Summary
Background
The spread of emerging SARS-CoV-2 immune escape sublineages, especially JN.1 and KP.2, has resulted in new waves of COVID-19 globally. The evolving memory B cell responses elicited by the parental Omicron variants to subvariants with substantial antigenic drift remain incompletely investigated.
Methods
Using the single B cell antibody cloning technology, we isolated single memory B cells, delineated the B cell receptor repertoire and conducted the pseudovirus-based assay for recovered neutralizing antibodies (NAb) screening. We analyzed the cryo-EM structures of top broadly NAbs (bnAbs) and evaluated their in vivo efficacy (golden Syrian hamster model).
Findings
By investigating the evolution of human B cell immunity, we discovered a new panel of bnAbs arising from vaccinees after Omicron BA.2/BA.5 breakthrough infections. Two lead bnAbs neutralized major Omicron subvariants including JN.1 and KP.2 with IC50 values less than 10 ng/mL, representing ultrapotent receptor binding domain (RBD)-specific class I bnAbs. They belonged to the IGHV3-53/3-66 clonotypes instead of evolving from the pre-existing vaccine-induced IGHV1-58/IGKV3-20 bnAb ZCB11. Despite sequence diversity, they targeted previously unrecognized, highly conserved conformational epitopes in the receptor binding motif (RBM) for ultrapotent ACE2 blockade. The lead bnAb ZCP3B4 not only protected the lungs of hamsters intranasally challenged with BA.5.2, BQ.1.1 and XBB.1.5 but also prevented their contact transmission.
Interpretation
Our findings demonstrated that class I bnAbs have evolved an ultrapotent mode of action protecting against highly transmissible and broad Omicron escape variants, and their epitopes are potential targets for novel bnAbs and vaccine development.
Funding
A full list of funding bodies that contributed to this study can be found in the Acknowledgements section.
Keywords: SARS-CoV-2, Omicron breakthrough infection, Broadly neutralizing antibody, Structural fitness of antibody, Prevention of XBB.1.5 transmission
Research in context.
Evidence before this study
During the early phase of pandemic, protective neutralizing antibodies (NAbs) against SARS-CoV-2 could be effectively elicited by the natural infection or WT-based vaccines, mainly targeting the receptor binding domain (RBD) of the spike protein. RBD epitopes that overlap the ACE2 receptor binding site (RBS) are immunodominant and concentrate most potent NAbs, especially class I NAbs. Antibodies in this class, which are mostly encoded by VH3-53/VH3-66 germlines, neutralize SARS-CoV-2 by directly blocking the spike protein binding to ACE2. However, the efficacy of class I NAbs has been challenged since the outbreak of variants of concern (VOCs). Members of the same class with similar breadth and potency against the emerging escape variants have rarely been documented in current literature and the CoV-AbDab.
Added value of this study
Our lead class I bnAbs were induced by Omicron BA.2/BA.5 breakthrough infections through reactivating memory B cell responses. They accumulated high levels of somatic hypermutations and displayed structural fitness to cross-neutralize both WT and variants ranging from 2020 to 2024. These bnAbs enhance the broad spectrum mainly by targeting the conserved conformational 'Jing' epitopes in RBM, which remained unchanged despite convergent mutations found in XBB, EG.5.1 and BA.2.86 sublineages. Such a unique mode of action for ultrapotent bnAbs allows them to overcome the Omicron antigenic shift from BA.1 to KP.2 variants.
Implications of all the available evidence
Our bnAbs were capable of cross-neutralizing emerging Omicron sublineages circulating 1–2 years after the biological sample collection, indicating a potential resistance to future escape. They have value as drug candidates in situations where prophylaxis or treatment are warranted. Moreover, the structural insights into bnAbs and antibody–antigen interaction provide new values for next-generation vaccine development and deep learning-guided antibody optimization.
Introduction
Since the beginning of COVID-19 pandemic, neutralizing antibodies (NAbs) against wild-type (WT) SARS-CoV-2 have been readily elicited during the natural course of infection.1, 2, 3, 4 Subsequently, NAbs were induced by WT-based vaccines, especially by mRNA-vectored vaccines,5,6 which were associated with reduced hospitalization and mortality.7,8 The evolution of SARS-CoV-2, however, resulted in the emergence of immune-escaped variants that outcompeted NAbs in the upper respiratory tract, leading to continuous viral transmission and pandemic.9, 10, 11, 12 Understanding how human memory B cells would respond to continuously evolving pandemic variants for generating broadly NAbs (bnAbs) is essential for the development of next generation antibody drugs and vaccines.
Prior ultrapotent NAbs that display IC50 values less than 10 ng/mL often target the immunodominant epitopes on the receptor binding domain (RBD) of SARS-CoV-2 spike protein with supersites overlapping or around the ACE2 receptor binding motif (RBM).1,13,14 The efficacies of these NAb drugs and WT-based vaccines have been severely reduced due to the global outbreak of highly mutated Omicron variants since November 2021.9, 10, 11, 12,15, 16, 17 The emerged sublineages BA.2, BA.2.75, BA.4/5 and BF7 displayed higher transmissibility and more significant antibody evasion than previous variants of concern (VOCs).9,11,18,19 Notably, their subsequent descendants such as BQ.1, XBB and EG.5.1 sublineages carry a series of convergent mutations that endow them with enormous growth advantages over their predecessors and even stronger resistance to antibody responses.12,20,21 BA.2.86 evolved with 34 additional mutations in the spike protein compared with BA.2 without showing more resistance to human sera than XBB.1.5 and EG.5.1.22 However, the JN.1 and KP.2 arising from BA.2.86 displayed increased fitness and outcompeted the previous dominant XBB lineage.23 The ongoing spread of these convergent variants has severely impaired the efficacy of NAbs from convalescents, fully vaccinated individuals and even vaccinees boosted with the bivalent mRNA vaccine.7,24, 25, 26 Although thousands of monoclonal NAbs were found targeting multiple spike epitopes, few retained ultrapotency and breadth against the current circulating JN.1 and KP.2 sublineages.22,27
Four classes of NAbs have been identified against multiple RBD epitopes, including the RBM face (class I), the outer faces (class II/III), and the cryptic but conserved inner surface (class IV).14,28, 29, 30 The reduced susceptibility of XBB, EG.5.1 and BA.2.86 sublineages to neutralization by such NAbs resulted from resistant mutations such as N460K, L455 F/S, F456L and F486 V/S/P evading class I/II NAbs, and R346T, K356T, K444T, V455P/H, G446S as well as F490S evading class III NAbs, respectively.12,18,22,31 Despite the NAb escape, these prevalent variants showed increased binding affinity to the host receptor ACE2 compared with WT (D614G).12,18,19 Given the huge diversity of B cell receptor (BCR) repertoire, we continued the search for ultrapotent bnAbs.32,33 In this study, we discovered a new panel of ultrapotent class I bnAbs with fitness for blockade of viral binding to receptor ACE2 and efficacy for preventing the contact transmission of BA.5.2, BQ.1.1 and XBB.1.5 sublineages.
Methods
Experimental model and subject details
Human subject
11 Omicron BA.2 case-patients7 receiving 3 doses of Sinovac (n = 5) or BNT162b2 (n = 6), and one Omicron BA.5 case-patient13 who received 3 doses of BNT162b2 before symptom onset were recruited for voluntary blood donation. Clinical and laboratory findings were entered into a predesigned database. Written informed consent was obtained from all study subjects. Blood samples were collected by professional clinical doctors and separated into plasma and peripheral blood mononuclear cells (PBMCs) by Ficoll-Hypaque gradient centrifugation. All plasma samples were heat-inactivated at 56 °C for at least 30 min before the test.
Live viruses
Authentic SARS-CoV-2 included Omicron BA.5.2 (hCoV-19/Hong Kong/HKU-220712-005/2022; GISAID: EPL_ISL_13777658), Omicron BQ.1.1 (hCoV-19/Hong Kong/HKU; GISAID: EPL_ISL_16342297), and Omicron XBB.1.5 (hCoV-19/Hong Kong/HKU; GISAID: EPL_ISL_17205250) were isolated from the combined nasopharyngeal-throat swabs of patients with COVID-19 in Hong Kong, respectively.34 All experiments involving authentic SARS-CoV-2 followed the approved standard operating procedures in Biosafety Level 3 (BSL-3).
Cell lines
Cell lines were maintained as previously described.35 HEK293T cells were cultured in DMEM containing 10% FBS+100 U/mL penicillin and incubated at 37 °C in a 5% CO2 setting. The above medium added with puromycin (1 μg/mL) was used for culturing HEK293T-hACE2, respectively. Expi293FTM cells were cultured in Expi293TM Expression Medium (Thermo Fisher Scientific) at 37 °C in an incubator with 80% relative humidity and a 5% CO2 setting on an orbital shaker platform at 125 ± 5 rpm/min (New Brunswick Innova™ 2100) according to the manufacturer's instructions.
Method details
Isolation of SARS-CoV-2 RBD/spike-specific IgG + single memory B cells by FACS
RBD/spike-specific single memory B cells were sorted as previously described.13,35 In brief, PBMCs from vaccinated convalescent donors were collected and incubated with an antibody cocktail and a His-tagged SARS-CoV-2 RBD/spike protein mixture (Sino Biological) for identification of RBD/spike-specific B cells. WT/BA.1/BA.2 RBDs combined with WT/BA.1/BA.2 spikes were used as sorting antigens for donor CUs, and BA.1/BA.2/BA.2.12.1/BA.5 spikes were used for donor ZC (5 μg/protein), respectively. The cocktail consisted of the Zombie viability dye (Biolegend), CD19-Percp-Cy5.5, CD3-Pacific Blue, CD14-Pacific Blue, CD56-Pacific Blue, IgM-Pacific Blue, IgD-Pacific Blue, IgG-PE, CD27-PE-Cy7 (2 μL/test, BD Biosciences or Biolegend). The mixture consisted of His-tagged WT, Omicron BA.1, BA.2, BA.2.12.1, or BA.5 RBD/spike. Two consecutive staining steps were conducted: the first one used an antibody and RBD/spike cocktail incubation of 30 min at 4 °C; the second staining involved staining with anti-His-APC (10 μL/test Abcam) and anti-His-FITC antibodies (2 μL/test Abcam) at 4 °C for 30 min to detect the His-tag of RBD/spike. The stained cells were washed and resuspended in PBS containing 2% FBS before being strained through a 70-μm cell mesh filter (BD Biosciences). SARS-CoV-2 RBD/spike-specific single B cells were gated as CD19+CD27+CD3-CD14-CD56-IgM-IgD-IgG + RBD/spike+ and sorted by FACSAria III cell sorter (BD) into 96-well PCR plates containing 10 μL of RNAase inhibiting RT-PCR catch buffer (1 M Tris–HCl pH 8.0, RNase inhibitor, DEPC-treated water, Thermo Scientific). Plates were then snap-frozen on dry ice and stored at −80 °C until the reverse transcription reaction. The population analysis of antigen-specific memory B cells was performed by FlowJo V10. RT-PCR was performed on isolated memory B cells, followed by antibody cloning, genetic analysis of the BCR repertoire and antibody production. For detailed information, see the Supplementary Methods.
ELISA analysis of antibody binding to trimeric spike
ELISA was performed as previously described.35 The recombinant trimeric spike proteins derived from SARS-CoV-2 (Sino Biological) were diluted to final concentrations of 1 μg/mL, coated onto 96-well plates (Corning 3690) and incubated at 4 °C overnight. Plates were washed with PBST (PBS containing 0.05% Tween-20) and blocked with blocking buffer (PBS containing 4% skim milk) at 37 °C for 1 h. Serially diluted isolated monoclonal antibodies were added to the plates and incubated at 37 °C for 1 h. Wells were then incubated with a secondary goat anti-human IgG labeled with horseradish peroxidase (HRP) (1:5000 Invitrogen). TMB substrate (SIGMA). Optical density (OD) at 450 nm was measured by SkanIt RE6.1 with VARIOSKAN Lux (Thermo Scientific). Competition ELISA was performed on antibodies for epitope mapping. For detailed information, see the Supplementary Methods.
Pseudovirus-based neutralization assay
The neutralizing activity of NAbs was determined using a pseudotype-based neutralization assay as previously described.35 Briefly, the pseudovirus was generated by co-transfection of HEK 293 T cells with pVax-1-S-COVID19/pCMV3-SARS-CoV-2-S_Δ18 and pNL4-3Luc_Env_Vpr, carrying the optimized gene of spike (S) and the human immunodeficiency virus type 1 backbone, respectively.3 The viral supernatant was collected 48 h post-transfection and frozen at −80 °C until use. The serially diluted monoclonal antibodies or sera were incubated with 200 TCID50 of pseudovirus at 37 °C for 1 h. The antibody-virus mixtures were subsequently added to pre-seeded HEK 293 T-ACE2 cells. 48 h later, infected cells were lysed to measure luciferase activity using a commercial kit (Promega, Madison, WI). Half-maximal inhibitory concentrations (IC50) of the evaluated antibody were determined by inhibitor vs. normalized response-3 Variable slope using GraphPad Prism 9.4.1.
Antibody binding kinetics of antibodies measured by surface plasmon resonance (SPR)
The binding kinetics and affinity of antibodies for the Omicron BA.5 spike trimer protein (Sino Biological) were analyzed by SPR (Biacore T200, Cytiva) as previously described.35 Specifically, the spike protein was covalently immobilized to a CM5 sensor chip via amine groups in 10 mM sodium acetate buffer (pH 5.0) for a final RU of around 250. SPR assays were run at a flow rate of 10 μL/min in HEPES buffer. For conventional kinetic/dose–response, serial dilutions of monoclonal antibodies were injected across the RBD protein surface for 180 s, followed by a 900 s dissociation phase using a multi-cycle method. The remaining analytes were removed in the surface regeneration step with the injection of 10 mM glycine-HCl (pH 1.5) for 60 s at a flow rate of 30 μL/min. Kinetic analysis of each reference subtracted injection series was performed using the Biacore Insight Evaluation Software (Cytiva). All sensorgram series were fit to a 1:1 (Langmuir) binding model of interaction.
Cryo-EM data collection and processing
Fab fragments of antibodies were prepared for cryo-EM data collection following the instruction of Pierce™ Fab Preparation Kit (Thermo). For detailed information, see the Supplementary Methods. Cryo-EM data collection was conducted using a Titan Krios G3 transmission electron microscope. It is equipped with a Gatan K3 direct electron detector and a Bio Quantum energy filter with a 20 eV slit width in the Biological Cryo-EM Center of HKUST. Automated data collection was performed using the EPU software manufactured by Thermo Fisher, with 40-frame movies collected at a total dose of about 50 e-/Å2 and 4.5 s exposure time. The counting mode of K3 was utilized to collect data, with the magnitude set at 81,000x and the defocus range set at −1.0 to −2.5 μm. The collected movies for ZCP3B4, ZCP4C9, ZCP4D5-1, and CUP2G3 were motion corrected and CTF estimated in cryoSPARC.36 The micrographs were manually checked. Particles were picked based on a reference template and extracted from the micrographs. The extracted particles were subjected to 2D classification. Particles from good 2D classes were selected for further 3D classification. For all four datasets, the data processing workflows were generally similar. Particles were picked based on the 2D average results, and the initial model was constructed by the picked particles with clear features and functioned as a reference for further heterogeneous refinement. CTF refinement and non-uniform refinement were used to obtain a higher-resolution map. For focused 3D classification, particles with clear density of bound Fabs were selected, and flexible refinement by cryoSPARC was utilized to resolve the flexible RBD-Fab region more clearly. For the ZCP4D5-1-Spike complex, we imported the particles into RELION37 for an additional focused 3D classification during the data processing.
Model building
The Omicron BA.5 RBD model was built based on the BA.1 RBD model (PDB code: 7U0N),38 and models for Fabs were predicted by AlphaFold.39 These models were used as initial models to dock into the cryo-EM density map, and the initial models were adjusted manually using Coot40 and refined in Coot and Phenix.41 Structural analysis was conducted in UCSF Chimera42 and UCSF ChimeraX.43
Hamster model and experiments
In vivo evaluation of monoclonal antibodies in the established golden Syrian hamster model of SARS-CoV-2 infection was performed as described previously.13,44,45 Female and male hamsters were purchased from the Chinese University of Hong Kong Laboratory Animal Service Centre through the HKU Centre for Comparative Medicine Research. 4–5 hamsters were housed per cage with sawdust bedding and environment enrichment. The hamsters were housed with access to standard pellet feed and water ad libitum until the live virus challenge in our BSL-3 animal facility. They were randomized from different litters into experimental groups. Experiments were performed in compliance with the relevant ethical regulations.44 For prophylaxis studies,13 1 day before the live virus challenge, three groups of hamsters were intraperitoneally injected with one dose of bnAbs ZCP3B4, ZCP4C9 or ZCP4D5-1 in PBS at 4.5 mg/kg, respectively. At day 0, each hamster was intranasally inoculated with a challenge dose of 100 μL of Dulbecco's Modified Eagle Medium containing 105 PFU of a mixture of Omicron BA.5.2, BQ.1.1 and XBB.1.5 variants (1:1:1 ratio of PFU, 1:1.2:1.9 ratio of RNA copies) under anesthesia with intraperitoneal ketamine (200 mg/kg) and xylazine (10 mg/kg). Accordingly, all hamsters were sacrificed for analysis at 4 days post infection (dpi) after virus challenge with high viral loads.44 The contact transmission studies were performed as previously described45 with slight modifications. Briefly, index hamsters were intranasally challenged with 105 PFU of a mixture of Omicron BA.5.2, BQ.1.1 and XBB.1.5 variants (1:1:1 ratio) at 0 dpi. Two groups of hamsters were intranasally administered with PBS and ZCP3B4 at 2 dpi, respectively. 8 h later, each virus-challenged index hamster was transferred to a new cage with two PBS- or antibody-treated hamsters as close contact. They were co-housed for 4 h before being transferred to separate new cages. The index and contact hamsters were sacrificed at 2 dpi and 2 days post-exposure, respectively. The hamsters were monitored daily for clinical signs of disease. Syrian hamsters typically clear viruses within one week after SARS-CoV-2 infection. Quantification of relative amounts of Omicron variants in tissues was performed by the next-generation sequencing. Quantification of the infectious virus was performed by plaque assay. Histopathological analysis or immunofluorescence (IF) staining was performed on harvested tissues. For detailed information, see the Supplementary Methods.
In vivo evaluation of the lead bnAb ZCP3B4 given at lower doses was performed as described above. For prophylaxis studies, two groups of hamsters were intraperitoneally injected with one dose of ZCP3B4 at 0.5 mg/kg and 1.5 mg/kg, respectively. For contact transmission studies, two groups of hamsters were intranasally administered with ZCP3B4 at 0.5 mg/kg and 1.5 mg/kg, respectively. The hamsters were monitored daily for clinical signs of disease. Quantification of the infectious virus was performed by plaque assay.
Ethics statement
This acquisition of blood samples from vaccinated convalescent donors for identification of broad neutralizing activities and isolation of potent monoclonal antibodies against COVID-19 received approval from the Institutional Review Board of The University of Hong Kong/Hospital Authority Hong Kong West Cluster (Ref No. UW 21-452). The research was conducted in strict accordance with the rules and regulations of the Hong Kong government for the protection of human subjects. The study subjects agreed and signed the written informed consents for research use of their blood samples and indirect identifiers.
All experimental procedures were approved by the Committee on the Use of Live Animals in Teaching and Research (CULATR 5518-20) of the University of Hong Kong.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 9.4.1 as previously described.35 Kolmogorov–Smirnov test was used for the normality test before group comparison analysis. For data passing the normality test, ordinary one-way ANOVA (parametric) and Tukey's multiple comparisons test were used to compare group means and differences between multiple groups. For data not passing the normality test, Kruskal–Wallis test (nonparametric) and Dunn's multiple comparisons test were used. Each p value is adjusted to account for multiple comparisons, and p < 0.05 was considered significant. The number of independent replicates performed, the number of animals in each group, and the specific details of statistical tests are reported in the figure legends and the methods section. Animal sample size was determined by the ‘E' value, where E = total number of animals-total number of groups. Based on the ANOVA, the animal size, which lies between 10 and 20, should be considered adequate. No randomization, blinding or inclusion/exclusion criteria were performed during the experiment or data analysis.
Role of funders
The funding source did not have any involvement in study design, data collection, data analyses, interpretation, writing of report, or decision to submit it for publication.
Results
Isolation of NAbs from convalescents after Omicron breakthrough infection
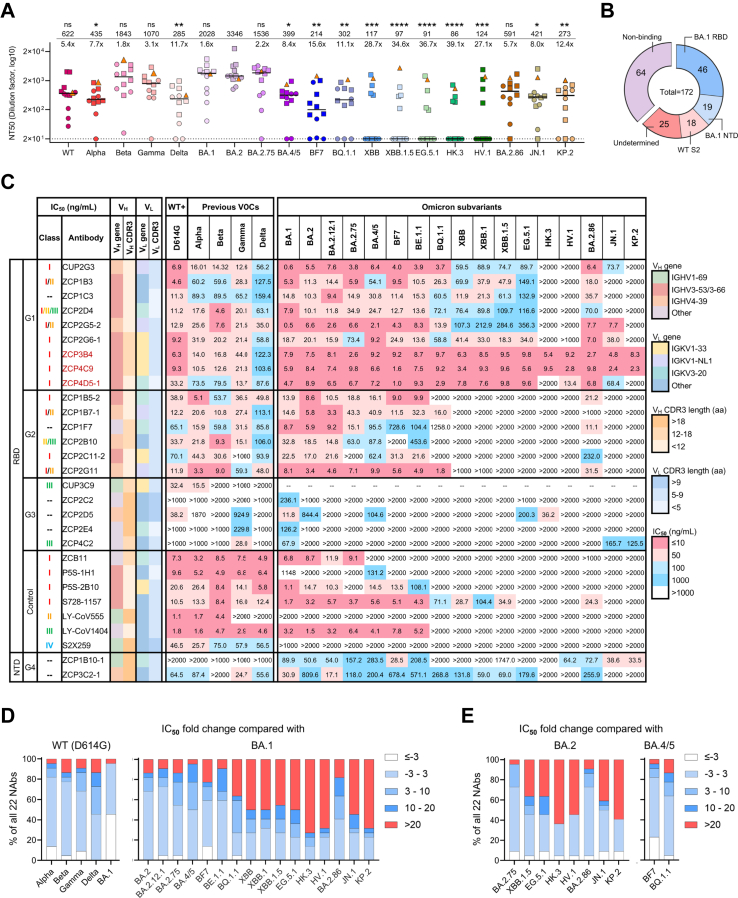
To search for ultrapotent bnAbs against SARS-CoV-2 variants, we screened plasma samples derived from a cohort of 12 convalescent people at median 142 days after vaccine breakthrough infection by Omicron BA.2 (n = 11, named CU) and BA.5 (n = 1, named ZC) in Hong Kong7,13 (Supplementary Table S1). The subjects in this cohort received 3 doses of vaccines (Sinovac, 5 CUs; BNT162b2, 6 CUs and ZC) at median 76 days prior to the infection. They displayed polyclonal neutralizing antibody responses to the pseudotyped WT and corresponding exposed variants (Fig. 1A). Other antigenically distinct variants XBB, EG.5.1 and their descendant lineages,12,22 however, displayed relative resistance to the serum neutralization. The geometric mean values of neutralization titers were significantly lower against these variants as compared to BA.2 with reductions of 11.7- to 39.1-fold. Only 4 CUs and ZC, who all received prior BNT162b2 vaccinations, had polyclonal NAbs cross-reactive to all variants tested (Supplementary Figure S1). To delineate the B cell receptor (BCR) repertoire, 150 WT/BA.1/BA.2 RBD/spike-specific and 210 BA.1/BA.2/BA.2.12.1/BA.5 spike-specific single memory B cells were sorted from CUs and ZC, respectively (Supplementary Figure S2). We then successfully obtained 172 monoclonal antibodies (mAbs) with naturally paired heavy chain (HC) and light chain (LC) sequences. The binding profile of antibody repertoire revealed that among mAbs specific to BA.1 spike (62.8%, 108/172), 42.6% (46/108) displayed binding activity to RBD as determined by ELISA (OD450 > 0.1; Fig. 1B and Supplementary Figure S3). Subsequently, 22 NAbs (20 from ZC and 2 from CUs) were identified based on their activity to neutralize pseudotyped WT or BA.1 (Supplementary Figure S3). Epitope mapping through competition with four previously defined RBD class I-IV NAbs (S2E1246 (class I), LY-CoV55547 (class II), LY-CoV140448 (class III) and S2X25949 (class IV)) indicated that most of the RBD-targeted NAbs belonged to class I-III (Supplementary Figure S4 and Supplementary Table S2). These results demonstrated consistently that vaccine breakthrough infections had activated diverse BCRs for generating multiple classes of NAbs as we and others previously described.18,20
Fig. 1.
Isolation of cross-reactive NAbs from convalescents after Omicron BA.2/BA.5 breakthrough infection. (A) Neutralization of pseudotyped SARS-CoV-2 WT and 18 variants by sera from a cohort of convalescents (n = 12). Neutralizing titer 50 (NT50) values represent the plasma dilution required to achieve 50% virus neutralization. The limit of detection is 20 (dash line). The triangle, squares and dots represent ZC (BNT162b), 6 CUs (BNT162b2) and 5 CUs (Sinovac), respectively, with a line indicating the median of each group. Geometric mean NT50 values are shown upon the symbols, and the fold reduction in geometric mean NT50 values for WT and each variant compared to BA.2 is also shown. Comparisons were made by Kruskal–Wallis test followed by Dunn's multiple comparisons test. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001; ns (not significant), p > 0.05. (B) Epitope specificity of 172 recovered mAbs as determined by ELISA. (C) Neutralization profiles of 22 newly identified NAbs against SARS-CoV-2 WT (D614G), previous VOCs and Omicron subvariants. IC50 values of all tested NAbs against the whole panel of pseudoviruses are summarized in the graph. The top three ultrapotent bnAbs are highlighted in red. Published NAb controls from class I-IV (ZCB11, P5S-1H1, P5S-2B10, S728-1157, LY-CoV555, LY-CoV1404 and S2X259) are included. The neutralizing potency is indicated according to the color bar. The antibody germline gene usage (IGHV and IGKV) and the length of CDR3 are also presented. aa, amino acid. (D) Fold changes in IC50 values of 22 NAbs against previous VOCs including Alpha, Beta, Gamma, Delta and Omicron BA.1 compared with the WT (D614G) (left), or against Omicron subvariants compared to BA.1 (right). (E) Fold changes in IC50 values of 22 NAbs against Omicron BA.2.75, XBB.1.5, HK.3, HV.1, EG.5.1, BA.2.86, JN.1 and KP.2, compared to ancestral BA.2 (left), or against BF7 and BQ.1.1 compared with ancestral BA.4/5 (right).
Ultrapotent bnAbs cross-neutralize SARS-CoV-2 VOCs and Omicron subvariants
Many NAbs isolated from convalescents after Omicron breakthrough infection to date mainly neutralize the exposed strain with limited breadth and few ultrapotent bnAbs have been found.18,20 We then evaluated the in vitro neutralizing activity of 22 newly identified NAbs. They showed diverse neutralizing profiles and some had great efficacy against a panel of pseudotyped WT (D614G) and 21 variants, including previously described variants of interest (VOIs) as well as Omicron subvariants prevalent in recent waves of COVID-19 (Supplementary Figures. S5A and S6A). Based on the epitope specificity and breadth of neutralization, these 22 NAbs were divided into four groups (Fig. 1C). Group 1 NAbs included 9 RBD-specific bnAbs (40.9%, 9/22) displaying the best breadth and potency against pseudoviruses tested, especially against XBB, EG.5.1 and BA.2.86 lineages compared with other NAbs in group 2–4. Of note, the top ultrapotent bnAbs were ZCP3B4 and ZCP4C9 based on their IC50 values consistently being less than 10 ng/mL against Omicron subvariants. Group 2 NAbs were particularly compromised by BQ.1.1, XBB, EG.5.1 or JN.1 sublineages, which might result from specific convergent mutations.12 Group 3 NAbs were effective for only a few variants. Their poor breadth was attributed to the weak binding activity to various spike trimers (Supplementary Figure S7). Finally, group 4 NAbs were NTD-specific, including a moderate bnAb ZCP3C2-1 and an Omicron-specific ZCP1B10-1. In previous studies on Omicron breakthrough infection, most pooled memory B cells from convalescent individuals were cross-reactive, demonstrating that the breakthrough infection mainly recalled memory B cells elicited by WT-based vaccines, but rarely recruited Omicron-restricted naive B cells.20,50, 51, 52, 53 Consistently, all group 1–4 NAbs except for CUP3C9 cross-bound to the spikes of both WT (D614G) and the Omicron variants (Supplementary Figure S7), probably derived from recalled responses.
We also compared the fold changes in IC50 values of these 22 NAbs against WT (D614G) and BA.1, respectively. There was no significant waning of neutralizing activity for most NAbs (>3-fold change in IC50 values) against Alpha, Beta, Gamma and BA.1 compared with WT (D614G). More than 50% of NAbs, however, were compromised against Delta (Fig. 1D, left). When recently emerged Omicron variants were compared with BA.1, about 50% of NAbs neutralizing BA.2, BA.4/5 and BE.1.1 lineages were of comparable potency. However, 60–86% of NAbs showed partially or completely diminished activity against BQ.1.1, XBB, EG.5.1 as well as recently circulating JN.1 and KP.2 sublineages (Fig. 1D, right). Similar results were obtained when descendent lineages were compared with ancestral BA.2 (Fig. 1E). Overall, these results indicated that several ultrapotent class I bnAbs were elicited after the Omicron BA.2/BA.5 breakthrough infection by recalling memory B cell responses.
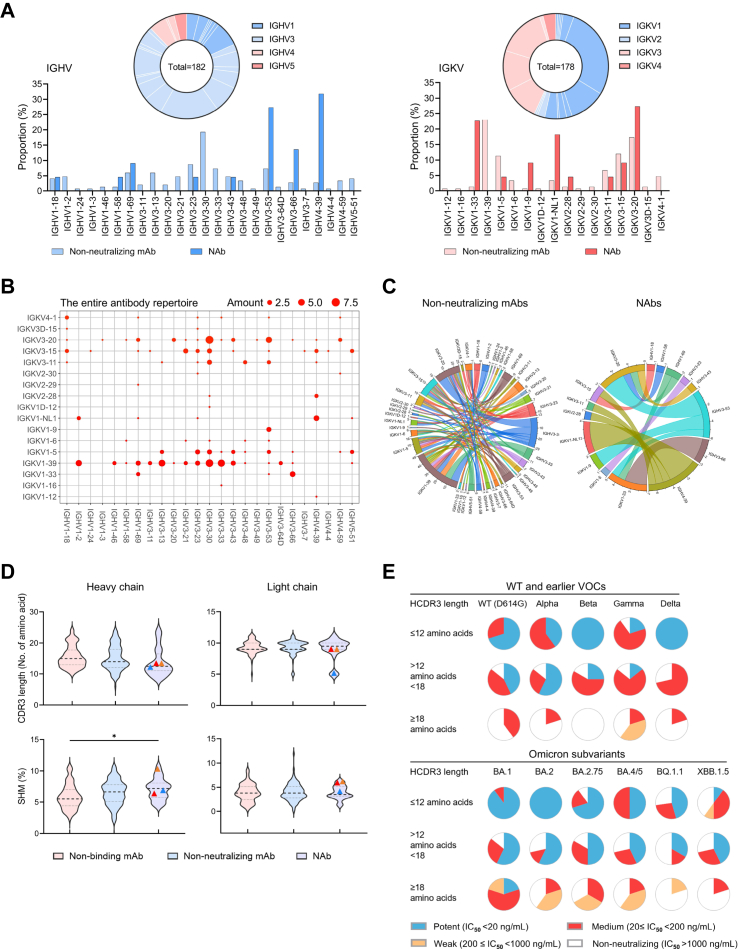
Ultrapotent class I bnAbs favor IGHV3-53/3–66 genes
Previously reported NAbs isolated from naturally infected or vaccinated individuals showed distinct patterns of BCR germline gene usage in HC and LC.6,54 Based on our analysis of all cloned antibody V genes, the HC V genes of 22 newly identified NAbs were enriched with 72.7% IGHV3-53/3–66/4-39 among 25 sub-families, and the LC V genes involved preferentially 68.2% IGKV1-33/1-NL1/3–20 among 19 sub-families (Fig. 2A). While the entire antibody repertoire displayed a pool of diverse clonotypes (Fig. 2B), NAbs belonged to IGHV3-53/3–66 (41%) and IGHV4-39/IGKV1-NL1 (18.2%) antibodies, respectively, as compared with non-NAbs (Fig. 2C). The predominance of IGHV3-53/3–66 paired with various LC genes has previously been documented for class I NAbs.55,56 Our three ultrapotent bnAbs ZCP3B4, ZCP4C9 and ZCP4D5-1 belonged to IGHV3-53/3–66 (Supplementary Tables S3 and S4). In contrast, the usage of IGHV4-39/IGKV1-NL1 by four NAbs obtained from two individuals (ZCP1B7-1, ZCP2G5-2, ZCP2G11 and CUP2G3) has rarely been found among previously published NAbs.57 Their over 98% identical VH and VL sequences (Supplementary Figure S8A) suggested a potential public antibody response.58,59 Subsequently, we analyzed clonal relatedness among all recovered mAbs with shared germline usage to understand their evolutionary relationship. ZCP2E4 was the only NAb identified from expanded clones (Supplementary Figure S8B). Despite the diverse neutralizing activity, there were no significant differences between non-NAbs and NAbs in the length of complementarity determining region 3 (CDR3) or the rate of somatic hypermutation (SHM) in both HC and LC (Fig. 2D). The ultrapotent bnAbs ZCP3B4, ZCP4C9 and ZCP4D5-1 consistently had short CDR3 lengths55,60 around or below the median of all NAbs (Fig. 2D, symbols). Apparently, NAbs with relatively longer CDR3 did not necessarily result in enhanced potency against SARS-CoV-2 variants (Fig. 2E). Overall, these results demonstrated that ultrapotent bnAbs elicited by Omicron breakthrough infection in the study subject ZC were preferentially encoded by IGHV3-53/3–66 with necessary SHM improving fitness for antigen interaction.
Fig. 2.
Immunogenetic properties of the antibody repertoire induced by Omicron breakthrough infection. (A) Antibody gene repertoire analysis of reactive memory B cells derived from 12 convalescent individuals. In pie charts, the number of all cloned antibody V genes is shown in the center for the heavy (left) or light chains (right). The colors represent variable gene families, and each fragment of the same color stands for one specific sub-family. The histograms summarize the IGHV (left) and IGKV (right) gene usage of 172 recovered mAbs, including 150 non-neutralizing mAbs (non-NAbs) and 22 NAbs, labeled in light and dark colors, respectively. (B and C) Parings of germline heavy and light V genes display preference among NAbs. (B) IGHV and IGKV pairings of 172 recovered mAbs are presented in the bubble diagram and (C) the diversity between non-NAbs and NAbs is indicated in chord diagrams. The outer circle border indicates the number of each pairing. (D) Comparison of CDR3 lengths and SHM rates between recovered non-binding mAbs (n = 64), non-NAbs (n = 86) and NAbs (n = 22). Amino acid lengths and SHM rates compared to germline sequences are presented in violin plots with kernel density estimation curves of the distribution. A dash line and two dotted lines indicate the median and quartiles of each group, respectively. bnAbs ZCP3B4 (red), ZCP4C9 (orange) and ZCP4D5-1 (blue) are presented by symbols. Comparisons were made by one-way ANOVA followed by Tukey's multiple comparisons test. ∗p < 0.05. (E) Neutralizing potency distribution of 22 NAbs against WT (D614G), earlier VOCs and Omicron subvariants. The NAbs are grouped according to their overall CDR-H3 amino acid (aa) lengths as follows: short (≤12 aa, upper panel), intermediate (12–18 aa, mid panel), or long (≥18 aa, lower panel).
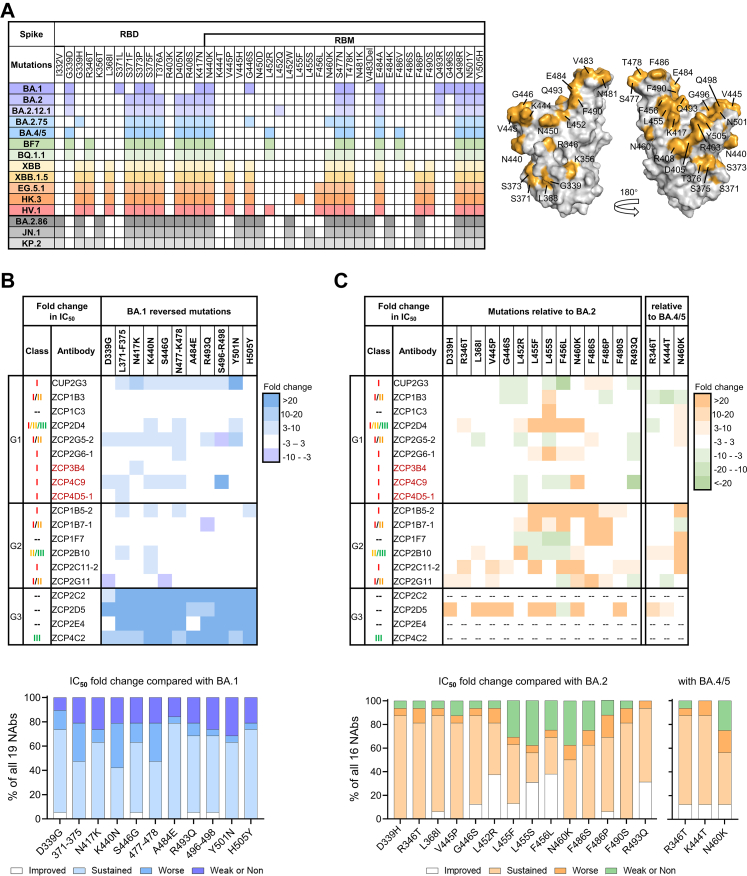
Inference of Omicron RBD mutation hotspots on antibody neutralization and evasion
Although immune imprinting was frequently observed in Omicron breakthrough infections, the recalled WT-primed memory B cells were likely to acquire increased affinity and neutralization potency against exposed viral variants.53,61 Expectably, we observed that compared to WT (D614G), more than 40% of newly identified NAbs had an improvement in neuralization potential against BA.1 (Fig. 1D), whose mutations are shared by BA.2, BA.4/5 and some other Omicron variants (Fig. 3A). To determine the possible impact of common mutations in Omicron RBD on antibody evolution, group 1–3 NAbs were screened for their neutralizing activities against a panel of 11 BA.1-derived pseudoviruses that carried mutations reversing to ancestral residues (Supplementary Figure S9). By measuring fold changes in IC50 values compared to parental BA.1 (Fig. 3B, top), we found that the activities of most NAbs in groups 1–2, including three ultrapotent bnAbs, were influenced moderately (<10-fold). In contrast, NAbs in group 3 displayed a >20-fold loss in IC50 values against the majority of pseudoviruses tested. These results suggested that most NAbs in group 1–2 likely directed at conserved epitopes of breakthrough Omicron variants. Moreover, it is possible that they have accommodated the evolved viral mutations. For example, S371F/L-S375F, N440K and S477N-T478K found in Omicron variants contributed to the increased neutralizing activity of over 50% of NAbs tested (Fig. 3B, bottom), probably by promoting antibody affinity maturation via SHM.
Fig. 3.
Inference of Omicron RBD mutation hotspots on NAb neutralization and evasion. (A) Specific RBD mutations carried by each Omicron subvariant. Mutation sites are colored on the WT RBD (PDB: 7B3O) (right). (B) Fold changes in IC50 values against BA.1 individual reversed mutations compared with BA.1. 19 anti-BA.1 NAbs in group1-3 were tested (top). Ranges are indicated according to the color bar. The top three ultrapotent bnAbs are highlighted in red. Impact of individual reversed mutations on tested NAbs potency as determined by proportions of improved (IC50 fold change < -3), sustained (−3≤ IC50 fold change ≤3), worse (3 < IC50 fold change ≤10) and weak/non (IC50 fold change >10) NAbs (bottom). (C) Fold changes in IC50 values against related convergent mutations compared with BA.2 or BA.4/5. 16 anti-BA.2/BA.4/5 NAbs in group1-3 were tested (top). Ranges are indicated according to the color bar. The top three ultrapotent bnAbs are highlighted in red. Impact of individual convergent mutations on tested NAbs potency as determined by proportions of improved (IC50 fold change < -3), sustained (−3≤ IC50 fold change ≤3), worse (3 < IC50 fold change ≤10) and weak/non (IC50 fold change >10) NAbs (bottom).
Considering that convergent Omicron RBD mutations cause the antibody-escape of emerging subvariants,12,20 we evaluated the activity of 16 anti-BA.2/BA.4/5 NAbs against corresponding pseudoviruses with individual mutations (Supplementary Figures S10 and S11). As indicated by fold changes in IC50 values compared with parental BA.2 or BA.4/5 (Fig. 3C, top), bnAbs in group 1, especially ultrapotent ZCP3B4 and ZCP4C9, were much less vulnerable to the whole panel tested. However, ultrapotent ZCP4D5-1 that tolerated individual mutations was escaped by the HK.3 and newly emerged KP.2 variant, probably owing to multiple synergetic mutations.17,23 The activity of group 2–3 NAbs, on the other hand, was significantly impaired by several mutations, leading to the complete loss of neutralization against BQ.1.1, XBB, EG.5.1 and BA.2.86 sublineages. L455F/S, F456L, N460K and F486S were the hotspots, conferring >3-fold resistance for 31–45% of NAbs tested (Fig. 3C, bottom). Notably, L455 F/S enhanced HV.1 and JN.1's ability to evade class I antibodies.27 The remaining mutations, including L368I, V445P and F490S found in XBB sublineages, showed limited impact on antibody evasion except for reversed R493Q. Interestingly, R493Q not only benefited ACE2 binding11,20 but also compensated for the activity loss of 31% of NAbs, including the ultrapotent bnAb ZCP4C9. These results suggested that epitopes of ultrapotent bnAbs ZCP3B4 and ZCP4C9 in group 1 might include residues critical for viral fitness and less likely to mutate as viral variants evolved.62
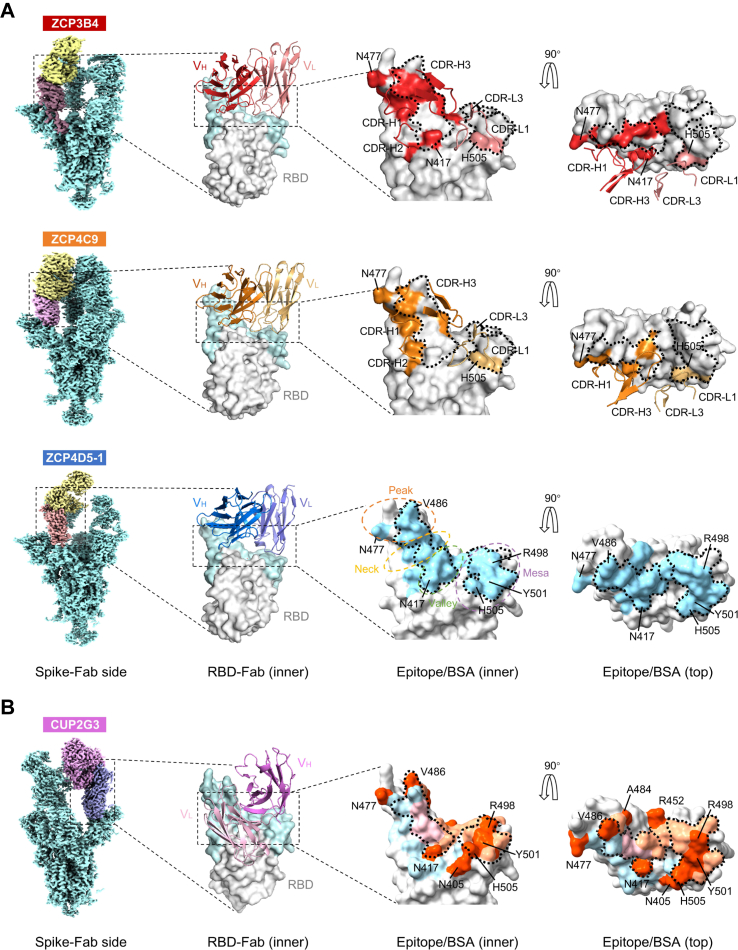
Structural insights into ultrapotent bnAbs
To reveal the molecular mechanisms of top ultrapotent bnAbs, we determined the structures of ZCP3B4, ZCP4C9 and ZCP4D5-1. Another class I bnAb CUP2G3 from a rarely reported clonotype probably adopted a unique binding mode, and thus its structure was also investigated. These bnAbs showed high-affinity binding to the BA.5 spike trimer at sub-nM concentrations (Supplementary Figure S12). Their Fabs in complex with BA.5 trimers were used for single particle cryo-EM analysis. ZCP3B4- and ZCP4C9-Spike complexes were determined at global resolutions of 3.88 Å and 3.68 Å (Supplementary Figures S13 and S14), respectively. To further explore the interactions between Fabs and RBDs, we conducted the flexible refinement to improve the local resolution of interactive regional maps at 4.08 Å (ZCP3B4) and 3.86 Å (ZCP4C9), allowing for atomic model building (Supplementary Figure S17 and Supplementary Table S5). Due to the relatively higher flexibility of the RBD-Fab regions, ZCP4D5-1- and CUP2G3-Spike complexes were determined at resolutions of 4.0 Å and 4.33 Å (Supplementary Figures S15-S17), respectively. We managed to identify their binding areas and describe features in broad terms since the allocation of side chains in predicted models was influenced by relatively low resolutions.
We found that BA.5 RBDs were all in the ‘up’ conformation after the binding of Fabs (Fig. 4A, Spike-Fab side), indicating that these bnAbs belonged to class I NAbs as previously described.28 However, their buried areas varied due to disparate mainchain distances between antibodies and RBD, as well as the angles of approach (Supplementary Video S1). ZCP3B4 buried a smaller surface area of 982.9 Å2, primarily contributed by VH (17 BSA residues) as its VL was further away from the top right of RBD (Supplementary Figure S18A and Supplementary Table S6, top). ZCP4C9 and ZCP4D5-1 relied on both VH and VL to bury surface areas of 1301.4 Å2 (29 BSA residues) and 1112.7 Å2 (22 BSA residues), respectively (Fig. 4A, Epitope/buried surface area (BSA), Supplementary Figure S19A and Supplementary Table S6, top). At the binding interfaces, ZCP3B4 and ZCP4C9 consistently utilized complementarity-determining region (CDR)-H1/2/3 and CDR-L1/3 to target RBD (Fig. 4A, Epitope/BSA) and formed tight atomic interactions (Supplementary Figures S18B and S19B). Their ‘Jing’ (a still principle to cope with changes) epitopes were conformational and primarily located on RBM, extensively overlapping with the binding site of ACE2 (Fig. 4A, Epitope/BSA). Moreover, the footprints of ZCP3B4 and ZCP4C9 avoided most of the mutation sites in RBD to minimize the impact of immune escape from Omicron subvariants. They did not contain any convergent mutations20 carried by BQ.1.1, XBB, EG.5.1 or BA.2.86 sublineages (Supplementary Table S6, bottom). ZCP4D5-1 shared most BSA residues with ZCP4C9 (Supplementary Table S6, top), but did not display comparable potency against HK.3, HV.1, JN.1 and KP.2, indicating their differences in binding epitopes. These results explained the molecular mechanism underlying ultrapotent class I bNAbs with fitness for breadth across a broad range of SARS-CoV-2 variants.
Fig. 4.
Structural basis for class I bnAbs ZCP3B4, ZCP4C9, ZCP4D5-1 and CUP2G3. (A) Binding modes and footprints of ZCP3B4, ZCP4C9 and ZCP4D5-1. Cryo-EM density maps of BA.5 spike trimers in complex with bnAb Fabs are shown in the side views. ‘Up’ RBDs of complexes are colored in dark brown (ZCP3B4), purple (ZCP4C9) and light orange (ZCP4D5-1), whereas bnAb Fabs are colored in yellow. The cartoons represent the structures of bnAb heavy chain and light chain variable regions (VH and VL) binding RBD (gray), viewed from the RBD inner face. The receptor binding motif (RBM) is colored in light cyan. HCDRs and LCDRs of ZCP3B4 and ZCP4C9 involved in the interaction are shown in the zoom-in figures. Epitopes of ZCP3B4 and ZCP4C9 and the buried surface area (BSA) of ZCP4D5-1 are shown in corresponding colors on the RBD surface viewed from the inner and top faces. BA.5 mutation sites involved in epitopes/BSA are also indicated. The RBM is topologically divided into ‘peak’, ‘neck’, ‘valley’ and ‘mesa’ subsections. The ACE2 binding site is outlined with dotted lines. PDB codes: 8K19 (BA.5 RBD-ZCP3B4 Fab) and 8K18 (BA.5 RBD-ZCP4C9 Fab). (B) Binding mode and footprint of CUP2G3. Cryo-EM density map of BA.5 spike trimer in complex with CUP2G3 Fabs is shown in the side view. The ‘up’ RBD of complex is colored in purple and Fab is in pink. The cartoon represents the structure of CUP2G3 heavy chain and light chain variable regions (VH and VL) binding RBD (gray), viewed from the RBD inner face. The receptor binding motif (RBM) is colored in light cyan. The buried surface areas (BSA) buried by VH (light orange), VL (light blue) and both (pink) are viewed from the inner and top faces. BA.5 mutation sites involved in the BSA are colored in orange. The ACE2 binding site is outlined with dotted lines.
ZCP3B4, ZCP4C9 and ZCP4D5-1 had better breadth and potency than recently isolated IGHV3-53/3–66 class I NAbs (e.g., P5S–2B10, P5S–1H154 and S728-115763) due to distinct binding epitopes (Supplementary Figure S20A). We characterized their genetic features compared to 103 CoV-AbDab57 NAbs against Omicron variants. Consistent with previous discoveries, the CDR-H3 lengths of ZCP3B4, ZCP4C9 and ZCP4D5-1 were relatively short and remained similar to published IGHV3-53/3–66 antibodies (Supplementary Figure S20B, left). Moreover, IGHV3-53/3–66 antibodies generally have limited mutations on HC.60 The geometric mean value of mutations was 8.5 for published NAbs, which were anti-BA.1 but escaped by XBB (n = 81). This value was slightly lower than 9.2 for published bnAbs, which remained cross-reactive to both BA.1 and XBB (n = 22) (Supplementary Figure S20B, right). In contrast, the geometric mean value of mutations among our IGHV3-53/3–66 antibodies was 13.9, which was significantly higher than those of previously published NAbs or bnAbs. Mutations accumulated by ZCP3B4, ZCP4C9 and ZCP4D5-1 were 12, 18 and 14, respectively. Although mutations such as A24V, G26E, F27I/L/V, T28I, S31R, S35N, V50I, I51M, S53P, S56T and Y58F have been found in previous IGHV3-53/3–66 antibodies60 (Supplementary Figure S20C) and single germline-reverted substitutions introduced in ZCP3B4, ZCP4C9 and ZCP4D5-1 (except for V27F and F58Y) did not significantly reduce their activity against XBB.1.5 (Supplementary Figure S20D), the combined mutations probably conferred the enhanced neutralization potency and breadth of our bnAbs.
The IGHV4-39/IGKV1-NL1-derived bnAb CUP2G3 adopted a previously unrecognized class I NAb binding mode in which VL and VH targeted the ‘neck’ and ‘mesa’ of RBM,29 respectively (Fig. 4B, Spike-Fab side and Supplementary Video S2). It buried a large RBD surface area (VH: 692.7 Å2; VL: 827.3 Å2) using both CDR1-3 and framework regions (FRs) (Fig. 4B, Epitope/BSA), involving 32 RBD residues that completely overlapped with ACE2 epitopes (Supplementary Table S6, top). As a result, the breadth of CUP2G3 was attributed to its expanded interaction with RBD for effective blocking of ACE2 binding. The buried area of CUP2G3 accommodated 15 SARS-CoV-2 mutation sites (RBD 403, 405, 417, 446, 452, 455, 456, 477, 484, 486, 493, 496, 498, 501 and 505), in which substitutions could impact the efficacy of previously reported class I NAbs.9,10,22,27,64 However, most of these mutations except for F486S/P did not diminish CUP2G3 neutralization (Fig. 3C), indicating that this bnAb was able to tolerate mutations within or close to epitopes. Due to this structural fitness, CUP2G3 maintained IC50 values below 10 ng/mL against most Omicron variants but with reduced potency against the XBB and EG.5.1 sublineages.
Efficacy of ZCP3B4, ZCP4C9 and ZCP4D5-1 against BA.5.2, BQ.1.1 and XBB.1.5
To determine the in vivo efficacy of ultrapotent ZCP3B4, ZCP4C9 and ZCP4D5-1, we tested these bnAbs in separate groups against live SARS-CoV-2 variants using our well-established golden Syrian hamster model13 (Fig. 5A). One day prior to the viral challenge, bnAbs were administered intra-peritoneally (i.p.) at 4.5 mg/kg in 3 groups of 15 hamsters (n = 5 per group), respectively. The control group (n = 6) was pretreated with PBS. Mixed viral challenge experiments have already been used to demonstrate the distinct replication fitness of co-circulating Omicron variants in animal models.65 We, therefore, sought to determine the protective potency of our lead bnAbs in hamsters challenged intranasally with 105 PFU of mixed BA.5.2, BQ.1.1 and XBB.1.5 at a 1:1:1 ratio to mimic the co-infection. Post–challenge, they were monitored for daily weight changes and euthanized at day 4 to harvest lungs and nasal turbinate (NT). XBB.1.5 had a competitive growth advantage over BA.5.2 and BQ.1.1 in both lungs and NT of hamsters without antibody pre-treatment (Fig. 5B), but both BA.5.2 and BQ.1.1 were still detected at low frequencies. The infection resulted in about 6% body weight loss over time in the PBS group, and no significant decrease was observed for bnAb-pretreated hamsters (Fig. 5C). Critically, pre-treatment with either ZCP3B4 or ZCP4C9 prevented the lung infection with undetectable infectious PFU, while ZCP4D5-1 reduced viral titers by more than 3 logs (Fig. 5D, left). In the following lower-dose studies, ZCP3B4 given at 1.5 mg/kg was equally effective (Supplementary Figure S21A and C, left). Minimal lung lesions were observed by histopathological analysis in hamsters pre-treated with bNAbs. Control hamsters, however, developed severe interstitial pneumonia with perivascular inflammatory cell infiltration, diffuse alveolar damage, alveolar septa thickness, edema of homogeneously pink materials and some hemorrhage foci (Fig. 5E, top). In the NT, lower infectious PFU were consistently observed in ZCP3B4- and ZCP4C9-pretreated hamsters (Fig. 5D, right), but tissue staining consistently revealed the substantial infiltration of submucosal immune cells as well as various amounts of damage to the respiratory and olfactory epithelium in each experimental group (Fig. 5E, bottom). These results demonstrated that ZCP3B4 and ZCP4C9 achieved significantly better protection than ZCP4D5-1 in preventing BA.5.2, BQ.1.1 and XBB.1.5 infections especially in the lungs.
Fig. 5.
Prophylactic efficacy of ZCP3B4, ZCP4C9 and ZCP4D5-1 against authentic Omicron BA.5.2, BQ.1.1 and XBB.1.5 in golden Syrian hamsters. (A) Experimental schedule and color coding for different treatment groups. Four groups of female hamsters received a single intraperitoneal injection of PBS (n = 6), 4.5 mg/kg of ZCP3B4 (n = 5), ZCP4C9 (n = 5) or ZCP4D5-1 (n = 5) one day before viral infection (−1 dpi). 24 h later (day 0), each group was challenged intranasally with a mixture of live Omicron BA.5.2, BQ.1.1 and XBB.1.5 (105 PFU/hamster). All animals were sacrificed on day 4 for final analysis. (B) Proportion of viral RNA copies in lungs and nasal turbinate (NT) homogenates of each group. The data is shown as mean ± SEM. (C) Daily body weight change of each group was measured after the viral infection. The data is shown as mean ± SEM. (D) Live viral plaque assay was used to quantify the number of infectious viruses in lungs and NT of each group. Log10-transformed plaque-forming unit (PFU) per mL was shown for each group. The dash line indicates the limit of detection. Each symbol represents an individual hamster with a line indicating the mean of each group. Statistics were generated using one-way ANOVA followed by Tukey's multiple comparisons test. ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001; ns (not significant), p > 0.05. (E) Representative histopathology of lung tissues and NT from pre-treated hamsters after viral challenge. Tissue sections were stained with hematoxylin and eosin (H&E). For PBS-treated hamsters, the infection could cause lung damage with alveolar septa thickening (black arrow), extensive inflammatory cell accumulation (blue arrow), homogeneously pink foci of edema (green arrow), and multifocal hemorrhage (red arrow). In NT, submucosal immune cell accumulation (blue arrow) as well as damage to the respiratory and olfactory epithelium (black arrow) are also indicated. The resolution is indicated by the scale bar.
Considering that ZCP3B4 and ZCP4C9 were able to reduce the amount of live infectious viruses in the NT more significantly than ZCP4D5-1, we sought to determine whether the lead bnAbs could prevent SARS-CoV-2 contact transmission. For this purpose, we designed and conducted an experiment of preventing natural co-housing transmission (Fig. 6A). Two days prior to the co-housing, index hamsters (n = 6) were inoculated intranasally with 105 PFU of the same stock of variant mixture containing BA.5.2, BQ.1.1 and XBB.1.5. Two days later, the treated group (n = 6) was administered intranasally with ZCP3B4 as a representative bnAb at 4.5 mg/kg, and the control group (n = 6) was treated by PBS 8 h before being co-housed with index hamsters at a 2:1 ratio. After 4 h of co-housing, index hamsters were euthanized immediately. Each ZCP3B4-treated and PBS-treated hamster was then separated, housed in individual cages, and euthanized at 4 dpi to harvest lungs and NT. We found that infectious viral titers were higher in the NT than in the lungs of control hamsters, modeling the situation of acute cases post-viral transmission.45 Importantly, ZCP3B4 prevented viral contact transmission without measurable live infectious virus in both lungs and NT of 5/6 animals except for one that had a low PFU number in NT (Fig. 6B), and even the lower doses (0.5 or 1.5 mg/kg) could achieve similar protection (Supplementary Figure S21D and E). Histopathological staining results of index and control hamsters showed consistently severe lung lesions, as well as inflammatory cell infiltration and damaged epithelium in the NT (Fig. 6C). Abundant infected cells (NP+) were also found in NT (Fig. 6D). Notably, less diffuse alveolar damage and inflammatory cell infiltration in the lungs of control hamsters were observed in this experiment than in the prophylactic study. We assumed that the discrepancy was likely due to the age difference because older hamsters were used in the first experiment. As expected, both lung and NT tissues derived from ZCP3B4-treated hamsters were normal (Fig. 6C). In addition, no NP+ cells were found in the entire NT tissue sections of 5/6 ZCP3B4-treated hamsters (Fig. 6D). These results demonstrated that ZCP3B4 could effectively prevent BA.5.2, BQ.1.1 and XBB.1.5 infections via contact transmission.
Fig. 6.
Efficacy of ZCP3B4 in preventing authentic Omicron BA.5.2, BQ.1.1 and XBB.1.5 transmission in golden Syrian hamsters. (A) Experimental schedule and color coding for different treatment groups. Index hamsters (male, n = 6) were challenged intranasally with a mixture of live Omicron BA.5.2, BQ.1.1 and XBB.1.5 (105 PFU/hamster) two days prior to the co-housing (day 0). On 2 dpi, two groups of male hamsters were administered intranasally with PBS (n = 6) or 4.5 mg/kg of ZCP3B4 (n = 6) 8 h before being co-housed with index hamsters at a 2:1 ratio. 4 h after the co-housing, index hamsters were sacrificed, whereas PBS- and ZCP3B4-treated hamsters were separated and sacrificed on day 4 for final analysis. (B) Live viral plaque assay was used to quantify the number of infectious viruses in lungs and nasal turbinate (NT) homogenates of each group. Log10-transformed plaque-forming unit (PFU) per mL was shown for each group. The dash line indicates the limit of detection. Each symbol represents an individual hamster with a line indicating the mean of each group. Statistics were generated using one-way ANOVA followed by Tukey's multiple comparisons test. ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. (C) Representative histopathology of lung tissues and NT from index and pre-treated hamsters after the co-housing. Tissue sections were stained with hematoxylin and eosin (H&E). For index and PBS-treated hamsters, the infection could cause lung damage with alveolar septa thickening (black arrow), extensive inflammatory cell accumulation (blue arrow), homogeneously pink foci of edema (green arrow), and multifocal hemorrhage (red arrow). In NT, submucosal immune cell accumulation (blue arrow) as well as damage to the respiratory and olfactory epithelium (black arrow) are also indicated. The resolution is indicated by the scale bar. (D) Representative images of infected cells in NT from index and pre-treated hamsters after the co-housing. Viral nucleocapsid protein (NP) was stained in green by immunofluorescence staining and cell nuclei were stained in blue with DAPI. The scale bar represents 100 μm.
Discussion
In this study, we isolated a total of 172 mAbs from convalescents after Omicron BA.2/BA.5 breakthrough infections by single B cell cloning technique. We found that 10 out of 22 NAbs were bnAbs with cross-neutralizing activities against major Omicron lineages including BA.5, XBB, and BA.2.86. Among them, the lead ultrapotent ZCP3B4 and ZCP4C9 were IGHV3-53/3–66 antibodies and showed IC50 values less than 10 ng/mL. Such ultrapotency and breadth have rarely been documented with NAbs cloned from the same family by analyzing current literatures and the CoV-AbDab.57 Mechanistically, instead of targeting the cryptic and conserved RBD epitopes like class IV bnAbs, these ultrapotent neutralizers belonged to class I bnAbs by structural analysis. They had undergone somatic hypermutations with fitness to interact with the RBM and avoid most documented mutations in Omicron sublineages. This resulted in remarkable breadth and effective protection against contact transmission of live BA.5.2, BQ.1.1 and XBB.1.5 strains.
Omicron breakthrough infection mainly induced cross-reactive NAb responses primarily against both WT and Omicron rather than Omicron-specific NAbs.7,50,51,66 Consistently, we found that 99.5% of newly identified NAbs could cross-bind to WT and Omicron, which were likely derived from memory B cells previously elicited by vaccinations (Supplementary Figure S7). During the breakthrough infection, NAbs often underwent maturation driven by somatic hypermutations after Omicron antigen exposure.61 In support of this notion, 66.7% of RBD-targeted NAbs, including ultrapotent ZCP3B4, ZCP4C9 and ZCP4D5-1, displayed better neutralization efficacy against BA.1 than BA.1 with reversed mutations (Fig. 3B, top). This process could potentially improve their interaction with mutated sites. Notably, WT-based vaccination alone was insufficient to induce such bnAbs with fitness because our previous study demonstrated that two doses of BNT162b2 vaccinations only induced one potent class I NAb ZCB11 against BA.1 from the same person.13 Surprisingly, after the BA.5 breakthrough infection in this person, newly cloned ultrapotent class I bnAbs were not further matured from IGHV1-58/IGKV3-20 of ZCB11. Only 1 of the 166 newly isolated ZC mAbs namely ZCP3G6-2 was clonally related58 to ZCB11, but they did not display significant clonal expansion (Supplementary Figures S3 and S8B). Instead, ZCP3B4, ZCP4C9 and ZCP4D5-1 were encoded by IGHV3-53/IGHV3-66 paired with various germline LC genes (Supplementary Table S4). These results demonstrated that ultrapotent bnAbs ZCP3B4, ZCP4C9 and ZCP4D5-1 encoded by IGHV3-53/3–66 likely possessed unique biochemical and structural features that rendered them natural fitness in binding and highly complementary in shape to the RBM of both WT and Omicron variants.30
Currently, few existing NAbs exhibit similar ultrapotency and breadth to our newly identified class I bnAbs against presently circulating JN.1 and KP.2 sublineages.23,27 The cryo-EM structural analysis revealed the neutralization mechanisms of ZCP3B4, ZCP4C9, ZCP4D5-1 and CUP2G3 (Fig. 4). These new bnAbs consistently targeted the top RBM surface of the ‘up’ RBD like previously reported class I NAbs. This class of antibodies included several ultrapotent NAbs against previous VOCs.13 Their potency and breadth, however, have reduced significantly attributed to K417 N/T, E484 K/A, L455 F/S, F456L, N501Y and F486 V/S/P mutations found in Omicron escape sublineages.9,12,27,67 Some studies, therefore, emphasized outer-face-targeting antibodies categorized in class II/III as a research focus for identifying ultrapotent bnAbs.29 We and others reported that class II/III NAbs might exhibit greater breadth,18,35,68 but few could neutralize BA.5 lineage and later dominant subvariants with ultrapotency.20 The footprints of ZCP3B4 and ZCP4C9, however, were almost retained throughout viral evolution and did not contain any convergent mutations and the specific mutations found in predominant XBB, EG.5.1 or BA.2.86 lineages22 (Supplementary Table S6, bottom). These results demonstrated that ZCP3B4 and ZCP4C9 have gained structural fitness to cross-neutralize both WT and Omicron sublineages by mainly targeting the conserved ‘Jing’ epitopes in RBD and accommodating the mutation sites. Although RBDs of the Omicron spike trimer prefer to maintain a stable ‘down’ state,18 competing with ACE2 for binding RBDs in an ‘up’ state remains a major mode of action for ultrapotent bnAbs as we demonstrated here.
The unique evolution of our ultrapotent bnAbs for targeting RBM has not been well investigated. For existing class I IGHV3-53/3-66 NAbs, such as P5S-2B10, P5S-1H1 and S728-1157, it has been well-documented that their binding to RBM relied significantly on the hydrogen-bond network mediated by 32NY33 in CDR-H1 and 53SGGS56 in CDR-H2, combined with the relatively short CDR-H3.56 Similarly, our bnAbs did not develop long CDR-H3, but instead accumulated higher levels of mutations during affinity maturation (Supplementary Figure S20B). Identified mutations were not unique among IGHV3-53/3-66 NAbs and independently had limited impact on the neutralizing activity (Supplementary Figure S20D), indicating that the breadth and ultrapotency of our bnAbs were owing to the extent of somatic hypermutations. To induce similar bnAbs for protection, vaccine design may consider the expression of stable RBDs of Omicron spike trimer in ‘up’ conformation for reactivating the family of IGHV3-53/3–66 antibody clonotypes. To control the RBD up/down equilibrium, double or quadruple mutations could be introduced into the spike subdomains.69 For CUP2G3, there were no prior NAb structures for comparison as it was from a rarely reported clonotype IGHV4-39/IGKV1-NL1 that accounted for <0.08% as documented in CoV-AbDab for mAb repertories.57 Unlike typical class I NAbs, CUP2G3 primarily relied on VL rather than VH to target the ‘peak’ and ‘neck’ of RBM,29 extensively overlapping with the major ACE2 epitopes (RBD 417-493)70 (Fig. 4B). Its distinct binding angle to RBD allowed both CDRs and FR to bury surface area mainly through germline-encoded residues (Supplementary Figure S20C). Since CUP2G3-like bnAbs have been rarely isolated from convalescent individuals, their implications for the rational design of vaccines remain to be investigated.
Sequentially emerged Omicron BA.1, BA.2 and BA.5, which have resulted in multiple widespread waves of SARS-CoV-2 infections in the world, exhibit great transmissibility in the upper airway.65 Our study continued to prove the in vivo replication advantage of later subvariant XBB.1.5 over BA.5 and BQ.1.1 (Fig. 5B), which may be associated with its enhanced transmissibility. The co-circulation of such highly transmissible strains outcompeted the development of clinical bnAb-based therapy to treat SARS-CoV-2 infection or co-infection, and most antibody drugs are no longer useful because of their reduced efficacy against recent Omicron escape mutants. For example, Bebtelovimab (LY-CoV1404) significantly lost cross-neutralizing activity against highly transmissible variants BQ.1.1 and XBB.1.520 (Fig. 1C). Since no therapeutic mAb is currently authorized for clinical use, significant concerns arise, particularly for immunocompromised individuals who fail to develop the antibodies needed for protection after vaccinations. The finding of several ultrapotent bnAbs in this study has shed light not only on understanding the fitness of memory B cells in response to vaccine breakthrough infections, but also on discovering promising antibody drug candidates. These bnAbs overcame the immune escape of major Omicron subvariants circulating in the past 2.5 years, indicating their potential against future VOIs. Furthermore, we demonstrated that ultrapotent bnAbs consistently displayed in vivo efficacy in reducing lung infection and damage against intranasal challenge by using BA.5, BQ.1.1 and XBB.1.5 in the hamster model (Fig. 5D and E). Since few existing NAbs show similar in vivo efficacy in preventing nasal infection and contact transmission against BA.5, BQ.1.1 and XBB.1.5,13,71,72 our bnAbs have the potential to be developed for future clinical use in treating immunocompromised patients and in healthcare facilities where the pre-exposure prophylaxis or post-exposure treatment is required. Finally, our understanding of bnAbs structural insights and the antibody–antigen interaction will help vaccine design and antibody engineering for enhanced potency and breadth.
Contributors
Z.C. conceived and supervised this study. M.L., R.Z. and Z.C. designed experiments. M.L., B.C., P.Z., Y.-L.L. and H.-O.M. did plasmid cloning, ELISA, and Western-blot. M.L., B.C. and Y.-L.L. expressed, purified and characterized antibodies. M.L., B.C. and N.L. performed pseudovirus neutralization assays. R.Z., N.L., Y.M. and Y.C. performed animal experiments. J.D.I, A.W.-H.C. and W.-M.C. conducted the Illumina sequencing. K.K.-W.T. isolated and provided authentic viruses and clinical specimens. K.-Y.Y. provided experimental resources. S.D. supervised the structural study. B.T., H.L. and S.D. solved the structures of antibodies.
Data sharing statement
Further information and requests for reagents and resources should be directed to and will be fulfilled by the lead contact, Z.C (zchenai@hku.hk).
Declaration of interests
The other authors declare no competing interests.
Acknowledgements
We thank Prof. David D. Ho, Lihong Liu and Linqi Zhang for providing various spike-expressing plasmids. We thank Hong Kong Customs and Excise Department for coordinating some subjects for participating in the study. This study was supported by the Hong Kong Research Grants Council Collaborative Research Fund (C7156-20G, C1134-20G and C5110-20G), and Shenzhen Science and Technology Program (JSGG20200225151410198); the Health@InnoHK, Innovation and Technology Commission; the National Program on Key Research Project of China (2020YFC0860600, 2020YFA0707500 and 2020YFA0707504), and donations from the Friends of Hope Education Fund in Hong Kong. Z.C.‘s team was also partly supported by the Hong Kong Theme-Based Research Scheme (T11-706/18-N, T11-702/24-N and T12-703/23-N) and Wellcome Trust P86433. This study was also partly supported by the Health and Medical Research Fund, the Food and Health Bureau, and the Government of the Hong Kong Special Administrative Region (Ref no.: COVID1903010-Project 4, COVID190123 and 20190572). Cryo-EM data were collected at the Biological Cryo-EM Centre at the Hong Kong University of Science and Technology (HKUST), generously supported by a donation from the Lo Kwee Seong Foundation. S.D. acknowledges support from the Research Grants Council (RGC) of Hong Kong (GRF16103321, GRF16102822, GRF16100223, C5033-19E, C6001-21E and C6012-22G) and HKUST start-up and initiation grants.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2024.105354.
Contributor Information
Shangyu Dang, Email: sdang@ust.hk.
Zhiwei Chen, Email: zchenai@hku.hk.
Appendix A. Supplementary data
References
- 1.Zhou D., Zhou R., Chen Z. Human neutralizing antibodies for SARS-CoV-2 prevention and immunotherapy. Immunother Adv. 2022;2(1) doi: 10.1093/immadv/ltab027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou R., To K.K., Wong Y.C., et al. Acute SARS-CoV-2 infection impairs dendritic cell and T cell responses. Immunity. 2020;53(4):864–877.e5. doi: 10.1016/j.immuni.2020.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu L., To K.K., Chan K.H., et al. High neutralizing antibody titer in intensive care unit patients with COVID-19. Emerg Microb Infect. 2020;9(1):1664–1670. doi: 10.1080/22221751.2020.1791738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wajnberg A., Amanat F., Firpo A., et al. Robust neutralizing antibodies to SARS-CoV-2 infection persist for months. Science. 2020;370(6521):1227–1230. doi: 10.1126/science.abd7728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Polack F.P., Thomas S.J., Kitchin N., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med. 2020;383(27):2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Z., Schmidt F., Weisblum Y., et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592(7855):616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou R., Liu N., Li X., et al. Three-dose vaccination-induced immune responses protect against SARS-CoV-2 Omicron BA.2: a population-based study in Hong Kong. Lancet Reg Health West Pac. 2023;32 doi: 10.1016/j.lanwpc.2022.100660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agrawal U., Katikireddi S.V., McCowan C., et al. COVID-19 hospital admissions and deaths after BNT162b2 and ChAdOx1 nCoV-19 vaccinations in 2.57 million people in Scotland (EAVE II): a prospective cohort study. Lancet Respir Med. 2021;9(12):1439–1449. doi: 10.1016/S2213-2600(21)00380-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu L., Iketani S., Guo Y., et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature. 2022;602(7898):676–681. doi: 10.1038/s41586-021-04388-0. [DOI] [PubMed] [Google Scholar]
- 10.Wang P., Nair M.S., Liu L., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593(7857):130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 11.Wang Q., Guo Y., Iketani S., et al. Antibody evasion by SARS-CoV-2 Omicron subvariants BA.2.12.1, BA.4 and BA.5. Nature. 2022;608(7923):603–608. doi: 10.1038/s41586-022-05053-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Q., Iketani S., Li Z., et al. Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. Cell. 2023;186(2):279–286.e8. doi: 10.1016/j.cell.2022.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou B., Zhou R., Tang B., et al. A broadly neutralizing antibody protects Syrian hamsters against SARS-CoV-2 Omicron challenge. Nat Commun. 2022;13(1):3589. doi: 10.1038/s41467-022-31259-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y., Zhao X., Zhou H., Zhu H., Jiang S., Wang P. Broadly neutralizing antibodies to SARS-CoV-2 and other human coronaviruses. Nat Rev Immunol. 2023;23(3):189–199. doi: 10.1038/s41577-022-00784-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andrews N., Stowe J., Kirsebom F., et al. Covid-19 vaccine effectiveness against the omicron (B.1.1.529) variant. N Engl J Med. 2022;386(16):1532–1546. doi: 10.1056/NEJMoa2119451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lau J.J., Cheng S.M.S., Leung K., et al. Real-world COVID-19 vaccine effectiveness against the Omicron BA.2 variant in a SARS-CoV-2 infection-naive population. Nat Med. 2023;29(2):348–357. doi: 10.1038/s41591-023-02219-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao Y., Wang J., Jian F., et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2022;602(7898):657–663. doi: 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao Y., Yisimayi A., Jian F., et al. BA.2.12.1, BA.4 and BA.5 escape antibodies elicited by Omicron infection. Nature. 2022;608(7923):593–602. doi: 10.1038/s41586-022-04980-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Q., Iketani S., Li Z., et al. Antigenic characterization of the SARS-CoV-2 Omicron subvariant BA.2.75. Cell Host Microbe. 2022;30(11):1512–1517.e4. doi: 10.1016/j.chom.2022.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao Y., Jian F., Wang J., et al. Imprinted SARS-CoV-2 humoral immunity induces convergent Omicron RBD evolution. Nature. 2023;614(7948):521–529. doi: 10.1038/s41586-022-05644-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang L., Kempf A., Nehlmeier I., et al. Neutralisation sensitivity of SARS-CoV-2 lineages EG.5.1 and XBB.2.3. Lancet Infect Dis. 2023;23(10):e391–e392. doi: 10.1016/S1473-3099(23)00547-9. [DOI] [PubMed] [Google Scholar]
- 22.Wang Q., Guo Y., Liu L., et al. Antigenicity and receptor affinity of SARS-CoV-2 BA.2.86 spike. Nature. 2023;624(7992):639–644. doi: 10.1038/s41586-023-06750-w. [DOI] [PubMed] [Google Scholar]
- 23.Kaku Y., Uriu K., Kosugi Y., et al. Virological characteristics of the SARS-CoV-2 KP.2 variant. Lancet Infect Dis. 2024;24:e416. doi: 10.1016/S1473-3099(24)00298-6. [DOI] [PubMed] [Google Scholar]
- 24.Zhang X., Chen L.L., Ip J.D., et al. Omicron sublineage recombinant XBB evades neutralising antibodies in recipients of BNT162b2 or CoronaVac vaccines. Lancet Microbe. 2023;4(3) doi: 10.1016/S2666-5247(22)00335-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Q., Bowen A., Tam A.R., et al. SARS-CoV-2 neutralising antibodies after bivalent versus monovalent booster. Lancet Infect Dis. 2023;23(5):527–528. doi: 10.1016/S1473-3099(23)00181-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Q., Bowen A., Valdez R., et al. Antibody response to omicron BA.4-BA.5 bivalent booster. N Engl J Med. 2023;388(6):567–569. doi: 10.1056/NEJMc2213907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang S., Yu Y., Xu Y., et al. Fast evolution of SARS-CoV-2 BA.2.86 to JN.1 under heavy immune pressure. Lancet Infect Dis. 2024;24(2):e70–e72. doi: 10.1016/S1473-3099(23)00744-2. [DOI] [PubMed] [Google Scholar]
- 28.Barnes C.O., Jette C.A., Abernathy M.E., et al. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588(7839):682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hastie K.M., Li H., Bedinger D., et al. Defining variant-resistant epitopes targeted by SARS-CoV-2 antibodies: a global consortium study. Science. 2021;374(6566):472–478. doi: 10.1126/science.abh2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qi H., Liu B., Wang X., Zhang L. The humoral response and antibodies against SARS-CoV-2 infection. Nat Immunol. 2022;23(7):1008–1020. doi: 10.1038/s41590-022-01248-5. [DOI] [PubMed] [Google Scholar]
- 31.Jian F., Feng L., Yang S., et al. Convergent evolution of SARS-CoV-2 XBB lineages on receptor-binding domain 455-456 synergistically enhances antibody evasion and ACE2 binding. PLoS Pathog. 2023;19(12) doi: 10.1371/journal.ppat.1011868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schaefer-Babajew D., Wang Z., Muecksch F., et al. Antibody feedback regulates immune memory after SARS-CoV-2 mRNA vaccination. Nature. 2023;613(7945):735–742. doi: 10.1038/s41586-022-05609-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tong P., Gautam A., Windsor I.W., et al. Memory B cell repertoire for recognition of evolving SARS-CoV-2 spike. Cell. 2021;184(19):4969–4980.e15. doi: 10.1016/j.cell.2021.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chan J.F., Yip C.C., To K.K., et al. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/hel real-time reverse transcription-PCR assay validated in vitro and with clinical specimens. J Clin Microbiol. 2020;58(5) doi: 10.1128/JCM.00310-20. e00310–e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luo M., Zhou B., Reddem E.R., et al. Structural insights into broadly neutralizing antibodies elicited by hybrid immunity against SARS-CoV-2. Emerg Microb Infect. 2023;12(1) doi: 10.1080/22221751.2022.2146538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods. 2017;14(3):290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 37.Zivanov J., Oton J., Ke Z., et al. A Bayesian approach to single-particle electron cryo-tomography in RELION-4.0. Elife. 2022;11 doi: 10.7554/eLife.83724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Geng Q., Shi K., Ye G., Zhang W., Aihara H., Li F. Structural basis for human receptor recognition by SARS-CoV-2 omicron variant BA.1. J Virol. 2022;96(8) doi: 10.1128/jvi.00249-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jumper J., Evans R., Pritzel A., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 41.Adams P.D., Gopal K., Grosse-Kunstleve R.W., et al. Recent developments in the PHENIX software for automated crystallographic structure determination. J Synchrotron Radiat. 2004;11(Pt 1):53–55. doi: 10.1107/s0909049503024130. [DOI] [PubMed] [Google Scholar]
- 42.Pettersen E.F., Goddard T.D., Huang C.C., et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 43.Pettersen E.F., Goddard T.D., Huang C.C., et al. UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 2021;30(1):70–82. doi: 10.1002/pro.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chan J.F., Zhang A.J., Yuan S., et al. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in a golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin Infect Dis. 2020;71(9):2428–2446. doi: 10.1093/cid/ciaa325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yuan S., Ye Z.W., Liang R., et al. Pathogenicity, transmissibility, and fitness of SARS-CoV-2 Omicron in Syrian hamsters. Science. 2022;377(6604):428–433. doi: 10.1126/science.abn8939. [DOI] [PubMed] [Google Scholar]
- 46.Tortorici M.A., Beltramello M., Lempp F.A., et al. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science. 2020;370(6519):950–957. doi: 10.1126/science.abe3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones B.E., Brown-Augsburger P.L., Corbett K.S., et al. The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates. Sci Transl Med. 2021;13(593):eabf1906. doi: 10.1126/scitranslmed.abf1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Westendorf K., Zentelis S., Wang L., et al. LY-CoV1404 (bebtelovimab) potently neutralizes SARS-CoV-2 variants. Cell Rep. 2022;39(7) doi: 10.1016/j.celrep.2022.110812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tortorici M.A., Czudnochowski N., Starr T.N., et al. Broad sarbecovirus neutralization by a human monoclonal antibody. Nature. 2021;597(7874):103–108. doi: 10.1038/s41586-021-03817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quandt J., Muik A., Salisch N., et al. Omicron BA.1 breakthrough infection drives cross-variant neutralization and memory B cell formation against conserved epitopes. Sci Immunol. 2022;7(75) doi: 10.1126/sciimmunol.abq2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park Y.J., Pinto D., Walls A.C., et al. Imprinted antibody responses against SARS-CoV-2 Omicron sublineages. Science. 2022;378(6620):619–627. doi: 10.1126/science.adc9127. [DOI] [PubMed] [Google Scholar]
- 52.Alsoussi W.B., Malladi S.K., Zhou J.Q., et al. SARS-CoV-2 Omicron boosting induces de novo B cell response in humans. Nature. 2023;617(7961):592–598. doi: 10.1038/s41586-023-06025-4. [DOI] [PubMed] [Google Scholar]
- 53.Sokal A., Barba-Spaeth G., Hunault L., et al. SARS-CoV-2 Omicron BA.1 breakthrough infection drives late remodeling of the memory B cell repertoire in vaccinated individuals. Immunity. 2023;56(9):2137–21351.e7. doi: 10.1016/j.immuni.2023.07.007. [DOI] [PubMed] [Google Scholar]
- 54.Ju B., Zhang Q., Wang Z., et al. Infection with wild-type SARS-CoV-2 elicits broadly neutralizing and protective antibodies against omicron subvariants. Nat Immunol. 2023;24(4):690–699. doi: 10.1038/s41590-023-01449-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yuan M., Liu H., Wu N.C., et al. Structural basis of a shared antibody response to SARS-CoV-2. Science. 2020;369(6507):1119–1123. doi: 10.1126/science.abd2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Q., Ju B., Ge J., et al. Potent and protective IGHV3-53/3-66 public antibodies and their shared escape mutant on the spike of SARS-CoV-2. Nat Commun. 2021;12(1):4210. doi: 10.1038/s41467-021-24514-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Raybould M.I.J., Kovaltsuk A., Marks C., Deane C.M. CoV-AbDab: the coronavirus antibody database. Bioinformatics. 2021;37(5):734–735. doi: 10.1093/bioinformatics/btaa739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Y., Yuan M., Lv H., Peng J., Wilson I.A., Wu N.C. A large-scale systematic survey reveals recurring molecular features of public antibody responses to SARS-CoV-2. Immunity. 2022;55(6):1105–11017.e4. doi: 10.1016/j.immuni.2022.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Setliff I., McDonnell W.J., Raju N., et al. Multi-donor Longitudinal antibody repertoire sequencing reveals the existence of public antibody clonotypes in HIV-1 infection. Cell Host Microbe. 2018;23(6):845–854.e6. doi: 10.1016/j.chom.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li L., Chen X., Wang Z., et al. Breakthrough infection elicits hypermutated IGHV3-53/3-66 public antibodies with broad and potent neutralizing activity against SARS-CoV-2 variants including the emerging EG.5 lineages. PLoS Pathog. 2023;19(12) doi: 10.1371/journal.ppat.1011856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kaku C.I., Starr T.N., Zhou P., et al. Evolution of antibody immunity following Omicron BA.1 breakthrough infection. Nat Commun. 2023;14(1):2751. doi: 10.1038/s41467-023-38345-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Starr T.N., Greaney A.J., Hilton S.K., et al. Deep mutational Scanning of SARS-CoV-2 receptor binding domain reveals Constraints on folding and ACE2 binding. Cell. 2020;182(5):1295–1310.e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Changrob S., Halfmann P.J., Liu H., et al. Site of vulnerability on SARS-CoV-2 spike induces broadly protective antibody against antigenically distinct Omicron subvariants. J Clin Invest. 2023;133(8) doi: 10.1172/JCI166844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang R., Zhang Q., Ge J., et al. Analysis of SARS-CoV-2 variant mutations reveals neutralization escape mechanisms and the ability to use ACE2 receptors from additional species. Immunity. 2021;54(7):1611–16121.e5. doi: 10.1016/j.immuni.2021.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shuai H., Chan J.F., Hu B., et al. The viral fitness and intrinsic pathogenicity of dominant SARS-CoV-2 Omicron sublineages BA.1, BA.2, and BA.5. eBioMedicine. 2023;95 doi: 10.1016/j.ebiom.2023.104753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Khan K., Karim F., Ganga Y., et al. Author Correction: omicron BA.4/BA.5 escape neutralizing immunity elicited by BA.1 infection. Nat Commun. 2022;13(1):6057. doi: 10.1038/s41467-022-33371-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhou T., Wang L., Misasi J., et al. Structural basis for potent antibody neutralization of SARS-CoV-2 variants including B.1.1.529. Science. 2022;376(6591) doi: 10.1126/science.abn8897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tuekprakhon A., Nutalai R., Dijokaite-Guraliuc A., et al. Antibody escape of SARS-CoV-2 Omicron BA.4 and BA.5 from vaccine and BA.1 serum. Cell. 2022;185(14):2422–2433.e13. doi: 10.1016/j.cell.2022.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Henderson R., Edwards R.J., Mansouri K., et al. Controlling the SARS-CoV-2 spike glycoprotein conformation. Nat Struct Mol Biol. 2020;27(10):925–933. doi: 10.1038/s41594-020-0479-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lan J., Ge J., Yu J., et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 71.Zhou D., Chan J.F., Zhou B., et al. Robust SARS-CoV-2 infection in nasal turbinates after treatment with systemic neutralizing antibodies. Cell Host Microbe. 2021;29(4):551–563.e5. doi: 10.1016/j.chom.2021.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhou B., Zhou R., Chan J.F., et al. SARS-CoV-2 hijacks neutralizing dimeric IgA for nasal infection and injury in Syrian hamsters. Emerg Microb Infect. 2023;12(2):2245921. doi: 10.1080/22221751.2023.2245921. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.