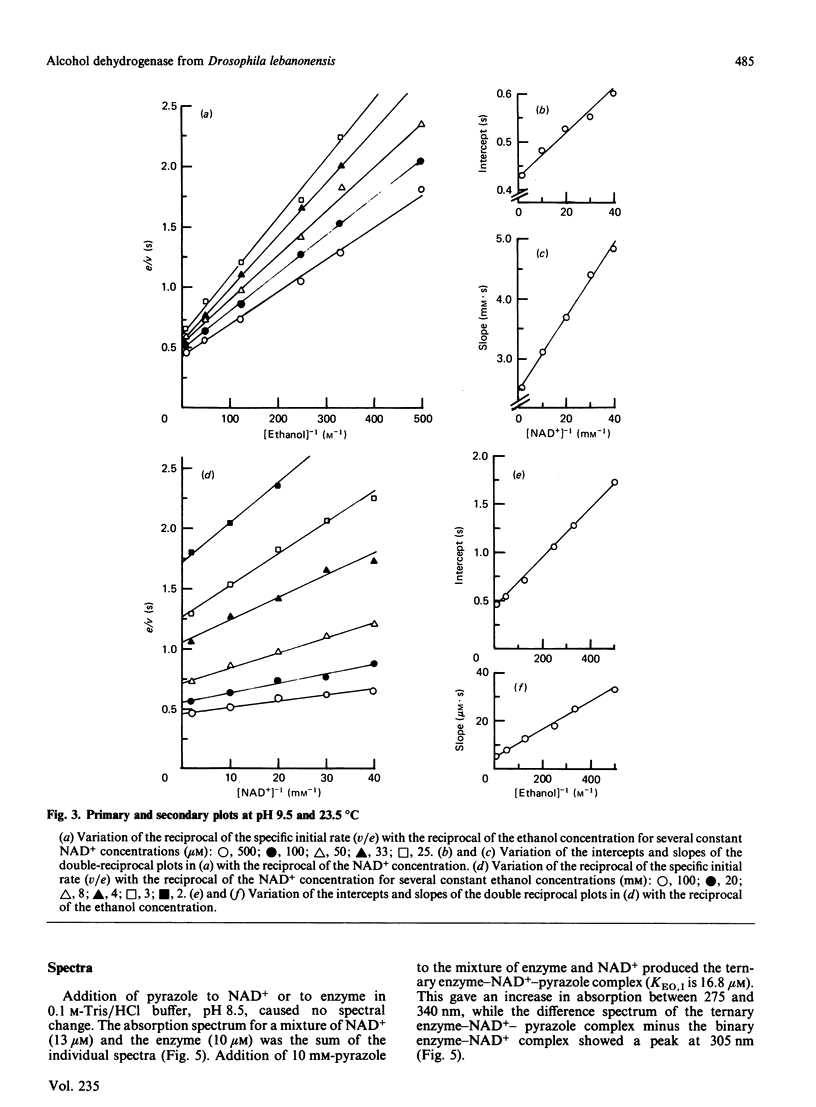
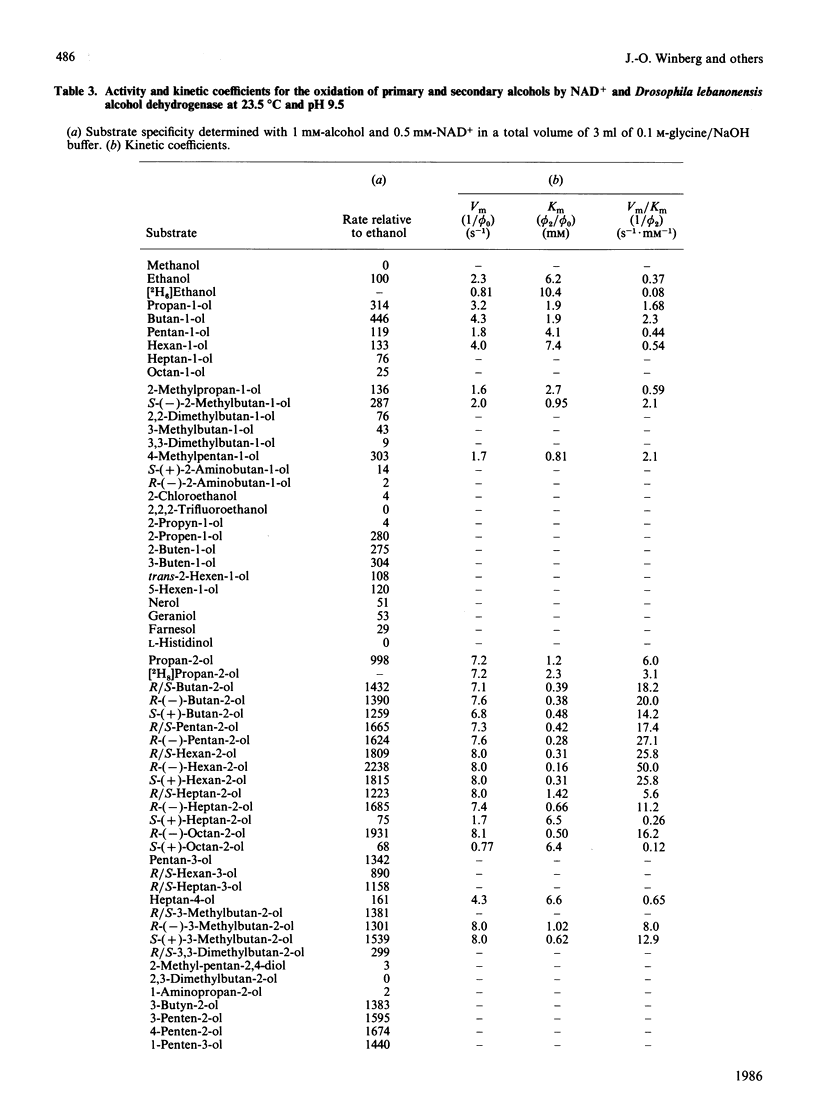
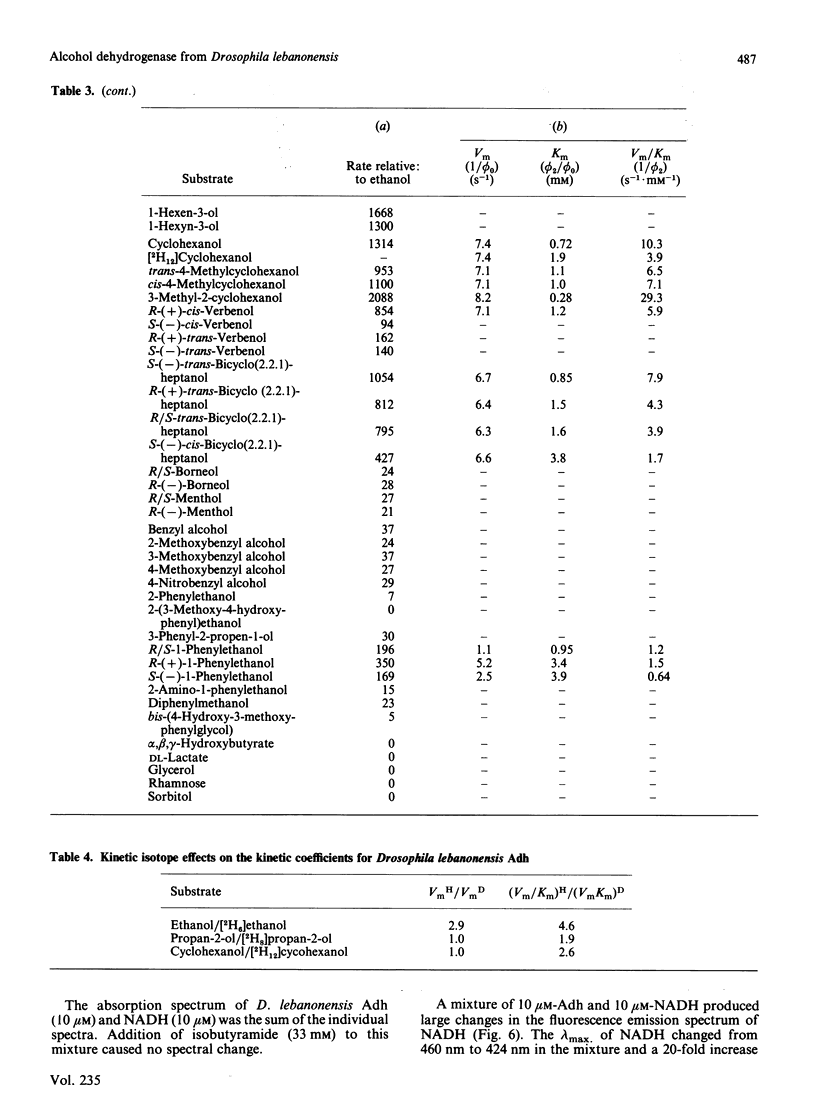
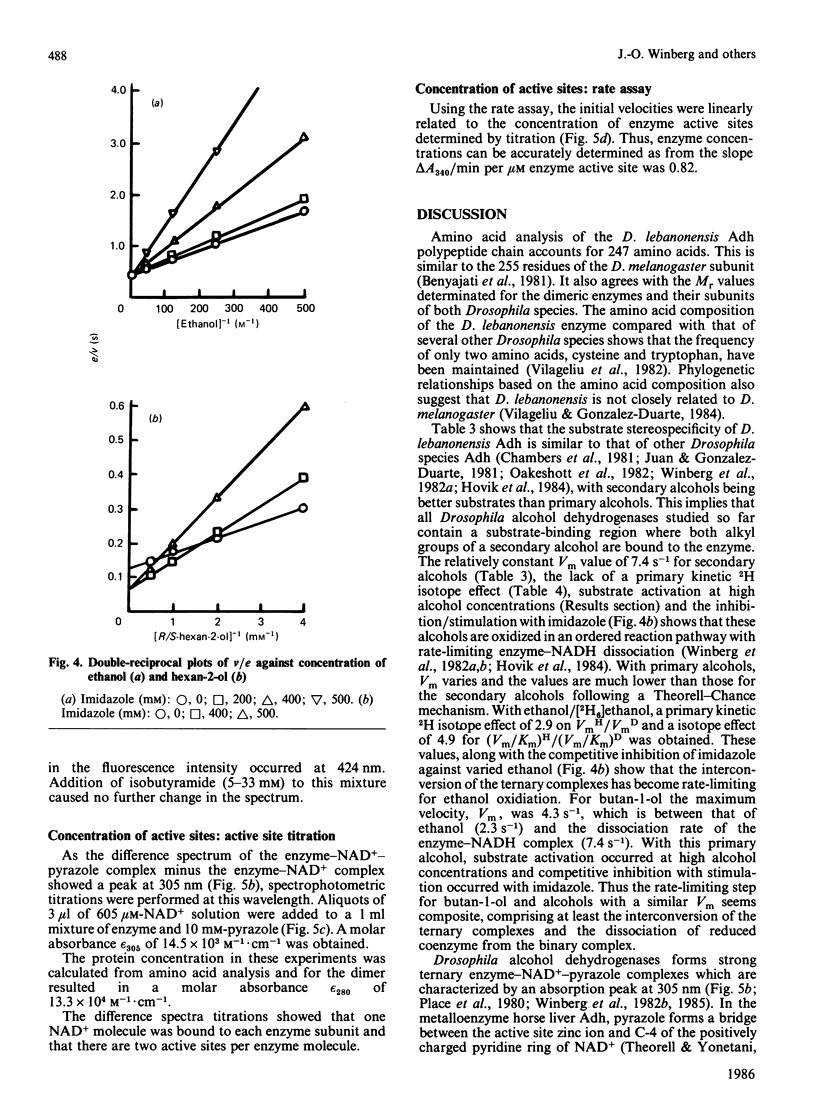
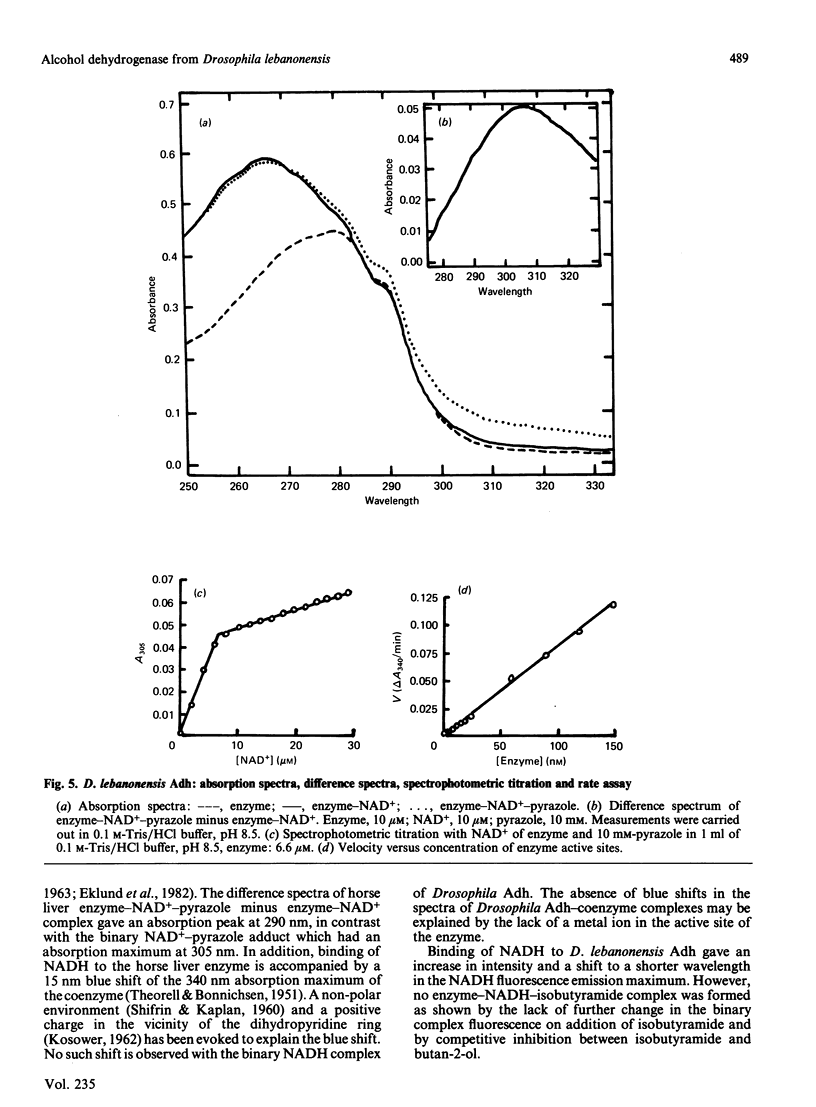
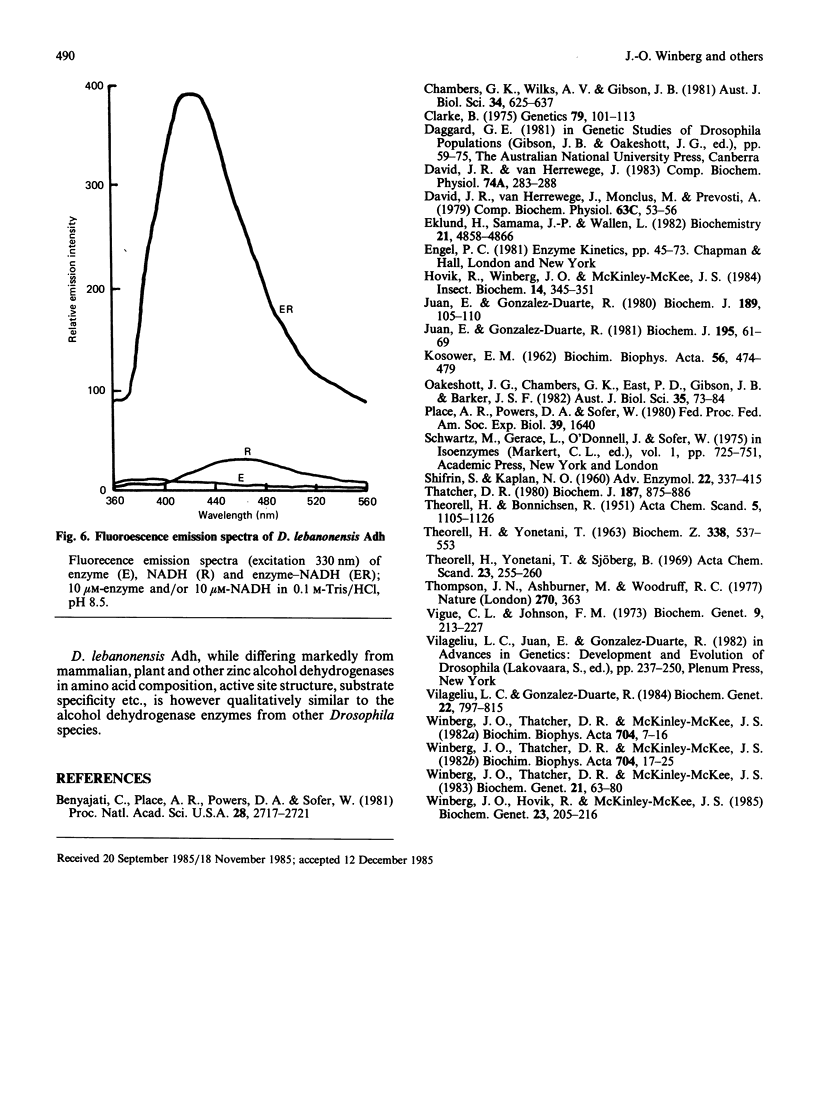
Abstract
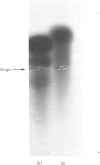
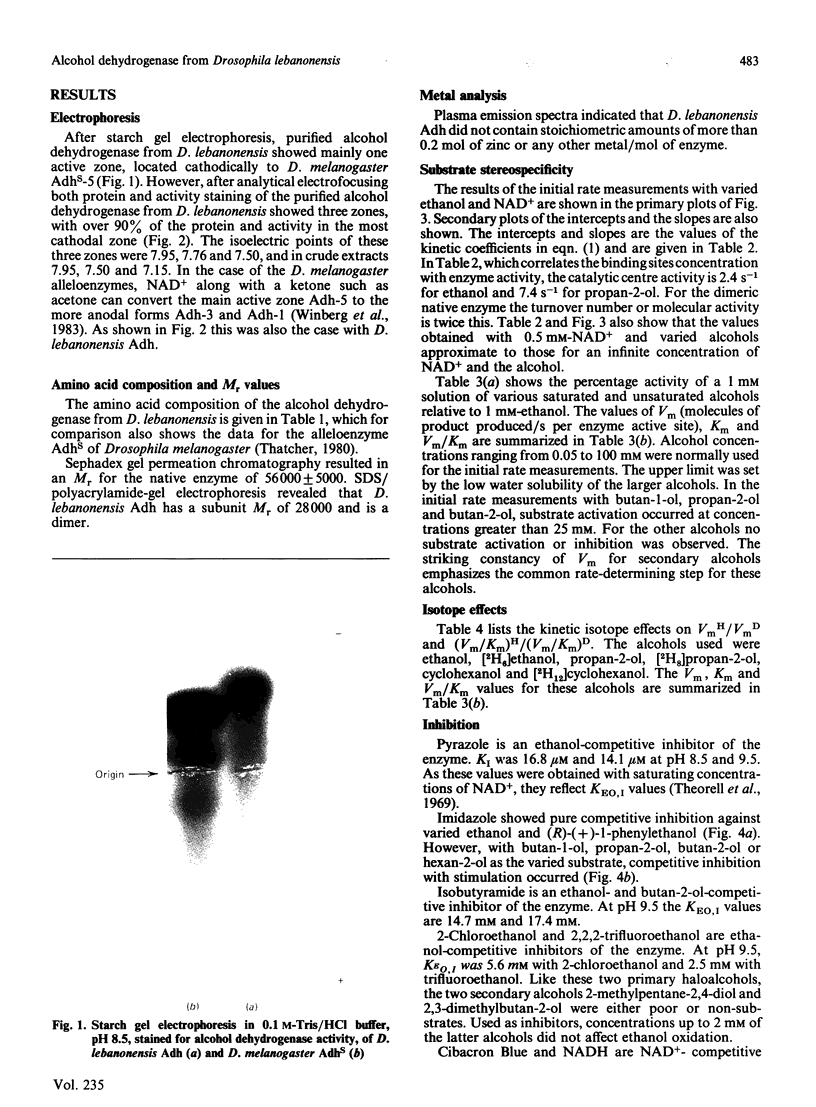
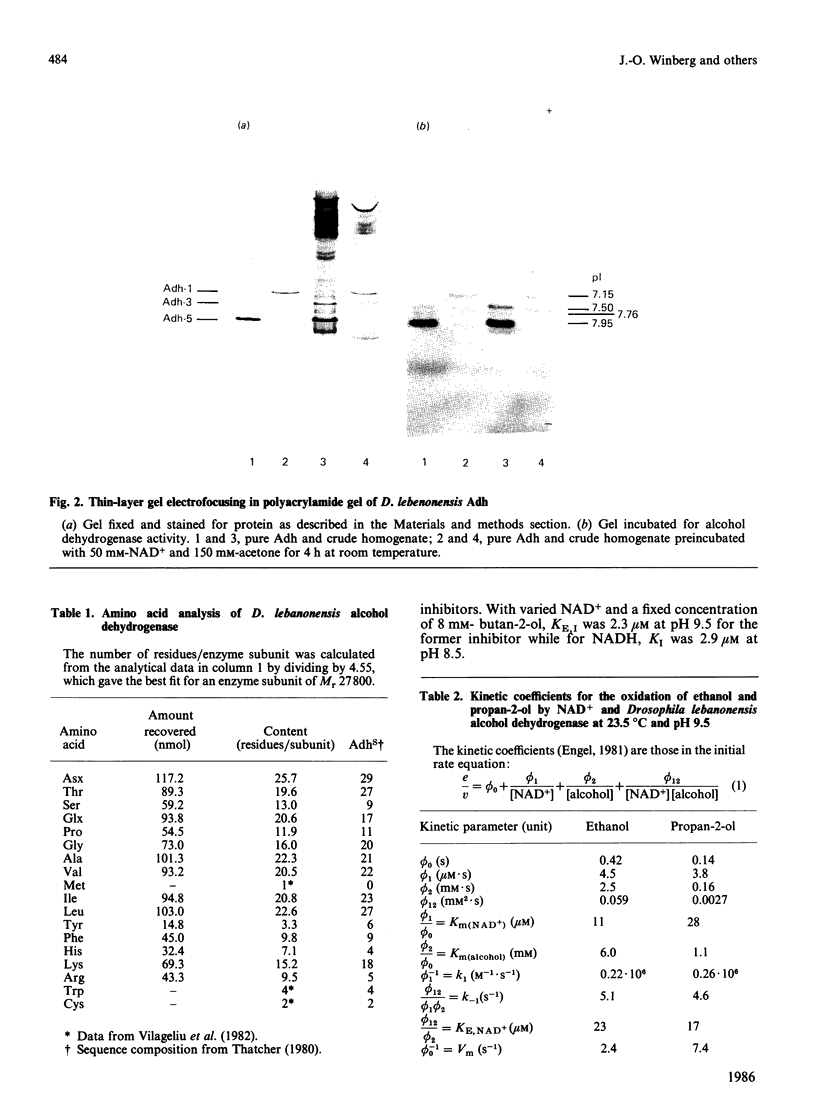
Purified Drosophila lebanonensis alcohol dehydrogenase (Adh) revealed one enzymically active zone in starch gel electrophoresis at pH 8.5. This zone was located on the cathode side of the origin. Incubation of D. lebanonensis Adh with NAD+ and acetone altered the electrophoretic pattern to more anodal migrating zones. D. lebanonensis Adh has an Mr of 56,000, a subunit of Mr of 28 000 and is a dimer with two active sites per enzyme molecule. This agrees with a polypeptide chain of 247 residues. Metal analysis by plasma emission spectroscopy indicated that this insect alcohol dehydrogenase is not a metalloenzyme. In studies of the substrate specificity and stereospecificity, D. lebanonensis Adh was more active with secondary than with primary alcohols. Both alkyl groups in the secondary alcohols interacted hydrophobically with the alcohol binding region of the active site. The catalytic centre activity for propan-2-ol was 7.4 s-1 and the maximum velocity of most secondary alcohols was approximately the same and indicative of rate-limiting enzyme-coenzyme dissociation. For primary alcohols the maximum velocity varied and was much lower than for secondary alcohols. The catalytic centre activity for ethanol was 2.4 s-1. With [2H6]ethanol a primary kinetic 2H isotope effect of 2.8 indicated that the interconversion of the ternary complexes was rate-limiting. Pyrazole was an ethanol-competitive inhibitor of the enzyme. The difference spectra of the enzyme-NAD+-pyrazole complex gave an absorption peak at 305 nm with epsilon 305 14.5 X 10(3) M-1 X cm-1. Concentrations and amounts of active enzyme can thus be determined. A kinetic rate assay to determine the concentration of enzyme active sites is also presented. This has been developed from active site concentrations established by titration at 305 nm of the enzyme and pyrazole with NAD+. In contrast with the amino acid composition, which indicated that D. lebanonensis Adh and the D. melanogaster alleloenzymes were not closely related, the enzymological studies showed that their active sites were similar although differing markedly from those of zinc alcohol dehydrogenases.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benyajati C., Place A. R., Powers D. A., Sofer W. Alcohol dehydrogenase gene of Drosophila melanogaster: relationship of intervening sequences to functional domains in the protein. Proc Natl Acad Sci U S A. 1981 May;78(5):2717–2721. doi: 10.1073/pnas.78.5.2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers G. K., Wilks A. V., Gibson J. B. An electrophoretically cryptic alcohol dehydrogenase variant in Drosophila melanogaster. III. Biochemical properties and comparison with common enzyme forms. Aust J Biol Sci. 1981;34(5-6):625–637. doi: 10.1071/bi9810625. [DOI] [PubMed] [Google Scholar]
- Clarke B. The contribution of ecological genetics to evolutionary theory: detecting the direct effects of natural selection on particular polymorphic loci. Genetics. 1975 Jun;79 (Suppl):101–113. [PubMed] [Google Scholar]
- Eklund H., Samama J. P., Wallén L. Pyrazole binding in crystalline binary and ternary complexes with liver alcohol dehydrogenase. Biochemistry. 1982 Sep 28;21(20):4858–4866. doi: 10.1021/bi00263a005. [DOI] [PubMed] [Google Scholar]
- Juan E., González-Duarte R. Determination of some biochemical and structural features of alcohol dehydrogenases from Drosophila simulans and Drosophila virilis. Comparison of their properties with the Drosophila melanogaster Adhs enzyme. Biochem J. 1981 Apr 1;195(1):61–69. doi: 10.1042/bj1950061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan E., González-Duarte R. Purification and enzyme stability of alcohol dehydrogenase from Drosophila simulans, Drosophila virilis and Drosophila melanogaster adhS. Biochem J. 1980 Jul 1;189(1):105–110. doi: 10.1042/bj1890105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- THEORELL H., YONETANI T. LIVER ALCOHOL DEHYDROGENASE-DPN-PYRAZOLE COMPLEX: A MODEL OF A TERNARY INTERMEDIATE IN THE ENZYME REACTION. Biochem Z. 1963;338:537–553. [PubMed] [Google Scholar]
- Thatcher D. R., Sawyer L. Secondary-structure prediction from the sequence of Drosophila melanogaster (fruitfly) alcohol dehydrogenase. Biochem J. 1980 Jun 1;187(3):884–886. doi: 10.1042/bj1870884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theorell H., Yonetani T., Sjöberg B. On the effects of some heterocyclic compounds on the enzymic activity of liver alcohol dehydrogenase. Acta Chem Scand. 1969;23(1):255–260. doi: 10.3891/acta.chem.scand.23-0255. [DOI] [PubMed] [Google Scholar]
- Thompson J. N., Jr, Ashburner M., Woodruff R. C. Presumptive control mutation for alcohol dehydrogenase in Drosophila melanogaster. Nature. 1977 Nov 24;270(5635):363–363. doi: 10.1038/270363a0. [DOI] [PubMed] [Google Scholar]
- Vigue C. L., Johnson F. M. Isozyme variability in species of the genus Drosophila. VI. Frequency-property-environment relationships of allelic alcohol dehydrogenases in D. melanogaster. Biochem Genet. 1973 Jul;9(3):213–227. doi: 10.1007/BF00485735. [DOI] [PubMed] [Google Scholar]
- Vilageliu L., González-Duarte R. Alcohol dehydrogenase from Drosophila funebris and Drosophila immigrans: molecular and evolutionary aspects. Biochem Genet. 1984 Oct;22(9-10):797–815. doi: 10.1007/BF00499474. [DOI] [PubMed] [Google Scholar]
- Winberg J. O., Hovik R., McKinley-McKee J. S. The alcohol dehydrogenase alleloenzymes AdhS and AdhF from the fruitfly Drosophila melanogaster: an enzymatic rate assay to determine the active-site concentration. Biochem Genet. 1985 Apr;23(3-4):205–216. doi: 10.1007/BF00504319. [DOI] [PubMed] [Google Scholar]
- Winberg J. O., Thatcher D. R., McKinley-McKee J. S. Alcohol dehydrogenase from the fruitfly Drosophila melanogaster. Inhibition studies of the alleloenzymes AdhS and AdhUF. Biochim Biophys Acta. 1982 May 21;704(1):17–25. doi: 10.1016/0167-4838(82)90126-1. [DOI] [PubMed] [Google Scholar]
- Winberg J. O., Thatcher D. R., McKinley-McKee J. S. Alcohol dehydrogenase from the fruitfly Drosophila melanogaster. Substrate specificity of the alleloenzymes AdhS and AdhUF. Biochim Biophys Acta. 1982 May 21;704(1):7–16. doi: 10.1016/0167-4838(82)90125-x. [DOI] [PubMed] [Google Scholar]
- Winberg J. O., Thatcher D. R., McKinley-McKee J. S. Drosophila melanogaster alcohol dehydrogenase: an electrophoretic study of the AdhS, AdhF, and AdhUF alleloenzymes. Biochem Genet. 1983 Feb;21(1-2):63–80. doi: 10.1007/BF02395392. [DOI] [PubMed] [Google Scholar]