Abstract
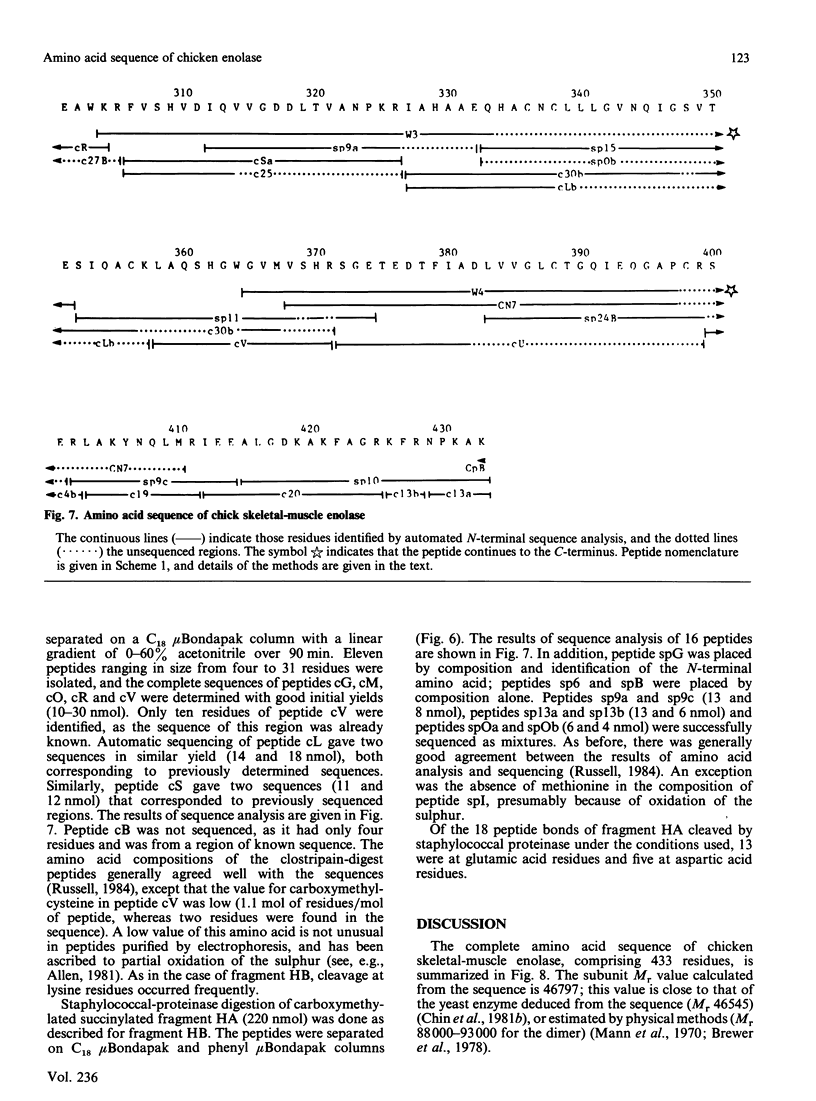
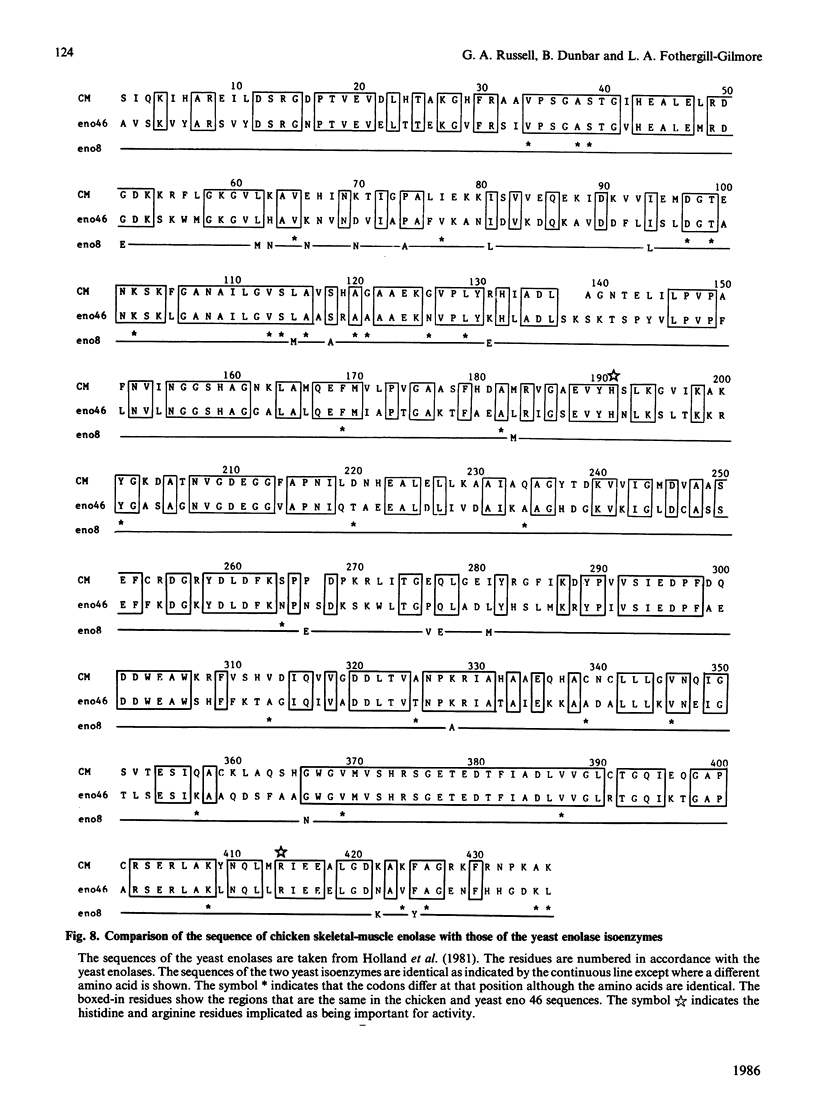
The complete amino acid sequence of chicken skeletal-muscle enolase, comprising 433 residues, was determined. The sequence was deduced by automated sequencing of hydroxylamine-cleavage, CNBr-cleavage, o-iodosobenzoic acid-cleavage, clostripain-digest and staphylococcal-proteinase-digest fragments. The presence of several acid-labile peptide bonds and the tenacious aggregation of most CNBr-cleavage fragments meant that a commonly used sequencing strategy involving initial CNBr cleavage was unproductive. Cleavage at the single Asn-Gly peptide bond with hydroxylamine proved to be particularly useful. Comparison of the sequence of chicken enolase with the two yeast enolase isoenzyme sequences shows that the enzyme is strongly conserved, with 60% of the residues identical. The histidine and arginine residues implicated as being important for the activity of yeast enolase are conserved in the chicken enzyme. Secondary-structure predictions are analysed in an accompanying paper [Sawyer, Fothergill-Gilmore & Russell (1986) Biochem. J. 236, 127-130].
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bornstein P., Balian G. Cleavage at Asn-Gly bonds with hydroxylamine. Methods Enzymol. 1977;47:132–145. doi: 10.1016/0076-6879(77)47016-2. [DOI] [PubMed] [Google Scholar]
- Brewer J. M. Yeast enolase: mechanism of activation by metal ions. CRC Crit Rev Biochem. 1981;11(3):209–254. doi: 10.3109/10409238109108702. [DOI] [PubMed] [Google Scholar]
- CRESTFIELD A. M., MOORE S., STEIN W. H. The preparation and enzymatic hydrolysis of reduced and S-carboxymethylated proteins. J Biol Chem. 1963 Feb;238:622–627. [PubMed] [Google Scholar]
- Carney D. N., Marangos P. J., Ihde D. C., Bunn P. A., Jr, Cohen M. H., Minna J. D., Gazdar A. F. Serum neuron-specific enolase: a marker for disease extent and response to therapy of small-cell lung cancer. Lancet. 1982 Mar 13;1(8272):583–585. doi: 10.1016/s0140-6736(82)91748-2. [DOI] [PubMed] [Google Scholar]
- Chin C. C., Brewer J. M., Eckard E., Wold F. The amino acid sequence of yeast enolase. Preparation and characterization of peptides produced by chemical and enzymatic fragmentation. J Biol Chem. 1981 Feb 10;256(3):1370–1376. [PubMed] [Google Scholar]
- Drapeau G. R., Boily Y., Houmard J. Purification and properties of an extracellular protease of Staphylococcus aureus. J Biol Chem. 1972 Oct 25;247(20):6720–6726. [PubMed] [Google Scholar]
- Elliott J. I., Brewer J. M. The inactivation of yeast enolase by 2,3-butanedione. Arch Biochem Biophys. 1978 Sep;190(1):351–357. doi: 10.1016/0003-9861(78)90285-0. [DOI] [PubMed] [Google Scholar]
- Elliott J. I., Brewer J. M. The relation of photooxidized histidines in yeast enolase to enzymatic activity. Arch Biochem Biophys. 1979 Jan;192(1):203–213. doi: 10.1016/0003-9861(79)90085-7. [DOI] [PubMed] [Google Scholar]
- Frank G. A cheap and simple method to achieve and maintain the necessary purity of reagents and solvents for automated amino acid sequence determination with the sequenator. Hoppe Seylers Z Physiol Chem. 1979 Jul;360(7):997–999. [PubMed] [Google Scholar]
- Freemont P. S., Dunbar B., Fothergill L. A. Human skeletal-muscle aldolase: N-terminal sequence analysis of CNBr- and o-iodosobenzoic acid-cleavage fragments. Arch Biochem Biophys. 1984 Jan;228(1):342–352. doi: 10.1016/0003-9861(84)90075-4. [DOI] [PubMed] [Google Scholar]
- Holland M. J., Holland J. P., Thill G. P., Jackson K. A. The primary structures of two yeast enolase genes. Homology between the 5' noncoding flanking regions of yeast enolase and glyceraldehyde-3-phosphate dehydrogenase genes. J Biol Chem. 1981 Feb 10;256(3):1385–1395. [PubMed] [Google Scholar]
- Hunkapiller M. W., Hood L. E. Direct microsequence analysis of polypeptides using an improved sequenator, a nonprotein carrier (polybrene), and high pressure liquid chromatography. Biochemistry. 1978 May 30;17(11):2124–2133. doi: 10.1021/bi00604a016. [DOI] [PubMed] [Google Scholar]
- Klapper D. G., Wilde C. E., 3rd, Capra J. D. Automated amino acid sequence of small peptides utilizing Polybrene. Anal Biochem. 1978 Mar;85(1):126–131. doi: 10.1016/0003-2697(78)90282-8. [DOI] [PubMed] [Google Scholar]
- Landon Cleavage at aspartyl-prolyl bonds. Methods Enzymol. 1977;47:145–149. doi: 10.1016/0076-6879(77)47017-4. [DOI] [PubMed] [Google Scholar]
- MALMSTROM B. G. The purification of yeast enolase by zone electrophoresis and ion-exchange chromatography, and the existence of several active forms of the enzyme. Arch Biochem Biophys. 1957 Jul;70(1):58–69. doi: 10.1016/0003-9861(57)90080-2. [DOI] [PubMed] [Google Scholar]
- Mahoney W. C., Hermodson M. A. High-yield cleavage of tryptophanyl peptide bonds by o-iodosobenzoic acid. Biochemistry. 1979 Aug 21;18(17):3810–3814. doi: 10.1021/bi00584a026. [DOI] [PubMed] [Google Scholar]
- Mahoney W. C., Smith P. K., Hermodson M. A. Fragmentation of proteins with o-iodosobenzoic acid: chemical mechanism and identification of o-iodoxybenzoic acid as a reactive contaminant that modifies tyrosyl residues. Biochemistry. 1981 Jan 20;20(2):443–448. doi: 10.1021/bi00505a033. [DOI] [PubMed] [Google Scholar]
- Mann K. G., Castellino F. J., Hargrave P. A. Molecular weight and subunit structure of yeast enolase. Biochemistry. 1970 Sep 29;9(20):4002–4007. doi: 10.1021/bi00822a020. [DOI] [PubMed] [Google Scholar]
- Rider C. C., Taylor C. B. Enolase isoenzymes. II. Hybridization studies, developmental and phylogenetic aspects. Biochim Biophys Acta. 1975 Sep 9;405(1):175–187. [PubMed] [Google Scholar]
- Rider C. C., Taylor C. B. Evidence for a new form of enolase in rat brain. Biochem Biophys Res Commun. 1975 Sep 16;66(2):814–820. doi: 10.1016/0006-291x(75)90582-3. [DOI] [PubMed] [Google Scholar]
- Riordan J. F., McElvany K. D., Borders C. L., Jr Arginyl residues: anion recognition sites in enzymes. Science. 1977 Mar 4;195(4281):884–886. doi: 10.1126/science.190679. [DOI] [PubMed] [Google Scholar]
- Rose I. A., O'Connell E. L. Inactivation and labeling of triose phosphate isomerase and enolase by glycidol phosphate. J Biol Chem. 1969 Dec 10;244(23):6548–6550. [PubMed] [Google Scholar]
- Sawyer L., Fothergill-Gilmore L. A., Russell G. A. The predicted secondary structure of enolase. Biochem J. 1986 May 15;236(1):127–130. doi: 10.1042/bj2360127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scopes R. K. Multiple enzyme purifications from muscle extracts by using affinity-elution-chromatographic procedures. Biochem J. 1977 Feb 1;161(2):265–277. doi: 10.1042/bj1610265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scopes R. K. Purification of glycolytic enzymes by using affinity-elution chromatography. Biochem J. 1977 Feb 1;161(2):253–263. doi: 10.1042/bj1610253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootton J. C., Baron A. J., Fincham J. R. The amino acid sequence of Neurospora NADP-specific glutamate dehydrogenase. Peptides from digestion with a staphylococcal proteinase. Biochem J. 1975 Sep;149(3):749–755. doi: 10.1042/bj1490749. [DOI] [PMC free article] [PubMed] [Google Scholar]


