ABSTRACT
In the era of antimicrobial resistance, phage-antibiotic combinations offer a promising therapeutic option, yet research on their synergy and antagonism is limited. This study aims to assess these interactions, focusing on protein synthesis inhibitors and cell envelope-active agents against multidrug-resistant bacterial strains. We evaluated synergistic and antagonistic interactions in multidrug-resistant Staphylococcus aureus, Enterococcus faecium, and Pseudomonas aeruginosa strains. Phages were combined with protein synthesis inhibitors [linezolid (LZD), minocycline (MIN), gentamicin (GEN), and azithromycin (AZM)] or cell envelope-active agents [daptomycin (DAP), ceftaroline (CPT), and cefepime (FEP)]. Modified checkerboard minimum inhibitory concentration assays and 24-h time-kill analyses were conducted, alongside one-step growth curves to analyze phage growth kinetics. Statistical comparisons used one-way analysis of variance (ANOVA) and the Tukey test (P < 0.05). In the checkerboard and 24-h time-kill analyses (TKA) of S. aureus and E. faecium, phage-LZD and phage-MIN combinations were antagonistic (FIC > 4) while phage-DAP and phage-CPT were synergistic (FIC 0.5) (ANOVA range of mean differences 0.52–2.59 log10 CFU/mL; P < 0.001). For P. aeruginosa, phage-AZM was antagonistic (FIC > 4), phage-GEN was additive (FIC = 1), and phage-FEP was synergistic (ANOVA range of mean differences 1.04–1.95 log10 CFU/mL; P < 0.001). Phage growth kinetics were altered in the presence of LZD and MIN against S. aureus and in the presence of LZD against a single E. faecium strain (HOU503). Our findings indicate that select protein synthesis inhibitors may induce phage-antibiotic antagonism. However, this antagonism may not solely stem from changes in phage growth kinetics, warranting further investigation into the complex interplay among strains, phage attributes, and antibiotic mechanisms affecting bacterial inhibition.
IMPORTANCE
In the face of escalating antimicrobial resistance, combining phages with antibiotics offers a promising avenue for treating infections unresponsive to traditional antibiotics. However, while studies have explored synergistic interactions, less attention has been given to potential antagonism and its impact on phage growth kinetics. This research evaluates the interplay between phages and antibiotics, revealing both synergistic and antagonistic patterns across various bacterial strains and shedding light on the complex dynamics that influence treatment efficacy. Understanding these interactions is crucial for optimizing combination therapies and advancing phage therapy as a viable solution for combating antimicrobial resistance.
KEYWORDS: bacteriophage, antibiotic, antagonism, Staphylococcus aureus, Enterococcus faecium, Pseudomonas aeruginosa
INTRODUCTION
Infections caused by antimicrobial-resistant (AMR) bacteria pose a grave global public health emergency, resulting in approximately 700,000 deaths yearly (1). Experts predict that this number will likely increase through 2050 (2). Reports from the World Health Organization suggest that we are on the brink of a post-antibiotic era in this century, while the Centers for Disease Control and Prevention declared that we have already entered this ominous era in their 2019 Antibiotic Resistance Threat Report (3, 4). Amidst the ongoing crisis, bacteriophage (phage) therapy is often proposed as part of the solution. Utilizing a combination of phages and antibiotics in phage-antibiotic combinations could address bacterial infections that are resistant or unresponsive to conventional antibiotic treatments (5–7). This approach applies two selective pressures on bacteria, offering a promising avenue for therapeutic intervention (5, 6).
Research on phage-antibiotic combinations and subsequent synergistic activity remains limited. Regrettably, the exploration of phage-antibiotic antagonism, where the combined activity of these agents is less effective than either agent alone, has received relatively less attention (8–11). While a comprehensive understanding of the underlying mechanisms of synergy and antagonism is currently lacking, researchers have identified a few possible explanations. First, antibiotics that target the protein synthesis pathway in bacteria, such as macrolides, tetracyclines, or aminoglycosides, can exhibit antagonistic interactions with phages compared to antibiotics that primarily target the bacterial cell envelope (8, 12). It is reasonable to hypothesize that this is because phages rely on host ribosomes for their own replication and protein synthesis. Thus, when protein synthesis inhibitors are present, they can potentially disrupt phage replication and reduce the production of phage progeny within the infected bacterial cells, leading to reduced overall phage efficacy (8). This has been demonstrated previously in in vitro and in vivo experiments wherein reduced phage efficacy was observed when combined with macrolides and tetracyclines (13, 14). In contrast, cell envelope-active antibiotics, including beta-lactams, primarily disrupt the integrity of the bacterial cell envelope, leading to cell lysis and bacterial death (15). Therefore, when cell envelope-active antibiotics are combined with phages, their modes of action can synergistically enhance bacterial cell lysis and overall antimicrobial activity as demonstrated in both planktonic and biofilm conditions (16, 17). However, not all observations match these hypotheses. For example, Liu et al. (18) observed synergy between Staphylococcus aureus phages and subinhibitory concentrations of protein synthesis inhibitors but not cell envelope-active agents in planktonic cells; synergy with protein synthesis inhibitors was maintained against in vitro and in vivo biofilms and could occur with otherwise phage-resistant strains. This might suggest that the subinhibitory antibiotic disadvantages cells sufficiently to allow phage-directed takeover without completely preventing phage replication. Clearly, however, the specific effects of phage-antibiotic combinations can vary widely and are not easily predictable based on antibiotic mechanisms of action. Therefore, the aim of this study was to evaluate in vitro synergistic and antagonistic activity among phage-antibiotic combinations with cell envelope-active agents and protein synthesis inhibitors against clinical strains of multidrug-resistant (MDR) S. aureus, Enterococcus faecium, and Pseudomonas aeruginosa, with evaluations of phage-antibiotic combination impacts on phage growth kinetics.
RESULTS
Bacterial isolates
Minimum inhibitory concentrations (MICs) for evaluated S. aureus, E. faecium, and P. aeruginosa against select protein synthesis pathway inhibitors and cell envelope-active agents are displayed in Table 1.
TABLE 1.
Minimum inhibitory concentrations (in mg/L) for evaluated S. aureus, E. faecium, and P. aeruginosa strains
| Staphylococcus aureus | Daptomycin (DAP) (MIC ≤ 1) | Ceftaroline (CPT) (MIC ≤ 1) | Linezolid (LZD) (MIC ≤ 4) | Minocycline (MIN) (MIC ≤ 4) |
|---|---|---|---|---|
| D712 | 4 | 1 | 1 | 0.125 |
| 684 | 4 | 0.5 | 1 | 2 |
Antibiotic class may not predict phage-antibiotic interactions
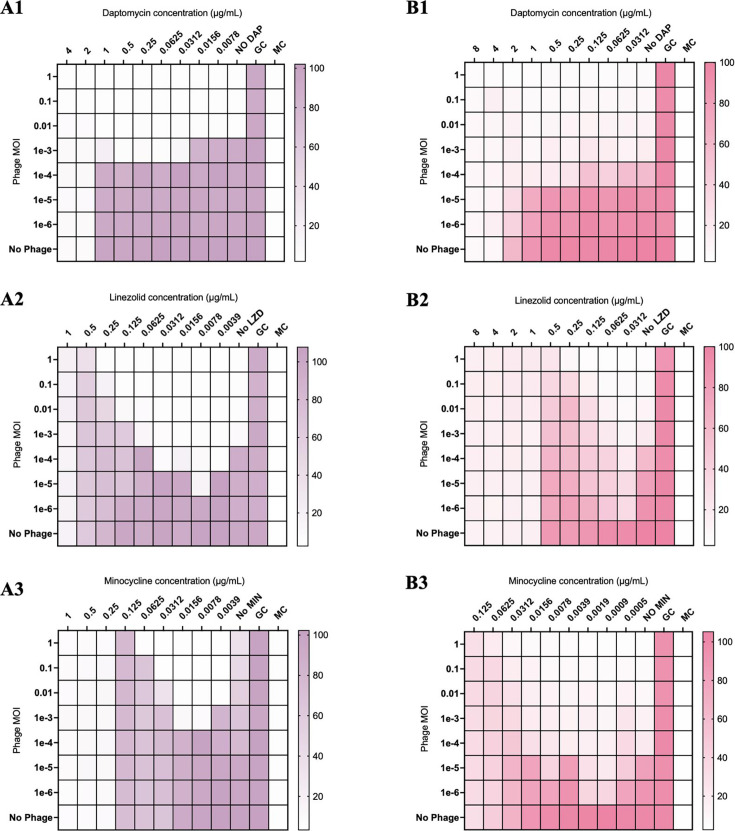
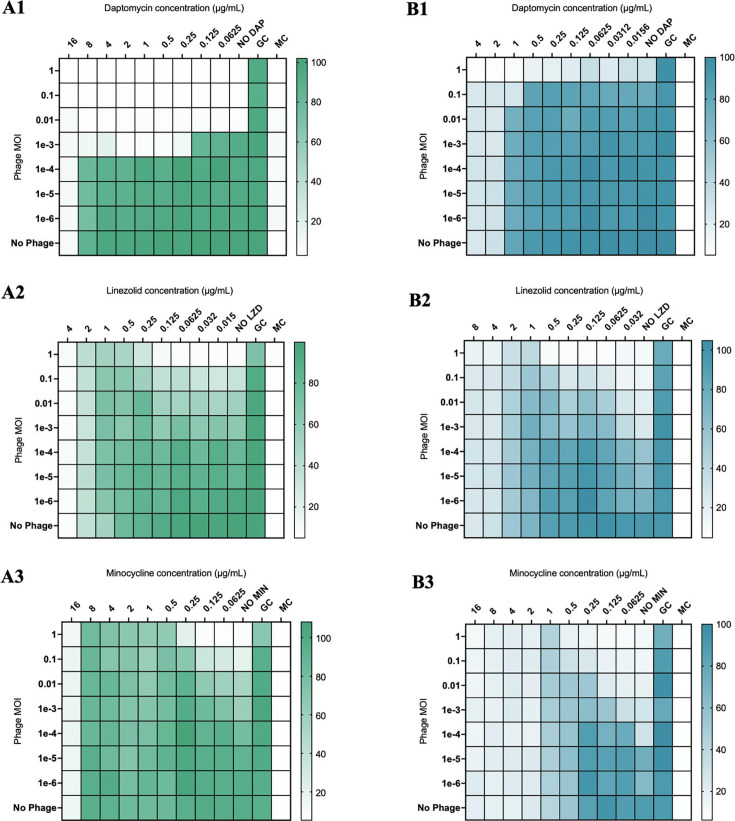
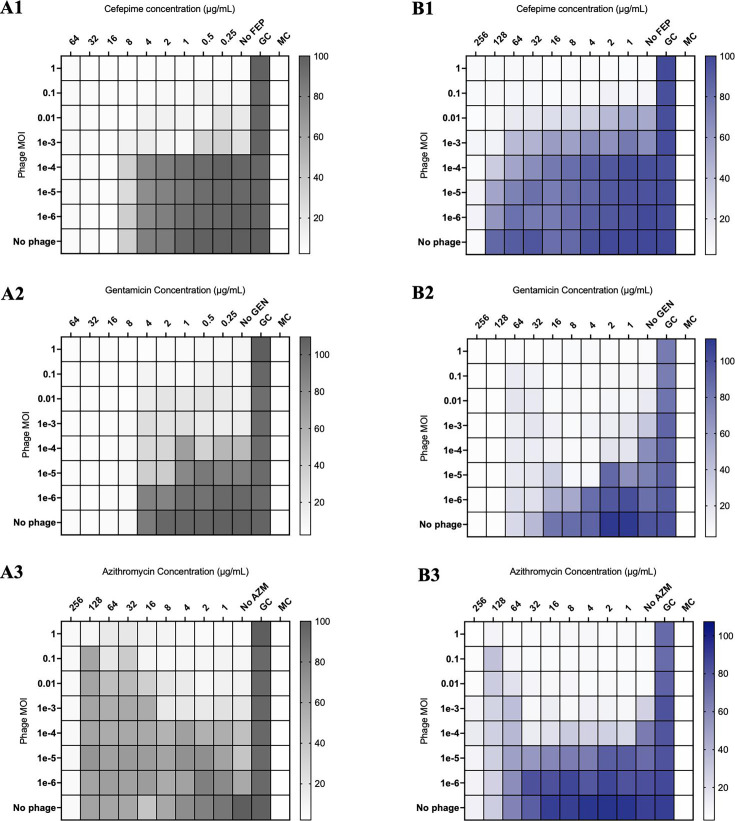
In modified checkerboard against S. aureus, phage-antibiotic combinations containing daptomycin (DAP) or ceftaroline (CPT) demonstrated synergistic killing of S. aureus strains D712 and 684, compared to DAP, CPT, and phage monotherapy at 0.5× minimum inhibitory concentration (MIC) [FIC 0.5, analysis of variance (ANOVA) range of mean difference 0.98–6.7 log10 CFU/mL; P < 0.001]; however, phage-antibiotic combinations with linezolid (LNZ) or minocycline (MIN) demonstrated antagonism at 0.5× MIC (FIC > 4, ANOVA range of mean difference 3.2–6.4 log10 CFU/mL; P < 0.001) (Fig. 1, 4A1, and A2). Similar findings were identified in E. faecium strains R497 and HOU503 with DAP- or CPT-containing combinations demonstrating synergy at 0.5× and 0.25× MIC (FIC 0.5, ANOVA range of mean difference 0.9–3.2 log10 CFU/mL; P < 0.001); while combinations with LNZ or MIN demonstrated antagonism at 0.5× MIC (FIC > 4, ANOVA range of mean difference 1.6–4.7 log10 CFU/mL; P < 0.001) (Fig. 2, 4B1, and B2). Against P. aeruginosa strains 9010 and 10266 in checkerboard and TKA, combinations with cefepime (FEP) or gentamicin (GENT) at 0.5× MIC demonstrated synergistic killing (FIC 0.5, ANOVA range of mean difference 1.8–5.3 log10 CFU/mL; P < 0.001); however, those with azithromycin (AZM) demonstrated antagonism at 0.5× and 0.25× MIC (FIC > 4, ANOVA range of mean difference 1.4–5.1 log10 CFU/mL; P < 0.001) (Fig. 3, 4C1, and C2).
Fig 1.
Modified checkerboard MIC of S. aureus strains D712 (A1–A3) and 684 (B1–B3) against phage-antibiotic combinations containing cell envelope-active agents or protein synthesis pathway inhibitors. GC, untreated growth control; MC, uninoculated media control. Comparisons are versus growth control and are depicted by the color gradient as a percentage of growth (right side of the figure).
Fig 2.
Modified checkerboard MIC of E. faecium strains R497 (A1–A3) and HOU503 (B1-B3) against phage-antibiotic combinations containing cell envelope-active agents or protein synthesis pathway inhibitors. GC, untreated growth control; MC, uninoculated media control. Comparisons are versus growth control and are depicted by the color gradient as a percentage of growth (right side of the figure).
Fig 3.
Modified checkerboard MIC of P. aeruginosa strains 9010 (A1-A3) and 10266 (B1-B3) against phage-antibiotic combinations containing cell envelope-active agents or protein synthesis pathway inhibitors. GC, untreated growth control; MC, uninoculated media control. Comparisons are versus growth control and are depicted by the color gradient as a percentage of growth (right side of the figure).
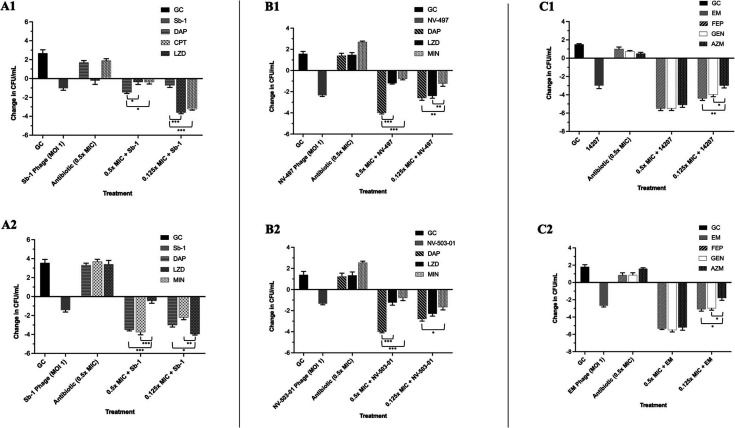
Fig 4.
Twenty-four-hour time-kill analyses of S. aureus strains D712 (A1) and 684 (A2), E. faecium strains R497 (B1) and HOU503 (B2), and P. aeruginosa strains 9010 (C1) and 10266 (C2) against phage-antibiotic combinations containing cell envelope-active agents or protein synthesis pathway inhibitors. All are completed in duplicate. P values were determined using a one-way ANOVA and Tukey’s post hoc test. *P < 0.05; **P < 0.005; and ***P < 0.001.
Disruption of bacteriophage kinetics is not antibiotic class-dependent
Standardized one-step phage growth curves were then conducted with select phage-antibiotic combinations to evaluate whether the demonstrated synergy and antagonism in checkerboard and TKA may be the result of altered phage growth kinetics in the presence of each cell envelope-active or protein synthesis inhibitor antibiotic (Fig. 5).
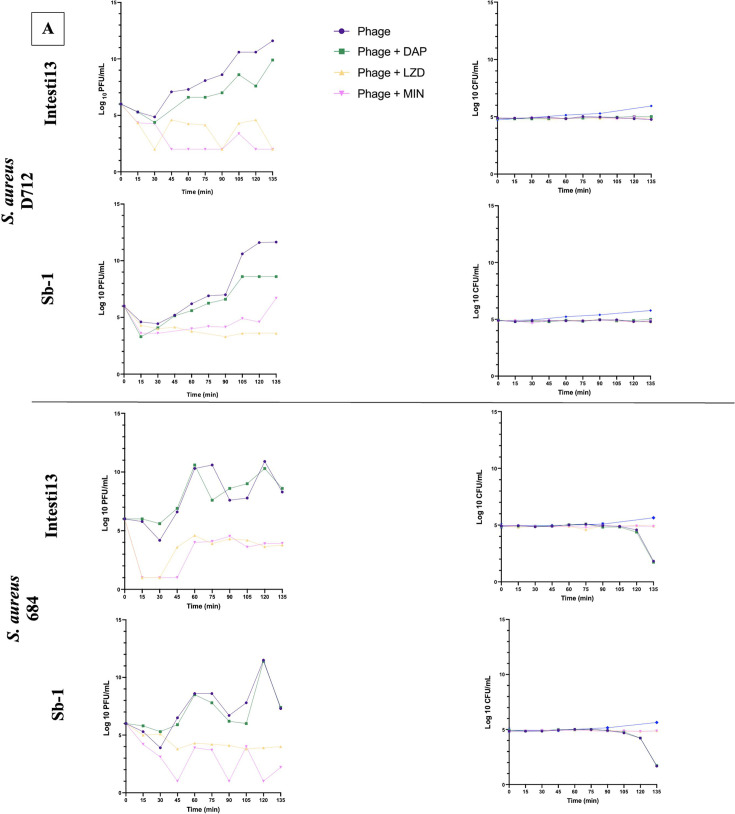
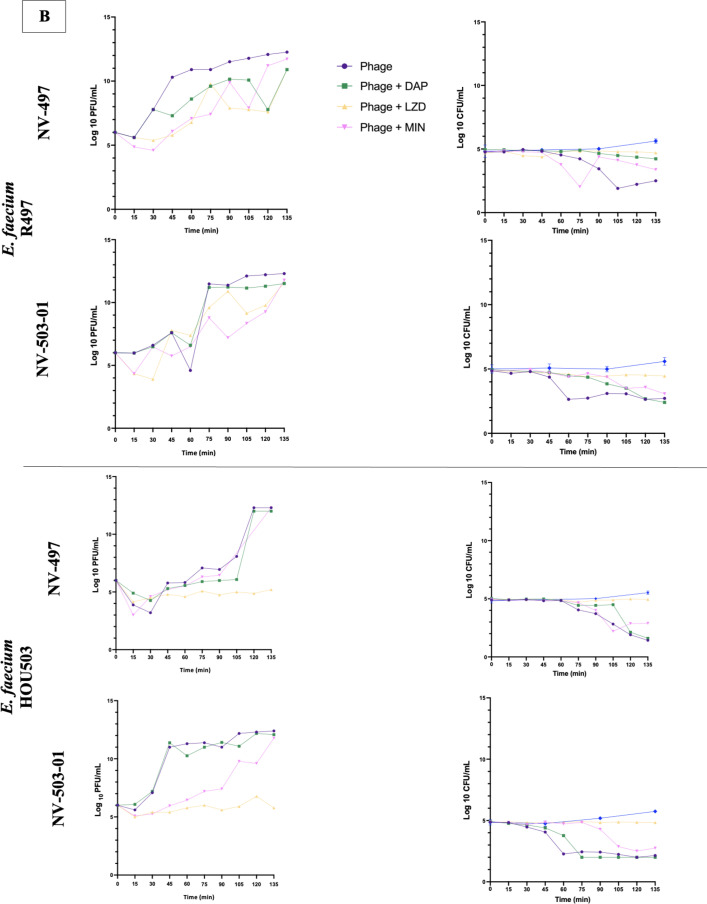
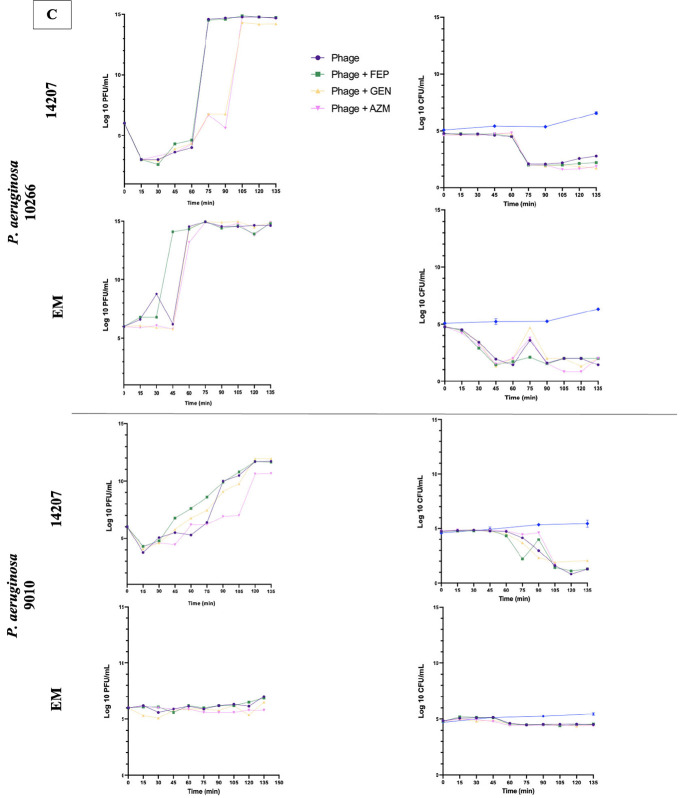
Fig 5.
Phage one-step growth curves, in duplicate, and corresponding CFU/mL for S. aureus phages Sb-1 and Intesti13 against S. aureus D712 and 684 (A), E. faecium phages NV-497 and NV-503–01 against E. faecium R497 and HOU503 (B), and P. aeruginosa phages EM and 14207 against P. aeruginosa 9010 and 10266 (C) with protein synthesis inhibitors or cell envelope-active agents. Limit of detection: 1 PFU/mL.
Against S. aureus strains D712 and 684, the addition of LZD or MIN to Intesti13 or Sb-1 significantly decreased phage burst size (−Δ 9.6–4.38 PFU/mL; P < 0.001) and resulted in substantial CFU decreases (Fig. 5A). Similarly, against E. faecium strain HOU503, the addition of LZD significantly decreased the burst size of phages NV-497 and NV-503–01 (−Δ 6.82–5.62 PFU/mL; P < 0.001); however, no significant difference in phage burst size was identified for combinations against E. faecium strain R497 (−Δ 1.4–0.52) (Fig. 5B). In addition to this variability between the E. faecium strains, there was evidence of variability by phage, with MIN inhibiting the burst size and antibacterial activity of phage NV-503-01, but not of NV-497. Against P. aeruginosa strains 9010 and 10266, no significant difference in phage burst size was identified for phages 14207 and EM in the presence of each antibiotic (−Δ 1.24–0.04) (Fig. 5C). Not all combinations that resulted in substantial phage replication also led to a detectable drop in bacterial concentration during the 135-min observation period, suggesting that replication of uninfected bacterial cells balanced the rate of phage-mediated lysis despite the presence of antibiotics. It is unknown whether the bacterial concentration would have declined at later time points.
DISCUSSION
The emergence of AMR bacteria presents a severe global public health crisis, necessitating the exploration of alternative treatment strategies (3, 19). Phage therapy, particularly when used in combination with standard-of-care antibiotics, offers a potential avenue for addressing bacterial infections that are resistant or unresponsive to conventional antibiotics (5, 6). In this study, we aimed to investigate the potential for synergistic or antagonistic interactions among cell envelope-active or protein synthesis pathway inhibitor antibiotics when combined with well-performing phages against clinically relevant MDR strains of S. aureus, E. faecium, and P. aeruginosa.
Our modified checkerboard assays (Fig. 1 to 3) and 24 h TKA (Fig. 4A1 through C2) revealed two general trends in bacterial killing against selected antibiotics (17, 20). First, synergistic killing by phage-antibiotic combinations was usually observed when increasing concentrations of certain bactericidal antibiotics (e.g., DAP, FEP, and GEN) were combined with increasing phage MOI (Fig.1A1, B1, 2A1, B1, 3A1, A2, B1 and B2). This synergism might be attributable to the complementary modes of action between the bactericidal antibiotics, which disrupt the bacterial cell envelope, and phage, which induce cell lysis during their life cycle (9, 21). In contrast, antagonistic interactions were usually observed when decreasing antibiotic concentrations were combined with increasing phage MOI, particularly with protein synthesis inhibitors (e.g., LZD, MIN, and AZM) (Fig. 1, 2A2, A3, B2, B3, 3A3 and B3). Notably, GEN, a bactericidal protein synthesis inhibitor demonstrated synergistic activity against P. aeruginosa isolates, which differs from the antagonistic interactions observed with the other protein synthesis inhibitors (22).
The observed synergistic and antagonistic patterns may be linked to the differential effects of bactericidal and bacteriostatic antibiotics on bacterial mechanisms and metabolism. Bactericidal antibiotics, such as aminoglycosides, disrupt essential bacterial processes, leading to irreversible damage and bacterial death (15). Phages, which replicate within bacterial cells, can benefit from increased cellular resources due to antibiotic-induced bactericidal action, potentially enhancing their replication and efficacy (9). Additionally, bactericidal antibiotics (e.g., beta-lactam antibiotics and gentamicin) primarily affect actively dividing bacterial cells, which can impact phage replication within the infected cells (23). In contrast, an antibiotic that demonstrates a bacteriostatic effect slows down bacterial growth and reduces metabolic activity without causing immediate cell death (24). This difference in metabolic state may affect phage replication and their ability to complete their life cycle within infected cells (25, 26). However, it is worth noting that the interactions between phages and antibiotics can be complex and context-dependent, influenced by various factors, including antibiotic type, phage characteristics, bacterial strains, and experimental conditions, including antibiotic concentration, media, stationary phase versus logarithmic dividing cells, and sequential administration versus administrating the phage and antibiotic at the same time (27–30). For example, the combination of minocycline and phage Sb-1 with S. aureus 684 showed a unique outcome: while phage replication was poor overall, the phage concentration fluctuated rapidly between 101 and 104 PFU/mL. The timing of these fluctuations is not consistent with the Sb-1 replication cycle in the strain and varied slightly between replicate experiments (data not shown). One hypothesis is that the data might reflect a low level of asynchronous phage infection and lysis, unlike the synchronous infection typical of one-step growth curves, perhaps due to a particular effect of minocycline on the cellular metabolism of this S. aureus strain.
One significant factor that might affect these observations is the timing of phage administration relative to antibiotic exposure. In our experiments, both the phage and antibiotic were administered simultaneously. If the phage was introduced prior to the antibiotic, allowing a window of, for example, 2 h, the observed antagonistic interactions might be reduced or altered (18, 28, 31, 32). This aspect warrants further investigation in future studies to determine the optimal sequencing of treatment for maximum efficacy.
In comparing our results with findings from previous studies on phage-antibiotic interactions, notable contrasts and similarities emerge. Gu et al. (9) demonstrated varying effects of antibiotics on Escherichia coli phages, including both antagonistic and synergistic interactions with different antibiotics such as ciprofloxacin, colistin, and ceftazidime. Similarly, our study revealed varied interactions between antibiotics and phages, with some combinations showing synergy while others exhibited antagonism. For instance, we observed synergistic killing with cell envelope-active antibiotics, such as daptomycin and ceftaroline, against S. aureus and E. faecium, aligning with the findings of Gu et al. (9) regarding synergy with ceftazidime. Conversely, we found antagonism with protein synthesis inhibitors like linezolid and minocycline, consistent with Gu et al.’s (9) observations of antagonism with certain antibiotics. Furthermore, our results align with Zuo et al.’s study (10), which showed antagonism between aminoglycosides and phages, particularly with kanamycin, similar to our findings of antagonism with gentamicin against P. aeruginosa. Additionally, our study’s findings of synergy with cefepime against P. aeruginosa correspond with Nicholls et al.’s findings (11) of synergy with cefepime and reduction in biofilm mass. These comparisons highlight the complex and context-dependent nature of phage-antibiotic interactions, influenced by bacterial strain, antibiotic class, and experimental conditions, emphasizing the importance of further research to elucidate these interactions for optimizing combination therapies.
One-step growth curves were used to assess the impact of antibiotics on phage growth kinetics (Fig. 5) (8). We observed that the addition of certain antibiotics to phage cultures resulted in a decrease in phage burst size. This suggests that antibiotics, particularly protein synthesis inhibitors like LZD and MIN, can interfere with phage replication and reduce the production of phage progeny within infected bacterial cells (33, 34).
As clinical trials evaluating phage-antibiotic combinations become more common, they will hopefully provide insight into how these complex interactions manifest in clinical settings. If combination therapies become more widespread, understanding those interactions will become an important part of both treatment selection in medical practice and antimicrobial stewardship to preserve the efficacy of available therapies (6, 26, 35, 36).
Despite the valuable insights gained from this study, there are notable limitations. The in vitro analyses were conducted over short 24-h durations using non-humanized antimicrobial concentrations. Based on our prior research, we believe that these methods are useful screening tools to identify synergistic and antagonistic interactions, but extended evaluations of these combinations and their impact on phage-antibiotic interactions are warranted, including testing additional antibiotic concentrations and phage MOI, administration order and timing of phage and antibiotics, and bacterial inoculum, all of which could influence whether synergy, additivity, or antagonism is observed. Additionally, this study focused on a specific set of antibiotics, and further research should explore the effects of other antibiotics within the same classes. The use of single phages in this study leaves room for investigating how combining phage cocktails with the same antibiotics may impact bacterial killing patterns. Another limitation of our study was that we did not conduct specific phage or antibiotic resistance testing. Due to the high volume of assays conducted and the primary focus on changes in bacterial killing, we opted to prioritize the exploration of synergistic or antagonistic interactions over resistance profiling. While our results provide valuable insights into potential interactions, it is essential to consider that the development of resistance, either to phages or antibiotics, could influence the outcomes of combined treatments. Future research should aim to elucidate the underlying mechanisms of phage-antibiotic interactions, incorporate resistance testing for a comprehensive understanding of the long-term implications, and develop strategies to mitigate their impact.
In conclusion, this study highlights the importance of understanding and addressing phage-antibiotic interactions in the context of phage therapy. The observed antagonistic interactions between some protein synthesis inhibitors and phages emphasize the need for careful consideration when designing phage-antibiotic combinations (PAC). Further research is warranted to elucidate the underlying mechanisms of phage-antibiotic interactions and develop strategies to optimize their impact, ultimately advancing the field of phage therapy as a viable therapeutic option for the future. While the field awaits the results of randomized clinical trials, in vitro investigations of phage-antibiotic combinations are crucial to advancing our understanding of potential interactions. Future studies, both in vitro and in vivo, should explore the combined action of phages and antibiotics, considering various factors that can influence these interactions.
MATERIALS AND METHODS
Bacterial isolates
Well-characterized clinical patient isolates of S. aureus (D712 and 684) (29, 30, 37), E. faecium (R497 and HOU503) (17, 20), and P. aeruginosa (9010 and 10266) (20, 38) were used in this study.
Antimicrobial agents and media
Antibiotics daptomycin, linezolid, minocycline, cefepime, gentamicin, and azithromycin were purchased commercially from Sigma Chemical Company (St. Louis, MO, USA), and CPT analytical powder was provided by AbbVie, Inc. (North Chicago, IL, USA). Mueller-Hinton broth II (Difco, Detroit, MI, USA) with 25 mg/L calcium and 12.5 mg/L magnesium was used for minimum inhibitory concentration testing and modified checkerboard analyses. For TKA, Mueller-Hinton broth II was used for S. aureus and Brain Heart Infusion (BHI) broth (Difco, Detroit, MI, USA) with 50 mg/L calcium and 12.5 mg/L magnesium for E. faecium and P. aeruginosa. Heart Infusion Broth (HIB) was used for all one-step growth curves. S. aureus was plated on trypticase soy agar, while E. faecium and P. aeruginosa were plated on BHI agar. For all experiments with DAP, an additional 25 mg/L of calcium was added to the broth due to the dependency of DAP on calcium for antimicrobial activity (39).
Antibiotic susceptibility testing
MICs of study antimicrobials were determined in duplicate by manual broth microdilution at approximately 106 CFU/mL according to the Clinical and Laboratory Standards Institute (39).
Bacteriophage source, propagation, and sensitivity assay
S. aureus phages Sb-1 and Intesti13 are myophages belonging to the Kayvirus genus that were isolated through plaque purification from bacteriophage solutions purchased from Eliava Institute (Tbilisi, Georgia); they were grown on S. aureus D712 and ATCC 19685, respectively (30). E. faecium phages NV-497 and NV-503-01 were isolated via plaque purification from wastewater in Maryland and grown on E. faecium host strains R497 and HOU503, respectively (20). P. aeruginosa phages EM and 14207 were isolated via plaque purification from wastewater in Orlando, FL, USA, and provided by J. Alexander (AdventHealth Orlando, Wintersprings, FL, USA) or purchased commercially from ATCC (Manassas, VA, USA) and grown on host strains EM-T3762627-2 or ATCC 14207-B, respectively (38, 40).
Phages were propagated to obtain high-titer stocks to use in resistance testing and time-kill analyses, as previously described (41). To begin, an underlay layer of 1.5% HIB agar was poured into square petri plates. A 6 mL overlay of 0.7% HIB agar was immediately combined with 100 µL of an overnight host S. aureus bacterial culture containing approximately 109 CFU/mL and poured atop the underlay layer. The overlay was briefly allowed to set, and following this, 750 µL of purified bacteriophage was spread over the top and incubated in a 37°C incubator overnight. The overlay agar was scraped into 3 mL of phosphate-buffered saline + 10 mM magnesium sulfate and centrifuged at 1,000 rpm for 25 minutes at 4°C. The supernatant was filtered and stored covered at 2°C–8°C for experimental use. All phage stocks have been sequenced to confirm their identities.
Phage activity was tested using a plate-based method (efficiency of plating), as reported previously (41). Briefly, 100 µL of a 16–18 h HIB culture (109 CFU/mL) was transferred to a 14 mL snap-cap tube, 6 mL of molten HIB (melted then held at 50°C) was added to the tube, and then the tube contents were poured evenly over a 100 × 100 mm square HIBA plate. Then, 10-fold serial dilutions of each phage were spotted onto the overlays. Plaques were counted after 20–24 h of incubation at 37°C.
Modified checkerboard assays
The activity of phages and antibiotic in combination, as well as their individual activities, was assessed to determine antibiotic and bacteriophage synergy in bacterial isolates. This was accomplished using a modified checkerboard assay with each bacterial isolate at 106 CFU/mL, as previously described (17, 20). Initially, a 96-well round-bottom microtiter tray was used for serially diluting a single antibiotic at a twofold concentration, similar to MIC testing, starting at 2×MIC for each antibiotic. In a separate 96-well tray, a second antibiotic was serially diluted twofold in a perpendicular direction. The process of diluting antibiotics in checkerboards containing phages followed the previously described approach. However, phage dilutions were performed at a 10-fold ratio instead of twofold to achieve a broader range of MOI. Once dilutions were completed, 50 µL of broth and isolate bacterial stock were added, and the plate was incubated at 37°C for 18–24 h before measuring the absorbance at OD600. The absorbance values were then used to generate a heat map representing the percentage of the bacterial population relative to the growth control standard. The equation used to calculate the FIC index is as follows using OD600 readings: FIC = [(ODdrug A in combination A+B)/ODagentA) + (OD drug B in combination A+B/ODagentB)]. Synergy, additive activity, and antagonism were defined based on the calculated FIC index values: ≤0.5, FIC of 1–4, and >4, respectively.
Time-kill analyses
Time-kill analyses were performed in duplicate in microwell plates to further evaluate phage-antibiotic interactions. In brief, media broth was inoculated with each strain at a targeted bacterial inoculum of 106 CFU/mL. Antibiotics were added at 0.5×, 0.25×, and 0.125× the MIC value to simulate typical and sub-inhibitory concentrations. Phage dosing was added to wells at an MOI of 1 (ratio of phage to target organism). In wells containing both phage and antibiotic, the antibiotic was added first directly followed by phage. Time-kill growth curves were constructed from samples removed at time points 0 (prior to the addition of drug and/or phage), 4, 8, and 24 h. Each 100 µL sample was aliquoted into a sterile Eppendorf containing 0.9 mL of 0.9% saline, as appropriate, and stored at freezing conditions of −40°C for 24 h. To neutralize phage during sample preparation, we used 40 mM sodium citrate buffer. Each sample was then thawed, and antibiotic carryover was accounted for by diluting samples in 0.9% saline, as appropriate. Diluted samples were plated on agar (easySpiral, Interscience for Microbiology, Saint Nom la Breteche, France, a detection limit of 102 CFU/mL), and incubated at 37°C for 24 h followed by counting of bacterial colonies (Scan 1200, Interscience for Microbiology, Saint Nom la Breteche, France). Synergy and bactericidal activity were defined as a ≥2 log10 CFU/mL kill compared to the most effective single agent and a ≥3 log10 CFU/mL reduction from the starting inoculum at 24 h. Antagonism was defined as worse killing with the PAC compared to the best single agent.
Statistical analysis was carried out using SPSS version 21.0 (IBM Corp., Armonk, NY, USA) software. One-way analysis of variance with Tukey’s multiple comparison test was used to compare changes in CFU/mL between regimens, where significance was considered at P < 0.05.
Analysis of phage infection activity via one-step growth curve
One-step growth experiments were conducted in duplicate similar to that previously reported (42, 43). A 10 mL volume of bacterial stock (2 × 109 CFU/mL) was centrifuged for 15 minutes with the resultant supernatant replaced with 1 mL of HIB. Next, 100 µL of phage (2 × 1010 PFU/mL) was added to the bacterial culture and left at 37°C for a duration of time equal to one-tenth of the doubling time of the bacterial strain. Antibiotic at 0.5× MIC was then added to corresponding one-compartment model flasks containing 200 mL HIB. The phage-bacteria mixture was then added to each one-compartment model to achieve starting bacterial and phage counts of 105 and 104 (MOI 0.1), respectively. At the time of transfer (T0), 1 mL was removed from each flask and centrifuged for 4 minutes at 14,000 RPM twice. The supernatant was removed for phage quantification. To the remaining bacterial pellet, 0.9 mL of normal saline was added, samples were plated on agar (easySpiral, Interscience for Microbiology, Saint Nom la Breteche, France, detection limit of 102 CFU/mL), and incubated at 37°C for 24 h followed by counting of bacterial colonies (Scan 1200, Interscience for Microbiology, Saint Nom la Breteche, France).
ACKNOWLEDGMENTS
The authors thank Cesar A. Arias, MD, PhD (Methodist Hospital, Houston, TX, USA), for providing E. faecium strains R497 and HOU503 for the work herein.
This research received no external funding. M.J.R. is supported by NIH grants R21 AI163726. This work was supported by work unit number A1417.
A.J.K.C., M.E., S.V., C.B., B.B., M.W., M.V.D., J.A., S.M.L., and M.J.R. were involved in the conceptualization, methodology, validation, formal analysis, investigation, writing—original draft preparation, and writing—review and editing—of this manuscript. All authors have read and agreed to the published version of the manuscript.
A.J.K.C., M.E., S.V., C.B., B.B., M.W., M.V.D., J.A., S.M.L., and M.J.R. have nothing to declare.
AFTER EPUB
[This article was published on 31 July 2024 with part of Figure 5 missing. Figure 5 was corrected in the current version, posted on 20 September 2024.]
The views expressed in this article reflect the results of research conducted by the authors and do not necessarily reflect the official policy or position of the Department of the Navy, Department of Defense, nor the United States Government.
Contributor Information
Ashlan J. Kunz Coyne, Email: ashlan.kunz.coyne@uky.edu.
Michael J. Rybak, Email: m.rybak@wayne.edu.
Stefano Pagliara, University of Exeter, Exeter, United Kingdom.
REFERENCES
- 1. Mancuso G, Midiri A, Gerace E, Biondo C. 2021. Bacterial antibiotic resistance: the most critical pathogens. Pathogens 10:1310. doi: 10.3390/pathogens10101310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. O’Neil J. 2016. Tackling drug-resistant infections globally: final report and recommendations, p 84. In Ro A, Ro A (ed), Resistance. Vol. 1. London, United kingdom. [Google Scholar]
- 3. Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, Ouellette M, Outterson K, Patel J, Cavaleri M, Cox EM, Houchens CR, Grayson ML, Hansen P, Singh N, Theuretzbacher U, Magrini N, WHO Pathogens Priority List Working Group . 2018. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 18:318–327. doi: 10.1016/S1473-3099(17)30753-3 [DOI] [PubMed] [Google Scholar]
- 4. Centers for Disease Control and Prevention . 2019. Centers for disease control and prevention: antibiotic resistance threats in the United States. Centers for Disease Control and Prevention, Atlanta, GA. Available from: https://www.cdc.gov/drugresistance/pdf/threatsreport/2019-ar-threats-report-508.pdf [Google Scholar]
- 5. Morrisette T, Kebriaei R, Lev KL, Morales S, Rybak MJ. 2020. Bacteriophage therapeutics: a primer for clinicians on phage-antibiotic combinations. Pharmacotherapy 40:153–168. doi: 10.1002/phar.2358 [DOI] [PubMed] [Google Scholar]
- 6. Suh Gina A, Lodise TP, Tamma PD, Knisely JM, Alexander J, Aslam S, Barton KD, Bizzell E, Totten KMC, Campbell JL, Chan BK, Cunningham SA, Goodman KE, Greenwood-Quaintance KE, Harris AD, Hesse S, Maresso A, Nussenblatt V, Pride D, Rybak MJ, Sund Z, van Duin D, Van Tyne D, Patel R, Antibacterial Resistance Leadership Group . 2022. Considerations for the use of phage therapy in clinical practice. Antimicrob Agents Chemother 66:e0207121. doi: 10.1128/AAC.02071-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Suh G.A, Patel R. 2023. Clinical phage microbiology: a narrative summary. Clin Microbiol Infect 29:710–713. doi: 10.1016/j.cmi.2023.02.006 [DOI] [PubMed] [Google Scholar]
- 8. Abedon ST. 2019. Phage-antibiotic combination treatments: antagonistic impacts of antibiotics on the pharmacodynamics of phage therapy? Antibiotics (Basel) 8:182. doi: 10.3390/antibiotics8040182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gu Liu C, Green SI, Min L, Clark JR, Salazar KC, Terwilliger AL, Kaplan HB, Trautner BW, Ramig RF, Maresso AW. 2020. Phage-antibiotic synergy is driven by a unique combination of Antibacterial mechanism of action and Stoichiometry. mBio 11:e01462–20. doi: 10.1128/mBio.01462-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zuo P, Yu P, Alvarez PJJ. 2021. Aminoglycosides antagonize bacteriophage proliferation, attenuating phage suppression of bacterial growth, biofilm formation, and antibiotic resistance. Appl Environ Microbiol 87:e0046821. doi: 10.1128/AEM.00468-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nicholls P, Clark JR, Gu Liu C, Terwilliger A, Maresso AW. 2023. Class-driven synergy and antagonism between a Pseudomonas phage and antibiotics. Infect Immun 91:e0006523. doi: 10.1128/iai.00065-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu C, Hong Q, Chang RYK, Kwok PCL, Chan HK. n.d. Phage-antibiotic therapy as a promising strategy to combat multidrug-resistant infections and to enhance antimicrobial efficiency. Antibiotics 11:570. doi: 10.3390/antibiotics11050570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rahman M, Kim S, Kim SM, Seol SY, Kim J. 2011. Characterization of induced Staphylococcus aureus bacteriophage SAP-26 and its anti-biofilm activity with rifampicin. Biofouling 27:1087–1093. doi: 10.1080/08927014.2011.631169 [DOI] [PubMed] [Google Scholar]
- 14. Valério N, Oliveira C, Jesus V, Branco T, Pereira C, Moreirinha C, Almeida A. 2017. Effects of single and combined use of bacteriophages and antibiotics to inactivate Escherichia coli. Virus Res 240:8–17. doi: 10.1016/j.virusres.2017.07.015 [DOI] [PubMed] [Google Scholar]
- 15. Epand RM, Walker C, Epand RF, Magarvey NA. 2016. Molecular mechanisms of membrane targeting antibiotics. Biochimica et Biophysica Acta (BBA) - Biomembranes 1858:980–987. doi: 10.1016/j.bbamem.2015.10.018 [DOI] [PubMed] [Google Scholar]
- 16. Lev K, Kunz Coyne AJ, Kebriaei R, Morrisette T, Stamper K, Holger DJ, Canfield GS, Duerkop BA, Arias CA, Rybak MJ. 2022. Evaluation of bacteriophage-antibiotic combination therapy for biofilm-embedded MDR Enterococcus faecium. Antibiotics (Basel) 11:392. doi: 10.3390/antibiotics11030392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kunz Coyne AJ, Stamper K, Kebriaei R, Holger DJ, El Ghali A, Morrisette T, Biswas B, Wilson M, Deschenes MV, Canfield GS, Duerkop BA, Arias CA, Rybak MJ. n.d. Phage cocktails with daptomycin and ampicillin eradicates biofilm-embedded multidrug-resistant Enterococcus faecium with preserved phage susceptibility. Antibiotics 11:1175. doi: 10.3390/antibiotics11091175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu S, Zhao Y, Hayes A, Hon K, Zhang G, Bennett C, Hu H, Finnie J, Morales S, Shearwin L, Psaltis AJ, Shearwin K, Wormald PJ, Vreugde S. 2021. Overcoming bacteriophage insensitivity in Staphylococcus aureus using clindamycin and azithromycin at subinhibitory concentrations. Allergy 76:3446–3458. doi: 10.1111/all.14883 [DOI] [PubMed] [Google Scholar]
- 19. Office of the Press Secretary . 2015. Fact sheet: obama administration releases national action plan to combat antibiotic-resistant bacteria. Washington, DC. Available from: https://www.whitehouse.gov/the-press-office/2015/12/22/fact-sheet-obama-administration-releases-national-action-plan-combating [Google Scholar]
- 20. Kunz Coyne AJ, Stamper K, El Ghali A, Kebriaei R, Biswas B, Wilson M, Deschenes MV, Tran TT, Arias CA, Rybak MJ. 2023. Phage-antibiotic cocktail rescues daptomycin and phage susceptibility against daptomycin-nonsusceptible Enterococcus faecium in a simulated endocardial vegetation ex vivo model. Microbiol Spectr 11:e0034023. doi: 10.1128/spectrum.00340-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Akturk E, Oliveira H, Santos SB, Costa S, Kuyumcu S, Melo LDR, Azeredo J. 2019. Synergistic action of phage and antibiotics: parameters to enhance the killing efficacy against mono and dual-species biofilms. Antibiotics (Basel) 8:103. doi: 10.3390/antibiotics8030103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ali L, Khambaty F, Diachenko G. 2006. Investigating the suitability of the calgary biofilm device for assessing the antimicrobial efficacy of new agents. Bioresour Technol 97:1887–1893. doi: 10.1016/j.biortech.2005.08.025 [DOI] [PubMed] [Google Scholar]
- 23. Baran A, Kwiatkowska A, Potocki L. 2023. Antibiotics and bacterial resistance-A short story of an endless arms race. Int J Mol Sci 24:5777. doi: 10.3390/ijms24065777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lin J, Zhou D, Steitz TA, Polikanov YS, Gagnon MG. 2018. Ribosome-targeting antibiotics: modes of action, mechanisms of resistance, and implications for drug design. Annu Rev Biochem 87:451–478. doi: 10.1146/annurev-biochem-062917-011942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Podlacha M, Grabowski Ł, Kosznik-Kawśnicka K, Zdrojewska K, Stasiłojć M, Węgrzyn G, Węgrzyn A. 2021. Interactions of bacteriophages with animal and human organisms-safety issues in the light of phage therapy. Int J Mol Sci 22:8937. doi: 10.3390/ijms22168937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hitchcock NM, Devequi Gomes Nunes D, Shiach J, Valeria Saraiva Hodel K, Dantas Viana Barbosa J, Alencar Pereira Rodrigues L, Coler BS, Botelho Pereira Soares M, Badaró R. 2023. Current clinical landscape and global potential of bacteriophage therapy. Viruses 15:1020. doi: 10.3390/v15041020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Howard-Varona C, Lindback MM, Bastien GE, Solonenko N, Zayed AA, Jang H, Andreopoulos B, Brewer HM, Glavina Del Rio T, Adkins JN, Paul S, Sullivan MB, Duhaime MB. 2020. Phage-specific metabolic reprogramming of virocells. ISME J 14:881–895. doi: 10.1038/s41396-019-0580-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kumaran D, Taha M, Yi Q, Ramirez-Arcos S, Diallo JS, Carli A, Abdelbary H. 2018. Does treatment order matter? Investigating the ability of bacteriophage to augment antibiotic activity against Staphylococcus aureus biofilms. Front Microbiol 9:127. doi: 10.3389/fmicb.2018.00127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kebriaei R, Lev KL, Shah RM, Stamper KC, Holger DJ, Morrisette T, Kunz Coyne AJ, Lehman SM, Rybak MJ. 2022. Eradication of biofilm-mediated methicillin-resistant Staphylococcus aureus infections in vitro: bacteriophage-antibiotic combination. Microbiol Spectr 10:e0041122. doi: 10.1128/spectrum.00411-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kebriaei R, Lehman SM, Shah RM, Stamper KC, Kunz Coyne AJ, Holger D, El Ghali A, Rybak MJ. 2023. Optimization of phage-antibiotic combinations against Staphylococcus aureus biofilms. Microbiol Spectr 11:e0491822. doi: 10.1128/spectrum.04918-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hall AR, De Vos D, Friman V-P, Pirnay J-P, Buckling A. 2012. Effects of sequential and simultaneous applications of bacteriophages on populations of Pseudomonas aeruginosa in vitro and in wax moth larvae. Appl Environ Microbiol 78:5646–5652. doi: 10.1128/AEM.00757-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mukhopadhyay S, Zhang P, To KKW, Liu Y, Bai C, Leung SSY. 2023. Sequential treatment effects on Phage-antibiotic synergistic application against multi-drug-resistant Acinetobacter Baumannii. Int J Antimicrob Agents 62:106951. doi: 10.1016/j.ijantimicag.2023.106951 [DOI] [PubMed] [Google Scholar]
- 33. Swaney SM, Aoki H, Ganoza MC, Shinabarger DL. 1998. The oxazolidinone linezolid inhibits initiation of protein synthesis in bacteria. Antimicrob Agents Chemother 42:3251–3255. doi: 10.1128/AAC.42.12.3251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Asadi A, Abdi M, Kouhsari E, Panahi P, Sholeh M, Sadeghifard N, Amiriani T, Ahmadi A, Maleki A, Gholami M. 2020. Focus on mechanisms of resistance, antibacterial activity, and clinical effectiveness: back to the future. J Glob Antimicrob Resist 22:161–174. doi: 10.1016/j.jgar.2020.01.022 [DOI] [PubMed] [Google Scholar]
- 35. Aslam S, Courtwright AM, Koval C, Lehman SM, Morales S, Furr C-LL, Rosas F, Brownstein MJ, Fackler JR, Sisson BM, Biswas B, Henry M, Luu T, Bivens BN, Hamilton T, Duplessis C, Logan C, Law N, Yung G, Turowski J, Anesi J, Strathdee SA, Schooley RT. 2019. Early clinical experience of bacteriophage therapy in 3 lung transplant recipients. American J Trans 19:2631–2639. doi: 10.1111/ajt.15503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nir-Paz R, Gelman D, Khouri A, Sisson BM, Fackler J, Alkalay-Oren S, Khalifa L, Rimon A, Yerushalmy O, Bader R, Amit S, Coppenhagen-Glazer S, Henry M, Quinones J, Malagon F, Biswas B, Moses AE, Merril G, Schooley RT, Brownstein MJ, Weil YA, Hazan R. 2019. Successful treatment of antibiotic-resistant, poly-microbial bone infection with bacteriophages and antibiotics combination. Clin Infect Dis 69:2015–2018. doi: 10.1093/cid/ciz222 [DOI] [PubMed] [Google Scholar]
- 37. Parra-Ruiz J, Vidaillac C, Rose WE, Rybak MJ. 2010. Activities of high-dose daptomycin, vancomycin, and moxifloxacin alone or in combination with Clarithromycin or rifampin in a novel in vitro model of Staphylococcus aureus biofilm. Antimicrob Agents Chemother 54:4329–4334. doi: 10.1128/AAC.00455-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Holger DJ, Lev KL, Kebriaei R, Morrisette T, Shah R, Alexander J, Lehman SM, Rybak MJ. 2022. Bacteriophage-antibiotic combination therapy for multidrug-resistant Pseudomonas aeruginosa: in vitro synergy testing. J Appl Microbiol 133:1636–1649. doi: 10.1111/jam.15647 [DOI] [PubMed] [Google Scholar]
- 39. CLSI . 2024. M07: dilution AST for aerobically grown bacteria. Available from: https://clsi.org/standards/products/microbiology/documents/m07/
- 40. Holger DJ, El Ghali A, Bhutani N, Lev KL, Stamper K, Kebriaei R, Kunz Coyne AJ, Morrisette T, Shah R, Alexander J, Lehman SM, Rojas LJ, Marshall SH, Bonomo RA, Rybak MJ. 2023. Phage-antibiotic combinations against multidrug-resistant Pseudomonas Aeruginosa in in vitro static and dynamic Biofilm models. Antimicrob Agents Chemother 67:e0057823. doi: 10.1128/aac.00578-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hyman P. 2019. Phages for phage therapy: isolation, characterization, and host range breadth. Pharmaceuticals (Basel) 12:35. doi: 10.3390/ph12010035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hyman P, Abedon ST. 2010. Bacteriophage host range and bacterial resistance. Adv Appl Microbiol 70:217–248. doi: 10.1016/S0065-2164(10)70007-1 [DOI] [PubMed] [Google Scholar]
- 43. Kropinski AM. 2018. Practical advice on the one-step growth curve. Methods Mol Biol 1681:41–47. doi: 10.1007/978-1-4939-7343-9_3 [DOI] [PubMed] [Google Scholar]