Abstract
Immune system and vaccine responses vary across geographical locations worldwide, not only between high and low-middle income countries (LMICs), but also between rural and urban populations within the same country. Lifestyle factors such as housing conditions, exposure to microorganisms and parasites and diet are associated with rural-and urban-living. However, the relationships between these lifestyle factors and immune profiles have not been mapped in detail. Here, we profiled the immune system of 100 healthy Tanzanians living across four rural/urban areas using mass cytometry. We developed a lifestyle score based on an individual's household assets, housing condition and recent dietary history and studied the association with cellular immune profiles. Seventeen out of 80 immune cell clusters were associated with living location or lifestyle score, with eight identifiable only using lifestyle score. Individuals with low lifestyle score, most of whom live in rural settings, showed higher frequencies of NK cells, plasmablasts, atypical memory B cells, T helper 2 cells, regulatory T cells and activated CD4+ T effector memory cells expressing CD38, HLA-DR and CTLA-4. In contrast, those with high lifestyle score, most of whom live in urban areas, showed a less activated state of the immune system illustrated by higher frequencies of naïve CD8+ T cells. Using an elastic net machine learning model, we identified cellular immune signatures most associated with lifestyle score. Assuming a link between these immune profiles and vaccine responses, these signatures may inform us on the cellular mechanisms underlying poor responses to vaccines, but also reduced autoimmunity and allergies in low- and middle-income countries.
1. Introduction
Variation in the immune system have been observed across populations in low and middle-income countries (LMICs) in Africa and Asia and those living in high-income countries (HICs) in Europe and the USA (de Ruiter et al., 2020; Mbow et al., 2014; Wager et al., 2019; Muyanja et al., 2014; Smolen et al., 2014; de Jong et al., 2021). In addition, immune system variation has been observed within countries, such as in rural compared to urban areas in Senegal (Mbow et al., 2014), Tanzania (Temba et al., 2021) and Indonesia (de Ruiter et al., 2020). The immune system of rural-living individuals in LMICs shows higher memory, activated and regulatory immune profiles, characterized by among others regulatory T cells and T helper 2 cells (Th2 cells), compared to urban-living individuals (de Ruiter et al., 2020; Mbow et al., 2014; Anuradha et al., 2015; Kemp et al., 2001). At the same time, reduced vaccine performance has been observed in populations living in LMICs, in particular in rural areas (Muyanja et al., 2014; Domingo et al., 2019; van Dorst et al., 2023). Moreover, it is known that in these same populations, there are less diseases of affluence, such as allergies or auto-immunities, where unchecked inflammation is a strong contributor (Muyanja et al., 2014; van Dorst et al., 2023; Tsang et al., 2014, 2020; Nehar-Belaid et al., 2023; Shannon et al., 2020; Avey et al., 2017; Okada et al., 2010; Murdaca et al., 2021; Bach, 2002).
Several factors determine the immune profile of an individual, including genetic and demographic factors, such as age and sex, as well as environmental factors, including exposure to microorganisms and parasites, type of housing and dietary history (Brodin et al., 2015; Klein and Flanagan, 2016). While genetics plays an important role in immune system variation during early childhood, this influence wanes with age due to cumulative exposure to environmental factors, including pathogens (Brodin et al., 2015; Brodin and Davis, 2017; Liston et al., 2021). This has been illustrated in individuals chronically infected with helminths, who exhibit skewed baseline immune profiles, characterized by higher frequencies of Th2, regulatory T cells and higher expression of activation and inhibitory markers such as cytotoxic T lymphocyte-associated protein 4 (CTLA-4), HLA-DR and programmed cell death protein 1 (PD-1) on T cells (Lubyayi et al., 2021; Wammes et al., 2016; Labuda et al., 2020). Furthermore, individuals infected with cytomegalovirus (CMV) show a disproportionately higher activation state of the immune system and an increased frequency of memory cells (Kaczorowski et al., 2017; Yan et al., 2021).
Socioeconomic status (SES) is intertwined with housing quality, nutritional status and access to healthcare (Carr et al., 2016; Chakraborty et al., 2016). These factors contribute to infection risk and, therefore, propel the vicious circle of infection/infestation, which strongly impacts the immune system (Murdaca et al., 2021; Carr et al., 2016; Chakraborty et al., 2016; Wikel, 1999; DHS, 2016; Fisk et al., 2010). The type of diet can also be linked to variation in immune profile, as was demonstrated in a recent study in Tanzania (Temba et al., 2021). In this study, rural-living Tanzanians harbored a more anti-inflammatory immune profile that correlated with higher levels of plant-derived flavonoid apigenin found in food mostly eaten in rural settings (Temba et al., 2021). Therefore, taken together, there is evidence for links between living environments such as housing, exposure to microorganisms and parasites, SES including individual assets and diet and immune system variation in LMICs.
Although the immune profiles of urban- and rural-living individuals have been directly compared, a more granular assessment of lifestyles irrespective of living location is lacking, as individuals living in rural areas may exhibit an urban lifestyle and vice versa. We hypothesized that a more refined measurement of lifestyle including housing status, assets (e.g. car, bicycle motorcycle or radio), and dietary history (i.e. frequency of consumption of common dietary products) will allow us to better explain immune variation previously related to rural or urban-living location. Especially, we aim to more precisely define immune signatures in individuals exhibiting immune hypo-responsiveness. Such information can have an impact on both communicable and non-communicable diseases, as a poor immune response to vaccines will affect susceptibility to vaccine-preventable infections, while poor responses to (self-)antigens can lead to fewer allergies or autoimmune diseases in rural-living individuals.
Therefore, we not only used mass cytometry to obtain a highly granular immune profile but also surveyed lifestyle variation among Tanzanian adults recruited from two rural and two urban locations to maximize lifestyle variation using a detailed questionnaire of housing conditions, assets and recent dietary history. We present a lifestyle score based on these questionnaire data, which places individuals on the spectrum ranging from rural to urban lifestyle. We used this lifestyle score to explain immune profile variation in Tanzanian adults living in rural and urban areas and contrasted this with immune signatures from urban-living Europeans. In addition, we utilized a machine learning model to define combined immune signatures most strongly associated with the lifestyle score.
2. Results
2.1. Characteristics of the study population
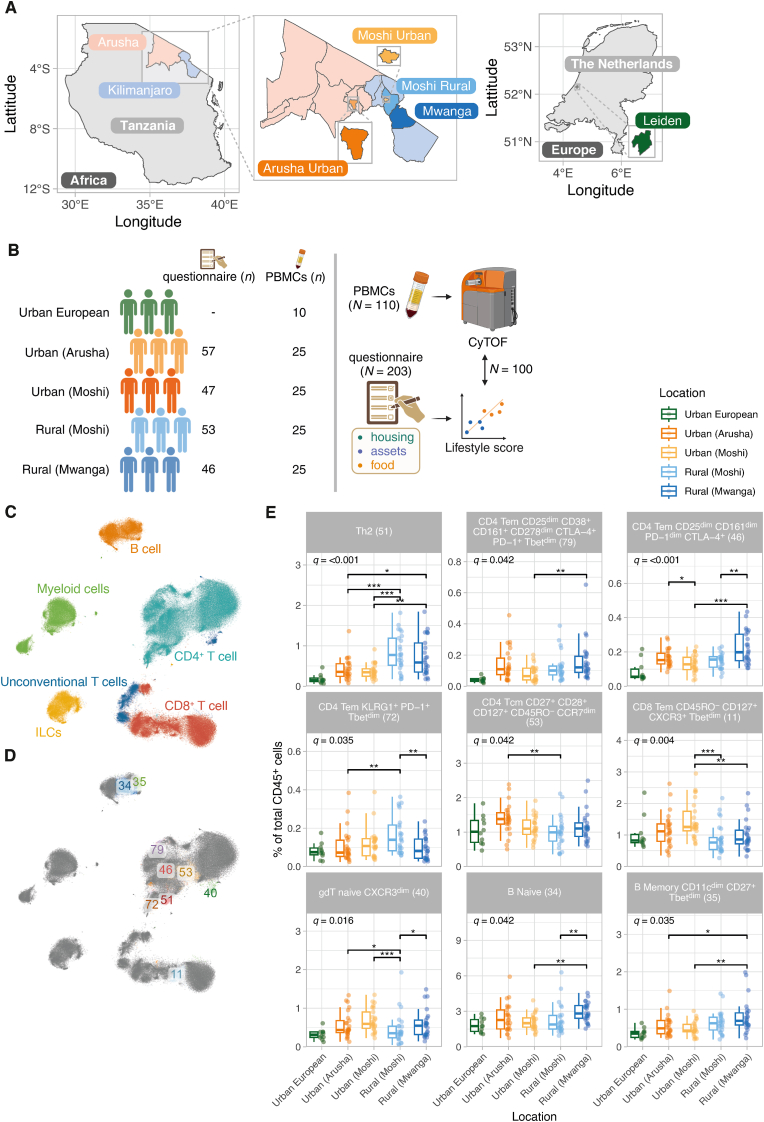
The Tanzanian study population consisted of 203 adults recruited from four geographical locations in northern Tanzania, including two urban locations, Arusha and Moshi Urban and two rural locations, Moshi Rural and Mwanga (Fig. 1A). These four locations were categorized as rural and urban based on the National Bureau of Statistics and the 2022 Census (TNBS, 2022). Detailed information on housing, assets and food history was collected using questionnaires (Temba et al., 2021; TDHS-, 2016) (Fig. 1B).
Fig. 1.
Mass cytometry immune profiles differ across individuals living in rural (Moshi Rural and Mwanga) and urban (Arusha and Moshi Urban) regions. A) Map of study sites in Tanzania and in The Netherlands. B) Graphical representation of sample numbers and the study design. C-D) t-distributed Stochastic Neighbor Embedding (t-SNE) visualizations (n = 1,500 random cells/individual); cells are coloured according to lineage (C) or significant cell cluster (D). E) Differential cell frequencies between rural and urban Tanzanian regions. Boxplots represent the 25th and 75th percentiles (lower and upper boundaries of boxes, respectively), the median (middle horizontal line) and measurements that fall within 1.5 times the interquartile range (IQR; distance between 25th and 75th percentiles; whiskers). Only clusters showing a significant effect of ‘location’ across Tanzanian sites were shown. The significance of ‘location’ was assessed using analysis of variance (ANOVA)-tests comparing a full (location, age [scaled] and sex [fixed effects] and sample ID [random effect]) and a simpler model, which was the same as the full model, except that we removed ‘location’ from the model. ANOVA p-values were corrected for multiple testing using the Benjamini-Hochberg method and referred to as q-values. Asterisks denote statistical significance (∗, q ≤ 0.05; ∗∗, q ≤ 0.01; ∗∗∗, q ≤ 0.001). The statistical significance of differences between each location was assessed using the emmeans-function (Tukey post hoc test). Urban Europeans were included in the figure for visual comparisons and were not included in statistical tests.
From these 203 individuals (Table S1), PBMC samples of 100 individuals were included for mass cytometry analyses (n = 100; n = 25 from each site in four sites) (Table 1). The median age was 25.0 years (interquartile range [IQR], 23–29 years). The prevalence of parasitic infections was 7% and these infections were detected only in individuals from rural areas (Table 1). As a comparator cohort, PBMC samples from ten Dutch individuals recruited in Leiden, The Netherlands (median age 29 [IQR 27–30], 50% female) were acquired using mass cytometry (referred to as ‘urban European’).
Table 1.
Baseline characteristics of the study population (N = 100).
| Variable | Overall, N = 100 | Urban Arusha, N = 25 | Urban Moshi, N = 25 | Rural Moshi, N = 25 | Rural Mwanga, N = 25 | p-value |
|---|---|---|---|---|---|---|
| Sex, female | 53 (53%) | 14 (56%) | 14 (56%) | 13 (52%) | 12 (48%) | 0.932 |
| Age | 25.0 (23.0, 29.0) | 25.0 (23.0, 30.0) | 25.0 (24.0, 27.0) | 24.0 (22.0, 27.0) | 25.0 (22.0, 31.0) | 0.686 |
| Age categories | 0.955 | |||||
| 18-25 | 56 (56%) | 13 (52%) | 14 (56%) | 15 (60%) | 14 (56%) | |
| 26-36 | 44 (44%) | 12 (48%) | 11 (44%) | 10 (40%) | 11 (44%) | |
| BMI | 22.8 (20.5, 26.0) | 21.8 (19.0, 26.8) | 24.1 (22.9, 28.4) | 22.3 (20.3, 26.7) | 22.4 (21.3, 24.6) | 0.243 |
| Missing | 1 | 1 | 0 | 0 | 0 | |
| BMI classification | 0.591 | |||||
| <18.5 | 7 (7.1%) | 3 (13%) | 2 (8.0%) | 1 (4.0%) | 1 (4.0%) | |
| 18.5–24.9 | 60 (61%) | 14 (58%) | 13 (52%) | 15 (60%) | 18 (72%) | |
| 25.0–29.9 | 16 (16%) | 2 (8.3%) | 5 (20%) | 4 (16%) | 5 (20%) | |
| >30 | 16 (16%) | 5 (21%) | 5 (20%) | 5 (20%) | 1 (4.0%) | |
| Missing | 1 | 1 | 0 | 0 | 0 | |
| Systolic blood pressure (mmHg) | 119 (110, 125) | 110 (109, 120) | 110 (100, 119) | 121 (112, 130) | 123 (119, 128) | <0.001 |
| Missing | 1 | 1 | 0 | 0 | 0 | |
| Diastolic blood pressure (mmHg) | 73 (70, 79) | 70 (70, 77) | 69 (64, 72) | 78 (70, 80) | 78 (74, 80) | <0.001 |
| Missing | 1 | 1 | 0 | 0 | 0 | |
| Hemoglobin level g/dl | 14.35 (13.30, 16.50) | 14.00 (13.30, 16.60) | 13.80 (12.40, 15.60) | 14.20 (13.70, 16.00) | 15.20 (13.80, 16.60) | 0.223 |
| Random blood sugar, mmol-1ˆˆ | 5.20 (4.60, 5.95) | 4.90 (4.40, 5.50) | 5.20 (4.70, 6.23) | 5.20 (4.10, 5.50) | 5.80 (4.90, 6.50) | 0.053 |
| Missing | 1 | 0 | 1 | 0 | 0 | |
| Highest level of education | <0.001 | |||||
| Primary | 30 (30%) | 0 (0%) | 0 (0%) | 13 (52%) | 17 (68%) | |
| Secondary | 24 (24%) | 6 (24%) | 0 (0%) | 10 (40%) | 8 (32%) | |
| College | 15 (15%) | 12 (48%) | 1 (4.0%) | 2 (8.0%) | 0 (0%) | |
| University | 31 (31%) | 7 (28%) | 24 (96%) | 0 (0%) | 0 (0%) | |
| Malaria | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | |
| Missing | 1 | 0 | 1 | 0 | 0 | |
| Helminth infectiona | 7 (7.0%) | 0 (0%) | 0 (0%) | 2 (8.0%) | 5 (20%) | 0.015 |
| Schistosomiasisb | 3 (3.0%) | 0 (0%) | 0 (0%) | 0 (0%) | 3 (12%) | 0.057 |
| Missing | 1 | 1 | 0 | 0 | 0 | |
| Insurance status | 31 (31%) | 13 (52%) | 15 (60%) | 3 (12%) | 0 (0%) | <0.001 |
| Occupation | <0.001 | |||||
| Farming | 20 (20%) | 0 (0%) | 1 (4.0%) | 5 (20%) | 14 (56%) | |
| Elementary occupation | 28 (28%) | 5 (20%) | 2 (8.0%) | 16 (64%) | 5 (20%) | |
| Student | 23 (23%) | 5 (20%) | 15 (60%) | 2 (8.0%) | 1 (4.0%) | |
| Employed/business owner | 20 (20%) | 10 (40%) | 5 (20%) | 2 (8.0%) | 3 (12%) | |
| Not employed | 9 (9.0%) | 5 (20%) | 2 (8.0%) | 0 (0%) | 2 (8.0%) | |
N = 100 participants. Values represent number of participants (percentage of total) and median (interquartile range [IQR]) for categorical and continuous variables, respectively. Comparisons between locations were performed using Fisher's exact, chi-squared and Mann–Whitney U test for categorical and continuous variables, respectively. a Stool was tested for helminths using the Kato-Katz method, testing for Schistosoma haematobium, Schistosoma mansoni, Ascaris lumbricoides, hookworm and Trichuris trichuria. b Tested for schistosomiasis using the POC-CCA method, testing for Schistosoma haematobium and Schistosoma mansoni.
2.2. Cellular immune profiles differ between rural- and urban-living Tanzanian adults
To characterize the cellular immune profiles between rural- and urban-living individuals, peripheral blood mononuclear cells (PBMCs) were stained with a panel of 37 metal-tagged antibodies. The processed single-cell level dataset contained 69.6 million live CD45+ cells, which allowed the identification of six major immune lineages, including B cells, CD4+ T cells, CD8+ T cells, innate lymphoid cells (ILCs), myeloid cells and unconventional T cells (including γδ T cells) (Fig. 1C). Clustering analyses using self-organizing maps (SOM), followed by hierarchical clustering resulted in 80 distinct immune cell clusters (Fig. S1 and Table S2). Cell clusters were annotated at subset-level by an expert immunologist. Cell labels were further refined by incorporating markers that exhibit variability within a given subset in the cell label. Using Generalized Linear Mixed Models (GLMMs), we identified nine clusters which were significantly different between the four locations, after adjusting for age and sex (Fig. 1D–E).
The CD4+ T cell lineage was composed of 28 cell clusters, of which 5 significantly differed across locations. Th2 cells (cluster 51) represented the strongest rural signal, where we observed significantly higher frequencies in rural-living locations (especially rural Moshi) compared to urban-living individuals (median 0.7% of total CD45+ cells across rural sites compared to 0.3% and 0.2% in urban Tanzanians and Europeans, respectively). Rural-living individuals additionally showed a significantly higher frequency of three cell clusters of CD4+ T cells. These clusters included CD161dim PD-1dim CTLA-4+ CD4+ T effector memory (Tem) cells (cluster 46), CD4+ Tem cells expressing CD38, CD161, CTLA-4 and PD-1 (cluster 79) and HLA-DRdim PD-1+ KLRG-1+ CD4+ Tem cells (cluster 72). In contrast, the CD27+ CD28+ CD45RO+ CD127+ CD4+ T central memory (Tcm) cell cluster (cluster 53) was higher in urban compared to rural-living individuals (Fig. 1E).
Within the CD8+ T cell lineage, 1 out of 15 CD8+ T cell clusters significantly differed across locations. This cluster was characterized by recently activated CD8+ Tem cells expressing CXCR3 and T-bet (cluster 11), which showed higher frequencies in urban compared to both rural locations (Fig. 1E). Furthermore, within the gamma delta (γδ) T cell lineage (containing 7 clusters), naïve γδ T cells expressing CXCR3 (cluster 40) were significantly higher in frequency in urban-living compared to both rural-living individuals. Finally, within the B cell lineage, we observed significantly higher frequencies of classical naive B cells (cluster 34) and atypical memory B cells expressing CD11c and Tbet (cluster 35) in rural- compared to urban-living locations (Fig. 1E). Six out of seven rural-associated clusters showed visual evidence of a rural-urban-European gradient, where cell frequencies showed a stepwise decrease from rural-to-urban and urban-to-European sites, except for cluster 40 (naïve γδ T cells). On the other hand, gradients were less clear for clusters enriched in urban Tanzanians.
2.3. Questionnaire data reveal differences in lifestyle between locations
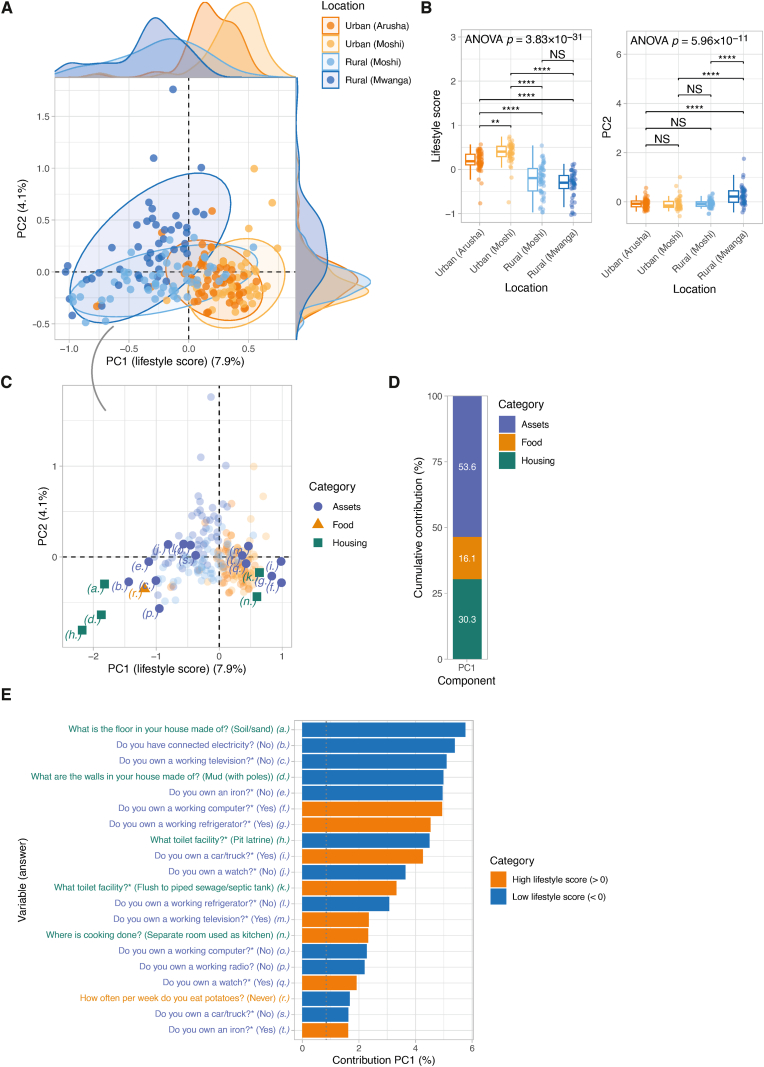
Within living locations, considerable variation in immune signatures was observed. Therefore, to better capture immune variation across locations, we developed a lifestyle score, which incorporates detailed questionnaire data on assets (e.g. possession of a watch, television or car), housing (i.e. materials used to construct the house) and food history (i.e. frequency of consumption of dietary products) into a single score. To obtain the lifestyle score, we applied Multiple Correspondence Analysis (MCA), a dimensionality reduction method similar to Principle Component Analysis (PCA), but for categorical data, which was applied to 38 questions (118 variable categories) collected from all 203 participants (Table S3 and Fig. S2). MCA clearly separated individuals based on living location, especially across principal component (PC) 1. Since the MCA was based on lifestyle questionnaire data and PC1 per definition explains most variance, PC1 was referred to as ‘lifestyle score’, explaining 7.8% of the variation in the questionnaire data (Fig. 2A). Across the first two principal components, we found that spread was highest in rural-compared to urban-living individuals (variance 6.1%/5.1% and 11.3%/11.2% for PC1/PC2 scores across urban and rural sites, respectively), indicating rural people have more heterogeneous lifestyles (Fig. 2B). Sensitivity analyses on condensed questionnaire data (collapsing rare categories and removing uninformative variables) showed that the relatively low percentage of variance explained by lifestyle score and other high-ranking principle components (Fig. S3A) is caused by the inclusion of rarer variable categories. Despite that, removing these had no important effect on the lifestyle score (Pearson r = 0.97, p-value <2.2 × 10−16).
Fig. 2.
Multiple Correspondence Analysis (MCA) based on questionnaire data to generate lifestyle score. A) MCA was applied to categorical questionnaire data (38 manually curated questions; 21 on assets, 11 on food and 6 on housing) (N = 203 individuals). Data points are coloured based on location. Ellipses reflect the data spread at a level of confidence of 95%. Density plots show the distribution of PC1 (lifestyle score) (x-axis) and PC2 (y-axis) score. B) Comparisons of PC1 (lifestyle score) and PC2 across locations. Global significance was assessed using analysis of variance (ANOVA) and post hoc tests between locations were performed using Tukey HSD tests. Asterisks denote statistical significance (NS, non-significant; ∗, p ≤ 0.05; ∗∗, p ≤ 0.01; ∗∗∗, p ≤ 0.001, p ≤ 0.0001). C) Coordinates of each variable category (a.-t.; see E) across dimensions 1 and 2. Variable categories with similar profiles are grouped together. D) Cumulative contributions (in percentage) of the variable categories by questionnaire data category (i.e. housing, assets and food). E) Contributions (in percentage) of variable categories to PC1 or lifestyle score. Bars are coloured based on whether a variable was associated with a high (>zero) or low (<zero) lifestyle score.
We found that the lifestyle score (PC1) was significantly associated with thirteen of 80 cell clusters, while none of the other principal components (PC2-PC5) showed any statistically significant associations with cell cluster frequencies (Fig. S3B), underscoring the validity and biological relevance of the lifestyle score.
Next, we explored the most strongly contributing lifestyle score variables across questionnaire categories, including housing conditions, assets and food history. Overall, assets showed the highest cumulative contribution to the lifestyle score (53.6%), followed by housing (30.3%) and food variables (16.1%) (Fig. 2D). Among the top 20 variables most strongly contributing to PC1, factors such as having a house with an earth/sand floor, a mud wall, no household electricity and a pit latrine as toilet were associated with low lifestyle score. Additionally, the lack of assets such as an ironing tool, refrigerator, computer, radio, car, television, or watch and not consuming potatoes was associated with a low lifestyle score. Factors associated with a high lifestyle score were a house with a flush toilet connected to a sewage/septic tank, a separate room used as a kitchen and possessing assets such as a car, a working computer and a refrigerator (Fig. 2E).
Besides lifestyle score (PC1), we found that PC2 explained 4.1% of the variance (Fig. S3A) and showed the highest spread across individuals living in rural Mwanga (variance across PC2 scores 15.0% compared to 2.9%–7.0% in other sites) (Fig. 2B). Similar to PC1, variables related to assets were most important (cumulative contribution 66.0%), particularly those related to livestock farming (Fig. S3C). PC3 through PC5 explained 3.2–3.5% of the variance (Fig. S3A), generally showing a higher cumulative contribution of food variables (40.3–49.4%) (Fig. S3C) compared to PC1 and PC2.
2.4. Lifestyle score association tests reveal additional immune cell clusters not previously linked to living location
We next assessed the association between lifestyle score and immune cell frequencies using GLMMs, adjusting for age and sex. We first verified that lifestyle score in individuals with matching mass cytometry data (n = 100), which was not significantly different from individuals without mass cytometry data available (Fig. S4).
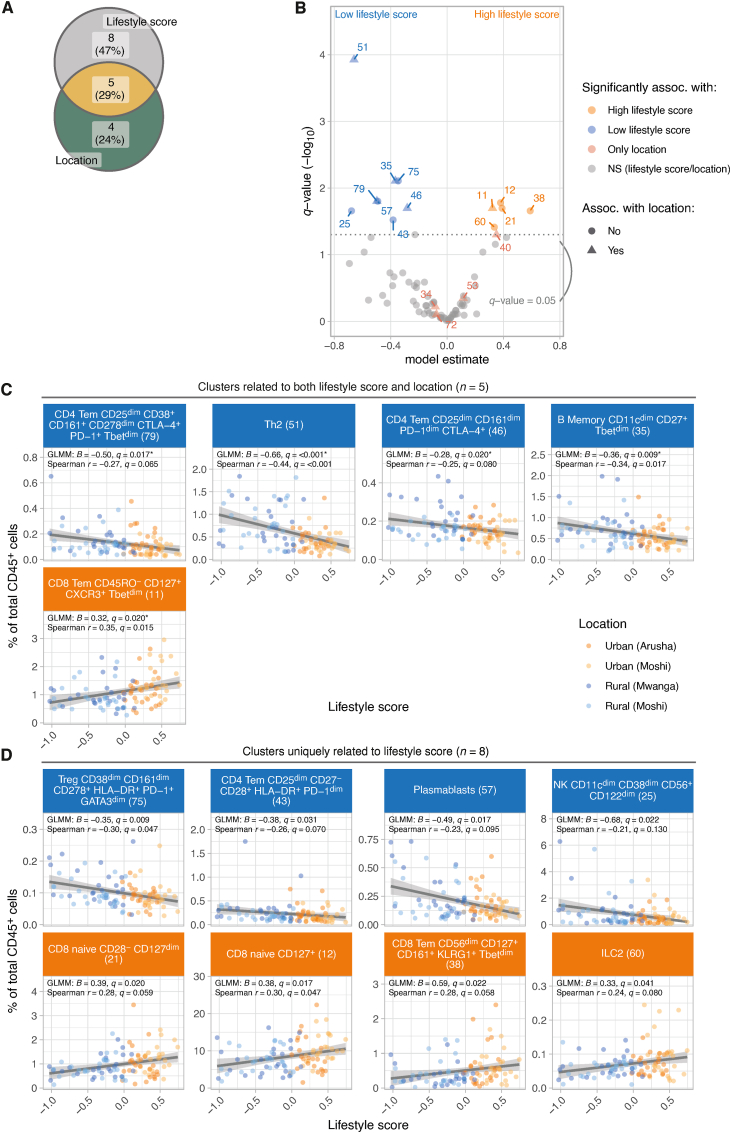
Overall, 13 cell clusters were associated with lifestyle score, of which 8 clusters were not identified by previous analyses where we assessed differences in immune profile between locations (Fig. 3A and B). Indeed, only one of these clusters (cluster 12; CD8+ naïve) showed a trend towards significance across locations (q = 0.055; Fig. S5). In addition, we confirmed 5 out of 9 clusters which were previously found to significantly differ across locations, which were Th2 cells (cluster 51; GLMM; β = −0.66), two CD4+ Tem clusters that were CTLA-4+ and/or CD161+ (cluster 79 and 46; β = −0.50 and −0.28, respectively), atypical memory B cells (cluster 35; β = −0.37) (rural-living location and low lifestyle score) and a CD8+ Tem cluster (cluster 11; β = 0.32) (urban-living location and high lifestyle score) (Fig. 3C). The additional clusters identified using the lifestyle score were two CD4+ Tem cell clusters that were associated with low lifestyle score: HLA-DR+ PD-1+ CD4+ Tem (cluster 43; β = −0.38) and regulatory T cells (cluster 75; β = −0.35). Furthermore, we identified a cluster of plasmablasts (cluster 57; β = −0.49), which was enriched in those with low lifestyle score. Last, an innate immune cell cluster of NK cells (cluster 25; β = −0.68) was also linked to a low lifestyle score (Fig. 3D).
Fig. 3.
Lifestyle score is associated with specific immune cell clusters not identified by comparisons across locations. A) Venn diagram indicating the number of cell clusters that show differences in cell frequencies 1) across locations [Fig. 1E], 2) both across locations and lifestyle score [Fig. 3C] and 3) only with lifestyle score [Fig. 3D]. Eight cell clusters were uniquely associated with lifestyle score and were not identified by comparisons across sampling locations. B) Volcano plot showing differential frequency results. Results were derived from a GLMM with cell frequency as outcome variable, lifestyle score, age (scaled) and sex as fixed effects and sample ID as a random effect. Model estimates and corresponding Benjamini-Hochberg (BH)-adjusted p-values (-log10(q-value)) were shown. Each point represents a cluster, clusters with q-values<0.05 are coloured by association (high or low lifestyle score, or only significantly associated with location). Shapes indicate whether lifestyle-associated clusters were also detected by comparisons across sampling locations. Each point is labelled with a cluster identifier. C-D) Scatter plots showing the association between lifestyle score and cell frequency for C) clusters significantly related to both location as well as lifestyle score and D) clusters uniquely related to lifestyle score (i.e. clusters not identified as differentially abundant between locations). Data points are coloured based on location. Lines represent linear fits to the data and are included for visualization purposes only. Statistical significance was assessed using a linear mixed model including lifestyle score, age (scaled) and sex as fixed effects and sample ID as random effect. Additionally, we ran univariable Spearman correlation tests, p-values were corrected for multiple testing using the Benjamini-Hochberg method (q-value). Asterisks indicate clusters that significantly differed between locations. Only cell clusters significant in GLMMs are shown.
In contrast, within the CD8+ T cell lineage, we identified three clusters of CD8+ T cells that were associated with high lifestyle score. These included two CD8+ naïve T cell clusters (cluster 12 and 21; β = 0.38 and 0.39, respectively) and a cluster of CD8+ Tem cells expressing CD161 and KLRG1 (cluster 38; β = 0.59). In addition, we found a positive association between higher frequencies of ILC2 (cluster 60; β = 0.33) and a high lifestyle score (Fig. 3D). Sensitivity analyses, where we jointly modelled lifestyle score and location and compared the model fit to simpler models (excluding either lifestyle score or location), indicated that indeed using lifestyle score we can detect an additional group of clusters which we could not have detected with location alone (Fig. S6).
2.5. Machine learning modelling links a combined immune endotype with a lifestyle score
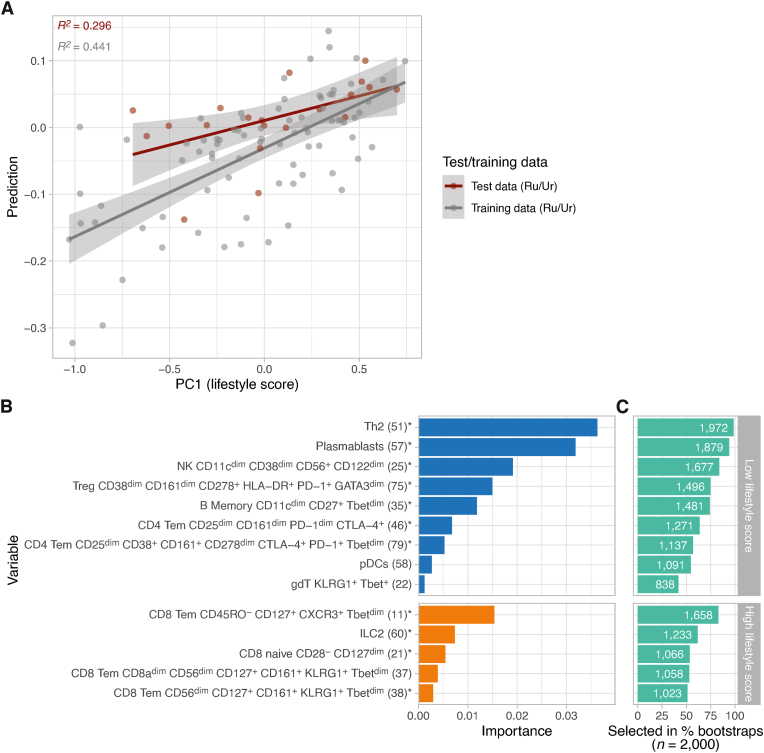
To investigate if a combination of immune cell clusters could be identified that together is associated with a lifestyle score (‘immune endotype’), a machine learning model (elastic net) was trained with lifestyle score as an outcome and cell cluster frequencies, age and sex as the predictor variables. Model training and hyperparameter tuning were performed on 80% of the data (n = 80 individuals; 2000 bootstrapped datasets) and the model was tested on the remaining 20% of the data (n = 20 individuals) (Fig. 4A). The model was able to predict 44.1% and 29.6% of the variance in the training and test data, respectively. Using feature importance analysis, we verified 11 of the 14 clusters that were previously associated with living location and/or lifestyle score. Compared to previous analyses, the current model is a multivariable model, estimating the contribution of each cell cluster to the prediction of lifestyle score while adjusting for all other cluster cell frequencies. Therefore, using this complementary approach, we identified three additional clusters, including CD8+ Tem cells expressing CD161 and KLRG1 (cluster 37) associated with high lifestyles score, pDCs (cluster 58) and γδ T-cells (cluster 22) related to low lifestyle score (Fig. 4B).
Fig. 4.
Machine learning model based on cell cluster frequencies can partly reconstruct lifestyle score. A) Performance of an elastic net machine learning model based on cell cluster frequencies (n = 80), age and sex trained to predict lifestyle score. Observed compared to predicted lifestyle score based on training (80%) and test data (20%; n = 5 samples per location) are shown. Using cell frequency data, we can explain ∼30% of the variance in lifestyle scores (leave-out test data). B) Feature importance of all features that remained in the model after feature shrinkage/regularization. Clusters previously associated with either location or lifestyle score (n = 17) are indicated (∗). Three clusters have not been associated with location nor lifestyle score in previous analyses. C) Feature stability across bootstraps. All features from the models fitted with the optimized tuning parameters (penalty/mixture) were extracted. The number of times a feature was selected across bootstrap samples serves as a score for stability of that feature (maximum score = 2000).
Taken together the elastic net model unveiled a fairly stable (Fig. 4C) immune endotype characterized by Th2 cells, regulatory T cells, atypical B memory cells, plasmablasts, NK cells, CTLA-4+ CD161+ CD4+ Tem, KLRG1+ γδ T-cells and plasmacytoid dendritic cells (pDCs) associated with a low lifestyle score. Inversely, the immune profile characterized by CD8+ naïve T cells, CXCR3+ CD127+ CD8+ Tem, two CD8+ Tem CD161+ CD56dim KLRG1+ and ILC2 is associated with a high lifestyle score (Fig. 4B).
3. Discussion
Here, we assessed the associations between location and/or lifestyle score and cellular immune profiles measured by mass cytometry. We found that seventeen of 80 clusters were associated with location or lifestyle score, with eight identifiable only when using lifestyle score, illustrating the ability of lifestyle score to capture immune variation. Indeed, individuals living in rural areas may exhibit an urban lifestyle and vice versa. This was further substantiated by applying a machine learning model, which identified a combined immune signature associated with lifestyle score.
We found an association between low lifestyle score and expression of activation markers such as CD38, HLA-DR and CTLA-4 on CD4+ Tem cells, along with expansion of Th2 and an increased frequency of regulatory T cells expressing CTLA-4. An increase in a specific memory T cell subsets might indicate that fewer naïve T cells are available for activation and expansion upon encounter with a new antigen. Furthermore, expression of activation/inhibitory markers on T cells can result in a reduced response to vaccines and allergens but may also explain a lower prevalence of autoimmune diseases in LMICs (Bach, 2002; Lubyayi et al., 2021; Maizels, 2016). Indeed, in rural Senegalese, immune profiles were enriched for HLA-DR-expressing CD4+ T cells compared to urban-living individuals (Mbow et al., 2014). Previous studies comparing rural and urban populations in Indonesia (de Ruiter et al., 2020; Wammes et al., 2016) and Gabon (Labuda et al., 2020; van Riet et al., 2007) found that immune profiles in rural-living individuals, characterized by high frequencies of Th2 cells, regulatory T cells expressing CTLA-4, HLA-DR, ICOS or CD161 and atypical memory B cells, were strongly linked to (chronic) helminth infections (de Ruiter et al., 2020; Wammes et al., 2016; Labuda et al., 2020).
In contrast to these previous studies, none of our participants tested positive for malaria and the prevalence of current helminth infections was very low. Therefore, we speculate that increased activation of CD4+ Tem cells, along with expansion of Th2 and higher regulatory T cell frequencies, may represent an immune footprint left behind by parasitic infection in the past or even during childhood, as have been suggested by others (Lubyayi et al., 2021; Djuardi et al., 2011; Mpairwe et al., 2014). Indeed, in 2005, the prevalence of schistosomiasis among school-aged children in two different schools located in one of the rural areas included in this study ranged between 34 and 70% with evidence for the presence of other soil-transmitted infections in the same setting (Poggensee et al., 2005). Thus, based on their age, our study participants likely experienced a high burden of helminth infections during childhood.
Alternatively, housing conditions related to a low lifestyle score (e.g. sand or earth floors and mud-wall houses) may predispose to different commensals or exposure to bacteria and fungi and their metabolites (McCall et al., 2020), some of which have immunomodulatory properties. Poor housing conditions also attract vectors like flies, lice, ticks, mites and mosquitoes, which may directly activate the immune system through components present in their saliva, even in the absence of disease transmission (Wikel, 1999; Vogt et al., 2018). Furthermore, rural-living individuals closely live with livestock and as such are exposed to an additional reservoir of micro-organisms and (zoonotic) pathogens (Libera et al., 2022). Taken together, past (parasitic) infections or unmeasured variables, such as the microbiome or exposure to vectors, are tightly linked to housing conditions. These factors may drive lifestyle-related immune variation, resulting in enrichment of Th2, regulatory T cells and activated T cells.
We found that individuals with low lifestyle score most of whom live in rural settings, display a higher frequency of plasmablasts. Plasmablasts are differentiated B cells with a short lifespan, which initiate early antibody responses during infections (Nutt et al., 2015; Wrammert et al., 2012; Fink, 2012). However, due to their high metabolic activity, the rapid development of short-lived plasmablasts can paradoxically impair humoral immunity by slowing down germinal centre formation. This, in turn may impair responsiveness to vaccines and reduce risk of developing allergies and autoimmunity by limiting the generation of long-lived plasma and memory B cells. Although this has been shown in the context of malaria infection (Vijay et al., 2020), which is not endemic in northern Tanzania, other infectious diseases endemic in the area, may similarly induce high levels of plasmablasts, including dengue (Hertz et al., 2012).
Last, we identified an association between both naïve CD8+ T cells and CD8+ Tem expressing CD161 and high lifestyle score. Although we lack immune markers to confirm, CD161+CD8+ Tem encompasses mucosal-associated invariant T cells (MAIT) cells. MAIT cells are abundant in blood and at mucosal sites and can activate dendritic cells that promote T follicular helper cells to induce mucosal antigen-specific IgA (Pankhurst et al., 2023). Therefore, the presence of such cells in urban-living individuals might indicate the propensity to react more strongly to antigens in a vaccine, allergens, or autoantigens. This aligns with the results of an earlier study indicating that healthy individuals residing in urban Moshi had a higher pro-inflammatory cytokine response upon pathogen challenge in an ex vivo PBMC stimulation assay compared to those living in rural areas (Temba et al., 2021; TDHS-, 2016). Regarding the naïve CD8+ T cells being enriched in urban-living, it has been noted that they allow new immune responses to be mounted to both infections and vaccines (Jongo et al., 2018). Their higher frequency in urban areas is in line with previous studies in Bangladeshi compared to (urban-living) North American children within the first three years of life (Godfrey et al., 2019) as well as in Malawian compared to UK adults (Ben-Smith et al., 2008). Reduced numbers of naïve CD8+ T cells was associated with a higher burden of intestinal worms and viral infections (e.g. CMV) in children from Bangladesh compared to those from the USA (Wager et al., 2019) and higher burden of CMV among Malawian adults (Ben-Smith et al., 2008). Similarly, we speculate that the association between high life score and naïve CD8+ T cells in our study is driven by reduced pathogen exposure in people living in urban settings due to differences in daily activities and hygiene practices compared to rural-living individuals.
The strengths of this study include the use of mass cytometry data in combination with the availability of detailed information on housing, assets and food history. Condensing this information into a single score allowed us to train a machine learning model to identify a distinct group of cell clusters (termed ‘immune endotype’), which was strongly associated with lifestyle score variation. Previous studies in HICs indicated that baseline (gene-expression-based) immune endotypes exhibiting a strong pro-inflammatory profile are predictive of improved vaccine responses in young adults across multiple vaccines (Fourati et al., 2022). In a similar fashion, we speculate the immune endotypes identified in this study are linked to vaccine responses in populations living in rural or urban Africa. As such, further phenotyping of immune endotypes in varied populations, not limited to HIC, using protein-based single-cell modalities such as mass cytometry, may deepen our understanding of variation in vaccine responses or reactivity to allergens or autoantigens and their underlying mechanisms. At the same time, using lifestyle scores opens opportunities for public health experts to screen individuals prone to, for example, vaccine hypo-responsiveness, informing policymakers on preventative measures, such as repeated vaccination. These interventions could target these high-risk individuals, potentially improving vaccine efficacy and public health outcomes. Since those mounting reduced vaccine responses are the very same individuals that also show lower responses to allergens and auto-antigens, immune phenotyping may also unveil new ways to prevent non-communicable diseases in urban-living individuals. Our study also has limitations. Among others, we did not assess cellular immune function through stimulation assays. In addition, future studies establishing direct links between low lifestyle score and responses to vaccines, allergens and autoantigens would be of great value.
In conclusion, in this study we comprehensively assessed the association between immune profiles and location and lifestyle variables in a LMIC. Additional cell clusters were detected through a more refined measurement of lifestyle. Follow-up studies should therefore focus on the links between lifestyle score, immune signature and functional immune responses, particularly in populations where vaccine responses are expected to be reduced and in populations with the highest prevalence of diseases linked to exaggerated immune responses to allergens and autoantigens.
4. Materials and methods
4.1. Study design
This observational study was conducted between September and October 2022 as part of the CapTan study. A total of 203 healthy Tanzanian participants aged between 18 and 35 years were included from two urban locations (Urban Arusha and Urban Moshi) and two rural locations (Rural Moshi and Mwanga) in northern Tanzania (Fig. 1A).
The study was approved both at a local level by the Ethical Board of the Kilimanjaro Christian Medical University College (No. 2588) and at the national level by the Tanzania National Ethical Committee Board (NIMR/HQ/R.8a/Vol.IX/4089). In addition, samples collected from ten Dutch 18 to 30-year-old adults enrolled between January 2022 and September 2022 were included in the TINO study (ClinicalTrials.gov, reference no. NCT06039527). The study was approved by the Ethics Committee of Leiden University Medical Center (NL77841.058.21).
4.2. Description of study areas
Arusha City (1400m above sea level; 617,631 inhabitants (TNBS, 2022)) is the administrative, business, commercial and educational centre of the Arusha region, as it accommodates most diplomatic and international activities. Due to these important regional functions, there is high diversity in ethnicity, economic status and lifestyle. Maasai, Meru and Chagga are the most common ethnicities. Most people living in Arusha City have access to good sanitation with the availability of clean, treated water. However, some people are slum dwellers, i.e. living in the city but practicing a rural lifestyle. Most people are self-employed or office employees in the government and private sectors (TNBS, 2022)
Kilimanjaro region has about 1.9 million inhabitants (TNBS, 2022) across seven different districts, three of which are included in this study (Moshi City, Rural Moshi and Mwanga). Moshi City (referred to as Urban Moshi) (700–950m above sea level; 331,733 inhabitants (TNBS, 2022)) is the administrative, commercial and educational center of the Kilimanjaro region. Most people live a Western lifestyle and have good general sanitation and access to clean water. The main ethnicities are Chagga and Pare. Formal business is the main activity, followed by government and public employment, while few people are involved in agricultural and entrepreneurial activities (TNBS, 2022).
People in Rural Moshi (535,803 inhabitants (TNBS, 2022)) are mainly involved in agricultural activities. Some people have access to clean water, while few use borehole water sources. People live in large family units and their main economic activities are subsistence farming and animal husbandry. The main ethnicity is Chagga and people follow Chagga traditions, such as drinking local brew from banana/plantain.
The population of Mwanga district (684m above sea level; 148,763 inhabitants (TNBS, 2022)) is mainly active in irrigation, subsistence farming and animal husbandry. The primary water sources are boreholes, rivers and dams, with only few people having access to tap water. Like Rural Moshi, people live in large family units. The main ethnicity is Pare, with few Chagga.
Europeans were recruited in the area around Leiden, an urban centre in The Netherlands. European individuals were Dutch.
4.3. Participant screening and enrollment
In rural communities, study information was given through community leaders and announcements during mass gatherings in mosques, churches and during village meetings. In urban communities, study information was distributed using leaflets and through community leaders, office announcements and university gatherings. Eligible participants (age 18–35 years and permanent residency of a given location) were asked to enroll in the study. Following informed consent, 230 participants were voluntarily screened for in- and exclusion criteria. Exclusion criteria were pregnancy, lactation, having acute or chronic diseases, being HIV-positive, recent use of antibiotics, use of antimalarials and use of tuberculostatic drugs. Participants were screened for HIV infection (SDBIOLINE HIV-1/2 3.0kit, LOT:03ADG020A), malaria (Malaria Ag p.f/Pan, Ref: 05FK60, LOT:05EDG018A) and soil-transmitted helminth such as hookworms (Ancylostoma duodenale and Necator americanus), Trichuris trichiura, Ascaris lumbricoides, Strongyloides stercoralis and Schistosoma mansoni using Kato-Katz or Schistosoma haematobium (POC-CCA, batch no:220701075). Furthermore, hemoglobin levels were measured (HemoCue Hb 301(CE:1450820055) and random blood glucose was assessed (ACCU-CHECK glucose test strips, Roche Diabetic care,06993761001). Weight and height were measured using a well-calibrated machine (RGZ-160, made from China), and last, blood pressure was measured using OMRON(SN:202111007949V). After nurse counseling, HIV-positive individual, individuals who had low or high blood pressure (≤90/60 mmHg and ≥140/90 mmHg, respectively) or had high blood glucose (≥7.1 mmol/L fasting or ≥11.1 mmol/L random glucose) were excluded and guided for further actions. People diagnosed with schistosomiasis or soil-transmitted helminth infections were treated with praziquantel and albendazole, respectively according to Tanzanian treatment guidelines. Based on exclusion criteria, 27 of 230 participants were excluded.
All questionnaires and clinical samples were collected by a trained study team, consisting of medical doctors, nurses and laboratory scientists. Data from Tanzanian individuals were collected using the cloud-based electronic data collection system REDCap, with a server hosted at the Kilimanjaro Clinical Research Institute in Tanzania. Data from Dutch participants were collected in a Castor database, with a server hosted in The Netherlands.
4.4. Lifestyle questionnaire
Questionnaires adopted from the Tanzania Demographic and Health Survey and Malaria Indicator Survey (TDHS-MIS) and previously published work conducted in Tanzania, focused on diet in relation to metabolic profiles and inflammatory status (Temba et al., 2021; TDHS-MIS, 2022) were used to collect data on basic demographics, wealth (house construction, general hygiene, land/animal/livestock/non-productive asset ownership) and (recent) food history. Combined, the collected information on wealth and food history was considered reflective of one's ‘lifestyle’. Among others, our questionnaire included questions on the material used to construct the house's floor, roof and walls, the source of water, the type of toilet and available cooking facilities. We assessed the number of milk cows, cattle, goats, sheep, horses and poultry owned and inquiries were made on land ownership and possession of non-productive assets, such as radios, televisions, computers, refrigerators and ironing tools (whether powered by charcoal or electricity), watches, motorcycles, trucks, animal-drawn carts, generators and motorboats. As diet was recently found to shape immune responses in a Tanzanian population (Temba et al., 2021), we additionally collected data on recent food history. We specifically focused on the frequency of various food types participants consume per week, including ugali (stiff porridge), plantain, rice, potatoes, meat, fish, beans/peas, green vegetables, cabbage, fruits and local beer.
4.5. PBMC isolation and cryopreservation
Blood was collected in sodium heparin tubes from 189 of 203 participants. PBMC isolation and cryopreservation were performed as previously described (de Ruiter et al., 2020). 27 Samples were excluded due to low blood quality, technical problems during PBMC isolation or low cell counts. The remaining 162 cryopreserved PBMC samples were transported from Moshi, Tanzania, to Leiden, The Netherlands, using a liquid nitrogen dry vapor shipper. Out of these samples, we selected 100 individuals (25 per location) for immune phenotyping based on age, sex and educational level. Apart from these variables, baseline demographics for the total cohort and the mass cytometry cohort were comparable (Table 1 and Table S1).
4.6. Mass cytometry antibody staining
Antibody panels were designed to phenotype immune cells ex vivo. Details on antibodies used are listed in Table S4. Antibodies were conjugated to metal using 100 μg of purified antibody combined with either the Maxpar X8 or MCP9 Antibody Labelling Kit (Fluidigm), as per the manufacturer's instructions. Conjugated antibodies were then stored in 200 μl of Antibody Stabilizer PBS (CANDOR Bioscience GmbH) at 4 °C. Titration of all antibodies was conducted on PBMC samples.
On the day of staining, cryopreserved PBMCs were thawed with 20% FCS/2 mM Mg2+/1:10,000 benzonase/RPMI medium at 37 °C and washed twice with 10% FCS/RPMI medium. For phenotyping, 3 × 106 cells per sample were prepared according to the Maxpar Nuclear Antigen Staining Protocol V2 (Fluidigm). PBMCs were washed with Maxpar staining buffer and centrifuged at 400g for 5 min in 5-ml Eppendorf tubes. Study samples were randomized over seven batches and for each batch up to 17 samples were barcoded. To barcode the samples, the cells were resuspended in 50 μl of Maxpar staining buffer and 50 μl of a barcode mix targeting β2-microglobulin (B2M) was added to each sample, employing a 6-choose-3 scheme using 106cadmium (Cd), 110Cd, 111Cd, 112Cd, 114Cd and 116Cd. After a 30-min room temperature incubation and a wash with Maxpar Staining Buffer, the cells were centrifuged, the supernatant was removed and the cells were resuspended in Maxpar staining buffer and pooled into one tube for each batch.
Subsequently, cells were treated with 5 ml (about 0.17 oz) of 500 × diluted Cell-ID Intercalator-103Rh (Fluidigm) for 15 min to identify dead cells. After washing with staining buffer, cells were incubated with 20 μl Human TruStain FcX Fc receptor blocking solution (BioLegend) and 130 μl of staining buffer at room temperature for 5 min. Next, 150 μl of a freshly prepared surface antibody cocktail was added for another 30-min room-temperature incubation. After a double wash with staining buffer, cells were fixed with 1.6% PFA in 5 ml PBS for 10 min. Post-centrifugation, cells underwent fixation and permeabilization using the eBioscience Foxp3/Transcription Factor Staining Buffer Set from eBioscience, followed by incubation with Human TruStain FcX receptor blocker. An intranuclear antibody cocktail was then added and the cells were incubated for an additional 30 min. After washing with permeabilization buffer and staining buffer, cells were fixed with 1.6% PFA in 5 ml PBS for 10 min. Finally, cells are stained with 1000 × diluted Cell-ID Intercalator-Ir (Fluidigm) in Maxpar Fix and Perm Buffer at room temperature for 1h and stored in RPMI 20% FCS 10% DMSO at −80 °C until acquisition.
4.7. Mass cytometry data acquisition
All barcoded samples within one batch were acquired simultaneously. Cells were measured using a Helios mass cytometer (Fluidigm) and calibrated as per Fluidigm's guidelines. Before measurement, cells underwent counting, washing with Milli-Q water, straining and then were suspended at a concentration of 1.0 × 106 cells/ml in a solution containing 10% EQ Four Element Calibration Beads from Fluidigm and Milli-Q water. Data acquisition in mass cytometry was performed using dual-count mode and with noise reduction. Various channels were used, including those for antibody detection, intercalators (103Rh, 191Ir, 193Ir), calibration beads (140Ce, 151Eu, 153Eu, 165Ho, 175Lu) and for tracking background/contamination (133Cs, 138Ba, 206Pb). Post-acquisition, the mass bead signal was used to standardize short-term signal variations, using the EQ passport P13H2302 as a reference throughout each experiment. When necessary, normalized FCS files were merged using Helios software, while retaining the beads.
4.8. Data analysis
All data preprocessing and statistics were performed in R v4.2.2 and RStudio Server v2022.03.999. All p-values were corrected for multiple testing using the Benjamini-Hochberg procedure (and referred to as q-values). P-/q-values<0.05 were considered statistically significant.
4.8.1. Data preprocessing
First, cells were automatically gated based on Gaussian parameters (CyTOFClean R-package; v1.03beta; https://github.com/JimboMahoney/cytofclean). Next, automatic gating was applied to select for intact/DNA+-(191Ir and 193Ir channels), CD45+- (89Y) and live cells (live/dead staining) (openCyto v2.10.1 R-package). All automatically set gates were manually inspected. Samples were compensated and debarcoded (CATALYST v1.22.0 R-package). Data were transformed using a hyperbolic arcsinh-transformation with a cofactor of 5 for downstream processing. Next, reference samples collected from healthy European adults included in each individual batch were used to train a CytoNorm-model (CytoNorm v0.0.17 R-package; CytoNorm.train-function; nQ = 101; goal = ‘mean’; k = 10; limit = 0–8). The trained model was applied to all samples, adjusting for batch effects (CytoNorm.normalize-function).
4.8.2. Cell clustering
Cells were subjected to flowSOM-clustering (15 × 15 hexagonal grid; rlen = 100; kohonen v3.0.11 R-package), followed by metaclustering at k = 80 clusters using the hierarchical clustering (factoextra v1.0.7 R-package, hcut-function, distance = ‘ward.D2’). The clustering map was trained on 100k cells per sample, the remaining cells were mapped to the trained map (predict.kohonen-function). Cell clusters were annotated at subset-level by an expert immunologist. Cell labels were further refined by incorporating markers that exhibit variability within a given subset in the cell label.
4.8.3. Lifestyle score
Multiple correspondence analysis (MCA) was applied to categorical questionnaire data (38 manually curated lifestyle-related questions; 21 on assets, 11 on food and 6 on housing) for all 203 Tanzanian participants (FactoMineR v2.7 R-package, MCA-function). Missing values are imputed using mode imputation. Principle component (PC) 1 was defined as ‘lifestyle score’, as this component, per definition, explained most variance across lifestyle questionnaire data. Coordinates of samples and variable categories were visualized in biplots. In addition, (cumulative) variable category contributions for lifestyle score were extracted and shown.
4.8.4. Statistical analyses
To understand the overall structure of the data, cells were placed on a two-dimensional t-distributed Stochastic Neighbor Embedding (t-SNE) map using the Fit-SNE algorithm v1.2.1 (https://github.com/KlugerLab/Fit-SNE/blob/master/fast_tsne.R). Fit-SNE was performed on a down-sampled dataset including 1500 cells per sample (max_iter = 1000; learning rate = n cells/12; perplexity = n cells/100).
To compare the frequency of cell clusters across rural and urban Tanzanian locations, we employed a generalized linear mixed model (binomial = ‘family’; link = ‘logit’; lme4 R-package v1.1-31). The number of cells in each cell cluster (as a fraction of total CD45+ cells per sample) was considered the dependent variable. We fit two models to assess the overall effect of location. Model 1 included (scaled) age and sex as fixed explanatory variables and ‘sample ID’ as a random intercept. ‘Sample ID’ was included as a random effect to deal with any under- or overdispersion due to the binomial model. Model 2 was the same as model 1, except that ‘location’ was added as a fixed explanatory variable. ANOVA tests were used to assess whether location (model 2) significantly improved model fit compared to model 1. Significant models (after correction for multiple testing using Benjamini-Hochberg) were subjected to pairwise comparisons between locations using the emmeans v1.8.5 R-package (model 2; Tukey post hoc test). The associations between cell cluster frequency and lifestyle score were also assessed using GLMMs, including lifestyle score, (scaled) age and sex as fixed explanatory variables and ‘sample ID’ as a random intercept. For sensitivity analyses, we fitted an additional ‘combined’ GLMM, including both location and lifestyle (LS) (as well as age (scaled) and sex) as fixed effects and sample ID as random effect. Model fit (using Akaike Information Criterion [AIC]) of the ‘combined’ GLMM was compared to same model, after removing either location or lifestyle score, to assess the relative importance of these variables to performance cluster-specific models.
4.8.5. Elastic net machine learning modelling
To identify a combined immune ‘endotype’ most associated with variation in lifestyle score, we fit an elastic net machine learning model (tidymodels v1.1.1 R-package, glmnet-engine). Scaled age, sex and cell frequencies of all 80 clusters were included as predictors and lifestyle score was included as an outcome variable. Data was randomly split into train (80%) and test (20%) data (stratified for living location). Model tuning was performed on training data using 2000 bootstrapped data samples, optimizing penalty and mixture parameters. The best model was identified based on the highest explained variance (R2) between observed and predicted lifestyle score (penalty = 0.788, mixture = 0.1). The final model was applied to both training and testing data to generate final estimates of model fit (R2). Variable importance was assessed using the vip v0.4.1 R-package. Feature stability was assessed by extracting all features from the models fitted with the optimized tuning parameters across bootstrap datasets (n = 2000). The number of times a feature was selected was used as a measure for feature stability.
CRediT authorship contribution statement
Jeremia J. Pyuza: Writing – original draft, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Marloes M.A.R. van Dorst: Writing – original draft, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Koen Stam: Writing – review & editing, Formal analysis. Linda Wammes: Writing – review & editing, Conceptualization. Marion König: Investigation. Vesla I. Kullaya: Writing – review & editing, Conceptualization. Yvonne Kruize: Investigation. Wesley Huisman: Writing – review & editing, Investigation. Nikuntufya Andongolile: Investigation. Anastazia Ngowi: Investigation. Elichilia R. Shao: Writing – review & editing. Alex Mremi: Writing – review & editing. Pancras C.W. Hogendoorn: Writing – original draft, Methodology. Sia E. Msuya: Writing – review & editing, Supervision, Methodology, Conceptualization. Simon P. Jochems: Writing – review & editing, Methodology, Formal analysis, Conceptualization. Wouter A.A. de Steenhuijsen Piters: Writing – review & editing, Supervision, Formal analysis, Data curation. Maria Yazdanbakhsh: Writing – review & editing, Supervision, Methodology, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by grants from the Dutch Research Organization (NWO) through the Spinoza prize awarded to Maria Yazdanbakhsh, the European Research Council (ERC) via the ERC Advanced Grant ‘REVERSE’ awarded to Maria Yazdanbakhsh (Grant No: 101055179), the LUMC Excellent Student Fellowship awarded to Marloes M.A.R. van Dorst and the LUMC Global PhD Fellowship awarded to Jeremia J. Pyuza from LUMC, The Netherlands. We would to like acknowledge all clinical and research staff at KCRI and KCMC in Tanzania who helped to make this study possible. We would also like to acknowledge the LUMC core facility for providing mass cytometry services. Finally, we would like to thank all volunteers who participated in this study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbih.2024.100863.
Contributor Information
Wouter A.A. de Steenhuijsen Piters, Email: w.a.a.de_steenhuijsen_piters@lumc.nl.
Maria Yazdanbakhsh, Email: m.yazdanbakhsh@lumc.nl.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
Data availability
Data will be made available on request.
References
- Anuradha R., Munisankar S., Dolla C., Kumaran P., Nutman T.B., Babu S. Parasite antigen-specific regulation of Th1, Th2, and Th17 responses in infection. J. Immunol. 2015;195:2241–2250. doi: 10.4049/jimmunol.1500745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avey S., Cheung F., Fermin D., Frelinger J., Gaujoux R., Gottardo R., Khatri P., Kleinstein S.H., Kotliarov Y., Meng H.L., et al. Multicohort analysis reveals baseline transcriptional predictors of influenza vaccination responses. Sci Immunol. 2017;2 doi: 10.1126/sciimmunol.aal4656. ARTN eaal465610.1126/sciimmunol.aal4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach J.F. Mechanisms of disease: the effect of infections on susceptibility to autoimmune and allergic diseases. N. Engl. J. Med. 2002;347:911–920. doi: 10.1056/NEJMra020100. [DOI] [PubMed] [Google Scholar]
- Ben-Smith A., Gorak-Stolinska P., Floyd S., Weir R.E., Lalor M.K., Mvula H., Crampin A.C., Wallace D., Beverley P.C.L., Fine P.E.M., Dockrell H.M. Differences between naive and memory T cell phenotype in Malawian and UK adolescents: a role for Cytomegalovirus? BMC Infect. Dis. 2008;8 doi: 10.1186/1471-2334-8-139. Artn 13910.1186/1471-2334-8-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodin P., Davis M.M. Human immune system variation. Nat. Rev. Immunol. 2017;17:21–29. doi: 10.1038/nri.2016.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodin P., Jojic V., Gao T., Bhattacharya S., Angel C.J., Furman D., Shen-Orr S., Dekker C.L., Swan G.E., Butte A.J., et al. Variation in the human immune system is largely driven by non-heritable influences. Cell. 2015;160:37–47. doi: 10.1016/j.cell.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr E.J., Dooley J., Garcia-Perez J.E., Lagou V., Lee J.C., Wouters C., Meyts I., Goris A., Boeckxstaens G., Linterman M.A., Liston A. The cellular composition of the human immune system is shaped by age and cohabitation. Nat. Immunol. 2016;17:461. doi: 10.1038/ni.3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty N.M., Fry K., Behl R., Longfield K. Simplified asset indices to measure wealth and equity in health programs: a reliability and validity analysis using Survey data from 16 countries. Glob Health-Sci Prac. 2016;4:141–154. doi: 10.9745/Ghsp-D-15-00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DHS . 2016. Wealth Index. [Google Scholar]
- Djuardi Y., Wammes L.J., Supali T., Sartono E., Yazdanbakhsh M. Immunological footprint: the development of a child's immune system in environments rich in microorganisms and parasites. Parasitology. 2011;138:1508–1518. doi: 10.1017/S0031182011000588. [DOI] [PubMed] [Google Scholar]
- Domingo C., Fraissinet J., Ansah P.O., Kelly C., Bhat N., Sow S.O., Mejia J.E. Long-term immunity against yellow fever in children vaccinated during infancy: a longitudinal cohort study. Lancet Infect. Dis. 2019;19:1363–1370. doi: 10.1016/S1473-3099(19)30323-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dorst M., Pyuza J.J., Nkurunungi G., Kullaya V.I., Smits H.H., Hogendoorn P.C.W., Wammes L.J., Everts B., Elliott A.M., Jochems S.P., Yazdanbakhsh M. Immunological factors linked to geographical variation in vaccine responses. Nat. Rev. Immunol. 2023 doi: 10.1038/s41577-023-00941-2. [DOI] [PubMed] [Google Scholar]
- Fink K. Origin and function of circulating plasmablasts during acute viral infections. Front. Immunol. 2012;3 doi: 10.3389/fimmu.2012.00078. ARTN 7810.3389/fimmu.2012.00078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk W.J., Eliseeva E.A., Mendell M.J. Association of residential dampness and mold with respiratory tract infections and bronchitis: a meta-analysis. Environ Health-Glob. 2010;9 doi: 10.1186/1476-069X-9-72. Artn 7210.1186/1476-069x-9-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourati S., Tomalin L.E., Mulè M.P., Chawla D.G., Gerritsen B., Rychkov D., Henrich E., Miller H.E.R., Hagan T., Diray-Arce J., et al. Pan-vaccine analysis reveals innate immune endotypes predictive of antibody responses to vaccination. Nat. Immunol. 2022;23:1777. doi: 10.1038/s41590-022-01329-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey D.I., Koay H.F., McCluskey J., Gherardin N.A. The biology and functional importance of MAIT cells. Nat. Immunol. 2019;20:1110–1128. doi: 10.1038/s41590-019-0444-8. [DOI] [PubMed] [Google Scholar]
- Hertz J.T., Munishi O.M., Ooi E.E., Howe S., Lim W.Y., Chow A., Morrissey A.B., Bartlett J.A., Onyango J.J., Maro V.P., et al. Chikungunya and dengue fever among hospitalized febrile patients in northern Tanzania. Am. J. Trop. Med. Hyg. 2012;86:171–177. doi: 10.4269/ajtmh.2012.11-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong S.E., van Unen V., Manurung M.D., Stam K.A., Goeman J.J., Jochems S.P., Höllt T., Pezzotti N., Mouwenda Y.D., Ongwe M.E.B., et al. Systems analysis and controlled malaria infection in Europeans and Africans elucidate naturally acquired immunity. Nat. Immunol. 2021;22:654. doi: 10.1038/s41590-021-00911-7. [DOI] [PubMed] [Google Scholar]
- Jongo S.A., Shekalaghe S.A., Church L.W.P., Ruben A.J., Schindler T., Zenklusen I., Rutishauser T., Rothen J., Tumbo A., Mkindi C., et al. Safety, immunogenicity, and protective efficacy against controlled human malaria infection of sporozoite vaccine in Tanzanian adults. Am. J. Trop. Med. Hyg. 2018;99:338–349. doi: 10.4269/ajtmh.17-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczorowski K.J., Shekhar K., Nkulikiyimfura D., Dekker C.L., Maecker H., Davis M.M., Chakraborty A.K., Brodin P. Continuous immunotypes describe human immune variation and predict diverse responses. P Natl Acad Sci USA. 2017;114:E6097–E6106. doi: 10.1073/pnas.1705065114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp K., Akanmori B.D., Hviid L. West African donors have high percentages of activated cytokine producing T cells that are prone to apoptosis. Clin. Exp. Immunol. 2001;126:69–75. doi: 10.1046/j.1365-2249.2001.01657.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein S.L., Flanagan K.L. Sex differences in immune responses. Nat. Rev. Immunol. 2016;16:626–638. doi: 10.1038/nri.2016.90. [DOI] [PubMed] [Google Scholar]
- Labuda L.A., Adegnika A.A., Rosa B.A., Martin J., Ateba-Ngoa U., Amoah A.S., Lima H.M., Meurs L., Mbow M., Manurung M.D., et al. A praziquantel treatment study of immune and transcriptome profiles in -infected Gabonese schoolchildren. J. Infect. Dis. 2020;222:2103–2113. doi: 10.1093/infdis/jiz641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libera K., Konieczny K., Grabska J., Szopka W., Augustyniak A., Pomorska-Mol M. Selected livestock-associated zoonoses as a growing challenge for public health. Infect. Dis. Rep. 2022;14:63–81. doi: 10.3390/idr14010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liston A., Humblet-Baron S., Duffy D., Goris A. Human immune diversity: from evolution to modernity. Nat. Immunol. 2021;22:1479–1489. doi: 10.1038/s41590-021-01058-1. [DOI] [PubMed] [Google Scholar]
- Lubyayi L., Mpairwe H., Nkurunungi G., Lule S.A., Nalwoga A., Webb E.L., Levin J., Elliott A.M. Infection-exposure in infancy is associated with reduced allergy-related disease in later childhood in a Ugandan cohort. Elife. 2021;10 doi: 10.7554/eLife.66022. ARTN e6602210.7554/eLife.66022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizels R.M. Parasitic helminth infections and the control of human allergic and autoimmune disorders. Clin. Microbiol. Infect. 2016;22:481–486. doi: 10.1016/j.cmi.2016.04.024. [DOI] [PubMed] [Google Scholar]
- Mbow M., de Jong S.E., Meurs L., Mboup S., Dieye T.N., Polman K., Yazdanbakhsh M. Changes in immunological profile as a function of urbanization and lifestyle. Immunology. 2014;143:569–577. doi: 10.1111/imm.12335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall L.I., Callewaert C., Zhu Q.Y., Song S.J., Bouslimani A., Minich J.J., Ernst M., Ruiz-Calderon J.F., Cavallin H., Pereira H.S., et al. Home chemical and microbial transitions across urbanization. Nat Microbiol. 2020;5:108–115. doi: 10.1038/s41564-019-0593-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mpairwe H., Tweyongyere R., Elliott A. Pregnancy and helminth infections. Parasite Immunol. 2014;36:328–337. doi: 10.1111/pim.12101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdaca G., Greco M., Borro M., Gangemi S. Hygiene hypothesis and autoimmune diseases: a narrative review of clinical evidences and mechanisms. Autoimmun. Rev. 2021;20 doi: 10.1016/j.autrev.2021.102845. ARTN 10284510.1016/j.autrev.2021.102845. [DOI] [PubMed] [Google Scholar]
- Muyanja E., Ssemaganda A., Ngauv P., Cubas R., Perrin H., Srinivasan D., Canderan G., Lawson B., Kopycinski J., Graham A.S., et al. Immune activation alters cellular and humoral responses to yellow fever 17D vaccine (vol 124, pg 3147, 2014) J. Clin. Invest. 2014;124:4669. doi: 10.1172/JCI75429. 4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehar-Belaid D., Sokolowski M., Ravichandran S., Banchereau J., Chaussabel D., Ucar D. Baseline immune states (BIS) associated with vaccine responsiveness and factors that shape the BIS. Semin. Immunol. 2023;70 doi: 10.1016/j.smim.2023.101842. ARTN 10184210.1016/j.smim.2023.101842. [DOI] [PubMed] [Google Scholar]
- Nutt S.L., Hodgkin P.D., Tarlinton D.M., Corcoran L.M. The generation of antibody-secreting plasma cells. Nat. Rev. Immunol. 2015;15:160–171. doi: 10.1038/nri3795. [DOI] [PubMed] [Google Scholar]
- Okada H., Kuhn C., Feillet H., Bach J.F. The 'hygiene hypothesis' for autoimmune and allergic diseases: an update. Clin. Exp. Immunol. 2010;160:1–9. doi: 10.1111/j.1365-2249.2010.04139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankhurst T.E., Buick K.H., Lange J.L., Marshall A.J., Button K.R., Palmer O.R., Farrand K.J., Montgomerie I., Bird T.W., Mason N.C., et al. MAIT cells activate dendritic cells to promote T(FH) cell differentiation and induce humoral immunity. Cell Rep. 2023;42 doi: 10.1016/j.celrep.2023.112310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poggensee G., Krantz I., Nordin P., Mtweve S., Ahlberg B., Mosha G., Freudenthal S. A six-year follow-up of schoolchildren for urinary and intestinal schistosomiasis and soil-transmitted helminthiasis in Northern Tanzania. Acta Trop. 2005;93:131–140. doi: 10.1016/j.actatropica.2004.10.003. [DOI] [PubMed] [Google Scholar]
- van Riet E., Adegnika A.A., Retra K., Vieira R., Tielens A.G.M., Lell B., Issifou S., Hartgers F.C., Rimmelzwaan G.F., Kremsner P.G., Yazdanbakhsh M. Cellular and humoral responses to influenza in Gabonese children living in rural and semi-urban areas. J. Infect. Dis. 2007;196:1671–1678. doi: 10.1086/522010. [DOI] [PubMed] [Google Scholar]
- de Ruiter K., Jochems S.P., Tahapary D.L., Stam K.A., König M., van Unen V., Laban S., Höllt T., Mbow M., Lelieveldt B.P.F., et al. Helminth infections drive heterogeneity in human type 2 and regulatory cells. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.aaw3703. ARTN eaaw370310.1126/scitranslmed.aaw3703. [DOI] [PubMed] [Google Scholar]
- Shannon C.P., Blimkie T.M., Ben-Othman R., Gladish N., Amenyogbe N., Drissler S., Edgar R.D., Chan Q., Krajden M., Foster L.J., et al. Multi-omic data integration allows baseline immune signatures to predict hepatitis B vaccine response in a small cohort. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.578801. ARTN 57880110.3389/fimmu.2020.578801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolen K.K., Ruck C.E., Fortuno E.S., Ho K., Dimitriu P., Mohn W.W., Speert D.P., Cooper P.J., Esser M., Goetghebuer T., et al. Pattern recognition receptor-mediated cytokine response in infants across 4 continents. J. Allergy Clin. Immunol. 2014;133:818. doi: 10.1016/j.jaci.2013.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TDHS M.I.S. Tanzania demographic and health Survey and malaria indicator Survey (TDHS-MIS) 2015-16. 2016. https://dhsprogram.com/publications/publication-FR321-DHS-Final-Reports.cfm
- TDHS-MIS The 2022 Tanzania demographic and health Survey and malaria indicator Survey (2022 TDHS-MIS) 2022. https://dhsprogram.com/pubs/pdf/FR382/FR382.pdf
- Temba G.S., Kullaya V., Pecht T., Mmbaga B.T., Aschenbrenner A.C., Ulas T., Kibiki G., Lyamuya F., Boahen C.K., Kumar V., et al. Urban living in healthy Tanzanians is associated with an inflammatory status driven by dietary and metabolic changes. Nat. Immunol. 2021;22:287. doi: 10.1038/s41590-021-00867-8. [DOI] [PubMed] [Google Scholar]
- TNBS Tanzania population and housing census- Tanzania National Bureau of statistics(TNBS) 2022. https://www.nbs.go.tz/index.php/en/census-surveys/population-and-housing-census
- Tsang J.S., Schwartzberg P.L., Kotliarov Y., Biancotto A., Xie Z., Germain R.N., Wang E., Olnes M.J., Narayanan M., Golding H., et al. Global analyses of human immune variation reveal baseline predictors of postvaccination responses (vol 157, pg 499, 2014) Cell. 2014;158:226. doi: 10.1016/j.cell.2014.06.015. 226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang J.S., Dobaño C., VanDamme P., Moncunill G., Marchant A., Ben Othman R., Sadarangani M., Koff W.C., Kollmann T.R. Improving vaccine-induced immunity: can baseline predict outcome? Trends Immunol. 2020;41:457–465. doi: 10.1016/j.it.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay R., Guthmiller J.J., Sturtz A.J., Surette F.A., Rogers K.J., Sompallae R.R., Li F.Y., Pope R.L., Chan J.A., Rivera F.D., et al. Infection-induced plasmablasts are a nutrient sink that impairs humoral immunity to malaria. Nat. Immunol. 2020;21:790. doi: 10.1038/s41590-020-0678-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt M.B., Lahon A., Arya R.P., Kneubehl A.R., Clinton J.L.S., Paust S., Rico-Hesse R. Mosquito saliva alone has profound effects on the human immune system. PLoS Neglected Trop. Dis. 2018;12 doi: 10.1371/journal.pntd.0006439. ARTN e000643910.1371/journal.pntd.0006439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wager L.E., Bolen C.R., Sigel N., Angel C.J.L., Guan L., Kirkpatrick B.D., Haque R., Tibshirani R.J., Parsonnet J., Petri W.A., Davis M.M. Increased T cell differentiation and cytolytic function in Bangladeshi compared to American children. Front. Immunol. 2019;10 doi: 10.3389/fimmu.2019.02239. ARTN 223910.3389/fimmu.2019.02239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wammes L.J., Hamid F., Wiria A.E., May L., Kaisar M.M.M., Prasetyani-Gieseler M.A., Djuardi Y., Wibowo H., Kruize Y.C.M., Verweij J.J., et al. Community deworming alleviates geohelminth-induced immune hyporesponsiveness. P Natl Acad Sci USA. 2016;113:12526–12531. doi: 10.1073/pnas.1604570113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikel S.K. Modulation of the host immune system by ectoparasitic arthropods - blood-feeding and tissue-dwelling arthropods manipulate host defenses to their advantage. Bioscience. 1999;49:311–320. doi: 10.2307/1313614. [DOI] [Google Scholar]
- Wrammert J., Onlamoon N., Akondy R.S., Perng G.C., Polsrila K., Chandele A., Kwissa M., Pulendran B., Wilson P.C., Wittawatmongkol O., et al. Rapid and massive virus-specific plasmablast responses during acute dengue virus infection in humans. J. Virol. 2012;86:2911–2918. doi: 10.1128/Jvi.06075-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z., Maecker H.T., Brodin P., Nygaard U.C., Lyu S.C., Davis M.M., Nadeau K.C., Andorf S. Aging and CMV discordance are associated with increased immune diversity between monozygotic twins. Immun. Ageing. 2021;18 doi: 10.1186/s12979-021-00216-1. ARTN 510.1186/s12979-021-00216-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.