Summary
This study aims to characterize dysregulation of phosphorylation for the 5XFAD mouse model of Alzheimer disease (AD). Employing global phosphoproteome measurements, we analyze temporal (3, 6, and 9 months) and sex-dependent effects on mouse hippocampus tissue to unveil molecular signatures associated with AD initiation and progression. Our findings reveal consistent phosphorylation of known AD biomarkers APOE and GFAP in 5XFAD mice, alongside candidates BIG3, CLCN6, and STX7, suggesting their potential as biomarkers for AD pathology. In addition, we identify PDK1 as a significantly dysregulated kinase at 9 months in females, and the regulation of gap junction activity as a key pathway associated with Alzheimer disease across all time points. AD-Xplorer, the interactive browser of our dataset, enables exploration of AD-related changes in phosphorylation, protein expression, kinase activities, and pathways. AD-Xplorer aids in biomarker discovery and therapeutic target identification, emphasizing temporal and sex-specific nature of significant phosphoproteomic signatures. Available at: https://yilmazs.shinyapps.io/ADXplorer.
Subject areas: molecular biology, Neuroscience, cell biology
Graphical abstract
Highlights
-
•
Phosphorylation-level dysregulation surpasses protein expression
-
•
Higher phospho-dysregulation in females, starting as early as 3-month time point
-
•
Candidate biomarkers BIG3, CLCN6, and STX7 exhibit consistent phospho-dysregulation
-
•
Developed AD-Xplorer: online tool to explore Alzheimer disease phosphoproteome
Molecular biology; Neuroscience; Cell biology
Introduction
Alzheimer disease (AD), a major cause of dementia, exhibits a high degree of heterogeneity and complexity in terms of risk factors, progression, and response to treatment.1,2,3,4 However, the conventional postmortem diagnosis of AD based on histopathological examination of amyloid-beta (Aβ) has limitations, as abnormal Aβ deposition occurs before neurodegeneration and cognitive decline.5,6 Despite these limitations, most proteome-wide investigations in the literature have focused on postmortem brain samples, which cannot capture the dynamic molecular changes occurring during the progression of AD.2,3 Elucidating the molecular mechanisms underlying abnormal Aβ deposition, as well as the timing and arrangement of the initiating and propagating pathways, could provide valuable insights into biomarkers and potential intervention strategies for predicting and managing AD progression in individual patients.7
The regulation of AD progression occurs at various levels, including DNA, RNA, and protein. The intricate nature of AD regulation is exemplified by transcriptomic and proteomic studies, which have shown a correlation of 0.45 between mRNA and protein levels.8 This suggests that these omics data are not individually sufficient to fully understand the important signaling events in AD. Besides translational control, post-translational modifications (PTMs), such as phosphorylation, also play a crucial role in determining protein levels and variations.9 Although abnormal protein phosphorylation in AD has been well documented10 and Cdk5-tau hyperphosphorylation axis is a hallmark of AD,11 there is a lack of comprehensive studies investigating PTMs at a proteome-wide scale in brain tissue. Therefore, the exploration of phosphoproteomics holds significant potential in providing additional insights and identifying further pathways implicated in the progression of AD.12
Mouse models offer an excellent opportunity to study the temporal aspects of AD progression. For instance, a longitudinal study using a well-established amyloid-driven AD model (5XFAD mice) analyzed the perfusates from the hippocampus and revealed dysregulation in glucose and lipid metabolism prior to AD pathogenesis, demonstrating the feasibility of identifying early molecular events in disease progression.13 Another study characterized 5- and 10-month-old 5XFAD and detected an astonishing 1,411 DEPs, of which vitamin-K-dependent protein S (PROS1) was identified as a candidate biomarker.14 Our previous study15,16 utilized the same set of 5XFAD mice to better understand the temporal variation in protein expression in mouse brain for wild-type (WT) compared to the AD mouse model at various time points relevant to the key hallmarks of disease progression in humans.
In this study, we expand on our previous work16 and characterize the phosphoproteomic changes in 5XFAD mice at different time points relevant to the key hallmarks of disease progression in humans. The 5XFAD mice express five familial AD mutations (amyloid precursor protein and presenilin 1 genes), leading to the accumulation of amyloidogenic Aβ42 and a cascade of pathological changes that closely resemble human AD. Notably, these mice exhibit a significant increase in Aβ42 levels, resulting in the early onset of plaque deposition (around 2 months of age) that progressively spreads throughout the brain along with astrocytosis and microgliosis (around 4 months of age). Subsequently, neuronal loss occurs in the cortex and subiculum at around 9 months of age. It is worth mentioning that, unlike the known human pathology, the Aβ plaques in the 5XFAD mouse model trigger the formation of neuritic tau plaque aggregates rather than neurofibrillary tangles, which are a crucial neuropathological hallmark of AD.17,18 Nevertheless, due to the overall similarities observed with human pathology, we designed a comprehensive study to explore the temporal and sex-related variations in protein phosphorylation, aiming to uncover new insights into known biomarkers and potentially identify targets for AD. In addition, we provide the interactive data mining tool AD-Xplorer with this manuscript, which is poised to become a versatile resource widely used by the AD research community.
Results
Experimental design
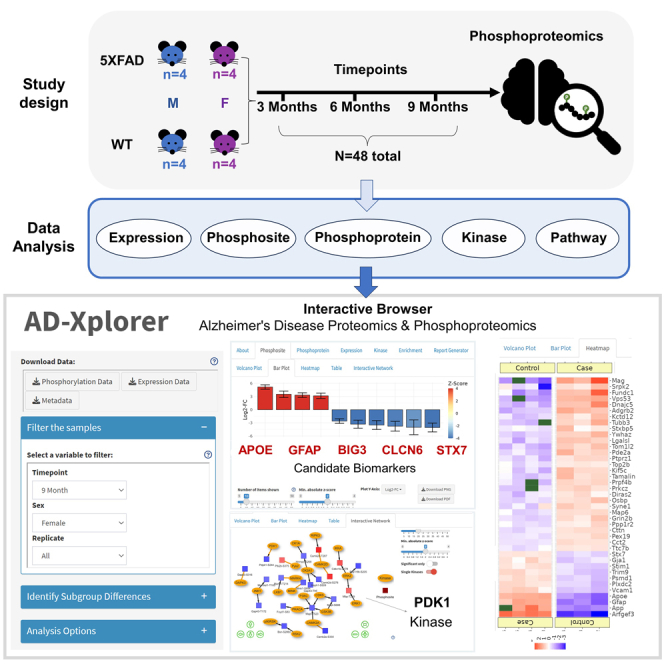
The study aimed to monitor the global phosphoproteome changes in the hippocampus of 5XFAD mice throughout different stages of AD progression. We chose to characterize the hippocampus due to its involvement in cognitive function, memory, and neurogenesis.19,20 Furthermore, the hippocampus is known for its early involvement in AD, characterized by its degeneration and abnormal protein accumulation in AD patients,21,22 making it an interesting tissue for us to probe. To achieve this, label-free liquid chromatography-tandem mass spectrometry (LC-MS/MS) was employed. Due to our complex experimental design, we made the decision of processing samples quickly in a fresh state comparable at specific time points. This made label-free the most convenient choice, as it does not require additional steps during sample preparation and expedites the analysis process as opposed to other approaches such as tandem mass tag (TMT).
A total of 48 samples were analyzed, comprising sets of 16 male and female 5XFAD mice, as well as wild-type (WT) mice, which were run contemporaneously. The hippocampi were collected and processed at three time points: 3, 6, and 9 months. Each time point consisted of eight samples for 5XFAD mice and eight samples for WT mice. Furthermore, within each group, the samples were subdivided based on sex, resulting in four samples per subgroup (Data S1; Figure S1). This comprehensive approach allowed for the investigation of the phosphoproteome changes in the hippocampus of male and female 5XFAD mice and male and female WT mice at each time point.
In our study, we conducted several analyses to gain a comprehensive understanding of the phosphoproteome changes in the hippocampus of 5XFAD mice during the progression of AD. First, we characterized phosphorylation levels as a function of phenotype and compared them to protein expression levels to identify any correlations and to assess the complementarity between these two measures. Next, we investigated the temporal and sex-linked patterns in phosphorylation with a statistical analysis to identify specific dysregulated phosphopeptides between the WT and 5XFAD mice groups, revealing distinct molecular signatures associated with AD progression. In addition, we performed a biomarker analysis, identifying proteins that are consistently phosphorylated across different time points that could potentially serve as markers for AD. For these identified proteins, we performed a computational prevalence analysis to quantify the stoichiometry of phosphorylation. In addition, we conducted kinase inference analysis to gain insights into the regulatory mechanisms involved in phosphorylation events. Finally, a pathway analysis was conducted to identify the biological pathways and networks impacted by the observed phosphoproteome changes. Together, these analyses provided a multi-faceted approach to uncovering the complex dynamics of phosphorylation and its implications in AD progression.
We validated our findings using independent validation data with a smaller number of samples, applying the same quantification method and statistical pipeline described. To enable interactive exploration of our findings and datasets, we developed an online tool named AD-Xplorer, which is available at https://yilmazs.shinyapps.io/ADXplorer.
Exploring complementarity: Dysregulation in phosphorylation vs. protein expression
We started our investigation by analyzing the trends in peptides identified through phosphorylation and comparing them with peptides identified from protein expression data of a previous study on the same set of mice.15 To ensure consistency, we applied the same thresholds that were used in the prior protein expression analysis. Specifically, we used 0.5 ≥ FC ≥ 2 and p ≤ 0.1 thresholds to screen phosphopeptides for differential phosphorylation, comparing 5XFAD mice with WT mice. Our initial observations revealed a gradual increase in peptides identified based on protein expression as time progressed (Figure S2). Similar trends were observed in the number of peptides identified through phosphorylation, with a slight decrease at the 6-month time point. Notably, although protein expression yielded a higher number of peptides that pass the cutoffs (around 2–5 times more), this is mainly due to higher numbers of peptides in the protein expression data (around 4–8 times more). However, when considering the percentage of peptides that satisfy the screening criteria relative to all peptides, phosphorylation exhibited higher prevalence compared to protein expression. In mixed-sex analysis, phosphorylation resulted in 2.26%, 1.96%, and 2.56% of peptides identified for 3-, 6-, and 9-month time points, whereas protein expression resulted in 0.51%, 0.73%, and 1.36%. This corresponds to 4.4×, 2.7×, and 1.9× higher phosphorylation prevalence compared to protein expression.
Note that our goal in this analysis is not to characterize the relationship between protein expression and phosphorylation at the peptide or individual protein level. Instead, we aim to evaluate the extent of dysregulation in protein expression vs. phosphorylation in AD and examine any correspondence between these changes. To achieve this, we identified proteins with differential expression and phosphorylation separately, comparing the numbers and overlap of these proteins.
Additionally, our sex-specific analysis showed a notably higher percentage of differentially phosphorylated peptides that pass the cutoffs in females for 3 and 9 months (6.67% and 6.56% respectively) and 1.85% for 6 months. These percentages correspond to 4.4×, 1.7×, and 5.2× higher prevalence of phosphorylation in females compared to protein expression and 4.7×, 1.0×, and 4.4× higher prevalence compared to males, respectively for 3-, 6-, and 9-month time points. These findings suggest that phosphorylation undergoes significant changes in the 5XFAD hippocampus at an early stage, potentially surpassing protein expression changes in magnitude.
Next, to assess the complementarity of phosphorylation information to protein expression, we examined their correlation at each time point. Comparing phosphorylation at the peptide-level and protein expression, we observed only a minor correlation (R2 = 0.09, 0.02, and 0.12 for 3, 6, and 9 months; Figure S3). However, when comparing protein expression with phosphorylation at the protein level (mean phosphorylation), we found a higher, yet still modest, correlation for all time points (R2 = 0.20, 0.07, and 0.27 for 3, 6, and 9 months). On the other hand, as a reference point, investigating the correlation between phosphorylation at the phosphosite level and phosphorylation at the protein level (mean phosphorylation), we observed a much stronger correlation (R2 = 0.56, 0.44, and 0.56 for 3, 6, and 9 months). These findings highlight that changes in phosphorylation and protein expression are not redundant but rather complementary to each other, as the lack of a strong correlation indicates they both provide distinct and valuable information.
Temporal dynamics: Phosphorylation profile of 5XFAD mouse model over time
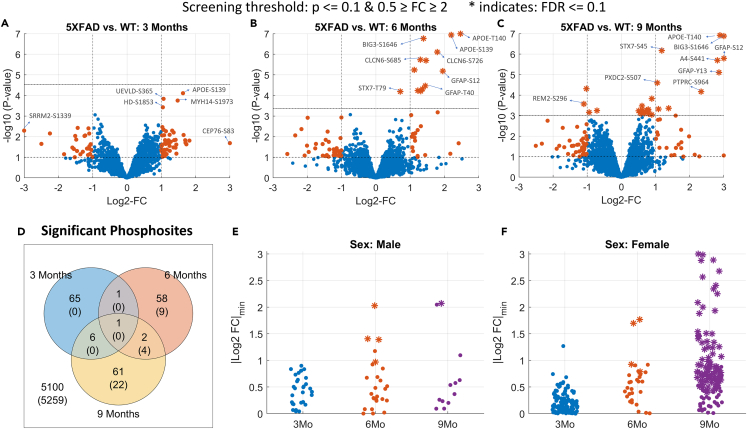
After the filtering and quality control steps, we identify 3,287; 3,198; and 2,748 phosphopeptides in the 3-month, 6-month, and 9-month datasets, respectively, all contributing to a total of hippocampal coverage of 1,782 phosphorylated proteins. The volcano plots for each dataset highlight a dramatic accumulation of differentially phosphorylated peptides at the 6-month time (screening threshold: 5XFAD/WT 0.5 ≥ FC ≥ 2 and p ≤ 0.1; Figures 1A–1C). Respectively in the 3-, 6-, and 9-month time points, 48, 28, and 30 phosphopeptides were upregulated, whereas 25, 34, and 40 phosphopeptides were downregulated. Thus, half of these differentially phosphorylated peptides are upregulated in the 5XFAD mice.
Figure 1.
Phosphosites identified as differentially phosphorylated for Alzheimer disease 5XFAD model at different time points
(A–C) Volcano plots indicating the screened phosphosites with high or low phosphorylation at 3-/6-/9-month time points. The dashed horizontal lines in each plot indicate the statistical significance threshold (p < 0.1 and FDR ≤ 0.1) and the vertical line the threshold on fold changes (>2). The statistically significant phosphosites at FDR ≤ 0.1 level are marked as stars (∗). The top differentially expressed phosphosites are highlighted in each panel.
(D) Venn diagram showing the number of phosphosites that pass the screening (p ≤ 0.1 and 0.5 ≥ FC ≥ 2) for 3/6/9 months data. Whereas, the numbers in parentheses indicate the significant ones at FDR ≤ 0.1 level.
(E and F) Identified phosphosites that pass the screening in sex-specific analysis. The y axis indicates a lower-bound of the 95% confidence intervals for each phosphosite.
To characterize the temporal persistence of these phosphorylation signatures, we analyzed the overlap among the three time points. The Venn diagram highlights the transitory nature of phosphorylation signaling in 5XFAD mice, with less than 5% of all phosphopeptides being dysregulated in two or more time points (Figure 1D). Indeed, only glial fibrillary acidic protein (GFAP) passed the screening thresholds across all three time points, exhibiting consistent hyperphosphorylation across time. This complements the proteome changes of the same set of mice that found GFAP protein expression to be significantly upregulated at all time points.15 This is expected, as GFAP is a glial and endothelial cell phenotypic marker and doubles as a classical astrogliosis marker,23,24 which in turn plays a central role in AD neuroinflammation.25,26
Sex differences in frequency, disease burden, and risk factors for AD are major long-standing questions in the field. Here, we exploited our sex-balanced experimental design to characterize the sex-dependent variations in phosphorylation across AD progression. We identified the differentially phosphorylated peptides separately for female and male groups at each time point (5XFAD male vs. WT male, and 5XFAD female vs. WT female, p ≤ 0.1 and 0.5≥FC ≥ 2). Male mice exhibit a consistent phosphopeptide signature throughout all time points, respectively 33, 38, and 23 differentially phosphorylated peptides at 3, 6, and 9 months (Figure 1E). In contrast, female 5XFAD mice display a distinct phosphopeptide profile with 169, 36, and 126 phosphopeptides at 3, 6, and 9 months, respectively (Figure 1F). In total, the number of differentially phosphorylated peptides identified for female mice is more than three times greater than that for male mice (Figure S4).
Temporal and sex-specific analysis: Phosphopeptide discoveries in 5XFAD mouse model
To control for false discoveries, we employed a moderated t test and applied the Benjamini-Hochberg procedure with a false discovery rate (FDR) threshold of 0.1. We imposed this significance threshold to identify individual phosphopeptides that showed significant dysregulation in each of the 3-/6-/9-month time points separately. In Figure 1, the phosphopeptides that met this significance threshold (FDR ≤ 0.1) are denoted by asterisks in panels A to C. Our analysis revealed an increasing number of dysregulated phosphopeptides identified at 3, 6, and 9 months, specifically 0, 13, and 26, respectively (Figure 1D). Among these dysregulated phosphopeptides, four were found to be common between 6- and 9-month time points: apolipoprotein E (APOE)-T140, brefeldin-A-inhibited guanine-nucleotide-exchange protein 3 (BIG3)-S1646, syntaxin 7 (STX7)-T79, and GFAP-S12 (Figures S5 and S6).
Next, among the phosphopeptides that we identify (FDR ≤ 0.1), we investigated which of these also met the fold-change threshold (5XFAD/WT 0.5 ≥ FC ≥ 2). We observed a total of 12 phosphopeptides for the 6-month time point and 12 phosphopeptides for the 9-month time point. Notable ones among these include APOE-S139, chloride voltage-gated channel 6 (CLCN6)-S726 and S685, and GFAP-T40, which exhibit significant upregulation exclusively in the 6-month time point, whereas CLCN6-S774, STX7-S45, GFAP-Y13, and amyloid-beta precursor protein (A4)-S441 are among phosphopeptides exhibiting significant upregulation only in the 9-month time point. Overall, we observe two phosphopeptides that are identified as significant in both 6- and 9-month time points, which are BIG3-S1646 and GFAP-S12. Apart from the known AD hallmarks, we detect the upregulation of STX7 and CLCN6, which implies the involvement of AD in the dysregulation of the late endocytic pathway.27 For a comprehensive list of the phosphopeptides identified as significant in this analysis, please refer to Data S2.
When we explore the sex-specific differences, we observe respectively 0, 4, and 142 phosphopeptides for females and 0, 4, and 1 phosphopeptides for males that pass the significance threshold (FDR ≤ 0.1) for 3-, 6-, and 9-month data. At 6 months, APOE-T140, APOE-S139, CLCN6-S685, and BIG3-S1646 are the discoveries for the female group, whereas APOE-T140, Presenilin-1 (PSN1)-S353, Brevican core protein (PGCB)-T544, and GFAP-S12 are for the male group. In females, there are two phosphopeptides that are identified as significant in both 6- and 9-month time points, which are BIG3-S1646 and APOE-T140. Whereas, in males, only APOE-T140 overlaps between 6- and 9-month time points.
Next, to further filter the 142 phosphopeptides we identified in the 9-month female group (FDR ≤ 0.1) and uncover sites of biological interest, we investigated those that exhibit a large difference in fold changes (5XFAD/WT 0.5 ≥ FC ≥ 2). We observed 88 out of these 142 findings also passed this fold-change threshold. Among these, the peptides that exhibit larger differences in fold changes (0.25 ≥ FC ≥ 4) are BIG3-S1646, GFAP-Y13, A4-S441, myelin-associated glycoprotein (MAG)-S547, microtubule-associated protein 2 (MTAP2)-S739, vacuolar-protein-sorting-associated protein 53 homolog (VPS53)-S377, oxysterol-binding protein 1 (OSBP1)-S349, cGMP-dependent 3′,5′-cyclic phosphodiesterase (PDE2A)-S911, and microtubule-associated protein 6 (MAP6)-S905. An STRING network analysis of these top phosphorylated proteins reveals an enrichment for Golgi apparatus proteins (Figure S7). This suggests that a dysregulation of the Golgi apparatus in female 5XFAD mice might contribute to the higher disease burden in female mice, especially in late AD stages.
To identify phosphopeptides that are differentially regulated between males and females, we compare 5XFAD/WT fold changes in females vs. the fold changes in males. We identify four phosphopeptides that are significantly different in the 9-month time point (FDR ≤ 0.1); these are MTAP2-S739, adhesion G protein-coupled receptor B2 (AGRB2)-S1277, 14-3-3 protein epsilon (1433E)-S210, and BTB/POZ-domain-containing protein KCTD12 (KCD12)-S178. Next, we investigate phosphopeptides that were significant in females, but not in males (FDR ≤ 0.1), that also exhibit a large fold-change difference between females and males (0.25 ≥ FC ≥ 4). Based on this analysis, we identify 23 peptides. Notable ones among these include BIG3-S1646, OSBP1-S349, VPS53-S377, vacuolar-protein-sorting-associated protein 35 (VPS35)-S786, MAG-S547, EH-domain-containing protein 3 (EHD3)-S456, receptor-type tyrosine-protein phosphatase zeta (PRPTZ)-S576, synapsin-1 (SYN1)-S67, NMDA 2B glutamate receptor ionotropic (NMDE2)-S948, and 14-3-3 protein zeta/delta (1433Z)-S207. We provide the rest of the significant findings in Data S2, and they can also be browsed from the AD-Xplorer application.
Potential Alzheimer’s disease markers: Consistent phospho-dysregulation across time
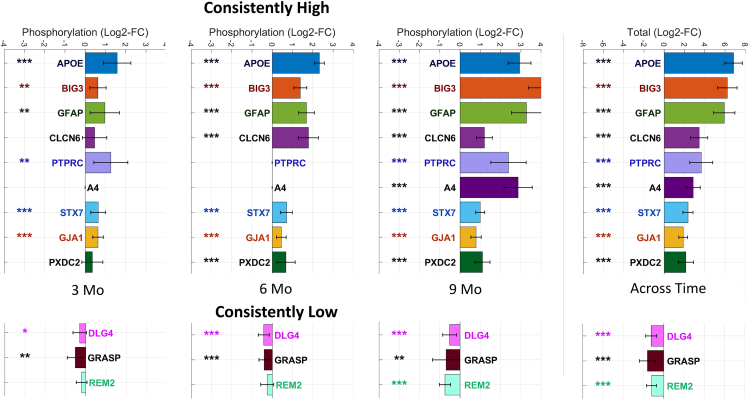
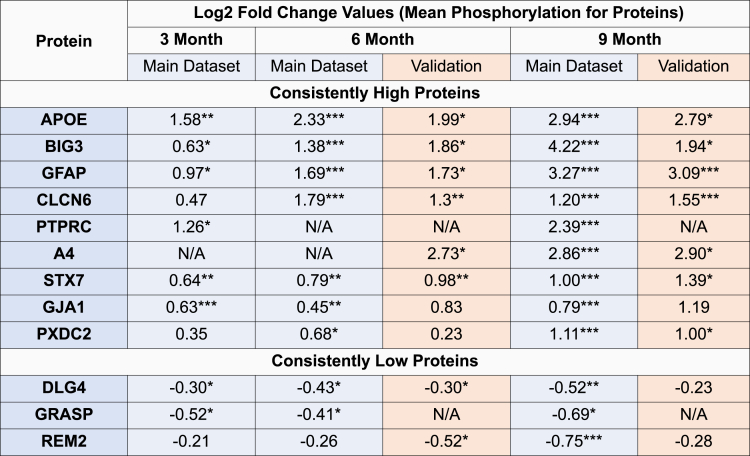
To identify potential biomarkers of disease progression, we conducted a longitudinal analysis of phosphoproteome profiling. We examined the dysregulation of proteins across all time points and calculated a consistency score based on their mean log2 fold change at the protein level and the total fold change across the three time points. Based on this analysis, we identified 31 proteins that were consistently upregulated and 18 proteins that were consistently downregulated, all at a FDR of 0.1 or lower. Among these proteins, several proteins emerged as candidate biomarkers. The nine proteins exhibiting consistently increased phosphorylation levels are APOE, GFAP, BIG3, CLCN6, protein tyrosine phosphatase receptor type C (PTPRC), A4, STX7, Gap junction alpha-1 protein (GJA1), plexin domain containing protein 2 (PXDC2). On the other hand, three proteins exhibiting consistently decreased phosphorylation are disks large homolog 4 (DLG4), Rad and gem-like GTP-binding protein 2 (REM2), and general receptor for phosphoinositide-1-associated scaffold protein (GRASP) (see Figure 2).
Figure 2.
Proteins identified as differentially phosphorylated for Alzheimer disease 5XFAD model across different time points
(Left) The bars indicate the mean phosphorylation for each protein identified as consistently phosphorylated, and the error bars indicate 95% confidence intervals. Proteins are marked according to their statistical significance levels: ∗p ≤ 0.1, ∗∗FDR ≤ 0.1, and ∗∗∗FDR ≤ 0.01.
In our sex-specific analysis, we observe distinct patterns between females and males. Specifically, we found 41 consistently high and 42 consistently low proteins in females, compared to 11 consistently high and 1 consistently low proteins in males. Interestingly, we observe 10 proteins that are identified as consistent in both female- and male-specific analysis, including APOE, BIG3, CLCN6, GFAP, STX7, PSN1, GJA1, serine incorporator 1 (SERC1), actin-binding LIM protein 1 (ABLM1), and brain acid soluble protein 1 (BASP1). Among these, only BASP1 exhibits decreased phosphorylation.
Moreover, we observe two proteins, neural proliferation differentiation and control protein 1 (NPDC1) and ubiquitin-conjugating enzyme E2 variant 3 (UEVLD), that are identified as consistently hyper-phosphorylated in males but not in females. On the other hand, when we examine the proteins identified exclusively in female-specific analysis, we observe a large number of proteins that exhibit a considerable total fold change across the three time points (0.25 ≥ total FC ≥ 4). These proteins include PXDC2, GRASP, MAG, electrogenic sodium bicarbonate cotransporter (S4A4), DnaJ homolog subfamily C member 5 (DNJC5), Src substrate cortactin (SRC8), synaptotagmin-2 (SYT2), GRIP1-associated protein 1 (GRAP1), SRSF protein kinase 2 (SRPK2), syntaxin-binding protein 5 (STXB5), galectin-1 (LEGL), synaptophysin (SYPH), basigin (BASI), and peroxisomal biogenesis factor 19 (PEX19). Among these, only PXDC2, S4A4, SRC8, DNJC5, and GRASP are also identified as consistently phosphorylated in the mixed sex analysis. Overall, our analysis reveals 19 consistently high and 27 consistently low proteins that are specific to the female-specific analysis and not in the mixed sex or male-specific analyses. For more detailed findings, please see Data S3.
Assessing functional significance: Stoichiometry of candidate phosphorylation markers
To assess phosphorylation stoichiometry, we conducted a computational analysis, computing the ratio of the top phosphopeptide to the top unmodified peptide for each protein, separately for each sample in both AD and control groups. Due to the limited ability of available technology to directly measure small intensities in unenriched samples, we utilized the phospho-enriched data to establish a scaling factor for each sample, estimating the intensities in unenriched data. The average phospho-enrichment factor was approximately 500 times (range: 244–912; Figure S8). As a result of this analysis, we report the mean phosphorylation rate across AD samples, along with the corresponding minimum and maximum values for each protein (shown in brackets). Notably, APOE and GFAP showed minimal phosphorylation rate (<1%) across all samples and time points, calling into question the biological significance of these modifications. In contrast, STX7 and GJA1 exhibited considerable phosphorylation rate, particularly at 9 months (STX7: 26.2% [18.0%–43.5%]; GJA1: 22.8% [14.5%–38.4%] at 9 months). We also observed a high phosphorylation rate of BIG3 at 6 months (33.5% [26.1%–43.1%]), while it was not identified at 3 months or 9 months. Detailed results are provided in Data S4.
Inferred kinase activities: Temporal and sex-specific analysis
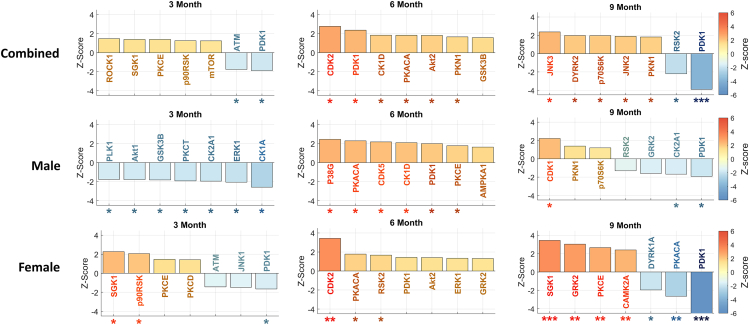
Phosphorylation, regulated by kinases, plays a crucial role in various cellular mechanisms and is of significant interest in AD research. To understand the impact of phosphorylation on AD, we focused on characterizing kinase activity. Using RoKAI, a tool for inferring kinase activity, we identified the top kinases at each time point. We performed separate analyses for each sex (male and female) as well as a combined, mixed-sex analysis (Figure 3).
Figure 3.
Inferred kinase activities across 3-/6-/9-month time points and different sexes
For each subplot, the top seven most significant kinases are shown. The error bars indicate 95% confidence intervals. Kinases are marked according to their statistical significance levels: ∗p ≤ 0.1, ∗∗FDR ≤ 0.1, and ∗∗∗FDR ≤ 0.01.
Investigating significant kinase dysregulation at FDR ≤ 0.1 level in our mixed sex analysis, we observe that phosphoinositide-dependent kinase 1 (PDK1) exhibits downregulation at 9 months. In the sex-specific analysis, we find a higher level of dysregulation in females compared to males, identifying several dysregulated kinases in females but none in males at the FDR ≤ 0.1 level. Specifically, we identify upregulation of cyclin-dependent kinase 2 (CDK2) in the 6-month female group, as well as upregulation of serum/glucocorticoid-regulated kinase 1 (SGK1), G-protein-coupled receptor kinase 2 (GRK2), protein kinase C epsilon (PKCE), and calcium/calmodulin-dependent protein kinase 2 alpha (CAMK2A) in 9-month female group. Additionally, we identify downregulation of PDK1 and protein kinase CAMP-activated catalytic subunit alpha (PKACA) in the 9-month female group. These results underscore the distinctive roles of kinases in AD progression between female and male mice. For detailed results, please refer to Data S5.
Dysregulation of PDK1: Phosphorylation, protein expression, and mRNA analysis
In the 5XFAD mouse model, we observed downregulation of PDK1 activity at 3 and 9 months, but a short-lived upregulation at the 6-month time point. Specifically, we observe divergent patterns of PDK1 regulation between female and male 5XFAD mice. The male dataset demonstrated significant upregulation of PDK1 activity at 6 months (p value ≤ 0.1), whereas the female dataset shows significant downregulation as early as the 3-month time point and identifies PDK1 upregulation (p value ≤ 0.1) as a top kinase (Figure 3).
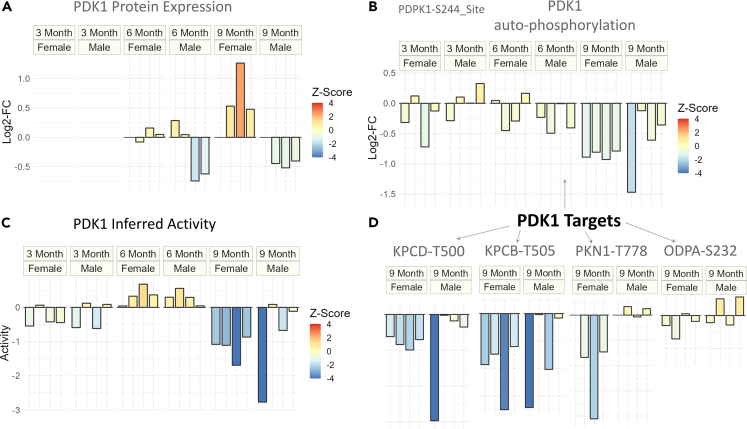
To further investigate the regulation of PDK1 at the protein expression level, we examined a previous study that analyzed global proteomic changes in the same set of 5XFAD mice15 (Figure 4A). The data revealed distinct expression patterns of PDK1 between male and female 5XFAD mice at 6 and 9 months. Male 5XFAD mice showed downregulation of PDK1 at both time points, whereas female 5XFAD mice exhibited little change at six months followed by significant upregulation at 9 months (Figure 4A). These findings indicate that changes in protein expression and phosphorylation of PDK1 are not entirely concordant. Specifically, at 6 months, male mice showed a trend of PDK1 downregulation, but inferred PDK1 activity was significantly upregulated (p value ≤ 0.1) (Figure 3). At 9 months, the upregulation of PDK1 expression in female mice was counteracted by a significant downregulation of inferred PDK1 activity (FDR ≤ 0.01). Conversely, at 9 months, male mice exhibited a downregulation trend in both PDK1 protein expression and inferred activity. These findings suggest that PDK1 is under complex regulation that changes throughout the progression of AD in both male and female mice.
Figure 4.
Dysregulation of PDK1 at protein expression and phosphorylation level for different sexes and time points of Alzheimer disease 5XFAD model
(A) Protein expression (log2-FC) of PDK1 for all time points and sexes.
(B) The auto-phosphorylation (log2-FC) of PDK1 for all time points and sexes.
(C) Inferred activity score of PDK1 for all time points and sexes.
(D) Phosphorylation (log2-FC) of known targets of PDK1 for 9 months’ data. For all plots, the coloring is done to reflect the degree of statistical significance based on Z scores.
To gain insights into the regulation of PDK1 at the transcriptional level, we examined a longitudinal 5XFAD transcriptomic study.28 We found that PDK1 mRNA levels remained relatively stable with a fold change (5XFAD/WT) from 4 to 18 months (Figure S9). These results indicate that the changes in PDK1 detected in 5XFAD mice are primarily regulated at the protein level and involve complex mechanisms.
To further investigate the regulation of PDK1 in 5XFAD mice, we analyzed the phosphorylation changes at PDK1’s constitutively activated site, S244 (Figure 4B). At 3 and 9 months, we observed that inferred activity and autophosphorylation levels for PDK1 are concordant (Figures 4B and 4C). However, at 6 months, although PDK1 inferred activity was upregulated, phosphorylation at the autophosphorylation site was not, suggesting that constitutive activation of PDK1 alone does not explain its regulation at 6 months. We observe a significant downregulation of the PDK1-S244 site only at 9 months (p value ≤ 0.1).
To explore the downstream signaling consequences of PDK1 at advanced stages of AD, we examine the phosphorylation levels of PDK1’s known targets that are quantified in our dataset (Figure 4D). We observe that the phosphorylation levels of protein kinase C beta (PRKCB) and protein kinase C delta (PRKCD) are particularly concordant with the inferred downregulation of PDK1 at 9 months.
Interpreting biological significance: Pathway analysis of phosphoproteome changes
To understand the relation between AD and various pathways, we conducted an analysis of phosphoproteomic datasets at different time points and performed a quantitative enrichment analysis using Reactome pathways.
Based on this analysis, we identify 5, 16, and 67 pathways that were significantly enriched at 3, 6, and 9 months respectively (FDR ≤ 0.1). Although most of these pathways are specific to particular time points, the regulation of gap junction activity was consistently enriched across all time points. Additionally, the scavenging by Class A receptors pathway showed significant enrichment at both 6 and 9 months (Table 1). Notable pathways unique to specific time points include the following: cation-coupled chloride cotransporters, RHO GTPases activate PAKs, DARPP-32 events, and RAF/MAP kinase cascade at 3 months; EPH-ephrin-mediated repulsion of cells, noncanonical activation of NOTCH3, nuclear signaling by ERBB4, NRIF signals cell death from the nucleus, and CS/DS degradation at 6 months; and GABA synthesis, release, reuptake, and degradation, insertion of tail-anchored proteins into the endoplasmic reticulum membrane, MTOR signaling, norepinephrine neurotransmitter release cycle, and dopamine neurotransmitter release cycle at 9 months (Table 1). For additional significant pathways, please refer to Data S6.
Table 1.
Reactome pathways identified as significant in AD phospho-proteome for 3-/6-/9-month time points
| Pathway | # of Proteins | Z score | FDR | |
|---|---|---|---|---|
| 3 Month | Regulation of gap junction activity | 2/3 | 4.60 | 0.0019 |
| Cation-coupled chloride cotransporters | 2/7 | 4.13 | 0.0053 | |
| RHO GTPases activate PAKs | 10/20 | 3.34 | 0.08 | |
| DARPP-32 events | 7/16 | 3.31 | 0.08 | |
| RAF/MAP kinase cascade | 26/91 | −4.28 | 0.004 | |
| 6 Month | Regulation of gap junction activity | 2/3 | 3.59 | 0.021 |
| Scavenging by class A receptors | 2/7 | 8.64 | 0 | |
| EPH-ephrin-mediated repulsion of cells | 5/30 | 4.90 | 0.00021 | |
| Noncanonical activation of NOTCH3 | 2/5 | 4.62 | 0.00054 | |
| Nuclear signaling by ERBB4 | 2/9 | 4.21 | 0.0028 | |
| NRIF signals cell death from the nucleus | 2/10 | 4.10 | 0.029 | |
| CS/DS degradation | 2/7 | −3.37 | 0.0033 | |
| 9 Month | Regulation of gap junction activity | 2/3 | 5.41 | 1.4e-05 |
| Scavenging by class A receptors | 2/7 | 5.66 | 6.4e-06 | |
| Insertion of tail-anchored proteins into the endoplasmic reticulum membrane | 4/12 | 4.83 | 0.00015 | |
| MTOR signaling | 5/14 | 3.71 | 0.006 | |
| Norepinephrine neurotransmitter release cycle | 8/15 | −4.10 | 0.0017 | |
| Dopamine neurotransmitter release cycle | 13/20 | −4.25 | 0.0015 | |
| GABA synthesis, release, reuptake, and degradation | 8/12 | −4.74 | 0.00018 |
Each row corresponds to a different pathway. Up to seven significant pathways are shown for each time point. “# of proteins” column indicates the number of proteins in the pathway; the first number is for proteins identified in our data, and the second one (after/) is for total number of proteins.
Validation on independent dataset: Confirming phosphoproteomic findings
To validate our findings and confirm the significance of candidate proteins, we analyzed an independent validation dataset for the 6- and 9-month time points, each consisting of a smaller sample size (n = 4, 2 5XFAD and 2 control). In the 6-month dataset, we identified 2,469 phosphopeptides in the main dataset and 3,274 in the validation dataset, with 1,330 phosphopeptides found to be common between the two. Using the same screening threshold and statistical analysis pipeline as described in previous sections (screening threshold: 5XFAD/WT 0.5 ≥ FC ≥ 2 and p ≤ 0.1), we investigated the significantly phosphorylated peptides. Among 12 peptides that passed the screening cutoffs in the main dataset and were present in the validation dataset, our analysis revealed 4 peptides that pass the same cutoffs in the validation dataset. These peptides include CLCN6-S726, GFAP-S12, GFAP-T40, and stromal interaction molecule 2 (STIM2)-S28. Notably, the observed overlap of four peptides is statistically significant, as it exceeds the expected overlap of approximately 0.3 peptides, corresponding to an 11.7-fold increase compared to the expected overlap (Fisher p value = 2.4e-4).
At the 9-month time point, we identified 2,576 phosphopeptides in the main dataset and 2,806 in the validation dataset, with 1,326 phosphopeptides found to be common between the two. Applying the same analysis, among 28 peptides that passed the cutoffs in the main dataset, 7 of them passed the cutoffs in the validation dataset as well: CLCN6-S774, GFAP-S12, A4-S441, PXDC2-S507, Ras-related protein Rab-3A (RAB3A)-S190, CaM kinase-like vesicle-associated protein (CAMKV)-T446, and protein kinase C and casein kinase substrate in neurons protein 1 (PACN1)-S427. The observed overlap of these seven peptides is statistically significant, exceeding the expected overlap of approximately 0.7 peptides, corresponding to a 10-fold increase compared to the expected overlap (Fisher p value = 2.5e-6).
Next, we conducted a protein-level analysis of mean phosphorylation using the same thresholds (screening threshold: 5XFAD/WT 0.5 ≥ FC ≥ 2 and p ≤ 0.1). At the 6-month time point, out of the 1,192 proteins in the main dataset and 1,181 proteins in the validation dataset, 785 were common between the two. Among these, seven proteins in the main dataset passed the cutoffs and four of them passed the cutoffs in both datasets: CLCN6, GFAP, APOE, and BIG3. Similarly, at the 9-month time point, we identified 1,227 proteins in the main dataset and 1,101 proteins in the validation dataset, with 788 proteins found to be common between the two. Among these, 11 proteins passed the cutoffs in the main dataset and 7 proteins in both: CLCN6, BIG3, GFAP, APOE, A4, PXDC2, and DNJC5. Notably, at 6 months, we also observe that CLCN6 and STX7 passed the FDR cutoff of 0.1 in the validation dataset despite STX7 exhibiting fold changes less than 2. Similarly, at 9 months, GFAP and CLCN6 passed the FDR cutoff of 0.1. Overall, the validation results for candidate proteins are presented in Figure 5. For more detailed results, please refer to Data S7.
Figure 5.
Validation data results for candidate proteins
Proteins are marked according to their statistical significance levels: ∗p ≤ 0.1, ∗∗FDR ≤ 0.1, and ∗∗∗FDR ≤ 0.01.
Discussion
We started our analysis by investigating the interplay between protein expression and phosphorylation, which revealed only a minor association between the two omic datasets. This is on par with previously published data29 that found only modest correlation between different omic datasets in brain tissues, advocating for multi-level omics approaches to studying AD. Furthermore, we found that phosphorylation exhibited a 1.9×-4.4× higher percentage of peptides passing the screening criteria than protein expression across all time points. Remarkably, our sex-specific analysis revealed that females exhibit a substantially higher phosphorylation prevalence (4.4×) compared to protein expression in our earliest 3-month time point. Overall, these results suggest that dysregulation in protein phosphorylation is more prevalent and complementary to changes in protein expression, underscoring the importance of phosphorylation regulation in comprehending AD, particularly in its early stages.
Our analysis on the global phosphoproteomic differences revealed a balanced distribution of up- and downregulated phosphopeptides in all time points, contrasting the proteomic data where more peptides were found to be upregulated than downregulated (greater than two-thirds).15 This dual regulation suggests that phosphorylation perpetuates the disease progression by disrupting homeostatic signals as much as it does by amplifying molecular signatures that exacerbate the disease. Surprisingly, a closer inspection revealed that the overlap among these phosphopeptides remained below 5% for all pairs of time points, implying distinct phosphorylation targeting at different AD stages. Additionally, our sex-specific analysis shows approximately 4.5× higher phospho-prevalence in 5XFAD females compared to 5XFAD males at 3 and 9 months, indicating that females exhibit higher phospho-dysregulation in both earlier and later stages of AD. This underscores the importance of developing sex-specific phosphorylation biomarkers that can be of potential clinical value for customized interventions in each sex.
Subsequently, we examined individual phosphosites displaying significant dysregulation at 3, 6, and 9 months (FDR ≤ 0.1; 0.5 ≥ FC ≥ 2). Remarkably, both temporal and sex-specific analyses consistently pinpointed the same top dysregulated proteins: APOE (S139, T140), BIG3 (S1646), CLCN6 (S726, S685, S774), GFAP (S12, Y13, T40), and STX7 (S45). In addition to the well-known proteins APOE and GFAP, we uncovered STX7 and CLCN6 as targets of phospho-dysregulation, suggesting AD’s involvement in the dysregulation of the late endocytic pathway.27 Sex-specific analysis highlighted that the top dysregulated proteins in females are notably enriched in the Golgi apparatus. Moreover, enrichment analysis based on sexual differences revealed enriched GO terms encompassing endocytic recycling, neuron development, and neuron death.
Our biomarker analysis revealed several proteins that exhibit consistent phosphorylation signatures across the time points, including well-known hallmarks of AD such as APOE, GFAP, and A4. Furthermore, our analysis unveiled the biomarker potential of additional proteins, including BIG3, STX7, PTPRC, CLCN6, GJA1, PXDC2, and GRASP. These proteins play diverse biological roles. For example, STX7 is involved in the SNARE complex and plays a crucial role in the late endocytic pathway,30,31 whereas BIG3 is known to regulate GABA neurotransmitter release.32 CLCN6, abundant in late endosomes of the nervous system, is found in amyloid plaques33 and associated with frontotemporal degeneration and dementia biomarkers.34,35 These findings suggest that CLCN6’s impact on autophagosome-mediated protein degradation might be critical in neurodegeneration.
Through our stoichiometry analysis, we examined the rate of phosphorylation among the top candidates identified in our biomarker analysis. Interestingly, we found that, although APOE and GFAP exhibit minimal phosphorylation rate (<1%) across all time points and samples, other candidates like STX7, GJA1, and BIG3 show a substantial rate of phosphorylation ranging from 20% to 40%. These contrasting profiles raise questions on the biological significance of the minor phosphorylation observed in APOE and GFAP, highlighting the potential relevance of the more pronounced phosphorylation events in STX7, GJA1, and BIG3. This prompts further exploration into the functional relevance of these phosphorylation events and their potential contributions to AD pathology.
Our kinase inference analysis indicated that PDK1 is significantly dysregulated, particularly at the 9-month time point in females. This observation is aligned with PDK1’s crucial involvement in synaptic plasticity and neuronal survival in the central nervous system (CNS), achieved by phosphorylating Akt, PKC, and p70S6, which are all pivotal for these processes.36 In AD, changes in PDK1 activity impact both the expression of Aβ and CDK5 through the PI3K/PDK1/Akt pathway.37,38,39 Our closer inspection into the PDK1 results at mRNA, protein expression, and phosphorylation level suggested PDK1 as a potentially dysregulated kinase throughout different stages of AD in the 5XFAD mouse model. Our results on PDK1 exhibited complex regulation at the protein expression and phosphorylation levels, which differ between male and females, with females exhibiting more pronounced changes, whereas the transcriptional regulation of PDK1 appears to be relatively stable over time. Further investigation into the downstream signaling pathways and targets of PDK1 may provide valuable insights into its role in AD progression.
One of the significant findings in our Reactome pathway analysis was the regulation of the gap junction activity. The related gap junction alpha-1 protein, GJA1 was also a significant finding in our biomarker analysis, exhibiting consistent hyperphosphorylation across all time points. Gap junctions play a crucial role in the cell cycling, migration, and survival of neurons and glial cells.40 Phosphorylation of gap junction proteins has been reported to regulate their assembly, trafficking, and stability.41 Whereas another significant pathway that we identify, scavenging by class A receptors, has been associated with microglia’s ability to bind and phagocytize amyloid-beta (Aβ), and studies have shown a positive correlation between class A scavenger receptors and the accumulation of Aβ in the brain of transgenic AD mice.42
Finally, AD-Xplorer, our online tool developed using the interactive data browser feature of RokaiXplorer43 enables interactive exploration of our AD-related proteomics datasets and is accessible at https://yilmazs.shinyapps.io/ADXplorer. This platform enables researchers to investigate changes in phosphorylation, protein expression, and inferred kinase activities, complemented by a pathway enrichment analysis based on significant findings. It provides several interactive visualizations, including volcano plots, bar plots, boxplots, heatmaps, and network views. The tool offers easy customization for focused subgroup analysis, such as selecting “9 Month” and “Female” to tailor the analysis to specific samples. To cater to diverse analytical needs, adjustable statistical cutoffs (based on p values, fold changes, or FDR) and various preprocessing options for quality control are provided. The tool supports exporting results as a formatted Excel report with detailed statistics and downloadable figure data. Overall, with an emphasis on temporal and sex-specific phosphoproteomic signatures, AD-Xplorer offers a user-friendly platform and facilitates in-depth exploration and interpretation of our dataset. It can aid in unraveling complexities of AD and serve as a valuable resource for biomarker discovery and therapeutic target identification.
Limitations of the study
Although 5XFAD is widely regarded as the gold standard among familial mouse models for AD, there are three primary concerns regarding its suitability as a stand-in for human AD: (1) although 5XFAD mice exhibit phenotypic traits based on single mutations in the APP and PSEN1 genes, familial AD in humans does not always stem from a straightforward inheritance of a single gene. Instead, it often manifests within families through a more complex genetic interplay, where the combined effects of multiple genes may contribute to an earlier onset of AD. (2) The Aβ42/Aβ40 ratio, a significant diagnostic indicator for AD, appears notably elevated in 5XFAD mice compared to human AD patients. This suggests a potential overestimation by the 5XFAD model regarding the impact of this toxicity on downstream targets.44 (3) Although dysregulation of Tau phosphorylation is a critical hallmark of AD, 5XFAD mice show minimal or no phosphorylation in several common phosphosites of Tau, such as S199, S202, and T205.45,46 This suggests that the 5XFAD model does not fully replicate the Tau biology observed in human AD patients. Overall, these differences between the 5XFAD model and human AD present challenges for uncovering biomarkers and the development of effective therapies.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Serhan Yilmaz (serhan.yilmaz@case.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE47 partner repository with the dataset identifier PXD044648 and https://doi.org/10.6019/PXD044648. The data and code to reproduce the analysis results are deposited to Figshare and available at: https://doi.org/10.6084/m9.figshare.23903715. Additionally, the data and findings presented in this study can be explored interactively from AD-Xplorer application (https://yilmazs.shinyapps.io/ADXplorer). Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Acknowledgments
This work was supported in part by the National Library of Medicine (NLM), National Institutes of General Medical Sciences (NIGMS), and the Office of the Director, National Institutes of Health (NIH), United States grants under R01-LM12980, R01-GM117208, R01-GM117208-03S1, S10-OD026882-01, and S10-OD028614-01. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NLM, NIH, or NIGMS.
Author contributions
D.S., X.Q., M.K., and M.R.C., conceptualization; S.Y. and F.B.T.P.L., formal analysis; S.Y. and F.B.T.P.L., writing—original draft; S.Y., F.B.T.P.L., D.S., X.Q., M.K., and M.R.C., writing—review and editing; F.B.T.P.L., D.S., and R.W., investigation; S.Y. and F.B.T.P.L., data curation; S.Y. and F.B.T.P.L., software; S.Y., visualization; S.Y., F.B.T.P.L., D.S., X.Q., M.K., and M.R.C., methodology; S.Y., F.B.T.P.L., and D.S., validation; X.Q. and M.R.C., resources; M.K. and M.R.C., supervision; M.R.C., project administration; M.K. and M.R.C. funding acquisition.
Declaration of interests
The authors declare no competing interests.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| sodium dodecyl sulfate | Sigma | Cat#L4509-10G |
| protease inhibitor cocktail | Sigma | Cat#P2714 |
| PhosphoSTOP | Roche | Cat#04906837001 |
| LysC | Wako | Cat#125-05061 |
| Trypsin | Thermo Fisher | Cat#90057 |
| Bio-Rad Protein Assay Kit – BSA | Bio-rad | Cat#5000002 |
| Amicon Ultra MWCO 10K filters | Millipore | Cat#UFC501096 |
| HLB 1cc (10 mg) extraction cartridges | Oasis | Lot#149A37130B |
| TiO2 phosphopeptide enrichment spin tips | Thermo Fisher | Cat#88303 |
| Deposited data | ||
| Phosphorylation - Mass Spectrometry data | PRIDE repository | PXD044648 |
| Protein Expression - Mass Spectrometry data (data from previous study) | PRIDE repository | PXD030161 |
| Experimental models: Organisms/strains | ||
| 5XFAD mice (B6SJL-Tg(APPSwFlLon, PSEN1∗M146L∗L286V) 6799Vas/Mmjax) | Jackson Laboratories | RRID:MMRRC_034840-JAX |
| WT mice (C57BL/6J) | Jackson Laboratories | RRID:IMSR_JAX:000664 |
| Software and algorithms | ||
| AD-Xplorer | https://yilmazs.shinyapps.io/ADXplorer | https://github.com/serhan-yilmaz/AD-Xplorer |
| Self-contained package (code and data) to reproduce the analysis results | Figshare | https://doi.org/10.6084/m9.figshare.23903715 |
| Other | ||
| NanoAcquity UPLC chromatography | Waters | 176016000 |
| Thermo Scientific Orbitrap Elite mass spectrometer | Thermo Fisher | FSN40129 |
Experimental model and study participant details
The study involved tracking alterations in the global phosphoproteome at distinct stages of Alzheimer’s Disease progression in the hippocampus of 5XFAD mice. For this purpose, label-free LC-MS/MS was employed. A total of 48 samples were analyzed, including sets of 16 male and female 5XFAD mice, as well as WT mice. The hippocampi were collected and processed at three time points: 3 months, 6 months, and 9 months. Each time point comprised 8 samples for both 5XFAD and WT mice. Additionally, within each group, samples were divided based on sex, resulting in 4 samples per subgroup.
All animal experiments in this study were conducted in accordance with protocols approved by the Institutional Animal Care and Use Committee of Case Western Reserve University and performed according to the National Institutes of Health Guide for the Care and Use of Laboratory Animals. The mice were mated, bred, and genotyped in the animal facility of Case Western Reserve University. Sufficient procedures were employed to reduce the pain and discomfort of the mice during the experiments. All mice were maintained under a 12 h/12 h light/dark cycle (light on at 6 a.m. and off at 6 p.m.) with ad libitum access to food and water under the ambient temperature at 23°C and humidity at 40–60%. 5XFAD transgenic mice overexpress known familial AD (FAD) mutations in APP (K670N/M671L + I716V + V717I) and PS1 (M146L + L286V) genes, under the control of the Thy1 promoter. All mice used in this study were maintained on a C57BL/6J (Strain #000664, The Jackson Laboratory) background. 5XFAD transgenic mice [Tg(APPSwFlLon, PSEN1∗M146L∗L286V)6799Vas, strain #034840-JAX] breeders were purchased from Jackson Laboratory. Wild-type control mice used in this study were 5xFAD littermates.
Method details
Hippocampus tissue samples were collected from 5XFAD and wild-type mice at three different time points (three, six, and nine months). The samples were lysed using a 2% SDS solution supplemented with a protease inhibitor cocktail (Cat#P2714, Sigma) and PhosphoSTOP (Cat#04906837001, Roche). The protein concentration in the lysates was determined using the Bio-Rad Protein Assay Kit – BSA (Cat#5000002, Bio-Rad).
To process the lysates, we followed the FASP protocol48 and performed digestion using dual LysC (Cat#125–05061, Wako) and trypsin (Cat#90057, Thermo Fisher) endoproteinase. The digested samples were then desalted using C18 cartridges. For phosphopeptide enrichment, TiO2 enrichment spin tips were used. Samples were normalized to 300 ng of digest, and a blind and randomized approach was employed for LC-MS/MS acquisition. We used a Waters NanoAcquity UPLC chromatography system coupled to a Thermo Scientific Orbitrap Elite mass spectrometer (Thermo Fisher Scientific, CA).
The raw LC-MS/MS data was processed using Peaks v10.0 Software (Bioinformatics Solutions, ON, CA). Peptide identification was performed within Peaks using the UNIPROT database (UNIPROT_MOUSE_091219, # of entries = 17026). The PEAKS search parameters included a mass error tolerance of 10 ppm for precursor ions, a mass tolerance of 0.6Da for fragment ions, trypsin enzyme specificity, and fixed variations of carbamidomethylation, as well as variable modifications of methionine oxidation and phosphorylation at serine, threonine, and tyrosine residues. The search allowed for one missed cleavage. Label-free phosphopeptide identification followed the default target decoy approach, with a PEAKS peptide score (−10logP) threshold of ≥15 and an FDR threshold of 1%. The abundance of individual peptides was determined by calculating the area under the curve (AUC). Data S1 contains the output excel files for all three time points, along with the corresponding sample information.
Quantification and statistical analysis
To ensure data quality, we performed a number of additional preprocessing steps. First, we filtered the peptides for monophosphorylated peptides. Next, we excluded quantifications with small intensities (<10ˆ4) from the analysis by treating them as missing values. Additionally, we filtered out peptides that have numerous missing values. Specifically, a peptide was included in the analysis only if it is identified in a minimum of 3 samples for both the WT and 5XFAD groups. We also filtered out 2 samples in the 9 months time point (one female and one male sample in the WT group) that exhibited substantially low mean intensities across all peptides (Figure S1). In addition, to alleviate potential imbalances between samples, we applied a preprocessing step that centers the log2 transformed intensities of all samples. This process ensures that the mean value across all peptides is the same for every sample, irrespective of their WT/5XFAD group. We investigated the differences between 5XFAD and WT samples by performing a moderated t-test, which utilizes an empirical Bayes method to shrink the sample variances toward a common value and to augment the degrees of freedom for the individual variances.49 In addition, we investigated potential sex differences based on a moderated t-test by incorporating sex as a covariate in the linear model (please see supplement for details).
To identify potential phosphorylation-based biomarkers that are consistent across time points, we conducted an analysis at the protein level. This approach was chosen due to the high amount of missingness at the phosphopeptide level, particularly when comparing different time point experiments (e.g., 3 months, 6 months, 9 months). The rate of missingness at the protein level was considerably less compared to the number of phosphopeptides identified across all three time points. By focusing the analysis at the protein level, we aimed to address this challenge and facilitate the assessment of consistency across various time points or groups. For this purpose, we computed the mean log-fold changes at the protein level and used a moderated t-test with the Satterthwaite approximation50 to compare 5XFAD and WT groups (see supplement for details). To assess consistency across time points, we introduced a simple statistic called the consistency score, which combined the log2-fold change results of individual time points. This score represents the total log2-fold change across time points, considering any missing values as log2-fold change of 0. This approach allowed us to account for changes in the same direction while canceling out those with opposite directions, facilitating the identification of proteins with consistent phosphorylation patterns.
To quantify the stoichiometry of phosphorylation, we conducted a computational analysis, calculating the ratio of the top phospho-peptide to the top unmodified peptide with highest intensities for each protein, separately for each sample in both 5XFAD and control groups. For this purpose, given the technological constraints in directly measuring small phosphorylation intensities in unenriched data,15 we used the phospho-enriched data to derive a scaling factor for each sample. This scaling factor enabled us to estimate the phosphorylation intensities in the unenriched data using the phospho-enriched data. To achieve this, we first identified common proteins between the two datasets and determined the highest phospho-peptide intensity for each protein. Then, we assessed how many times, on average, the intensities in the phospho-enriched data was higher compared to the unenriched data for each sample using the geometric mean (Figure S8). By dividing the intensities in the phospho-enriched data by this scaling factor, we obtained the estimated phosphorylation intensities for each protein in the unenriched samples. Finally, we calculated the prevalence of phosphorylation for each protein and sample, reporting mean, minimum and maximum values across samples in both AD and control groups.
For the kinase analysis, we considered kinases with at least two known targets identified in our dataset and performed the kinase activity inference using the RoKAI algorithm, which makes use of a functional network to improve the accuracy and robustness of the inference.51 To identify biological pathways and networks impacted by the observed phosphoproteome changes, we performed a quantitative pathway enrichment analysis based on the mean phosphorylation (log2-FC) of proteins. This approach helped us overcome the limitations of relying on arbitrary significance thresholds and provided a more interpretable measure of pathway enrichment, enabling us to quantify the magnitude of enrichment in addition to identifying the significant pathways affected by the observed changes in the phosphoproteome. We conducted the pathway analysis using Reactome pathways52 that met the following inclusion criteria. We excluded pathways with a large portion of missing data, considering only those with at least two proteins identified in our dataset. Additionally, we required that these proteins accounted for at least 10% of all proteins in the pathway. To avoid redundancy, we filtered out highly similar pathways sharing the same protein sets, retaining the smaller pathway in cases of duplication.
In our phosphopeptide analysis, to determine phosphopeptides that are of significance, we applied two sets of thresholds. The first set, which includes a p-value cutoff of p ≤ 0.1 followed by a fold change cutoff of 0.5 ≥ FC ≥ 2, enables us to compare results across different time points and/or different sexes. This simple criteria provides a consistent framework for making comparisons between different groups. However, to address the multiple comparisons issue and to enable individual discoveries, we also employed a second set of thresholds. This involves utilizing the Benjamini-Hochberg procedure to apply a false discovery rate (FDR) cutoff of less than 0.1, ensuring statistically significant findings that are unlikely to be due to random noise or false positives.
Additional resources
To aid in the interpretation of our findings, we used the interactive data browser feature of RokaiXplorer43 application (https://rokai.io/explorer) to develop the online tool AD-Xplorer. In this tool, the users can interactively explore the AD phospho-proteomic dataset that we present in this paper, as well as the proteomic dataset that is a result of our previous study on the same set of mice.15 In addition to the previous analyses, AD-Xplorer includes a gene ontology (GO) term enrichment performed with an over-representation analysis (chi-squared test with Yates’ correction) based on a screened set of peptides or proteins (default cutoffs: p ≤ 0.1 and 0.5 ≥ FC ≥ 2).
Published: September 13, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.110941.
Contributor Information
Serhan Yılmaz, Email: serhan.yilmaz@case.edu.
Filipa Blasco Tavares Pereira Lopes, Email: filipa@case.edu.
Mehmet Koyutürk, Email: koyuturk@case.edu.
Mark R. Chance, Email: mark.chance@case.edu.
Supplemental information
References
- 1.GBD 2016 Dementia Collaborators Global, regional, and national burden of Alzheimer's disease and other dementias, 1990–2016 a systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2019;18:88–106. doi: 10.1016/S1474-4422(18)30403-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferreira D., Wahlund L.-O., Westman E. The heterogeneity within Alzheimer's disease. Aging (Albany NY) 2018;10:3058–3060. doi: 10.18632/aging.101638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van der Flier W.M. Clinical heterogeneity in familial Alzheimer's disease. Lancet Neurol. 2016;15:1296–1298. doi: 10.1016/S1474-4422(16)30275-7. [DOI] [PubMed] [Google Scholar]
- 4.Devi G., Scheltens P. Heterogeneity of Alzheimer’s disease: consequence for drug trials? Alzheimers Res. Ther. 2018;10:122. doi: 10.1186/s13195-018-0455-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Troncoso J.C., Martin L.J., Dal Forno G., Kawas C.H. Neuropathology in controls and demented subjects from the Baltimore longitudinal study of aging. Neurobiol. Aging. 1996;17:365–371. doi: 10.1016/0197-4580(96)00028-0. [DOI] [PubMed] [Google Scholar]
- 6.Troncoso J.C., Cataldo A.M., Nixon R.A., Barnett J.L., Lee M.K., Checler F., Fowler D.R., Smialek J.E., Crain B., Martin L.J., Kawas C.H. Neuropathology of preclinical and clinical late-onset. Ann. Neurol. 1998;43:673–676. doi: 10.1002/ana.410430519. [DOI] [PubMed] [Google Scholar]
- 7.Jack C.R., Knopman D.S., Jagust W.J., Petersen R.C., Weiner M.W., Aisen P.S., Shaw L.M., Vemuri P., Wiste H.J., Weigand S.D., et al. Tracking pathophysiological processes in Alzheimer's disease: an updated hypothetical model of dynamic biomarkers. Lancet Neurol. 2013;12:207–216. doi: 10.1016/S1474-4422(12)70291-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharma K., Schmitt S., Bergner C.G., Tyanova S., Kannaiyan N., Manrique-Hoyos N., Kongi K., Cantuti L., Hanisch U.K., Philips M.A., et al. Cell type- and brain region-resolved mouse brain proteome. Nat. Neurosci. 2015;18:1819–1831. doi: 10.1038/nn.4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Needham E.J., Parker B.L., Burykin T., James D.E., Humphrey S.J. Illuminating the dark phosphoproteome. Sci. Signal. 2019;12:eaau8645. doi: 10.1126/scisignal.aau8645. [DOI] [PubMed] [Google Scholar]
- 10.Kovacech B., Zilka N., Novak M. New age of neuroproteomics in Alzheimer's disease research. Cell. Mol. Neurobiol. 2009;29:799–805. doi: 10.1007/s10571-009-9358-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noble W., Olm V., Takata K., Casey E., Mary O., Meyerson J., Gaynor K., LaFrancois J., Wang L., Kondo T., et al. Cdk5 is a key factor in tau aggregation and tangle formation in vivo. Neuron. 2003;38:555–565. doi: 10.1016/S0896-6273(03)00259-9. [DOI] [PubMed] [Google Scholar]
- 12.Marttinen M., Paananen J., Neme A., Mitra V., Takalo M., Natunen T., Paldanius K.M.A., Mäkinen P., Bremang M., Kurki M.I., et al. A multiomic approach to characterize the temporal sequence in Alzheimer's disease-related pathology. Neurobiol. Dis. 2019;124:454–468. doi: 10.1016/j.nbd.2018.12.009. [DOI] [PubMed] [Google Scholar]
- 13.Gurel B., Cansev M., Koc C., Ocalan B., Cakir A., Aydin S., Kahveci N., Ulus I.H., Sahin B., Basar M.K., Baykal A.T. Proteomics Analysis of CA1 Region of the Hippocampus in Pre-Progression and Pathological Stages in a Mouse Model of the Alzheimer’s Disease. Curr. Alzheimer Res. 2019;16:613–621. doi: 10.2174/1567205016666190730155926. [DOI] [PubMed] [Google Scholar]
- 14.Kim D.K., Han D., Park J., Choi H., Park J.-C., Cha M.Y., Woo J., Byun M.S., Lee D.Y., Kim Y., Mook-Jung I. Deep proteome profiling of the hippocampus in the 5XFAD mouse model reveals biological process alterations and a novel biomarker of Alzheimer's disease. Exp. Mol. Med. 2019;51:1–17. doi: 10.1038/s12276-019-0326-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blasco Tavares Pereira Lopes F., Schlatzer D., Wang R., Li X., Feng E., Koyutürk M., Qi X., Chance M.R. Temporal and Sex-Linked Protein Expression Dynamics in a Familial Model of Alzheimer’s Disease. Mol. Cell. Proteomics. 2022;21:100280. doi: 10.1016/j.mcpro.2022.100280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blasco Tavares Pereira Lopes F., Schlatzer D., Wang R., Li X., Feng E., Koyutürk M., Qi X., Chance M.R. Temporal and Sex-Linked Protein Expression Dynamics in a Familial Model of Alzheimer's Disease. Mol. Cell. Proteomics. 2022;21:100280. doi: 10.1016/j.mcpro.2022.100280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li T., Braunstein K.E., Zhang J., Lau A., Sibener L., Deeble C., Wong P.C. The neuritic plaque facilitates pathological conversion of tau in an Alzheimer’s disease mouse model. Nat. Commun. 2016;7:12082. doi: 10.1038/ncomms12082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo J.L., Narasimhan S., Changolkar L., He Z., Stieber A., Zhang B., Gathagan R.J., Iba M., McBride J.D., Trojanowski J.Q., Lee V.M.Y. Unique pathological tau conformers from Alzheimer's brains transmit tau pathology in nontransgenic mice. J. Exp. Med. 2016;213:2635–2654. doi: 10.1084/jem.20160833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu K., Li Y., Yu H., Hu Y. CTBP1 Confers Protection for Hippocampal and Cortical Neurons in Rat Models of Alzheimer's Disease. Neuroimmunomodulation. 2019;26:139–152. doi: 10.1159/000500942. [DOI] [PubMed] [Google Scholar]
- 20.Poo M.M., Pignatelli M., Ryan T.J., Tonegawa S., Bonhoeffer T., Martin K.C., Rudenko A., Tsai L.H., Tsien R.W., Fishell G., et al. What is memory? The present state of the engram. BMC Biol. 2016;14:40. doi: 10.1186/s12915-016-0261-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Josephs K.A., Dickson D.W., Tosakulwong N., Weigand S.D., Murray M.E., Petrucelli L., Liesinger A.M., Senjem M.L., Spychalla A.J., Knopman D.S., et al. Rates of hippocampal atrophy and presence of post-mortem TDP-43 in patients with Alzheimer's disease: a longitudinal retrospective study. Lancet Neurol. 2017;16:917–924. doi: 10.1016/s1474-4422(17)30284-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rao Y.L., Ganaraja B., Murlimanju B.V., Joy T., Krishnamurthy A., Agrawal A. Hippocampus and its involvement in Alzheimer's disease: a review. 3 Biotech. 2022;12:55. doi: 10.1007/s13205-022-03123-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pixley S.K., de Vellis J. Transition between immature radial glia and mature astrocytes studied with a monoclonal antibody to vimentin. Brain Res. 1984;317:201–209. doi: 10.1016/0165-3806(84)90097-X. [DOI] [PubMed] [Google Scholar]
- 24.Levin E.C., Acharya N.K., Sedeyn J.C., Venkataraman V., D'Andrea M.R., Wang H.Y., Nagele R.G. Neuronal expression of vimentin in the Alzheimer's disease brain may be part of a generalized dendritic damage-response mechanism. Brain Res. 2009;1298:194–207. doi: 10.1016/j.brainres.2009.08.072. [DOI] [PubMed] [Google Scholar]
- 25.McColl T.J., Brady R.D., Shultz S.R., Lovick L., Webster K.M., Sun M., McDonald S.J., O'Brien T.J., Semple B.D. Mild Traumatic Brain Injury in Adolescent Mice Alters Skull Bone Properties to Influence a Subsequent Brain Impact at Adulthood: A Pilot Study. Front. Neurol. 2018;9:372. doi: 10.3389/fneur.2018.00372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bilbao A., Rieker C., Cannella N., Parlato R., Golda S., Piechota M., Korostynski M., Engblom D., Przewlocki R., Schütz G., et al. CREB activity in dopamine D1 receptor expressing neurons regulates cocaine-induced behavioral effects. Front. Behav. Neurosci. 2014;8:212. doi: 10.3389/fnbeh.2014.00212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jentsch T.J., Pusch M. CLC Chloride Channels and Transporters: Structure, Function, Physiology, and Disease. Physiol. Rev. 2018;98:1493–1590. doi: 10.1152/physrev.00047.2017. [DOI] [PubMed] [Google Scholar]
- 28.Forner S., Kawauchi S., Balderrama-Gutierrez G., Kramár E.A., Matheos D.P., Phan J., Javonillo D.I., Tran K.M., Hingco E., da Cunha C., et al. Systematic phenotyping and characterization of the 5xFAD mouse model of Alzheimer’s disease. Sci. Data. 2021;8:270. doi: 10.1038/s41597-021-01054-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dammer E.B., Ping L., Duong D.M., Modeste E.S., Seyfried N.T., Lah J.J., Levey A.I., Johnson E.C.B. Multi-platform proteomic analysis of Alzheimer's disease cerebrospinal fluid and plasma reveals network biomarkers associated with proteostasis and the matrisome. Alzheimers Res. Ther. 2022;14:174. doi: 10.1186/s13195-022-01113-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu P., Hankins H.M., MacDonald C., Erlinger S.J., Frazier M.N., Diab N.S., Piper R.C., Jackson L.P., MacGurn J.A., Graham T.R. COPI mediates recycling of an exocytic SNARE by recognition of a ubiquitin sorting signal. Elife. 2017;6:e28342. doi: 10.7554/eLife.28342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sogorb-Esteve A., Nilsson J., Swift I.J., Heller C., Bocchetta M., Russell L.L., Peakman G., Convery R.S., van Swieten J.C., Seelaar H., et al. Differential impairment of cerebrospinal fluid synaptic biomarkers in the genetic forms of frontotemporal dementia. Alzheimers Res. Ther. 2022;14:118. doi: 10.1186/s13195-022-01042-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu T., Li H., Hong W., Han W. Brefeldin A-inhibited guanine nucleotide exchange protein 3 is localized in lysosomes and regulates GABA signaling in hippocampal neurons. J. Neurochem. 2016;139:748–756. doi: 10.1111/jnc.13859. [DOI] [PubMed] [Google Scholar]
- 33.Drummond E., Kavanagh T., Pires G., Marta-Ariza M., Kanshin E., Nayak S., Faustin A., Berdah V., Ueberheide B., Wisniewski T. The amyloid plaque proteome in early onset Alzheimer’s disease and Down syndrome. Acta Neuropathol. Commun. 2022;10:53. doi: 10.1186/s40478-022-01356-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sassi C., Capozzo R., Hammer M., Zecca C., Federoff M., Blauwendraat C., Bernstein N., Ding J., Gibbs J.R., Price T., et al. Exploring dementia and neuronal ceroid lipofuscinosis genes in 100 FTD-like patients from 6 towns and rural villages on the Adriatic Sea cost of Apulia. Sci. Rep. 2021;11:6353. doi: 10.1038/s41598-021-85494-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Del Greco M F., Pattaro C., Luchner A., Pichler I., Winkler T., Hicks A.A., Fuchsberger C., Franke A., Melville S.A., Peters A., et al. Genome-wide association analysis and fine mapping of NT-proBNP level provide novel insight into the role of the MTHFR-CLCN6-NPPA-NPPB gene cluster. Hum. Mol. Genet. 2011;20:1660–1671. doi: 10.1093/hmg/ddr035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bayascas J.R. PDK1: the major transducer of PI 3-kinase actions. Curr. Top. Microbiol. Immunol. 2010;346:9–29. doi: 10.1007/82_2010_43. [DOI] [PubMed] [Google Scholar]
- 37.Pietri M., Dakowski C., Hannaoui S., Alleaume-Butaux A., Hernandez-Rapp J., Ragagnin A., Mouillet-Richard S., Haik S., Bailly Y., Peyrin J.-M., et al. PDK1 decreases TACE-mediated α-secretase activity and promotes disease progression in prion and Alzheimer's diseases. Nat. Med. 2013;19:1124–1131. doi: 10.1038/nm.3302. [DOI] [PubMed] [Google Scholar]
- 38.Wen Y., Planel E., Herman M., Figueroa H.Y., Wang L., Liu L., Lau L.F., Yu W.H., Duff K.E. Interplay between cyclin-dependent kinase 5 and glycogen synthase kinase 3 beta mediated by neuregulin signaling leads to differential effects on tau phosphorylation and amyloid precursor protein processing. J. Neurosci. 2008;28:2624–2632. doi: 10.1523/jneurosci.5245-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nava P., Capaldo C.T., Koch S., Kolegraff K., Rankin C.R., Farkas A.E., Feasel M.E., Li L., Addis C., Parkos C.A., Nusrat A. JAM-A regulates epithelial proliferation through Akt/β-catenin signalling. EMBO Rep. 2011;12:314–320. doi: 10.1038/embor.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zlomuzica A., Viggiano D., Degen J., Binder S., Ruocco L.A., Sadile A.G., Willecke K., Huston J.P., Dere E. Behavioral alterations and changes in Ca/calmodulin kinase II levels in the striatum of connexin36 deficient mice. Behav. Brain Res. 2012;226:293–300. doi: 10.1016/j.bbr.2011.08.028. [DOI] [PubMed] [Google Scholar]
- 41.Boassa D., Solan J.L., Papas A., Thornton P., Lampe P.D., Sosinsky G.E. Trafficking and recycling of the connexin43 gap junction protein during mitosis. Traffic. 2010;11:1471–1486. doi: 10.1111/j.1600-0854.2010.01109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bornemann K.D., Wiederhold K.H., Pauli C., Ermini F., Stalder M., Schnell L., Sommer B., Jucker M., Staufenbiel M. Abeta-induced inflammatory processes in microglia cells of APP23 transgenic mice. Am. J. Pathol. 2001;158:63–73. doi: 10.1016/s0002-9440(10)63945-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yılmaz S., Tavares Pereira Lopes F.B., Schlatzer D., Ayati M., Chance M.R., Koyutürk M. Making Proteomics Accessible: RokaiXplorer for interactive analysis of phospho-proteomic data. Bioinformatics Adv. 2024:vbae077. doi: 10.1093/bioadv/vbae077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oakley, H., Cole, S.L., Logan, S., Maus, E., Shao, P., Craft J., Guillozet-Bongaarts A., Ohno M., Disterhoft J., Van Eldik L., et al. Intraneuronal β-Amyloid Aggregates, Neurodegeneration, and Neuron Loss in Transgenic Mice with Five Familial Alzheimer's Disease Mutations: Potential Factors in Amyloid Plaque Formation. J. Neurosci. 2006;26:10129–10140. doi: 10.1523/JNEUROSCI.1202-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bennett D.A. Mixed pathologies and neural reserve: Implications of complexity for Alzheimer disease drug discovery. PLoS Med. 2017;14:e1002256. doi: 10.1371/journal.pmed.1002256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kovacs G.G., Milenkovic I., Wöhrer A., Höftberger R., Gelpi E., Haberler C., Hönigschnabl S., Reiner-Concin A., Heinzl H., Jungwirth S., et al. Non-Alzheimer neurodegenerative pathologies and their combinations are more frequent than commonly believed in the elderly brain: a community-based autopsy series. Acta Neuropathol. 2013;126:365–384. doi: 10.1007/s00401-013-1157-y. [DOI] [PubMed] [Google Scholar]
- 47.Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M., et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wiśniewski J.R., Zougman A., Nagaraj N., Mann M. Universal sample preparation method for proteome analysis. Nat. Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 49.Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 50.Satterthwaite F.E. An approximate distribution of estimates of variance components. Biometrics. 1946;2:110–114. [PubMed] [Google Scholar]
- 51.Yılmaz S., Ayati M., Schlatzer D., Çiçek A.E., Chance M.R., Koyutürk M. Robust inference of kinase activity using functional networks. Nat. Commun. 2021;12:1177. doi: 10.1038/s41467-021-21211-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jassal B., Matthews L., Viteri G., Gong C., Lorente P., Fabregat A., Sidiropoulos K., Cook J., Gillespie M., Haw R., et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2020;48:D498–D503. doi: 10.1093/nar/gkz1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE47 partner repository with the dataset identifier PXD044648 and https://doi.org/10.6019/PXD044648. The data and code to reproduce the analysis results are deposited to Figshare and available at: https://doi.org/10.6084/m9.figshare.23903715. Additionally, the data and findings presented in this study can be explored interactively from AD-Xplorer application (https://yilmazs.shinyapps.io/ADXplorer). Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.