virus vector system to show that increased expression of one of these genes, PBR (encoding peripheral benzodiazepine receptor), plays a protective role for the host in Sindbis virus encephalitis.
MATERIALS AND METHODS
Recombinant virus strains.
Previously described recombinant strains of double-subgenomic Sindbis viruses, dsTE12H (formerly referred to as dsTE12) (37) and dsTE12Q (29) were used for most experiments. To construct recombinant chimeric Sindbis viruses expressing mouse PBR, a 509-bp fragment containing the open reading frame of the mouse PBR gene was amplified by PCR from cDNA prepared from dsTE12H-infected mouse brains, adding BstEII restriction sites to the upstream and downstream primer and a flag epitope sequence to the upstream primer. The mouse PBR gene was cloned into the BstEII sites of dsTE12Q to generate plasmid dsTE12Q/PBR, and the correct sequence of the mouse PBR gene insert was confirmed by sequencing. A control chimeric viral cDNA, dsTE12Q/PBR-control, was also constructed in which the start codon of the PBR gene was deleted. Stock viruses (dsTE12Q, dsTE12H, dsTE12Q/PBR, and dsTE12Q/PBRcontrol) were produced from viral cDNA clones as previously described (37) and stock titers were determined by plaque assay titer determination on BHK-21 cells.
Animal studies.
The right cerebral hemispheres of 1- and 10-day-old CD1 mice (Charles River) were inoculated with 1,000 PFU of virus in 0.03 ml of Hanks' balanced salt solution. A 0.03-ml volume of Hanks' balanced salt solution was used for mock infections. For mortality studies, three to five separate litters were inoculated with each virus and the mice were observed daily for 21 days to monitor survival. For RNA isolation, brains were dissected at 1, 2, and 3 days (24, 48, and 72 h) after inoculation (for reverse transcription-PCR [RT]-PCR analysis) and at 3 days (66 h) after inoculation (for GeneChip expression analysis). The left hemisphere of the brain was rapidly frozen and stored at −80°C. The right cerebral hemisphere was fixed by immersion in 4% paraformaldehyde. For virus titer determination experiments, the left cerebral hemisphere was used to make a freeze-thaw 10% homogenate in Hanks' balanced salt solution for subsequent plaque assay on BHK-21 cells.
Histopathology.
The right cerebral hemispheres of paraformaldehyde-fixed mouse brains were embedded in paraffin, and a series of 4-μm-thick sagittal sections were cut from medial to lateral. For each dsTE12Q- and dsTE12H-infected brain, sequential sections were examined by hematoxylin and eosin (H & E) staining to detect histopathology, terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) staining to detect apoptosis, and immunoperoxidase staining to detect Sindbis virus antigen, mouse PBR, mouse IRF-7, and mouse MCP-1. For each dsTE12Q/PBR- and dsTE12Q/PBR control-infected brain, sequential sections were examined by H & E staining, TUNEL, and immunoperoxidase staining to detect Sindbis virus antigen and flag-epitope-tagged mouse PBR. TUNEL staining was performed using the ApopTag peroxidase in situ apoptosis detection kit (Intergen). Immunostaining to detect Sindbis virus antigen was performed using a polyclonal anti-Sindbis virus antibody (1:400 dilution) and the avidin-biotin peroxidase method (Vectastain ABC kit; Vector Laboratories). A similar method was used to detect mouse PBR with a polyclonal rabbit anti-PBR antibody (1:150 dilution; Trevigen), IRF-7 with a polyclonal rabbit anti-murine IRF-7 antibody (1:50 dilution; Zymed Laboratories), MCP-1 with a polyclonal rabbit anti-murine MCP-1 antibody (1:50 dilution; PeproTech), and the flag epitope with monoclonal anti-flag antibody M2 (20 μg/ml; Sigma). The number of TUNEL-positive cells in each sagittal section of dsTE12Q/PBR- and dsTE12H/PBRcontrol-infected brains was counted at ×20 magnification by an observer blinded to the treatment group. Results are expressed as the mean number of apoptotic cells per ×20 magnification field.
GeneChip expression analysis sample preparation and hybridization.
RNA was prepared for GeneChip expression analysis as specified by the manufacturer (Affymetrix) and as described in previously published protocols (14, 35, 64). Briefly, total RNA was isolated from the left hemispheres of brains dissected 3 days after virus infection using TRIzol reagent (Gibco/BRL). RNA from six mice per treatment group was pooled and used for GeneChip expression analysis. Poly(A)+ mRNA was isolated from 200 μg of pooled total RNA using Oligotex Direct mRNA kit (Qiagen). cDNA was synthesized from 1 μg of poly(A)+ mRNA using a T7 (dT)24 high-pressure liquid chromatography-purified primer (GENSET) and SuperScript choice system (Gibco/BRL). Biotin-labeled cRNA was generated from 1 μg of cDNA using the RNA transcript labeling kit (ENZO). RNeasy mini-kits (Qiagen) were used to remove the unincorporated nucleoside triphosphates. The samples were precipitated in ethanol and resuspended in diethylpyrocarbonate-treated H2O at a concentration of 0.5 to 1 μg/μl. Samples were fragmented to 50 to 200 bp in 5× fragmentation buffer (200 mM Tris acetate [pH 8.1], 500 mM potassium acetate, 150 mM magnesium acetate) for 35 min at 95°C. The samples were hybridized to the Murine 6500 GeneChip subarrays A, B, C, and D (Affymetrix) overnight, washed, and stained as previously described (63, 64).
Data analysis.
The data collected after hybridization of biotin-labeled cRNA from each treatment group was analyzed using GeneChip Suite software. Internal controls for hybridization intensity and cRNA synthesis were included on each chip and have been discussed elsewhere (14). Samples were normalized to glyceraldehyde phosphate dehydrogenase, actin, and 18S rRNA levels. The Absolute Analysis algorithm was used to calculate whether genes were “absent” or “present” based on previously described parameters (64). To calculate fold changes in genes in two different samples, the Comparison Analysis algorithm (Affymetrix) was used (64). Three different comparison analyses were performed, including one with cRNA from mock-infected brains as the baseline sample and cRNA from dsTE12Q-infected brains as the experimental sample (QvM), one with cRNA from mock-infected brains as the baseline sample and cRNA from dsTE12H-infected brains as the experimental sample (HvM), and one with cRNA from dsTE12Q-infected brains as the baseline sample and cRNA from dsTE12Q-infected brains as the experimental sample (HvQ). Genes which were “absent” in the absolute analysis of the baseline sample and “decreasing” in the experimental sample on comparison analysis were considered false positives. Genes which were “absent” in the absolute analysis of the experimental sample and also “increasing” in the experimental sample upon comparison analysis were also considered false positives.
Analysis of CNS gene expression by RT-PCR.
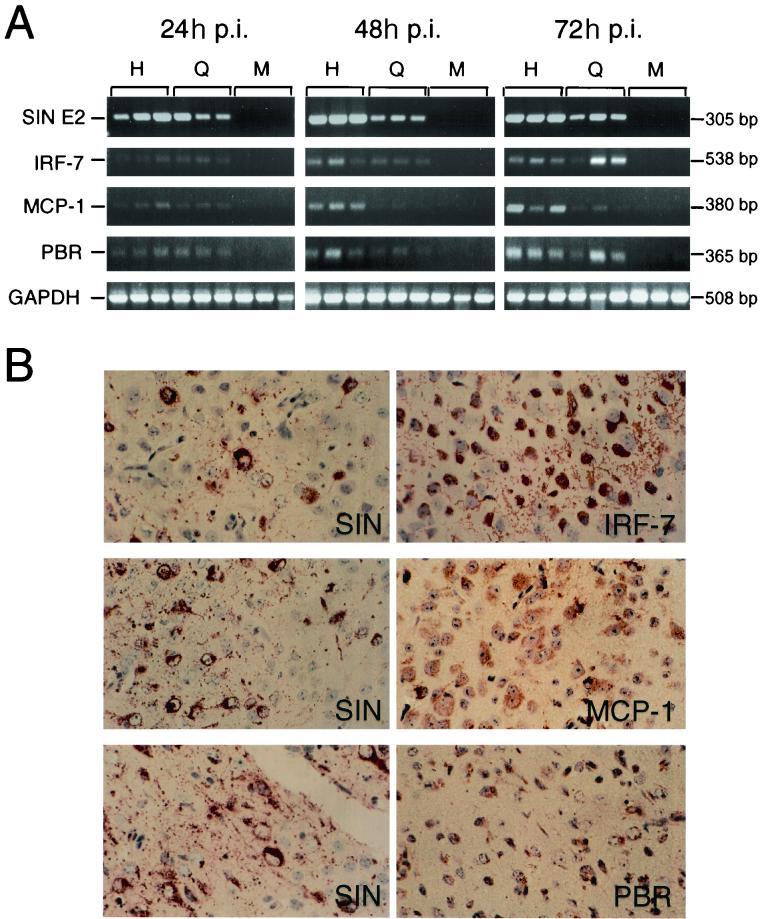
Total RNA was extracted using TRIzol Reagent, and poly(A) RNA was isolated using an Oligotex Direct mRNA kit (Qiagen) from triplicate mouse brains on days 1, 2, and 3 after mock infection or infection with 1,000 PFU of dsTE12H or dsTE12Q, intracerebrally. A 1-μg portion of poly(A)+ RNA was used to synthesize cDNA with avian myeloblastosis virus reverse transcriptase (Boehringer Mannheim) and an oligo(dT) primer (Boehringer Mannheim), and PCR was performed as described previously (62) to detect the Sindbis virus E2 glycoprotein gene (plus-strand primer [8607–8626], 5′-GGATCGTCTGGCAGAAGCAA-3′; minus-strand primer [8893–8912], 5′-AAGCCTTCTACACGGTCCTG-3′), murine MCP-1 (plus-strand primer, 5′-ACCAAGCTCAAGAGAGAGGT-3′; minus-strand primer, 5′-CTGGATTCACAGAGAGGGAA-3′), murine IRF-7 (plus-strand primer, 5′-CAGCGAGTGCTGTTTGGAGA-3′; minus-strand primer, 5′-ACTGCAGAACCTGAAGCAAGAG-3′), murine PBR (plus-strand primer, 5′-TCATGGGAGCCTACTTTGTG-3′; minus-strand primer, 5′-CAGGTAAGGATACAGCAAGC-3′), and the constituitively expressed cellular gene, glyceraldehyde phosphate dehydrogenase (plus-strand primer, 5′-ACCACCATGGAGAAGGCTGG-3′; minus-strand primer, 5′-CTCAGTGTAGCCCAGGATGC-3′). Serial dilutions of positive controls for each gene of interest were amplified at 20, 25, 30, and 35 cycles to determine the optimal number of amplification cycles that resulted in a linear relationship between the initial RNA input and PCR product. Twenty-five cycles of amplification was used in each PCR in the data shown in Fig. 8A.
FIG. 8.
Expression of IRF-7, MCP-1, and PBR in Sindbis virus-infected mouse brains. (A) RT-PCR analysis of RNA samples from triplicate mouse brains harvested at serial time points after infection with dsTE12H or dsTE12Q or mock infection. H, dsTE12H infection; Q, dsTE12Q infection; M, mock infection; p.i., postinfection. (B) Immunoperoxidase staining demonstrating expression in a similar brain region of Sindbis virus antigen (left column) and IRF-7, MCP-1, and PBR in adjacent sections (right column) 3 days after infection with dsTE12H. Top and middle panels are from the brain stem: bottom panel is from the posterior neocortex. Magnification, ×776.
RESULTS
dsTE12H produces fatal disease and dsTE12Q produces asymptomatic disease in 10-day-old CD1 mice.
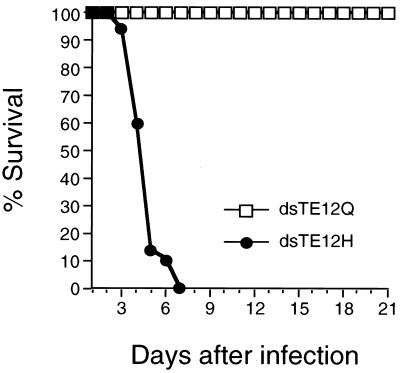
To compare the host gene response during virulent and avirulent CNS infections, we studied the pathogenesis of two different strains of Sindbis virus, dsTE12H and dsTE12Q. dsTE12H and dsTE12Q are both recombinant strains of Sindbis virus that contain duplicated internal subgenomic promoters. Although viral replication is somewhat attenuated by the presence of a duplicated subgenomic promoter, we chose to use these strains because of their utility as vectors for expressing candidate host pathogenesis regulatory genes in neurons in vivo. The strains dsTE12Q and dsTE12H differ by only 1 amino acid at position 55 in the E2 envelope glycoprotein; dsTE12Q contains a wild-type glutamine at E2-55 and dsTE12H contains a histidine substitution. The histidine substitution at E2-55 has been previously shown to be an important determinant of neurovirulence for older mice in non-double-subgenomic recombinant strains of Sindbis virus (59, 60). Similarly, we found that intracerebral inoculation of 1,000 PFU of dsTE12H resulted in 100% mortality in 10-day-old mice within 7 days after infection whereas no mortality or clinically apparent disease was observed in mice infected with dsTE12Q (Fig. 1). These data demonstrate that in the double-subgenomic strain of Sindbis virus, a histidine mutation at position E2-55 confers neurovirulence in 10-day-old mice.
FIG. 1.
Survival curve of 10-day-old mice infected with 1,000 PFU dsTE12H or dsTE12Q. Data represent combined survival probabilities for three independent litters with 10 to 12 mice per litter.
dsTE12H replicates to higher levels and causes increased apoptosis in the brains of 10-day-old mice.
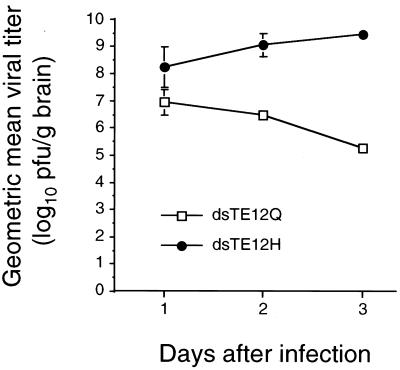
In parallel with increased mortality, we found higher levels of Sindbis virus replication in the brains of mice infected with dsTE12H compared to the brains of mice infected with dsTE12Q (Fig. 2). By day 3 after infection (when most dsTE12H-infected mice are moribund), mean viral titers were greater than 109 PFU per g of brain in mice infected with dsTE12H. In dsTE12Q-infected mice, viral titers peaked at 1 day postinfection at 107 PFU per g of brain and declined gradually thereafter. More extensive viral antigen staining was also observed in the brains of mice infected with dsTE12H than in the brains of mice infected with dsTE12Q (representative photomicrographs are shown in Fig. 3A and B). Despite greater numbers of viral antigen-positive cells in the brains of dsTE12H-infected mice, the tropism of the two Sindbis virus strains did not differ. In both dsTE12H- and dsTE12Q-infected mouse brains, neurons were the most prominent cell type infected and virus-infected foci were identified most frequently in the posterior neocortex, hippocampus, colliculus, and striatum.
FIG. 2.
Viral growth of dsTE12H and dsTE12Q in mouse brain. Each data point represents the geometric mean viral titer and standard error of the mean for three mouse brains.
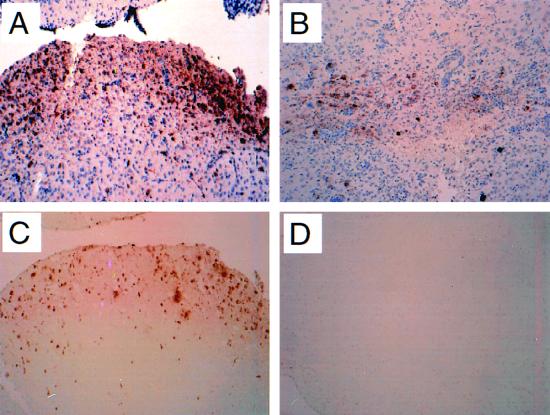
FIG. 3.
Immunoperoxidase labeling to detect Sindbis virus antigen (A and B) and TUNEL labeling to detect apoptotic nuclei (C and D) in the brains of dsTE12H-infected (A and C) and dsTE12Q-infected (B and D) mice. Representative sections are shown from the superior colliculus region 3 days after virus infection. Magnification, ×243.
We analyzed adjacent mouse brain sections by immunohistochemistry to detect Sindbis virus antigen, by H & E staining to detect neuronal pathology, and by TUNEL staining to detect apoptotic nuclei on days 1, 2, and 3 after infection (Fig. 3). In dsTE12Q-infected brains, pathology and TUNEL-positive cells were rarely observed in the regions of viral antigen-positive cells (Fig. 3B and D and data not shown). In contrast, in dsTE12H-infected brains, virus-infected foci displayed numerous pyknotic neuronal nuclei and numerous TUNEL-positive neuronal nuclei (Fig. 3A and C and data not shown). This observation is consistent with a previous report indicating that a histidine mutation at E2-55 in a similar background strain of a non-double-subgenomic Sindbis virus results in increased neuronal apoptosis in 2-week-old mice (40). Surprisingly, very little perivascular cuffing or other evidence of inflammation was observed in the brains of both dsTE12Q- and dsTE12H-infected mice.
Thus, the neurovirulent strain, dsTE12H replicates to higher titers in the CNS than does the avirulent strain, dsTE12Q, and induces more neuronal death but does not elicit a greater detectable inflammatory response.
dsTE12H infection increases the expression of more host genes than does dsTE12Q infection.
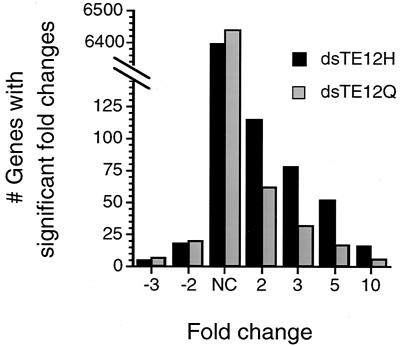
To investigate differences in the host response to infection with the avirulent dsTE12Q strain and the virulent dsTE12H strain, we performed cDNA microarray analysis of pooled RNA harvested from mouse brains 3 days after infection using the Mu6500 GeneChip (Affymetrix), which includes probe pairs that hybridize with approximately 6,500 different murine genes. In dsTE12Q-, dsTE12H-, and mock-infected brains, approximately 25% of the genes were judged to be “present” by GeneChip software absolute algorithm analysis. The overall number of host genes that had increased expression was greater in response to dsTE12H infection than in response to dsTE12Q infection (Fig. 4). Compared to mock-infected brains, the mRNA expression of 115 genes increased by at least twofold in dsTE12H-infected brains and the mRNA expression of only 62 genes increased by at least twofold in dsTE12Q-infected brains. In contrast, the number of genes with decreased expression did not significantly differ in dsTE12H- and dsTE12Q-infected brains (n = 18 and 20, respectively).
FIG. 4.
Graph showing the distribution of fold changes in gene expression on comparison of dsTE12H- or dsTE12Q-infected brains with mock-infected brains. Each category includes all genes that have at least the fold change indicated on the x axis. NC, no change. See Materials and Methods for an explanation of the data analysis.
dsTE12H infection results in higher levels of expression of many different cellular genes.
The expression of many cellular genes increased in both dsTE12H- and dsTE12Q-infected compared to mock-infected brains (Table 1). The majority of these genes fell into functional categories that are known or likely to be important in the host response to viral infections, including chemokines, extracellular matrix and cell adhesion molecules, hematopoietic cell surface molecules, and genes involved in antigen presentation, protein degradation, interferon (IFN)-related, and complement-related pathways. While many genes had between 2- and 10-fold-increased expression, the expression of a limited number of genes increased more than 10-fold in dsTE12H-infected brains; these included chemokine genes (MCP-5 and MCP-1), complement-related genes (complement component C4 and factor B), and genes encoding tissue inhibitor of metalloprotease, lymphocyte antigen LY-6E.1, PBR, cyclophilin C, and interferon response factor 7, and several IFN-inducible genes of unknown function (GARG-39, IFN-induced 15-kDa protein, IFN-α-induced 11.5-kDa protein, and GARG49).
TABLE 1.
Genes whose expression is increased twofold or more in dsTE12H- and dsTE12Q-infected mouse brains
| Gene family and gene namea | GenBank no. | Fold increasea
|
||
|---|---|---|---|---|
| HvMb | QvMc | HvQd | ||
| Actin/cytoskeleton | ||||
| Vimentin | M26251 | 7 | 2.9 | 2.9 |
| Antigen presentation | ||||
| MHC class III region | W90837 | 2.6 | 1.9 | NC |
| TAP2 | U60087 | 4.8 | 3.8 | 3.8 |
| HAM1 | M55637 | 6 | 5.6 | NC |
| MHC class I D-region cell surface Ag | M27034 | 7.7 | 5.8 | NC |
| β2-Microglobulin | AA059700 | 22.3 | 13.9 | 1.6 |
| Chemokines | ||||
| MIG | M34815 | 2.8 | 2 | NC |
| Crg-2/IP-10 | M86829 | 5 | 2.2 | 3.1 |
| FIC/MCP-3 | X70058 | 7.9 | 4.1 | 4.7 |
| MCP-5 | U50712 | 10.1 | 2.6 | NC |
| JE/MCP-1 | M19681 | 18.8 | 4.8 | 5.2 |
| CNS specific | ||||
| Synaptobrevin/endobrevin | AA049140 | 3.2 | 2.7 | NC |
| Glial fibrillary acidic protein | K01347 | 12.5 | 7.2 | 2.5 |
| Complement | ||||
| Complement receptor C3 β (CD18) | X14951 | 3.3 | 1.9 | 2.5 |
| C1q C chain | X66295 | 4 | 2 | NC |
| Complement C1q B chain | M22531 | 5.3 | 2.9 | 1.8 |
| C1Q α chain | X58861 | 5.3 | 3.1 | NC |
| Complement component C2 | M57891 | 5.4 | 4.6 | NC |
| Complement component C4 | M17440 | 11.2 | 9.1 | 1.9 |
| Factor B/serine protease | M57890 | 14 | 10.5 | 3.6 |
| DNA replication | ||||
| Histone H1 | J03482 | 2 | 2 | NC |
| DNA replication licensing factor | D86726 | 2.2 | 1.8 | NC |
| Thymidylate kinase homolog | L32973 | 3.9 | 2 | NC |
| Extracellular matrix and cell adhesion | ||||
| Extracellular matrix protein 1 | L33416 | 2.7 | 1.7 | NC |
| VCAM-1 | L22355 | 5.2 | 2 | 2.7 |
| Galectin-1 | W13002 | 5.2 | 2.6 | 2 |
| Mac-2 (galectin-3) | X16834 | 7.4 | 1.9 | 3.9 |
| TIMP | M28312 | 12.4 | 3.8 | 3.2 |
| Granule associated | ||||
| Granzyme B | X04072 | 4 | 3.2 | 2.4 |
| Serglycin | X16133 | 2.8 | 2.1 | 2 |
| Growth factor | ||||
| Granulin precursor | M86736 | 3.2 | 1.8 | 1.9 |
| Hematopoietic cell surface molecules | ||||
| CD63/ME491 | D16432 | 2.5 | 1.4 | NC |
| Thymic shared antigen 1 | U47737 | 3.3 | 2.8 | NC |
| Mast cell surface glycoprotein Gp49B | U05265 | 3.4 | 1.7 | 3 |
| Leukocyte common Ag Ly-5 | M14343 | 3.5 | 3 | NC |
| Monocyte diff. Ag (CD14) | M34510 | 3.7 | 2.1 | 3.1 |
| Lymphocyte diff. Ag B7 (CD52) | M55561 | 4.6 | 2.4 | NC |
| IgE receptor γ subunit | W41745 | 5 | 2.7 | 1.9 |
| Lymphocyte Ag Ly-6E.1 | X04653 | 32.4 | 15.1 | 1.9 |
| IFN pathway related | ||||
| 204 IFN-activatable protein | M31419 | 2.4 | 1.9 | NC |
| G protein like LRG-47 | U19119 | 2.9 | 1.9 | 2.4 |
| GTP binding protein (GTP2) | U15636 | 4.6 | 2.9 | NC |
| GBP3 | U44731 | 6.8 | 4.1 | 2.6 |
| Nmi | AA013783 | 6.4 | 6.6 | NC |
| Sim to IFN-inducible 1-8D | W29434 | 7.3 | 2.2 | 3.3 |
| IFN-induced GBP-2 | AA153021 | 7.6 | 5.2 | 2.9 |
| IRG-47 | M63630 | 7.6 | 5.3 | 2.8 |
| IRF-1 | M21065 | 8 | 4.3 | 3.5 |
| Stat1 | U06924 | 8.3 | 2.6 | NC |
| GARG-39 | U43085 | 10.9 | 1.7 | 3.7 |
| IFN-induced 15 kDa protein | X56602 | 13.5 | 10.4 | 2.2 |
| IFN-α induced 11.5-kDa protein | AA145371 | 15.1 | 8.4 | 1.9 |
| IRF-7 | U73037 | 25.1 | 4.9 | 2 |
| GARG49/irg2 | L32974 | 26.3 | 9 | 2.7 |
| Lipocalins | ||||
| Apollipoprotein D | L39123 | 6.4 | 2.9 | 2.2 |
| Lipocalin 2 (24p3) | W13166 | 7.2 | 2.8 | 6.1 |
| Mitochondrial function | ||||
| Uncoupling protein 2 (UCP2) | U69135 | 5.2 | 2 | NC |
| Peripheral benzodiazepine receptor | D21207 | 10.6 | 2.4 | 2.6 |
| Prostaglandin synthesis | ||||
| G/H synthase (COX-2) | M88242 | 2.6 | 2.1 | 2.1 |
| Protein degradation | ||||
| Cathepsin S precursor | AA146437 | 4.7 | 6.1 | NC |
| Cathepsin C | AA144887 | 5.2 | 4.8 | NC |
| Ubiquitin protein ligase E1 | AA170444 | 5.3 | 2.9 | 1.8 |
| Proteasome (Lmp2) gene | L11613 | 7 | 2.1 | NC |
| Cathepsin S precursor | AA089333 | 7.7 | 6.6 | NC |
| 20S proteasome Lmp7 | U22031 | 9 | 5 | 2.3 |
| Protein synthesis | ||||
| 40S ribosomal protein S16 | AA034714 | 2.5 | 2.8 | NC |
| Suit homolog | W11169 | 5.7 | 32.4 | NC |
| Proto-oncogenes | ||||
| GTP-binding protein rhoG | AA063858 | 2.4 | 3 | NC |
| Tyrosine kinase (HCK) | J03023 | 5.7 | 4.8 | NC |
| Transcription factors | ||||
| NK10 | X79828 | 2.7 | 3.2 | NC |
| C/EBP δ | X61800 | 3.1 | 2.1 | NC |
| C/EBP β | X62600 | 3.1 | 2.7 | NC |
| Chop-10 | X67083 | 4.2 | 2.4 | 2.9 |
| Miscellaneous | ||||
| Arnyloid β (A4) | W34965 | 2.3 | 1.7 | NC |
| Tissue plasminogen activator | J03520 | 2.8 | 1.5 | NC |
| EST sim to sec61b subunit | AA097018 | 3.9 | 4.2 | NC |
| Cyclophilin C | L16894 | 13.6 | 13.1 | 2.1 |
Genes in bold are significantly increased twofold or greater in dsTE12H-infected compared to dsTE120-infected brains. Abbreviations: diff., differentiation; sim, similar; Ag, antigen; EST, expressed sequence tag.
dsTE12H- compared to mock-infected brains.
dsTE12Q- compared to mock-infected brains.
dsTE12H- compared to dsTE12Q-infected brains. NC, no change.
For about 55% of the genes that were induced in both dsTE12Q- and dsTE12H-infected brains, the magnitude of increase was at least twofold higher in dsTE12H-infected than dsTE12Q-infected brains. The expression of only two of these genes, encoding MCP-1, a CC chemokine, and lipocalin 2 (44), an acute-phase protein, increased by more than fivefold between dsTE12H- and dsTE12Q-infected brains. The cellular mRNAs whose levels increased by a greater magnitude in dsTE12H- than dsTE12Q-infected brains represent candidate genes that may play a role in the host protective or pathologic response to the more virulent Sindbis virus infection. There were no cellular mRNAs whose levels increased by a greater magnitude in dsTE12Q- than in dsTE12H-infected brains and which could be identified as candidate host factors that might contribute to increased host resistance to the avirulent dsTE12Q strain.
Some cellular genes have increased expression uniquely in dsTE12H-infected brains.
To further identify candidate host genes that may play a role in the host protective or pathologic response during lethal CNS infection, we identified genes that had increased expression in dsTE12H-infected brains but not in dsTE12Q-infected brains (Table 2). In general, these genes fell into the same functional groups as cellular mRNAs whose levels were increased in both dsTE12H- and dsTE12Q-infected brains. However, low levels of increased expression (two- to fourfold range) were seen in unique functional groups of genes including those encoding annexins (e.g., annexin 2, annexin 1, annexin 1 ligand), antiapoptotic Bcl-2 family members (e.g., Mcl-1 and Bfl-1), growth arrest/DNA damage-inducible proteins (e.g., GADD45γ and MyD118), and macrophage inflammatory proteins (MRP14, MRP8, and chitotriosidase). Of the genes that had increased expression uniquely in dsTE12H-infected brains, the highest levels of increase were observed for the CC chemokine RANTES and the regIII-γ-proteasome activator (7.5- and 6.4-fold, respectively, in comparison to dsTE12Q-infected brain).
TABLE 2.
Genes which are uniquely induced in dsTE12H-infected brains and which exhibit significant fold increases compared to their induction in dsTE12Q-infected brains
| Gene family and gene name | GenBank no. | Fold increase
|
||
|---|---|---|---|---|
| HvMa | QvMb | HvQc | ||
| Annexins | ||||
| Annexin 2 (calpactin I heavy chain) | D10024 | 2.9 | NCd | 1.9 |
| Annexin I | M69260 | 3 | NC | 2.5 |
| Annexin I ligand | M16465 | 3.2 | NC | 2.3 |
| Bcl-2 family members | ||||
| Mcl-1 | U35623 | 2.1 | NC | 1.8 |
| Bfl-1 | L16462 | 2.2 | NC | 2.1 |
| Cysteine protease | ||||
| Caspase-11 | U59463 | 2.9 | NC | 2.3 |
| Cysteine protease inhibitor | ||||
| Cystatin B | AA015331 | 2.5 | NC | 2.7 |
| CNS specific | ||||
| Mouse brain testican | X92864 | 2.2 | NC | 1.8 |
| Chemokines | ||||
| MIP-1β | M23503 | 7 | NC | 3.3 |
| RANTES | U02298 | 10.3 | NC | 7.5 |
| Extracellular matrix and cell adhesion | ||||
| α-Catenin | X59990 | 2.4 | NC | 2.3 |
| Matrix Gla protein (MGP) | D00613 | 2.5 | NC | 2.6 |
| ICAM-1 | M31585 | 3 | NC | 2.8 |
| Growth arrest and DNA damage inducible | ||||
| GADD 45γ | AA138777 | 3.1 | NC | 2.4 |
| MyD118 | X54149 | 3.4 | NC | 3.4 |
| Hematopoietic cell surface receptors and ligands | ||||
| CD53 | Z16078 | 2.2 | NCd | 1.9 |
| CD30 ligand | L09754 | 2.3 | NC | 2.2 |
| G-CSF receptor | M58288 | 2.4 | NC | 2 |
| Macrosialin (similar to human CD68) | X68273 | 2.6 | NC | 2.3 |
| Lyb-2.1 | J04170 | 2.7 | NC | 2.5 |
| Ly-6C | D86232 | 7.3 | NC | 3.6 |
| IFN related | ||||
| Mx1 protein | M21117 | 2.6 | NC | 2.8 |
| Similar to IFN-inducible 1-8U | AA144469 | 4.5 | NC | 2.1 |
| GARG-16 | U43084 | 6.6 | NC | 2.6 |
| Macrophage inflammatory proteins | ||||
| MRP14 | M83219 | 2.8 | NC | 1.7 |
| MRP8 | X87966 | 2.8 | NC | 1.8 |
| Chitotriosidase | D87757 | 4.1 | NC | 2.5 |
| Protein degradation | ||||
| PA28 β subunit | U60328 | 3.7 | NC | 1.6 |
| Cathepsin Z | AA116604 | 5.9 | NC | 1.9 |
| PA28 β subunit | U60329 | 6 | NC | 3 |
| Reg III-γ-proteasome activator | AA123026 | 6.3 | NC | 6.4 |
| Lysozyme M gene | M21050 | 9.2 | NC | 3.5 |
| Transcriptional activator | ||||
| α-NAC (Naca) | U48363 | 2 | NC | 2 |
| Miscellaneous | ||||
| Tumor-associated membrane protein | U25633 | 2.2 | NC | 1.5 |
| Serum amyloid A4 | M17790 | 2.4 | NC | 2.1 |
| Metallothionein-I | V00835 | 2.6 | NC | 2.9 |
| Heme oxygenase | X56824 | 2.6 | NC | 2.7 |
| ob/ob haptoglobin | M96827 | 2.9 | NC | 3 |
| Adipophilin | M93275 | 8.3 | NC | 4.5 |
dsTE12H- compared to mock-infected brains.
dsTE12Q- compared to mock-infected brains.
dsTE12H- compared to dsTE12Q-infected brains.
NC, no change.
dsTE12Q and dsTE12H infection results in decreased expression of a limited number of cellular genes.
Compared to the increases in gene expression observed in dsTE12Q- and dsTE12H-infected brains, the expression of a smaller number of host genes decreased in response to Sindbis virus infection and the magnitude of the decreases tended to be small, ranging mostly between 2- and 3.5-fold (Table 3). This observation may in part reflect decreases in gene expression occurring primarily in virally infected neurons (which constitute only a small subpopulation of the total RNA analyzed), whereas increases in gene expression probably occur in both infected neurons and other uninfected cells in the brain. Interestingly, the largest category of genes with decreases in expression encoded proteins that are localized in synaptic nerve terminals and involved in synaptic function. Decreases in expression were also observed for other gene categories including those involved in cholesterol and steroid metabolism, cytoskeletal proteins, nuclear hormone receptors, neuronal receptors, and signal transduction molecules.
TABLE 3.
Genes whose expression is decreased twofold or greater in dsTE12H- or dsTE12Q-infected mouse brains
| Gene family and gene namea | GenBank no. | Fold changea
|
||
|---|---|---|---|---|
| HvMb | QvMc | HvQd | ||
| Cholesterol and steroid metabolism | ||||
| Hydroxysteroid sulfotransferase | L27121 | −2.0 | −2.6 | −1.6 |
| Similar to human sterol reductase | W15994 | −3.2 | −1.6 | −2.0 |
| Cytoskeletal | ||||
| Cytokeratin 10 | L00193 | −2.3 | −4.6 | −1.5 |
| Laminin B2 chain | J03484 | NC | −3.6 | 3.0 |
| Laminin receptor | J02870 | NC | −7.2 | 5.5 |
| DNA replication | ||||
| Similar to human thymidylate kinase | AA089117 | −2.7 | NC | −2.4 |
| Granule associated | ||||
| RNA binding protein TIAR | U55861 | NC | −2.0 | 2.2 |
| Mitochondrial function | ||||
| Adenine nucleotide translocase 1 | U27315 | NC | −2.3 | 3.0 |
| Nuclear hormone receptors | ||||
| c-erbA α2 thyroid hormone receptor | X07751 | −2.4 | NC | −2.0 |
| EAR2 (v-erbA-related protein) | X76654 | −2.4 | NC | −2.1 |
| Neuronal receptors | ||||
| GABA-α receptor subunit | M86567 | −2.0 | −2.1 | NC |
| Neurotensin receptor 2 | U51908 | −3.1 | −2.2 | NC |
| Other receptor | ||||
| Endothelin B receptor | U32329 | −3.1 | NC | −2.5 |
| Protein degradation | ||||
| Polyubiquitin 4 | W13646 | −2.2 | −2.5 | NC |
| Proteasome Z subunit | D83585 | NC | −4.1 | 4.4 |
| Protein synthesis | ||||
| EF1α2 | W30609 | −3.9 | NC | −3.1 |
| Signal transduction | ||||
| P21-activated kinase 3 | W50708 | −2.2 | −2.0 | NC |
| Similar to Rap1 GTPase-activating protein | W11780 | −2.3 | NC | −2.2 |
| Synaptic function | ||||
| Endophilin | U58886 | NC | −2.2 | 2.3 |
| α-SNAP | AA037945 | −2.4 | NC | −2.2 |
| Similar to human ankyrin G | AA030658 | −2.4 | NC | −2.4 |
| Complexin II (presynaptic protein) | D38613 | −2.9 | −3.0 | NC |
| Heat shock cognate protein 70 | AA137580 | −3.4 | −2.1 | NC |
| Transcription factor | ||||
| Ets-related protein 81 (ER81) | L10426 | NC | −2.1 | 2.1 |
| Unknown function | ||||
| FGF-inducible gene 14 (FIN14) | U42386 | NC | −2.1 | 2.5 |
| Brain protein I3 | AA060064 | −2.8 | −1.5 | −2.3 |
| EST similar to HBV X interacting protein | AA051557 | NC | −3.4 | 3.3 |
| EST similar to CGI-69 | AA124273 | NC | −4.1 | 2.8 |
| G protein γ3 subunit | AA049022 | −1.6 | −4.3 | 2.8 |
Abbreviations: EST, expressed sequence tag; HBV, hepatitis B virus; NC, no change.
dsTE12H- compared to mock-infected brains.
dsTE12Q- compared to mock-infected brains.
dsTE12H- compared to dsTE12Q-infected brains.
A subset of genes that are involved in IFN signaling or regulated by IFNs have increased expression in dsTE12H- and dsTE12Q-infected brains.
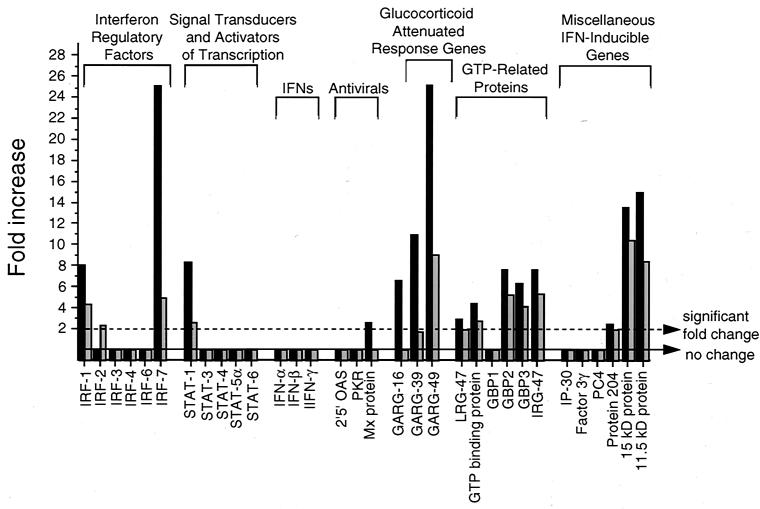
IFN-α/β are known to play a critical role in defense against Sindbis virus infections (52; reviewed in reference 21). To help define the specific molecules that may be important in mediating these effects, we compared the levels of gene expression in infected brains of all genes included in the Mu6500 GeneChip that either play a role in IFN signaling or are known to be induced by IFN (14) (Fig. 5). IFN-regulatory factors, especially IRF-1, IRF-3, and IRF-7, are important transcriptional activators of IFN-α/β gene expression during viral infections (reviewed in references 45 and 51), and studies with targeted mutant mice have demonstrated a role for both IRF-1 and IRF-2 in protection against Venezuelan equine encephalitis virus (20). IRF-1 and IRF-7, but not IRF-2, IRF-3, IRF-4, IRF-5, or IRF-6, gene expression increased in dsTE12H-infected (8-fold for IRF-1; 25.1-fold for IRF-7) and in dsTE12Q-infected (4.3-fold for IRF-1; 4.9-fold for IRF-7) brains compared to mock-infected brains. Stat 1, a signaling molecule that mediates most biologic responses induced by both IFN-α/β and IFN-γ (reviewed in reference 23), but not Stat 3, 4, 5a, and 6, also increased in dsTE12H-infected (8.3-fold) and dsTE12Q-infected (2.6-fold) brains. The expression of IFN-α, IFN-β, and well-characterized IFN-α/β-inducible, antiviral genes such as those encoding 2,5-oligoadenylate synthetase, double-stranded-RNA-activated protein kinase (PKR), and Mx protein did not change significantly following Sindbis virus infection. However, a number of other IFN-α/β-inducible genes (14) with less well-characterized roles in antiviral defense were induced strongly in dsTE12H-infected brains and to a lesser extent in dsTE12Q-infected brains, including glucocorticoid-attenuated response genes (GARG-16, GARG-39, and GARG-49) and genes encoding GTP-related proteins (GBP2, GBP3, and IRG47), a ubiquitin-like molecule (IFN-induced 15-kDa protein), and an 11.5-kDa protein of unknown function.
FIG. 5.
mRNA expression of genes involved in IFN signaling or induced by IFNs in dsTE12H-infected (solid bars) and dsTE12Q-infected brains (shaded bars). Mock-infected brains were used as the baseline for comparison analyses.
A subset of chemokines is induced in dsTE12H-infected and to a lesser extent in dsTE12Q-infected brains.
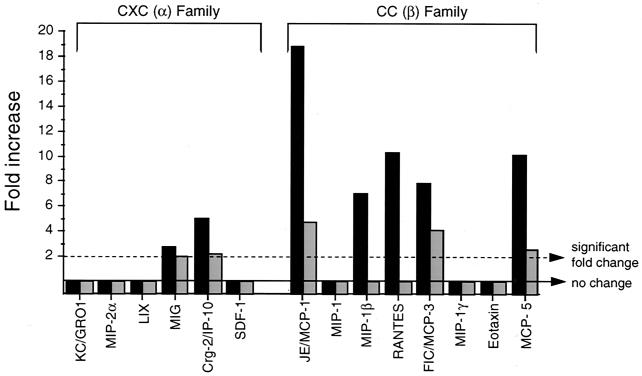
Chemokines are an important functional subgroup of IFN-inducible cytokines that play a role in both normal CNS development as well as in protective and pathologic responses during CNS viral infections (reviewed in references 1 and 6). To investigate which chemokines may be important in the host response to neurovirulent Sindbis virus infection, we compared the levels of expression of all known chemokines included in the Affymetrix 6500 GeneChip (Fig. 6). Small increases in gene expression were observed in dsTE12H-infected brains for the CXC family members MIG and Crg-2 but not for other CXC family members such as KC/GRO1, MIP-2α, LIX, and SDF-1. Among the CC chemokine family members, the highest levels of increases in expression were observed in dsTE12H-infected brains for JE/MCP-1, RANTES, MCP-5, FIC/MCP-3, and MIP-1β. In dsTE12Q-infected brains, smaller increases were also observed for MCP-1, MCP-3, and MCP-5 but not for MIP-1β or RANTES. No increases were observed in either dsTE12H- or dsTE12Q-infected brains for the CC chemokines MIP-1, MIP-1γ, and eotaxin.
FIG. 6.
mRNA expression of chemokines in dsTE12H-infected (solid bars) and dsTE12Q-infected (shaded bars) brains. Mock-infected brains were used as the baseline for comparison analyses.
Potential apoptotic mediators are induced in the brains of Sindbis virus-infected mice.
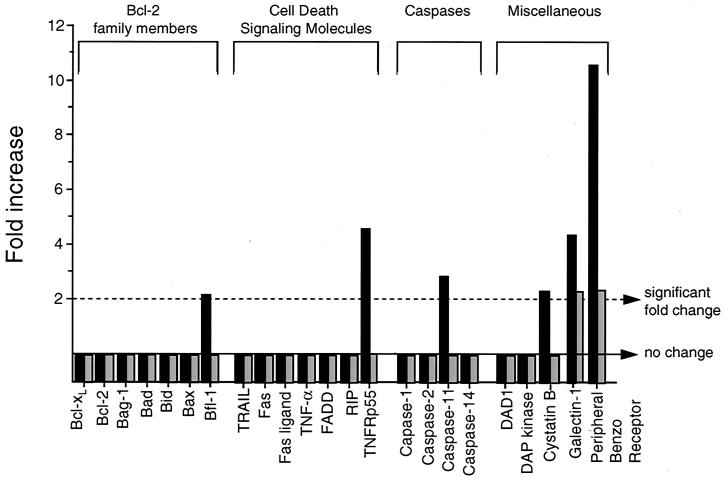
Several lines of evidence suggest that apoptosis plays an important role in the pathogenesis of fatal Sindbis virus encephalitis (reviewed in reference 36). However, the specific host molecules that are important in the regulation of Sindbis virus-induced apoptosis in vivo are unknown. To gain insight into potential host pathways that may be important in either the induction or prevention of apoptosis in Sindbis virus-infected brains, we compared the levels of gene expression of all known and putative apopototic mediators included on the Mu6500 GeneChip (Fig. 7). Among the Bcl-2 family members, the only change observed was a minimal increase in the expression of the antiapoptotic family members bfl-1 and mcl-1 in dsTE12H-infected brains. Among molecules involved in cell death receptor signaling pathways, the expression of TNFRp55, but not other molecules, increased in dsTE12H-infected brains. The physiologic role of this increase in apoptosis induction in dsTE12H infection is unclear, since mice lacking TNFRp55 are not protected against disease induced by dsTE12H (data not shown). The Mu6500 GeneChip contained only four caspases, C-1, C-2, C-11, and C-14, and of these, only C-11 (a caspase involved in inflammation but not apoptosis) showed an increase in the level in dsTE12H-infected compared to mock-infected brains.
FIG. 7.
mRNA expression of pro- and antiapototic genes in dsTE12H-infected (solid bars) and dsTE12Q-infected (shaded bars) brains. Mock-infected brains were used as the baseline for comparison analyses.
While few significant increases were observed in gene expression in dsTE12Q-and dsTE12H-infected brains among well-characterized apoptotic mediators such as Bcl-2 family members, cell death receptor-signaling molecules, and caspases, there was more marked induction of a gene encoding PBR, a mitochondrial protein with putative anti-apoptotic function. The expression of PBR increased 10.6-fold in dsTE12H- and 2.4-fold in dsTE12Q-infected brains. PBR associates with the voltage-dependent anion channel (VDAC) and the adenine nucleotide transporter (ANT) in the mitochondrial membrane (47) and has been shown to have antiapoptotic activity in Jurkat cells (5). The expression of another gene encoding a protein with putative proapoptotic function, galectin 1, is also mildly increased in dsTE12H-infected brains. However, the proapoptotic activity of galectin 1 has been found only in T lymphocytes (reviewed in reference 57), and in peripheral nerves, galectin 1 promotes axonal regeneration following axotomy (25).
RT-PCR analysis of selected mRNAs and immunohistochemical analysis of selected proteins in dsTE12H- and dsTE12Q-infected brains.
To validate our findings from GeneChip expression analyses, we performed RT-PCR (Fig. 8A) and immunohistochemistry of mouse brain sections (Fig. 8B) to detect the expression of selected representative genes, IRF-7, MCP-1, and PBR, from three major categories (IFN signaling, chemokines, and apoptotic mediators, respectively) that are likely to be important in the host response to neurovirulent Sindbis virus infection. Increased levels of IRF-7, MCP-1, and PBR mRNA (but not the constituitively expressed GAPDH mRNA) were detected by RT-PCR in Sindbis virus-infected compared to mock-infected brains on days 1, 2, and 3 after infection. In general, the magnitude of the increases for MCP-1, PBR, and, to a lesser extent, IRF-7 was proportional to the amount of Sindbis virus RNA. Thus, these findings confirm that for selected genes, mRNA expression as measured by RT-PCR analysis of individual mouse brain RNA samples yields results that are biologically similar to those obtained using GeneChip expression analysis of pooled RNA samples.
In parallel with the increased mRNA expression, we also found that there was increased IRF-7, MCP-1, and PBR protein expression in Sindbis virus-infected brains. No detectable expression of these proteins was observed in mock-infected brains (data not shown), and the highest levels of expression were observed in dsTE12H-infected brains, specifically in regions of the brain that also displayed Sindbis virus antigen immunoreactivity (see representative photomicrographs in Fig. 8B). Within such regions, there was expression of IRF-7, MCP-1, and PBR in cells that morphologically appeared to be neurons, as well as in some nonneuronal cells. These findings demonstrate that at least for some of the genes that had increased mRNA expression in GeneChip studies (Table 1) of Sindbis virus-infected brains, there is a corresponding increase in CNS protein expression.
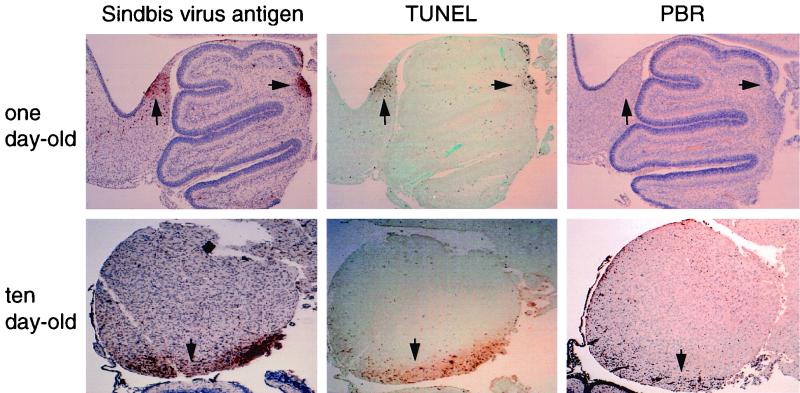
Neuronal PBR expression exerts a protective effect in Sindbis virus encephalitis.
Our GeneChip expression analysis identified a number of host genes that have altered expression in Sindbis virus-infected brains. Although such genes represent candidate mediators of CNS viral pathogenesis, the exact significance of alterations in the expression of given genes is unknown. Given the importance of neuronal apoptosis in Sindbis virus pathogenesis (reviewed in reference 36), we tested the hypothesis that increased expression of PBR, a putative antiapoptotic molecule (5, 8, 49), represents a protective host response in Sindbis virus-infected neurons. While dsTE12H but not dsTE12Q is neurovirulent in 10-day-old mice, both dsTE12H and dsTE12Q result in 100% mortality in 1-day-old mice. In contrast to 10-day-old mice, where PBR expression increased in both dsTE12H- and dsTE12Q-infected brains, we found that no PBR expression was detected by RT-PCR analysis (data not shown) or immunohistochemistry (Fig. 9) in the brains of 1-day-old mice infected with either dsTE12H or dsTE12Q. Therefore, we used the Sindbis virus vector system to evaluate whether the expression of PBR in neurons of Sindbis virus-infected 1-day-old mice (which do not normally upregulate PBR expression in response to Sindbis virus infection) would alter the natural history of fatal Sindbis virus encephalitis. We cloned flag epitope-tagged PBR and a control construct of PBR lacking the start codon into dsTE12Q to generate the recombinant chimeric viruses, dsTE12Q/PBR and dsTE12Q/PBRcontrol.
FIG. 9.
PBR induction in apoptotic areas of 10-day-old mouse brain but not 1-day-old mouse brain infected with Sindbis virus. Adjacent mouse brain sections were stained to detect Sindbis virus antigen, TUNEL-positive nuclei, or PBR expression 3 days after infection. Shown are representative sections that include regions of the colliculus and cerebellum. Arrows denote specific regions of each section that demonstrate both viral antigen staining and TUNEL-positive nuclei. Magnification, ×85.
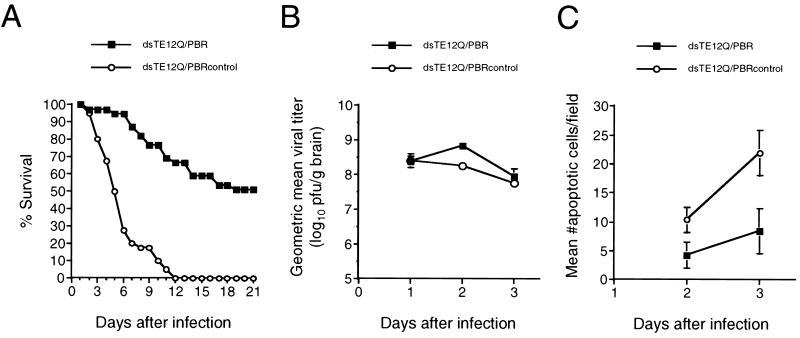
Following dsTE12Q/PBR infection, flag-PBR could be detected in regions of the brain that demonstrated Sindbis virus immunoreactivity (data not shown). Infection with 1,000 PFU of dsTE12Q/PBRcontrol resulted in 100% mortality, whereas 51% of mice infected with dsTE12Q/PBR survived (Fig. 10A). (A similar survival rate was also observed following infection with a chimeric virus expressing PBR lacking a flag epitope [data not shown].) Viral replication as measured by plaque assay titer determination did not differ significantly in the brains of dsTE12Q/PBR- and dsTE12Q/PBRcontrol-infected mice, except that the brains of dsTE12Q/PBR-infected mice had slightly higher mean viral titers on day 2 after infection (Fig. 10B). However, on both days 2 and 3 after infection, the brains of dsTE12Q/PBR-infected mice had significantly fewer cells that demonstrated TUNEL positivity (Fig. 10C). Thus, neuronal PBR expression in virally infected neurons can decrease neuronal apoptosis and protect 1-day-old mice against fatal disease without inhibiting viral replication.
FIG. 10.
Enforced PBR expression protects neonatal mice against lethal Sindbis virus infection. (A) Survival curves of 1-day-old mice infected intracerebrally with 1,000 PFU of dsTE12Q/PBR or dsTE12Q/PBRcontrol. The results represent combined survival curves for five independent litters with 10 to 12 mice per litter. (B) Viral growth in mouse brains infected with dsTE12Q/PBR or dsTE12Q/PBRcontrol. Each data point represents the mean and standard error of the mean for three mouse brains. (C) Quantitation of apoptotic cells in mouse brains infected with dsTE12Q/PBR or dsTE12Q/PBRcontrol. Each data point represents the mean and standard error of the mean for sagittal sections from three to five different mouse brains. Quantitation was performed as described in Materials and Methods.
DISCUSSION
In this study, we compared the CNS gene expression profiles in 10-day-old mice infected intracerebrally with two strains of Sindbis virus, dsTE12Q and dsTE12H, that are closely related genetically but differ dramatically in neurovirulence. dsTE12Q contains a wild-type glutamine at E2-55, replicates to lower titers in mouse brain, results in less neuronal death, and produces no clinically apparent disease, whereas dsTE12H contains a neurovirulent histidine substition at E2-55, replicates to higher titers in mouse brain, results in more neuronal death, and invariably produces fatal disease. In parallel with the increased neurovirulence of dsTE12H, the increases in host gene expression were more marked in dsTE12H-infected brains. The expression levels of more cellular mRNAs were increased in dsTE12H-infected compared to dsTE12Q-infected brains, and for cellular mRNAs that had increased levels in both dsTE12H- and dsTE12Q-infected brains, the magnitude of increase was usually greater in dsTE12H-infected brains. This increased host gene expression in mice infected with a more neurovirulent strain may play a role in both increased antiviral defense and increased virus-induced injury in the brain.
Our results identify several host genes with altered CNS expression that may be involved in the host response to Sindbis virus infection. The majority of these genes can be classified into functional groups that are already known to be involved in the host response to infection with Sindbis virus and other neurotropic viruses, including genes encoding antigen presentation molecules, immune cell activation markers, chemokines, and IFN-inducible gene products. The identification of specific genes within each of these functional groups that have increased expression in Sindbis virus-infected brains may help elucidate the molecular details of important host response pathways. For example, the expression levels of two CC chemokines, RANTES and MCP-1, which have both neuroprotective and proinflammatory effects in the CNS, both increased significantly in dsTE12H-infected compared to in dsTE12Q-infected brains, and these two chemokines represent candidate regulators of Sindbis virus neuropathogenesis. RANTES and MCP-1 are upregulated in several other CNS viral infections (2, 11, 12, 33, 34, 46, 53, 54), including in genetically susceptible but not genetically resistant strains of mice infected with mouse adenovirus type 1 (10). The selective increase in the level of RANTES in dsTE12H- but not in dsTE12Q-infected brains is interesting since RANTES has been directly implicated in the pathogenesis of mouse hepatitis virus-induced demyelination (34). The upregulation of MCP-1 is also noteworthy in light of recent findings by Gil et al. (18a). They demonstrated that IFN-α/β/γ receptor-deficient mice were much more susceptible than Stat1-deficient mice to Sindbis virus infection and that MCP-1 was one of the few genes that is induced by IFN in a Stat-1-independent manner. This suggests that MCP-1 may be an IFN-regulated gene that is important in protection against Sindbis virus infection.
Surprisingly, no increases were observed in IFN-α or IFN-β in brains infected with either dsTE12H or dsTE12Q, despite induction of IFN-α/β transcriptional activators (e.g., IRF-1 and IRF-7), IFN-α/β signaling molecules (e.g., Stat1), and a number of known IFN-α/β-inducible genes (see above). This may reflect the lower sensitivity of the GeneChip analysis compared to bioassays previously used to measure induction of IFN-α/β activity in Sindbis virus-infected brains (56, 61). Alternatively, since IFN production is an early response in CNS Sindbis virus infections, our microarray analysis may have been performed at a time point that was too late to detect increases in IFN-α or IFN-β mRNA levels. These considerations raise the possibility that IFNs and other important genes that have increased expression in Sindbis virus-infected brains may have been missed in our screen, either due to the intrinsic sensitivity of the GeneChip assay or to the kinetics of host gene expression. Despite these limitations, the specific IFN pathway-related genes that are increased in Sindbis virus-infected brains represent potential mediators of the IFN-α/β antiviral host response to Sindbis virus infection.
The increased expression of one class of inflammatory genes, complement-related proteins, was a somewhat unexpected finding in our study. Complement-related proteins are upregulated in brain microglia in response to cerebral ischemia (55), and complement-mediated lysis has been postulated to play a role in neurodegenerative diseases (16). However, despite the importance of complement as part of the innate immune response to microbial invasion, very little attention has been devoted in the past two decades to the role of complement in CNS viral infections. In addition, there are no previous reports of altered gene expression of complement proteins within the brain during viral infection. However, Hirsch et al. found that Sindbis virus-infected mice treated with cobra venom factor to deplete the third component of complement had 1,000-fold-greater viral titers in the brain (25). Although the mechanism was postulated to reflect systemic effects of complement on viral replication in the bloodstream (26), our observation that expression of complement genes increases in Sindbis virus-infected mouse brains raises the possibility that complement may also play a more direct role in antiviral responses in the CNS.
It is also interesting that the marked increases in the expression of inflammatory response genes in 10-day-old Sindbis virus-infected mouse brains were observed in the absence of histologic evidence of frank “encephalitis” (i.e., brain inflammation). Similar increases in chemokine gene expression in the absence of inflammation have also been found in neonatal rodents infected with Borna disease virus (54). Although we cannot rule out the possibility that some of the changes in inflammatory gene expression reflect infiltrating inflammatory cells that we failed to detect on H & E analysis of mouse brain sections, most of the alterations in inflammatory gene expression are probably occurring within intrinsic brain cells. This hypothesis is consistent with increasing evidence demonstrating that different types of brain cells, including neurons, microglia, and astrocytes, are capable of producing cytokines, chemokines, and other immunomoregulatory molecules (reviewed in reference 6). In support of our hypothesis, we found that expression of IRF-7 and the CC chemokine MCP-1 were detected in the same regions of the brain and in the same cell types which expressed Sindbis virus antigen (i.e., neurons). Similarly, Wesselingh et al. reported production of numerous cytokines by intrinsic brain cells in scid mice infected with Sindbis virus (62). Given the important effects of immune mediators in the CNS (neuroprotective as well as neurotoxic), the host gene response profile in a CNS infection may be a more accurate reflection of pathogenetically relevant “inflammation” than traditional histologic criteria.
In addition to identifying specific candidate host response genes within functional groups that are already known to be important in CNS viral infections, our findings identify novel groups of genes that are not yet known to play a role in the host response to Sindbis virus infection. We observed significant increases in the expression of several host genes involved in protein degradation, particularly in the brains of mice infected with the more neurovirulent strain of Sindbis virus, dsTE12H. Although some of these genes such as the proteasomal components are involved in antigen processing, the upregulated expression of genes involved in lysosomal, ubiquitin-mediated, and other forms of proteolysis could have additional, as yet undefined effects on the pathogenesis of CNS viral infections. For example, a role for upregulation of the gene encoding lysosomal M (the protein degradation gene with the greatest increase in expression in dsTE12H-infected brains) has been postulated to play a role in microglia-induced neurotoxic injury and the generation of spongiform changes in the brains of mice affected with an experimental prion disease (32). We also observed decreases in expression of several host genes encoding proteins that localize to synaptic nerve terminals and are involved in synaptic function (e.g., endophilin, α-SNAP, ankyrin G, complexin II, and heat shock cognate protein 70), although these decreases were modest and it is difficult to speculate on their biological significance. Nonetheless, in view of previous reports that lymphocytic choriomeningitis virus selectively decreases the transcription of a neuronal gene, GAP-43, involved in synaptic plasticity (7, 13) and that Borna disease virus selectively depresses certain neurotransmitter mRNA levels (42, 43), the decreased expression of these genes in Sindbis virus-infected brains may be of potential relevance to understanding the effects of Sindbis virus infection on neuronal function.
While our findings identify candidate regulators of Sindbis virus neuropathogenesis, the significance of alterations in expression of individual genes is unclear without further investigation, including direct tests of gene function in vivo. First, it is possible that the GeneChip assay produced “false-positive” results. Our RT-PCR and immunohistochemical analyses of IRF-7, MCP-1, and PBR expression provide confirmation of our GeneChip findings by independent methods and argue against this possibility. In addition, we have performed RNase protection assays to detect apoptotic regulatory gene expression in dsTE12Q- and dsTE12H-infected brains from 10-day-old mice and our results are consistent with the findings of our GeneChip analysis; there is increased expression of bfl-1 mRNA (but not other bcl-2 family members) and of TNFRp55 mRNA (C. Johnston and B. Levine, unpublished data). Second, a more major limitation is that one cannot ascertain from data related to the expression level of a gene whether the gene plays a role in viral pathogenesis. For example, higher levels of expression of any individual gene in dsTE12H-infected compared to mock- or dsTE12Q-infected brains could (i) be involved in the increased neuropathology of dsTE12H infection, (ii) represent a more vigorous host protective response induced by higher levels of viral replication, or (iii) have no beneficial or detrimental role in dsTE12H pathogenesis.
To address this latter limitation, we chose to use the Sindbis virus vector system as a functional screen to evaluate a novel candidate regulator of pathogenesis, PBR, which was identified in our GeneChip expression analysis. We selected PBR for further analysis based on several considerations. First, cellular mRNA levels were higher in dsTE12H- than in dsTE12Q-infected brains, raising the possibility that PBR upregulation might either play a role in neuropathology or represent a more vigorous host antiviral response induced by higher levels of viral replication. Second, our immunohistochemical staining confirmed that PBR protein expression was upregulated in virally infected apoptotic neuronal foci in the brains of 10-day-old dsTE12H-infected mice. This enabled us to examine the effects of enforced neuronal PBR expression by a recombinant chimeric Sindbis virus in 1-day-old mice that lack detectable increases in endogenous PBR following Sindbis virus infection. Third, PBR was of particular interest because it is a potentially novel mitochondrial regulator of apoptosis and Sindbis virus-induced apoptosis plays an important role in CNS viral pathogenesis. Therefore, we hypothesized that upregulation of PBR in virally infected neurons may be a host defense mechanism designed to block Sindbis virus-induced apoptosis.
Our findings demonstrate that enforced neuronal PBR expression protects 1-day-old mice against lethal Sindbis virus encephalitis and decreases the number of apoptotic cells in Sindbis virus-infected brains. To our knowledge, our microarray analysis results represent the first demonstration of upregulation of PBR in a viral infection and our studies with the Sindbis virus vector system represent the first demonstration of a protective role for PBR upregulation in CNS disease. PBR expression has been reported to increase in the brain in response to a variety of other, nonviral, neurotoxic insults including excitotoxic injury, ischemia, and chemical sympathetectomy, although in most studies, the increased expression was thought to be in reactive glial cells (reviewed in references 4 and 17). However, PBR is thought to mediate the electrophysiological actions of a PBR agonist on cerebellar Purkinje neurons (3), indicating a functional role for PBR in at least some neuronal populations. The significance of upregulation of PBR expression in CNS injury has been unclear, but increased PBR expression has been postulated to play a role in protecting human hematopoietic cells against oxygen radical damage (8), rat corpora lutea against gonadotropin releasing hormone-induced apoptosis (49), and human epidermal cells against free radical damage from UV exposure (58). The mechanism of protective action of upregulated PBR expression is postulated to involve an increase in the level of mitochondrial membrane cholesterol, which prevents the release of cytochrome c through the mitochondrial permeability membrane transition pore.
The results of our study suggest that the upregulation of PBR may be an important host protective response against Sindbis virus infection, as well as other CNS insults. The apparent failure of 1-day-old mice to increase PBR expression following Sindbis virus infection may contribute to their increased susceptibility to fatal disease, and CNS developmental changes in PBR upregulation may contribute to the resistance of older mice to Sindbis virus-induced neuronal injury. The ability of enforced neuronal expression of PBR, a mitochondrial membrane protein that associates with VDAC and ANT, to reduce the number of apoptotic neurons in Sindbis virus-infected brains suggests that Sindbis virus-induced neuronal apoptosis may be triggered through a mitochondrial pathway. This hypothesis is consistent with the findings of Jan et al. (28) indicating that Sindbis virus-induced apoptosis in neuroblastoma cells involves the release of ceramide, a known inducer of mitochondrial cytochrome c (reviewed in reference 19). Regardless of the mechanism by which PBR exerts its neuroprotective actions, our findings raise the possibility that modulation of this receptor by the administration of previously characterized, selective pharmacologist PBR agonists (reviewed in references 4 and 17) may be beneficial in the treatment of CNS viral diseases.
The identification of PBR as a novel molecule involved in the host protective response to Sindbis virus infection illustrates the potential utility of combining microarray analysis with functional screens to study host genes that regulate viral pathogenesis. Further analysis of additional host genes that were found to have increased expression in Sindbis virus-infected brains will likely yield new insights into the role of specific host responses in CNS alphavirus pathogenesis.
ACKNOWLEDGMENTS
We thank the Microarray Facility of the Albert Einstein College of Medicine for assistance in performing Affymetrix GeneChip expression analysis and James Goldman and Carol Troy for assistance in performing neuropathology analysis.
This work was supported by NIH grants R29A140246 and RO1 AI44157 and an Irma T. Hirschl Trust Scholar Award to B.L.
REFERENCES
- 1.Ascensio V C, Campbell I L. Chemokines in the CNS: plurifunctional mediators in diverse states. Trends Neurosci. 1999;22:504–512. doi: 10.1016/s0166-2236(99)01453-8. [DOI] [PubMed] [Google Scholar]
- 2.Asensio V C, Campbell I L. Chemokine gene expression in the brains of mice with lymphocytic choriomeningitis. J Virol. 1997;71:7832–7840. doi: 10.1128/jvi.71.10.7832-7840.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basile A S, Bolger G T, Lueddens H W, Skolnick P. Electrophysiological actions of Ro5–4864 on cerebellar Purkinje neurons: evidence for “peripheral” benzodiazepine receptor-mediated depression. J Pharmacol Exp Ther. 1989;248:463–469. [PubMed] [Google Scholar]
- 4.Beurdeley-Thoma A, Miccoli L, Oudard S, Dutrillaux B, Poupon M F. The peripheral benzodiazepine receptors: a review. J Neuro-Oncol. 2000;46:45–56. doi: 10.1023/a:1006456715525. [DOI] [PubMed] [Google Scholar]
- 5.Bono F, Lamarche I, Prabonnaud V, LeFur G, Herbert J M. Peripheral benzodiazepine receptor agonists exhibit potent antiapoptotic activities. Biochem Biophys Res Commun. 1999;265:457–461. doi: 10.1006/bbrc.1999.1683. [DOI] [PubMed] [Google Scholar]
- 6.Campbell I L. Cytokines and chemokines in defense and damage in the intact CNS. In: Peterson P K, Remington J S, editors. New concepts in the immunopathogenesis of CNS infections. Oxford, United Kingdom: Blackwell Science; 2000. [Google Scholar]
- 7.Cao W, Oldstone M B, de la Torre J C. Viral persistent infection affects both transcriptional and posttranscriptional regulation of neuron-specific molecule GAP43. Virology. 1997;230:147–154. doi: 10.1006/viro.1997.8458. [DOI] [PubMed] [Google Scholar]
- 8.Carayon P, Portier M, Dussossoy D, Bord A, Petitpretre G, Canat X, LeFur G, Casellas P. Involvement of peripheral benzodiazepine receptors in the protection of hematopoietic cells against oxygen radical damage. Blood. 1996;87:3170–3178. [PubMed] [Google Scholar]
- 9.Chang Y E, Laimans L A. Microarray analysis identified interferon-inducible genes and Stat-1 as major transcriptional targets of human papillomavirus type 31. J Virol. 2000;74:4174–4182. doi: 10.1128/jvi.74.9.4174-4182.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charles P C, Chen X, Horwitz M S, Brosnan C F. Differential chemokine induction by the mouse adenovirus type-1 in the central nervous system of susceptible and resistant strains of mice. J Neurovirol. 1999;5:55–64. doi: 10.3109/13550289909029746. [DOI] [PubMed] [Google Scholar]
- 11.Chen C J, Liao S L, Kuo M D, Wang Y M. Astrocytic alteration induced by Japanese encephalitis virus infection. Neuroreport. 2000;11:1933–1937. doi: 10.1097/00001756-200006260-00025. [DOI] [PubMed] [Google Scholar]
- 12.Conant K, Garzino A, Nath A, McArthur J C, Halliday W, Power C, Gallo R C, Major E O. Induction of monocyte chemoattract protein-1 in HIV-1 Tat-stimulated astrocytes and elevation in AIDS dementia. Proc Natl Acad Sci USA. 1998;95:3117–3121. doi: 10.1073/pnas.95.6.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de la Torre J C, Mallory M, Brot M, Gold L, Koob G, Oldstone M B, Masliah E. Viral persistence in neurons alters synaptic plasticity and cognitive functions without destruction of brain cells. Virology. 1996;220:508–515. doi: 10.1006/viro.1996.0340. [DOI] [PubMed] [Google Scholar]
- 14.Der S D, Zhou A, Williams B R G, Silverman R H. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci USA. 1998;95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Derocg J-M, Jbilo O, Bouaboula M, Sequi M, Clere C, Casellas P. Genomic and functional changes induced by the activation of the peripheral cannabinoid receptor CB2 in the promyelocytic cells HL-60. J Biol Chem. 2000;275:15621–15628. doi: 10.1074/jbc.275.21.15621. [DOI] [PubMed] [Google Scholar]
- 16.Gasque P, Dean Y D, McGreal E P, VanBeek J, Morgan B P. Complement components of the immune system in health and disease in the CNS. Immunopharmacology. 2000;49:171–186. doi: 10.1016/s0162-3109(00)80302-1. [DOI] [PubMed] [Google Scholar]
- 17.Gavish M, Bachman I, Shoukrun R, Katz Y, Veenman L, Weisinger G, Weizman A. Enigma of the peripheral denzodiazepine receptor. Pharmacol Rev. 1991;51:629–650. [PubMed] [Google Scholar]
- 18.Geiss G K, Bumgarner R E, An M C, Agy M B, van ter Wout A B, Hammersmark E, Carter V S, Upchurch D, Mullins J I, Katze M G. Large-scale monitoring of host cell gene expression during HIV-1 infection using cDNA microarrays. Virology. 2000;266:8–16. doi: 10.1006/viro.1999.0044. [DOI] [PubMed] [Google Scholar]
- 18a.Gil M L, Bohn E, O'Guin A K, Ramana C V, Levine B, Stark G R, Virgin H W, Schreiber R D. Biological consequences of Stat1-independent IFN signaling. Proc Natl Acad Sci USA. 2001;98:6680–6685. doi: 10.1073/pnas.111163898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Green D, Reed J. Mitochondria and apoptosis. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 20.Grieder F B, Vogel S N. Role of interferon and interferon regulatory factors in early protection against Venezuelan equine encephalitis virus infection. Virology. 1999;257:106–118. doi: 10.1006/viro.1999.9662. [DOI] [PubMed] [Google Scholar]
- 21.Griffin D E. Alphavirus pathogenesis and immunity. In: Schlesinger S, Schlesinger M J, editors. The Togaviridae and Flaviviridae. New York, N.Y: Plenum Press; 1986. pp. 209–236. [Google Scholar]
- 22.Griffin D E. Molecular pathogenesis of Sindbis virus encephalitis in experimental animals. Adv Virus Res. 1989;36:255–271. doi: 10.1016/s0065-3527(08)60587-4. [DOI] [PubMed] [Google Scholar]
- 23.Hague S J, Williams B R. Signal transduction in the interferon system. Semin Oncol. 1998;25:14–22. [PubMed] [Google Scholar]
- 24.Hardwick J M, Levine B. Sindbis virus vector system for functional analysis of apoptosis regulators. Methods Enzymol. 2000;322:492–508. doi: 10.1016/s0076-6879(00)22045-4. [DOI] [PubMed] [Google Scholar]
- 25.Hirsch R L, Griffin D E, Winkelstein J A. The effect of complement depletion on the course of Sindbis virus infection in mice. J Immunol. 1978;121:1276–1278. [PubMed] [Google Scholar]
- 26.Hirsch R L, Griffin D E, Winkelstein J A. The role of complement in viral infections. II. The clearance of Sindbis virus from the bloodstream and central nervous system of mice depleted of complement. J Infect Dis. 1980;141:212–217. doi: 10.1093/infdis/141.2.212. [DOI] [PubMed] [Google Scholar]
- 27.Horie H, Inagaki Y, Sohma Y, Nozaw R, Okawa K, Hasegawa M, Muramatsu N, Kawano H, Horie M, Koyama H, Sakai I, Takeshita K, Kowada Y, Takano M, Kadova T. Galectin-1 regulates initial axonal growth in peripheral nerves after axotomy. J Neurosci. 1999;19:9964–9974. doi: 10.1523/JNEUROSCI.19-22-09964.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jan J-T, Chatterjee S, Griffin D. Sindbis virus entry into cells triggers apoptosis by activating sphingomyelinase, leading to the release of ceramide. J Virol. 2000;74:6425–6432. doi: 10.1128/jvi.74.14.6425-6432.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joe A, Ferrari G, Jiang H, Levine B. Dominant inhibitory Ras expression delays Sindbis virus-induced apoptosis in neuronal cells. J Virol. 1996;68:7744–7751. doi: 10.1128/jvi.70.11.7744-7751.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson R T, McFarland H F. Age-dependent resistance to viral encephalitis: sutides of infections due to Sindbis virus in mice. J Infect Dis. 1972;125:257–262. doi: 10.1093/infdis/125.3.257. [DOI] [PubMed] [Google Scholar]
- 31.Kerr D A, Nery J P, Traystman R T, Chau B N, Hardwick J M. Survival motor neuron protein modulates neuron-specific apoptosis. Proc Natl Acad Sci USA. 2000;97:13312–13317. doi: 10.1073/pnas.230364197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kopacek J, Sakaguchi S, Shigematsu K, Nishida N, Atarashi R, Nakaoke R, Moriuchi R, Niwa M, Katamine S. Upregulation of the genes encoding lysosomal hydrolases, a perforin-like protein, and peroxidases in the brains of mice affected with an experimental prion disease. J Virol. 2000;74:411–417. doi: 10.1128/jvi.74.1.411-417.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lane T E, Asensio V C, Yu N, Paoletti A D, Campbell I L, Buchmeier M J. Dynamic regulation of alpha- and beta-chemokine expression in the central nervous system during mouse hepatitis virus-induced demyclinating disease. J Immunol. 1998;160:970–978. [PubMed] [Google Scholar]
- 34.Lane T E, Lieu M T, Chen B P, Asensio V C, Samawi R M, Paoletti A D, Campbell I L, Kunkel S L, Fox H S, Buchmeier M J. A central role for CD4+ T cells and RANTES in virus-induced central nervous system inflammation and demyelination. J Virol. 2000;74:1415–1424. doi: 10.1128/jvi.74.3.1415-1424.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee C-K, Klopp R G, Weinruch R, Prolla T A. Gene expression profile of aging and its retardation by caloric restriction. Science. 1999;285:1390–1393. doi: 10.1126/science.285.5432.1390. [DOI] [PubMed] [Google Scholar]
- 36.Levine, B. Apoptosis in viral infections of neurons: a protective or pathologic host response? Curr. Top. Microbiol. Immunol., in press. [DOI] [PubMed]
- 37.Levine B, Goldman J E, Jiang H H, Griffin D E, Hardwick J M. Bcl-2 protects mice against fatal alphavirus encephalitis. Proc Natl Acad Sci USA. 1996;93:4810–4815. doi: 10.1073/pnas.93.10.4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levine B, Griffin D E. Molecular analysis of neurovirulent strains of Sindbis virus that evolve during persistent infection of scid mice. J Virol. 1993;67:6872–6875. doi: 10.1128/jvi.67.11.6872-6875.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lewis J, Oyler G, Ueno K, Fannjiang Y, Chau B, Vornov J, Korsmeyer S, Zou S, Hardwick J. Inhibition of virus-induced neuronal apoptosis by Bax. Nat Med. 1999;5:832–835. doi: 10.1038/10556. [DOI] [PubMed] [Google Scholar]
- 40.Lewis J, Wesslingh S L, Griffin D E, Hardwick J M. Alphavirus-induced apoptosis in mouse brains correlates with neurovirulence. J Virol. 1996;70:1828–1835. doi: 10.1128/jvi.70.3.1828-1835.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang X H, Kleeman L, Jiang H H, Gordon G, Goldman J E, Berry G, Herman B, Levine B. Protection against fatal Sindbis virus encephalitis by Beclin, a novel Bcl-2 interacting protein. J Virol. 1998;72:8586–8596. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lipkin W I, Battenberg E L, Bloom F E, Oldstone M B. Viral infection of neurons can depress neurotransmitter mRNA levels without histologic injury. Brain Res. 1988;451:333–339. doi: 10.1016/0006-8993(88)90779-2. [DOI] [PubMed] [Google Scholar]
- 43.Lipkin W I, Carbone K M, Wilson M C, Duchala C S, Narayan O, Oldstone M B. Neurotransmitter abnormalities in Borna disease. Brain Res. 1988;475:366–370. doi: 10.1016/0006-8993(88)90627-0. [DOI] [PubMed] [Google Scholar]
- 44.Liu Q, Nilsen-Hamiliton M. Identification of a new acute phase protein. J Biol Chem. 1995;70:22565–22570. doi: 10.1074/jbc.270.38.22565. [DOI] [PubMed] [Google Scholar]
- 45.Mamane Y, Heylbroeck C, Genin P, Algarte M, Servant M J, LePage C, DeLuca C, Kwon H, Lin R, Hiscott J. Interferon regulatory factors: the next generation. Gene. 1999;237:1–14. doi: 10.1016/s0378-1119(99)00262-0. [DOI] [PubMed] [Google Scholar]
- 46.Manchester M, Eto D, Oldstone M. Characterization of the inflammatory response during acute measles encephalitis in NSE-CD46 transgenic mice. J Neuroimmunol. 1999;96:207–217. doi: 10.1016/s0165-5728(99)00036-3. [DOI] [PubMed] [Google Scholar]
- 47.McEnery M W, Snowman A M, Trifiletti R R, Snyder S H. Isolation of the mitochondrial benzodiazepine receptor: association with the voltage-dependent anion channel and the adenine nucleotide carrier. Proc Natl Acad Sci USA. 1992;89:3170–3174. doi: 10.1073/pnas.89.8.3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mossman K L, Macgregor P F, Rozmus J J, Goryachev A B, Edwards A M, Smiley J M. Herpes simplex virus triggers and then disarms a host antiviral response. J Virol. 2001;75:750–758. doi: 10.1128/JVI.75.2.750-758.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Papadopoulos V, Dharmarajan A M, Li H, Culty M, Lemay M, Sridaran R. Mitochondrial peripheral-type benzodiazepine receptor expression. Correlation with gonadotropin-releasing hormone (GnRH) agonist-induced apoptosis in the corpus luteum. Biochem Pharmacol. 1999;58:1389–1393. doi: 10.1016/s0006-2952(99)00215-4. [DOI] [PubMed] [Google Scholar]
- 50.Peterson P K, Remington J S, editors. New concepts in the immunopathogenesis of CNS infections. Malden, Mass: Blackwell Science, Inc.; 2000. [Google Scholar]
- 51.Pitha P M, Au W C, Lowther W, Juang Y T, Schafer S L, Burysek L, Hiscott J, Moore P A. Role of the interferon regulatory factors (IRFs) in virus-mediated signaling and regulation of cell growth. Biochemie. 1998;80:651–658. doi: 10.1016/s0300-9084(99)80018-2. [DOI] [PubMed] [Google Scholar]
- 52.Ryman K D, Klimstra W B, Nguyen H B, Biron C A, Johnston R E. Alpha/beta interferon protects adult mice from fatal Sindbis virus infection and is an important determinant of cell and tissue tropism. J Virol. 2000;74:3366–3378. doi: 10.1128/jvi.74.7.3366-3378.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sasseville V G, Smith M M, Mackay C R, Pauley D R, Mansfield K G, Ringler D J, Lackner A A. Chemokine expression in simian immunodeficiency virus-induced AIDS encephalitis. Am J Pathol. 1996;149:1459–1467. [PMC free article] [PubMed] [Google Scholar]
- 54.Sauder C, Hallensleben W, Pagenstecher A, Schneckenburger S, Biro L, Pertlik D, Hausmann J, Suter M, Staeheli P. Chemokine gene expression in astrocytes of Borna disease virus-infected rats and mice in the absence of inflammation. J Virol. 2000;74:9267–9280. doi: 10.1128/jvi.74.19.9267-9280.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schafer M K, Schwaeble W J, Post C, Salvati P, Calabresi M, Sim R B, Petry F, Loos M, Weihe E. Complement C1q is dramatically up-regulated in brain microglia in response to transient global cerebral ischemia. J Immunol. 2000;164:5446–5452. doi: 10.4049/jimmunol.164.10.5446. [DOI] [PubMed] [Google Scholar]
- 56.Sherman L A, Griffin D E. Pathogenesis of encephalitis induced in newborn mice by virulent and avirulent strains of Sindbis virus. J Virol. 1990;64:2041–2046. doi: 10.1128/jvi.64.5.2041-2046.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sotomayor C E, Rabinovich G A. Galectin-1 induces central and peripheral death: implications in T-cell physiology. Dev Immunol. 2000;7:117–129. doi: 10.1155/2000/36321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stoebner P E, Carayon P, Penarier G, Frechin N, Barneon G, Casellas P, Cano J P, Meynadier J, Meunier L. The expression of peripheral benzodiazepine receptors in human skin: the relationship with epidermal cell differentiation. Br J Dermatol. 1999;140:1010–1016. doi: 10.1046/j.1365-2133.1999.02896.x. [DOI] [PubMed] [Google Scholar]
- 59.Tucker P C, Strauss E G, Kuhn R J, Strauss J H, Griffin D E. Viral determinants of age-dependent virulence of Sindbis virus for mice. J Virol. 1993;67:4605–4610. doi: 10.1128/jvi.67.8.4605-4610.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ubol S, Tucker P, Griffin D, Hardwick J. Neurovirulent strains of alphavirus induce apoptosis in bcl 2-expressing cells: role of a single amino acid change in the E2 glycoprotein. Proc Natl Acad Sci USA. 1994;91:5202–5206. doi: 10.1073/pnas.91.11.5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vilcek J. Production of interferon by newborn and adult mice infected with Sindbis virus. Virology. 1964;22:651–652. doi: 10.1016/0042-6822(64)90091-1. [DOI] [PubMed] [Google Scholar]
- 62.Wesselingh S L, Levine B, Fox R J, Choi S, Griffin D E. Intracerebral cytokine mRNA expression during fatal and nonfatal alphavirus encephalitis suggests a predominant type 2 T cell response. J Immunol. 1994;152:1289–1297. [PubMed] [Google Scholar]
- 63.Wodicka L, Dong H, Mittmann M, Ho M H, Lockhart D J. Genome-wide expression monitoring in Saccharomyces cerevisiae. Nat Biotechnol. 1997;15:1359–1367. doi: 10.1038/nbt1297-1359. [DOI] [PubMed] [Google Scholar]
- 64.Zhu H, Cong J-P, Mamtora G, Gineras T, Shenk T. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc Natl Acad Sci USA. 1998;95:14470–14475. doi: 10.1073/pnas.95.24.14470. [DOI] [PMC free article] [PubMed] [Google Scholar]