Abstract
Envelope glycoproteins (Envs) of human immunodeficiency virus type 2 (HIV-2) are frequently able to use chemokine receptors, CXCR4 or CCR5, in the absence of CD4. However, while these Envs are commonly dual-tropic, no isolate has been described to date that is CD4 independent on both CXCR4 and CCR5. In this report we show that a variant of HIV-2/NIHz, termed HIV-2/vcp, previously shown to utilize CXCR4 without CD4, is also CD4 independent on rhesus (rh) CCR5, but requires CD4 to fuse with human (hu) CCR5. The critical determinant for this effect was an acidic amino acid at position 13 in the CCR5 N terminus, which is an asparagine in huCCR5 and an aspartic acid in rhCCR5. Transferring the huCCR5 N terminus with an N13D substitution to CCR2b or CXCR2 was sufficient to render these heterologous chemokine receptors permissive for CD4-independent fusion. Chimeric Envs between HIV-2/vcp and a CD4-dependent clone of HIV-2/NIHz as well as site-directed Env mutations implicated a positively charged amino acid (lysine or arginine) at position 427 in the C4 region of the HIV-2/vcp env gene product (VCP) gp120 as a key determinant for this phenotype. Because CD4-independent use of CCR5 mapped to a negatively charged amino acid in the CCR5 N terminus and a positively charged amino acid in the gp120 C4 domain, an electrostatic interaction between these residues or domains is likely. Although not required for CD4-dependent fusion, this interaction may serve to increase the binding affinity of Env and CCR5 and/or to facilitate subsequent conformational changes that are required for fusion. Because the structural requirements for chemokine receptor use by HIV are likely to be more stringent in the absence of CD4, CD4-independent viruses should be particularly useful in dissecting molecular events that are critical for viral entry.
Human (HIV) and simian (SIV) immunodeficiency viruses enter cells by fusing with the cellular membrane in a reaction triggered by an interaction between the viral envelope glycoprotein (Env) and two cellular molecules, CD4 and a chemokine receptor, generally either CCR5 or CXCR4 (1, 11, 14, 18, 19, 28). All HIVs described to date have been shown to utilize CCR5, CXCR4, or both for entry. Although additional members of the chemokine receptor family and several other seven transmembrane domain receptors can serve as coreceptors for HIV and SIV in vitro, their importance in vivo is currently unclear (3, 32).
The HIV Env is composed of gp120 and gp41 subunits that are noncovalently associated on virions in trimeric complexes (8, 69). Binding of CD4 to gp120 causes a conformational change that exposes and/or induces the formation of a highly conserved domain in gp120 that is important for binding to CCR5 (50, 58). This region, termed the bridging sheet, consists largely of a four-stranded, antiparallel β-sheet formed by the V1/V2 stem and components of the fourth conserved region (C4) of gp120 (42, 58). Conservation of this region among different HIV-1 isolates and, to some extent, HIV-2 suggests that it may also be important for interactions with CXCR4 and thus represents a generic chemokine receptor binding site. Exposure of the bridging sheet as a consequence of CD4 binding is thought to involve movement of the V1/V2 and V3 hypervariable loops (49, 68, 72, 73) that flank this domain (42, 58) and determine specificity for which chemokine receptor is utilized (10–12, 34, 66). Subsequent binding of gp120 to the chemokine receptor leads to final conformational changes in gp41 that ultimately result in fusion between the viral and cellular membranes (8, 15, 69).
Several laboratory-adapted HIV-1 isolates as well as many primary HIV-2 and SIV strains are able to bypass the requirement for CD4 (20, 23, 25, 33, 39, 43, 54, 55). Env proteins from these CD4-independent isolates have been shown to interact directly with chemokine receptors, indicating that their chemokine receptor binding site is formed and exposed without the need for CD4 triggering (21, 31, 33, 39, 46). These findings and the fact that some feline immunodeficiency virus strains also use CXCR4 as a sole receptor (71) suggest that chemokine receptors are the primordial receptors for lentiviruses and that primate lentiviruses subsequently evolved to bind to CD4 as well (23, 25, 39). Interestingly, CD4-independent R5- and X4-tropic isolates exhibit enhanced susceptibility to antibody-mediated neutralization by anti-gp120 monoclonal antibodies as well as sera from HIV-1-infected patients, suggesting that these variants are strongly selected against in vivo (24, 33, 38).
Despite the many CD4-independent isolates identified thus far, none have been able to utilize both CXCR4 and CCR5 independently of CD4. One CD4-independent, X4-tropic variant of HIV-1, termed 8x, could fuse using CCR5 in the absence of CD4 when it contained the V3 loop of an R5-tropic virus, suggesting exposure of a region that could interact with both CXCR4 and CCR5 (33, 43). However, this Env lost the ability to fuse with CXCR4-expressing cells (33, 43), suggesting additional structural constraints on chemokine receptor usage. Moreover, in an analysis of primary HIV-2 isolates, which are characteristically dualtropic for CXCR4 and CCR5 in the presence of CD4, CD4 independence was observed for one but not both of these receptors (54). It has been proposed that structural differences between CXCR4 and CCR5 could preclude their ability to function independently of CD4 for a single Env protein and that CD4 binding may allow greater tolerance for variations in Env-chemokine receptor interactions (54).
In this study, we demonstrate that an HIV-2 Env, termed HIV-2/vcp, that was previously shown to be CD4 independent on CXCR4 (25), could also utilize rhesus (rh) CCR5 in the absence of CD4, while it required CD4 to fuse with human (hu) CCR5. Determinants on both CCR5 and gp120 that were responsible for CD4-independent use of rhCCR5 were identified. We found that a negatively charged amino acid at position 13 in the rhCCR5 N terminus and a positively charged amino acid at position 427 in the C4 domain of gp120 were required for CD4-independent fusion on CCR5. This finding suggests that an electrostatic interaction between these residues or domains occurs and likely modulates the binding affinity and/or subsequent conformational changes that are required for fusion.
MATERIALS AND METHODS
Plasmids and plasmid construction.
The env genes from HIV-2/ROD/A and HIV-2/ROD/B were cloned identically into a modified pCR3.1 vector from full-length infectious proviral molecular clones pACR23 and ROD/B.14 (55; G. Lin and J. A. Hoxie, submitted for publication). The HIV-2/vcp env gene (designated VCP) was recloned from the original pCR3.1 expression vector (25) to generate an env clone with the 5′ untranslated region from ROD/A, producing an expression vector identical to the ROD/A and ROD/B env clones except for the open reading frame (Lin and Hoxie, submitted). An env clone designated NIHzc8 was generated by Josephine Romano (University of Pennsylvania, Philadelphia) from a viral swarm of HIV-2/NIHz-infected CEM cells, provided by Celia C. LaBranche (Duke University, Durham, N.C.) in a manner similar to that described for the original VCP env clone (25). This clone was subsequently recloned into the HIV2-/vcp env clone equalized to ROD/A with BsmI 5′ and BamHI 3′, in essence exchanging the open reading frames.
Chimeras were generated between VCP and NIHzc8 with restriction enzymes HindIII in the vector and combinations of BstXI, PstI, and MaeI to generate exchanges between the V1/V2, V3, and V4/C4 regions, respectively. The QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.) was used to generate point mutations in VCP and NIHzc8 env clones. The HIV-2/vcp env clone with a deleted V1/V2 hypervariable loop (residues 110 to 194 deleted and replaced with a glycine-alanine-glycine motif) was constructed with QuikChange with the following pair of complementary primers: 5′CAAATTAACACCCTTATGTGTAGGTGCCGGCCATTGCAATACATCAGTC3′ and 5′GACTGATGTATTGCAATGGCCGGCACCTACACATAAGGGTGTTAATTTG3′.
Human and rhesus CCR5 with the corresponding position 13 change (huCCR5/N13D and rhCCR5/D13N) have been described (21). QuikChange was used to generate human and rhesus CCR5 with either a glutamic acid or an alanine at position 13. Constructs for huCCR2b and chimeras between huCCR5 and huCCR2b have been described (62). The ones used in this study were C25-01, C25-06, and C25-28. Constructs for huCXCR2 and chimeras between huCCR5 and huCXCR2 have been described (17). QuikChange was also used to introduce an aspartic acid at position 13 for chimeras containing the huCCR5 N terminus.
Rhesus CD4, CXCR4, and CCR5 in pcDNA3 were provided by Bridget A. Puffer (University of Pennsylvania). Human CD4, CXCR4, and CCR5 in pcDNA3 and the reporter plasmid encoding luciferase under the control of a T7 promoter (T7.luciferase), used in a cell-cell fusion assay, have been described previously (22, 33, 43, 61).
Cell-cell fusion assay.
A cell-cell fusion assay (22, 61) was modified as described (Lin and Hoxie, submitted for publication). Briefly, to generate effector env-expressing cells, QT6 quail fibrosarcoma cells in T-25 flasks were infected with vaccinia virus strain WR at a multiplicity of infection of 10 for 1 h at 37°C and transfected for 4 h by the standard calcium phosphate method with 6 μg of the desired env-expressing plasmid and 6 μg of pSP64.vE/L.T7 RNAP (Lin and Hoxie, submitted), a plasmid containing T7 RNA polymerase under the regulation of the synthetic vaccinia virus early and late promoter (6). Cells were incubated overnight with rifampin at a concentration of 100 μg/ml. To generate target receptor cells, QT6 quail cells in 24-well plates were transfected with the desired receptors and the T7.luciferase reporter plasmid in a total of 1 μg by the standard calcium phosphate method for 4 h and expressed overnight. Effector cells were mixed with receptor target cells and cell-cell fusion was assessed 7.5 h later by lysing with 0.5% NP-40–phosphate-buffered saline (PBS). After addition of luciferase substrate (Promega, Madison, Wis.), luciferase activity was quantified with a Wallac 1450 Microbeta Plus luminometer.
Analysis of CCR5 expression and tyrosine sulfation levels.
QT6 quail cells were transfected with huCCR5 and huCCR5 mutants by the standard calcium phosphate method. After overnight expression, cells were detached with ice-cold 1 mM EDTA-PBS and analyzed by fluorescence-activated cell sorting (FACS) (43) using a monoclonal antibody to CCR5 (2D7) directly conjugated to phycoerythrin (BD PharMingen, San Diego, Calif.). Cells were also labeled in six-well plates for 4 h with either 500 μCi total of [35S]cysteine and [35S]methionine or 500 μCi of [35S]sulfate (NEN Life Science Products, Boston, Mass.) and analyzed for CCR5 tyrosine sulfation as described (48). After radiolabeling, cells were lysed in Cymal-5 (Anatrace, Maumee, Ohio) lysis buffer containing 1% detergent, 100 mM (NH4)2SO4, 20 mM Tris-HCl (pH 7.5), 10% glycerol, and Complete protease inhibitor cocktail (Roche Molecular Biochemicals, Indianapolis, Ind.) and immunoprecipitated with 2D7 and protein A/G beads overnight. Beads were pelleted and washed with Cymal-5 lysis buffer supplemented with 500 mM NaCl. Sodium dodecyl sulfate (SDS) sample buffer was added to the beads and heated at 55°C for 1 h, and samples analyzed by SDS-polyacrylamide gel electrophoresis (PAGE). Radioactivity corresponding to either [35S]cysteine- and [35S]methionine- or [35S]sulfate-labeled CCR5 was quantified on a PhosphorImager.
RESULTS
HIV-2 exhibits CD4-independent use of rhCCR5 but not huCCR5.
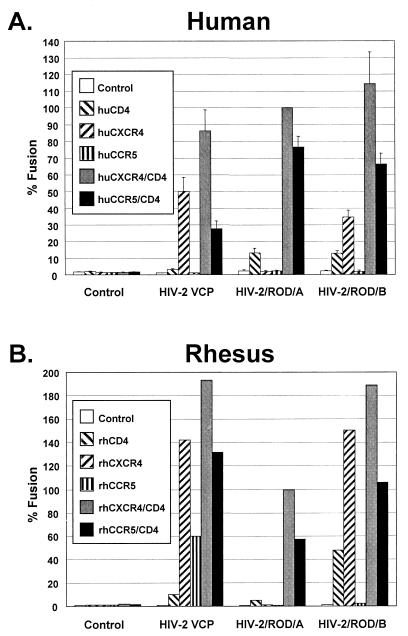
HIV-2/vcp has been shown to utilize human CXCR4 in the absence of CD4 (25). Although HIV-2 isolates are commonly dual-tropic for CXCR4 and CCR5 and are frequently CD4 independent for one of these coreceptors, no HIV-2 Envs described to date have been able to use both CXCR4 and CCR5 in the absence of CD4 (54). Coreceptor usage of VCP was further evaluated on human and rhesus CXCR4 and CCR5 in the presence and absence of CD4. We found that this Env was able to use both human and rhesus CXCR4 independently of CD4. Remarkably, it was also able to use rhCCR5 but not huCCR5 in the absence of CD4, although it could use both receptors when CD4 was present (Fig. 1A and 1B). We also evaluated HIV-2/ROD/A Env, which is CD4 dependent on CXCR4, and HIV-2/ROD/B Env, which was derived from ROD/A and is CD4 independent on CXCR4 (55). Although ROD/A and ROD/B utilized huCCR5 and rhCCR5 in the presence of CD4, neither clone could use these receptors in the absence of CD4 (Fig. 1A and 1B). Both VCP and particularly ROD/B were able to fuse when the QT6 quail target cells expressed CD4 alone, raising the possibility of an endogenous CD4-dependent coreceptor on this nonmammalian cell type. Nonetheless, these findings indicated that the HIV-2/vcp Env must contain determinants that enable it to use both CXCR4 and rhCCR5 without CD4 and that rhCCR5 must differ from huCCR5 in a way that supports CD4-independent fusion.
FIG. 1.
Evaluation of HIV-2 Env proteins with human and rhesus receptors. HIV-2 env clones VCP, ROD/A, and ROD/B were evaluated in a cell-cell fusion assay on QT6 target cells expressing (A) human or (B) rhesus receptors CXCR4 and CCR5 in the presence or absence of human or rhesus CD4. Values are represented as percent fusion, calculated using luciferase activity normalized to ROD/A fusion on CXCR4 with CD4. Mean values + standard error of the mean (SEM) are represented.
Role of CCR5 amino terminus in CD4-independent fusion.
Previous reports have shown differential use of human and rhesus CCR5 by SIV Envs. SIVmac239 gp120 binds to rhCCR5 independently of CD4 but requires soluble CD4 to bind to huCCR5 (46). SIVmac/CP-MAC can use rhCCR5 for infection in the absence of CD4 but requires CD4 to use huCCR5 (21). Both studies mapped the determinant for CD4 independence to residue 13 in the CCR5 N terminus, which is an asparagine (N) for huCCR5 and an aspartic acid (D) for rhCCR5. Thus, changing position 13 in huCCR5 to a D (huCCR5/N13D) conferred CD4 independence, while changing position 13 in rhCCR5 to an N (rhCCR5/D13N) abrogated CD4 independence (21, 46).
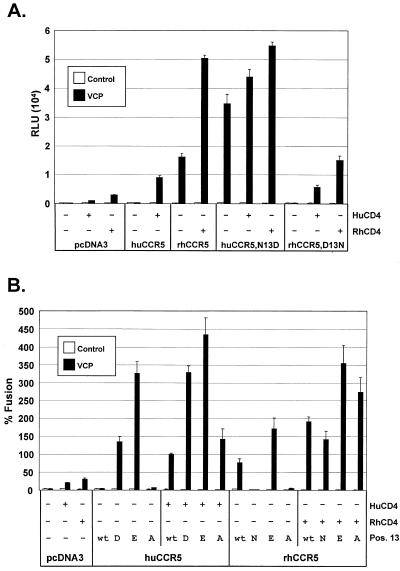
To determine if this amino acid had a similar effect for HIV-2/vcp, huCCR5/N13D and rhCCR5/D13N were evaluated in fusion assays with and without CD4 (Fig. 2A). In the presence of human or rhesus CD4, VCP could fuse with both hu- and rhCCR5, although fusion was typically greater when rhCD4 was used with rhCCR5. Although fusion with huCCR5 was CD4 dependent, it became CD4 independent with the N13D substitution. CD4-independent use of rhCCR5 was abrogated when it contained the D13N substitution, although this receptor remained functional in the presence of CD4. Thus, a D at position 13 was critical for CD4-independent fusion on CCR5 by VCP.
FIG. 2.
Human and rhesus CCR5 N-terminal mutations at position 13. (A) HIV-2/vcp was evaluated in cell-cell fusion on QT6 target cells expressing huCCR5, rhCCR5, or CCR5 mutants containing either an aspartic acid (D) or an asparagine (N) at position 13, as indicated. The ability of these CCR5 N-terminal mutants to support membrane fusion by VCP Env was assessed in the presence or absence of both human and rhesus CD4. Results are expressed as luciferase activity in relative light units (RLU). (B) VCP Env fusion was also assessed with CCR5 mutants containing either a glutamic acid (E) or an alanine (A) at position 13, as indicated. Values are represented as percent fusion, calculated using RLU normalized to VCP fusion on huCCR5 with huCD4. Mean values + SEM are represented.
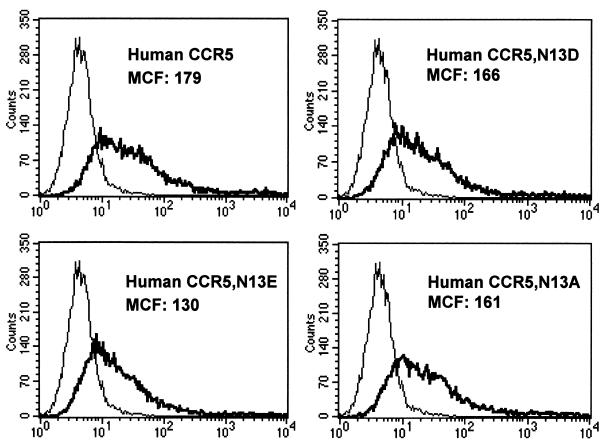
To determine if CD4-independent use of rhCCR5 was due to the presence of the aspartic acid, the mere presence of a negative charge, or the specific loss of an asparagine, hu- and rhCCR5 mutants that contained either a glutamic acid (E) or an alanine (A) at position 13 were created. FACS analysis using a monoclonal antibody (2D7) that binds to a determinant in the second extracellular loop of CCR5 (44, 63) demonstrated that all huCCR5 mutants were expressed at levels comparable to wild-type huCCR5 (Fig. 3). Although the HIV-2/vcp Env was able to fuse with each construct in the presence of CD4, it was only CD4 independent on CCR5 molecules that contained a D or an E, but not an N or an A (Fig. 2B). Therefore, the ability of rhCCR5 to support CD4-independent fusion by HIV-2/vcp was due to the presence of a negative charge in the N terminus at position 13.
FIG. 3.
Cell surface expression of CCR5 mutants. QT6 quail cells were transiently transfected with huCCR5 or huCCR5 mutants by the standard calcium phosphate method, and after overnight expression, cells were detached with ice-cold 1 mM EDTA-PBS and analyzed by FACS using a monoclonal antibody to CCR5 (2D7) directly conjugated to phycoerythrin. Histograms are labeled with mean channel fluorescence (MCF). Cells expressing CCR5 are shown with a bold line, while cells transfected with vector alone are shown with a thin line.
CCR5 N terminus can confer CD4-independent fusion on CCR5 by HIV-2/vcp to heterologous chemokine receptors.
Interactions of HIV with CCR5 are conformationally complex, involving to at least some degree all extracellular domains of this coreceptor. However, the N-terminal domain of CCR5 and the second extracellular loop are particularly important regions for Env interactions (4). In addition to being CD4 dependent on huCCR5, VCP also exhibited CD4-dependent fusion with cells expressing huCCR2b or huCXCR2 (Fig. 4A, data not shown), which are 76 and 34% homologous to huCCR5, respectively (17, 62). To determine if the N terminus of CCR5 with a negative charge at position 13 could confer CD4 independence on other receptors, we analyzed usage of CCR2b and CXCR2 chimeras containing the N terminus of CCR5 or other domains of CCR5. These receptor chimeras were previously shown to be functional for other CCR5-tropic HIV-1 Envs (17).
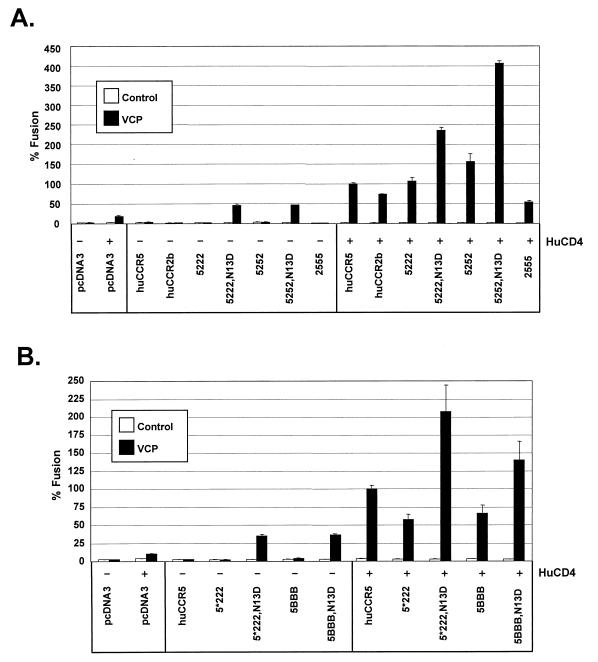
FIG. 4.
Role of CCR5 N terminus in CD4-independent use of CCR2b and CXCR2. HIV-2/vcp was evaluated in cell-cell fusion on QT6 target cells expressing (A) human CCR2b/CCR5 chimeras with the entire N terminus as well as the first transmembrane domain exchanged, or (B) human CCR2b/CCR5 and CXCR2/CCR5 chimeras containing a minimal N-terminal exchange made at the conserved cysteine at position 20. Nomenclature shows the first number representing the N terminus and the second, third, and fourth numbers representing the second, third, and fourth extracellulat loops, respectively. 5 refers to CCR5, 2 refers to CCR2b, and B refers to CXCR2. An asterisk (5*222) indicates that this construct differs from 5222 in that the N-terminal exchange was made at cysteine 20. CCR5 N-terminal chimeras denoted N13D contained an aspartic acid at residue 13. Values are represented as percent fusion, calculated using luciferase activity normalized to VCP fusion on huCCR5 with huCD4. Mean values + SEM are represented.
CCR2b chimeras that contained either the entire N-terminal domain of CCR5 or only the first 20 residues supported CD4-dependent fusion with VCP (Fig. 4A and 4B). However, these chimeras supported CD4-independent fusion when residue 13 was changed to an aspartic acid (Fig. 4A and 4B). The same observation was made with the more heterologous CXCR2/CCR5 N-terminal chimeras (Fig. 4B). Notably, the second extracellular loop of CCR5 was not critical for CD4 independence, since on the CCR5/CCR2b chimeras, VCP used 5222/N13D without CD4 just as well as it used 5252/N13D (Fig. 4A). Therefore, the N-terminal domain of huCCR5 containing a negatively charged residue at position 13 was sufficient to confer CD4-independent fusion by HIV-2/vcp onto heterologous chemokine receptors.
Tyrosine sulfation levels of CCR5 mutants.
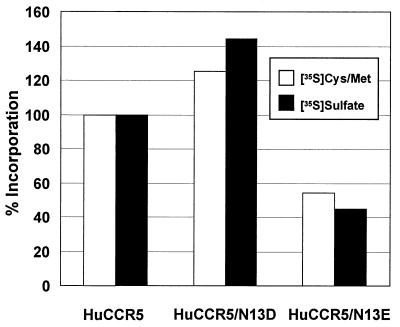
Tyrosines in the N terminus of CCR5 at positions 3, 10, 14, and 15 are sulfated and play an important role in gp120 binding and efficient HIV entry (13, 26, 27). Because acidic residues surrounding tyrosines serve as tyrosine sulfation signals, especially acidic residues at the −1 position from a tyrosine (5, 36, 45, 60), we considered the possibility that a D or an E at position 13 could improve the efficiency of CCR5 utilization and permit CD4 independence by increasing the level of tyrosine sulfation. Therefore, we examined the relative levels of tyrosine sulfation between huCCR5 and huCCR5 containing a D or an E at position 13 by radiolabeling with either [35S]sulfate or [35S]cysteine and [35S]methionine as previously described (48). By using [35S]cysteine and [35S]methionine incorporation as a means to normalize for CCR5 expression, we were able to determine the relative levels of sulfation. We found that [35S]sulfate incorporation into huCCR5/N13D and huCCR5/N13E relative to wild-type CCR5 was 115 and 82%, respectively (Fig. 5), indicating that differences in tyrosine sulfation could not account for the CD4-independent fusion. Thus, a negatively charged amino acid at position 13 may be more directly involved in CD4-independent use of rhCCR5.
FIG. 5.
Tyrosine sulfation levels of CCR5 mutants. QT6 quail cells were transiently transfected with huCCR5 or huCCR5 mutants by the standard calcium phosphate method, radiolabeled with [35S]cysteine and [35S]methionine or [35S]sulfate, lysed, immunoprecipitated with 2D7, and analyzed by SDS-PAGE as described in the text. Radioactivity corresponding to either [35S]cysteine- and [35S]methionine- or [35S]sulfate-labeled CCR5 was quantified on a PhosphorImager. Background radioactive counts from pcDNA3 control-transfected cells were subtracted, and results are expressed as the relative number of cpm normalized to huCCR5. huCCR5/N13D and N13E ratios for [35S]sulfate to [35S]cysteine and [35S]methionine labeling were 115 and 82%, respectively.
Derivation and sequence analysis of a CD4-dependent clone from HIV-2/NIHz compared to HIV-2/vcp.
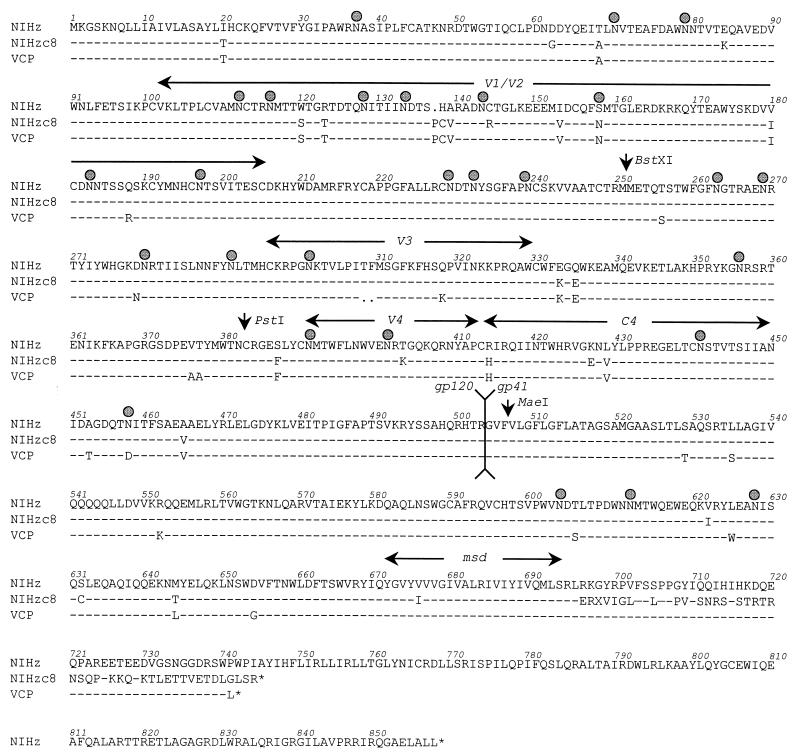
To identify regions within the HIV-2/vcp Env that determined its CD4 independence on CCR5, we derived an env clone from an early passage of the parental HIV-2/NIHz isolate. This clone, designated NIHzc8, utilized both CCR5 and CXCR4 in fusion assays, but only when CD4 was present (see Fig. 7; data not shown). The sequences of the NIHzc8 and VCP Env clones are shown and compared to the published sequence of NIHz (Fig. 6) (75). VCP differed from NIHzc8 by 11 amino acids in gp120 and 9 amino acids in the gp41 ecto- and membrane-spanning domains. Both NIHzc8 and NIHz contain an unpaired cysteine in the V1/V2 domain, which in HIV-2 and SIV contains four additional cysteines compared to HIV-1 (35). NIHzc8 also acquired a cysteine at position 632 in the gp41 ectodomain. In addition, relative to NIHz, NIHzc8 and VCP contained mutations that resulted in the loss of a predicted N-linked glycosylation site at position 401 and 458, respectively. NIHzc8 also has a frameshift mutation in the TM cytoplasmic domain at position 696 that results in an aberrant and prematurely truncated tail of 34 amino acids. Similar to many laboratory-adapted HIV-2 and SIV isolates (37, 41), VCP contained a premature stop codon in its cytoplasmic tail.
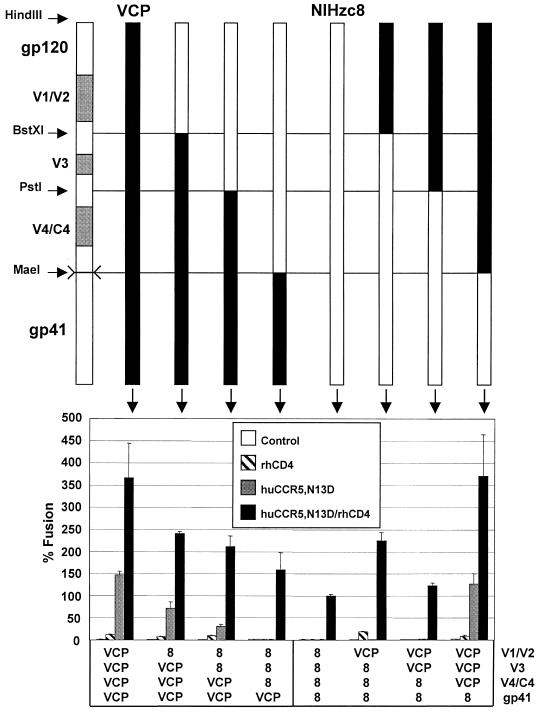
FIG. 7.
Mapping determinants for CD4-independent use of CCR5 with progressive N- to C-terminal exchanges. HIV-2/vcp and HIV-2/NIHzc8 chimeras were constructed using the indicated restriction sites (see also Fig. 6) and evaluated in cell-cell fusion on QT6 target cells expressing huCCR5/N13D with and without rhesus CD4. Values are represented as percent fusion, calculated using luciferase activity normalized to NIHzc8 fusion on huCCR5/N13D with rhCD4. Mean values + SEM are represented.
FIG. 6.
Sequence analysis of HIV-2 env clones NIHzc8 and VCP in comparison to NIHz. Sequences are shown for the HIV-2/vcp env in comparison to HIV-2/NIHz (75) and an env clone derived from HIV-2/NIHz-infected cells (NIHzc8). NIHzc8 is CD4 dependent for rhesus and human CCR5 and CXCR4 (not shown). Positions of variable loops, conserved domain 4, the gp120/gp41 cleavage site, and the TM membrane-spanning domain (msd) are indicated. Predicted N-linked glycosylation sites are represented with a shaded circle. NIHzc8 contains a frameshift mutation at position 696, resulting in a prematurely truncated cytoplasmic tail at position 745. The VCP cytoplasmic tail is also prematurely truncated at position 742. NIHzc8 and VCP have each lost a consensus N-linked glycosylation site at positions 401 and 458, respectively. Restriction sites (BstXI, PstI, and MaeI) used to construct NIHzc8 and VCP Env chimeras are indicated with a vertical arrow.
Mapping Env determinants for CD4 independence using Env chimeras.
To identify the structural determinants in HIV-2/vcp Env that enabled it to use rhCCR5 independently of CD4, chimeras were made between VCP and the CD4-dependent NIHzc8 clone. Env chimeras were constructed using available restriction sites to exchange the V1/V2, the V3, and the V4/C4 loops in various combinations, as well as the entire gp120 and gp41 subunits (Fig. 6, 7, and 8). Fusion assays were performed on cells expressing huCCR5/N13D, since we hoped to identify determinants on Env that would likely interact with residue 13 in the CCR5 N terminus. All constructs were competent for fusion when CD4 was present, although the levels of activity varied. The CD4-independent activity of VCP clearly resided within gp120, since the NIHzc8 gp120 paired with VCP gp41 lost CD4 independence, while a chimera containing VCP gp120 with the NIHzc8 gp41 was fully CD4 independent (Fig. 7). Thus, the marked differences in the cytoplasmic domains of these proteins had no obvious effect on the CD4-independent phenotype, nor did other changes in the gp41 ectodomain.
FIG. 8.
Mapping determinants for CD4-independent use of CCR5 with exchanges of gp120 subdomains. HIV-2/vcp and HIV-2/NIHzc8 chimeras were constructed using the indicated restriction sites and evaluated in cell-cell fusion on QT6 target cells expressing huCCR5/N13D with and without rhesus CD4. Values are represented as percent fusion, calculated using luciferase activity normalized to NIHzc8 fusion on huCCR5/N13D with rhCD4. Mean values + SEM are represented.
Within gp120, the V4/C4 domain appeared to be particularly important, since VCP chimeras containing the NIHzc8 V4/C4 domain alone or in combination with the V1/V2 or V3 domain became CD4 dependent on huCCR5/N13D (Fig. 8). However, the VCP V4/C4 domain could not by itself confer CD4 independence on NIHzc8 (Fig. 8), although introducing the VCP V4/C4 region onto an NIHzc8 background in combination with either V1/V2 or V3 did confer CD4 independence (Fig. 8). Therefore, the VCP V4/C4 domain was necessary but not sufficient to confer CD4 independence to NIHzc8.
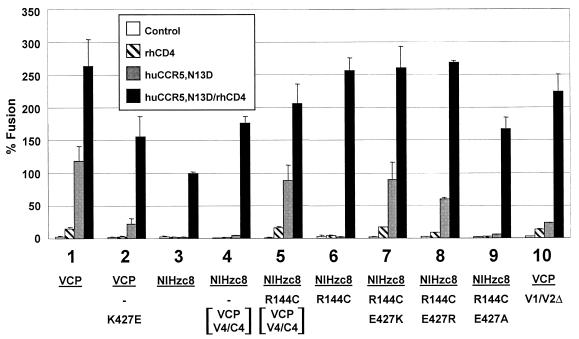
In analyzing sequence differences between NIHzc8 and VCP, the loss of a cysteine in the NIHzc8 V1/V2 loop was intriguing because unpaired cysteine residues in the ectodomains of HIV and SIV glycoproteins are uncommon. Although not sufficient to confer CD4 independence on NIHzc8, the VCP V1/V2 clearly contributed since this domain, as noted above, in conjunction with V4/C4 rendered NIHzc8 fully CD4 independent (Fig. 8). We therefore considered the possibility that the absence of C144 in NIHzc8 disrupted the V1/V2 domain and adversely affected Env function. When we restored a cysteine at position 144 in the NIHzc8 chimera containing the VCP V4/C4 region, we found that the resulting construct was fully CD4 independent (Fig. 9, compare numbers 4, 5, and 6). These findings indicated that differences between the NIHzc8 and VCP V4/C4 domains are important for CD4-independent use of CCR5.
FIG. 9.
Fine mapping of determinants of CD4 independence. Point mutations were introduced into HIV-2/vcp and HIV-2/NIHzc8 env clones and chimeras as depicted at positions 144 and 427 through QuikChange site-directed mutagenesis and evaluated in cell-cell fusion on QT6 target cells expressing huCCR5/N13D with and without rhesus CD4. Constructs 4 and 5 contain the VCP V4/C4 domains (PstI to MaeI fragment) on an NIHzc8 background. An HIV-2/vcp env construct containing a deleted V1/V2 hypervariable loop that was replaced with a glycine-alanine-glycine motif was also evaluated. Values are represented as percent fusion, calculated using luciferase activity normalized to NIHzc8 fusion on huCCR5/N13D with rhCD4. Mean values + SEM are represented.
Identification of lysine 427 within V4/C4 domain as a determinant for CD4-independent use of CCR5.
VCP differed from NIHzc8 at four positions in the V4/C4 domain (403, 427, 453, and 458) (Fig. 6). Among these changes, we were particularly interested in the difference at position 427, since that resulted in a change to a positively charged lysine in VCP from the negatively charged glutamic acid in NIHzc8 (Fig. 6). Given the importance of a negatively charged residue at position 13 in the CCR5 N terminus for CD4-independent fusion, we hypothesized that a gain in positive charges would facilitate VCP CD4 independence. Indeed, by changing residue 427 to a lysine in the NIHzc8/R144C background, CD4 independence was conferred (Fig. 9, compare numbers 6 and 7). Moreover, CD4 independence was also conferred if position 427 was changed to a different positively charged residue (arginine), but was lost if an alanine was substituted (Fig. 9, numbers 8 and 9). Conversely, when position 427 in VCP was changed to a glutamic acid, a considerable degree of CD4 independence was lost (Fig. 9, number 2). Despite its role in CD4 independence, VCP residue C144 was not believed to contribute to the interaction with residue 13 in the CCR5 N terminus because VCP Env with the entire V1/V2 hypervariable loop deleted retained some degree of CD4 independence (Fig. 9, number 10). Therefore, similar to the analysis of the CCR5 receptor, what was critical for CD4 independence on Env was the presence of a positively charged residue at position 427 in the C4 region and not a specific basic residue.
DISCUSSION
The fusion activity of HIV and SIV Envs is typically triggered by sequential interactions with CD4 and a coreceptor. For primary HIV-1 strains, CD4 binding induces structural alterations in gp120 that enable it to interact with CCR5 and/or CXCR4 (67, 72). While coreceptor binding is needed for membrane fusion, it is not clear exactly how coreceptors bind to Env, nor are the structural consequences of this interaction well understood. CD4-independent virus strains provide a means to dissect the roles played by CD4 and coreceptor binding in the virus entry process, since coreceptor binding alone is sufficient to elicit membrane fusion for these viruses (20, 23, 25, 33, 39, 43, 54, 55).
We have previously shown that HIV-2/vcp can efficiently use CXCR4 independently of CD4 (25). In the present study we found that VCP could also use rhesus but not human CCR5 independently of CD4. Of the four extracellular differences between huCCR5 and rhCCR5 (9), the critical determinant for CD4-independent use of rhCCR5 by VCP mapped solely to residue 13 in the CCR5 N terminus. Specifically, an acidic amino acid at this position enabled either huCCR5 or rhCCR5 to support CD4-independent fusion. Potential indirect effects of a negatively charged residue at position 13, such as an increase in coreceptor expression or tyrosine sulfation of the CCR5 N terminus, which would have confounded our interpretation, were ruled out. Previous studies have shown that SIVmac239 gp120 binding to rhCCR5 and CD4-independent SIVmac/CP-MAC infection are likewise dependent on an acidic residue at position 13 in the CCR5 N terminus (21, 46). Moreover, the N-terminal domain of huCCR5 with a negatively charged residue at position 13 was sufficient to confer CD4-independent use by VCP on CCR2b and CXCR2, underscoring the importance of the CCR5 N terminus for Env binding and efficient coreceptor use. Consistent with this view, CCR5 N-terminal peptides containing sulfated tyrosines at position 10 and 14 have been shown to bind gp120-CD4 complexes from HIV-1 CCR5-tropic isolates and to inhibit virus entry, suggesting that this domain by itself adopts a conformation that binds directly to gp120 (13, 27).
By using chimeras between VCP and a closely related CD4-dependent Env, we identified a critical determinant on Env for CD4-independent use of CCR5 as a positively charged residue in the C4 domain of gp120 at position 427. By homology analysis, VCP residue 427 corresponds to HIV-1 HXBc2 residue 432, which lies in the highly conserved four-stranded, antiparallel β-sheet implicated in forming the CCR5 binding site (Fig. 10) (42, 57). The requirement for a positively charged residue at this position in gp120 coupled with the need for a negatively charged residue at position 13 in CCR5 suggests that an electrostatic interaction between these residues is important for the CD4-independent phenotype. A number of other basic residues are located in the CCR5 binding site, including several that have been shown to be important for CCR5 binding (Fig. 10) (57). These basic residues could potentially interact with negatively charged residues in the CCR5 N terminus, including the sulfated tyrosines. However, our results with VCP are clearly context dependent, since there are other R5-tropic, CD4-independent isolates that can use human CCR5, which lacks the aspartic acid at position 13 (21, 23, 33, 39, 58). In addition, the V3 loop of Env is also implicated in interacting with the CCR5 N terminus and plays a critical role in coreceptor choice (10–12, 27, 66). Finally, the CCR5 extracellular loops also play a role in HIV entry (17, 62), although it is not clear how they interact with Env.
FIG. 10.
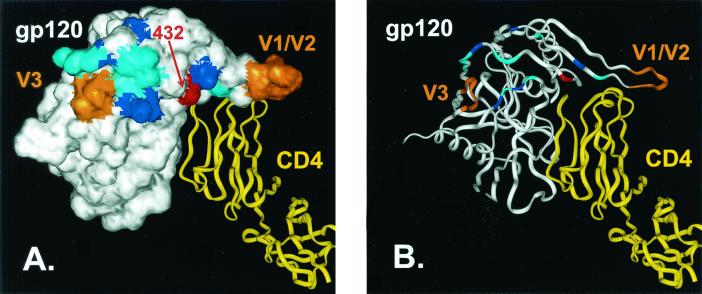
Positively charged residues in the putative CCR5 binding site on gp120. A space-filling model (A) and a ribbon diagram (B) of the HIV-1 gp120 core crystal structure (white) is depicted complexed with a ribbon diagram of CD4 (yellow) (42). HIV-1 amino acid 432, which corresponds to residue 427 of NIHzc8 (Glu), is shown in red. Amino acid positions at which mutations produced a ≥50% decrease in HIV-1/YU2 gp120 binding to CCR5 are shown in cyan. Positively charged residues included within this region (K121, K207, R419, K421, and R440) are shown in blue (58). Positions of the V1/V2 stem and the base of the V3 loop are shown in orange. As shown in Fig. 9, E427K and E427R mutations in NIHzc8 enabled this clone to use huCCR5/N13D independently of CD4.
How does a single amino acid change in gp120 result in a CD4-independent phenotype? While it could be argued that the lysine at position 427 is involved in exposing the coreceptor binding site on gp120, a VCP Env containing a lysine to glutamic acid substitution at position 427 remained highly CD4 independent on CXCR4 (data not shown). Thus, we consider it less likely that a basic residue at this position simply exposes the coreceptor binding site. However, it is possible that the region in the bridging sheet responsible for CXCR4 binding is at least somewhat different from the region implicated in CCR5 binding. Thus, the acquisition of a positively charged amino acid at 427 could result in enhanced exposure of that portion of the coreceptor binding site needed to interact with CCR5.
It is also possible that the electrostatic interactions that we have implicated enhance the affinity and/or avidity of Env-coreceptor binding. Recent work indicates that multiple coreceptor binding events are needed for optimal fusion activity of Env trimers (40). Thus, Envs that bind coreceptor with high affinity will engage multiple coreceptors more quickly than Envs that bind with low affinity, resulting in more efficient triggering. While our studies have not yet addressed the affinity of the VCP Env for CCR5, studies by Cormier et al. (13) have shown that gp120 binding to sulfated peptides from the CCR5 N terminus can be quantitated. It will be of great interest to determine if CD4-dependent and CD4-independent use of hu- and rhCCR5, respectively, corresponds to differences in affinity of VCP gp120 binding to sulfated peptides from these receptors.
Finally, it is possible that an electrostatic interaction between residue 427 in VCP gp120 and residue 13 in CCR5 allows more efficient triggering of postbinding, fusion-inducing conformational changes in Env. Thermodynamic analysis of the gp120-CD4 binding reaction indicates that extensive structural rearrangements in gp120 occur upon CD4 binding that likely involve movements of the V1/V2 and V3 hypervariable loops as well as formation of the bridging sheet, which connects the inner and outer domains of gp120 (49, 50, 72, 73). However, the exact structural consequences of coreceptor binding to Env are unknown, and it is unclear how this interaction leads to conformational changes in gp41 and formation of the six α-helical bundle that ultimately drives membrane fusion (7, 47, 59, 64, 70). VCP Env can interact with huCCR5, since fusion occurs when CD4 is present. CD4, in addition to Env binding, may serve to facilitate the coreceptor-induced structural changes in Env that huCCR5 cannot induce alone. Without CD4, the more favorable electrostatic interaction provided by a negatively charged residue at position 13 in rhCCR5 may serve to transmit signals to gp41 that enable subsequent conformational changes to occur.
It is likely that CD4 binding allows more genetic variation in the Env-chemokine receptor interaction to be tolerated. In this context, CD4 may serve not only to induce favorable conformational changes for coreceptor binding, but could also contribute to the net avidity of Env with a cellular CD4-chemokine receptor complex (74). However, our demonstration that VCP can use both CXCR4 and CCR5 independently of CD4 indicates that these divergent coreceptors clearly can adopt a conformation that permits a direct interaction with a single gp120. Nonetheless, coreceptor binding may itself be a complex and cooperative process involving several Env chemokine receptor subdomains. While this study has implicated an interaction between the CCR5 N terminus and the gp120 C4 domain, another study of an X4-tropic, CD4-independent HIV-2 implicated an interaction with the CXCR4 second extracellular loop (53). Studies to identify the determinants and mechanisms for CD4-independent use of CXCR4 by HIV-2/vcp may help to identify additional coreceptor domains that have been optimized by this Env.
Given the relatively broad expression pattern of CXCR4 in human tissues and the more restricted expression of CD4, CD4-independent viruses would be expected to exhibit expanded tropism in vivo. However, primary CD4-independent HIV-1 isolates have not yet been identified, perhaps because of their markedly enhanced sensitivity to neutralizing antibodies, resulting in selective immune pressure in vivo against their emergence (24, 33, 38). A CD4-independent HIV-1 gp120 protein that we have described has been shown to have a stably exposed coreceptor binding site, raising the possibility that such genetically triggered Env proteins may also elicit antibodies against this highly conserved region (31, 33). The feasibility of targeting cryptic epitopes on Env or fusion intermediates is evidenced by the ability of strategically deglycosylated Envs to generate antibody responses with increased neutralizing activity (56) and the success of peptide-based pharmaceuticals in preventing membrane fusion (7, 47, 59, 64, 70). Combining immunologic and pharmacologic strategies that target concealed epitopes and fusion intermediates may be highly synergistic in inhibiting HIV infection (16).
In summary, our findings have implicated an electrostatic interaction between an HIV-2 gp120 and the CCR5 N terminus as playing a key role in events that lead to viral entry. Although CD4 likely modulates the exposure, affinity, and/or orientation of Env that enables it to engage coreceptors, an accumulation of favorable charge-charge interactions can apparently obviate the need for CD4, at least in the context of cell-cell fusion used in our analysis. In addition, accessory molecules such as the newly discovered C-type lectins DC-SIGN and DC-SIGNR, which also bind HIV Env, are generating increased interest as factors that can enhance HIV transmission to T cells via a trans mechanism (2, 29, 30, 51, 52, 65). Whether these other attachment mediators also induce structural changes in Env that bear on interactions with CD4 and coreceptors or simply serve to bind virus remains to be determined. The development of a molecular model that takes into account all components of viral entry will lead to the more rational design of pharmacologic and immunologic inhibitors that target Env domains critical to the viral entry process. CD4-independent isolates that interact with both CXCR4 and CCR5 may be particularly useful in this regard.
ACKNOWLEDGMENTS
We thank Bridget A. Puffer, Jacqueline D. Reeves, and Josephine Romano for plasmids; George J. Leslie for technical assistance; Michael Farzan for technical advice; and Celia C. LaBranche for providing the HIV-2/NIHz virus stock.
G.L. was supported by the National Institutes of Health Medical Scientist Training Program and the Cell and Molecular Biology Training Grant. B.L. was supported by NIH grant KO8 HL03923 and a Burroughs Wellcome Fund Career Development Award. J.A.H. and R.W.D. were supported by NIH grant RO1 AI45378 and by an Elizabeth Glaser Scientist Award from the Pediatric AIDS Foundation and a Burroughs Wellcome Fund Translational Research Award to R.W.D. Support was also provided by the Penn Center for AIDS Research, NIH grant P30 AI45008.
REFERENCES
- 1.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 2.Bashirova A A, Geijtenbeek T B, van Duijnhoven G C, van Vliet S J, Eilering J B, Martin M P, Wu L, Martin T D, Viebig N, Knolle P A, KewalRamani V N, van Kooyk Y, Carrington M. A dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin (DC-SIGN)-related protein is highly expressed on human liver sinusoidal endothelial cells and promotes hiv-1 infection. J Exp Med. 2001;193:671–678. doi: 10.1084/jem.193.6.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger E A. HIV entry and tropism: when one receptor is not enough. Adv Exp Med Biol. 1998;452:151–157. [PubMed] [Google Scholar]
- 4.Berson J F, Doms R W. Structure-function studies of the HIV-1 coreceptors. Semin Immunol. 1998;10:237–248. doi: 10.1006/smim.1998.0130. [DOI] [PubMed] [Google Scholar]
- 5.Bundgaard J R, Vuust J, Rehfeld J F. New consensus features for tyrosine O-sulfation determined by mutational analysis. J Biol Chem. 1997;272:21700–21705. doi: 10.1074/jbc.272.35.21700. [DOI] [PubMed] [Google Scholar]
- 6.Chakrabarti S, Sisler J R, Moss B. Compact, synthetic, vaccinia virus early/late promoter for protein expression. Biotechniques. 1997;23:1094–1097. doi: 10.2144/97236st07. [DOI] [PubMed] [Google Scholar]
- 7.Chan D C, Chutkowski C T, Kim P S. Evidence that a prominent cavity in the coiled coil of HIV type 1 gp41 is an attractive drug target. Proc Natl Acad Sci USA. 1998;95:15613–15617. doi: 10.1073/pnas.95.26.15613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan D C, Fass D, Berger J M, Kim P S. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 9.Chen Z, Zhou P, Ho D D, Landau N R, Marx P A. Genetically divergent strains of simian immunodeficiency virus use CCR5 as a coreceptor for entry. J Virol. 1997;71:2705–2714. doi: 10.1128/jvi.71.4.2705-2714.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho M W, Lee M K, Carney M C, Berson J F, Doms R W, Martin M A. Identification of determinants on a dualtropic human immunodeficiency virus type 1 envelope glycoprotein that confer usage of CXCR4. J Virol. 1998;72:2509–2515. doi: 10.1128/jvi.72.3.2509-2515.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 12.Cocchi F, DeVico A L, Garzino-Demo A, Cara A, Gallo R C, Lusso P. The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat Med. 1996;2:1244–1247. doi: 10.1038/nm1196-1244. [DOI] [PubMed] [Google Scholar]
- 13.Cormier E G, Persuh M, Thompson D A, Lin S W, Sakmar T P, Olson W C, Dragic T. Specific interaction of CCR5 amino-terminal domain peptides containing sulfotyrosines with HIV-1 envelope glycoprotein gp120. Proc Natl Acad Sci USA. 2000;97:5762–5767. doi: 10.1073/pnas.97.11.5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 15.Doms R W. Beyond receptor expression: the influence of receptor conformation, density, and affinity in HIV-1 infection. Virology. 2000;276:229–237. doi: 10.1006/viro.2000.0612. [DOI] [PubMed] [Google Scholar]
- 16.Doms R W, Moore J P. HIV-1 membrane fusion: targets of opportunity. J Cell Biol. 2000;151:F9–F14. doi: 10.1083/jcb.151.2.f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doranz B J, Lu Z H, Rucker J, Zhang T Y, Sharron M, Cen Y H, Wang Z X, Guo H H, Du J G, Accavitti M A, Doms R W, Peiper S C. Two distinct CCR5 domains can mediate coreceptor usage by human immunodeficiency virus type 1. J Virol. 1997;71:6305–6314. doi: 10.1128/jvi.71.9.6305-6314.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the beta- chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 19.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 20.Dumonceaux J, Nisole S, Chanel C, Quivet L, Amara A, Baleux F, Briand P, Hazan U. Spontaneous mutations in the env gene of the human immunodeficiency virus type 1 NDK isolate are associated with a CD4-independent entry phenotype. J Virol. 1998;72:512–519. doi: 10.1128/jvi.72.1.512-519.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edinger A L, Blanpain C, Kunstman K J, Wolinsky S M, Parmentier M, Doms R W. Functional dissection of CCR5 coreceptor function through the use of CD4-independent simian immunodeficiency virus strains. J Virol. 1999;73:4062–4073. doi: 10.1128/jvi.73.5.4062-4073.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edinger A L, Doms R W. A cell-cell fusion assay to monitor HIV-1 Env interactions with chemokine receptors. In: Michael N L, Kim J H, editors. Methods in molecular medicine. Vol. 17. Totowa, N.J: Humana Press, Inc.; 1999. pp. 41–49. [DOI] [PubMed] [Google Scholar]
- 23.Edinger A L, Mankowski J L, Doranz B J, Margulies B J, Lee B, Rucker J, Sharron M, Hoffman T L, Berson J F, Zink M C, Hirsch V M, Clements J E, Doms R W. CD4-independent, CCR5-dependent infection of brain capillary endothelial cells by a neurovirulent simian immunodeficiency virus strain. Proc Natl Acad Sci USA. 1997;94:14742–14747. doi: 10.1073/pnas.94.26.14742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edwards T G, Hoffman T L, Baribaud F, Wyss S, LaBranche C C, Romano J, Adkinson J, Sharron M, Hoxie J A, Doms R W. Relationships between CD4 independence, neutralization sensitivity, and exposure of a CD4-induced epitope in a human immunodeficiency virus type 1 envelope protein. J Virol. 2001;75:5230–5239. doi: 10.1128/JVI.75.11.5230-5239.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Endres M J, Clapham P R, Marsh M, Ahuja M, Turner J D, McKnight A, Thomas J F, Stoebenau-Haggarty B, Choe S, Vance P J, Wells T N, Power C A, Sutterwala S S, Doms R W, Landau N R, Hoxie J A. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell. 1996;87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 26.Farzan M, Mirzabekov T, Kolchinsky P, Wyatt R, Cayabyab M, Gerard N P, Gerard C, Sodroski J, Choe H. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell. 1999;96:667–676. doi: 10.1016/s0092-8674(00)80577-2. [DOI] [PubMed] [Google Scholar]
- 27.Farzan M, Vasilieva N, Schnitzler C E, Chung S, Robinson J, Gerard N P, Gerard C, Choe H, Sodroski J. A tyrosine-sulfated peptide based on the N terminus of CCR5 interacts with a CD4-enhanced epitope of the HIV-1 gp120 envelope glycoprotein and inhibits HIV-1 entry. J Biol Chem. 2000;275:33516–33521. doi: 10.1074/jbc.M007228200. [DOI] [PubMed] [Google Scholar]
- 28.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 29.Geijtenbeek T B, Kwon D S, Torensma R, van Vliet S J, van Duijnhoven G C, Middel J, Cornelissen I L, Nottet H S, KewalRamani V N, Littman D R, Figdor C G, van Kooyk Y. DC-SIGN, a dendritic cell-specific HIV-1-binding protein that enhances trans-infection of T cells. Cell. 2000;100:587–597. doi: 10.1016/s0092-8674(00)80694-7. [DOI] [PubMed] [Google Scholar]
- 30.Geijtenbeek T B, Torensma R, van Vliet S J, van Duijnhoven G C, Adema G J, van Kooyk Y, Figdor C G. Identification of DC-SIGN, a novel dendritic cell-specific ICAM-3 receptor that supports primary immune responses. Cell. 2000;100:575–585. doi: 10.1016/s0092-8674(00)80693-5. [DOI] [PubMed] [Google Scholar]
- 31.Hoffman T L, Canziani G, Jia L, Rucker J, Doms R W. A biosensor assay for studying ligand-membrane receptor interactions: binding of antibodies and HIV-1 Env to chemokine receptors. Proc Natl Acad Sci USA. 2000;97:11215–11220. doi: 10.1073/pnas.190274097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoffman T L, Doms R W. Chemokines and coreceptors in HIV/SIV-host interactions. AIDS. 1998;12:S17–S26. [PubMed] [Google Scholar]
- 33.Hoffman T L, LaBranche C C, Zhang W, Canziani G, Robinson J, Chaiken I, Hoxie J A, Doms R W. Stable exposure of the coreceptor-binding site in a CD4-independent HIV-1 envelope protein. Proc Natl Acad Sci USA. 1999;96:6359–6364. doi: 10.1073/pnas.96.11.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoffman T L, Stephens E B, Narayan O, Doms R W. HIV type I envelope determinants for use of the CCR2b, CCR3, STRL33, and APJ coreceptors. Proc Natl Acad Sci USA. 1998;95:11360–11365. doi: 10.1073/pnas.95.19.11360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoxie J A. Hypothetical assignment of intrachain disulfide bonds for HIV-2 and SIV envelope glycoproteins. AIDS Res Hum Retroviruses. 1991;7:495–499. doi: 10.1089/aid.1991.7.495. [DOI] [PubMed] [Google Scholar]
- 36.Kehoe J W, Bertozzi C R. Tyrosine sulfation: a modulator of extracellular protein-protein interactions. Chem Biol. 2000;7:R57–R61. doi: 10.1016/s1074-5521(00)00093-4. [DOI] [PubMed] [Google Scholar]
- 37.Kodama T, Wooley D P, Naidu Y M, Kestler H W, 3rd, Daniel M D, Li Y, Desrosiers R C. Significance of premature stop codons in env of simian immunodeficiency virus. J Virol. 1989;63:4709–4714. doi: 10.1128/jvi.63.11.4709-4714.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kolchinsky P, Kiprilov E, Sodroski J. Increased neutralization sensitivity of CD4-independent human immunodeficiency virus variants. J Virol. 2001;75:2041–2050. doi: 10.1128/JVI.75.5.2041-2050.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kolchinsky P, Mirzabekov T, Farzan M, Kiprilov E, Cayabyab M, Mooney L J, Choe H, Sodroski J. Adaptation of a CCR5-using, primary human immunodeficiency virus type 1 isolate for CD4-independent replication. J Virol. 1999;73:8120–8126. doi: 10.1128/jvi.73.10.8120-8126.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuhmann S E, Platt E J, Kozak S L, Kabat D. Cooperation of multiple CCR5 coreceptors is required for infections by human immunodeficiency virus type 1. J Virol. 2000;74:7005–7015. doi: 10.1128/jvi.74.15.7005-7015.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuiken C, Foley B, Hahn B, Marx P, McCutchan F, Mellors J, Mullins J, Wolinsky S, Korber B. Human retroviruses and AIDS. Los Alamos, N. Mex: Theoretical Biology and Biophysics, Los Alamos National Laboratory; 1999. [Google Scholar]
- 42.Kwong P D, Wyatt R, Robinson J, Sweet R W, Sodroski J, Hendrickson W A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.LaBranche C C, Hoffman T L, Romano J, Haggarty B S, Edwards T G, Matthews T J, Doms R W, Hoxie J A. Determinants of CD4 independence for a human immunodeficiency virus type 1 variant map outside regions required for coreceptor specificity. J Virol. 1999;73:10310–10319. doi: 10.1128/jvi.73.12.10310-10319.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee B, Sharron M, Blanpain C, Doranz B J, Vakili J, Setoh P, Berg E, Liu G, Guy H R, Durell S R, Parmentier M, Chang C N, Price K, Tsang M, Doms R W. Epitope mapping of CCR5 reveals multiple conformational states and distinct but overlapping structures involved in chemokine and coreceptor function. J Biol Chem. 1999;274:9617–9626. doi: 10.1074/jbc.274.14.9617. [DOI] [PubMed] [Google Scholar]
- 45.Lin W H, Larsen K, Hortin G L, Roth J A. Recognition of substrates by tyrosylprotein sulfotransferase. Determination of affinity by acidic amino acids near the target sites. J Biol Chem. 1992;267:2876–2879. [PubMed] [Google Scholar]
- 46.Martin K A, Wyatt R, Farzan M, Choe H, Marcon L, Desjardins E, Robinson J, Sodroski J, Gerard C, Gerard N P. CD4-independent binding of SIV gp120 to rhesus CCR5. Science. 1997;278:1470–1473. doi: 10.1126/science.278.5342.1470. [DOI] [PubMed] [Google Scholar]
- 47.Melikyan G B, Markosyan R M, Hemmati H, Delmedico M K, Lambert D M, Cohen F S. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J Cell Biol. 2000;151:413–424. doi: 10.1083/jcb.151.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mirzabekov T, Bannert N, Farzan M, Hofmann W, Kolchinsky P, Wu L, Wyatt R, Sodroski J. Enhanced expression, native purification, and characterization of CCR5, a principal HIV-1 coreceptor. J Biol Chem. 1999;274:28745–28750. doi: 10.1074/jbc.274.40.28745. [DOI] [PubMed] [Google Scholar]
- 49.Moore J P, Sattentau Q J, Wyatt R, Sodroski J. Probing the structure of the human immunodeficiency virus surface glycoprotein gp120 with a panel of monoclonal antibodies. J Virol. 1994;68:469–484. doi: 10.1128/jvi.68.1.469-484.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Myszka D G, Sweet R W, Hensley P, Brigham-Burke M, Kwong P D, Hendrickson W A, Wyatt R, Sodroski J, Doyle M L. Energetics of the HIV gp120-CD4 binding reaction. Proc Natl Acad Sci USA. 2000;97:9026–9031. doi: 10.1073/pnas.97.16.9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pohlmann S, Baribaud F, Lee B, Leslie G J, Sanchez M D, Hiebenthal-Millow K, Munch J, Kirchhoff F, Doms R W. DC-SIGN interactions with human immunodeficiency virus type 1 and 2 and simian immunodeficiency virus. J Virol. 2001;75:4664–4672. doi: 10.1128/JVI.75.10.4664-4672.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pohlmann S, Soilleux E J, Baribaud F, Leslie G J, Morris L S, Trowsdale J, Lee B, Coleman N, Doms R W. DC-SIGNR, a DC-SIGN homologue expressed in endothelial cells, binds to human and simian immunodeficiency viruses and activates infection in trans. Proc Natl Acad Sci USA. 2001;98:2670–2675. doi: 10.1073/pnas.051631398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reeves J D, Heveker N, Brelot A, Alizon M, Clapham P R, Picard L. The second extracellular loop of CXCR4 is involved in CD4-independent entry of human immunodeficiency virus type 2. J Gen Virol. 1998;79:1793–1799. doi: 10.1099/0022-1317-79-7-1793. [DOI] [PubMed] [Google Scholar]
- 54.Reeves J D, Hibbitts S, Simmons G, McKnight A, Azevedo-Pereira J M, Moniz-Pereira J, Clapham P R. Primary human immunodeficiency virus type 2 (HIV-2) isolates infect CD4-negative cells via CCR5 and CXCR4: comparison with HIV-1 and simian immunodeficiency virus and relevance to cell tropism in vivo. J Virol. 1999;73:7795–7804. doi: 10.1128/jvi.73.9.7795-7804.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reeves J D, Schulz T F. The CD4-independent tropism of human immunodeficiency virus type 2 involves several regions of the envelope protein and correlates with a reduced activation threshold for envelope-mediated fusion. J Virol. 1997;71:1453–1465. doi: 10.1128/jvi.71.2.1453-1465.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reitter J N, Means R E, Desrosiers R C. A role for carbohydrates in immune evasion in AIDS. Nat Med. 1998;4:679–684. doi: 10.1038/nm0698-679. [DOI] [PubMed] [Google Scholar]
- 57.Rizzuto C, Sodroski J. Fine definition of a conserved CCR5-binding region on the human immunodeficiency virus type 1 glycoprotein 120. AIDS Res Hum Retroviruses. 2000;16:741–749. doi: 10.1089/088922200308747. [DOI] [PubMed] [Google Scholar]
- 58.Rizzuto C D, Wyatt R, Hernandez-Ramos N, Sun Y, Kwong P D, Hendrickson W A, Sodroski J. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 59.Root M J, Kay M S, Kim P S. Protein design of an HIV-1 entry inhibitor. Science. 2001;11:11. doi: 10.1126/science.1057453. [DOI] [PubMed] [Google Scholar]
- 60.Rosenquist G L, Nicholas H B. Analysis of sequence requirements for protein tyrosine sulfation. Protein Sci. 1993;2:215–222. doi: 10.1002/pro.5560020210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rucker J, Doranz B J, Edinger A L, Long D, Berson J F, Doms R W. Cell-cell fusion assay to study role of chemokine receptors in human immunodeficiency virus type 1 entry. Methods Enzymol. 1997;288:118–133. doi: 10.1016/s0076-6879(97)88011-1. [DOI] [PubMed] [Google Scholar]
- 62.Rucker J, Samson M, Doranz B J, Libert F, Berson J F, Yi Y, Smyth R J, Collman R G, Broder C C, Vassart G, Doms R W, Parmentier M. Regions in beta-chemokine receptors CCR5 and CCR2b that determine HIV-1 cofactor specificity. Cell. 1996;87:437–446. doi: 10.1016/s0092-8674(00)81364-1. [DOI] [PubMed] [Google Scholar]
- 63.Siciliano S J, Kuhmann S E, Weng Y, Madani N, Springer M S, Lineberger J E, Danzeisen R, Miller M D, Kavanaugh M P, DeMartino J A, Kabat D. A critical site in the core of the CCR5 chemokine receptor required for binding and infectivity of human immunodeficiency virus type 1. J Biol Chem. 1999;274:1905–1913. doi: 10.1074/jbc.274.4.1905. [DOI] [PubMed] [Google Scholar]
- 64.Sodroski J G. HIV-1 entry inhibitors in the side pocket. Cell. 1999;99:243–246. doi: 10.1016/s0092-8674(00)81655-4. [DOI] [PubMed] [Google Scholar]
- 65.Soilleux E J, Barten R, Trowsdale J. DC-SIGN; a related gene, DC-SIGNR; and CD23 form a cluster on 19p13. J Immunol. 2000;165:2937–2942. doi: 10.4049/jimmunol.165.6.2937. [DOI] [PubMed] [Google Scholar]
- 66.Speck R F, Wehrly K, Platt E J, Atchison R E, Charo I F, Kabat D, Chesebro B, Goldsmith M A. Selective employment of chemokine receptors as human immunodeficiency virus type 1 coreceptors determined by individual amino acids within the envelope V3 loop. J Virol. 1997;71:7136–7139. doi: 10.1128/jvi.71.9.7136-7139.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Trkola A, Dragic T, Arthos J, Binley J M, Olson W C, Allaway G P, Cheng-Mayer C, Robinson J, Maddon P J, Moore J P. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 68.Trkola A, Ketas T, Kewalramani V N, Endorf F, Binley J M, Katinger H, Robinson J, Littman D R, Moore J P. Neutralization sensitivity of human immunodeficiency virus type 1 primary isolates to antibodies and CD4-based reagents is independent of coreceptor usage. J Virol. 1998;72:1876–1885. doi: 10.1128/jvi.72.3.1876-1885.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weissenhorn W, Dessen A, Harrison S C, Skehel J J, Wiley D C. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 70.Wild C T, Shugars D C, Greenwell T K, McDanal C B, Matthews T J. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc Natl Acad Sci USA. 1994;91:9770–9774. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Willett B J, Hosie M J, Neil J C, Turner J D, Hoxie J A. Common mechanism of infection by lentiviruses. Nature. 1997;385:587. doi: 10.1038/385587a0. [DOI] [PubMed] [Google Scholar]
- 72.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A A, Desjardin E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 73.Wyatt R, Thali M, Tilley S, Pinter A, Posner M, Ho D, Robinson J, Sodroski J. Relationship of the human immunodeficiency virus type 1 gp120 third variable loop to a component of the CD4 binding site in the fourth conserved region. J Virol. 1992;66:6997–7004. doi: 10.1128/jvi.66.12.6997-7004.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xiao X, Wu L, Stantchev T S, Feng Y R, Ugolini S, Chen H, Shen Z, Riley J L, Broder C C, Sattentau Q J, Dimitrov D S. Constitutive cell surface association between CD4 and CCR5. Proc Natl Acad Sci USA. 1999;96:7496–7501. doi: 10.1073/pnas.96.13.7496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zagury J F, Franchini G, Reitz M, Collalti E, Starcich B, Hall L, Fargnoli K, Jagodzinski L, Guo H G, Laure F, Arya S K, Josephs S, Zagury D, Wong-Staal F, Gallo R C. Genetic variability between isolates of human immunodeficiency virus (HIV) type 2 is comparable to the variability among HIV type 1. Proc Natl Acad Sci USA. 1988;85:5941–5945. doi: 10.1073/pnas.85.16.5941. [DOI] [PMC free article] [PubMed] [Google Scholar]