Abstract
Background
The hippocampus, with its complex subfields, is linked to numerous neuropsychiatric traits. While most research has focused on its global structure or a few specific subfields, a comprehensive analysis of hippocampal substructures and their genetic correlations across a wide range of neuropsychiatric traits remains underexplored. Given the hippocampus’s high heritability, considering hippocampal and subfield volumes (HASV) as endophenotypes for neuropsychiatric conditions is essential.
Methods
We analyzed MRI-derived volumetric data of hippocampal and subfield structures from 41,525 UK Biobank participants. Genome-wide association studies (GWAS) on 24 HASV traits were conducted, followed by genetic correlation, overlap, and Mendelian randomization (MR) analyses with 10 common neuropsychiatric traits. Polygenic risk scores (PRS) based on HASV traits were also evaluated for predicting these traits.
Results
Our analysis identified 352 independent genetic variants surpassing a significance threshold of 2.1 × 10−9 within the 24 HASV traits, located across 93 chromosomal regions. Notably, the regions 12q14.3, 17q21.31, 12q24.22, 6q21, 9q33.1, 6q25.1, and 2q24.2 were found to influence multiple HASVs. Gene set analysis revealed enrichment of neural differentiation and signaling pathways, as well as protein binding and degradation. Of 240 HASV-neuropsychiatric trait pairs, 75 demonstrated significant genetic correlations (P < 0.05/240), revealing 433 pleiotropic loci. Particularly, genes like ACBD4, ARHGAP27, KANSL1, MAPT, ARL17A, and ARL17B were involved in over 50 HASV-neuropsychiatric pairs. Leveraging Mendelian randomization analysis, we further confirmed that atrophy in the left hippocampus, right hippocampus, right hippocampal body, and right CA1-3 region were associated with an increased risk of developing Parkinson’s disease (PD). Furthermore, PRS for all four HASVs were significantly linked to a higher risk of Parkinson’s disease (PD), with the highest hazard ratio (HR) of 1.30 (95% CI 1.18–1.43, P = 6.15 × 10⁻⁸) for right hippocampal volume.
Conclusions
These findings highlight the extensive distribution of pleiotropic genetic determinants between HASVs and neuropsychiatric traits. Moreover, they suggest a significant potential for effectively managing and intervening in these diseases during their early stages.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12916-024-03682-8.
Keywords: Hippocampus, Neuropsychiatric, Pleiotropic, Parkinson’s disease
Background
The human hippocampus, situated in the medial temporal lobe, is crucial for fundamental cognitive functions such as learning, memory [1, 2], and stress regulation [3]. This intricate structure consists of histologically distinct subfields, each of which exhibits structural and functional changes associated with various complex neurological and psychiatric conditions, including Alzheimer’s disease (AD) [4], Parkinson’s disease (PD) [5], bipolar disorder (BIP) [6], and schizophrenia (SCZ) [7]. Despite the hippocampus’ significance, current research has predominantly focused on either its global architecture or on the association between specific subfields and a limited number of neuropsychiatric traits. A comprehensive understanding of the hippocampal substructures’ correlations and impacts across a broader spectrum of disorders remains largely unexplored.
As a recognized biomarker for AD, hippocampal volume has an estimated heritability exceeding 75% based on twin studies [8]. This high heritability, therefore, underscores the importance of considering hippocampal and subfield volumes (HASV) as endophenotypes for a spectrum of neuropsychiatric conditions. Among imaging modalities, magnetic resonance imaging (MRI) is favored for its superior soft tissue contrast, positioning it as the non-invasive examination tool of choice for studying the human brain in vivo. Moreover, recent advancements in MRI technology, paired with progress in the adaptive segmentation of hippocampal subregions, have significantly enhanced our ability to rapidly and accurately estimate the volume of the entire hippocampus and its individual subregions from MRI data [9]. Consequently, this presents an opportunity to integrate large-scale brain MRI and genetic data, thereby increasing our understanding of the genetic architecture underlying HASV and their association with a range of neuropsychiatric traits.
Importantly, neuropsychiatric traits, notably AD and PD, exhibit relentless progression once diagnosed. Hence, proactive identification of high-risk subpopulations in their early stages is imperative for timely interventions. Currently, there is a scarcity of genetic or biological markers for risk prediction in these diseases. We propose leveraging polygenic risk scores (PRS) derived from HASV traits for this purpose, as the computation of PRS requires only partial genetic information, which is essentially fixed at birth. PRS have been extensively investigated for predicting disease onset and have proven invaluable in identifying high-risk populations and guiding decision-making in other diseases [10]. However, to our knowledge, there is limited research assessing the predictive role of PRS related to hippocampal volume in the occurrence of these diseases.
In the current study, we initially conducted genome-wide association studies (GWAS) on 24 well-segmented and quantified HASV in a cohort of 41,525 individuals. Subsequently, we performed variants and genes annotation analyses to investigate the biological significance of the GWAS findings, with a particular focus on the genomic distribution of these loci and the genes that significantly impact multiple HASV traits. Then, we assessed the genetic associations between the 24 HASVs and 97 other regional brain volumes, along with a range of neuropsychiatric traits using genetic correlation and genetic overlap analyses. For the HASV-neuropsychiatric trait pairs that showed significant correlations, we sequentially investigated the pleiotropic associations through various statistical genetic approaches, from the genome-wide to the variant and gene levels, to disentangle the underlying shared genetic etiology. Following this, we utilized Mendelian randomization (MR) to further confirm the causal connections across these significant genetic associations. Lastly, to facilitate clinical application and early intervention, we established PRS derived from HASV traits and validated their predictive ability for these neuropsychiatric traits in approximately 450,000 individuals without brain imaging data. An overview of the study design and analyses is provided in Fig. 1.
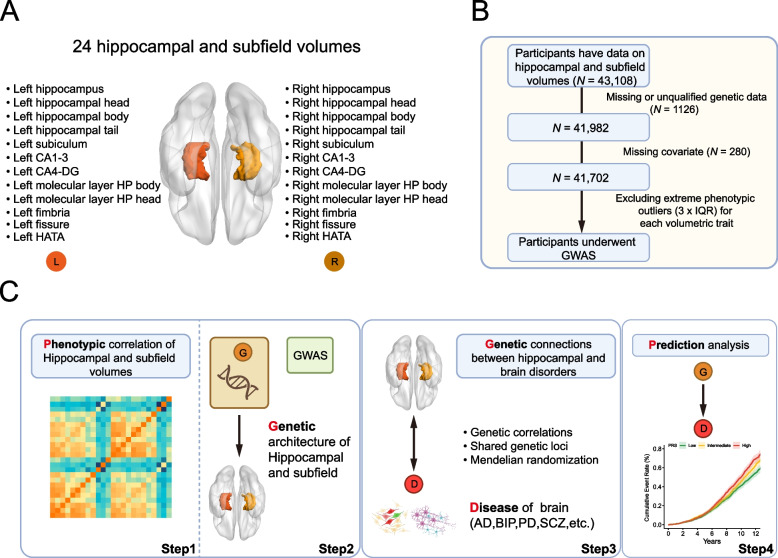
Fig. 1.
Study overview and workflow.A Schematic illustration of the hippocampus regions. B Sample selection flowchart. C A brief description of the overall workflow and major analyses
Methods
Study population
The UKBB is a large prospective cohort study of over 500,000 participants recruited at 22 assessment centers across the UK between 2006 and 2010 [11]. It has gathered a wealth of information on participants, including health and lifestyle data, physical measurements, biological samples, imputed genome-wide genotypes, and a portion of participants had brain MRI data [12]. All participants provided informed consent. The ethical committees from the North West Multi-Center Research Ethics approved the study.
Sample selection
In this study, we initially downloaded volumetric data for 44 hippocampal and subfield structures from the UK Biobank (UKB) official website, which had been segmented using Freesurfer (Field ID 26620–26663). The initial dataset included hippocampal segmentation information for 43,108 individuals who had not withdrawn consent as of December 2020. To ensure data quality, we excluded individuals with missing genetic data or those whose data did not meet quality standards (1126 individuals), as well as those lacking covariate information (280 individuals) (see Fig. 1B). Ultimately, our sample consisted of 41,702 individuals, with an average age of 55.0 years at the time of enrollment, and 47.5% were male (for detailed demographic information, refer to (Additional file 1: Table S1). It is important to note that to minimize the partial volume effect [13], we combined the volumes of certain subfields: CA1 and CA2/3 were merged as CA1-3, CA4, and GC-ML-DG were merged as CA4-DG, and parasubiculum, presubiculum, and subiculum were merged as subiculum. As a result, we obtained phenotypic data for 24 merged hippocampal and subfield volumes (HASV), with these subfields symmetrically distributed in the left and right cerebral hemispheres, as illustrated in Fig. 1Aand detailed in Additional file 1: Table S2. It is worth emphasizing that our subsequent GWAS analyses were conducted based on these 24 merged HASV phenotypes. Prior to performing GWAS for each HASV trait, we further excluded individuals with extreme phenotype outliers (identified using the three times the interquartile range, IQR, criterion). And the largest sample size among these HASV traits is 41,525. The means and standard deviations of these 24 HASV traits can be found in Additional file 1: Table S2 for reference in subsequent research and analysis. The UKB Data-Fields of covariates were listed in Additional file 1: Table S3. The definitions were used for GWAS participant exclusion and PRS assessment. And the Data Field in UKBB of the first in-patient diagnosis of relevant brain disorders was provided in Additional file 1: Table S4.
Genotyping and imputation
Detailed information on genotyping and imputation in the UKB has been described previously1. Briefly, participants were genotyped based on UK BiLEVE Axiom™ Array by Affymetrix (807,411 markers for 49,950 participants) and UKB Axiom Array by Affymetrix (825,927 markers for 438,427 participants). Genotype imputation was based on merged UK10K sequencing and 1000 Genomes phase3 reference panels with SHAPEIT3 and IMPUTE32. Variant positions were keyed to the GRCh37 human genome reference.
Genome-wide association study
We computed residuals for each HASV trait by regressing them on covariates such as age, sex, BMI, and imaging center. Subsequently, after rank-based inverse normal transformation of these residuals (Additional file 2: Fig. S1-2), we performed GWAS for the transformed HASV traits. This analysis utilized approximately 8.5 million well-imputed variants, each with a minor allele frequency (MAF) of ≥ 1%, and an imputation quality (INFO) score > 0.3 and was conducted using BOLT-LMM v2.3.6 [14]. GWAS analysis models were adjusted for age, sex, BMI, and principal component (PC) 1–10. BOLT-LMM accounts for ancestral heterogeneity, cryptic population structure, and sample relatedness by fitting a linear mixed model with a Bayesian mixture prior as a random effect [15–17]. Previous evidence supports the use of LMM approaches to perform GWAS of admixed populations, which may provide favorable statistical power [16, 18, 19], and similar approaches have been taken previously [15–17]. As expected, our GWAS analyses did not reveal any evidence of confounding arising from population stratification or cryptic relatedness in our 24 GWASs. The genomic inflation factor ranged from 1.10 to 1.20, while the linkage disequilibrium (LD)-score regression intercept [20, 21] consistently remained below 1.03. Moreover, (intercept − 1)/(mean(χ2) − 1) was less than 0.12, further supporting the conclusion that HASV traits were influenced more by polygenicity than population structure (Additional file 1: Table S3). Observed scale heritability (h2) was estimated using the slope of LDSC regression. To identify genetic loci, we uploaded this summary statistic to the FUMA platform v1.5.03. Using the 1000GPhase3 EUR as a reference panel, we identified independent significant SNPs at the statistical significance threshold P < 5 × 10–8. All SNPs at r2 < 0.6 with each other were considered as independent significant SNPs and a fraction of the independent significant SNPs in approximate linkage equilibrium with each other at r2 < 0.1 were considered as lead SNPs.
Functional follow-up with FUMA
We utilized two main approaches to map genome-wide significant loci to genes via FUMA default settings and specialized datasets, as described as follows: (1) positional mapping of variants, whereby variants within a 10kB window from known protein-coding genes in the human reference assembly (GRCh37/hg19) are mapped and (2) eQTL mapping whereby allelic variations at a variant is significantly linked to expression of a gene, where we considered eQTLs within heart atrial appendage and heart left ventricle from GTEx v8.
We also performed a generalized gene set analysis using MAGMA within FUMA. Variants within exonic, intronic, and untranslated regions were chosen for each gene. The 18,888 protein-coding genes were used in MAGMA. The mean of the summary statistic (χ2) of GWAS for the variants in a gene was used to determine the gene-based P-value4. The Bonferroni method was used to calculate the P-value significance threshold, which is 2.64 × 10–6 when 0.05 is divided by the total number of genes (18,888).
Transcriptome-wide association study
For each of the 24 HASV traits, we performed a TWAS to identify the most strongly associated gene at each locus based on imputed cis-regulated gene expression. We used FUSION with eQTL data from GTEx v8. Precomputed transcript expression reference weights for the brain–hippocampus (3457 genes) were obtained from the FUSION authors, website (http://gusevlab.org/projects/fusion/). A significance threshold of P < 6.03 × 10−7 was applied, accounting for the number of genes and HASV traits. FUSION was then run with its default settings.
Pathway enrichment and tissue expression analyses
Functional enrichment and pathway characterization of the candidate genes associated with each HASV trait were performed using the clusterProfiler package [22]. Tissue expression analyses were obtained from GTEx which were also integrated in FUMA. Average gene expression per tissue type was utilized as a gene covariate to test for a positive link between gene expression in a given tissue type and genetic correlations.
Genetic correlation analysis
Using summary statistics, we applied LDSC software [20, 21] to estimate the genetic correlations (1) between 24 HASV traits, (2) between 24 HASV traits and 97 other regional brain volumes [23], and (3) between 24 HASV traits and 10 common brain disease: AD (Alzheimer’s disease) [24], attention-deficit hyperactivity disorder (ADHD) [25], anorexia nervosa (AN) [26], anxiety disorder (ANX) [27], bipolar disorder (BIP) [28], epilepsy [29], insomnia, PD (Parkinson’s disease) [30], post-traumatic stress disorder (PTSD) [31], and schizophrenia (SCZ) [32]. These analyses were performed according to the standard analysis process of LDSC. We performed LDSC using well-imputed HapMap3 variants (http://ldsc.broadinstitute.org/static/media/w_hm3.noMHC.snplist.zip) and pre-computed LD scores of European ancestry from the 1000 Genomes Project Phase3 (https://data.broadinstitute.org/alkesgroup/LDSCORE/eur_w_ld_chr.tar.bz2). We did not constrain the intercepts in LDSC analysis, which could not only account for residual confounding but also indicate whether there was potential sample overlap between two GWAS studies.
Genetic overlap analysis
Given that the genetic correlation analysis only reflects the overall correlation across the genome between traits, we further applied GPA (genetic analysis incorporating pleiotropy and annotation) [33] to explore the overall genetic overlap. For each trait pair, GPA relies on four distinct models to classify SNPs into four categories, aims to estimate the proportions of SNPs in each model, and uses a likelihood ratio test to assess the statistical significance for overall genetic overlap [33]. GPA assumes that P-values from null SNPs (not associated with the trait) follow the uniform distribution and non-null SNPs (associated with the trait) follow the Beta distribution, then extends the assumption to two GWASs and proposes four models (M00, M10, M01, and M11) to classify these SNPs into four categories: (i) SNPs associated with neither of traits, (ii) SNPs only associated with the first trait, (iii) SNPs only associated with the second trait, and (iv) SNPs associated with both traits. GPA aims to estimate the proportions of SNPs in these models (PM) and uses likelihood ratio test (LRT) to assess the statistical significance for overall genetic overlap. Note that the proportion of risk SNPs should not be extremely small to enable GPA to work well [33]. To alleviate the influence of LD on GPA, we performed LD pruning based on the 1000 Genomes Phase 3 European-ancestry genotypes using PLINK1.9 to obtain relatively independent SNPs.
Pairwise pleiotropic analysis using PLACO
For the union set of pairwise traits with significant genetic correlation or genetic overlap, we used the recently developed pleiotropic analysis under composite null hypothesis (PLACO), which could account for potential correlation between two traits, to identify pleiotropic SNPs [34]. For a given variant, PLACO detects pleiotropic associations by considering a composite null hypothesis, where the null hypothesis H0 is a composite of the global null {beta trait1 = beta trait2 = 0} and the sub-null hypotheses are {beta trait1 = 0, beta trait2 ≠ 0} and {beta trait1 ≠ 0, beta trait2 = 0}. That is, PLACO tests H0: beta trait1 × beta trait2 = 0 vs H1: beta trait1 × beta trait2 ≠ 0, and the test statistic of PLACO is TPLACO = Ztrait1 Ztrait2 [34]. For each trait pair, we denote trait1 and trait2 as HASV trait and brain disease, beta trait1 and beta trait2 as the effect sizes of a SNP on two traits, Ztrait1 and Ztrait2 as the observed Z-scores of a SNP from corresponding GWAS summary data, respectively. The rejection of H0 statistically suggests that the SNP would be a potential pleiotropic variant shared between two traits. Overlapped SNPs between GWASs of each pairwise traits were included and the summary statistics were harmonized to align to the same effect allele. SNPs with squared Z-scores above 80 were removed since extremely large effect sizes could produce spurious signals [34]. We de-correlated the Z-scores using the correlation matrix estimated from GWAS summary statistics to account for potential sample overlap. SNPs with P < 6.67 × 10−10 (5 × 10−8/75, Bonferroni correction) were declared as significant pleiotropic variants.
Mendelian randomization (MR) analysis
The TwoSampleMR [35] and MendelianRandomization [36] R packages were primarily used to perform two-sample MR. Effect allele coding was harmonized across phenotypes using the harmonise_data function. Strand ambiguous SNPs were excluded. Genome-wide significant SNPs were LD clumped (P < 5 × 10–8, r2 ≤ 0.001 in 1000 Genomes Phase 3 European data, over a 10 megabase window) to ensure independence. SNPs within highly pleiotropic regions, the MHC region (hg19 coordinates: Chromosome 6, 28,477,797–33,448,354 base pairs) was excluded. To further mitigate the impact of pleiotropy, we refined our instrument variables (IVs) by removing SNPs associated with confounding factors such as socioeconomic status, education, drinking, and smoking behavior. Additionally, we excluded IVs identified as pleiotropic in our PLACO analysis. Finally, we assessed the strength of genetic associations of instrumental SNPs and addressed the issue of weak instrument bias by calculating phenotype variance explained (PVE) by genetic variants and F statistics. The primary MR analysis was conducted using the inverse variance weighted (IVW) estimator with multiplicative random effects. Additional MR analysis was performed using MR-Egger [37], weighted median [38], weighted mode [39], IVW method using robust regression (MR-Robust) [40], and MR robust adjusted profile score (MR-RAPS) [41].
MR sensitivity analyses
Several sensitivity analyses were conducted to assess the robustness of findings and account for pleiotropy. MR PRESSO [42] was used to identify heterogeneity (global test) and outliers (outlier test) and to determine if the outlier-adjusted IVW estimate significantly differed from the unadjusted. The MR-Egger intercept, Cochran’s Q statistic, and MR-PRESSO Global Test were used to confirm that pleiotropic effects were not driving the observed associations. To evaluate correlated horizontal pleiotropy, the CAUSE method [43] was applied. This method fits a series of nested models: a “null” model where only uncorrelated horizontal pleiotropy (defined as direct effects of genes on the outcome with net zero effect) is modeled (parameter q), a “sharing” model where an additional parameter (parameter eta) is fit to account for correlated horizontal pleiotropy, and a “causal” model where a causal effect parameter (parameter gamma) is fit in addition to the sharing parameter. To test the hypothesis that a causal model explained the relationship better than a sharing model, the causal and sharing model fits were compared using the difference in the expected log pointwise posterior density. Specifically, if the causal model fits better than the sharing model, this implies that the additional complexity needed to model a causal effect is justified and thus is evident that data are consistent with a causal effect. If, however, there is not significant evidence that the causal model fits better than the sharing model, this implies that shared pleiotropy alone is sufficient to explain the observed association.
Polygenic risk score development
We used the C + T (clumping + thresholding) method [44] to construct the polygenic risk score (PRS) of each HCAS trait based on the effect sizes derived from the HASV GWASs. The PRS was calculated through a weighted model, as shown below.
where β values (the log of odds ratio) is the summary statistic for the effective allele and G is the number of the effective allele observed. We used variants with genome-wide significant (P < 5 × 10–7) and clumping window (r2 < 0.1, kb = 250) to derive PRS. We categorized participants into three genetic risk levels: low (lowest tertile), intermediate (second tertile), and high (highest tertile).
Results
Genome-wide association studies of 24 hippocampal and subfield volumes
To understand the common genetic basis for variation in hippocampal volumes, we performed a series of GWAS on 24 HASV with a maximum sample size of 41,525 individuals. The baseline characteristics of the study population are detailed in Additional file 1: Table S1, whereas Additional file 1: Table S2, Additional file 2: Fig. S1A and Fig. S2 provide a summary of the 24 HASV traits. Subsequently, we aimed to assess the phenotypic correlations among these HASV traits. Approximately one-third of the phenotype pairs exhibited correlation values (r2) greater than 0.70, while half showed correlations ranging from 0.30 to 0.70. Additionally, we observed weak negative correlations between certain phenotype pairs, such as the correlation between left hippocampal fissure and right fimbria (r2 = − 0.10, P < 0.05/276, Bonferroni corrected), as illustrated in Additional file 2: Fig. S1B.
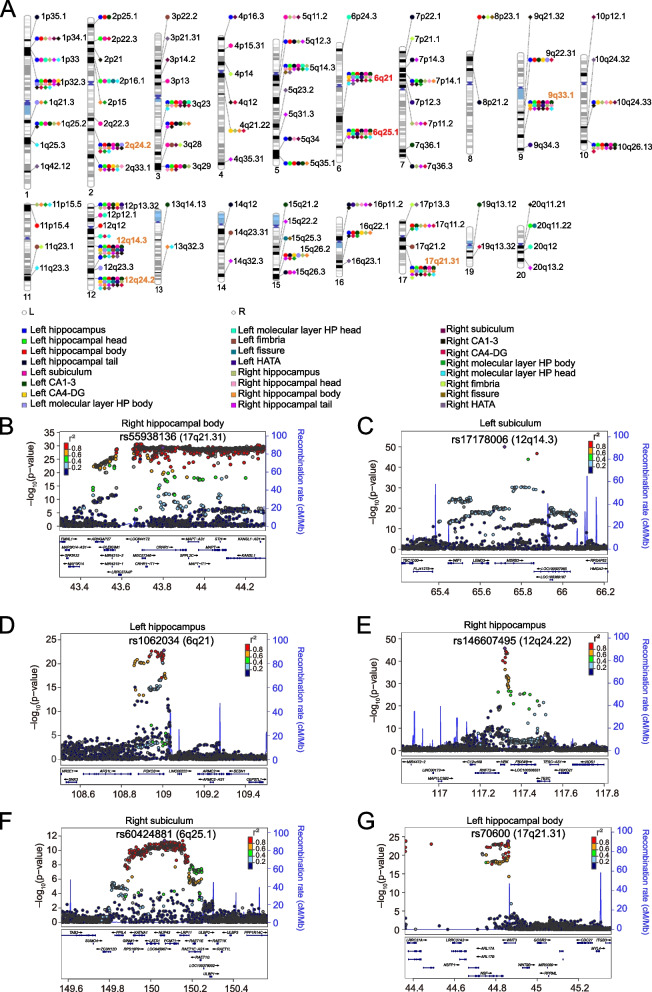
We identified 578 significant variant–trait associations at P < 5.0 × 10−8, of which 352 associations survived P < 2.1 × 10−9 (Additional file 1: Table S5-6, Additional file 2: Fig. S3-4). Notably, among the 578 variants, 93 were found to be associated with at least two HASV traits, resulting in a total of 317 unique variants associated with the 24 HASV traits, distributed across 93 distinct chromosomal regions, including specific regions like12q14.3 [45, 46], 17q21.31 [45], 12q24.22 [45, 46], 6q21, 9q33.1 [47], 6q25.1, and 2q24.2 [46, 47] (Fig. 2A). Particularly, the lead single-nucleotide polymorphism (SNP) rs55938136 on chromosome 17 within the LINC02210-CRHR1 (Fig. 2B) was associated with 19 HASV traits. Similarly, rs17178006 within the MSRB3 on chromosome 12 (Fig. 2C) was associated with 18 HASV traits. In addition, rs1062034 on chromosome 6 within the FOXO3 (Fig. 2D) was linked to 12 HASV traits, while rs146607495 within the HRK on chromosome 12 (Fig. 2E) demonstrated an association with 12 HASV traits as well. Notably, 6q21 and 6q25.1 were recently reported to influence multiple HASVs [48].
Fig. 2.
Genomic loci associated with 24 hippocampal and subfield volumes (HASVs). A Ideogram of 93 genomic regions associated with 24 hippocampal and subfield volumes. Orange name labels denote genomic regions that have been widely reported to be associated with hippocampal volume, while red represents newly identified loci in this study. B–G These regional plots illustrate an instance of corresponding loci that exerted influence on multiple HASV traits
Functional characterization of risk variants
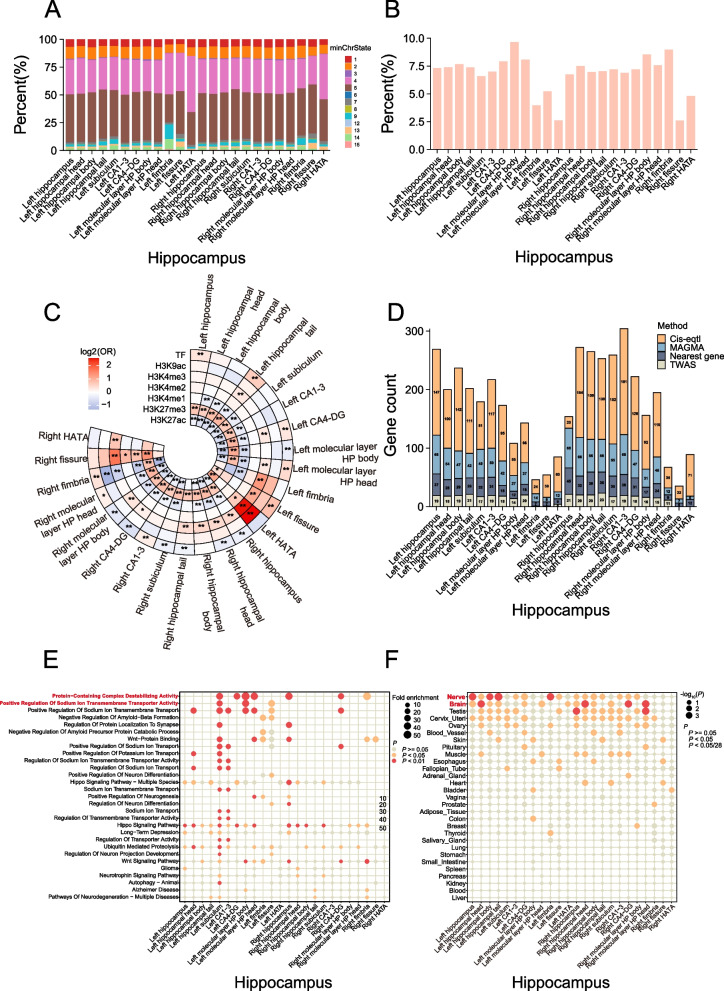
We functionally annotated candidate variants that were in linkage disequilibrium (r2 ≥ 0.6) with one of the independent significant SNPs for each HASV traits using functional mapping and annotation of GWAS (FUMA) [49] (Additional file 1: Table S7). The number of candidate variants ranged from 539 in the right hippocampal fissure to 8589 in the right hippocampus. We next observed that approximately 90% of these candidates in each HASV trait variants were localized within accessible chromatin regions (which is less than 1% of the total number of approximately 8.5 million GWAS SNPs), represented by chromatin states with scores ranging from 1 to 7 (Fig. 3A), which suggests their potential functional significance [50, 51]. Additionally, around 6.8% of the candidate SNPs were classified under regulomeDB categories 1 or 2 (Fig. 3B), indicating their potential involvement in regulatory functions [50]. Subsequently, we investigated whether these candidate variants were enriched among genetic regulatory elements. We generated control variants in a 1:1 ratio using vSampler [52]. The enrichment analysis revealed that candidate variants exhibited a positive association with H3K4 trimethylation marks (H3K4me3) when compared to control variants. Conversely, candidate SNPs showed a negative enrichment for H3K27 acetylation marks (H3K27ac) (Fig. 3C). Notably, H3K27me3 is a well-known mark associated with gene silencing and downregulation [53], while H3K27ac is linked to enhancer regions and can promote gene transcription and expression [54]. Our findings lead us to reasonably infer that these candidate SNPs could be implicated in gene regulation, potentially modulating gene expression levels.
Fig. 3.
Annotation of risk variants and genes. A The minimum chromatin state across 127 tissue and cell types for candidate SNPs for each of the 24 HASV traits, with lower states indicating higher accessibility and states 1–7 referring to open chromatin states. B The bar charts represent the proportions of RegulomeDB scores 1 or 2 among the risk variants for each of the 24 HASV traits. A lower score suggests a higher likelihood of having a regulatory function. C Histone modifications and transcription factor (TF) peaks were primarily sourced from hippocampus samples provided by ENCODE. When hippocampus-specific data was unavailable, brain data was used instead. The enrichment of candidate SNPs in these epigenomic marks was assessed relative to control variants, which were generated in a 1:1 ratio using vSampler. Statistical significance was determined using a Fisher test. An asterisk denotes statistically significant differences (*P < 0.05; **P < 2.08 × 10−4, Bonferroni corrected). D The stacked bar charts depict the number of genes mapped using four distinct strategies: physical position, eQTL association, transcriptome-wide association study (TWAS), and multi-marker analysis of genomic annotation (MAGMA), for each of the 24 HASV traits. E Pathway analysis of genes associated with each of the 24 HASV traits based on the molecular signatures database. F Tissue expression results across 29 specific tissue types from GTEx v8 in FUMA. The chromatin states are 1 = active transcription start site (TSS); 2 = flanking active TSS; 3 = transcription at gene 5’ and 3’; 4 = strong transcription; 5 = weak transcription; 6 = genic enhancers; 7 = enhancers; 8 = zinc finger genes and repeats; 9 = heterochromatic; 10 = bivalent/poised TSS; 11 = flanking bivalent/poised TSS/Enh; 12 = bivalent enhancer; 13 = repressed PolyComb; 14 = weak repressed PolyComb; 15 = quiescent/low
Identification and functional annotation of susceptible genes associated with HASVs
We further sought to identify candidate genes influencing HASV traits variation by combining evidence from physical position, eQTL association, transcriptome-wide association study (TWAS), and multi-marker analysis of genomic annotation (MAGMA). Our analysis revealed a total of 4184 mapped genes associated with the 24 HASV traits (Fig. 3D). Specifically, taking into account the physical position within ± 1 Mb of the lead variant, we pinpointed 578 genes (Additional file 1: Table S6). Additionally, eQTL mapping led to the discovery of 2289 genes associated with HASV traits (Additional file 1: Table S8). Meanwhile, TWAS analysis contributed to the identification of 351 genes (P < 6.03 × 10–7) (Additional file 1: Table S9). Furthermore, MAGMA analyses yielded 996 significant genes (mean χ2 statistics, P < 2.64 × 10−6) (Additional file 1: Table S10). Notably, our findings revealed that 323 genes shared at least two HASV traits or were identified by multiple methods, resulting in a total of 694 unique genes associated with the 24 HASV traits (Additional file 1: Table S11). Among these genes, several notable ones, including MSRB3, HRK, CRHR1, FOXO3, NUP43, ASTN2, GINM1, LEMD3, WNT3, PCMT1, SSBP3, LRP11, and MAPT, exerted significant pleiotropic on nearly 24 HASV traits (Additional file 2: Fig. S5).
Subsequently, we conducted gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses to gain a comprehensive understanding of the functions of these susceptible genes. As expected, these genes showed significant enrichment in several functional categories, including (1) ion transmembrane transport processes (e.g., regulation of sodium ion transport, positive regulation of potassium ion transport), (2) neural differentiation and signaling pathways (e.g., regulation of neuron differentiation, hippo signaling pathway, Wnt signaling pathway), (3) protein binding and degradation (e.g., protein-containing complex destabilizing activity, negative regulation of amyloid-beta formation, Wnt-protein binding), and (4) brain-related diseases (e.g., AD, long-term depression, glioma) (Fig. 3E and Additional file 1: Table S12). The tissue enrichment analysis revealed that these genes were predominantly expressed in nerve and brain tissues, aligning with our expectations (Fig. 3F and Additional file 1: Table S13). In summary, these findings represent a substantial expansion of our understanding of the genetic basis of HASV traits and underscore their role in the development of brain-related diseases.
Heritability and genetic correlation of HASVs
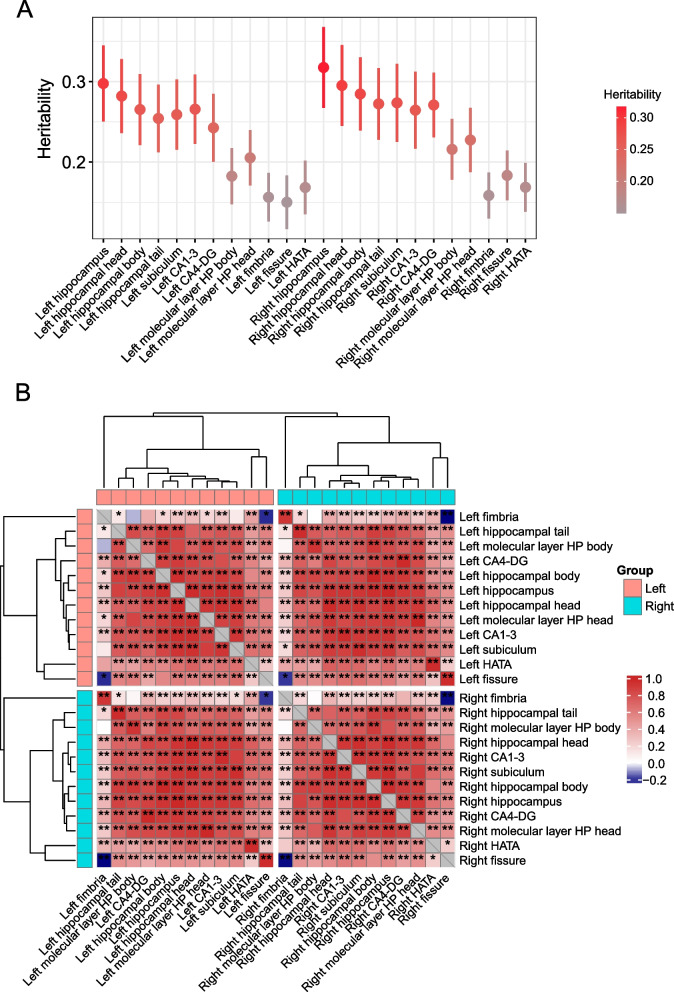
Using summary statistics, we applied LD score regression (LDSC software) [20, 21] to estimate the heritability and genetic correlation among these 24 HASV traits. The mean heritability (h2) was 0.24 for the 24 traits (ranging from h2 = 0.15 of the left hippocampal fissure to h2 = 0.32 for the right hippocampus trait; Fig. 4A). Of significant note were the robust genetic correlations observed within these traits (Fig. 4B and Additional file 1: Table S14). Particularly, the genetic correlations between corresponding regions of the left and right hippocampus consistently exceeded 0.95 (P < 0.05/276, Bonferroni corrected). Additionally, when focusing on individual cerebral hemispheres, moderate genetic correlations were found between the hippocampus fimbria, hippocampal fissure, and HATA subregions with the remaining nine regions, indicating moderate influences. In contrast, stronger genetic correlations emerged among the remaining nine regions ranging from 0.80 to 0.90. These intriguing observations suggest these traits are under pleiotropic genetic control.
Fig. 4.
SNP heritability and genetic correlations of 24 HASV traits. A SNP heritability of 24 HASV traits. B Genetic correlations between 24 HASV traits. One asterisk denotes the nominal level (0.05), while two asterisks indicate genetic correlations that have survived multiple testing adjustments using the Bonferroni correction (P < 0.05/276). The colors represent the magnitude of genetic correlations
Building on this groundwork, we extended our inquiry to explore the genetic connections between the HASV traits and 97 other regional brain volumes [23]. Employing a significance threshold of P < 2.15 × 10–5 (0.05/2328, Bonferroni corrected), we unearthed noteworthy associations between HASV traits and various brain regions, encompassing total brain volume, amygdalae, white matter, thalami, ventricles, and more, as visually presented in Additional file 2: Fig. S6 and comprehensively detailed in Additional file 1: Table S15. As expected, we found clear genetic correlations between the amygdala and multiple HASV traits, particularly with the HATA (hippocampal amygdala transition area) subregion (genetic correlation > 0.47, P < 5.15 × 10−10). This reinforces the functional interplay between the amygdala and the hippocampal complex, two components nestled within the medial temporal lobe, each tied to discrete memory systems. In emotional contexts, these systems intersect, with the amygdala regulating the encoding and retention of hippocampal-dependent memories, while the hippocampal complex shapes the amygdala's reaction to emotional stimuli [55].
Genetic correlations and genetic overlaps between HASVs and neuropsychiatric traits
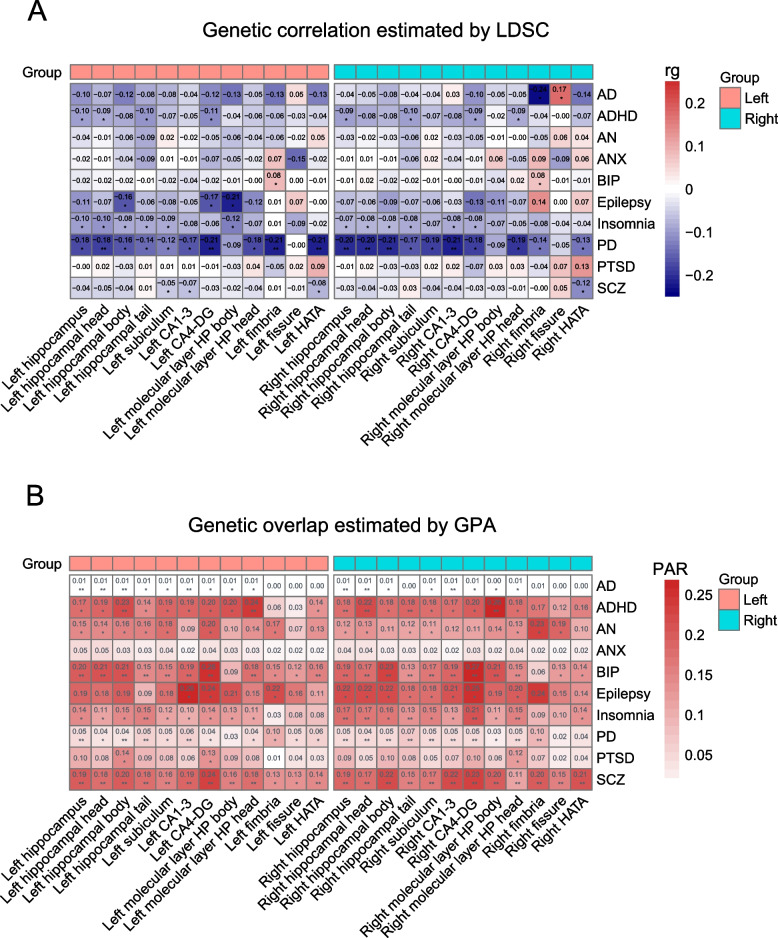
Previous research has revealed that the hippocampus is a critical brain region involved in memory formation, spatial navigation and other higher-level cognitive functions [56–58], and structural abnormalities in the hippocampus have been connected to a variety of neurological and psychiatric diseases [4, 5, 59–61]. We performed genetic correlation analyses between HASVs and the ten most frequent neuropsychiatric traits: AD, attention-deficit hyperactivity disorder (ADHD), anorexia nervosa (AN), anxiety disorder (ANX), BIP, epilepsy, insomnia, PD, post-traumatic stress disorder (PTSD), and schizophrenia (SCZ). As expected, the majority of HASV traits displayed significant positive genetic associations with PD from LDSC (ranging from 0.18 to 0.21, P < 0.05/240, Bonferroni corrected), as depicted in Fig. 5A and Additional file 1: Table S16. We further applied genetic analysis incorporating pleiotropy and annotation (GPA) [33] to explore the overall genetic overlap between those traits. We discovered genetic overlap not only just with PD but also with other diseases that had previously shown no clear genetic link in genetic correlation analysis, such as AD, ADHD, AN, BIP, insomnia, PTSD, and SCZ (Fig. 5B and Additional file 1: Table S17). By integrating the results of genetic correlation and genetic overlap analyses, we finally produced a set of 75 significant pairwise traits (P < 0.05/240, Bonferroni corrected, Table 1). These findings imply that the genetic foundation of HASVs is complex and involves multiple neuropsychiatric traits.
Fig. 5.
Genetic connections between 24 HASV traits and 10 brain disorders.A Genetic correlations between 24 HASV traits (X-axis) and 10 brain disorders (Y-axis). Genetic correlation was estimated using the LDSC method. Asterisk denotes statistically significant differences, *P < 0.05; **P < 2.08 × 10−4 (0.05/240, Bonferroni corrected). B Genetic overlaps between 24 HASV traits (X-axis) and 10 brain disorders (Y-axis). Genetic overlap was estimated using the GPA method. We introduced PAR as PM 11/(PM10 + PM01 + PM11) to represent the proportion of pleiotropic SNPs associated with both traits against the proportion of SNPs associated with at least 1 trait. Asterisk denotes statistically significant differences, *P < 0.05; **P < 2.08 × 10−4 (0.05/240, Bonferroni corrected). LDSC, linkage disequilibrium score regression; GPA, genetic analysis incorporating pleiotropy and annotation method; PAR, pleiotropy association ratio; PM11, proportion of genetic variants associated with both traits; AD, Alzheimer’s disease; ADHD, attention-deficit hyperactivity disorder; AN, anorexia nervosa; ANX, anxiety disorder; BIP, bipolar disorder; PD, Parkinson’s disease; PTSD, post-traumatic stress disorder; SCZ, schizophrenia
Table 1.
Seventy-five significant trait pairs with genetic correlations or overlaps
| Hippocampus | Neuropsychiatric traits | rg | se | P value for LDSC | PM11 | PAR | P value for GPA | Either significant |
|---|---|---|---|---|---|---|---|---|
| Left hippocampus | AD | − 0.1021 | 0.0766 | 1.83E-01 | 0.0005 | 0.0090 | 2.45E-06 | GPA |
| Left hippocampus | BIP | − 0.0232 | 0.0285 | 4.14E-01 | 0.0415 | 0.2023 | 5.64E-11 | GPA |
| Left hippocampus | PD | − 0.1757 | 0.0499 | 4.00E-04 | 0.0034 | 0.0467 | 1.24E-05 | GPA |
| Left hippocampus | SCZ | − 0.0428 | 0.0272 | 1.16E-01 | 0.0437 | 0.1944 | 2.24E-17 | GPA |
| Left hippocampal head | AD | − 0.0720 | 0.0733 | 3.26E-01 | 0.0005 | 0.0093 | 3.19E-05 | GPA |
| Left hippocampal head | BIP | − 0.0175 | 0.0288 | 5.43E-01 | 0.0410 | 0.2056 | 5.11E-11 | GPA |
| Left hippocampal head | PD | − 0.1808 | 0.0481 | 2.00E-04 | 0.0027 | 0.0404 | 5.48E-04 | LDSC |
| Left hippocampal head | SCZ | − 0.0460 | 0.0267 | 8.57E-02 | 0.0399 | 0.1807 | 9.26E-16 | GPA |
| Left hippocampal body | AD | − 0.1235 | 0.0798 | 1.22E-01 | 0.0005 | 0.0080 | 1.46E-04 | GPA |
| Left hippocampal body | ADHD | − 0.0825 | 0.0467 | 7.77E-02 | 0.0486 | 0.2256 | 1.20E-04 | GPA |
| Left hippocampal body | BIP | − 0.0175 | 0.0313 | 5.76E-01 | 0.0439 | 0.2127 | 9.57E-10 | GPA |
| Left hippocampal body | PD | − 0.1635 | 0.0563 | 3.70E-03 | 0.0031 | 0.0396 | 9.87E-05 | GPA |
| Left hippocampal body | SCZ | − 0.0414 | 0.0297 | 1.64E-01 | 0.0465 | 0.2041 | 8.62E-15 | GPA |
| Left hippocampal tail | BIP | − 0.0088 | 0.0308 | 7.75E-01 | 0.0296 | 0.1464 | 2.29E-06 | GPA |
| Left hippocampal tail | Insomnia | − 0.0866 | 0.0400 | 3.05E-02 | 0.0343 | 0.1511 | 1.40E-04 | GPA |
| Left hippocampal tail | PD | − 0.1441 | 0.0518 | 5.40E-03 | 0.0031 | 0.0545 | 3.99E-05 | GPA |
| Left hippocampal tail | SCZ | 0.0101 | 0.0305 | 7.40E-01 | 0.0385 | 0.1798 | 5.40E-15 | GPA |
| Left subiculum | BIP | − 0.0213 | 0.0297 | 4.72E-01 | 0.0310 | 0.1507 | 2.68E-06 | GPA |
| Left subiculum | SCZ | − 0.0548 | 0.0277 | 4.83E-02 | 0.0357 | 0.1611 | 1.08E-12 | GPA |
| Left CA1-3 | AD | − 0.0443 | 0.0737 | 5.47E-01 | 0.0005 | 0.0099 | 5.41E-06 | GPA |
| Left CA1-3 | BIP | − 0.0379 | 0.0300 | 2.06E-01 | 0.0376 | 0.1924 | 1.76E-10 | GPA |
| Left CA1-3 | PD | − 0.1684 | 0.0505 | 9.00E-04 | 0.0036 | 0.0597 | 7.17E-06 | GPA |
| Left CA1-3 | SCZ | − 0.0651 | 0.0283 | 2.12E-02 | 0.0404 | 0.1867 | 1.06E-15 | GPA |
| Left CA4-DG | BIP | − 0.0233 | 0.0311 | 4.53E-01 | 0.0533 | 0.2578 | 2.35E-11 | GPA |
| Left CA4-DG | PD | − 0.2104 | 0.0532 | 7.67E-05 | 0.0029 | 0.0359 | 1.03E-03 | LDSC |
| Left CA4-DG | SCZ | − 0.0289 | 0.0297 | 3.32E-01 | 0.0535 | 0.2359 | 1.40E-16 | GPA |
| Left molecular layer HP body | SCZ | − 0.0245 | 0.0363 | 5.00E-01 | 0.0380 | 0.1577 | 1.17E-04 | GPA |
| Left molecular layer HP head | ADHD | − 0.0596 | 0.0434 | 1.70E-01 | 0.0502 | 0.2439 | 1.44E-04 | GPA |
| Left molecular layer HP head | BIP | − 0.0037 | 0.0320 | 9.08E-01 | 0.0355 | 0.1756 | 5.18E-06 | GPA |
| Left molecular layer HP head | SCZ | − 0.0383 | 0.0316 | 2.26E-01 | 0.0409 | 0.1795 | 3.00E-08 | GPA |
| Left HATA | BIP | − 0.0050 | 0.0365 | 8.91E-01 | 0.0305 | 0.1577 | 8.65E-06 | GPA |
| Left HATA | PD | − 0.2102 | 0.0562 | 2.00E-04 | 0.0030 | 0.0619 | 4.72E-02 | LDSC |
| Left HATA | SCZ | − 0.0779 | 0.0352 | 2.70E-02 | 0.0306 | 0.1414 | 1.37E-06 | GPA |
| Right hippocampus | AD | − 0.0393 | 0.0721 | 5.86E-01 | 0.0005 | 0.0088 | 7.05E-05 | GPA |
| Right hippocampus | ADHD | − 0.0926 | 0.0421 | 2.77E-02 | 0.0375 | 0.1824 | 2.07E-04 | GPA |
| Right hippocampus | BIP | − 0.0080 | 0.0288 | 7.81E-01 | 0.0379 | 0.1866 | 3.25E-11 | GPA |
| Right hippocampus | Insomnia | − 0.0745 | 0.0366 | 4.16E-02 | 0.0401 | 0.1748 | 5.91E-06 | GPA |
| Right hippocampus | PD | − 0.2042 | 0.0509 | 6.00E-05 | 0.0034 | 0.0505 | 8.64E-07 | LDSC, GPA |
| Right hippocampus | SCZ | − 0.0296 | 0.0268 | 2.70E-01 | 0.0414 | 0.1889 | 1.09E-21 | GPA |
| Right hippocampal head | AD | − 0.0488 | 0.0735 | 5.07E-01 | 0.0005 | 0.0096 | 8.46E-06 | GPA |
| Right hippocampal head | ADHD | − 0.0815 | 0.0423 | 5.40E-02 | 0.0431 | 0.2172 | 8.80E-06 | GPA |
| Right hippocampal head | BIP | 0.0200 | 0.0301 | 5.07E-01 | 0.0345 | 0.1722 | 3.08E-10 | GPA |
| Right hippocampal head | Insomnia | − 0.0760 | 0.0359 | 3.41E-02 | 0.0383 | 0.1657 | 1.51E-05 | GPA |
| Right hippocampal head | PD | − 0.2002 | 0.0488 | 4.13E-05 | 0.0028 | 0.0448 | 2.43E-05 | LDSC, GPA |
| Right hippocampal head | SCZ | − 0.0409 | 0.0289 | 1.57E-01 | 0.0372 | 0.1727 | 4.82E-19 | GPA |
| Right hippocampal body | BIP | − 0.0244 | 0.0306 | 4.25E-01 | 0.0480 | 0.2334 | 1.15E-11 | GPA |
| Right hippocampal body | PD | − 0.2080 | 0.0568 | 2.00E-04 | 0.0036 | 0.0475 | 2.45E-05 | LDSC, GPA |
| Right hippocampal body | SCZ | − 0.0280 | 0.0272 | 3.04E-01 | 0.0493 | 0.2238 | 1.89E-19 | GPA |
| Right hippocampal tail | ADHD | − 0.0980 | 0.0452 | 3.00E-02 | 0.0355 | 0.1844 | 1.09E-04 | GPA |
| Right hippocampal tail | BIP | − 0.0195 | 0.0301 | 5.17E-01 | 0.0267 | 0.1337 | 6.62E-07 | GPA |
| Right hippocampal tail | Insomnia | − 0.0833 | 0.0398 | 3.62E-02 | 0.0294 | 0.1342 | 1.09E-04 | GPA |
| Right hippocampal tail | PD | − 0.1666 | 0.0503 | 9.00E-04 | 0.0039 | 0.0703 | 4.95E-06 | GPA |
| Right hippocampal tail | SCZ | 0.0282 | 0.0295 | 3.38E-01 | 0.0332 | 0.1549 | 6.86E-15 | GPA |
| Right subiculum | BIP | − 0.0052 | 0.0321 | 8.71E-01 | 0.0325 | 0.1655 | 1.13E-09 | GPA |
| Right subiculum | PD | − 0.1851 | 0.0508 | 3.00E-04 | 0.0029 | 0.0510 | 4.07E-06 | GPA |
| Right subiculum | SCZ | − 0.0411 | 0.0274 | 1.34E-01 | 0.0365 | 0.1702 | 5.41E-19 | GPA |
| Right CA1-3 | AD | 0.0252 | 0.0766 | 7.42E-01 | 0.0005 | 0.0086 | 5.31E-05 | GPA |
| Right CA1-3 | BIP | − 0.0034 | 0.0320 | 9.15E-01 | 0.0381 | 0.1930 | 7.12E-11 | GPA |
| Right CA1-3 | PD | − 0.2136 | 0.0554 | 1.00E-04 | 0.0031 | 0.0481 | 1.62E-04 | LDSC, GPA |
| Right CA1-3 | SCZ | − 0.0383 | 0.0282 | 1.75E-01 | 0.0454 | 0.2184 | 1.43E-26 | GPA |
| Right CA4-DG | BIP | − 0.0122 | 0.0307 | 6.92E-01 | 0.0547 | 0.2689 | 5.25E-14 | GPA |
| Right CA4-DG | Insomnia | − 0.0822 | 0.0353 | 1.97E-02 | 0.0507 | 0.2147 | 1.35E-05 | GPA |
| Right CA4-DG | PD | − 0.1838 | 0.0527 | 5.00E-04 | 0.0040 | 0.0511 | 1.68E-05 | GPA |
| Right CA4-DG | SCZ | − 0.0279 | 0.0304 | 3.58E-01 | 0.0524 | 0.2307 | 1.56E-17 | GPA |
| Right molecular layer HP body | ADHD | − 0.0155 | 0.0480 | 7.47E-01 | 0.0496 | 0.2597 | 1.49E-05 | GPA |
| Right molecular layer HP body | BIP | − 0.0380 | 0.0348 | 2.75E-01 | 0.0416 | 0.2054 | 3.86E-08 | GPA |
| Right molecular layer HP body | SCZ | − 0.0250 | 0.0333 | 4.53E-01 | 0.0447 | 0.2002 | 1.16E-10 | GPA |
| Right molecular layer HP head | BIP | 0.0225 | 0.0315 | 4.75E-01 | 0.0299 | 0.1477 | 1.52E-05 | GPA |
| Right molecular layer HP head | Insomnia | − 0.0529 | 0.0378 | 1.62E-01 | 0.0324 | 0.1518 | 3.37E-06 | GPA |
| Right molecular layer HP head | PD | − 0.1929 | 0.0576 | 8.00E-04 | 0.0027 | 0.0491 | 7.21E-05 | GPA |
| Right molecular layer HP head | SCZ | − 0.0149 | 0.0303 | 6.23E-01 | 0.0260 | 0.1140 | 9.11E-05 | GPA |
| Right fimbria | PD | − 0.1362 | 0.0606 | 2.46E-02 | 0.0053 | 0.0953 | 3.32E-06 | GPA |
| Right fimbria | SCZ | − 0.0011 | 0.0335 | 9.74E-01 | 0.0460 | 0.2035 | 3.05E-06 | GPA |
| Right hippocampal fissure | SCZ | 0.0523 | 0.0322 | 1.04E-01 | 0.0329 | 0.1539 | 8.21E-08 | GPA |
| Right HATA | SCZ | − 0.1155 | 0.0336 | 5.89E-04 | 0.0476 | 0.2070 | 6.45E-08 | GPA |
Genetic correlation and genetic overlap were estimated by LDSC and GPA methods, respectively. Bonferroni-corrected significance threshold was set at P < 2.08 × 10–3(0.05/240), producing a final union set of 75 pairwise traits with significant genetic correlation or genetic overlap for subsequent analysis. We introduced PAR as PM 11/(PM10 + PM01 + PM11) to represent the proportion of pleiotropic SNPs associated with both traits against the proportion of SNPs associated with at least 1 trait. Asterisk denotes statistically significant differences, *P < 0.05; **P < 2.08 × 10–4(0.05/240, Bonferroni corrected). LDSC linkage disequilibrium score regression, GPA genetic analysis incorporating pleiotropy and annotation method, PAR pleiotropy association ratio, PM11 proportion of genetic variants associated with both traits, AD Alzheimer’s disease, ADHD attention-deficit hyperactivity disorder, AN anorexia nervosa, ANX anxiety disorder, BIP bipolar disorder, PD Parkinson’s disease, PTSD post-traumatic stress disorder, SCZ schizophrenia
Shared loci between HASVs and neuropsychiatric traits
Our comprehensive examination extended to identifying shared genetic loci between HASVs and neuropsychiatric traits. Through a novel pleiotropic analysis under the composite null hypothesis (PLACO) [34] across 75 trait pairs, we identified a total of 133,100 variants (8567 unique) pleiotropic variants at the threshold of P < 6.67 × 10–10 (5 × 10−8/75, Bonferroni correction). After performing variant clumping (r2 < 0.1 and 6.67 < 5 × 10−10), we totally identified 433 pleiotropic lead SNPs across the 75 trait pairs (Additional file 1: Table S18). Notable, loci such as 17q21.31, 2q33.1, 12q14.3, 6q21, 16p11.2, 2q24.2, and 6p22.2 displayed associations with at least 20 trait pairs (Additional file 1: Table S19). To better understand the functional relevance of these pleiotropic variants, we conducted a comprehensive mapping of variants to genes by employing three distinct methodologies–positional analysis, eQTL analysis, and MAGMA analysis (Additional file 1: Table S20). We found that genes like ACBD4, ARHGAP27, KANSL1, MAPT, ARL17A, and ARL17B were implicated in no fewer than 50 trait pairs, across a range of diseases including AD, ADHD, BIP, insomnia, PD, and SCZ (Additional file 1: Table S21). In summary, these findings reinforce the strong associations between hippocampal and subfield structures and various neurological and psychiatric conditions, providing a foundation for a deeper understanding of the complex genetic factors influencing these diseases.
Causal hippocampal-brain disease relationships detected by Mendelian randomization
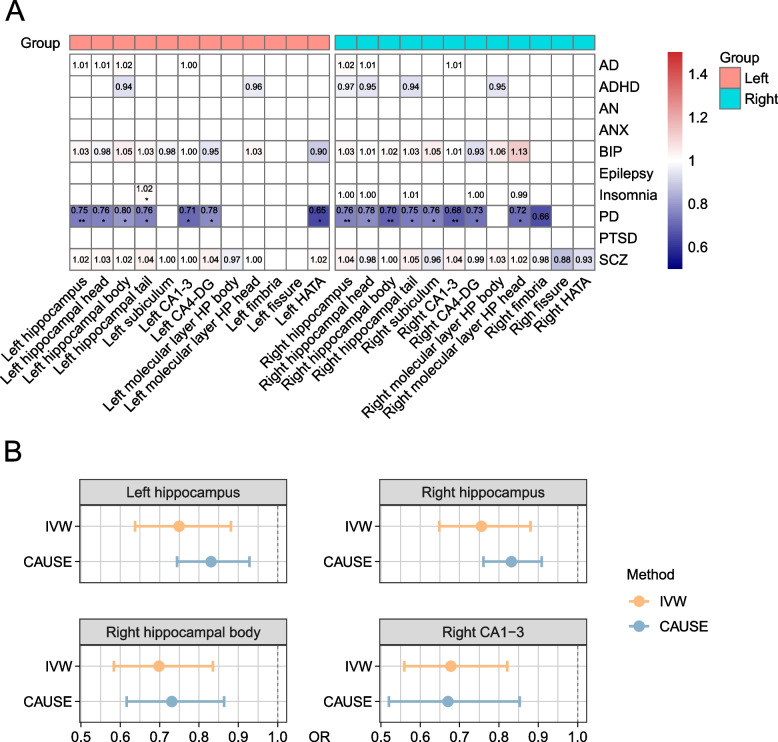
To further verify the potential causation of the 75 pairs (HASVs to neuropsychiatric traits), we performed two-sample Mendelian randomization (MR) analyses [62] using genetic instruments for neuropsychiatric traits among individuals of European ancestry. We employed the inverse variance weighted method (IVW) as our primary analysis approach. As shown in Fig. 6A and Additional file 1: Table S22, we observed that four HASVs (left hippocampus, right hippocampus, right hippocampal body, right CA1-3) were causally associated with PD a threshold of P < 6.67 × 10–4 (0.05/75, Bonferroni correction). For example, a decrease of 1 s.d. volume value of the CA1-3 in the right hemisphere was associated with 32% higher odds of PD (IVW OR = 0.68, 95% CI of 0.56 to 0.82, P = 7.28 × 10−5). Similar volumes decreased in the right hippocampal body were also associated with a higher risk of PD (IVW OR = 0.70, 95% CI of 0.58 to 0.84, P = 8.46 × 10−5). However, at a nominal threshold of P < 0.05, we observed more hippocampal subfields that were suggestively causally associated with PD.
Fig. 6.
Causal effects between HASV traits and brain disorders using Mendelian Randomization. A This heatmap presents the results of two-sample Mendelian randomization (MR) analyses for the 75 trait pairs (direction is HASV to brain disorders) using the IVW method. Statistically significant differences are denoted by asterisks: *P < 0.05; **P < 6.67 × 10−4 (0.05/75, Bonferroni corrected). B Forest plots for the four HASV traits that were Bonferroni corrected significant, showing causal effects on PD using two MR methods: IVW and CAUSE. IVW inverse variance weighted, CAUSE causal analysis using summary effect estimates, OR odds ratio, AD Alzheimer’s disease, ADHD attention-deficit hyperactivity disorder, AN anorexia nervosa, ANX anxiety disorder, BIP bipolar disorder, PD Parkinson’s disease, PTSD post-traumatic stress disorder, SCZ schizophrenia
To ensure that these findings were robust and not influenced by pleiotropy, we evaluated several pleiotropy indicators, including the MR-Egger intercept, Cochran’s Q statistic, and MR-PRESSO Global Test, all of which confirmed that the results were not biased by horizontal pleiotropic effects (Additional file 1: Table S22). However, while these methods address horizontal pleiotropy, we also considered the possibility of correlated pleiotropy, where shared biological pathways might influence both HASV traits and PD. To account for this, we employed CAUSE (causal analysis using summary effect estimates) to compare nested competing models. Results were considered consistent with a causal effect if the model with a causal effect parameter (causal model) provided a significantly better fit than the reduced model fit with only a shared effect parameter for correlated horizontal pleiotropy (sharing model). Our analyses supported causal effects of HASV on PD, with the ΔELPDCausal vs. Sharingvalues for all four trait pairs were significantly negative (P < 0.05) (Additional file 1: Table S23). We note that the causal effect estimates of these models were comparable in magnitude (overlapping 95% CIs) to the IVW estimates (Fig. 6B). These findings align with previous research, underscoring the significant role of HASV traits in contributing to the pathogenesis of PD.
Polygenic score associated with neuropsychiatric traits
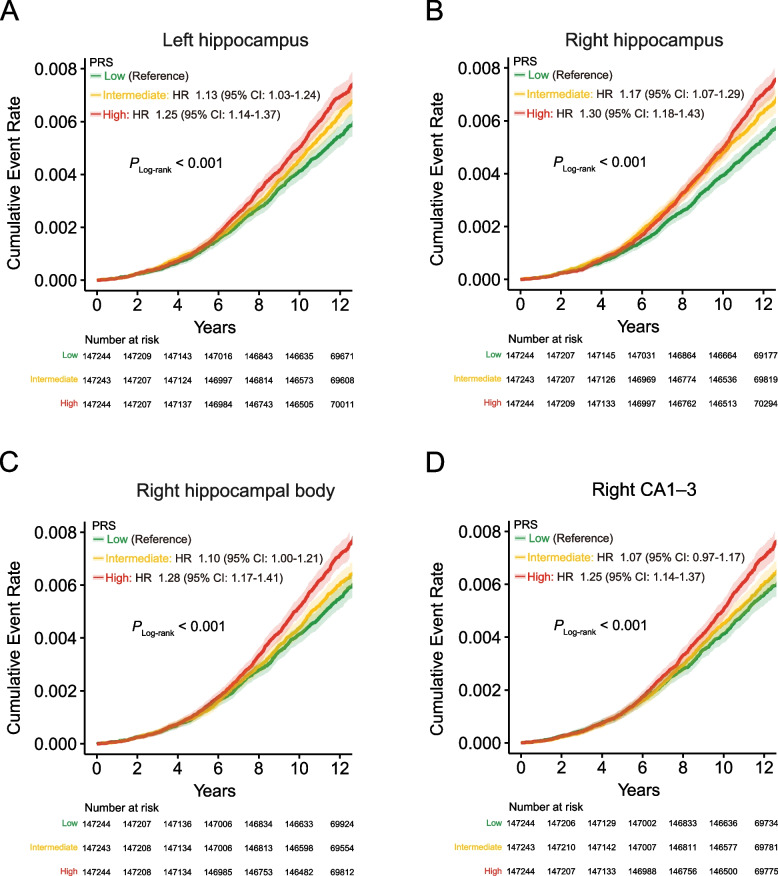
To validate the previously established causal associations involving the four HASVs, we formulated PRS by incorporating genetic dosage weights based on the effect sizes of independent genetic variants (P < 1 × 10–7, r2 = 0.1, kb = 250) derived from the corresponding GWAS results (Additional file 1: Table S24). We meticulously assessed the predictive abilities of these PRS on PD in a cohort of 441,731 UK Biobank participants, who had not undergone brain MRI scans and had no prior diagnoses of the respective diseases at the time of enrollment. As expected, PRS derived from all four HASVs demonstrated significant predictive capacity for PD incidence (Fig. 7 and Additional file 1: Table S25). The most pronounced results were observed in the right hippocampus, where individuals classified as high risk based on the PRS showed a notable 1.30-fold increased risk compared to their low-risk counterparts (95% CI: 1.18–1.43, P = 6.15 × 10–8; Fig. 7B). A similar trend was observed with the remaining HASV PRSs. In summary, our findings suggest that increased genetically determined HASVs are associated with elevated risks of both PD, offering the potential for the identification of high-risk individuals and enabling timely intervention strategies.
Fig. 7.
Cumulative incidence of Parkinson's disease stratified by PRS. A–D These survival curves include 441,731 individuals who had not undergone brain MRI scans and had no prior diagnosis of Parkinson’s disease at the time of enrollment. The y-axis represents the cumulative incidence (1 minus the Kaplan–Meier survival estimate) of a Parkinson’s disease diagnosis, while the x-axis indicates the number of years since enrollment in the UK Biobank. Individuals with a high polygenic score are depicted in red, those in the intermediate tertiles are in orange, and those in the low tertiles are in green. The 95% confidence intervals, derived from the cumulative hazard standard error, are represented with lighter shades
Discussion
By leveraging the finely delineated subfields of the hippocampus based on the rigorous quality control of brain MRI data from a cohort of over 40,000 individuals, we shed light on the shared biological mechanisms involved in neuropsychiatric traits. In our study, we identified several regions, including 12q14.3 [45, 46], 17q21.3 [45], 12q24.22 [45, 46], 9q33.11 [47], and 2q24.23 [46, 47], that have been associated with hippocampal volume and are known to play important roles in neuropsychiatric traits. These loci have been implicated in various biological processes, such as cell proliferation, synaptic plasticity, and neuronal apoptosis. For example, MSRB3, located in the chromosomal region 12q14.31, has been suggested to contribute to the reduction of methionine sulfoxide residues in proteins, potentially affecting processes related to AD and hippocampal atrophy [63–66]. HRK, situated in the chromosomal region 12q24.22, plays a crucial role in neuronal apoptosis [67] and exhibits the highest expression levels in hippocampal tissue, as observed in the genotype–tissue expression (GTEx) project [68]. These findings suggest the involvement of HRK in regulating hippocampal volume and may provide insights into the molecular mechanisms underlying neurodegenerative diseases. Furthermore, the 17q21.31 region, which has been widely recognized for its role in hippocampal development and neurodegenerative diseases, including PD and AD, was also confirmed to influence hippocampal volume in our study. We identified several genes within this region, including well-studied genes such as CRHR118, MAPT [45, 69], STH1, and KANSL1 [70], which have been extensively investigated for these diseases. Additionally, our study also unveiled a range of recently reported genes in the 17q21.31 region, such as ARL17A [71], ARL17B [71], LRRC37A2 [71, 72], NSF [71], PLEKHM1 [73], SPPL2C [71], and LRRC37A [71, 72], which may contribute to the pathogenesis of neurodegenerative diseases.
Furthermore, our study also validates two recently reported regions, 6q21 and 6q25.1, which have a significant impact on multiple HASVs [48]. At the 6q21 locus, we observed that the G allele of rs1062034 (minor allele frequency = 0.36), located in the UTR3 region within FOXO3, was associated with a decrease in hippocampal volume (β = − 0.08; P = 6.9 × 10−23). FOXO3 functions downstream of the insulin/IGF signaling pathway and plays a crucial role in maintaining adult neural precursor cell homeostasis [74–76]. Studies [77] have demonstrated that FOXO3 can trigger axonal degeneration upon the withdrawal of neurotrophic factors, suggesting its potential involvement in regulating hippocampal volume and establishing a link between neurotrophic signaling and structural changes in the hippocampus. In contrast, at the 6q25.1 locus, the C allele of rs60424881 (minor allele frequency = 0.36), which lies in an intronic region within NUP43, was associated with an increase in hippocampal volume (β = 0.05; P = 6.1 × 10−12). The NUP43 gene is a constituent of the nuclear pore complex (NPC). Recent studies have revealed a close association between abnormalities in the NPC and various neurodegenerative diseases [78], including AD, Huntington’s disease, and PD. Future research endeavors are expected to elucidate the potential role and underlying mechanisms of NUP43 in neurodegenerative diseases.
The heritability estimates we found for the HASVs, which ranged from 0.15 to 0.32, are in line with earlier large-scale studies [79, 80]. These findings highlight the significant contribution of genetic factors in shaping the variability of hippocampal subregions. In addition, we found weaker genetic relationships between the HATA subregions, hippocampal fissure, hippocampal fimbria, and the remaining regions of the hippocampus in our analysis. Stronger genetic relationships, on the other hand, were seen in the remaining hippocampal regions. This disparity reflects the complex interplay of genetic factors and the distinct physiological functions that gray matter and white matter play within the hippocampus. Moreover, we conducted a comprehensive exploration of the genetic associations between the hippocampal subregions and 97 other subregions of the brain. The results revealed widespread connections between the hippocampus and brain regions [23], particularly the amygdala. Building upon this, we further investigated the relationships between the HASVs and ten common neuropsychiatric traits.
Importantly, through the integration of genetic correlation, genetic overlap, and MR analysis, we confirmed that atrophy in the left hippocampus, right hippocampus, right hippocampal body, and right CA1-3 region is associated with an increased risk of developing PD, which aligns with previous clinical research. The hippocampus is crucial for cognitive functions, and its atrophy is linked to memory impairment and other cognitive deficits. Multiple clinical studies have observed whole hippocampal atrophy (either bilateral or unilateral) in PD patients with mild cognitive impairment or dementia [5, 81–83]. Additionally, studies in healthy individuals highlight that the CA1-3 subregions play essential roles in episodic memory recollection and are strongly correlated with learning and recognition scores [84]. This supports our finding that atrophy in the CA1-3 regions is associated with increased PD risk. Furthermore, Foo and colleagues [85] measured hippocampal subfield volumes in PD patients and examined their correlation with cognitive and motor decline over 18 months. They found reduced volumes in the right CA1 at baseline and observed further reduction in the right CA2-3 after 18 months. This volume reduction was accompanied by significant declines in episodic memory and executive function in PD converters (patients who transitioned from PD with normal cognition to PD with mild cognitive impairment), compared to PD-stable patients (those who did not experience cognitive decline). This longitudinal evidence further corroborates our MR findings, underscoring the role of hippocampal atrophy—particularly in the CA1-3 regions—in the progression of cognitive decline in PD.
The utility of PRS in forecasting disease onset has been extensively explored, serving as a valuable tool for identifying high-risk populations and aiding decision-making [10]. However, the enduring predictive capabilities of PRS derived from HASVs across neuropsychiatric traits have remained largely unexplored. In our investigation, extending from the insights garnered through MR analysis, we further assess the influence of PRS on the occurrence of PD in a follow-up cohort comprising nearly 450,000 individuals, observed over a median period of 11.1 years. Remarkably, our findings highlighted the substantial predictive potential of four PRSs for PD, distinctly showcasing their effectiveness in foreseeing the occurrence of this disease.
This study has several limitations. Our analyses rely on data from the ongoing UK Biobank brain imaging study, which includes only about 10% of all UK Biobank participants (as of 2020) and predominantly represents individuals of European ancestry. The UK Biobank is also known for its “healthy volunteer” selection bias, which may not fully capture the broader European population [86]. To better account for population-specific variations in genetic effects, future research should incorporate expansive and diverse imaging datasets from global populations, as recommended by more open and large-scale imaging studies [87]. Additionally, using overlapping UK Biobank samples for genetic correlation estimates of the 24 HASV traits may result in inflated correlations. Despite this, high-quality, large-sample brain MRI images are scarce elsewhere, making the UK Biobank dataset indispensable. Furthermore, while identifying specific genes through multiple strategies provides a strong indication, further experimental studies using gene-editing techniques in cellular and animal models are needed.
Conclusions
In conclusion, our study sheds light on 352 independent significant (P < 2.1 × 10−9) variants intricately linked to the 24 HASVs. Notably, the regions 12q14.3, 17q21.31, 12q24.22, 6q21, 9q33.1, 6q25.1, and 2q24.2 were found to influence multiple HASVs. Furthermore, our exploration delves deeper, revealing an expansive and intricate genetic interconnection that binds HASV traits to a spectrum of brain disorders. Significantly, through meticulous observation of a cohort comprising nearly 450,000 individuals, we unveil the potential of utilizing PRS derived from HASVs as a potent tool for risk stratification in PD. This approach has the potential to significantly enhance our ability to effectively manage and intervene in PD in early stages.
Supplementary Information
Acknowledgements
We are grateful to all members who participated in the study, as well as all individuals who helped us complete the research.
Abbreviations
- AD
Alzheimer’s disease
- ADHD
Attention-deficit hyperactivity disorder
- AN
Anorexia nervosa
- ANX
Anxiety disorder
- BMI
Body mass index
- BIP
Bipolar disorder
- CI
Confidence interval
- GPA
Genetic analysis incorporating pleiotropy and annotation
- GWAS
Genome-wide association studies
- HASV
Hippocampal and subfield volumes
- HR
Hazard ratio
- IQR
Interquartile range
- IVW
Inverse variance weighted
- MRI
Magnetic resonance imaging
- MR
Mendelian randomization
- PD
Parkinson’s disease
- PLACO
Pleiotropic analysis under composite null hypothesis
- PRS
Polygenic risk scores
- PVE
Phenotype variance explained
- PTSD
Post-traumatic stress disorder
- SCZ
Schizophrenia
- TWAS
Transcriptome-wide association study
- UKB
UK Biobank
Authors’ contributions
JT, XM, and XL were the overall principal investigators in this study who conceived the study and obtained financial support. JT and XM were responsible for the study design and supervised the entire study. CN, MJ, and YC organized the data, carried out the statistical analysis, and participated in writing the first draft of the manuscript. CN designed and drew the figures. LF, KH, ZL, MZ, CC, Y.M.L, NH, DZ, Y.Z.L, SC, YJ, CH, ZW, ZC, HL, GL, QM, HG, WT, HZ, XY, CH, YW, BL and YZ analyzed the data. All authors approved the final report for publication.
Funding
This work was supported by National Science Fund for Excellent Young Scholars (NSFC-82322058), Program of National Natural Science Foundation of China (NSFC-82103929, NSFC-82273713), Young Elite Scientists Sponsorship Program by CAST (2022QNRC001), National Science Fund for Distinguished Young Scholars of Hubei Province of China (2023AFA046), and Knowledge Innovation Program of Wuhan (whkxjsj011, 2023020201010073), Fundamental Research Funds for the Central Universities (2042024kf1012) for Jianbo Tian; National Key R&D Program of China (2022YFA0806601), National Science Fund for Distinguished Young Scholars of China (NSFC-81925032), Key Program of National Natural Science Foundation of China (NSFC-82130098), the Leading Talent Program of the Health Commission of Hubei Province (EWT2173), Knowledge Innovation Program of Wuhan (2023020201010060) and Fundamental Research Funds for the Central Universities (2042022rc0026, 2042023kf1005) for Xiaoping Miao; Program of National Natural Science Foundation of China (NSFC-82003547, NSFC-82373663), Program of Health Commission of Hubei Province (WJ2023M045), and Knowledge Innovation Program of Wuhan (2023020201020244) for Ying Zhu.
Availability of data and materials
Data from the UKB (www.ukbiobank.ac.uk/register-apply) are available to all researchers upon making an application. This research has been conducted using the UKB Resource under Application 63454. Source data are provided with this paper. Data sources of publicly available GWAS results were listed in Additional file 1: Table S4. The GWAS summary statistics generated in this study have been deposited in the Human Genome Research Institute GWAS Catalog under accession codes GCST90296069-GCST90296092. All bioinformatics and statistical analysis tools used in the present study are open source, and details about them are available in the “Methods” section. No customized code was used to process or analyze the data.
Data from the UKB (www.ukbiobank.ac.uk/register-apply) are available to all researchers upon making an application. This research has been conducted using the UKB Resource under Application 63,454. Source data are provided with this paper. Data sources of publicly available GWAS results were listed in Supplementary Table 24. The GWAS summary statistics generated in this study have been deposited in the Human Genome Research Institute GWAS Catalog under accession codes GCST90296069-GCST90296092. All bioinformatics and statistical analysis tools used in the present study are open source, and details about them are available in the Methods section. No customized code was used to process or analyze the data.
Declarations
Ethics approval and consent to participate
All participants provided written informed consent before enrolment in the UK Biobank, which was conducted in accordance with the Declaration of Helsinki. The UK Biobank study, and the sharing of anonymized data with the research community, was approved by the North West Multi-center Research Ethics Committee (REC reference: 12/NW/03820).
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Caibo Ning, Meng Jin, and Yimin Cai contributed equally to this work.
Contributor Information
Xiangpan Li, Email: rm001227@whu.edu.cn.
Xiaoping Miao, Email: xpmiao@whu.edu.cn.
Jianbo Tian, Email: tianjb@whu.edu.cn.
References
- 1.Maguire EA, Gadian DG, Johnsrude IS, Good CD, Ashburner J, Frackowiak RS, et al. Navigation-related structural change in the hippocampi of taxi drivers. Proc Natl Acad Sci U S A. 2000;97:4398–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burgess N, Maguire EA, O’Keefe J. The human hippocampus and spatial and episodic memory. Neuron. 2002;35:625–41. [DOI] [PubMed] [Google Scholar]
- 3.Snyder JS, Soumier A, Brewer M, Pickel J, Cameron HA. Adult hippocampal neurogenesis buffers stress responses and depressive behaviour. Nature. 2011;476:458–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moreno-Jiménez EP, Flor-García M, Terreros-Roncal J, Rábano A, Cafini F, Pallas-Bazarra N, et al. Adult hippocampal neurogenesis is abundant in neurologically healthy subjects and drops sharply in patients with Alzheimer’s disease. Nat Med. 2019;25:554–60. [DOI] [PubMed] [Google Scholar]
- 5.Calabresi P, Castrioto A, Di Filippo M, Picconi B. New experimental and clinical links between the hippocampus and the dopaminergic system in Parkinson’s disease. Lancet Neurol. 2013;12:811–21. [DOI] [PubMed] [Google Scholar]
- 6.Hibar DP, Westlye LT, van Erp TGM, Rasmussen J, Leonardo CD, Faskowitz J, et al. Subcortical volumetric abnormalities in bipolar disorder. Mol Psychiatry. 2016;21:1710–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Erp TGM, Hibar DP, Rasmussen JM, Glahn DC, Pearlson GD, Andreassen OA, et al. Subcortical brain volume abnormalities in 2028 individuals with schizophrenia and 2540 healthy controls via the ENIGMA consortium. Mol Psychiatry. 2016;21:547–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hibar DP, Stein JL, Renteria ME, Arias-Vasquez A, Desrivières S, Jahanshad N, et al. Common genetic variants influence human subcortical brain structures. Nature. 2015;520:224–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iglesias JE, Augustinack JC, Nguyen K, Player CM, Player A, Wright M, et al. A computational atlas of the hippocampal formation using ex vivo, ultra-high resolution MRI: Application to adaptive segmentation of in vivo MRI. Neuroimage. 2015;115:117–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Torkamani A, Wineinger NE, Topol EJ. The personal and clinical utility of polygenic risk scores. Nat Rev Genet. 2018;19:581–90. [DOI] [PubMed] [Google Scholar]
- 11.Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miller KL, Alfaro-Almagro F, Bangerter NK, Thomas DL, Yacoub E, Xu J, et al. Multimodal population brain imaging in the UK Biobank prospective epidemiological study. Nat Neurosci. 2016;19:1523–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shahid SS, Wen Q, Risacher SL, Farlow MR, Unverzagt FW, Apostolova LG, et al. Hippocampal-subfield microstructures and their relation to plasma biomarkers in Alzheimer’s disease. Brain. 2022;145:2149–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loh P-R, Kichaev G, Gazal S, Schoech AP, Price AL. Mixed-model association for biobank-scale datasets. Nat Genet. 2018;50:906–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pirruccello JP, Bick A, Wang M, Chaffin M, Friedman S, Yao J, et al. Analysis of cardiac magnetic resonance imaging in 36,000 individuals yields genetic insights into dilated cardiomyopathy. Nat Commun. 2020;11:2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wojcik GL, Graff M, Nishimura KK, Tao R, Haessler J, Gignoux CR, et al. Genetic analyses of diverse populations improves discovery for complex traits. Nature. 2019;570:514–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Page GP, Kanias T, Guo YJ, Lanteri MC, Zhang X, Mast AE, et al. Multiple-ancestry genome-wide association study identifies 27 loci associated with measures of hemolysis following blood storage. J Clin Invest. 2021;131(e146077):146077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lloyd-Jones LR, Robinson MR, Moser G, Zeng J, Beleza S, Barsh GS, et al. Inference on the genetic basis of eye and skin color in an admixed population via Bayesian linear mixed models. Genetics. 2017;206:1113–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caliebe A, Tekola-Ayele F, Darst BF, Wang X, Song YE, Gui J, et al. Including diverse and admixed populations in genetic epidemiology research. Genet Epidemiol. 2022;46:347–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bulik-Sullivan BK. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bulik-Sullivan B. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao B, Luo T, Li T, Li Y, Zhang J, Shan Y, et al. Genome-wide association analysis of 19,629 individuals identifies variants influencing regional brain volumes and refines their genetic co-architecture with cognitive and mental health traits. Nat Genet. 2019;51:1637–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jansen IE, Savage JE, Watanabe K, Bryois J, Williams DM, Steinberg S, et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat Genet. 2019;51:404–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Demontis D. Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat Genet. 2019;51:63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watson HJ, Yilmaz Z, Thornton LM, Hübel C, Coleman JRI, Gaspar HA, et al. Genome-wide association study identifies eight risk loci and implicates metabo-psychiatric origins for anorexia nervosa. Nat Genet. 2019;51:1207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Forstner AJ, Awasthi S, Wolf C, Maron E, Erhardt A, Czamara D, et al. Genome-wide association study of panic disorder reveals genetic overlap with neuroticism and depression. Mol Psychiatry. 2021;26:4179–90. [DOI] [PubMed] [Google Scholar]
- 28.Mullins N, Forstner AJ, O’Connell KS, Coombes B, Coleman JRI, Qiao Z, et al. Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat Genet. 2021;53:817–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.International League Against Epilepsy Consortium on Complex Epilepsies. Genome-wide mega-analysis identifies 16 loci and highlights diverse biological mechanisms in the common epilepsies. Nat Commun. 2018;9:5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nalls MA, Blauwendraat C, Vallerga CL, Heilbron K, Bandres-Ciga S, Chang D, et al. Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 2019;18:1091–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nievergelt CM, Maihofer AX, Klengel T, Atkinson EG, Chen C-Y, Choi KW, et al. International meta-analysis of PTSD genome-wide association studies identifies sex- and ancestry-specific genetic risk loci. Nat Commun. 2019;10:4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pardiñas AF, Holmans P, Pocklington AJ, Escott-Price V, Ripke S, Carrera N, et al. Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet. 2018;50:381–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chung D, Yang C, Li C, Gelernter J, Zhao H. GPA: a statistical approach to prioritizing GWAS results by integrating pleiotropy and annotation. PLoS Genet. 2014;10:e1004787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ray D, Chatterjee N. A powerful method for pleiotropic analysis under composite null hypothesis identifies novel shared loci between type 2 diabetes and prostate cancer. PLoS Genet. 2020;16:e1009218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7:e34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yavorska OO, Burgess S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int J Epidemiol. 2017;46:1734–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44:512–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40:304–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hartwig FP, Davey Smith G, Bowden J. Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int J Epidemiol. 2017;46:1985–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Slob EAW, Burgess S. A comparison of robust Mendelian randomization methods using summary data. Genet Epidemiol. 2020;44:313–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao Q, Wang J, Hemani G, Bowden J, Small DS. Statistical inference in two-sample summary-data Mendelian randomization using robust adjusted profile score. Ann Stat. 2020;48:1742–69. [Google Scholar]
- 42.Verbanck M, Chen C-Y, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50:693–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morrison J, Knoblauch N, Marcus JH, Stephens M, He X. Mendelian randomization accounting for correlated and uncorrelated pleiotropic effects using genome-wide summary statistics. Nat Genet. 2020;52:740–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choi SW, Mak TS-H, O’Reilly PF. Tutorial: a guide to performing polygenic risk score analyses. Nat Protoc. 2020;15:2759–72. [DOI] [PMC free article] [PubMed]
- 45.Bahrami S, Nordengen K, Shadrin AA, Frei O, Van Der Meer D, Dale AM, et al. Distributed genetic architecture across the hippocampal formation implies common neuropathology across brain disorders. Nat Commun. 2022;13:3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bis JC, DeCarli C, Smith AV, van der Lijn F, Crivello F, Fornage M, et al. Common variants at 12q14 and 12q24 are associated with hippocampal volume. Nat Genet. 2012;44:545–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hibar DP, Adams HHH, Jahanshad N, Chauhan G, Stein JL, Hofer E, et al. Novel genetic loci associated with hippocampal volume. Nat Commun. 2017;8:13624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu N, Zhang L, Tian T, Cheng J, Zhang B, Qiu S, et al. Cross-ancestry genome-wide association meta-analyses of hippocampal and subfield volumes. Nat Genet. 2023;55(7):1126–37. [DOI] [PubMed]
- 49.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8:1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang D, Wang Z, Zhou Y, Liang Q, Sham PC, Yao H, et al. vSampler: fast and annotation-based matched variant sampling tool. Bioinformatics. 2021;37:1915–7. [DOI] [PubMed] [Google Scholar]
- 53.Cai Y, Zhang Y, Loh YP, Tng JQ, Lim MC, Cao Z, et al. H3K27me3-rich genomic regions can function as silencers to repress gene expression via chromatin interactions. Nat Commun. 2021;12:719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, et al. Transcriptional super-enhancers connected to cell identity and disease. Cell. 2013;155:10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed]
- 55.Phelps EA. Human emotion and memory: interactions of the amygdala and hippocampal complex. Curr Opin Neurobiol. 2004;14:198–202. [DOI] [PubMed] [Google Scholar]
- 56.Scoville WB, Milner B. Loss of recent memory after bilateral hippocampal lesions. J Neurol Neurosurg Psychiatry. 1957;20:11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Morris RG, Garrud P, Rawlins JN, O’Keefe J. Place navigation impaired in rats with hippocampal lesions. Nature. 1982;297:681–3. [DOI] [PubMed] [Google Scholar]
- 58.Lisman J, Buzsáki G, Eichenbaum H, Nadel L, Ranganath C, Redish AD. Viewpoints: how the hippocampus contributes to memory, navigation and cognition. Nat Neurosci. 2017;20:1434–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mattai A, Hosanagar A, Weisinger B, Greenstein D, Stidd R, Clasen L, et al. Hippocampal volume development in healthy siblings of childhood-onset schizophrenia patients. Am J Psychiatry. 2011;168:427–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Treadway MT, Waskom ML, Dillon DG, Holmes AJ, Park MTM, Chakravarty MM, et al. Illness progression, recent stress, and morphometry of hippocampal subfields and medial prefrontal cortex in major depression. Biol Psychiatry. 2015;77:285–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pitman RK, Rasmusson AM, Koenen KC, Shin LM, Orr SP, Gilbertson MW, et al. Biological studies of post-traumatic stress disorder. Nat Rev Neurosci. 2012;13:769–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Burgess S, Dudbridge F, Thompson SG. Combining information on multiple instrumental variables in Mendelian randomization: comparison of allele score and summarized data methods. Stat Med. 2016;35:1880–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.González S, Huerta JM, Alvarez-Uría J, Fernández S, Patterson AM, Lasheras C. Serum selenium is associated with plasma homocysteine concentrations in elderly humans. J Nutr. 2004;134:1736–40. [DOI] [PubMed] [Google Scholar]
- 64.Kalmijn S, Launer LJ, Lindemans J, Bots ML, Hofman A, Breteler MM. Total homocysteine and cognitive decline in a community-based sample of elderly subjects: the Rotterdam Study. Am J Epidemiol. 1999;150:283–9. [DOI] [PubMed] [Google Scholar]
- 65.Prins ND, Den Heijer T, Hofman A, Koudstaal PJ, Jolles J, Clarke R, et al. Homocysteine and cognitive function in the elderly: the Rotterdam Scan Study. Neurology. 2002;59:1375–80. [DOI] [PubMed] [Google Scholar]
- 66.den Heijer T, Vermeer SE, Clarke R, Oudkerk M, Koudstaal PJ, Hofman A, et al. Homocysteine and brain atrophy on MRI of non-demented elderly. Brain. 2003;126(Pt 1):170–5. [DOI] [PubMed] [Google Scholar]
- 67.Towers E, Gilley J, Randall R, Hughes R, Kristiansen M, Ham J. The proapoptotic dp5 gene is a direct target of the MLK-JNK-c-Jun pathway in sympathetic neurons. Nucleic Acids Res. 2009;37(9):3044–60. [DOI] [PMC free article] [PubMed]
- 68.Aguet F, Brown AA, Castel SE, Davis JR, He Y, Jo B, et al. Genetic effects on gene expression across human tissues. Nature. 2017;550:204–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Le Guennec K, Quenez O, Nicolas G, Wallon D, Rousseau S, Richard A-C, et al. 17q21.31 duplication causes prominent tau-related dementia with increased MAPT expression. Mol Psychiatry. 2017;22:1119–25. [DOI] [PubMed] [Google Scholar]
- 70.Jun G, Ibrahim-Verbaas CA, Vronskaya M, Lambert J-C, Chung J, Naj AC, et al. A novel Alzheimer disease locus located near the gene encoding tau protein. Mol Psychiatry. 2016;21:108–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Soto-Beasley AI, Walton RL, Valentino RR, Hook PW, Labbé C, Heckman MG, et al. Screening non-MAPT genes of the Chr17q21 H1 haplotype in Parkinson’s disease. Parkinsonism Relat Disord. 2020;78:138–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bowles KR, Pugh DA, Liu Y, Patel T, Renton AE, Bandres-Ciga S, et al. 17q21.31 sub-haplotypes underlying H1-associated risk for Parkinson’s disease are associated with LRRC37A/2 expression in astrocytes. Mol Neurodegener. 2022;17:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cooper YA, Teyssier N, Dräger NM, Guo Q, Davis JE, Sattler SM, et al. Functional regulatory variants implicate distinct transcriptional networks in dementia. Science. 2022;377:eabi8654. [DOI] [PubMed] [Google Scholar]
- 74.Paik J, Ding Z, Narurkar R, Ramkissoon S, Muller F, Kamoun WS, et al. FoxOs cooperatively regulate diverse pathways governing neural stem cell homeostasis. Cell Stem Cell. 2009;5:540–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–12. [DOI] [PubMed] [Google Scholar]
- 76.Renault VM, Rafalski VA, Morgan AA, Salih DAM, Brett JO, Webb AE, et al. FoxO3 regulates neural stem cell homeostasis. Cell Stem Cell. 2009;5:527–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Santo EE, Paik J. FOXO in Neural Cells and Diseases of the Nervous System. Curr Top Dev Biol. 2018;127:105–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu J, Hetzer MW. Nuclear pore complex maintenance and implications for age-related diseases. Trends Cell Biol. 2022;32:216–27. [DOI] [PubMed] [Google Scholar]
- 79.van der Meer D, Rokicki J, Kaufmann T, Córdova-Palomera A, Moberget T, Alnæs D, et al. Brain scans from 21,297 individuals reveal the genetic architecture of hippocampal subfield volumes. Mol Psychiatry. 2020;25:3053–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Franke B, Stein JL, Ripke S, Anttila V, Hibar DP, van Hulzen KJE, et al. Genetic influences on schizophrenia and subcortical brain volumes: large-scale proof of concept. Nat Neurosci. 2016;19:420–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Das T, Hwang JJ, Poston KL. Episodic recognition memory and the hippocampus in Parkinson’s disease: A review. Cortex. 2019;113:191–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kandiah N, Zainal NH, Narasimhalu K, Chander RJ, Ng A, Mak E, et al. Hippocampal volume and white matter disease in the prediction of dementia in Parkinson’s disease. Parkinsonism Relat Disord. 2014;20:1203–8. [DOI] [PubMed] [Google Scholar]
- 83.Segura B, Baggio HC, Marti MJ, Valldeoriola F, Compta Y, Garcia-Diaz AI, et al. Cortical thinning associated with mild cognitive impairment in Parkinson’s disease. Mov Disord. 2014;29:1495–503. [DOI] [PubMed] [Google Scholar]
- 84.Chadwick MJ, Bonnici HM, Maguire EA. CA3 size predicts the precision of memory recall. Proc Natl Acad Sci U S A. 2014;111:10720–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Foo H, Mak E, Chander RJ, Ng A, Au WL, Sitoh YY, et al. Associations of hippocampal subfields in the progression of cognitive decline related to Parkinson’s disease. NeuroImage Clinical. 2017;14:37–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fry A, Littlejohns TJ, Sudlow C, Doherty N, Adamska L, Sprosen T, et al. Comparison of sociodemographic and health-related characteristics of UK Biobank participants with those of the general population. Am J Epidemiol. 2017;186:1026–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Laird AR. Large, open datasets for human connectomics research: considerations for reproducible and responsible data use. Neuroimage. 2021;244:118579. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data from the UKB (www.ukbiobank.ac.uk/register-apply) are available to all researchers upon making an application. This research has been conducted using the UKB Resource under Application 63454. Source data are provided with this paper. Data sources of publicly available GWAS results were listed in Additional file 1: Table S4. The GWAS summary statistics generated in this study have been deposited in the Human Genome Research Institute GWAS Catalog under accession codes GCST90296069-GCST90296092. All bioinformatics and statistical analysis tools used in the present study are open source, and details about them are available in the “Methods” section. No customized code was used to process or analyze the data.
Data from the UKB (www.ukbiobank.ac.uk/register-apply) are available to all researchers upon making an application. This research has been conducted using the UKB Resource under Application 63,454. Source data are provided with this paper. Data sources of publicly available GWAS results were listed in Supplementary Table 24. The GWAS summary statistics generated in this study have been deposited in the Human Genome Research Institute GWAS Catalog under accession codes GCST90296069-GCST90296092. All bioinformatics and statistical analysis tools used in the present study are open source, and details about them are available in the Methods section. No customized code was used to process or analyze the data.