ABSTRACT
Neisseria meningitidis serogroups A, B, C, W, X, and Y cause invasive meningococcal disease (IMD) worldwide. Factor H binding protein (FHbp), a key meningococcal virulence factor, is an antigen included in both licensed meningococcal serogroup B (MenB) vaccines. This review examines the biology and epidemiology of FHbp and assesses the ability and potential of FHbp vaccine antigens to protect against IMD. Using evidence from the literature and the contemporary PubMLST database, we discuss analyses of MenB genotypes on the representation of the most prevalent multilocus sequence typing (MLST)/clonal complexes, FHbp subfamily distribution, and FHbp and porin A (PorA) variants. We further discuss that the similar genotypes, distribution, and diversity of FHbp variant types have remained stable over long time periods, supporting the potential for FHbp-containing, protein-based vaccines to protect against IMD, including MenB-FHbp (Trumenba®), which contains two lipidated FHbp antigens (one each from both FHbp subfamilies: A and B).
KEYWORDS: Vaccines, invasive meningococcal disease, factor H binding protein, genotypic diversity, clonal complex, Neisseria meningitidis
Introduction
Neisseria meningitidis is an obligate human bacterium that after droplet-borne transmission can, under certain circumstances, penetrate the mucosal layer of the nasopharynx to cause invasive meningococcal disease (IMD).1–4 The incidence of IMD is low, with recent estimates between 0 and 10.2 per 100,000, depending on country/region.5 However, IMD can cause sequelae that include high morbidity and mortality5; additionally, IMD outbreaks are unpredictable and cause devastating effects.6
Of the 12 known meningococcal serogroups, six (A, B, C, W, X, Y) are primarily associated with IMD.2 The predominant disease-causing meningococcal serogroups have varied by geography and over time, and are influenced by interventions such as the introduction of vaccines.7,8 In various countries/regions, a high proportion of IMD cases are caused by meningococcal serogroup B (MenB); therefore, the development of a broadly protective MenB vaccine has been considered an important public health initiative.9–13
Protection against meningococcal infection is provided by complement-dependent, antibody-mediated bactericidal activity.3 The natural mechanism of protection can be mimicked and quantified in vitro by the serum bactericidal antibody (SBA) assay, which comprises meningococcal isolates, serum antibodies recognizing bacterial surface structures, and a complement source.14 Given the low incidence of IMD, traditional clinical endpoints in vaccine efficacy studies would require unrealistically large enrollment numbers; therefore, licensure of meningococcal vaccines has depended on the demonstration of functional bactericidal immune responses as a correlate of protection from disease, measured via the SBA assay.14
Vaccines based on capsular polysaccharide protein conjugates are used to protect against meningococcal serogroups A, C, W, and Y (MenA, MenC, MenW, and MenY, respectively) disease.1 However, because the MenB capsular polysaccharide consists of polysialic acid units similar to molecules present on human neuronal cells, it has poor functional immunogenicity and is not a suitable vaccine antigen.15
Two independent discovery efforts identified factor H binding protein (FHbp) as a candidate MenB vaccine antigen.16,17 The FHbp antigen is a component of both licensed protein-based MenB vaccines: MenB-FHbp (Trumenba,® bivalent rLP2086; Pfizer Inc, Philadelphia, PA) and MenB-4C (Bexsero,® 4CMenB; GSK Vaccines srl, Siena, Italy).18,19 FHbp is a 27-kDa lipoprotein present on the meningococcal surface that binds human factor H (FH),16,20 thereby functioning as a virulence factor enabling the meningococcus to evade killing by human complement.21 The FHbp gene is found in most meningococcal strains regardless of serogroup; therefore, MenB vaccines containing FHbp antigens have potential to prevent non-MenB IMD.22–25
Critical examination of FHbp as a vaccine antigen may contribute to an enhanced understanding of the most effective use and assessment of MenB vaccines. Thus, the objectives of this review are to (1) assess the biochemical, genetic, and functional characteristics of FHbp; (2) discuss considerations for the development and bactericidal activity assessment of MenB vaccines containing FHbp antigens; (3) evaluate the epidemiology of FHbp in MenB and non-MenB strains as reported in the scientific literature and at the Neisseria Multi Locus Sequence Typing website (PubMLST; http://pubmlst.org/neisseria/); and (4) assess the potential of FHbp as a vaccine antigen to protect against non-MenB IMD.
Biochemical, genetic, and functional characteristics of FHbp
The synthesis of FHbp and its transport from the cytosol to cell surface have been elegantly described by da Silva and colleagues.26 FHbp is synthesized as a preprolipoprotein with a cleavable N-terminal signal peptide for transportation across the inner membrane by the Sec translocon. The signal peptide carries a conserved lipobox at its C terminus, and the thiol group of the invariant cysteine residue therein is triacylated in the inner membrane. First, a preprolipoprotein diacylglyceryl transferase, Lgt, diacylates the lipobox, forming a prolipoprotein. The lipoprotein signal peptidase, Lsp, then cleaves the signal peptide from the prolipoprotein, rendering the diacylated cysteine as the N-terminal residue. Subsequently, the apolipoprotein N-acyl transferase, Lnt, adds an amide-linked fatty acid to this residue, generating a triacylated protein. This mature lipoprotein is transported by the lipoprotein outer membrane localization apparatus, Lol, to the outer membrane.
da Silva and colleagues demonstrated that apolipoprotein N-acyl transferase Lnt activity was not necessary for surface expression because both diacylated and triacylated FHbp could be transported to the outer membrane.26 However, the efficiency of transport and level of FHbp surface expression were substantially reduced in the absence of Lnt activity. It was also found that levels of transcripts coding for FHbp in Lnt-deficient bacteria were significantly reduced. The authors hypothesized that diacylated FHbp accumulation in the periplasm may lead to feedback inhibition of FHbp expression via down-regulation of FHbp transcription and potentially also because of FHbp protein translation down-regulation or increased proteolysis.26
Subsequently, da Silva and colleagues found that the majority of a UK MenB isolate collection displays nonsynonymous single nucleotide polymorphisms (SNPs) in the FHbp signal peptide.27 One of the SNPs alters a polar amino acid affecting lipidation and signal peptide cleavage. MenB isolates with signal peptide sequences including this SNP displayed partial FHbp retention in the cytoplasm because of reduced binding to SecA. However, some FHbp translocated and localized to the outer membrane despite the processing defect.
Lipidation is critical for FHbp surface expression, which is required for the role of FHbp as a virulence factor and target for antibody-dependent, complement-mediated bactericidal activity. As evidence, preclinical studies clearly showed lipidated FHbp elicits higher and broader bactericidal activity than nonlipidated forms.28 In mice vaccinated with lipidated FHbp variants, SBA using human complement (hSBA) titers against heterologous MenB test strains were consistently ten-fold higher than those in mice vaccinated with the corresponding nonlipidated FHbp.16 Similarly, protective hSBA responses in rhesus macaques were considerably more robust after vaccination with a lipidated subfamily A FHbp variant compared with the analogous nonlipidated FHbp antigen (100% vs 50% responders [response based on ≥4-fold increase in hSBA titer compared with pre-vaccination titer] against a test strain expressing homologous FHbp, and 60% vs 0% responders against a test strain expressing heterologous subfamily A FHbp).28 On the basis of these preclinical data, the MenB-FHbp vaccine was developed using lipidated forms of FHbp as described below.
The key function of FHbp is binding of FH. Although sequence diversity across FHbp variant types is considerable, critical FH binding regions are conserved across both FHbp subfamilies (A and B).29,30 To prevent unintended host cell damage, FH down-regulates the alternative complement pathway by acting as a cofactor for factor I–mediated cleavage of C3b, thereby increasing the production of inactive C3b and the C3bBb decay rate.21,31 By binding FH at the meningococcal cell surface, FHbp reduces complement-mediated killing of the bacteria.21,32 Experiments illustrating the role of FHbp as a meningococcal virulence factor included the observation that laboratory-generated MenB FHbp deletion mutant strains were highly susceptible to killing by human sera and whole blood, whereas isogenic wild-type parent strains were considerably less susceptible.21,33
Given that FHbp specifically binds FH, this may dampen immune responses to FHbp antigens in humans.20 Because murine factor H does not bind to FHbp, preclinical studies in mice and rabbits may not be good surrogates to monitor clinical immune responses to FHbp antigens34; this suggestion is consistent with the observation that immune responses to FHbp in transgenic mice expressing high levels of FH were not as robust as the immune response in wild-type mice.35 Moreover, mutant FHbp antigens that do not bind FH elicit an enhanced immune response compared with wild-type FHbp in FH transgenic mice.34,35 Accordingly, modified FHbp that minimizes binding of FH has been proposed as a next-generation vaccine antigen.
Development and bactericidal activity assessment of MenB vaccines containing FHbp antigens
Discovery
Pfizer used a combined biochemical and immunologic screening approach to discover surface-expressed proteins with broad MenB distribution and sufficient amino acid sequence conservation to induce hSBA responses against endemic, epidemic, and outbreak strains.28 The outer membrane lipoprotein LP2086, subsequently identified as FHbp, was recognized as an antigen able to elicit functional hSBA responses in assays targeting MenB clinical isolates expressing FHbp variants heterologous to the vaccine antigens.16,28
FHbp possesses several characteristics contributing to its suitability as a broadly effective MenB vaccine antigen. As an outer membrane lipoprotein expressed at the bacterial surface, FHbp is accessible to the host immune system.30 Mature FHbp variants range from 253 to 266 amino acids in length, with a characteristic tri-Pam-Cys–modified N-terminus serving to anchor the protein to the outer membrane. A full-length gene capable of coding for FHbp is present in >99% of invasive MenB isolates36–38 and in the majority of carriage isolates across meningococcal serogroups.39–41 N meningitidis is an organism noted for genomic plasticity;42 therefore, widespread retention of the gene suggests FHbp plays a critical role in the physiology of this pathogen.36
It should be noted that because FHbp was independently discovered by two institutions, different nomenclature for FHbp classification has been adopted: the Pfizer nomenclature (used throughout this article) and the Novartis nomenclature, with no consensus on which should be regarded as standard.30 FHbp can be classified into two subfamilies or three variants using sequence-based clustering; subfamilies can be further divided into groups based on the N- and C-terminal domains of the protein.30 Individual protein subvariants are assigned a numerical identifier based on sequence homology, alongside a prefix indicating the unique variant designation (eg, A01 or 1.1).43 Each subfamily/variant member is more closely related to other FHbps in that subfamily/variant by sequence than to members of other subfamily/variant groups.16 For example, analysis of 1837 invasive MenB strains from the United States, Europe, New Zealand, and South Africa revealed that within each of the two subfamilies, FHbp variants share 83% to 99% pairwise amino acid sequence identity but only 60% to 75% identity between subfamilies.37
Consistent with the amino acid sequence of FHbp variants, functional immune responses to lipidated FHbp antigens are largely subfamily-restricted.16,44 Immunization with subfamily A FHbp antigens elicits high functional immune responses to MenB strains expressing subfamily A variants but reduced or very low responses to strains expressing subfamily B variants. Similarly, robust serum bactericidal activity elicited in response to subfamily B FHbp antigens is largely restricted to MenB strains expressing subfamily B FHbp variants. MenB-FHbp, the bivalent vaccine comprising an FHbp variant from subfamily A and a second from subfamily B, elicits broad-spectrum functional immune responses against strains expressing either subfamily A or B variants.18,44
The recognition of FHbp as an important virulence factor and its attributes as a vaccine antigen contributed to the development of two recombinant MenB vaccines, which are either directed to FHbp as the sole target or containing FHbp as one of the antigenic components.30 MenB-FHbp includes two variants of FHbp, one from subfamily A (variant A05) and the second from subfamily B (variant B01).18 As in the native protein, both vaccine antigens are expressed as recombinant proteins with an amino terminal lipid tail.18,26 A vaccine containing an FHbp from each of the two FHbp subfamilies is predicted to provide broad coverage against diverse MenB disease strains.28 Specifically, MenB-FHbp, which comprises an FHbp variant from subfamily A and a second from subfamily B, elicits broad-spectrum functional immune responses against strains expressing either subfamily A or B variants.18,44 In contrast, MenB-4C contains three recombinant-expressed MenB antigens: neisserial heparin-binding antigen (NHBA) fused to genome-derived neisserial antigen 1030 (GNA1030), nonlipidated FHbp from subfamily B (variant B24) fused to GNA2091, and neisserial adhesin A (NadA).19,30 In addition to the recombinant proteins, outer membrane vesicles prepared from the NZ98/254 MenB outbreak strain are included as the fourth component of the vaccine.19
SBA assays as surrogates of vaccine efficacy
The SBA assay is established as an in vitro surrogate of vaccine efficacy and routinely used to support licensure of meningococcal vaccines, and it was used for clinical evaluation and approval of MenB-FHbp and MenB-4C.45 The SBA assay, performed using human or rabbit complement (hSBA or rSBA, respectively), assesses sera antibody levels capable of killing the target meningococcal strain in a complement-dependent fashion.14 Based on the pioneering work of Goldschneider and colleagues,46 the presence of serum bactericidal activity measured by the SBA assay is recognized to correlate with protection from IMD occurring in vivo, provided that meningococcal test strains are representative of circulating IMD isolates and that human complement sources allow for measurement of antibody-mediated, as opposed to intrinsic, bactericidal activity against a diverse set of strains.14
Because the bacterial capsular polysaccharide antigen is conserved across MenA, MenC, MenW, and MenY strains, only one test strain expressing an invariant polysaccharide capsule is required to assess coverage for each of these serogroups.45 Testing of MenB vaccines composed of protein antigens that can exhibit sequence diversity across disease-causing isolates provides a unique challenge, whereby hSBA assays using multiple test strains that represent the antigenic diversity of disease-causing isolates must be used.45 Broad coverage of protein-based MenB vaccines against a diverse set of MenB test strains has been demonstrated in clinical trials and various analyses.47–49
Determinants of the bactericidal activity of anti-FHbp antibodies
The interaction of FHbp on the bacterial surface with serum antibody is fundamental to the bactericidal activity measured in hSBA assays as a correlate of protective immunity.32,33 Antibodies elicited in response to an FHbp vaccine antigen potentially provide two functional outcomes. Anti-FHbp antibodies can block FH-mediated down-regulation of the alternative complement pathway by interfering with the binding of FH to FHbp at the meningococcal cell surface.14,21,33 Binding of anti-FHbp antibodies at the bacterial surface could also serve as an initial step of the classical complement pathway, leading to phagocytosis and bactericidal activity. As discussed earlier, FHbp is capable of eliciting a bactericidal immune response against diverse IMD isolates.28 Early studies showed that FHbp subfamily A or B vaccine antigens evoke subfamily-restricted immune responses that are cross-protective against MenB strains expressing heterologous FHbp variants from the same subfamily;16,17 it was concluded that a bivalent FHbp vaccine representing both subfamilies may provide broad coverage against most MenB strains.16
In a set of 100 invasive MenB isolates selected to represent FHbp, porin A (PorA), and multilocus sequence typing (MLST) diversity, rabbit sera and human sera raised against a bivalent FHbp vaccine representing both subfamilies killed 87% (87/100) and 80% (36/45) of strains, respectively.44 The FHbp sequence variant did not predict killing in the hSBA assay, with strains representing a range of different sequence types being killed; rather, the best predictor of bactericidal activity was the level of FHbp surface expression.44 To this end, the absolute quantification of FHbp on diverse MenB strains showed 15-fold expression level variation among strains and indicated that a minimum of 757 molecules separated by ≤130 nm is necessary to engage C1q, the first component of the classical complement pathway (note, this quantification was not specific for surface-localized FHbp).50 To determine the relative in vitro FHbp surface expression levels on MenB strains, a flow cytometry assay (MEASURE) was developed using intact target bacteria prepared under the same culture conditions used to measure functional immune responses in the hSBA assay.51 Binding of a murine monoclonal antibody (MN86-994-11) that recognizes an epitope common to nearly all FHbp variants is followed by secondary detection reagents. The mean fluorescence intensity (MFI) in this assay represents an indirect measure of FHbp expression levels on the surface of the target bacteria. Importantly, an evaluation of 1814 MenB IMD isolates in the MEASURE assay demonstrated that >91% of these isolates exhibited sufficient FHbp surface expression to be susceptible to bactericidal killing by MenB-FHbp vaccine-induced antibodies.51
da Silva and colleagues postulated that allelic variation in the FHbp signal peptide may impact lipidation levels, efficiency of signal peptide cleavage, and FHbp surface expression among naturally occurring isolates, potentially resulting in differential bactericidal activity.27 This suggests that appropriate FHbp lipidation impacts surface expression levels and therefore bactericidal activity levels in functional antibody assays (ie, hSBA). In their study, a reduction in FHbp surface expression in the absence of Lnt substantially affected N meningitidis binding to the anti-FHbp antibody, JAR4.26 The authors speculated an impact on the breadth of protective efficacy of FHbp-based vaccines against N meningitidis, specifically against a subset of isolates that may contain SNPs that would reduce surface FHbp expression and therefore bactericidal activity.27 However, each of the signal peptide variants was represented in an unbiased and comprehensively selected N meningitidis hSBA strain panel used in clinical trials to support MenB-FHbp licensure.52 Immune sera from MenB-FHbp−vaccinated participants displayed high levels of bactericidal activity (hSBA geometric mean titers for four primary test strains of 23.7–218.4 and for ten additional test strains of 21.4–93.6) against isolates containing each of the SNPs. Notwithstanding any differential surface expression and relative susceptibility to bactericidal activity between isolates, the effectiveness of MenB-FHbp against the strain collection containing each of the SNPs in question should allay concern stemming from the experimental observations of da Silva and colleagues.52
Epidemiology of FHbp: FHbp subfamily distribution among MenB serogroups
Studies before 2007
For MenB disease, the molecular epidemiology of FHbp has been the focus of numerous studies. The main import from the results of these studies is that FHbp subfamily distribution can vary geographically. In a study of 86 MenB strains collected during the 1960s to 1990s from Europe, the United States, and Chile, 30% of isolates coded for FHbp subfamily A variants and 70% for subfamily B variants.16 A very similar subfamily distribution was observed in a set of 1263 MenB strains systematically collected in the United States and Europe between 2000 and 2006, with 29% of isolates coding for subfamily A and 71% subfamily B FHbp variants.37 A modified and extended version of this prevalence-based (ie, reflective of IMD strain prevalence in a particular geographic location over a given time period) MenB strain collection that included 1841 MenB strains from the United States and Europe had 31% subfamily A variant strains overall. However, the percentage of subfamily A variant strains within this collection fluctuated by country, ranging from 21% in Germany to 43% in the United States.36 For comparison, 59% of MenB isolates in a smaller South African collection in 2005 coded for subfamily A variants.53 Similarly, 62% of MenB strains collected between 2009 and 2013 in western Canada coded for subfamily A variants.54
A smaller number of studies have reported on FHbp epidemiology in MenB carriage isolates. Unlike the majority of invasive MenB isolates, genes coding for FHbp subfamily A variants were most common among carriage isolates. For example, a longitudinal carriage study of adolescents and young adults between 14 and 22 years of age in Genoa, Italy, in 2011 reported that 80% of MenB isolates coded for FHbp subfamily A variants.39 Genes coding for subfamily A variants were also detected in the majority of carriage isolates (58.6%) collected from adolescents in 2012 in a second study from Italy.55 Similarly, in a longitudinal study conducted among adolescents and young adults (10–25 years old) from secondary schools and universities in the United Kingdom during 2011, 89% of MenB carriage isolates were genotyped to FHbp subfamily A.56 Finally, a study of carriage isolates from adolescents in the United States collected in 1998 and between 2006 and 2007 reported that 91% of MenB carriage isolates code for FHbp subfamily A variants.40
The coverage of vastly diverse FHbp variants has been examined previously. Individual sera from adolescents and young adults vaccinated with MenB-FHbp in clinical trials displayed substantial bactericidal activity against each of 27 MenB test strains (hSBA titers ≥ 1:8 were achieved by 60.0%–100% and 31.8%–84.6% of samples expressing FHbp subfamily A and B, respectively, after two doses and 61.7%–100% and 55.6%–100% after three doses).48 The 27 strains in this study included prevalent strains from 2000–2006 in the United States and Europe, as well as isolates from later outbreaks until 2014. Based on MLST clonal complex and the FHbp variant expressed, these strains were estimated to represent approximately 80% of invasive MenB isolates circulating in Europe and the United States. Of the 27 MenB strains tested, 25 expressed FHbp variants heterologous to those in the MenB-FHbp vaccine. Another set of 148 MenB isolates from Greece was evaluated using the Meningococcal Antigen Typing System (MATS) in 2014 for predicting coverage by the MenB-4C vaccine.57 Coverage of the highest number of isolates in this set was predicted by NHBA expression (78.4%), followed by FHbp (52.7%), PorA (8.1%), and NadA (0.7%). Among a collection of Spanish IMD isolates collected from 2015–2018 (108, 103, 87, and 112 isolates each year, respectively), it was predicted that 64.2% of strains would be covered by the MenB-FHbp vaccine based on hSBA data from clinical trials.58 In contrast, only 15.9% of strains would be predicted to be covered by MenB-4C via FHbp based on MATS analysis. This strain set was composed of 65.9% FHbp subfamily A variants and 32.4% FHbp subfamily B variants, which likely explains the differential coverage levels provided by the two vaccines; the bivalent MenB-FHbp vaccine contains two FHbp peptides, one from each subfamily, whereas the MenB-4C vaccine contains only one FHbp subfamily B peptide.18,19
Comparison of the Pfizer MenB collection (2000–2006) with the PubMLST dataset (2007–2021)
These earlier literature reports represented limited geographic areas and/or short time periods. Because FHbp strain diversity varies geographically and temporally, continuous molecular surveillance of strains is critical to evaluate ongoing vaccine coverage. Therefore, we conducted a comprehensive analysis of invasive MenB strains deposited at PubMLST.59,60 PubMLST entries were accessed on March 25, 2022, at which time the PubMLST database contained >81,000 meningococcal entries and included isolates originating on all five inhabited continents. Approximately one-third of isolates were collected during the past decade. Search criteria for disease-causing meningococcal isolates were (1) year of isolation is 2021 or earlier, (2) FHbp peptide designation is “confirmed” with a unique ID, (3) serogroup is defined as A, B, C, W, X, or Y, and (4) disease status is any of the following: invasive, meningitis, septicaemia, meningitis and/or septicaemia. Isolates from sexually transmitted infections and conjunctivitis were excluded. A total of 13,799 entries passed the selection criteria and were selected for further analysis.
Applying similar filters as above, we identified 6203 invasive MenB isolates in the PubMLST database (Table 1). These included 2104 (33.9%) subfamily A and 4088 (65.9%) subfamily B strains. The five most highly represented variants in MenB isolates were B16, B24, B09, A22, and B03 (Figure 1). These results indicate diversity of FHbp variants among IMD-causing MenB isolates in the human population. We further probed whether the degree, composition, and extent of diversity changed over time. MenB-FHbp was originally licensed based on correlates of efficacy (ie, bactericidal activity in SBA assays against a diverse set of strains).61,62 To predict the continued effectiveness of MenB-FHbp in protecting against a contemporary set of IMD-causing MenB strains, we compared the diversity described in our collection (ie, the Pfizer MenB Collection [2000–2006]) with a contemporary dataset from PubMLST (2007–2021). The MenB strain pool in the Pfizer MenB Collection comprised a prevalence-based dataset from select European countries and the United States collected between 2000–2006 and contained 1814 strains (with the caveat that only isolates that could be cultured from patients with IMD were represented); in comparison, the PubMLST dataset was assembled from 2007–2021 globally, was not prevalence based, and contained 5659 strains (Table 2). The contribution of different countries to each of these datasets indicates the PubMLST dataset is proportionately representative of the countries in the Pfizer MenB Collection and includes additional important data from other countries on different continents. As such, the PubMLST dataset, although not prevalence based, appears to provide a robust platform for analysis of global genotypic diversity in FHbp among IMD-causing MenB strains.
Table 1.
FHbp subfamily distribution among invasive disease isolates for each meningococcal serogroup obtained from the PubMLST dataset (2007–2021).
| Serogroup | Number (% Total) |
Number (% FHbp Subfamily A) |
Number (% FHbp Subfamily B) |
Number (% Subfamily A/B Hybrid) |
Number (% Null) |
Year of Isolation |
|---|---|---|---|---|---|---|
| MenA | 402 (2.9) | 7 (1.7) | 395 (98.3) | 0 (0) | 0 (0) | 1915–2021 |
| MenB | 6203 (45.0) | 2104 (33.9) | 4088 (65.9) | 7 (0.1) | 4 (0.03)a | 1940–2021 |
| MenC | 2125 (15.4) | 1220 (57.4) | 891 (41.9) | 4 (0.2) | 10 (0.5)a | 1965–2021 |
| MenW | 3291 (23.8) | 2391 (72.7) | 899 (27.3) | 0 (0) | 1 (0.03) | 1976–2021 |
| MenX | 105 (0.8) | 10 (9.5) | 95 (90.5) | 0 (0) | 0 (0) | 1986–2019 |
| MenY | 1673 (12.1) | 1595 (95.3) | 78 (4.7) | 0 (0) | 0 (0) | 1986–2021 |
Note: The PubMLST analysis was conducted on March 25, 2022, and retrieved 13,799 entries corresponding to unique identifications of FHbp for serogroups A, B, C, W, X, and Y. Hybrid strains have both FHbp subfamily A and subfamily B regions. Null strains lack FHbp expression.
aIncludes isolates (two MenB and one MenC) that are FHbp Pfizer “not defined” but which have an assigned FHbp peptide.
FHbp = factor H binding protein; MenA = meningococcal serogroup A; MenB = meningococcal serogroup B; MenC = meningococcal serogroup C; MenW = meningococcal serogroup W; MenX = meningococcal serogroup X; MenY = meningococcal serogroup Y.
Figure 1.
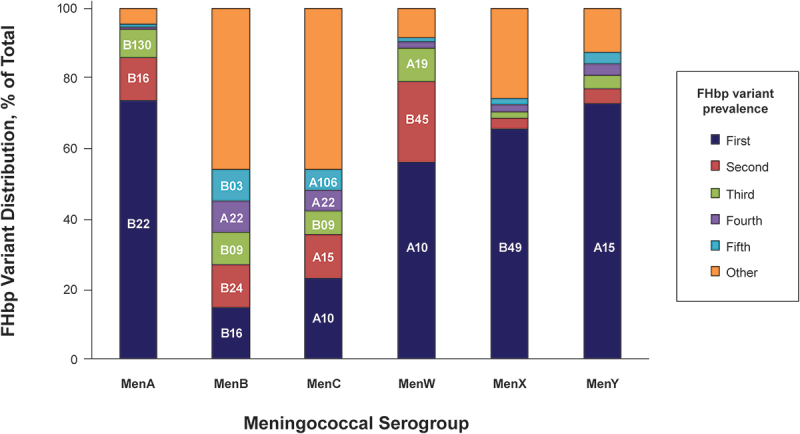
Most prevalent FHbp variants for each meningococcal serogroup.
The PubMLST analysis was conducted on March 25, 2022. FHbp variants were arranged from most to least represented in each serogroup. The five most common represented variants are shown (these are labeled where space allows). The remaining variants were pooled and shown under “other” category. Nomenclatures for each variant are shown in Supplementary Table S1. FHbp = factor H binding protein; MenA = meningococcal serogroup A; MenB = meningococcal serogroup B; MenC = meningococcal serogroup C; MenW = meningococcal serogroup W; MenX = meningococcal serogroup X; MenY = meningococcal serogroup Y.
Table 2.
Characteristics of the Pfizer MenB Collection and the contemporary PubMLST dataset.
| Pfizer MenB Collection (Extended MenB Strain Pool, N = 1814) |
Contemporary PubMLST Dataset (N = 5659) |
|||
|---|---|---|---|---|
| Number of Strains (% Total) |
Number of Strains (% Total) |
|||
| Country of Origin | (2000–2006) | Disease Years | (2007–2021) | Disease Years |
| United Kingdom France Norway Czech Republic United States Germany Spain Ireland Netherlands Poland South Africa |
536 (30) | 2001–2006 | 2731 (48) | 2007–2021 |
| 244 (13) | 2001–2006 | 603 (11) | 2007–2021 | |
| 23 (1) | 2001–2006 | 10 (0.2) | 2008–2012 | |
| 28 (2) | 2001–2006 | 66 (1) | 2007–2019 | |
| 432 (24) | 2000–2005 | 603 (11) | 2007–2021 | |
| 205 (11) | 2001–2006 | 357 (6) | 2008–2021 | |
| 346 (19) | 2001–2006 | 52 (1) | 2013–2018 | |
| NA | NA | 253 (4) | 2008–2019 | |
| NA | NA | 223 (4) | 2008–2019 | |
| NA | NA | 119 (2) | 2011–2017 | |
| NA |
NA |
109 (2) |
2016–2021 |
|
| Years of isolation | 2000–2006 | 2007–2021 | ||
| Prevalence based?a | Yes | No | ||
aThe Pfizer MenB Collection was systematically compiled to reflect the prevalence of IMD strains in circulation in different geographic regions (for details, see Zlotnick, et al.28); in contrast, the PubMLST dataset is global, constantly updated, and dependent on submission of new isolates for assessment. IMD = invasive meningococcal disease; MenB = meningococcal serogroup B; NA = not available.
The subfamily composition of the PubMLST dataset (66% subfamily B; 34% subfamily A) was comparable with that of the Pfizer MenB Collection strain pool (70% subfamily B; 30% subfamily A). Moreover, the ten most common represented FHbp variants were the same in both collections and had comparable cumulative representations (76.6% in the Pfizer MenB Collection strain pool; 72.5% in the PubMLST dataset; Figure 2). We further evaluated the diversity of clonal complexes and the FHbp diversity within each clonal complex. Figure 3 presents the seven most prevalent clonal complexes identified in the Pfizer MenB Collection and the corresponding clonal complexes from the PubMLST dataset. The seven most common clonal complexes in the Pfizer MenB Collection were all represented in the PubMLST dataset, and the level of FHbp diversity within these clonal complexes was similar in each strain pool (ie, in the Pfizer MenB Collection and in the PubMLST dataset, the same FHbp strains were highly represented within each of the seven most common clonal complexes).
Figure 2.
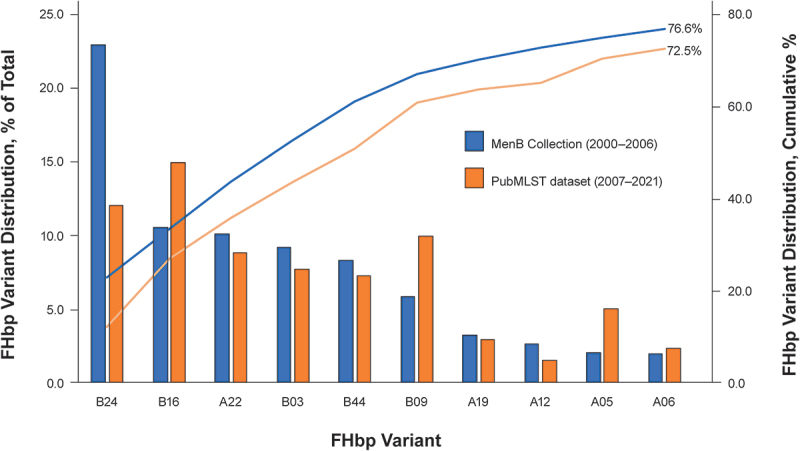
Comparative distribution of FHbp variants in the respective MenB strain pools.
The ten most prevalent FHbp variants identified in the Pfizer MenB Collection (2000–2006) are illustrated. The prevalence of these ten FHbp variants within the Pfizer MenB Collection is compared with prevalence of the same isolates among all isolates from the contemporary PubMLST dataset (2007–2021). Cumulative percentages of these ten FHbp variants within the two strain pools were similar: they represented 76.6% of all isolates in the Pfizer MenB Collection from 2000–2006 and 72.5% of all isolates in the contemporary PubMLST collection from 2007–2021. Similarly, the overall FHbp subfamily distribution was largely unchanged: 30% from subfamily A and 70% from subfamily B among Pfizer MenB Collection strains; 34% from subfamily A and 66% from subfamily B among contemporary PubMLST strains. Nomenclatures for each variant are shown in Supplementary Table S1. FHbp = factor H binding protein; MenB = meningococcal serogroup B.
Figure 3.
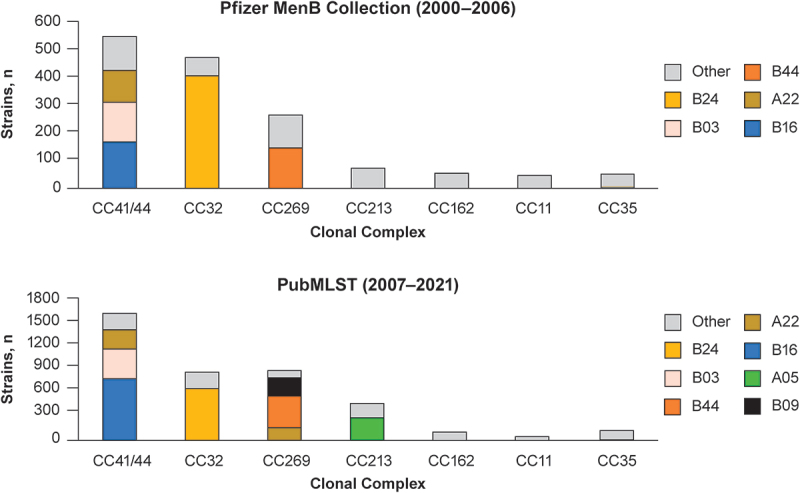
Comparative clonal complex distribution in the respective MenB strain pools.
The seven most prevalent CCs identified in the Pfizer MenB Collection (2000–2006) are shown in the upper panel. The prevalence of these CCs among all MenB disease isolates from the contemporary PubMLST dataset (2007–2021) are shown in the lower panel. Highly represented (n > 100) FHbp variants within each CC are shown with a non-gray color; less frequently expressed variants are categorized as “other” and shown in gray. Nomenclatures for each variant are shown in Supplementary Table S1. CC = clonal complex; FHbp = factor H binding protein; MenB = meningococcal serogroup B.
We also compared FHbp diversity with another dominant meningococcal surface antigen: PorA. As shown in Figure 4, the six most common PorA variants were represented in both the Pfizer MenB Collection and the PubMLST dataset, and there was comparable FHbp diversity within each PorA variant (ie, in the Pfizer MenB Collection and in the PubMLST dataset, the same co-expressed FHbp strains were highly represented within the six most common PorA variants). Collectively, these results suggest that the PubMLST database is a robust platform for analysis of FHbp strain diversity among IMD isolates from the population.
Figure 4.
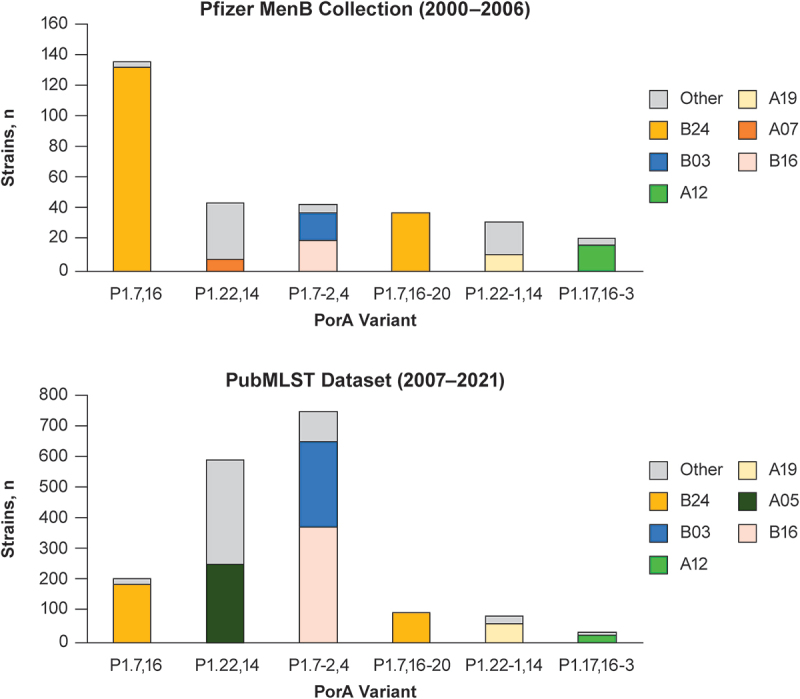
Comparative PorA variant type distribution and co-expression of highly represented FHbp variants in the respective MenB strain pools.
The six most prevalent PorA variants identified in the Pfizer MenB Collection (2000–2006) are shown in the upper panel. The prevalence of these PorA variants among all MenB disease isolates from the contemporary PubMLST collection (2007–2021) is shown in the lower panel. Highly represented (n > 100) or dominant FHbp variants co-expressed with each PorA variant are shown with a non-gray color; FHbp variants less frequently co-expressed with each PorA variant are categorized as “other” and shown in gray. Nomenclatures for each variant are shown in Supplementary Table S1. FHbp = factor H binding protein; MenB = meningococcal serogroup B; PorA = porin A.
Epidemiology of FHbp: FHbp subfamily distribution among non-MenB isolates
The epidemiology of FHbp in invasive non-MenB isolates has also been reported in the literature, albeit with smaller datasets compared with MenB.23,24,53,63–66 Therefore, we also leveraged the PubMLST database to evaluate the FHbp variant diversity within non-MenB serogroups.
Meningococcal serogroup A
Three studies in the literature reported FHbp subfamily distribution among MenA disease-causing strains. Mothibeli and colleagues determined the prevalence and sequence diversity of FHbp in 20 invasive MenA strains in South Africa during 2005.53 The FHbp variants coded for by these 20 MenA strains were all from subfamily B. Similarly, Pajon and colleagues collected and analyzed 31 MenA strains that were part of a collection obtained over a 45-year period from 17 African (mostly sub-Saharan) countries.63 All of the MenA strains in this study were found to code for FHbp subfamily B variants. One study assessed the prevalence and diversity of IMD isolates collected in the Netherlands between 1960 and 2009.64 Of the 11 IMD MenA strains collected, the majority (9/11) coded for FHbp subfamily B variants. The current PubMLST survey identified 402 invasive MenA strains, which accounted for 2.9% of all IMD isolates in the collection (Table 1). Consistent with literature reports, the vast majority of these (98.3% [395/402]) coded for subfamily B variants. The two most prevalent FHbp variants, B22 (74%) and B16 (12%), represent approximately 86% of the total (Figure 1).
Meningococcal serogroup C
In a study by Harris and colleagues, FHbp was detected in each of the 116 invasive MenC isolates from the US Active Bacterial Core Surveillance sites from 2000 to 2001.23 Nine of these, all genotypically typed as ST11 isolates, encoded a truncated protein due to a single nucleotide 356 deletion causing a frameshift and early termination. Of the 116 isolates, 57 (49%) coded for subfamily A variants (including 11 unique FHbp variants), and 59 (51%) were characterized as subfamily B isolates (32 unique variants). Of the 43 different FHbp variants detected, 23 had not been found among the MenB FHbp variants already described; none of these new variants differed from previously known variants by >7 amino acids. Examination of FHbp surface expression by flow cytometry, using a monoclonal antibody that previously detected surface FHbp for 98% of 1263 MenB isolates, detected the FHbp protein on 69% of MenC isolates. SBA assays were developed for six MenC isolates positive for FHbp surface expression (two subfamily A; four subfamily B); all six isolates were susceptible to killing by the sera of rabbits and macaques previously vaccinated with MenB-FHbp. In another publication, Tsang and colleagues examined the FHbp variant type in 50 MenC isolates from culture-confirmed IMD cases in Canada from 2009 to 2013.65 In this collection, five of the 50 isolates coded for a truncated version of FHbp (attributed to a frame-shift mutation), 34 coded for subfamily A variants, and 11 coded for subfamily B variants.
In the PubMLST analysis, the distribution of FHbp subfamilies was similar to literature reports. Specifically, 2125 FHbp sequences (15.4% of total) were found among invasive MenC strains; of these, 1220 (57.4%) were subfamily A and 891 (41.9%) were subfamily B (Table 1). There was notable diversity in FHbp variants among MenC isolates, with A10 (23%) and A15 (12%) representing the two most common variants, followed by B09, A22, and A106 (Figure 1).
Meningococcal serogroup W
One study in the literature analyzed FHbp variant type in MenW strains. In the previously described study by Pajon and colleagues assessing MenW strains from African countries, 34% (18/53) of MenW strains in the collection were found to code for FHbp subfamily B and 66% (35/53) for subfamily A.63 In the current PubMLST analysis, 3291 invasive MenW strains were identified, including 2391 (72.7%) subfamily A variants and 899 (27.3%) subfamily B variants (Table 1). The most common variants in this MenW collection were A10 (56%) and B45 (23%; Figure 1).
Meningococcal serogroup X
The study by Pajon and colleagues included 22 meningococcal serogroup X (MenX) strains from Africa.63 All but one of the MenX strains (95.5%) in this collection coded for FHbp subfamily B variants. In a study assessing potential coverage of MenX by the MenB-4C vaccine, nine MenX isolates from African countries (Chad, Niger, Burkina Faso) and two from France were characterized; all of the African isolates coded for subfamily B variants, whereas both isolates from France contained subfamily A sequences.24 Invasive MenX strains were represented to a limited extent in the PubMLST collection compared with other serogroups, with only 105 invasive MenX strains having an FHbp assignment. Ten (9.5%) MenX strains coded for FHbp subfamily A variants and 95 (90.5%) for subfamily B variants (Table 1); the most common variant was B49 (66%; Figure 1).
Meningococcal serogroup Y
Only one study reported on FHbp variant type in MenY strains. Law and colleagues collected 16 invasive MenY strains from Quebec, Canada, from 2009 to 2013.66 All but one MenY strain (93.8%) in this collection coded for a subfamily A variant. Consistent with this publication, PubMLST analysis identified 1673 invasive MenY strains with an FHbp variant assignment, of which 1595 (95.3%) coded for FHbp subfamily A variants and 78 (4.7%) for subfamily B variants (Table 1). The most common variant was A15, accounting for 73% of these MenY strains (Figure 1).
Potential of FHbp as a vaccine antigen to protect against non–serogroup B invasive meningococcal disease
As described in the previous section, Harris and colleagues examined the susceptibility of a panel of six diverse MenC isolates to sera from rabbits and macaques previously vaccinated with bivalent MenB-FHbp (ie, containing variants A05 and B01 as vaccine antigens).23 The FHbp variants of the MenC isolates evaluated were A19, A68, B149, B157, B151, and B24; flow cytometry FHbp MFI values of these isolates ranged from 1592 to 6532, indicating varying levels of FHbp surface expression. All six MenC isolates tested were susceptible to both the rabbit and the macaque immune sera as assessed by the hSBA assay, providing evidence that MenB-FHbp may protect across serogroups.
The study carried out by Hong and colleagues evaluated the ability of the MenB-4C vaccine to protect against MenX isolates.24 Eleven MenX isolates (nine from Africa and two from France) were serotyped, genotyped, and tested in hSBA assays using pooled sera from MenB-4C–vaccinated infants (n = 40), adolescents (n = 12), or adults (n = 23). Each of the nine MenX isolates from Africa coded for subfamily B FHbp variants (the same subfamily of the variant contained in MenB-4C), and the two French isolates coded for FHbp subfamily A variants. Data from MATS predicted that all nine African isolates would be covered by the MenB-4C FHbp antigen (ie, variant B24); this was confirmed by the hSBA assays, in which the nine African MenX strains were killed by pooled immune sera, but the two French MenX strains were not susceptible. As MenB-FHbp contains an FHbp variant from each subfamily, it is possible that it may provide broader coverage against MenX isolates.
Ladhani and colleagues have described the epidemiology, clinical presentation, and molecular characteristics of MenW disease in England and Wales.67,68 From 2009, MenW cases in England increased due to the spread of an endemic hypervirulent ST11 strain (MenW:cc11).67 MenB-4C was introduced into the routine infant immunization program in the United Kingdom in 2015. Comparison of MenW:cc11 isolates to the MenB-4C vaccine antigens revealed the predominance of alleles for non–cross-protective PorA (P1.5,2), non–cross-protective FHbp (variant 2 peptide 22 [ie, subfamily A]), potentially cross-protective NHBA (peptide 29), and highly cross-protective NadA variants (NadA-2/3).67,68 hSBA assays against six MenW:c11 isolates from patients with IMD were carried out using pooled infant/toddler serum samples from phase II MenB-4C studies. hSBA titers were ≥1:32 against all six isolates, with MenB-4C toddler booster sera giving titers comparable to MenACWY pooled adolescent sera.67 Subsequent real-world evidence indicates MenB-4C is providing protection against MenW disease in the United Kingdom; based on Poisson modeling, it is estimated that MenB-4C directly prevented 98 (95% CI, 34–201) MenW cases in vaccine-eligible children during the first 4 years of implementation in the national infant immunization program. As the MenB-FHbp vaccine contains both subfamily A and B FHbp antigens,18 it also has the potential to protect against the MenW:cc11 strain.
Harris and colleagues reported on the effect of MenB-FHbp–elicited antibodies against non-MenB isolates from human studies using hSBA assays.25 MenB-FHbp was found to elicit bactericidal responses greater than the recognized correlate of protection (hSBA titer ≥ 1:446,69) against non-MenB disease-causing strains (ie, MenC, MenY, MenW, and MenX serogroups) in adolescents, suggesting that the vaccine may provide broad protection against divergent serogroups and emergent strains.25 Bactericidal responses for the MenA strain were substantially lower, but the cause of this was not clear. Interestingly, nearly all participants achieved protective levels of bactericidal antibodies against a representative strain of the hypervirulent MenW:cc11 sublineage after two MenB-FHbp doses, and the majority had a four-fold response after three doses. MenB-FHbp also elicited substantial bactericidal responses against a MenX strain isolated from a Burkina Faso outbreak.25 In comparison, the study from Hong and colleagues using pooled sera found that MenB-4C elicited a bactericidal response against a subset of MenX strains from Africa but had low responses against two MenX isolates from France.24
Discussion
Meningococcal disease, and particularly MenB disease, remains an important cause of morbidity and mortality worldwide.1,2 Additionally, the dynamic nature of IMD epidemiology and the occurrence of outbreaks support the importance of having broadly protective vaccines as a key public health concern.6–8
FHbp is a key virulence factor and antigen, and a component of both licensed MenB vaccines.18–20 Several post-translational processes affect the biological functioning of FHbp. Lnt-mediated triacylation is required for efficient translocation and localization of FHbp molecules to the outer membrane,26 and lipidation is required for its cell surface expression and for its role as a virulence factor and target for antibody-dependent, complement-mediated bactericidal activity.16,28 FHbp is classified, on the basis of its amino acid sequence, into two subfamilies: A and B.23 The functional importance of FHbp to N meningitidis is evidenced by the presence of FHbp in almost all (>99%) strains in the United States and Europe and by the conservation in both FHbp subfamilies of the N-terminal region responsible for FH binding.29,36–38 Importantly, FHbp subfamily A and B vaccine antigens evoke subfamily-restricted immune responses, with the greatest bactericidal activity observed against members of the respective subfamily.16 Licensure of MenB-FHbp, which contains two lipidated FHbp antigens, one from each subfamily, was supported by the demonstration of bactericidal activity against a diverse group of subfamily A and B disease and outbreak test strains.45,47,48
To evaluate FHbp-based vaccine coverage against MenB and non-MenB strains, we conducted molecular surveillance of strains using a large, contemporary (2007–2021) PubMLST dataset. An important advantage of the PubMLST analysis examining the epidemiology of FHbp in non-MenB strains is the breadth of contemporary invasive isolates compared with that of earlier publications. In this regard, PubMLST allows institutions to upload disease isolate information rapidly; therefore, new sequence and vaccine antigen variant types are constantly added. The PubMLST database also expands on the countries and continents that served as a source of data for previous reports. However, limitations of the PubMLST data analysis should be noted. The PubMLST database is dependent on submission of isolates for assessment and therefore may not reflect all circulating isolates at a given time. Additionally, the quality and accuracy of the isolate information added to PubMLST are dependent on the uploading institution. The potential for redundancy between the PubMLST collection isolates and those in the literature is also noted. Lastly, bactericidal activity in the SBA assay is regarded as the gold standard correlate of protective immunity to support vaccine licensure,14 and it is inappropriate to attempt to use sequencing data to predict vaccine coverage. These limitations must be considered when interpreting the expanded insights gleaned from the PubMLST database.
Considering the largely comparable strain diversity between the historical and contemporary datasets discussed in this article, data used for vaccine licensure further support the broad-spectrum effectiveness of MenB vaccines, and MenB-FHbp in particular. There were minor differences between the two datasets; a subset of FHbp variants in the global 2007–2021 collection was not represented in the Pfizer 2000–2006 Collection, underscoring the value of the PubMLST database for continual molecular surveillance of FHbp strain diversity to assess ongoing coverage provided by licensed MenB vaccines.
Presence of the FHbp gene, which suggests the expression of a full-length protein, is a feature of nearly all IMD isolates including non-MenB IMD isolates. As such, various lines of evidence suggest the possibility of protection against non-MenB IMD using FHbp-containing vaccines. Our analysis of the PubMLST dataset reveals that, except for a subset of MenC strains, all MenB and non-MenB disease-causing strains carry a full-length FHbp gene capable of expressing functional FHbp. Subfamily A variants appear to be more common among MenC, MenW, and Men Y strains, whereas subfamily B variants predominated among MenA, MenB, and MenX strains. Moreover, FHbp variants in most non-MenB strains appear to be less genetically diverse than FHbp variants in MenB strains. However, it should be noted that although there are large prevalence-based collections of MenB strains available, the data for non-MenB strains are much less substantial, thereby limiting the ability to make comparative statements about genetic diversity.
In conclusion, we have shown the utility of PubMLST to study FHbp diversity among MenB and non-MenB strains. Because of the presence of FHbp antigens in both MenB and non-MenB strains, such studies are important to unravel the potential of MenB vaccines in providing protection across meningococcal serogroups and across the diversity of disease strains. However, limited strain collection has resulted in a paucity of data regarding the diversity of FHbp among non-MenB strains. We have discussed the potential utility of the PubMLST collection to overcome this lack of resources and to evaluate genotypic diversity among non-MenB disease strains, with particular emphasis on the potential for FHbp-containing vaccines to protect against IMD caused by non-MenB isolates.
Supplementary Material
Biography
Zhenghui Li has a PhD focused on genetics (bioinformatics) from Dartmouth College. He has 8 years of combined academic and industrial work experience, with a particular focus on vaccine and anti-infectives research and development. He is a subject matter expert in next-generation sequencing (DNA-Seq, RNA-Seq, scRNA-seq) experiment design, execution and data analysis. Zhenghui is also proficient in R, Python, and workflow system Airflow, as well as being skilled in ML/AI framework.
Funding Statement
This work was supported by Pfizer Inc. Editorial/medical writing support was provided by Tricia Newell, PhD, and James Currie, PhD, of ICON (Blue Bell, PA), and was funded by Pfizer Inc. Authors thank Professor Martin Maiden, Professor Angela Brueggemann, and Dr Keith Jolley for the development and management of PubMLST and Dr Paul Liberator for his review and early input into this project.
Author Contribution Statement
ZL and AKM were responsible for the conception and design of this study. All authors were responsible for analysis and interpretation of the data; the drafting of the paper, revising it critically for intellectual content; and the final approval of the version to be published. All authors agree to be accountable for all aspects of the work.
Ethical statement
This study did not involve human participants and therefore ethical approval was not required.
Disclosure statement
All authors are employees of Pfizer and may hold stock or stock options.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website at https://doi.org/10.1080/21645515.2024.2409502
References
- 1.Cohn AC, MacNeil JR, Clark TA, Ortega-Sanchez IR, Briere EZ, Meissner HC, Baker CJ, Messonnier NE.. Prevention and control of meningococcal disease: recommendations of the Advisory Committee on Immunization Practices (ACIP). MMWR Recomm Rep. 2013;62(RR–2):1–13. https://www.cdc.gov/mmwr/preview/mmwrhtml/rr6202a1.htm. [PubMed] [Google Scholar]
- 2.MacNeil J, Cohn A. Chapter 8: meningococcal disease. Manual for the Surveillance of Vaccine-Preventable Diseases. 5th ed. Atlanta (GA): Centers for Disease Control and Prevention; 2017. [Google Scholar]
- 3.Stephens DS, Greenwood B, Brandtzaeg P. Epidemic meningitis, meningococcaemia, and Neisseria meningitidis. Lancet. 2007;369(9580):2196–2210. doi: 10.1016/S0140-6736(07)61016-2. [DOI] [PubMed] [Google Scholar]
- 4.Read RC. Neisseria meningitidis and meningococcal disease: recent discoveries and innovations. Curr Opin Infect Dis. 2019;32(6):601–608. doi: 10.1097/QCO.0000000000000606. [DOI] [PubMed] [Google Scholar]
- 5.Pardo de Santayana C, Tin Tin Htar M, Findlow J, Balmer P. Epidemiology of invasive meningococcal disease worldwide from 2010–2019: a literature review. Epidemiol Infect. 2023;151:e57. doi: 10.1017/S0950268823000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soumahoro L, Abitbol V, Vicic N, Bekkat-Berkani R, Safadi MAP. Meningococcal disease outbreaks: a moving target and a case for routine preventative vaccination. Infect Dis Ther. 2021;10(4):1949–1988. doi: 10.1007/s40121-021-00499-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jafri RZ, Ali A, Messonnier NE, Tevi-Benissan C, Durrheim D, Eskola J, Fermon F, Klugman KP, Ramsay M, Sow S, et al. Global epidemiology of invasive meningococcal disease. Popul Health Metr. 2013;11(1):11–17. doi: 10.1186/1478-7954-11-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mbaeyi S, Pondo T, Blain A, Yankey D, Potts C, Cohn A, Hariri S, Shang N, MacNeil JR. Incidence of meningococcal disease before and after implementation of quadrivalent meningococcal conjugate vaccine in the United States. JAMA Pediatr. 2020;174(9):843–851. doi: 10.1001/jamapediatrics.2020.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention. Enhanced Meningococcal Disease Surveillance Report . 2016. [accessed 2024 Aug 19]. https://stacks.cdc.gov/view/cdc/49452.
- 10.European Centre for Disease Prevention and Control . Surveillance Atlas of Infectious Diseases: invasive meningococcal disease. [accessed 2024 Aug 19]. https://atlas.ecdc.europa.eu/public/index.aspx?Dataset=27&HealthTopic=36.
- 11.Public Health Agency of Canada . Vaccine preventable disease: surveillance report to December 31. 2015. [accessed 2024 Aug 19]. https://www.canada.ca/en/public-health/services/publications/healthy-living/vaccine-preventable-disease-surveillance-report-december-31-2015.html.
- 12.Australian Government Department of Health . Invasive meningococcal disease national surveillance report with a focus on MenW. 2017.
- 13.New Zealand Ministry of Health . Notifiable diseases in New Zealand annual report 2016. [accessed 2024 Aug 19]. https://sacnzs.org.nz/assets/1Reports/Public-Health-Surveillance-/Annual-notifiable-disease-report-and-data/esr-notifiable-diseases-annual-surveillance-summary-2016.pdf.
- 14.Findlow J, Lucidarme J, Taha MK, Burman C, Balmer P. Correlates of protection for meningococcal surface protein vaccines: lessons from the past. Expert Rev Vaccines. 2022;21(6):739–751. doi: 10.1080/14760584.2021.1940144. [DOI] [PubMed] [Google Scholar]
- 15.Finne J, Leinonen M, Makela PH. Antigenic similarities between brain components and bacteria causing meningitis. Implications for vaccine development and pathogenesis. Lancet. 1983;2(8346):355–357. doi: 10.1016/S0140-6736(83)90340-9. [DOI] [PubMed] [Google Scholar]
- 16.Fletcher LD, Bernfield L, Barniak V, Farley JE, Howell A, Knauf M, Ooi P, Smith RP, Weise P, Wetherell M, et al. Vaccine potential of the Neisseria meningitidis 2086 lipoprotein. Infect Immun. 2004;72(4):2088–2100. doi: 10.1128/IAI.72.4.2088-2100.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masignani V, Comanducci M, Giuliani MM, Bambini S, Adu-Bobie J, Arico B, Brunelli B, Pieri A, Santini L, Savino S, et al. Vaccination against Neisseria meningitidis using three variants of the lipoprotein GNA1870. J Exp Med. 2003;197(6):789–799. doi: 10.1084/jem.20021911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wyeth Pharmaceuticals LLC, a subsidiary of Pfizer Inc . Trumenba® (meningococcal group B vaccine). Philadelphia (PA): Wyeth Pharmaceuticals LLC, a subsidiary of Pfizer Inc.; 2021. [Google Scholar]
- 19.GSK Vaccines Srl . Bexsero (meningococcal group B vaccine [rDNA, component, adsorbed]). Summary of product characteristics. Siena, Italy: GSK Vaccines Srl; 2022. [Google Scholar]
- 20.Schneider MC, Prosser BE, Caesar JJ, Kugelberg E, Li S, Zhang Q, Quoraishi S, Lovett JE, Deane JE, Sim RB, et al. Neisseria meningitidis recruits factor H using protein mimicry of host carbohydrates. Nature. 2009;458(7240):890–893. doi: 10.1038/nature07769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Madico G, Welsch JA, Lewis LA, McNaughton A, Perlman DH, Costello CE, Ngampasutadol J, Vogel U, Granoff DM, Ram S. The meningococcal vaccine candidate GNA1870 binds the complement regulatory protein factor H and enhances serum resistance. J Immunol. 2006;177(1):501–510. doi: 10.4049/jimmunol.177.1.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beernink PT, Caugant DA, Welsch JA, Koeberling O, Granoff DM. Meningococcal factor H-binding protein variants expressed by epidemic capsular group A, W-135, and X strains from Africa. J Infect Dis. 2009;199(9):1360–1368. doi: 10.1086/597806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harris SL, Zhu D, Murphy E, McNeil LK, Wang X, Mayer LW, Harrison LH, Jansen KU, Anderson AS. Preclinical evidence for the potential of a bivalent fHBP vaccine to prevent Neisseria meningitidis serogroup C disease. Hum Vaccin. 2011;7(suppl):68–74. doi: 10.4161/hv.7.0.14564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong E, Giuliani MM, Deghmane AE, Comanducci M, Brunelli B, Dull P, Pizza M, Taha MK. Could the multicomponent meningococcal serogroup B vaccine (4CMenB) control Neisseria meningitidis capsular group X outbreaks in Africa? Vaccine. 2013;31(7):1113–1116. doi: 10.1016/j.vaccine.2012.12.022. [DOI] [PubMed] [Google Scholar]
- 25.Harris SL, Tan C, Andrew L, Hao L, Liberator PA, Absalon J, Anderson AS, Jones TR. The bivalent factor H binding protein meningococcal serogroup B vaccine elicits bactericidal antibodies against representative non-serogroup B meningococci. Vaccine. 2018;36(45):6867–6874. doi: 10.1016/j.vaccine.2018.05.081. [DOI] [PubMed] [Google Scholar]
- 26.da Silva RAG, Churchward CP, Karlyshev AV, Eleftheriadou O, Snabaitis AK, Longman MR, Ryan A, Griffin R. The role of apolipoprotein N-acyl transferase, Lnt, in the lipidation of factor H binding protein of Neisseria meningitidis strain MC58 and its potential as a drug target. Br J Pharmacol. 2017;174(14):2247–2260. doi: 10.1111/bph.13660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.da Silva RAG, Karlyshev AV, Oldfield NJ, Wooldridge KG, Bayliss CD, Ryan A, Griffin R. Variant signal peptides of vaccine antigen, FHbp, impair processing affecting surface localization and antibody-mediated killing in most meningococcal isolates. Front Microbiol. 2019;10:2847. doi: 10.3389/fmicb.2019.02847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zlotnick GW, Jones TR, Liberator P, Hao L, Harris S, McNeil LK, Zhu D, Perez J, Eiden J, Jansen KU, et al. The discovery and development of a novel vaccine to protect against Neisseria meningitidis serogroup B disease. Hum Vaccin Immunother. 2015;11(1):5–13. doi: 10.4161/hv.34293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brehony C, Wilson DJ, Maiden MC. Variation of the factor H-binding protein of Neisseria meningitidis. Microbiology. 2009;155(Pt 12):4155–4169. doi: 10.1099/mic.0.027995-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McNeil LK, Zagursky R, Shuo L, Murphy E, Zlotnick G, Hoiseth SK, Jansen KU, Andersen AS. The role of factor H binding protein in Neisseria meningitidis virulence and its potential as a vaccine candidate to broadly protect against meningococcal disease. Microbiol Mol Biol Rev. 2013;77(2):234–252. doi: 10.1128/MMBR.00056-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walport MJ. Complement. First of two parts. N Engl J Med. 2001;344(14):1058–1066. doi: 10.1056/nejm200104053441406. [DOI] [PubMed] [Google Scholar]
- 32.Giuntini S, Reason DC, Granoff DM. Complement-mediated bactericidal activity of anti-factor H binding protein monoclonal antibodies against the meningococcus relies upon blocking factor H binding. Infect Immun. 2011;79(9):3751–3759. doi: 10.1128/IAI.05182-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seib KL, Serruto D, Oriente F, Delany I, Adu-Bobie J, Veggi D, Arico B, Rappuoli R, Pizza M. Factor H-binding protein is important for meningococcal survival in human whole blood and serum and in the presence of the antimicrobial peptide LL-37. Infect Immun. 2009;77(1):292–299. doi: 10.1128/IAI.01071-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Granoff DM, Ram S, Beernink PT. Does binding of complement factor H to the meningococcal vaccine antigen, factor H binding protein, decrease protective serum antibody responses? Clin Vaccine Immunol. 2013;20(8):1099–1107. doi: 10.1128/cvi.00260-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Beernink PT, Shaughnessy J, Braga EM, Liu Q, Rice PA, Ram S, Granoff DM. A meningococcal factor H binding protein mutant that eliminates factor H binding enhances protective antibody responses to vaccination. J Immunol. 2011;186(6):3606–3614. doi: 10.4049/jimmunol.1003470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoiseth SK, Murphy E, Andrew L, Vogel U, Frosch M, Hellenbrand W, Abad R, Vazquez JA, Borrow R, Findlow J, et al. A multi-country evaluation of Neisseria meningitidis serogroup B factor H-binding proteins and implications for vaccine coverage in different age groups. Pediatr Infect Dis J. 2013;32(10):1096–1101. doi: 10.1097/INF.0b013e31829aa63b. [DOI] [PubMed] [Google Scholar]
- 37.Murphy E, Andrew L, Lee KL, Dilts DA, Nunez L, Fink PS, Ambrose K, Borrow R, Findlow J, Taha MK, et al. Sequence diversity of the factor H binding protein vaccine candidate in epidemiologically relevant strains of serogroup B Neisseria meningitidis. J Infect Dis. 2009;200(3):379–389. doi: 10.1086/600141. [DOI] [PubMed] [Google Scholar]
- 38.Lucidarme J, Tan L, Exley RM, Findlow J, Borrow R, Tang CM. Characterization of Neisseria meningitidis isolates that do not express the virulence factor and vaccine antigen factor H binding protein. Clin Vaccine Immunol. 2011;18(6):1002–1014. doi: 10.1128/CVI.00055-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gasparini R, Comanducci M, Amicizia D, Ansaldi F, Canepa P, Orsi A, Icardi G, Rizzitelli E, De Angelis G, Bambini S, et al. Molecular and serological diversity of Neisseria meningitidis carrier strains isolated from Italian students aged 14 to 22 years. J Clin Microbiol. 2014;52(6):1901–1910. doi: 10.1128/JCM.03584-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marsh JW, Shutt KA, Pajon R, Tulenko MM, Liu S, Hollick RA, Kiehlbauch JA, Clark TA, Stephens DS, Arnold KE, et al. Diversity of factor H-binding protein in Neisseria meningitidis carriage isolates. Vaccine. 2011;29(35):6049–6058. doi: 10.1016/j.vaccine.2011.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lemee L, Hong E, Etienne M, Deghmane AE, Delbos V, Terrade A, Berthelot G, Caron F, Taha MK. Genetic diversity and levels of expression of factor H binding protein among carriage isolates of Neisseria meningitidis. PLoS One. 2014;9(9):e107240. doi: 10.1371/journal.pone.0107240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rouphael NG, Stephens DS. Neisseria meningitidis: biology, microbiology, and epidemiology. Methods Mol Bio. 2012;799:1–20. doi: 10.1007/978-1-61779-346-2_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Seib KL, Scarselli M, Comanducci M, Toneatto D, Masignani V. Neisseria meningitidis factor H-binding protein fHbp: a key virulence factor and vaccine antigen. Expert Rev Vaccines. 2015;14(6):841–859. doi: 10.1586/14760584.2015.1016915. [DOI] [PubMed] [Google Scholar]
- 44.Jiang HQ, Hoiseth SK, Harris SL, McNeil LK, Zhu D, Tan C, Scott AA, Alexander K, Mason K, Miller L, et al. Broad vaccine coverage predicted for a bivalent recombinant factor H binding protein based vaccine to prevent serogroup B meningococcal disease. Vaccine. 2010;28(37):6086–6093. doi: 10.1016/j.vaccine.2010.06.083. [DOI] [PubMed] [Google Scholar]
- 45.Donald RG, Hawkins JC, Hao L, Liberator P, Jones TR, Harris SL, Perez JL, Eiden JJ, Jansen KU, Anderson AS. Meningococcal serogroup B vaccines: estimating breadth of coverage. Hum Vaccin Immunother. 2017;13(2):255–265. doi: 10.1080/21645515.2017.1264750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goldschneider I, Gotschlich EC, Artenstein MS. Human immunity to the meningococcus. I. The role of humoral antibodies. J Exp Med. 1969;129(6):1307–1326. doi: 10.1084/jem.129.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ostergaard L, Vesikari T, Absalon J, Beeslaar J, Ward BJ, Senders S, Eiden JJ, Jansen KU, Anderson AS, York LJ, et al. A bivalent meningococcal B vaccine in adolescents and young adults. N Engl J Med. 2017;347(24):2349–2362. doi: 10.1056/NEJMoa1614474. [DOI] [PubMed] [Google Scholar]
- 48.Harris SL, Donald RG, Hawkins JC, Tan C, O’Neill R, McNeil LK, Perez JL, Anderson AS, Jansen KU, Jones TR. Neisseria meningitidis serogroup B vaccine, bivalent rLP2086, induces broad serum bactericidal activity against diverse invasive disease strains including outbreak strains. Pediatr Infect Dis J. 2017;36(2):216–223. doi: 10.1097/INF.0000000000001399. [DOI] [PubMed] [Google Scholar]
- 49.Findlow J, Borrow R, Stephens DS, Liberator P, Anderson AS, Balmer P, Jodar L. Correlates of protection for meningococcal surface protein vaccines: current approaches for the determination of breadth of coverage. Expert Rev Vaccines. 2022;21(6):753–769. doi: 10.1080/14760584.2022.2064850. [DOI] [PubMed] [Google Scholar]
- 50.Biagini M, Spinsanti M, De Angelis G, Tomei S, Ferlenghi I, Scarselli M, Rigat F, Messuti N, Biolchi A, Muzzi A, et al. Expression of factor H binding protein in meningococcal strains can vary at least 15-fold and is genetically determined. Proc Natl Acad Sci USA. 2016;113(10):2714–2719. doi: 10.1073/pnas.1521142113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McNeil LK, Donald RGK, Gribenko A, French R, Lambert N, Harris SL, Jones TR, Li S, Zlotnick G, Vogel U, et al. Predicting the susceptibility of meningococcal serogroup B isolates to bactericidal antibodies elicited by bivalent rLP2086, a novel prophylactic vaccine. mBio. 2018;9(2):e00036‒00018. doi: 10.1128/mBio.00036-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liberator P, Donald RGK, Balmer P, Findlow J, Anderson AS. Variant signal peptides of vaccine antigen, FHbp, impair processing affecting surface localization and antibody-mediated killing in most meningococcal isolates [commentary]. Front Microbiol. 2020;11:538209. doi: 10.3389/fmicb.2020.538209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mothibeli KM, du Plessis M, von Gottberg A, Murphy E, Hoiseth SK, Zlotnick G, Klugman KP. Distribution of factor H binding protein beyond serogroup B: variation among five serogroups of invasive Neisseria meningitidis in South Africa. Vaccine. 2011;29(11):2187–2192. doi: 10.1016/j.vaccine.2010.11.072. [DOI] [PubMed] [Google Scholar]
- 54.Law DK, Zhou J, Deng S, Hoang L, Tyrrell G, Horsman G, Wylie J, Tsang RS. Determination of serotyping antigens, clonal analysis and genetic characterization of the 4CMenB vaccine antigen genes in invasive Neisseria meningitidis from Western Canada, 2009 to 2013. J Med Microbiol. 2014;63(Pt 11):1490–1499. doi: 10.1099/jmm.0.079921-0. [DOI] [PubMed] [Google Scholar]
- 55.Esposito S, Zampiero A, Terranova L, Montinaro V, Scala A, Ansuini V, Principi N. Genetic characteristics of Neisseria meningitidis serogroup B strains carried by adolescents living in Milan, Italy: implications for vaccine efficacy. Hum Vaccin Immunother. 2013;9(11):2296–2303. doi: 10.4161/hv.25800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jeppesen CA, Snape MD, Robinson H, Gossger N, John TM, Voysey M, Ladhani S, Okike IO, Oeser C, Kent A, et al. Meningococcal carriage in adolescents in the United Kingdom to inform timing of an adolescent vaccination strategy. J Infect. 2015;71(1):43–52. doi: 10.1016/j.jinf.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 57.Tzanakaki G, Hong E, Kesanopoulos K, Xirogianni A, Bambini S, Orlandi L, Comanducci M, Muzzi A, Taha MK. Diversity of Greek meningococcal serogroup B isolates and estimated coverage of the 4CMenB meningococcal vaccine. BMC Microbiol. 2014;14:111. doi: 10.1186/1471-2180-14-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abad R, Garcia-Amil C, Navarro C, Martin E, Martin-Diaz A, Vazquez JA. Molecular characterization of invasive serogroup B Neisseria meningitidis isolates from Spain during 2015-2018: evolution of the vaccine antigen factor H binding protein (FHbp). J Infect. 2021;82(4):37–44. doi: 10.1016/j.jinf.2021.01.030. [DOI] [PubMed] [Google Scholar]
- 59.Jolley KA, Maiden MC. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinf. 2010;11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018;3:124. doi: 10.12688/wellcomeopenres.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wyeth Pharmaceuticals LLC, a subsidiary of Pfizer Inc . Trumenba (meningococcal group B vaccine). Philadelphia (PA): Wyeth Pharmaceuticals LLC, a subsidiary of Pfizer Inc.; 2024. [Google Scholar]
- 62.Pfizer Europe MA EEIG . Trumenba (meningococcal group B vaccine). Summary of product characteristics. Bruxelles, Belgium: Pfizer Europe MA EEIG; 2022. [Google Scholar]
- 63.Pajon R, Fergus AM, Koeberling O, Caugant DA, Granoff DM. Meningococcal factor H binding proteins in epidemic strains from Africa: implications for vaccine development. PLoS Negl Trop Dis. 2011;5(9):e1302. doi: 10.1371/journal.pntd.0001302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bambini S, Piet J, Muzzi A, Keijzers W, Comandi S, De Tora L, Pizza M, Rappuoli R, van de Beek D, van der Ende A, et al. An analysis of the sequence variability of meningococcal fHbp, NadA and NHBA over a 50-year period in the Netherlands. PLoS One. 2013;8(5):e65043. doi: 10.1371/journal.pone.0065043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tsang RS, Hoang L, Tyrrell G, Horsman G, Wylie J, Jamieson FB, Lefebvre B, Taha MK. Genetic and antigenic characterization of Canadian invasive Neisseria meningitidis serogroup C (MenC) case isolates in the post-MenC conjugate vaccine era, 2009-2013. J Med Microbiol. 2015;64(Pt 2):174–179. doi: 10.1099/jmm.0.000006-0. [DOI] [PubMed] [Google Scholar]
- 66.Law DK, Lefebvre B, Gilca R, Deng S, Zhou J, De Wals P, Tsang RS. Characterization of invasive Neisseria meningitidis strains from Québec, Canada, during a period of increased serogroup B disease, 2009-2013: phenotyping and genotyping with special emphasis on the non-carbohydrate protein vaccine targets. BMC Microbiol. 2015;15(1):143. doi: 10.1186/s12866-015-0469-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ladhani SN, Giuliani MM, Biolchi A, Pizza M, Beebeejaun K, Lucidarme J, Findlow J, Ramsay ME, Borrow R. Effectiveness of meningococcal B vaccine against endemic hypervirulent Neisseria meningitidis W strain, England. Emerg Infect Dis. 2016;22(2):309–311. doi: 10.3201/eid2202.150369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ladhani SN, Beebeejaun K, Lucidarme J, Campbell H, Gray S, Kaczmarski E, Ramsay ME, Borrow R. Increase in endemic Neisseria meningitidis capsular group W sequence type 11 complex associated with severe invasive disease in England and Wales. Clin Infect Dis. 2015;60(4):578–585. doi: 10.1093/cid/ciu881. [DOI] [PubMed] [Google Scholar]
- 69.Borrow R, Balmer P, Miller E. Meningococcal surrogates of protection—serum bactericidal antibody activity. Vaccine. 2005;23(17–18):2222–2227. doi: 10.1016/j.vaccine.2005.01.051. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


