Abstract
Here we demonstrate replication of human herpesvirus 7 (HHV-7), a T-lymphotropic virus, in macrophages. Productive replication was lost after 2 weeks, but HHV-7 DNA was detected up to 1 month after infection. Thus, macrophages become infected by HHV-7 and might play an important role as a viral reservoir, as has been demonstrated for human immunodeficiency virus type 1.
Human herpesvirus 7 (HHV-7) was first isolated in 1990 from peripheral blood mononuclear cells (PBMC), under conditions required to promote T-cell activation, from a healthy individual and later on also from a patient with chronic fatigue syndrome (3, 5). HHV-7 utilizes CD4 as its main receptor to enter the target cell and has also been shown to interfere with human immunodeficiency virus type 1 infection in CD4+ T cells (6, 11). So far, only CD4+ T lymphocytes (isolated from PBMC and cord blood mononuclear cells) and SupT1 cells, immature T-cells derived from a non-Hodgkin's lymphoma, have proved sensitive to HHV-7 infection (2, 12). Lack of productive HHV-7 infection in CD4+ macrophages was observed by quantitative PCR and immunofluorescence assays to detect HHV-7 antigen in vitro (4). It has also been demonstrated that HHV-7 antigen is expressed in macrophages/monocytes of AIDS-associated Kaposi's sarcoma samples (8). Another, more recent study has shown that in lymph nodes of AIDS patients HHV-7 is mainly present in macrophages (9). This suggests that macrophages can be infected with HHV-7 and may be a site of latency and reactivation of HHV-7 in vivo.
In the present study, we evaluated HHV-7 infection of macrophages in vitro. Our results suggest that HHV-7 can infect macrophages and that this infection may persist in these cells for a long time. Macrophages were isolated by density gradient centrifugation from PBMC from healthy blood donors and further enriched, as described in detail elsewhere (1). More than 99% of the cells were CD14+ CD4+ CD3−, and no HHV-7 DNA was detected by PCR in these cell cultures. The stock of HHV-7 (strain KHR) was grown in CD8-depleted PBMC, as described previously (13). The CD4+ T-cell line SupT1, the CD4− cell line HSB-2, the CD4+ cell line MT-2, and the monocytic CD4+ THP-1 cell line were purchased from the American Type Culture Collection (Rockville, Md.).
To study the HHV-7 infection in macrophages, the cells were inoculated with virus stock for 4 h and then extensively washed. The SupT1, HSB-2, MT-2, and THP-1 cell lines were also used to determine HHV-7 infection. The mock-infected and HHV-7-infected macrophage cultures were evaluated regularly by light microscopy. PCR, reverse transcription (RT)-PCR, and electron microscopy were also used to detect HHV-7 DNA, mRNA, and HHV-7 particles.
DNA was isolated from mock-infected and HHV-7-infected macrophages at different time points after HHV-7 infection. Briefly, the cells were washed extensively with phosphate-buffered saline and collected, and total DNA of the cell pellet was isolated using QIAamp DNA blood Mini Kit (Qiagen, Hilden, Germany) following the protocol described in the product manual. The following primer pairs derived from U10, an HHV-7 α gene, were used: HV7, 5′-TATCCCAGCTGTTTTCATATAGTAAC-3′, and HV8, 5′-GCCTTGCGGTAGCACTAGATTTTTTG-3′. These oligonucleotide primers were proven to amplify only HHV-7 DNA and not DNA from other human herpesviruses (2). The hot-start PCR was carried out in a 25-μl reaction volume containing 1 μg of total DNA, 1.5 mM MgCl2, 0.5 μM concentrations of each primer, 100 μM concentrations of each deoxynucleoside triphosphate (Gibco BRL), and 1 U of SuperTaq DNA polymerase (HT Biotechnologies, Cambridge, United Kingdom). Thirty-five amplification cycles were used with the following cycling parameters: 94°C for 1 min, 57.5°C for 1 min, and 72°C for 1 min, with an increase of 3 s per cycle and an additional final extension at 72°C for 5 min. The resulting amplified fragment had a size of 186 bp and was analyzed using agarose gel electrophoresis and ethidium bromide staining.
In order to detect the productive HHV-7 infection in macrophages, total RNA of mock-infected and HHV-7-infected macrophages and SupT1 cells was isolated at day 4, day 7, day 14, and day 21 after infection using the RNeasy Mini kit (Qiagen). RNA concentrations were determined spectrophotometrically. DNA contamination was eliminated by digestion with 50 U of RNase-free DNase I (Roche, Mannheim, Germany) at room temperature for 30 min. Residual DNase activity was eliminated by adding 2.5 mM EDTA and heating at 65°C for 10 min. Reverse transcription was carried out in a 50-μl reaction volume, containing 10 μl of buffer, 250 μM concentrations of each dNTP, 2 μg of total RNA, 0.5 μg of reverse primer, 130 U of human placental RNase inhibitor (Amersham), and 1.8 U of RAV-2 reverse transcriptase (Amersham), and the reaction was carried out for 90 min at 45°C. Five microliters of cDNA thus obtained was used to perform hot-start PCR as described above.
β-Actin DNA and cDNA levels were measured as an internal standard and were found to be comparable for all samples (data not shown). In order to determine the sensitivity of the PCR, the amplicon was cloned into a T-vector (Invitrogen). After 35 amplification cycles, 1,000 copies of the HHV-7 U10 DNA fragment were detected (data not shown).
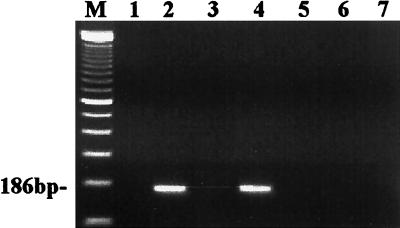
The PCR results of all the cell lines are shown in Fig. 1. As shown in this figure, neither HHV-7 DNA nor cDNA was detected in mock-infected SupT1 cells (Fig. 1, lanes 1 and 3). HHV-7 DNA and cDNA from the HHV-7-infected SupT1 cells could be detected at day 7 after infection (Fig. 1, lanes 2 and 4). This points to a productive HHV-7 infection in SupT1 cells. In contrast, no HHV-7 DNA could be detected in DNA extracts from the CD4− cell line HSB-2, the CD4+ cell line MT-2, and the monocytic cell line THP-1 at day 7 after HHV-7 infection (Fig. 1, lanes 5, 6, and 7). Thus, HSB-2, MT-2, and THP-1 cells could not become infected with HHV-7.
FIG. 1.
Agarose gel (ethidium bromide stained) showing the results of PCR and RT-PCR amplification of HHV-7 DNA and mRNA, obtained from mock- and HHV-7-infected cell lines at day 7 postinfection. A 35-cycle PCR amplification was performed on a 186-bp fragment from the U10 gene of HHV-7. Lane M, DNA size marker (100-bp ladder); lane 1, cDNA from mock-infected SupT1 cells; lane 2, cDNA from HHV-7-infected SupT1 cells at day 7 postinfection; lane 3, DNA from mock-infected SupT1 cells; lanes 4 to 7, DNA from HHV-7-infected SupT1 cells, HSB-2 cells, MT-2 cells, and THP-1 cells, respectively, at day 7 postinfection.
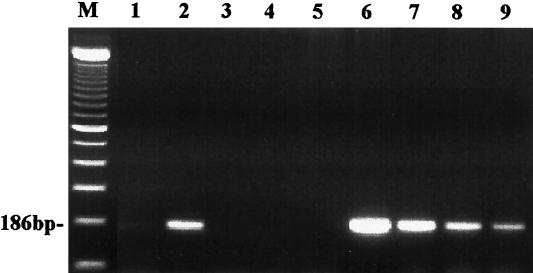
Then, by RT-PCR we analyzed RNA extracts from mock-infected and HHV-7-infected macrophages for the presence of HHV-7 transcription. No HHV-7 cDNA was detected in mock-infected macrophages at day 7 (Fig. 2, lane 1), whereas HHV-7 cDNA from HHV-7-infected macrophages at day 7 was clearly positive (Fig. 2; lane 2). This suggests that macrophages were able to produce new virus after HHV-7 infection. However, at days 14 and 21 postinfection, cDNA could no longer be detected by RT-PCR (Fig. 2, lanes 3 and 4). In parallel, DNA extracts were analyzed by PCR. No HHV-7 DNA was found in mock-infected macrophages (Fig. 2, lane 5). However, the DNA extracts from the HHV-7-infected macrophages from day 7 to day 30 were all positive by PCR (Fig. 2, lanes 6 to 9). This suggests that the viral DNA was maintained in macrophages for at least 30 days. The mock-infected macrophages remained always negative by PCR. Furthermore, all the DNA extracts from HHV-7-infected macrophages, which were isolated from several independent experiments, were always positive for HHV-7 DNA. This supports the notion of a latent HHV-7 infection in macrophages.
FIG. 2.
PCR and RT-PCR amplification of HHV-7 DNA and mRNA, isolated from mock-infected and HHV-7-infected macrophages. Lane M, DNA size marker (100-bp ladder); lane 1, cDNA from the mock-infected macrophages at day 7 postinfection; lanes 2 to 4, cDNA from the HHV-7-infected macrophages at day 7, day 14, and day 21, respectively, after HHV-7 infection; lane 5, DNA from the mock-infected macrophages at day 7; lanes 6 to 9, DNA from the HHV-7-infected macrophages at day 7, day 14, day 21, and day 30, respectively, postinfection.
The productive HHV-7 infection in macrophages was also detected by electron microscopy. The mock-infected and HHV-7-infected macrophages were detached, and the cell pellet was fixed in 2% glutaraldehyde for 15 min, pelleted, and resuspended in 2.5% glutaraldehyde in 0.1 M phosphate buffer (pH 7.2) at 4°C. The cells were centrifuged again and postfixed in 1% osmium tetroxide in 0.1 M phosphate buffer. The samples were embedded in epoxy resin, and 50-nm sections were cut, collected on copper grids, stained with uranyl acetate and lead citrate, and examined with a Philips CM10 electron microscope operated at 60 kV.
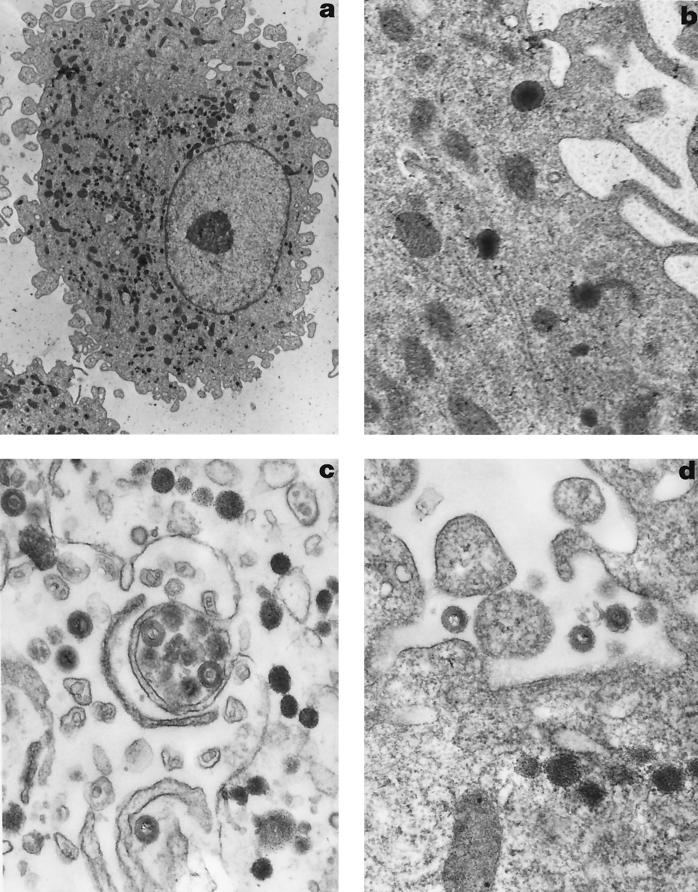
This electron microscopic analysis revealed the presence of HHV-7 viral particles in HHV-7-infected macrophages at day 7 postinfection (Fig. 3a) but not in mock-infected macrophages (data not shown). The mature HHV-7 particles were visible in the cytoplasm (Fig. 3b) and extracellular space at day 7 postinfection. Also, at day 9 postinfection, the mature virus particles were found in the cytoplasm and extracellular space (Fig. 3c and d). The mature HHV-7 virions were about 185 to 205 nm in diameter and resembled the virions in HHV-7-infected SupT1 cells described by Klussmann et al. (11). This reveals productive HHV-7 infection in macrophages. At day 12 postinfection, no virion particles were detected in the cytoplasm of HHV-7-infected macrophages anymore (data not shown).
FIG. 3.
Electron microscopic appearance of HHV-7-infected macrophages. (a) HHV-7-infected macrophages at day 7 postinfection (magnification, ×5,359). (b) HHV-7 virions in the cytoplasm at day 7 postinfection (magnification, ×31,935). (c) HHV-7 virus particles in the cytoplasm and extracellular space at day 9 postinfection (magnification, ×39,760). (d) Mature HHV-7 particles in the extracellular space at day 9 postinfection (magnification, ×39,760).
To confirm that productive HHV-7 replication had occurred in macrophages, we also tried to isolate the HHV-7 from the supernatant of HHV-7-infected macrophages. When cell-free supernatant from the HHV-7-infected macrophages was harvested at different time points (day 1 until day 30 postinfection) and inoculated on SupT1 cells, only the supernatant collected from days 5 to 10 (from eight different experiments) yielded infection in the SupT1 cells, as assessed by the appearance of cytopathic effect and HHV-7 antigen expression (data not shown). The supernatant harvested at day 1, day 14, day 21, and day 30 postinfection was not able to infect the SupT1 cells. These observations correspond to the results shown in Fig. 2 and 3. This demonstrates that infective HHV-7 particles were present in the supernatant of HHV-7-infected macrophage cultures at a given time point (7 days) but decreased thereafter.
HHV-7 is a T-lymphotropic virus which has been shown to replicate in CD4+ T lymphocytes and in the SupT1 cell line in vitro (3, 11). In vivo studies have shown that HHV-7 antigen can be found in a number of tissues, such as lung, skin, and mammary gland (7), and the salivary gland is proposed to be a major site for virus replication (10). A more recent study further showed that in AIDS patients the presence of HHV-7 in lymph nodes is mainly restricted to macrophages (9). Although a previous study could not find evidence for HHV-7 replication in macrophages in vitro (4), our findings clearly indicate that HHV-7 is able to replicate in macrophages. First, we showed that all the DNA extracts taken at different time points from HHV-7-infected macrophages were positive for HHV-7 DNA. Even 1 month after HHV-7 infection, HHV-7 DNA could be detected in the HHV-7-infected macrophages. This suggests that HHV-7 can enter the macrophages and may persist there in a latent state. Second, RNA extracts from HHV-7-infected macrophages taken at day 7 postinfection were positive for HHV-7 by RT-PCR, whereas the RNA extracts taken at day 14 and later were negative. This suggests that the productive HHV-7 infection lasted only for about 1 week. Third, HHV-7-infected macrophages produced infectious HHV-7, as demonstrated by their infectivity in SupT1 cells. The fact that no HHV-7 could be isolated from the supernatant at day 1 after HHV-7 infection indicates that the virus was derived from newly produced HHV-7 particles. Finally, electron microscopic analysis clearly revealed the presence of mature virus particles in the cytoplasm and extracellular space, thus indicating a productive infection.
In conclusion, we have demonstrated that, in addition to CD4+ T lymphocytes, macrophages can also be infected by HHV-7. Productive HHV-7 infection in macrophages occurs during 1 to 2 weeks after HHV-7 inoculation, but HHV-7 DNA can still be detected for 1 month. Thus, HHV-7 may remain in a latent state in macrophages, and if occurring in vivo these macrophages may serve a role as viral reservoir.
Acknowledgments
We thank Sandra Claes, Erik Fonteyn, Chris Armee, Rolande Renwart, and Michel Rooseleer for excellent technical assistance and K. Yamanishi for kindly providing the KHR strain of HHV-7.
This work was supported by the Fonds voor Wetenschappelijk Onderzoek (FWO) Vlaanderen (Krediet no. G.0104.98) and the Geconcerteerde Onderzoekacties Vlaamse Gemeenschap (project no. 2000/12).
REFERENCES
- 1.Aquaro S, Menten P, Struyf S, Proost P, Van Damme J, De Clercq E, Schols D. The LD78β isoform of MIP-1α is the most potent CC-chemokine in inhibiting CCR5-dependent human immunodeficiency virus type 1 replication in human macrophages. J Virol. 2001;75:4402–4406. doi: 10.1128/JVI.75.9.4402-4406.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berneman Z N, Ablashi D V, Li G, Eger-Fletcher M, Reitz M S, Jr, Hung C L, Brus I, Komaroff A L, Gallo R C. Human herpesvirus 7 is a T-lymphotropic virus and is related to, but significantly different from, human herpesvirus 6 and human cytomegalovirus. Proc Natl Acad Sci USA. 1992;89:10552–10556. doi: 10.1073/pnas.89.21.10552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berneman Z N, Gallo R C, Ablashi D V, Frenkel N, Katsafanas G, Kramarsky B, Brus I. Human herpesvirus 7 (HHV-7) strain JI: independent confirmation of HHV-7. J Infect Dis. 1992;166:690–691. doi: 10.1093/infdis/166.3.690. [DOI] [PubMed] [Google Scholar]
- 4.Crowley R W, Secchiero P, Zella D, Cara A, Gallo R C, Lusso P. Interference between human herpesvirus 7 and HIV-1 in mononuclear phagocytes. J Immunol. 1996;156:2004–2008. [PubMed] [Google Scholar]
- 5.Frenkel N, Schirmer E C, Wyatt L S, Katsafanas G, Roffman E, Danovich R M, June C H. Isolation of a new herpesvirus from human CD4+ T cells. Proc Natl Acad Sci USA. 1990;87:748–752. doi: 10.1073/pnas.87.2.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furukawa M, Yasukawa M, Yakushijin Y, Fujita S. Distinct effects of human herpesvirus 6 and human herpesvirus 7 on surface molecule expression and function of CD4+ T cells. J Immunol. 1994;152:5768–5775. [PubMed] [Google Scholar]
- 7.Kempf W, Adams V, Mirandola P, Menotti L, Di Luca D, Wey N, Muller B, Campadelli-Fiume G. Persistence of human herpesvirus 7 in normal tissues detected by expression of a structural antigen. J Infect Dis. 1998;178:841–845. doi: 10.1086/515339. [DOI] [PubMed] [Google Scholar]
- 8.Kempf W, Adams V, Wey N, Moos R, Schmid M, Avitabile E, Campadelli-Fiume G. CD68+ cells of monocyte/macrophage lineage in the environment of AIDS-associated and classic-sporadic Kaposi sarcoma are singly or doubly infected with human herpesviruses 7 and 6B. Proc Natl Acad Sci USA. 1997;94:7600–7605. doi: 10.1073/pnas.94.14.7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kempf W, Muller B, Maurer R, Adams V, Campadelli Fiume G. Increased expression of human herpesvirus 7 in lymphoid organs of AIDS patients. J Clin Virol. 2000;16:193–201. doi: 10.1016/s1386-6532(99)00083-9. [DOI] [PubMed] [Google Scholar]
- 10.Kidd I M, Clark D A, Ait-Khaled M, Griffiths P D, Emery V C. Measurement of human herpesvirus 7 load in peripheral blood and saliva of healthy subjects by quantitative polymerase chain reaction. J Infect Dis. 1996;174:396–401. doi: 10.1093/infdis/174.2.396. [DOI] [PubMed] [Google Scholar]
- 11.Klussmann J P, Krueger E, Sloots T, Berneman Z, Arnold G, Krueger G R F. Ultrastructural study of human herpesvirus-7 replication in tissue culture. Virchows Arch. 1997;430:417–426. doi: 10.1007/s004280050051. [DOI] [PubMed] [Google Scholar]
- 12.Lusso P, Secchiero P, Crowley R W, Garzino-Demo A, Berneman Z N, Gallo R C. CD4 is a critical component of the receptor for human herpesvirus 7: interference with human immunodeficiency virus. Proc Natl Acad Sci USA. 1994;91:3872–3876. doi: 10.1073/pnas.91.9.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, Schols D, De Clercq E. Selective activity of various antiviral compounds against HHV-7 infection. Antiviral Res. 1999;43:23–35. doi: 10.1016/s0166-3542(99)00031-5. [DOI] [PubMed] [Google Scholar]