Abstract
During replication of herpes simplex virus type 1 (HSV-1), viral DNA is synthesized in the infected cell nucleus, where DNA-free capsids are also assembled. Genome-length DNA molecules are then cut out of a larger, multigenome concatemer and packaged into capsids. Here we report the results of experiments carried out to test the idea that the HSV-1 UL6 gene product (pUL6) forms the portal through which viral DNA passes as it enters the capsid. Since DNA must enter at a unique site, immunoelectron microscopy experiments were undertaken to determine the location of pUL6. After specific immunogold staining of HSV-1 B capsids, pUL6 was found, by its attached gold label, at one of the 12 capsid vertices. Label was not observed at multiple vertices, at nonvertex sites, or in capsids lacking pUL6. In immunoblot experiments, the pUL6 copy number in purified B capsids was found to be 14.8 ± 2.6. Biochemical experiments to isolate pUL6 were carried out, beginning with insect cells infected with a recombinant baculovirus expressing the UL6 gene. After purification, pUL6 was found in the form of rings, which were observed in electron micrographs to have outside and inside diameters of 16.4 ± 1.1 and 5.0 ± 0.7 nm, respectively, and a height of 19.5 ± 1.9 nm. The particle weights of individual rings as determined by scanning transmission electron microscopy showed a majority population with a mass corresponding to an oligomeric state of 12. The results are interpreted to support the view that pUL6 forms the DNA entry portal, since it exists at a unique site in the capsid and forms a channel through which DNA can pass. The HSV-1 portal is the first identified in a virus infecting a eukaryote. In its dimensions and oligomeric state, the pUL6 portal resembles the connector or portal complexes employed for DNA encapsidation in double-stranded DNA bacteriophages such as φ29, T4, and P22. This similarity supports the proposed evolutionary relationship between herpesviruses and double-stranded DNA phages and suggests the basic mechanism of DNA packaging is conserved.
Herpes simplex virus type 1 (HSV-1) is the prototypical member of the herpesviruses, a virus family whose members infect vertebrate animals. Eight herpesviruses, including HSV-1, are able to infect humans. Like all herpesviruses, HSV-1 consists of a T=16 icosahedral capsid surrounded by a membrane envelope (14, 31, 39). The virus DNA is contained inside the capsid. During HSV-1 replication, viral DNA is synthesized in the infected cell nucleus, where a large, branched concatemer consisting of many genomes is formed (15, 21, 34, 54). Newly replicated DNA is then packaged into preformed capsids. Packaging is considered to begin with the procapsid, a spherical intermediate in assembly of the mature capsid (27, 36). Procapsids are transformed into the mature, angular, icosahedral morphology at approximately the same time as DNA is encapsidated. Once packaging is complete, DNA-containing capsids are enveloped and released from the host cell.
Studies to clarify the mechanism of HSV-1 DNA encapsidation have focused on specific, conserved, cis-acting DNA sequences (pac sites) (23, 38, 40, 49) and on trans-acting, virus-encoded proteins. Genetic analyses have identified a total of seven HSV-1-encoded proteins, products of the genes UL6, UL15, UL17, UL25, UL28, UL32, and UL33, that are demonstrated to be specifically involved in DNA packaging (14, 51). When cells are infected with HSV-1 mutants lacking the function of any of the seven genes, capsid formation and DNA replication occur normally, but no packaging takes place.
In its basic features, HSV-1 DNA encapsidation resembles that observed in double-stranded DNA (dsDNA) bacteriophages, such as P22, T7, and λ. Bacteriophage assembly proceeds through the formation of spherical precursor capsids (procapsids), and bacteriophages have a unique vertex, a dodecameric ring of 12 proteins, through which the DNA enters and exits the capsid (3, 45). The portal vertex is the docking site for the packaging proteins, such as terminase, the protein responsible for cleavage of monomeric units from concatemeric DNA and translocation of DNA into the capsid in an ATP-dependent fashion (5, 7, 8, 45). Portal proteins are found as integral capsid components in both procapsids and mature capsids, while terminase proteins are only associated with procapsids and are not present in mature capsids or virions (24).
By analogy to the better-studied phage systems, functions for the HSV-1 packaging proteins have been tentatively assigned on the basis of (i) limited amino acid sequence conservation with phage proteins and (ii) the behavior of mutant viruses. For example, amino acid sequence homology has been observed between the phage T4 terminase and the HSV-1 UL15 gene product, suggesting pUL15 may have terminase function (30). The proteins encoded by the UL15 and UL28 genes have been shown to interact with one another and have been proposed to form a two-subunit terminase (18, 19). Further support for this assignment comes from the finding that pUL28 or its homolog from HCMV can specifically recognize pac site DNA (1, 6). In order for packaging to occur, the terminase must associate at least transiently with the procapsid. Genetic and biochemical evidence indicates that pUL6 may provide a docking site for the terminase. For example, pUL6 is found in procapsids and mature capsids in conserved amounts, while pUL15 and pUL28 are only transiently associated with B capsids and procapsids (29, 36). Also, in cells infected with a UL6-null mutant virus, pUL15 is not found associated with B capsids (33, 53).
Here we report the results of experiments designed to test the idea that pUL6 is the HSV-1 portal protein. Studies have been carried out to determine the location of pUL6 in the capsid and to examine the structure of purified pUL6 in solution. Because DNA must enter the capsid at a single site, it is presumed that pUL6 will be found at a single, unique location in the capsid. In solution, pUL6 is expected to have a structure that is consistent with its presumptive role as a channel through which DNA can be translocated into the capsid.
MATERIALS AND METHODS
Cells and viruses.
Experiments were carried out with the KOS strain of HSV-1 which was grown at 34°C on Vero cells maintained in minimal essential medium containing 10% calf serum and antibiotics (35). HSV-1 mutant strain hr-74 (UL6 deletion), a strain with a lacZ cassette inserted at position 378 of the UL6 gene, was propagated on the complementing cell line UL6–31, where titers of 107 to 108 were obtained (20). Vero cells grown in 850-cm2 roller bottles were infected with hr74 at a multiplicity of infection of 10; infection was allowed to proceed for 24 h at 34°C, after which, infected cells were harvested and used for capsid isolation. Previously described procedures were employed for capsid purification from cells infected with either HSV-1 KOS or hr74 (35). Recombinant baculovirus Autographa californica nuclear polyhedrosis virus encoding the HSV-1 UL6 gene was grown as previously described on Spodoptera frugiperda (Sf9) cells (44). Cells were infected at a multiplicity of 5, and infection was allowed to proceed for 64 h at 28°C before cells were harvested by centrifugation.
Antibody labeling of HSV-1 capsids.
B capsids to be employed in antibody labeling experiments were purified (26) from cells infected with wild-type HSV-1 or with the UL6 deletion strain hr74 and suspended in TNE (20 mM Tris-HCl [pH 7.5], 0.5 M NaCl, 1 mM EDTA) at a concentration of 0.5 mg/ml. Capsids (5 μl) were adsorbed to Formvar-carbon-coated electron microscope grids (15 s at room temperature), and further operations were performed by floating grids, specimen side down, on solution drops (∼300 μl), which were placed on Parafilm and maintained at room temperature in a closed, humidified petri dish. Specimens were washed (twice) for 30 s with 0.5× TNE to remove unadsorbed capsids, for 1 h with blocker (0.5× TNE containing 5% goat serum, 5% bovine serum albumin, and 0.1% fish gelatin) to saturate nonspecific binding sites, and then for 1 h with preadsorbed (see below) rabbit polyclonal antibody anti-MBP (maltose binding protein)-UL6 (43) diluted 1:50 in 0.5× TNE. Excess anti-MBP-UL6 was removed by washing eight times (3 min each) with blocker, and specimens were exposed for 45 min to secondary antibody, goat anti-rabbit immunoglobulin G conjugated to 10-nm-diameter gold beads (EY Laboratories, San Mateo, Calif.), diluted 1:25 with blocker. Grids were then washed with blocker (five times), 0.5× TNE (three times), and phosphate-buffered saline (PBS; once). Capsids were fixed by immersing grids for 5 min in 500 μl of PBS containing 5% glutaraldehyde, washed (twice) in 0.5× TNE, stained by immersion for 30 s in 1% uranyl acetate, blotted, and allowed to dry at room temperature before examination in the electron microscope.
Anti-MBP-UL6 (raised against a fusion protein consisting of the C-terminal 298 amino acids of pUL6 and Escherichia coli MBP) (43) was absorbed with B capsids prepared from the UL6 deletion strain (hr74). Capsids (25 μl) were added to 5 μl of anti-MBP-UL6 and incubated for 1 h at room temperature. The solution volume was then adjusted to 250 μl with 0.5× TNE and centrifuged for 30 min at 30,000 rpm (32,000 × g) in a Beckman TL-100 ultracentrifuge (TLA-100 rotor) to pellet capsids. The supernatant was removed and used directly for capsid labeling as described above.
pUL6 purification.
Purification of pUL6 (molecular weight [MW], 74,087) was carried out beginning with Sf9 cells infected with BAC-UL6 as described above. Except as indicated, all purification steps were performed at 4°C. After being harvested by centrifugation, infected cells (∼2.0 ml of packed cells) were suspended in 2 volumes of PBS plus 0.1 volume of a protease inhibitor cocktail (aprotinin, leupeptin, and Pefabloc; stock solution prepared by dissolving one tablet of Boehringer Mannheim Complete EDTA-free in 5 ml of PBS) and lysed by three cycles of freezing and thawing. The lysate was then centrifuged for 5 min at 16,000 × g, and the supernatant was discarded. The pellet, which contains pUL6 in the form of inclusion bodies, was resuspended (by gentle sonication in a bath sonicator) in 1 ml of TNE plus 0.1 ml of protease inhibitors, adjusted to 10 mM dithiothreitol–2% Triton X-100, and incubated for 30 min on ice to solubilize cellular membranes. The resulting suspension was centrifuged at 16,000 × g for 5 min, and the supernatant was discarded. The pellet was resuspended in 1 ml of TNE (with protease inhibitors), adjusted to 20 mM MgSO4 plus 0.5 mg of DNase I per ml, incubated for 10 min at room temperature, and centrifuged for 5 min at 16,000 × g, and the supernatant was discarded. The pellet was resuspended in 1 ml of 1 M arginine (pH 7.4) and incubated for 10 min on ice to solubilize pUL6. The resulting solution was clarified by centrifugation at 30,000 rpm (32,000 × g) in the TLA-100 rotor and applied to the top of a 5-ml gradient of 10 to 30% sucrose containing 1 M arginine and 20 mM Tris-HCl (pH 7.5). After centrifugation for 48 h at 33,000 rpm (105,000 × g) in a Beckman SW50.1 rotor, the pUL6-containing band was identified by examining the gradient with a high-intensity lamp (top illumination) and isolated by fractionating the gradient. A yield of ∼0.25 mg of purified pUL6 was obtained in preparations beginning with ∼2 ml of packed cells.
Negative stain electron microscopy.
Electron microscopic examination of purified pUL6 was carried out beginning with specimens removed directly from sucrose density gradients (the last step in the purification procedure as described above). A drop (∼30 μl) of the sample was placed on a Parafilm sheet and allowed to stand for 0.5 to 1.0 min. A carbon-coated electron microscope grid was placed on top of the drop, and pUL6 was allowed to adsorb for 10 to 30 s. The grid was then washed twice in 20 mM Tris-HCl (pH 7.5), stained for 1 min with 1% uranyl acetate, blotted, and allowed to air dry. In some preparations, grids were glow discharged by exposure at 10 mA for 10 s in a Polaron E5100 vacuum evaporator operated at 100 mTorr. Specimens were observed and photographed in a Philips 400T electron microscope operated at 80 keV. Electron microscope negatives were digitized in a flatbed scanner, and images were measured with Photoshop 5.0.
STEM.
Purified pUL6 was examined by scanning transmission electron microscopy (STEM) beginning with pUL6 dissolved in 20 mM Tris-HCl (pH 7.5) containing 1 M arginine. A 3-μl aliquot was injected into a drop of 20 mM Tris-HCl (pH 7.5) on a thin (2 to 3 nm) carbon film supported by a holey carbon film on a titanium grid. Tobacco mosaic virus (TMV), an internal mass standard, had been previously applied to the grid. After incubation for 2 min to permit pUL6 to adsorb to the carbon, the grid was washed with 20 mM ammonium acetate, blotted, quick-frozen by being plunged into liquid nitrogen slush, and freeze-dried overnight. The grids were transferred under vacuum to the STEM and observed in the dark-field mode. Digital images were recorded in clean areas with an adequate number of pUL6 rings and with TMV as a standard. Masses of individual pUL6 rings were measured from the images by using software described previously (50).
Amino acid sequence analysis.
Amino acid sequences for the HSV-1, HSV-2, Kaposi's sarcoma-associated herpesvirus (KSHV [HHV-8]), and Epstein-Barr virus (EBV) putative portal proteins were obtained from the GenBank protein database under accession no. P10190, P89429, AAC57125, and MP_039880, respectively. These sequences were compiled into and aligned with MacVector version 7.0. Secondary structure predictions were performed with Ph.D. Predict (32). Multicoil was used to analyze the UL6 leucine zipper (52).
Other methods.
Previously described procedures were employed for sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis, staining of gels with Coomassie blue, and Western immunoblotting (25, 27, 36). Gels to be stained with Sypro orange (Molecular Probes, Eugene, Oreg.) were incubated overnight in the dark with 1:5,000 Sypro orange dye in 7.5% acetic acid and rinsed according to the manufacturer's instructions. A UV transilluminator was used to view stained gels and record them digitally. In immunoblotting experiments, unabsorbed anti-MBP-UL6 was used at a dilution of 1:1,000. Coomassie-stained protein bands in polyacrylamide gels and bands in immunoblot radioautographs were determined quantitatively by densitometric scanning in a flatbed scanner (transmission mode) followed by integration of band density with UN-SCAN-IT (version 5.1) software.
RESULTS
Capsid location of pUL6.
Experiments to determine the location of pUL6 in the capsid were performed with B capsids prepared from infected Vero cells. These were adsorbed to carbon-coated electron microscope grids and stained with a pUL6-specific antibody, anti-MBP-UL6 (43), that had been preabsorbed with HSV-1 capsids lacking pUL6. Specific staining was followed by treatment with an anti-antibody conjugated to gold beads, which are expected to mark the location of pUL6 in the capsid. Control experiments were performed with capsids prepared from cells infected with hr74, a pUL6 deletion mutant of HSV-1 (20).
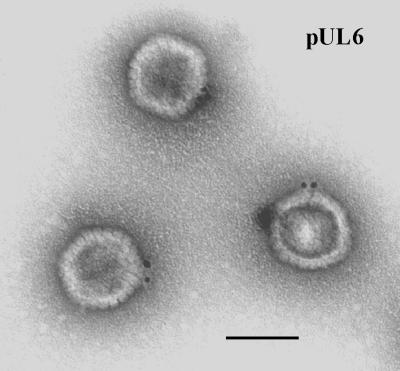
After examination in the electron microscope, specifically stained wild-type HSV-1 capsids demonstrated the presence of gold label at a single capsid vertex (Fig. 1). Specifically labeled capsid vertices were often found to contain more than one gold bead, as shown in Fig. 1 and Table 1, but only one vertex was labeled in most cases. The proportions of capsids labeled at a single vertex were 32 and 41%, respectively, in two experiments as shown in Table 2. The corresponding values were 1 and 2%, respectively, for capsids lacking pUL6 (Table 2). Background labeling, comparable to that found with UL6 deletion capsids, was observed when the anti-MPB-UL6 antibody was omitted or replaced with a nonspecific antiserum (data not shown).
FIG. 1.
Electron micrograph of HSV-1 B capsids after staining with antibody specific for pUL6 followed by an anti-antibody conjugated to gold beads. Note that gold beads are found at a single capsid vertex. Bar, 100 nm.
TABLE 1.
Immunogold staining of HSV-1 capsids for pUL6 and number of gold beads present at labeled capsid vertexa
| No. of gold beads at labeled vertex | No. of capsids
|
|||
|---|---|---|---|---|
| Expt 1
|
Expt 2
|
|||
| Wild-type HSV-1 | UL6 deletion | Wild-type HSV-1 | UL6 deletion | |
| 1 | 42 | 3 | 35 | 9 |
| 2 | 20 | 0 | 40 | 1 |
| 3 | 4 | 0 | 16 | 1 |
| 4 | 2 | 0 | 1 | 1 |
| 5 | 1 | 0 | 0 | 0 |
| Total no. of capsids counted | 69 | 3 | 92 | 12 |
Gold beads present at the labeled capsid vertex were counted in images such as Fig. 1.
TABLE 2.
Immunogold staining of pUL6 in HSV-1 B capsids
| Capsid type | Total no. of capsids counted | No. (%) of
capsids with gold beads at:
|
|
|---|---|---|---|
| 1 vertex | 2 vertices | ||
| Expt 1 | |||
| Wild-type HSV-1 | 219 | 69 (32) | 3 (1) |
| UL6 deletion | 211 | 3 (1) | 0 |
| Expt 2 | |||
| Wild-type HSV-1 | 222 | 92 (41) | 6 (3) |
| UL6 deletion | 502 | 12 (2) | 1 (<1) |
Gold label was observed at two capsid vertices in 1 and 3% of wild-type capsids in the two experiments, respectively (Table 2). We interpret this low level of labeling at a second vertex to be nonspecific, because it is similar in amount to the percentage of UL6 deletion capsids having a labeled vertex (i.e., 1 and 2% in the two experiments). Only background gold label was seen at nonvertex sites (data not shown).
Amount of pUL6 in the capsid.
The abundance of pUL6 in HSV-1 capsids was determined quantitatively by Western immunoblot experiments performed with known amounts of B capsids and purified pUL6. Identical capsid specimens were employed for SDS-polyacrylamide gel electrophoresis followed by (i) staining with Coomassie blue to determine the number of capsids present and (ii) immunoblot analysis and specific staining with anti-MBP-UL6 (43) to measure the amount of pUL6. The immunoblot signal corresponding to pUL6 in capsids was measured quantitatively by densitometric scanning of autoradiographs and interpreted with reference to a standard curve obtained by similar immunoblot analysis carried out on the same gel with pUL6 purified as described below. In a second study, the same measurements were made with capsid proteins stained in step i above with Sypro orange rather than Coomassie blue.
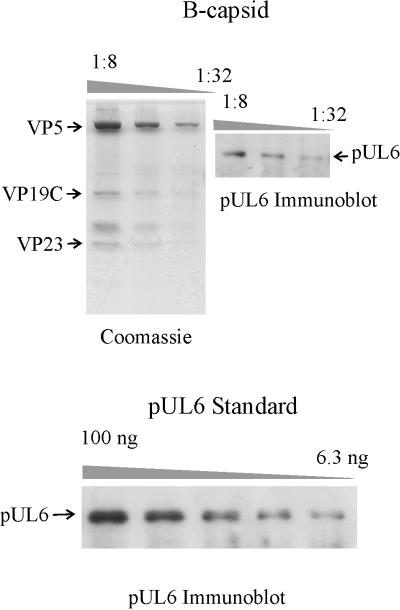
Figure 2 shows the results of immunoblot experiments performed on the same gel with capsids (top panel) and with purified pUL6 (bottom). The pUL6 copy number per capsid was calculated after quantitative determination of the capsid amount (based on staining of the VP5 band) and the immunoblot signal. In a representative experiment, for example, capsids containing 1.10 μg of VP5 (MW, 149,075) were found to have 8.0 ng of pUL6 (MW, 74,087). In all experiments, the pUL6 copy number per capsid was found to be 16.6 ± 1.0 (n = 3) and 12.5 ± 1.8 (n = 3) for gels in which capsid protein staining was with Coomassie blue and Sypro orange, respectively. Combination of the results of the Coomassie blue and Sypro orange experiments yields a copy number of 14.8 ± 2.6 (n = 6).
FIG. 2.
pUL6 content of HSV-1 B-capsids as determined by Western immunoblotting. (Top left) B capsid proteins separated by SDS-polyacrylamide gel electrophoresis and stained with Coomassie blue. (Top right) Same as the gel on the left, except that pUL6 is stained specifically by Western immunoblotting. (Bottom) Purified pUL6 stained by Western immunoblotting on the same gel as that shown in the top right. The pUL6 signal in the bottom panel was employed as a standard curve to determine the amount present in B capsids as shown at the top right.
pUL6 structure.
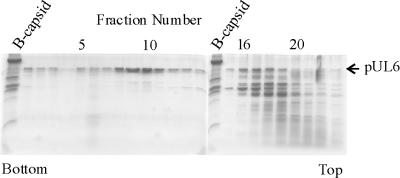
Studies of pUL6 structure were carried out with protein purified from insect cells containing pUL6 as a result of infection with a recombinant baculovirus, BAC-UL6 (22), expressing the UL6 gene. Purification was accomplished by a four-step procedure in which inclusion bodies, found to contain pUL6, were first isolated in crude form by extraction of cells with 2% Triton X-100 followed by 0.5 mg of DNase I per ml. pUL6 was then solubilized by treatment of inclusion bodies with 1 M arginine, an antiaggregation agent (41), and purified further by sucrose density gradient ultracentrifugation. Sucrose density gradients contained 1 M arginine because pUL6 was insoluble if arginine was removed (data not shown). During sucrose gradient centrifugation, much of the pUL6 was found to migrate more rapidly than contaminating insect cell proteins, as shown by SDS-polyacrylamide gel analysis of gradient fractions (Fig. 3). Whereas a substantial portion of pUL6 was found in fractions 8 to 11, most other proteins were found in fractions 15 to 21.
FIG. 3.
SDS-polyacrylamide gel electrophoresis of the fractions obtained during the sucrose density gradient ultracentrifugation step of pUL6 purification. The gel was stained with Coomassie blue. Fractions from the bottom of the gradient are shown to the left, and lanes containing protein standards (B capsid proteins) are indicated. Note that fractions highly enriched in pUL6 (fractions 8 to 11) are well separated from those (fractions 15 to 21) containing most contaminating insect cell proteins.
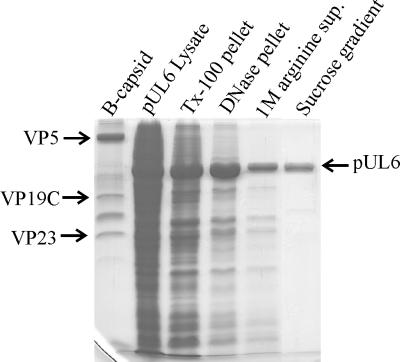
Figure 4 shows the results of an SDS-polyacrylamide gel analysis carried out with the pUL6-containing fraction at each step of the purification procedure. Densitometric scanning of stained gels demonstrated that pUL6 constituted 96% or more of the protein present in the peak sucrose gradient fractions (in four preparations). Purified pUL6 was found to be stained in immunoblots with specific antiserum (i.e., anti-MBP-UL6), as shown in Fig. 2 (bottom panel). The major contaminant in pUL6 preparations, a minor band accounting for 3 to 4% of the protein present and migrating between VP19C and VP23 (see Fig. 4, rightmost column), was not identified. It migrated coincidentally with pUL6 during sucrose density gradient centrifugation, suggesting it might be a degradation product of pUL6, but it did not react with pUL6-specific antiserum in immunoblots (data not shown).
FIG. 4.
SDS-polyacrylamide gel electrophoresis (Coomassie stain) of the pUL6-containing fraction at each stage of the purification procedure. HSV-1 B capsid proteins (leftmost lane) are included for reference, and the position of the pUL6 band is indicated. Tx-100, Triton X-100; sup., supernatant.
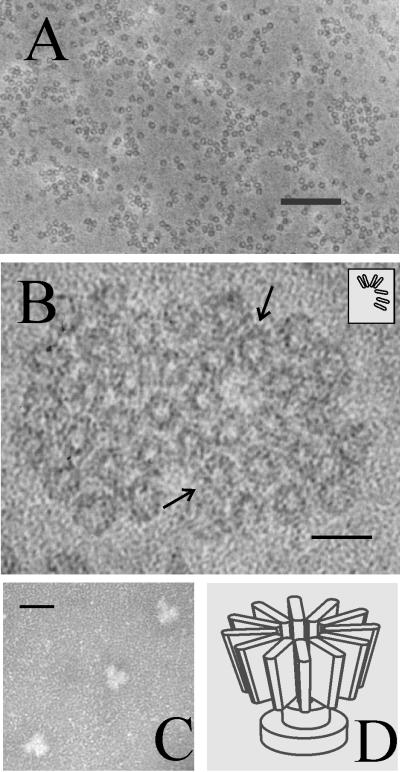
Structural characterization of pUL6 was performed by electron microscopy of negatively stained preparations. Specimens for this analysis were taken directly from the sucrose density gradient step of purification, diluted, and stained on standard or glow-discharged grids. Electron microscopic examination of specimens on standard grids showed a uniform population of small rings (Fig. 5A). No other structures were observed in significant amounts. Single rings were seen most often, but rings were also found to associate laterally to form small aggregates, including pairs, chains, and islands of rings, as shown in Fig. 5B. Measurement of the images showed the rings have outside and inside diameters of 16.4 ± 1.1 and 5.0 ± 0.7 nm (n = 56), respectively.
FIG. 5.
Electron microscopy of purified pUL6. Specimens were prepared by negative staining on standard (A and B) or glow-discharged (C) grids as described in Materials and Methods. Note that on standard grids, pUL6 is found in the form of small rings (A) and that at higher magnification the rings can be seen to be composed of subunits (B; arrows). The inset in panel B shows a schematic representation of the subunits present in the upper-arrowed ring. When adsorbed to glow-discharged grids, portal complexes were found to yield Y-shaped images, as shown in panel C. Shown in panel D is a diagrammatic representation of the portal complex structure designed to be consistent with the images shown in panels A, B, and C. The bars are 200 nm (A) and 20 nm (B and C).
In some images, pUL6 rings could be seen to be composed of uniformly sized subunits directed radially outward from the ring axis. The arrows in Fig. 5B, for instance, indicate rings where subunits are visible, and other such rings can be seen in the same micrograph. Subunits visible in the upper-arrowed ring are shown diagrammatically in the Fig. 5B inset. The dimensions of the subunits (∼2 by ∼6 nm in projection) suggest the subunits may be pUL6 monomers.
When electron microscopy of purified pUL6 was performed with glow-discharged grids, images were often found to be Y-shaped, as shown in Fig. 5C. We interpret such images to be lateral views of the portal complex as contrasted with the axial views shown in Figs. 5A and B. The lateral views suggest the complex may have the shape of a goblet, as shown schematically in Fig. 5D. According to this interpretation, the upper right, middle, and lower left images shown in Fig. 5C would correspond to clockwise rotations of the schematic representation by ∼160, 30, and 190°, respectively. Measurements made with images interpreted to be lateral views showed widths in the wide and narrow portions of 16.6 ± 2.3 nm (n = 39) and 7.5 ± 1.3 nm (n = 39), respectively. The length was 19.5 ± 1.9 nm (n = 65), and the narrow portion constituted 0.37 ± 0.05 nm (n = 29) of the total length.
STEM mass measurements.
The masses of individual pUL6 rings were measured by dark-field STEM and used to compute the pUL6 oligomeric state in rings. The method depends on the fact that the number of scattered electrons (collected in two annular detectors) from each pixel is directly proportional to the mass thickness of that pixel. Summing the scattered electrons over all the pixels for a particle, one obtains a value proportional to the particle mass. From the total mass, the ring oligomeric state was calculated with an MW of 74,087 for pUL6.
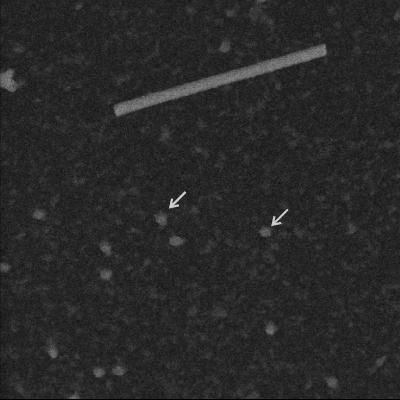
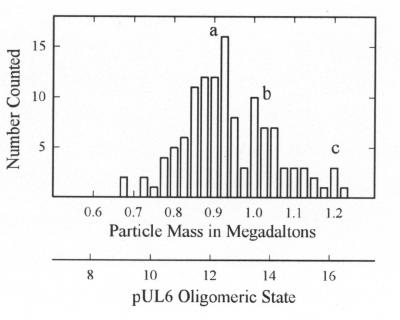
Examination of pUL6 in the STEM showed a uniform population of molecules that were circular or slightly irregular in profile, as shown in Fig. 6. Mass measurements were made with 127 such images derived from a total of 21 micrographs. The results showed pUL6 rings are somewhat heterogeneous, with a predominant population having a mass of ∼0.9 MDa corresponding to an oligomeric state of 12 (Fig. 7). Smaller populations were observed with mass values corresponding to oligomeric states of 14 and 16 (Fig. 7). For comparison, bacteriophage portal complexes or connectors are found to have oligomeric states of 12 or 13 (3, 45).
FIG. 6.
Dark-field STEM of purified pUL6. Mass measurements were made from pUL6 images such as those indicated by the arrows. TMV (top center; diameter, 18 nm) is included as a mass standard.
FIG. 7.
Histogram showing the masses determined from 127 individual pUL6 images such as those shown in Fig. 6. Note that the predominant population occurs at a mass of ∼0.9 MDa corresponding to a pUL6 oligomeric state of 12.
Amino acid sequence analyses.
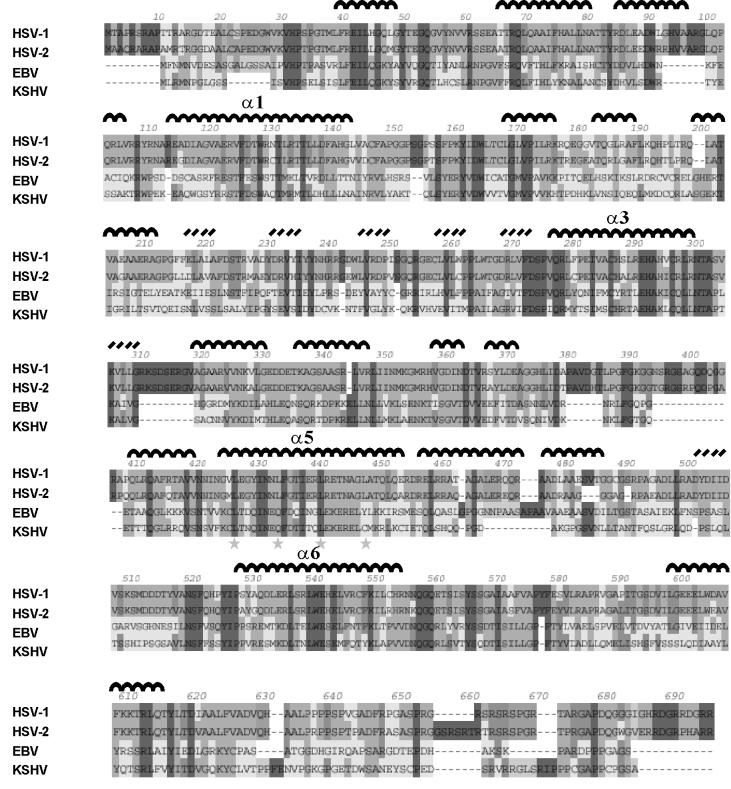
To attempt to identify conserved amino acid sequence elements that might relate to its function, we compared the pUL6 sequence with those of homologs encoded by other herpesviruses. The pUL6 sequence was aligned with the homologs of three representative human herpesviruses, HSV-2, EBV, and KSHV, and the alignment was shaded to indicate the degree of homology. Regions of amino acid identity in all proteins (darkest shading) were observed throughout the alignment, with 146 (21.5%) of the 676 pUL6 amino acids showing identity with all of the other herpesviruses having an amino acid at the relevant position. Identity with at least one of the three other herpesviruses was observed at 622 of 676 amino acids in the pUL6 sequence. All herpesvirus pUL6 homologs examined were found to contain a putative leucine zipper motif in a predicted α-helical region between residues 425 and 447 (marked with stars in Fig. 8). Other features that have been conserved include the MW, isoelectric point, and overall charge (data not shown).
FIG. 8.
Amino acid sequence alignment of HSV-1, HSV-2, EBV, and KSHV pUL6 protein homologs. The intensity of shading is indicative of the relative homology at each position, with dark gray shading representing amino acid identity. Predicted α-helical regions are marked with inverted black arcs above the HSV-1 protein sequence; black slanted lines indicate regions of predicted β structure. The stars between amino acids 425 and 447 mark the location of a presumptive leucine zipper found in the pUL6 homologs in all herpesviruses.
The program Ph.D. Predict was employed to identify presumptive secondary structural features in the amino acid sequences of pUL6 and its homologs in HSV-2, EBV, and KSHV. The results for pUL6 are shown in Fig. 8, where predicted α-helices and regions of β structure are indicated by inverted U's and angled dashes, respectively. The results show that α-helices are the predominant predicted structural feature accounting for ∼40% of the pUL6 sequence. Several long α-helices are predicted (e.g., those marked α1, α3, α5, and α6), and these may correspond to similar long α-helices present in the phage φ29 portal protein (37). Compared to α-helices, much less β structure is predicted in pUL6, with most occurring in a single region between amino acids 216 and 273.
DISCUSSION
Previous studies have indicated that HSV-1 DNA encapsidation may occur by mechanisms similar to those of the better-studied bacteriophages such as T7, T4, and λ (4, 8). Packaging in these phage systems begins when terminase makes a cut in the newly replicated, concatemeric phage DNA. The terminase-DNA end complex then attaches to a progeny procapsid by way of the portal or connector, a ring-shaped structure located at a unique site in the procapsid shell. DNA is injected through a hole in the portal oligomer with energy provided by terminase-catalyzed ATP hydrolysis. Recent studies of the phage φ29 portal complex suggest turning of the 12-subunit portal ring drives DNA translocation into the capsid (37). Packaging is completed when terminase makes a second cut in the DNA, and the portal is sealed. It was reasonable to assume that HSV-1 DNA packaging would conform to the phage model, because the mechanisms of capsid formation are similar in the two and because HSV-1 encodes a homolog of phage T4 terminase (30).
Location of pUL6 in the capsid.
In the studies described here, we tested the idea that the HSV-1 UL6 gene product is the structural subunit of the portal. We determined the location of pUL6 in the capsid, its copy number per capsid, and the structure of the purified protein in solution. In immunoelectron microscopic studies designed to determine the location of pUL6, 32 to 41% of capsids were found to have specific label at a single vertex and not at other sites. This observation is the first to demonstrate the presence of a unique capsid vertex in any member of the herpesvirus family and in any mammalian virus. A similar experimental method was employed to demonstrate that the phage T4 portal protein, gp20, is located at a unique capsid vertex (10). The location of pUL6 at a unique site in the capsid is compatible with the view that it functions as the portal, since the linear dsDNA genome presumably must enter through a single opening.
The proportion of capsids labeled in immunoelectron microscopic experiments (i.e., 32 to 41%) could not be increased by changes in the antibody concentration or in other experimental variables tested, including temperature and ionic strength. We suggest that inability to label pUL6 in some capsids may be due to (i) blocking of antibody access to capsids at or near the site of attachment to the electron microscope grid, (ii) preferential attachment of capsids to the grid by way of the pUL6 vertex, or (iii) masking of the portal complex (e.g., by steric blocking of antibody by the major capsid protein) on the surface of some capsids. Labeling of pUL6 capsid vertices with more than one gold bead as shown in Fig. 1 and Table 1 is expected to be possible, particularly if portal complexes contain more than a single pUL6 molecule.
pUL6 copy number per capsid.
The pUL6 copy number per capsid was found to be 14.8 ± 2.6 in Western immunoblot experiments in which purified pUL6 was employed as a standard. Calculation of the pUL6 copy number was carried out with the assumption that all capsids contain a portal complex, as suggested in previous reports (29, 36). The experimental value is in satisfactory agreement with the value of 12 expected if the HSV-1 portal is a 12-member ring like the portals of most dsDNA bacteriophage (3, 45). The higher value of the experimental compared to the expected copy number (i.e.,cf. 14.8 and 12) may result from an underestimate of the number of capsids analyzed due to uncertainties in determining VP5 quantitatively in stained SDS-polyacrylamide gels.
In a recent study, Ogasawara et al. (28) reported a value for the pUL6 copy number per capsid (44 ± 13) substantially higher than that observed here. The measurement was made by quantitative determination of a stained band in an SDS-polyacrylamide gel. We suggest the higher value may have resulted from the presence of non-UL6 proteins in the relevant region of the gel.
pUL6 purification.
Purification of pUL6 from recombinant baculovirus-infected Sf9 cells was complicated by the fact that it was present in insoluble inclusion bodies. The protein had to be dissolved before it could be purified and its structure examined in solution. Solubilization of pUL6 by treatment of inclusion bodies with 1 M arginine was therefore a crucial step in the purification procedure. Arginine was found to solubilize pUL6 after unsuccessful attempts with many other compounds, including salts, chaotropic agents, detergents, and reducing agents. Not all inclusion body-associated pUL6 was solubilized in 1 M arginine. The proportion was estimated to be ∼15% in most preparations (W. Newcomb, unpublished observation). Solubility of pUL6 required the continuous presence of 1 M arginine. If arginine was removed (e.g., by dialysis), pUL6 was found to aggregate and come out of solution. Such aggregates could be resolubilized, however, by further treatment with 1 M arginine.
pUL6 structure.
The structure of purified pUL6 was examined by electron microscopy of negatively stained specimens. Micrographs showed two views: a ring (Fig. 5A and 5B) and a Y-shaped structure (Fig. 5C) interpreted to be a lateral view of the ring. Together, the two images suggest the portal complex may have a goblet shape as shown schematically in Fig. 5D. According to this interpretation, the ring images would be axial views of the structure as seen from the wide end. It is consistent with this idea that the dimension of the Y images as measured at the wide end (16.6 ± 2.3 nm) is in satisfactory agreement with the measured diameter of the ring images (16.4 ± 1.1 nm). Rings presumptively corresponding to a view of the portal narrow end were observed in electron micrographs, but they were rare (data not shown). This observation suggests the narrow end may have a high affinity for the carbon-coated surface of electron microscope grids.
We suggest that the portal is oriented in the HSV-1 capsid with the narrow end pointing outward and the wide end toward the center of the capsid cavity. This orientation would correspond to that of the phage φ29 portal, the only portal whose orientation in the capsid has been determined (42). The structure of the HSV-1 portal complex, with a central channel able to accommodate DNA, is compatible with its proposed function as an opening through which DNA can enter the capsid.
The HSV-1 portal complex has important similarities to the portals of dsDNA bacteriophage. For example, the dimensions of the ring images, 16.4 ± 1.1 and 5.0 ± 0.7 nm for the outside and inside diameters respectively, are within the range of values observed for purified bacteriophage portals examined in comparable preparations. In six representative phage portals, the ranges of values were reported to be 14.5 to 17.5 nm for the outside diameter and 2.5 to 4.5 nm for the inside (3, 45).
In some micrographs, pUL6 rings could be seen to be composed of distinct subunits, which we interpret to be pUL6 monomers (Fig. 5B). Efforts to count the subunits yielded results that were in general agreement with determination of the ring oligomeric state by STEM. Distinct subunits are also observed in comparable electron micrographs of purified bacteriophage portal proteins, including those of phages T3, SPP1, T4, λ, and P22 (2, 11, 12, 17, 46). Efforts are currently under way to determine the structure of the pUL6 portal at higher resolution.
The length of the HSV-1 portal, 19.5 nm, is somewhat greater than the lengths reported for bacteriophage portals. For example, a range of 8.5 to 14.0 nm is reported for six phage portals (45). We suggest the difference may be related to the greater thickness of the capsid shell in HSV-1 (15 nm) compared to dsDNA bacteriophage (2 to 3 nm) (55).
Use of STEM to determine the mass of individual pUL6 portals showed that the population is heterogeneous with respect to the oligomeric state. A major population had an oligomeric state of 12, but there were significant minority populations corresponding to oligomeric states of 14 and 16. It is expected that the portal in the capsid will correspond to the majority population (i.e., the 12-mer), and we are now testing that expectation. The minor populations of 14- and 16-mers may indicate that the basic building block of the ring is a pUL6 dimer. This is consistent with the presence of a conserved putative leucine zipper (Fig. 8) that is predicted to be of the type that forms homodimeric interactions (52). In studies involving the use of electron cryomicroscopy and three-dimensional image reconstruction, the portal complex of phage φ29 was found to have 12-fold symmetry (42).
Structural similarities to other portal proteins.
A high degree of amino acid sequence conservation was observed between pUL6 and its homologs encoded in other human herpesviruses (Fig. 8). Regions of identity and homology were observed in all parts of the sequence. We interpret this observation to be consistent with the view that the homologs in other herpesviruses encode structurally similar portal complexes involved in DNA encapsidation.
α-Helices are the predominant secondary structural feature predicted to be present in the pUL6 molecule, accounting for ∼40% of the total amino acid sequence, as shown in Fig. 8. We note that α-helices are found to be the predominant elements of secondary structure in the phage φ29 portal protein (gp10), the only portal protein whose structure has been determined by X-ray crystallography (37). The gp10 structure includes three long α-helices that each traverse nearly the entire length of the funnel-shaped portal. The predicted α-helical regions of pUL6 include several long helices (i.e., those indicated by α1, α3, α5, and α6 in Fig. 8) that have the potential to have a similar orientation to those in the gp10 structure. Thus, the structures of the φ29 and HSV-1 portal proteins may be conserved despite amino acid sequence divergence, a situation that has been observed in functionally homologous proteins in a wide variety of organisms (9, 13, 16). We are currently using methods of genetic analysis to test the proposed structural homology between the φ29 and HSV-1 portal proteins.
The proposed evolutionary relationship between herpesviruses and dsDNA bacteriophage is supported by the results reported here showing a similarity in the structure of the portal protein and in its location in the capsid. The basic mechanism of DNA encapsidation appears to have been conserved over the period of time, probably 2 billion years or more, that links bacteria with vertebrate organisms, the hosts for herpesviruses.
In addition to serving as an entry site for DNA encapsidation, the phage T4 portal complex also functions as an initiator of capsid formation (5, 47, 48). Capsids are assembled at a membrane-bound site containing the portal protein (gp20) and gp40, another phage-encoded protein. The functioning of the portal in this way may ensure that only one portal complex is incorporated into each capsid. Although it is clear that morphologically normal HSV-1 capsids can form in the absence of pUL6 (20, 25, 44), it may nevertheless be the case that pUL6 is required for assembly of encapsidation-competent capsids in vivo. In the future, it may be productive, therefore, to examine how pUL6 becomes incorporated into capsids.
ACKNOWLEDGMENTS
We thank Martha Simon and Joe Wall for performing STEM mass analyses, Joel Baines for anti-MBP-UL6, and Jacob Nellissery for an important suggestion regarding the use of arginine.
This work was supported by NIH awards AI41644 (J.C.B.) and AI37549 (S.K.W.), and by NSF award MCB-9904879 (J.C.B).
REFERENCES
- 1.Adelman K, Salmon B, Baines J D. Herpes simplex virus DNA packaging sequences adopt novel structures that are specifically recognized by a component of the cleavage and packaging machinery. Proc Natl Acad Sci USA. 2001;98:3086–3091. doi: 10.1073/pnas.061555698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bazinet C, Benbasat J, King J, Carazo J M, Carrascosa J L. Purification and organization of the gene 1 portal protein required for phage P22 DNA packaging. Biochemistry. 1988;27:1849–1856. doi: 10.1021/bi00406a009. [DOI] [PubMed] [Google Scholar]
- 3.Bazinet C, King J. The DNA translocating vertex of dsDNA bacteriophage. Annu Rev Microbiol. 1985;39:109–129. doi: 10.1146/annurev.mi.39.100185.000545. [DOI] [PubMed] [Google Scholar]
- 4.Black L W. DNA packaging in dsDNA bacteriophages. Annu Rev Microbiol. 1989;43:267–292. doi: 10.1146/annurev.mi.43.100189.001411. [DOI] [PubMed] [Google Scholar]
- 5.Black L W, Showe M K, Steven A C. Morphogenesis of the T4 head. In: Karam J D, editor. Molecular biology of bacteriophage T4. Washington, D.C.: ASM Press; 1994. pp. 218–258. [Google Scholar]
- 6.Bogner E, Radsak K, Stinski M F. The gene product of human cytomegalovirus open reading frame UL56 binds the pac motif and has specific nuclease activity. J Virol. 1998;72:2259–2264. doi: 10.1128/jvi.72.3.2259-2264.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Catalano C E. The terminase enzyme from bacteriophage lambda: a DNA-packaging machine. Cell Mol Life Sci. 2000;57:128–148. doi: 10.1007/s000180050503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Catalano C E, Cue D, Feiss M. Virus DNA packaging: the strategy used by phage lambda. Mol Microbiol. 1995;16:1075–1086. doi: 10.1111/j.1365-2958.1995.tb02333.x. [DOI] [PubMed] [Google Scholar]
- 9.Doherty A J, Suh S W. Structural and mechanistic conservation in DNA ligases. Nucleic Acids Res. 2000;28:4051–4058. doi: 10.1093/nar/28.21.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Driedonks R A, Caldentey J. Gene 20 product of bacteriophage T4. II. Its structural organization in prehead and bacteriophage. J Mol Biol. 1983;166:341–360. doi: 10.1016/s0022-2836(83)80089-8. [DOI] [PubMed] [Google Scholar]
- 11.Driedonks R A, Engel A, ten Heggeler B, van Driel R. Gene 20 product of bacteriophage T4: its purification and structure. J Mol Biol. 1981;152:641–662. doi: 10.1016/0022-2836(81)90121-2. [DOI] [PubMed] [Google Scholar]
- 12.Dube P, Tavares P, Lurz R, van Heel M. The portal protein of bacteriophage SPP1: a DNA pump with 13-fold symmetry. EMBO J. 1993;12:1303–1309. doi: 10.1002/j.1460-2075.1993.tb05775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halaby D M, Poupon A, Mornon J. The immunoglobulin fold family: sequence analysis and 3D structure comparisons. Protein Eng. 1999;12:563–571. doi: 10.1093/protein/12.7.563. [DOI] [PubMed] [Google Scholar]
- 14.Homa F L, Brown J C. Capsid assembly and DNA packaging in herpes simplex virus. Rev Med Virol. 1997;7:107–122. doi: 10.1002/(sici)1099-1654(199707)7:2<107::aid-rmv191>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 15.Jacob R J, Morse L S, Roizman B. Anatomy of herpes simplex virus DNA. XII. Accumulation of head-to-tail concatemers in nuclei of infected cells and their role in the generation of the four isomeric arrangements of viral DNA. J Virol. 1979;29:448–457. doi: 10.1128/jvi.29.2.448-457.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katti M V, Sami-Subbu R, Ranjekar P K, Gupta V S. Amino acid repeat patterns in protein sequences: their diversity and structural-functional implications. Protein Sci. 2000;9:1203–1209. doi: 10.1110/ps.9.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kochan J, Carrascosa J L, Murialdo H. Bacteriophage lambda preconnectors. Purification and structure. J Mol Biol. 1984;174:433–447. doi: 10.1016/0022-2836(84)90330-9. [DOI] [PubMed] [Google Scholar]
- 18.Koslowski K M, Shaver P R, Casey II J T, Wilson T, Yamanaka G, Sheaffer A K, Tenney D J, Pederson N E. Physical and functional interactions between the herpes simplex virus UL15 and UL28 DNA cleavage and packaging proteins. J Virol. 1999;73:1704–1707. doi: 10.1128/jvi.73.2.1704-1707.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krosky P M, Underwood M R, Turk S R, Feng K W-H, Jain R K, Ptak R G, Westerman A C, Biron K K, Townsend L B, Drach J C. Resistance of human cytomegalovirus to benzimidazole ribonucleosides maps to two open reading frames: UL89 and UL56. J Virol. 1998;72:4721–4728. doi: 10.1128/jvi.72.6.4721-4728.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamberti C, Weller S K. The herpes simplex virus type 1 UL6 protein is essential for cleavage and packaging but not for genomic inversion. Virology. 1996;226:403–407. doi: 10.1006/viro.1996.0668. [DOI] [PubMed] [Google Scholar]
- 21.Martinez R, Sarisky R T, Weber P C, Weller S K. Herpes simplex virus type 1 alkaline nuclease is required for efficient processing of viral DNA replication intermediates. J Virol. 1996;70:2075–2085. doi: 10.1128/jvi.70.4.2075-2085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McNab A R, Desai P, Person S, Roof L L, Thomsen D R, Newcomb W W, Brown J C, Homa F L. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J Virol. 1998;72:1060–1070. doi: 10.1128/jvi.72.2.1060-1070.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mocarski E S, Roizman B. Structure and role of the herpes simplex virus DNA termini in inversion, circularization and generation of virion DNA. Cell. 1982;31:89–97. doi: 10.1016/0092-8674(82)90408-1. [DOI] [PubMed] [Google Scholar]
- 24.Murialdo H, Becker A. Head morphogenesis of complex double-stranded deoxyribonucleic acid bacteriophages. Microbiol Rev. 1978;42:529–576. doi: 10.1128/mr.42.3.529-576.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newcomb W W, Homa F L, Thomsen D R, Trus B L, Cheng N, Steven A C, Booy F, Brown J C. Assembly of the herpes simplex virus procapsid from purified components and identification of small complexes containing the major capsid and scaffolding proteins. J Virol. 1999;73:4239–4250. doi: 10.1128/jvi.73.5.4239-4250.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Newcomb W W, Trus B L, Booy F P, Steven A C, Wall J S, Brown J C. Structure of the herpes simplex virus capsid: molecular composition of the pentons and the triplexes. J Mol Biol. 1993;232:499–511. doi: 10.1006/jmbi.1993.1406. [DOI] [PubMed] [Google Scholar]
- 27.Newcomb W W, Trus B L, Cheng N, Steven A C, Sheaffer A K, Tenney D J, Weller S K, Brown J C. Isolation of herpes simplex virus procapsids from cells infected with a protease-deficient mutant virus. J Virol. 2000;74:1663–1673. doi: 10.1128/jvi.74.4.1663-1673.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogasawara M, Suzutani T, Yoshida I, Azuma M. Role of the UL25 gene product in packaging DNA into the herpes simplex virus capsid: location of UL25 product in the capsid and demonstration that it binds DNA. J Virol. 2001;75:1427–1436. doi: 10.1128/JVI.75.3.1427-1436.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patel A H, MacLean J B. The product of the UL6 gene of herpes simplex virus type 1 is associated with virus capsids. Virology. 1995;206:465–478. doi: 10.1016/s0042-6822(95)80062-x. [DOI] [PubMed] [Google Scholar]
- 30.Poon A P W, Roizman B. Characterization of a temperature-sensitive mutant of the UL15 open reading frame of herpes simplex virus 1. J Virol. 1993;67:4497–4503. doi: 10.1128/jvi.67.8.4497-4503.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rixon F J. Structure and assembly of herpesviruses. Semin Virol. 1993;4:135–144. [Google Scholar]
- 32.Rost B. PHD: predicting one-dimensional protein structure by profile-based neural networks. Methods Enzymol. 1996;266:525–539. doi: 10.1016/s0076-6879(96)66033-9. [DOI] [PubMed] [Google Scholar]
- 33.Salmon B, Baines J D. Herpes simplex virus DNA cleavage and packaging: association of multiple forms of UL15-encoded proteins with B capsids requires at least the UL6, UL17, and UL28 genes. J Virol. 1998;72:3045–3050. doi: 10.1128/jvi.72.4.3045-3050.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Severini A, Morgan A R, Tovell D R, Tyrrell D L. Study of the structure of replicative intermediates of HSV-1 DNA by pulsed-field gel electrophoresis. Virology. 1994;200:428–435. doi: 10.1006/viro.1994.1206. [DOI] [PubMed] [Google Scholar]
- 35.Sheaffer A K, Newcomb W W, Brown J C, Gao M, Weller S K, Tenney D J. Evidence for controlled incorporation of herpes simplex virus type 1 UL26 protease into capsids. J Virol. 2000;74:6838–6848. doi: 10.1128/jvi.74.15.6838-6848.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sheaffer A K, Newcomb W W, Gao M, Yu D, Weller S K, Brown J C, Tenney D J. Herpes simplex virus DNA cleavage and packaging proteins associate with the procapsid prior to its maturation. J Virol. 2001;75:687–698. doi: 10.1128/JVI.75.2.687-698.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simpson A A, Tao Y, Leiman P G, Badasso M O, He Y, Jardine P J, Olson N H, Morais M C, Grimes S, Anderson D L, Baker T S, Rossmann M G. Structure of the bacteriophage phi29 DNA packaging motor. Nature. 2000;408:745–750. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spaete R R, Frenkel N. The herpes simplex virus amplicon: analyses of cis-acting replication functions. Proc Natl Acad Sci USA. 1985;82:694–698. doi: 10.1073/pnas.82.3.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steven A C, Spear P G. Herpesvirus capsid assembly and envelopment. In: Burnett R, Chiu W, Garcea R, editors. Structural biology of viruses. New York, N.Y: Oxford University Press; 1996. pp. 312–351. [Google Scholar]
- 40.Stow N D, McMonagle E C, Davison A J. Fragments from both termini of the herpes simplex virus type 1 genome contain signals required for the encapsidation of viral DNA. Nucleic Acids Res. 1983;11:8205–8220. doi: 10.1093/nar/11.23.8205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suenaga M, Ohmae H, Tsuji S, Itoh T, Nishimura O. Renaturation of recombinant human neurotrophin-3 from inclusion bodies using a suppressor agent of aggregation. Biotechnol Appl Biochem. 1998;28:119–124. [PubMed] [Google Scholar]
- 42.Tao Y, Olson N H, Xu W, Anderson D L, Rossmann M G, Baker T S. Assembly of a tailed bacterial virus and its genome release studied in three dimensions. Cell. 1998;95:431–437. doi: 10.1016/s0092-8674(00)81773-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taus N S, Salmon B, Baines J D. The herpes simplex virus 1 UL17 gene is required for localization of capsids and major and minor capsid proteins to intranuclear sites where viral DNA is cleaved and packaged. Virology. 1998;252:115–125. doi: 10.1006/viro.1998.9439. [DOI] [PubMed] [Google Scholar]
- 44.Thomsen D R, Roof L L, Homa F L. Assembly of herpes simplex virus (HSV) intermediate capsids in insect cells infected with recombinant baculoviruses expressing HSV capsid proteins. J Virol. 1994;68:2442–2457. doi: 10.1128/jvi.68.4.2442-2457.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Valpuesta J M, Carrascosa J L. Structure of viral connectors and their function in bacteriophage assembly and DNA packaging. Q Rev Biophys. 1994;27:107–155. doi: 10.1017/s0033583500004510. [DOI] [PubMed] [Google Scholar]
- 46.Valpuesta J M, Fujisawa H, Marco S, Carazo J M, Carrascosa J L. Three-dimensional structure of T3 connector purified from overexpressing bacteria. J Mol Biol. 1992;224:103–112. doi: 10.1016/0022-2836(92)90579-9. [DOI] [PubMed] [Google Scholar]
- 47.van Driel R, Couture E. Assembly of bacteriophage T4 head-related structures. II. In vitro assembly of prehead-like structures. J Mol Biol. 1978;123:115–128. doi: 10.1016/0022-2836(78)90316-9. [DOI] [PubMed] [Google Scholar]
- 48.van Driel R, Couture E. Assembly of the scaffolding core of bacteriophage T4 proheads. J Mol Biol. 1978;123:713–719. doi: 10.1016/0022-2836(78)90217-6. [DOI] [PubMed] [Google Scholar]
- 49.Vlazny D A, Frenkel N. Replication of herpes simplex virus DNA: localization of replication recognition signals within defective virus genomes. Proc Natl Acad Sci USA. 1981;78:742–746. doi: 10.1073/pnas.78.2.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wall J S, Hainfeld J F, Simon M N. Scanning transmission electron microscopy of nuclear structures. Methods Cell Biol. 1998;53:139–164. doi: 10.1016/s0091-679x(08)60878-x. [DOI] [PubMed] [Google Scholar]
- 51.Weller S K. Herpes simplex virus DNA replication and genome maturation. In: Cooper G M, Temin R G, Sugden B, editors. The DNA provirus: Howard Temin's scientific legacy. Washington, D.C.: American Society for Microbiology; 1995. pp. 189–213. [Google Scholar]
- 52.Wolf E, Kim P S, Berger B. MultiCoil: a program for predicting two- and three-stranded coiled coils. Protein Sci. 1997;6:1179–1189. doi: 10.1002/pro.5560060606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu D, Weller S K. Herpes simplex virus type 1 cleavage and packaging proteins UL15 and UL28 are associated with B but not C capsids during packaging. J Virol. 1998;72:7428–7439. doi: 10.1128/jvi.72.9.7428-7439.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang X, Efstathiou S, Simmons A. Identification of novel herpes simplex virus replicative intermediates by field inversion gel electrophoresis: implications for viral DNA amplification strategies. Virology. 1994;202:530–539. doi: 10.1006/viro.1994.1375. [DOI] [PubMed] [Google Scholar]
- 55.Zhou Z H, Dougherty M, Jakana J, He J, Rixon F J, Chiu W. Seeing the herpesvirus capsid at 8.5 Å. Science. 2000;288:877–880. doi: 10.1126/science.288.5467.877. [DOI] [PubMed] [Google Scholar]