Abstract
Mice expressing the Fv-4 gene are resistant to infection by ecotropic murine leukemia viruses (MuLVs). The Fv-4 gene encodes an envelope (Env) protein whose putative receptor-binding domain resembles that of ecotropic MuLV Env protein. Resistance to ecotropic MuLVs appears to result from viral interference involving binding of the endogenously expressed Fv-4 env-encoded protein to the ecotropic receptor, although the immune system also plays a role in resistance. The Fv-4 env-encoded protein is processed normally and can be incorporated into virus particles but is unable to promote viral entry. Among the many sequence variations between the transmembrane (TM) subunit of the Fv-4 env-encoded protein and the TM subunits of other MuLV Env proteins, there is a substitution of an arginine residue in the Fv-4 env-encoded protein for a glycine residue (gly-491 in Moloney MuLV Env) that is otherwise conserved in all of the other MuLVs. This residue is present in the MuLV TM fusion peptide sequence. In this study, gly-491 of Moloney MuLV Env has been replaced with other residues and a mutant Env bearing a substitution for gly-487 was also created. G491R recapitulates the Fv-4 Env phenotype in cell culture, indicating that this substitution is sufficient for creation of an Env protein that can establish the interference-mediated resistance to ecotropic viruses produced by the Fv-4 gene. Analysis of the mutant MuLV Env proteins also has implications for an understanding of the role of conserved glycine residues in fusion peptides and for the engineering of organismal resistance to retroviruses.
The mouse Fv-4 gene controls susceptibility to infection by ecotropic murine leukemia viruses (MuLVs) but does not affect susceptibility to other MuLV subgroups (20, 32, 47, 51). The Fv-4 locus is an endogenous defective provirus that contains the end of the pol gene, a complete env gene, and a 3′ long terminal repeat that resembles the ecotropic MuLV genome (14). The env gene product shares amino acid sequence identity with the Env proteins of the ecotropic Cas-Br-E (90% identity) and Moloney (70% identity) MuLVs (Mo-MuLVs) (29). MuLV envelope (Env) proteins consist of two subunits, SU (surface protein) and TM (transmembrane protein). The sequence of the putative receptor-binding site of Fv-4 SU is highly similar to that of the ecotropic MuLVs (5, 27, 29), which bind to a receptor present on rodent cells (1). Cells carrying the Fv-4r-encoding allele produce cell surface antigens immunologically related to ecotropic MuLV Env proteins but do not produce ecotropic infectious MuLV (15, 16, 50).
The basis of the resistance to infection by ecotropic viruses conferred by the Fv-4 gene remains a controversial subject. The immune response has been suggested to play a subsidiary (25) or, more recently, a dominant (12, 51) role in resistance, although the mechanism is incompletely defined. It is nevertheless probable that viral interference is an important component of the lack of susceptibility of Fv-4r mice to infection by ecotropic MuLVs (16, 17, 25). The phenomenon of retroviral interference (40, 44, 45) dictates that a cell productively infected by a retrovirus is infected much less efficiently by the same retrovirus or by a retrovirus that utilizes the same receptor for entry, although it can be infected readily by retroviruses that employ a different receptor. This interference is the result of the binding of the endogenous retroviral Env protein to the cellular receptor so that it is unavailable for interaction with Env protein on exogenous virus that uses the same receptor to enter cells. The binding of ecotropic MuLV SU and the cellular receptor probably occurs intracellularly (23). An article that suggested that release of Fv-4 SU from a cell and its subsequent binding to the ecotropic receptor on cell surfaces was at least partially responsible for Fv-4-mediated interference (30) has been retracted (31).
We have approached these issues by investigating the amino acid sequence and biochemical basis of the Fv-4 phenotype in the context of our knowledge concerning the function of other ecotropic MuLV Env proteins. During the progress of the Mo-MuLV Env protein through the secretory system, it forms a trimer (21) and is subsequently proteolytically processed into two subunits (10, 49), gp70 (amino acids 34 to 468) and p15E (amino acids 470 to 665), that are linked through a disulfide bond (33, 35, 36). Once the Env protein has reached the surface of a cell, it is incorporated into a budding type C retroviral particle. The gp70 subunit (SU) is on the outside of the particle, whereas the p15E protein (TM) possesses an extraparticle domain, a membrane-spanning domain, and a 35-amino-acid domain that resides within the particle (34). The TM protein possesses, at its amino terminus, a sequence that is believed to encode the fusion peptide, which, upon exposure, promotes the first steps in the membrane fusion process that occurs during viral entry (19, 52). The membrane-spanning domain of TM has recently been shown also to participate in the process of membrane fusion (48). During a late stage of viral particle maturation, the carboxy-terminal 16 amino acid residues of the p15E protein are removed by the retroviral gag-pol-encoded protease, resulting in a 12-kDa Env TM protein (3, 9, 10, 22, 42, 43, 46). The Env protein present on the membrane of an infected cell is therefore structurally different from that found in a mature particle. Removal of the carboxy-terminal region activates the Env so that it is capable of fusing membranes in a receptor-dependent fashion (38, 39). It has been proposed that the binding of SU to the cellular receptor results in a thiol-disulfide exchange reaction leading to the elimination of the cystine bridge between the SU and TM subunits and consequent exposure of the TM fusion peptide that promotes membrane fusion and viral entry (35, 41).
Through the construction of a recombinant Mo-MuLV bearing the Fv-4 env gene, it was determined that the Fv-4 env-encoded protein is expressed, processed, and incorporated into Mo-MuLV particles at normal levels (29). However, recombinant virus bearing the Fv-4 env-encoded protein was not infectious (29). Investigations of the infectiousness of recombinant Mo-MuLVs bearing Env proteins that were chimeras of the Fv-4 and Mo-MuLV Env proteins indicated that the major source of the defect in the Fv-4 env-encoded protein was located in Fv-4 env TM, although there was a reduction in infectiousness when the carboxy-terminal half of the chimeric Env proteins originated from Fv-4 env SU (29).
Although there are 18 amino acid differences between the Fv-4 and MuLV TM proteins, we focused on an arginine residue encoded by the Fv-4 env gene that is conserved as a glycine residue (gly-491 in Mo-MuLV Env) in the TM proteins of all of the other MuLV TM proteins (Fig. 1). This residue lies near the amino terminus of TM in the putative fusion peptide (19, 52). The fusion peptides of viral glycoproteins are apolar, and those at the amino-terminal ends of the fusion protein also tend to be glycine rich. Although the actual biophysical role of the fusion peptide is controversial (2, 4), its apolar character is believed to be essential for interaction with the target cellular membrane and the consequent membrane perturbation that leads to fusion. Our hypothesis was that replacement of gly-491 with an arginine residue would disrupt the promotion of membrane fusion. Although other differences in the sequence of the Mo-MuLV and Fv-4 Env proteins might contribute to the Fv-4r phenotype, our analysis suggested that the single substitution would be sufficient to give the Mo-MuLV Env protein a defect similar to that of the Fv-4 Env protein in cell culture experiments.
FIG. 1.
Amino acid sequence of the Fv-4 fusion peptide at the amino terminus of TM compared to those of other MuLVs (GenBank accession numbers 387149, 58830, 119478, 332083, and 332027, respectively). The residues that were mutated are in light gray.
Our experiments also addressed the actual roles of gly-491 and the nearby conserved residue gly-487 in fusion peptide function. The role of the glycine residues in fusion peptides that are present at the amino termini of viral TM proteins that promote fusion is incompletely understood (4). In any representation of the Mo-MuLV TM fusion peptide as an α-helix, there is no segregation of the glycine residues to a particular face. This indicates that previous suggestions about the roles of the glycines in helix packing or about the role of the fusion peptide hydrophobic moment (4) may not be applicable to the TM fusion peptide.
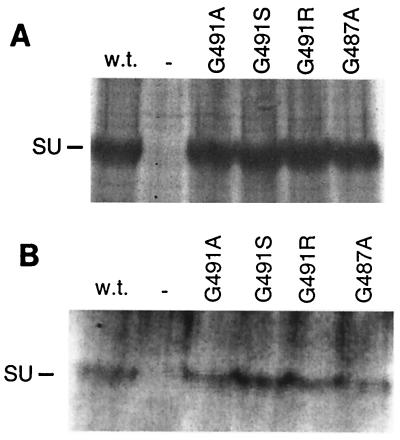
The processing and function of mutant Mo-MuLV Env proteins with gly-491 replaced with either an alanine, an arginine, or a serine residue and of one with gly-487 replaced with an alanine residue were studied. Mutagenesis of Mo-MuLV env was performed on env-containing subclones of pMOVΨ- (28) in pTZ18U by utilizing the Bio-Rad Muta-Gene Phagemid Mutagenesis Kit (48). The Mo-MuLV Env proteins were expressed from pLTRSDSA, a vector that contains the Mo-MuLV long terminal repeat enhancer and promoter and encodes the Mo-MuLV splice donor and acceptor sequences and the simian virus 40 polyadenylation addition signal (48). The vector expressing the wild-type Mo-MuLV Env protein is penv1min (48), whereas those expressing the mutant Env proteins are referred to as penv1G491A, penv1G491R, penv1G491R, and penv1G487A. Expression and processing of the G491A, G491R, and G491S Env proteins in gpGFP cells (ΦNX cells, a second-generation 293T-based retroviral packaging cell line transfected with MFG.S-GFP-S65T, a retroviral vector encoding the Aequorea victoria green fluorescent protein S65T mutant [48]) equivalent to that of the wild-type Env protein were observed through radioimmunoprecipitation assays, whereas slightly reduced levels of the G487A Env protein were detected (Fig. 2A). Normal incorporation of the G491A, G491R, and G491S Env proteins into recombinant retroviral particles produced by the gpGFP cells was also observed, whereas incorporation of G487A Env was reduced (Fig. 2B).
FIG. 2.
Analysis of processing (A) and incorporation into virus particles (B) of the various mutant Mo-MuLV Env proteins in gpGFP cells. gpGFP cells expressing the mutant Env proteins were labeled with [35S]cysteine-[35S]methionine, and the cell lysate (A) and viral supernatant (B) were immunoprecipitated with antibody against SU. An analysis of cell lysate and virus from gpGFP cells transfected with the wild-type (w.t.) Env plasmid or mock transfected (−) is also presented. At the left is indicated the position of SU (70 kDa).
The ability of the mutant Env proteins to bind to the cellular receptor was determined by testing their function in an interference assay utilizing stable clones of NIH 3T3 cells expressing the mutant env genes (48). Medium from E86 nlslacZ cells, which produce replication-defective Mo-MuLV retrovirus that confers nucleus-localized β-galactosidase expression on infected cells (48), was incubated with stable NIH 3T3 Env-expressing clones, which were established as previously described (48). The susceptibility of these cells to superinfection was measured by staining with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), a synthetic substrate of β-galactosidase. A reduction in the number of X-Gal-stained cells indicates that the expressed mutant Env protein is able to bind to the cellular receptor. Cells expressing G491R had a 13-fold reduction in susceptibility to transduction, similar to cells expressing the wild-type Env protein. Cells expressing G491A Env had a 10-fold reduction in susceptibility, whereas those expressing the G491S and G487A Env proteins had a fivefold reduction. The extent of interference roughly correlates with the level of Env protein expression in the transfected NIH 3T3 cells (Fig. 3). Of central importance is the fact that G491R Env can confer wild-type levels of interference.
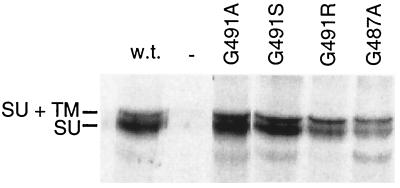
FIG. 3.
Normal processing of mutant Env proteins in NIH 3T3 cells. NIH 3T3 cells expressing mutant Env proteins labeled with [35S]cysteine-[35S]methionine and immunoprecipitated with antibody against SU. GP+E-86 cells were used as the positive control for wild-type (w.t.) Env protein expression, while NIH 3T3 cells were used as the negative control (−). At the left are indicated the positions of wild-type uncleaved SU plus TM (85 kDa) and SU (70 kDa). There was a cross-reactive protein in the cell lysate that migrated slightly slower than SU plus TM (85 kDa).
The transduction capacity of recombinant viruses bearing the mutant Env proteins produced by gpGFP cells was measured by flow cytometry as previously described (48). Virus particles bearing the G491A, G491S, or G487A Env protein were able to promote viral transduction at levels similar to those of virus bearing wild-type Env, whereas virus bearing the G491R Env protein was completely defective (Table 1).
TABLE 1.
Analysis of the abilities of mutant Env proteins to promote membrane fusion and viral entry
| Env protein or cell type | Syncytium formationa | Virus titerb |
|---|---|---|
| G491A | 69 ± 6 | (1.0 ± 0.1) × 105 |
| G491R | <0.2 | <5.0 × 102 |
| G491S | 91 ± 19 | (9.9 ± 5.0) × 104 |
| G487A | 46 ± 10 | (9.8 ± 1.9) × 104 |
| Wild type | 100c | 1.0 × 105 |
| Cells not expressing Env | <0.2 | <5.0 × 102 |
NIH 3T3 cells were transiently transfected with plasmids encoding Env proteins lacking the last 16 amino acids, and syncytia were counted 48 h posttransfection. Data are the averages of three independent experiments.
Virus titer is given as transducing units per milliliter. Data are the averages of three independent experiments.
The values shown were normalized to the percentage of nuclei in syncytia of cells expressing the wild-type Env protein lacking the last 16 amino acids (17.3%).
In order to examine the abilities of the various mutant Env proteins to promote receptor-mediated-membrane fusion, syncytium assays were carried out. NIH 3T3 cells, which possess the Mo-MuLV receptor and are susceptible to infection by Mo-MuLV, were transiently transfected with plasmids encoding the mutant Env proteins lacking the last 16 amino acid residues in the cytoplasmic domain (48). The last 16 amino acid residues in the cytoplasmic domain of TM (referred to as the R peptide) must be removed in order for fusion, that is, multinucleated cells, or syncytia, to be observed (38, 39). In this assay, the G491A and G491S Env proteins were able to promote membrane fusion at levels similar to those promoted by wild-type Env whereas G491R Env was completely incapable of promoting fusion (Table 1). The capacity of G487A Env to promote fusion was moderately reduced.
These results demonstrate that replacing G491 with alanine or serine has little effect on the ability of the mutant Env proteins to promote fusion or viral transduction. On the other hand, when G491 is replaced with an arginine, although the Env protein is able to bind to the cellular receptor and is processed and incorporated into virus particles at normal levels, it is incapable of promoting fusion and viral entry. These properties of G491R Env are similar to those of the Fv-4 Env protein, and this fact supports the idea that the natural resistance to ecotropic infection conferred by the Fv-4 Env protein is due to a combination of the ability of Fv-4 Env to produce interference and its inability to promote membrane fusion.
Another potential mechanism by which Fv-4 Env produces resistance to ecotropic viruses is dependent upon the high level of amino acid sequence identity between the TM proteins of the Env proteins of ecotropic viruses and that produced by the Fv-4r-encoding locus. The TM proteins are responsible for the trimerization of the MuLV Env proteins (6, 7). When cells expressing Fv-4 Env are infected with ecotropic virus (interference is not absolute), it would be expected that the new progeny virus would incorporate Env proteins that are mixed oligomers of the Fv-4 and wild-type ecotropic Env proteins. We conducted the following experiment to examine the consequences of expression of G491R Env, an Fv-4 Env surrogate, upon transduction mediated by wild-type Env. A constant amount of penv1min (0.5 μg), which encodes the wild-type Mo-MuLV Env protein, was transfected into cultures of gpGFP cells. Cotransfected into the cultures was a constant mass of DNA (3.5 μg) containing various proportions of penv1G491R and pLTRSDSA, the expression vector. Increasing amounts of G491R Env expression decreased the transduction titer of the recombinant virus (Fig. 4). These data indicate that it is likely that Fv-4 Env not only reduces ecotropic virus entry into cells through interference but also diminishes the infectiousness of the virus produced by the rare infected cell.
FIG. 4.
Effect of G491R mutant Env on the ability of wild-type Env to promote transduction. A constant amount of plasmid penv1min (0.5 μg) and various amounts of penv1G491R were transfected into gpGFP cells, and the virus present in the supernatant medium was used to transduce NIH 3T3 cells. The level of transduction was determined by flow cytometry.
The fact that replacing a glycine residue in the fusion peptide with an arginine could eliminate the ability of the Mo-MuLV Env protein to promote fusion is not unexpected. It has been found in a number of systems that placing a charged residue into viral fusion peptides by substitution adversely affects the ability of the viral glycoprotein to promote fusion (8, 11, 18, 24, 26, 37). For example, replacing AKV MuLV TM gly-17 (the equivalent of Mo-MuLV Env gly-486) with an arginine (19) resulted in an Env protein that could no longer promote membrane fusion and replacing Mo-MuLV Env leu-478 with a glutamate (52) produced an Env protein that could promote neither fusion nor infection but was processed and incorporated into virus particles at normal levels (19, 52).
The role of the glycine residues in viral amino-terminal fusion peptides is controversial (4). We hypothesize that the glycine residues may, in fact, play their most important roles not in membrane fusion per se but in the native uncleaved and/or cleaved conformation of Mo-MuLV Env. In this scenario, in the native cleaved conformation, the aliphatic side chains are buried in a cavity whereas the glycine residues are on the surface. For later function as a fusion peptide, the glycine residues cannot be replaced with polar or charged residues because that would interfere with membrane fusion. On the other hand, because the glycine residues are exposed in the native cleaved conformation, they cannot be replaced with hydrophobic residues. Under this scheme, the glycine residues are conserved because they are the only residues that are neither polar, charged, nor hydrophobic. They may also be conserved because they allow close packing of the fusion peptide on the face of the cavity. In our studies, substitutions of uncharged residues for the fusion peptide glycine residues had little (gly-487) or no (gly-491) effect on function. In contrast, Mo-MuLV Env proteins bearing substitutions of uncharged residues for gly-482 were defective in processing to SU and TM and incorporation into retrovirus particles (52). These data are consistent with a structural role for the fusion peptide glycine residues; substitutions for individual glycine residues have different effects because of variations in the criticalness of a given residue to the maintenance of the native conformation.
Although there are 18 amino acid residue differences between the Fv-4 and Mo-MuLV TMs, we chose to focus on the arginine-for-glycine substitution because, as our experiments have borne out, we anticipated that the placement of an arginine in the Mo-MuLV Env fusion peptide would be sufficient to give it a defect in cell culture experiments similar to that of Fv-4 Env. It is possible that other sequence differences between the Fv-4 and Mo-MuLV TMs contribute to the Fv-4 defects, but recent data suggest that this is unlikely. Fv-4 Env is closely related to the Env protein of the replication-competent Cas-Br-E MuLV (six differences in TM outside of a short divergent carboxy-terminal sequence [29]). Env proteins from endogenous replication-competent Mus musculus castaneus Mo-MuLVs that are even closer to Fv-4 Env have been recently cloned (13). The Env protein of one of these Cas-E-type MuLVs (Frg-3), which appears to be a prototype, differs from Fv-4 Env at only five residues (at two of which the Fv-4 Env sequence is identical to that of Cas-Br-E Env) and possesses an identical carboxy-terminal sequence (13). Of these differences, only the arginine-for-glycine substitution in the fusion peptide is not found in the Env proteins of any replication-competent Cas-E-type or other MuLV. It can be concluded that the defect in Fv-4 Env probably results from the substitution whose effects we have investigated upon its transfer to Mo-MuLV Env.
Our model for Fv-4 restriction of ecotropic virus replication and pathogenesis is that the Fv-4 Env protein binds to the ecotropic receptor and prevents it from interacting with Env protein on exogenous virus, perhaps through retention of the functional receptor molecules within the cell. The reduction of ecotropic virus entry and infection is dependent on the level of Fv-4 locus expression and is not absolute. The titer of virus produced by the rare infected cell is reduced through incorporation of Fv-4 Env into progeny virions, leading to inhibition of the function of the wild-type Env protein. This phenomenon may also underlie the failure to detect infectious ecotropic MuLV from Fv-4r M. musculus castaneus mice despite the fact that they possesses endogenous replication-competent MuLV proviruses (13). A functioning immune system may be unnecessary for elimination of the low level of virus produced. If, however, expression of the Fv-4 locus is reduced (in a heterozygote, for example), then lower levels of interference will result in more cells being infected by ecotropic viruses and there will be less inhibition of the function of wild-type Env protein in progeny virions. In the absence of a functioning immune system, there may be sufficient virus production for the recombination events that lead to disease to transpire. Such a scenario seems to be the most plausible explanation for the apparent role of the immune system in Fv-4 restriction when there is only one copy of the locus (51). It seems unlikely that Fv-4 Env itself stimulates the immune response to the Env proteins on ecotropic viruses.
The properties of the Fv-4 Env protein have been transferred to Mo-MuLV Env by the replacement of a single residue. Our results suggest an approach to the introduction of resistance to a retrovirus, such as human immunodeficiency virus, into an organism. Transfer of a gene encoding a normally processed but fusion-defective retroviral Env protein into susceptible cells would interfere with viral entry and potentially reduce the infectiousness of virus emerging from the cell. In the case of human immunodeficiency virus, it might be additionally desirable if the Env protein could bind to the chemokine receptor(s) without the necessity of binding to CD4.
Acknowledgments
This work was supported by the Purdue Research Foundation and American Cancer Society grant IRG-17-36 to D.A.S. This research was also supported by an NIH Biophysics Training Grant (GM08296) and a GAANN Fellowship to G.M.T.
REFERENCES
- 1.Albritton L M, Tseng L, Scadden D, Cunningham J M. A putative murine ecotropic retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell. 1989;57:659–666. doi: 10.1016/0092-8674(89)90134-7. [DOI] [PubMed] [Google Scholar]
- 2.Bonnafous P, Stegmann T. Membrane perturbation and fusion pore formation in influenza hemagglutinin-mediated membrane fusion. A new model for fusion. J Biol Chem. 2000;275:6160–6166. doi: 10.1074/jbc.275.9.6160. [DOI] [PubMed] [Google Scholar]
- 3.Crawford S, Goff S P. A deletion mutation in the 5′ part of the pol gene of Moloney murine leukemia virus blocks proteolytic processing of the gag and pol polyproteins. J Virol. 1985;53:899–907. doi: 10.1128/jvi.53.3.899-907.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Durell S R, Martin I, Ruysschaert J M, Shai Y, Blumenthal R. What studies of fusion peptides tell us about viral envelope glycoprotein-mediated membrane fusion. Mol Membr Biol. 1997;14:97–112. doi: 10.3109/09687689709048170. [DOI] [PubMed] [Google Scholar]
- 5.Fass D, Davey R A, Hamson C A, Kim P S, Cunningham J M, Berger J M. Structure of a murine leukemia virus receptor-binding glycoprotein at 2.0 Angstrom resolution. Science. 1997;277:1662–1666. doi: 10.1126/science.277.5332.1662. [DOI] [PubMed] [Google Scholar]
- 6.Fass D, Harrison S C, Kim P S. Retrovirus envelope domain at 1.7 Angstrom resolution. Nat Struct Biol. 1996;3:465–469. doi: 10.1038/nsb0596-465. [DOI] [PubMed] [Google Scholar]
- 7.Fass D, Kim P S. Dissection of a retrovirus envelope protein reveals structural similarity to influenza hemagglutinin. Curr Biol. 1995;5:1377–1383. doi: 10.1016/s0960-9822(95)00275-2. [DOI] [PubMed] [Google Scholar]
- 8.Freed E O, Myers D J, Risser R. Characterization of the fusion domain of the human immunodeficiency virus type 1 envelope glycoprotein gp41. Proc Natl Acad Sci USA. 1990;87:4650–4654. doi: 10.1073/pnas.87.12.4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Green N, Shinnick T M, Witte O, Ponticelli A, Sutcliffe J G, Lerner R A. Sequence-specific antibodies show that maturation of Moloney leukemia virus envelope polyprotein involves removal of a COOH-terminal peptide. Proc Natl Acad Sci USA. 1981;78:6023–6027. doi: 10.1073/pnas.78.10.6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henderson L E, Sowder R, Copeland T D, Smythers G, Oroszlan S. Quantitative separation of murine leukemia virus proteins by reversed-phase high-pressure liquid chromatography reveals newly described gag and env cleavage products. J Virol. 1984;52:492–500. doi: 10.1128/jvi.52.2.492-500.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hernandez L D, White J M. Mutational analysis of the candidate internal fusion peptide of the avian leukosis and sarcoma virus subgroup A envelope glycoprotein. J Virol. 1998;72:3259–3267. doi: 10.1128/jvi.72.4.3259-3267.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Higo K, Kubo Y, Iwatani Y, Ono T, Maeda M, Hiai H, Masuda T, Kuribayashi K, Zhang F, Lamin T Y, Adachi A, Ishimoto A. Susceptibility of nude mice carrying the Fv-4 gene to Friend murine leukemia virus infection. J Virol. 1997;71:750–754. doi: 10.1128/jvi.71.1.750-754.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ikeda H, Kato K, Kitani H, Suzuki T, Yoshida T, Inaguma Y, Yamamoto N, Suh J-G, Hyun B-H, Yamagata T, Namikawa T, Tomita T. Virological properties and nucleotide sequences of Cas-E-type endogenous ecotropic murine leukemia viruses in South Asian wild mice, Mus musculus castaneus. J Virol. 2001;75:5049–5058. doi: 10.1128/JVI.75.11.5049-5058.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ikeda H, Laigret F, Martin M A, Repaske R. Characterization of a molecularly cloned retroviral sequence associated with Fv-4 resistance. J Virol. 1985;55:768–777. doi: 10.1128/jvi.55.3.768-777.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ikeda H, Odaka T. A cell membrane “gp70” associated with Fv-4 gene: immunological characterization, and tissue and strain distribution. Virology. 1984;133:65–76. doi: 10.1016/0042-6822(84)90426-4. [DOI] [PubMed] [Google Scholar]
- 16.Ikeda H, Odaka T. Cellular expression of murine leukemia virus gp70-related antigen on thymocytes of uninfected mice correlates with Fv-4 gene-controlled resistance to Friend leukemia virus infection. Virology. 1983;128:127–139. doi: 10.1016/0042-6822(83)90324-0. [DOI] [PubMed] [Google Scholar]
- 17.Ikeda H, Sugimura H. Fv-4 resistance gene: a truncated endogenous murine leukemia virus with ecotropic interference properties. J Virol. 1989;63:5405–5412. doi: 10.1128/jvi.63.12.5405-5412.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ito H, Watanabe S, Sanchez A, Whitt M A, Kawaoka Y. Mutational analysis of the putative fusion domain of Ebola virus glycoprotein. J Virol. 1999;73:8907–8912. doi: 10.1128/jvi.73.10.8907-8912.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones J S, Risser R. Cell fusion induced by the murine leukemia virus envelope glycoprotein. J Virol. 1993;67:67–74. doi: 10.1128/jvi.67.1.67-74.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kai K, Ikeda H, Yuasa Y, Suzuki S, Odaka T. Mouse strain resistant to N-, B-, and NB-tropic murine leukemia viruses. J Virol. 1976;20:436–440. doi: 10.1128/jvi.20.2.436-440.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamps C A, Lin Y C, Wong P K. Oligomerization and transport of the envelope protein of Moloney murine leukemia virus-TB and of ts1, a neurovirulent temperature-sensitive mutant of MoMuLV-TB. Virology. 1991;184:687–694. doi: 10.1016/0042-6822(91)90438-h. [DOI] [PubMed] [Google Scholar]
- 22.Katoh I, Yoshinaka Y, Rein A, Shibuya M, Odaka T, Oroszlan S. Murine leukemia virus maturation: protease region required for conversion from “immature” to “mature” core form and for virus infectivity. Virology. 1985;145:280–292. doi: 10.1016/0042-6822(85)90161-8. [DOI] [PubMed] [Google Scholar]
- 23.Kim J W, Cunningham J M. N-linked glycosylation of the receptor for murine ecotropic retroviruses is altered in virus-infected cells. J Biol Chem. 1993;268:16316–16320. [PubMed] [Google Scholar]
- 24.Levy-Mintz P, Kielian M. Mutagenesis of the putative fusion domain of the Semliki Forest virus spike protein. J Virol. 1991;65:4292–4300. doi: 10.1128/jvi.65.8.4292-4300.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Limjoco T I, Dickie P, Ikeda H, Silver J. Transgenic Fv-4 mice resistant to Friend virus. J Virol. 1993;67:4163–4168. doi: 10.1128/jvi.67.7.4163-4168.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo Z, Weiss S R. Roles in cell-to-cell fusion of two conserved hydrophobic regions in the murine coronavirus spike protein. Virology. 1998;244:483–494. doi: 10.1006/viro.1998.9121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacKrell A J, Soong N W, Curtis C M, Anderson W F. Identification of a subdomain in the Moloney murine leukemia virus envelope protein involved in receptor binding. J Virol. 1996;70:1768–1774. doi: 10.1128/jvi.70.3.1768-1774.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mann R, Mulligan R C, Baltimore D. Construction of a retrovirus packaging mutant and its use to produce helper-free defective retrovirus. Cell. 1983;33:153–159. doi: 10.1016/0092-8674(83)90344-6. [DOI] [PubMed] [Google Scholar]
- 29.Masuda M, Yoshikura H. Construction and characterization of the recombinant Moloney murine leukemia viruses bearing the mouse Fv-4 env gene. J Virol. 1990;64:1033–1043. doi: 10.1128/jvi.64.3.1033-1043.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nihrane A, Fujita K, Willey R, Lyu M S, Silver J. Murine leukemia virus envelope protein in transgenic-mouse serum blocks infection in vitro. J Virol. 1996;70:1882–1889. doi: 10.1128/jvi.70.3.1882-1889.1996. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Nihrane A, Fujita K, Willey R, Lyu M S, Silver J. Murine leukemia virus envelope protein in transgenic-mouse serum blocks infection in vitro. J Virol. 1998;72:8462. doi: 10.1128/jvi.72.10.8462-8462.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Odaka T, Ikeda H, Yoshikura H, Moriwaki K, Suzuki S. Fv-4: gene controlling resistance to NB-tropic Friend murine leukemia virus. Distribution in wild mice, introduction into genetic background of BALB/c mice, and mapping of chromosomes. JNCI. 1981;67:1123–1127. [PubMed] [Google Scholar]
- 33.Pinter A, Fleissner E. The presence of disulfide-linked gp70–p15(E) complexes in AKR murine leukemia virus. Virology. 1977;83:417–422. doi: 10.1016/0042-6822(77)90187-8. [DOI] [PubMed] [Google Scholar]
- 34.Pinter A, Honnen W J. Topography of murine leukemia virus envelope proteins: characterization of transmembrane components. J Virol. 1983;46:1056–1060. doi: 10.1128/jvi.46.3.1056-1060.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pinter A, Kopelman R, Li Z, Kayman S C, Sanders D A. Localization of the labile disulfide bond between SU and TM of the murine leukemia virus envelope protein complex to a highly conserved CWLC motif in SU that resembles the active site sequence of thiol-disulfide exchange enzymes. J Virol. 1997;71:8073–8077. doi: 10.1128/jvi.71.10.8073-8077.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pinter A, Lieman-Hurwitz J, Fleissner E. The nature of the association between the murine leukemia virus envelope proteins. Virology. 1978;91:345–351. doi: 10.1016/0042-6822(78)90382-3. [DOI] [PubMed] [Google Scholar]
- 37.Qiao H, Armstrong R T, Melikyan G B, Cohen F S, White J M. A specific point mutant at position 1 of the influenza hemagglutinin fusion peptide displays a hemifusion phenotype. Mol Biol Cell. 1999;10:2759–2769. doi: 10.1091/mbc.10.8.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ragheb J A, Anderson W F. pH-independent murine leukemia virus ecotropic envelope-mediated cell fusion: implications for the role of the R peptide and p12E TM in viral entry. J Virol. 1994;68:3220–3231. doi: 10.1128/jvi.68.5.3220-3231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rein A, Mirro J, Haynes J G, Ernst S M, Nagashima K. Function of the cytoplasmic domain of a retroviral transmembrane protein: p15E-p2E cleavage activates the membrane fusion capability of the murine leukemia virus Env protein. J Virol. 1994;68:1773–1781. doi: 10.1128/jvi.68.3.1773-1781.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rein A, Schultz A. Different recombinant murine leukemia viruses use different cell surface receptors. Virology. 1984;136:144–152. doi: 10.1016/0042-6822(84)90255-1. [DOI] [PubMed] [Google Scholar]
- 41.Sanders D A. Sulfhydryl involvement in fusion mechanisms. In: Hilderson H, Fuller S, editors. Fusion of biological membranes and related problems. New York, N.Y: Kluwer Academic/Plenum Publishers; 2000. pp. 483–514. [Google Scholar]
- 42.Schultz A, Rein A. Maturation of murine leukemia virus env proteins in the absence of other viral proteins. Virology. 1985;145:335–339. doi: 10.1016/0042-6822(85)90168-0. [DOI] [PubMed] [Google Scholar]
- 43.Shinnick T M, Lerner R A, Sutcliffe J G. Nucleotide sequence of Moloney murine leukaemia virus. Nature. 1981;293:543–548. doi: 10.1038/293543a0. [DOI] [PubMed] [Google Scholar]
- 44.Steck F T, Rubin H. The mechanism of interference between an avian leukosis virus and Rous sarcoma virus. I. Establishment of interference. Virology. 1966;29:628–641. doi: 10.1016/0042-6822(66)90287-x. [DOI] [PubMed] [Google Scholar]
- 45.Steck F T, Rubin H. The mechanism of interference between an avian leukosis virus and Rous sarcoma virus. II. Early steps of infection by RSV of cells under conditions of interference. Virology. 1966;29:642–653. doi: 10.1016/0042-6822(66)90288-1. [DOI] [PubMed] [Google Scholar]
- 46.Sutcliffe J G, Shinnick T M, Green N, Liu F T, Niman H L, Lerner R A. Chemical synthesis of a polypeptide predicted from nucleotide sequence allows detection of a new retroviral gene product. Nature. 1980;287:801–805. doi: 10.1038/287801a0. [DOI] [PubMed] [Google Scholar]
- 47.Suzuki S. FV-4: a new gene affecting the splenomegaly induction by Friend leukemia virus. Jpn J Exp Med. 1975;45:473–478. [PubMed] [Google Scholar]
- 48.Taylor G M, Sanders D A. The role of the membrane-spanning domain sequence in glycoprotein-mediated membrane fusion. Mol Biol Cell. 1999;10:2803–2815. doi: 10.1091/mbc.10.9.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Witte O N, Tsukamoto A A, Weissman I L. Cellular maturation of oncornavirus glycoproteins: topological arrangement of precursor and product forms in cellular membranes. Virology. 1977;76:539–553. doi: 10.1016/0042-6822(77)90236-7. [DOI] [PubMed] [Google Scholar]
- 50.Yoshikura H, Naito Y, Moriwaki K. Unstable resistance of G mouse fibroblasts to ecotropic murine leukemia virus infection. J Virol. 1979;29:1078–1086. doi: 10.1128/jvi.29.3.1078-1086.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang F, Ya L T, Iwatani Y, Higo K, Suzuki Y, Tanaka M, Nakahara T, Ono T, Sakai H, Kuribayashi K, Ishimoto A. Resistance to Friend murine leukemia virus infection conferred by the Fv-4 gene is recessive but appears dominant from the effect of the immune system. J Virol. 2000;74:6193–6197. doi: 10.1128/jvi.74.13.6193-6197.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu N-L, Cannon P M, Chen D, Anderson W F. Mutational analysis of the fusion peptide of Moloney murine leukemia virus transmembrane protein p15E. J Virol. 1998;72:1632–1639. doi: 10.1128/jvi.72.2.1632-1639.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]