Abstract
Despite reported influences of the intratumoral microbiome on cancer progression, its role in this subtype remains unclear. This study aimed to characterize the microbial landscape and signatures of kidney renal clear cell carcinoma using RNA-Seq data from The Cancer Genome Atlas. Following microbial decontamination, differential microbial analysis was conducted between tumorous and adjacent non-tumorous samples. Compared to non-tumorous samples, tumorous microbiota exhibited reduced α and β diversity and distinct phylum-level communities. Differential microbial analysis between patients exhibiting long and short overall survival revealed ten significant differential microbial genera, with six genera correlating with a positive prognosis (Plasmodium, Babesia, Toxoplasma, Cytobacillus, Alicyclobacillus, Verrucomicrobium) and four with a negative prognosis (Colletotrichum, Leuconostoc, Gluconobacter, and Parabacteroides). Employing Cox regression analysis and support vector machines, a prognosis-related microbiome risk signature was developed, achieving an AUC of 0.809. Based on this risk signature, two microbiome-based subtypes were found to be significantly associated with distinct clinical prognoses and immune microenvironments. These findings were corroborated by significant correlations between prognostic-relevant microorganisms and 30 immune-related differentially expressed genes. Specifically, microbial genera associated with a negative prognosis were linked to a pro-tumor acute inflammatory immune response, whereas genera related to a positive prognosis were associated with an anti-tumor adaptive immune response. In conclusion, microbiome-based subtyping revealed correlations between tumor microbiome, clinical prognosis, and tumor microenvironment, indicating intratumoral microbiota as a promising prognostic biomarker for kidney renal clear cell carcinoma.
Keywords: Intratumor microbiota, Kidney renal clear cell carcinoma, Tumor microenvironment, Biomarkers
1. Introduction
To date, cancer remains a major global health issue, with renal cell carcinoma (RCC) being the tenth most prevalent of all cancer types [1]. Approximately 30 percent of patients with RCC are diagnosed at advanced stages, and the five-year (5 y) survival rate is only 10 percent [2]. Owing to the lack of sensitivity to conventional radiotherapy and chemotherapy, patients with metastatic kidney renal clear cell carcinoma (KIRC), the most common RCC subtype, are treated with immunotherapy and targeted therapy as a first line of treatment [[3], [4], [5]]. Despite the clinical benefits of these therapies, a deeper understanding of RCC pathogenesis is crucially needed to improve survival rates [6]. The microbiome, which is defined as all the microorganisms and their genomes within a specific environment, profoundly influences the metabolic, immunologic, and homeostatic functions of the host. Recent studies have shown that microbiotas play important roles in the progression and treatment responses of multiple cancers, including RCC [7,8].
The intratumoral microbiota, including bacteria [8,9], fungi [10], and viruses [11], plays an influential role in cancer initiation, tumor progression, and responses toward cancer therapies [12]. However, the importance of the tumor microbiome and its association with carcinogenesis remain controversial [13]. On the one hand, as a crucial component of the tumor microenvironment (TME), the tumor microbiome can facilitate tumorigenesis by inducing the suppression of innate and adaptive immunity [14]. On the other hand, intratumoral microorganisms can suppress carcinogenesis by altering immune cell activation [15]. This intricate interplay underscores the close relationship between tumors and their microbiomes, which is influenced by specific microbial species and individual tumor status, potentially affecting the sensitivity of the cancer cells to immunotherapy [16,17]. Given the potential strategy of improving patient prognosis by manipulating the composition of the intratumoral microbiota, further exploration of the interplay between the tumor microbiome and tumor immunology and treatment prognosis is needed.
Although intratumoral microbiomes have already been characterized in various cancers, those specifically associated with RCC remain understudied [18]. Urinary tract infections have been identified as a modifiable risk factor for RCC development [19]. A 2022 study revealed an inverse correlation between bacterial load and PU.1 macrophages, providing insights into how bacteria influence the phenotypes of RCC-related immune cells [20,21]. Although several attempts have been made to investigate the microbiomes within RCCs, the reliability of the results remains questionable owing to the small sample sizes and inadequate decontamination procedures used [22]. Moreover, the correlations among the intratumoral microbiomes, immune landscape of RCC, and prognostic implications require further exploration [23].
In this study, KIRC transcriptome sequencing data and patient clinical data was obtained from The Cancer Genome Atlas (TCGA) database. Subsequently, differentially abundant microorganisms were identified between patients with long-versus short-term overall survival (OS) and constructed a Cox regression-based risk score signature. Further analyses, including differential gene expression and gene set enrichment, were performed to elucidate the underlying immunogenic mechanisms by which these risk-associated microorganisms affect the tumor immune response and disease prognosis.
2. Material and methods
2.1. Design of the study
The primary objective of this study was to elucidate the impact of the intratumoral microbiota on tumor immunity and prognosis in KIRC. To achieve this, intratumoral microbial profiles, tumor transcriptome characteristics, and patient survival data need to be obtained simultaneously. A comprehensive analysis was conducted using RNA-Seq data from 532 patients with KIRC referenced on TCGA. First, the intratumoral microbiota information was extracted from the RNA-seq data. To ensure microbial profile extraction accuracy and minimize contamination, a validated analytical pipeline [9] was employed. Subsequently, patients were stratified into two groups based on OS and compared between patients with long and short survival for the differential intratumoral microbiota. Then, a survival-associated microbial score based on Cox regression modeling was constructed to categorize patients into two distinct risk groups. By analyzing the differences in tumor transcriptomes between groups, performing functional enrichment, and analyzing immune infiltration, we aimed to elucidate the specific mechanisms underlying how differential microbiota influence tumor immunity. The study was conducted between October 2022 and August 2023 at Peking Union Medical College, Beijing, China.
2.2. TCGA-sourced kidney renal clear cell carcinoma raw sequencing data
The Genomic Data Commons database provides gene sequencing data for 890 patients with RCC from the TCGA project, including 532 with KIRC, 290 with kidney papillary cell carcinoma, 66 with chromophobe RCC, and 2 with sarcomas. All 532 KIRC samples were selected, and 613 raw transcriptome sequencing data (binary alignment/map (bam) files) were downloaded. Of these, 541 cancerous (Tumor) and 72 paracancerous (Normal) samples were included.
2.3. TCGA-sourced kidney renal clear cell carcinoma RNA-seq data
The KIRC RNA-Seq data were obtained from raw sequencing data after comparison using STAR software (version 2.5.3a) [24]. The basic workflow of STAR is divided into two steps: generating a genome index file and posting the read segments back to the genome. This completed the alignment of the transcriptome data. The comparison generated gene expression data in count form, which can be read using the TCGAbiolinks package in R (version 4.1.1) [25].
2.4. Patient information and clinical characteristics
The basic information and clinical data of the 532 patients with KIRC that were downloaded from TCGA are presented in Table 1. Of the 532 patients participating in the study, 64.8 percent were male, and the median age was 61 y (the oldest patient was 90 y and the youngest was 26 y). In term of race, 86.7 percent were White, 10.5 percent were Black or African American, 1.5 percent were Asian, and 1.3 percent had no recorded race. Regarding clinical characteristics, the numbers of patients with tumor node metastasis (TNM) stage I, II, III, and IV disease were 266 (50.3 %), 57 (10.8 %), 123 (23.3 %), and 83 (15.7 %), respectively. In total, 285 (53.6 %) patients received chemotherapy and 247 (46.4 %) received radiation therapy, and the data did not show any patients who received other therapies (e.g., targeted therapy or immunotherapy). Based on the observed endpoints, 357 (67.1 %) patients survived and 175 (32.9 %) died (of various reasons). According to the computation based on length of observation, 96 (18.0 %) patients would survive for less than 1 y, 289 (54.3 %) would survive for 1–5 y, and 147 (27.6 %) would survive for over 5 y overall.
Table 1.
Information of patients with KIRC and the clinical features from TCGA data.
| Patient information | Number (%) |
|---|---|
| Gender | |
| Male | 345 (64.8) |
| Female | 187 (35.2) |
| Age (median), y | 61 (26–90) |
| TNM staging | |
| Ⅰ | 266 (50.3) |
| Ⅱ | 57 (10.8) |
| Ⅲ | 123 (23.3) |
| Ⅳ | 83 (15.7) |
| Treatment | |
| Chemotherapy | 285 (53.6) |
| Radiotherapy | 247 (46.4) |
| Survival status | |
| Deceased | 175 (32.9) |
| Surviving | 357 (67.1) |
| Overall survival | |
| <1 y | 96 (18.0) |
| 1–5 y | 289 (54.3) |
| >5 y | 147 (27.6) |
Abbreviations: KIRC, kidney renal clear cell carcinoma; TCGA, The Cancer Genome Atlas; TNM, tumor node metastasis; y, year.
2.5. Microbial extraction
The original bam files were converted to fastq format using SAMtools software (version 1.3.1) [26] and the data were subsequently classified into microorganisms using Kraken2 software (version 2.0.8) [27]. Microbial read counts were extracted from the fastq files. The results of the Kraken2 were read at the “Species” level, yielding 9886 reads in total.
2.6. Microbial contamination removement
Rigorous filtration and decontamination were performed. Microbial reads with low expression (i.e., those that did not meet the “expressed in ≥10 individuals” or “the total expression of a microorganism >100” criteria) as well as Human and Archaea reads were filtered out. Consequently, 42.2 percent of the reads were removed, leaving 5716 reads. Then, the decontam package in R (version 1.6.0) [28] was used to remove microorganisms with nonlinear variation in expression, resulting in 1181 remaining reads, with 88.1 percent of the original reads removed.
2.7. Microbial diversity analysis
The microbial read counts were read at the “Species” level, and each microorganism was identified as an operational taxonomic unit (OTU). The OTU table of patients with KIRC was analyzed using the vegan package in R (version 2.5–6) (https://cran.r-project.org/package=vegan); that is, the sequences of all samples were randomly selected to a uniform amount of data to ensure their homogeneity. The α-diversity indices were also calculated using vegan, and the Wilcoxon test was used to analyze the differences between groups. In this study, vegan was used to calculate the Bray–Curtis distance and Jaccard index distance using a weighted and an unweighted algorithm, respectively. Additionally, principal coordinate analysis was performed to demonstrate the differences in biological β-diversity between subgroups of KIRC samples and the significance of differences between groups was tested using the Adonis test. Linear discriminant analysis effect size (LEfSe) was performed using the online tool accessible at http://www.bic.ac.cn/BIC.
2.8. Analysis of variance by rank-sum test
The filtered and decontaminated microbial readings were standardized using VOOM and SNM and subsequently submitted to the Wilcoxon rank-sum test to search for differential microorganisms.
2.9. Survival analysis
Univariate and multivariate regression analyses with the Cox proportional hazards regression model were performed using the survival package in R (version 3.3-1) (https://CRAN.R-project.org/package=survival). The risk values were obtained on the basis of the scores of significant variables and used for survival prediction, and the survminer package in R (version 0.4.7) (https://cran.r-project.org/web/packages/survminer/index.html) was used to plot the Kaplan–Meier curves.
2.10. Support vector machine and receiver operating characteristic curves
A support vector machine (SVM) model was used to predict the survival of patients with KIRC based on the screened core microorganisms. Receiver operating characteristic (ROC) curves were plotted and the area under the ROC curve (AUC) values were calculated based on the predicted results.
2.11. Differential gene expression analysis
DESeq2 (version 1.32.0) [29] with patient gene counts was used as the input matrix. The data were normalized to compare gene expression in the two different groups of patients.
2.12. Correlation analysis
The Pearson and Spearman coefficients were used for establishing the correlation between microbial abundance and gene expression.
2.13. Gene functional enrichment analysis
The clusterProfiler package in R (version 4.0.5) [30] was used for gene functional and pathway enrichment analyses on the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases and for gene set enrichment analysis (GSEA).
2.14. Immune infiltration analysis
Immune infiltration analysis was performed using the xcell (version 1.1.0) [31] and CIBERSORT (version 0.1.0) packages in R [32].
3. Results
3.1. Microorganism profile of patients with KIRC
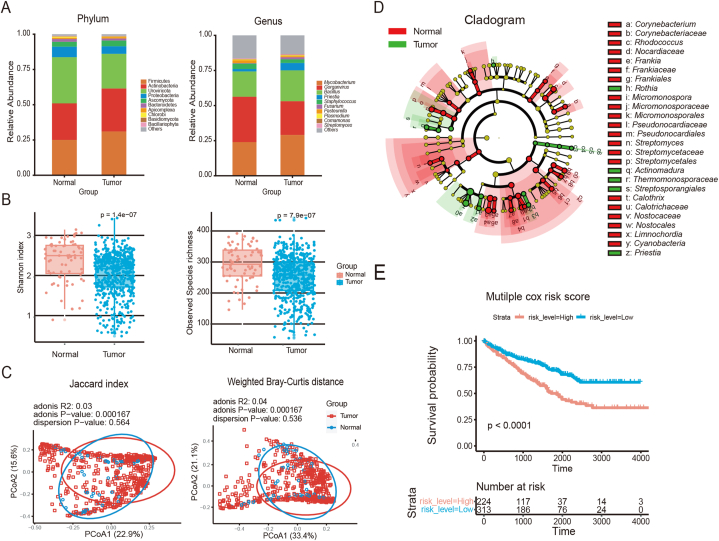
Using the filtered and decontaminated microbial readings, we mapped the tumor microbiomes of patients diagnosed with KIRC. Each microorganism was represented as an OTU, which is a measure of microbial diversity based on genetic similarity (Fig. 1A). This comparison allowed visualization of the microbial profiles of the cancerous (Tumor) and adjacent noncancerous (Normal) tissue samples. Among the identified microbial phyla, Firmicutes, Actinobacteria, Uroviricota, and Proteobacteria were present at the highest relative abundance. The microbial genera with the highest relative abundances were Mycobacterium, Gorganvirus, Bacillus, and Preistia.
Fig. 1.
Overall profile of the comparison between cancerous and paracancerous tissue microbiotas
Abbreviations: Tumor, cancerous samples; Normal, paracancerous samples; PCoA, principal coordinate analysis; KM curve, Kaplan–Meier survival curve; Cox analysis, Cox's proportional hazards regression model analysis.
(A) Overlay of the microbial compositions at the phylum and genus levels. The vertical coordinates represent the relative abundance, and colors represents the top 10 microorganisms. (B) Box plots showing the α-diversity (Shannon and Observed species indices) of cancerous and paracancerous samples, Wilcoxon test. (C) PCoA plots showing the β-diversity of cancerous and paracancerous samples, Adonis test. (D) Cladistic map of microbial taxa in cancerous and paracancerous tissues, marked with significantly different genus. (E) KM curves based on risk level. P values were determined using the Cox proportional hazards risk model.
To reveal the differences in microbial diversity between the tumorous and adjacent non-tumorous samples,α- and β-diversity analyses were performed. The results showed that microbial diversity was significantly reduced in the tumor samples (Fig. 1B) and a significant difference was observed in microbial composition between the tumorous and adjacent non-tumorous samples (Fig. 1C). These results suggest that the TME of KIRC harbors distinct microbial communities compared with the surrounding non-tumorous tissue.
To mitigate the impact of sequencing depth and batch effects on the LEfSe analysis, VOOM normalization together with SNM was performed [9]. Subsequently, the corrected results were tested with the Wilcoxon rank-sum test, leading to the identification of 10 differentially abundant genera between the tumorous and adjacent non-tumorous samples. These genera were Colletotrichum, Arthrobacter, Tumebacillus, Leuconostoc, Thermus, Azoarcus, Pseudochrobactrum, Gluconobacter, Parabacteroides, and Leptospirillum (Fig. 1D).
To further investigate the impact of these 10 KIRC-associated microbial genera on patient survival, a univariate Cox proportional hazards regression analysis was conducted for each genus. Initially, the cutoff values for microbial expression were determined and the optimal cutoff point was obtained using the surv_cutpoint function of the survminer package in R. On the basis of the cutoff point, data above the threshold were defined as high expression (high) and those below the threshold were defined as low expression (low). Univariate Cox regression analysis indicated that five of the 10 genera significantly affected survival. After excluding those not associated with human diseases, four genera (Colletotrichum, Leuconostoc, Gluconobacter, and Parabacteroides) remained significant predictors of patient survival (Table 2). The significance of these four genera persisted after the multifactorial Cox analysis. We calculated the risk score based on the scores of the significant variables and stratified the patients with KIRC into high- and low-risk groups according to the median risk value. As shown in Fig. 1E, patients with high-risk scores had significantly shorter survival outcomes than those with low-risk scores. These findings suggest that the unique microbial signature of KIRC has the potential to predict patient outcomes, providing a basis for the further stratification of patients into long- and short-term survival groups and subsequent analysis of their differential microbiotas.
Table 2.
Univariate Cox statistics for the most differentially abundant microorganisms between cancerous and paracancerous KIRC samples.
| Microorganism | Beta | HR (95 % CI) | Wald test | ap Value |
|---|---|---|---|---|
| Colletotrichum | −0.51 | 0.6 (0.41–0.88) | 6.9 | 0.0088 |
| Arthrobacter | −0.22 | 0.81 (0.58–1.1) | 1.6 | 0.21 |
| Tumebacillus | 0.25 | 1.3 (0.94–1.8) | 2.5 | 0.11 |
| Leuconostoc | −0.42 | 0.66 (0.46–0.94) | 5.4 | 0.02 |
| Thermus | 0.45 | 1.6 (0.91–2.7) | 2.6 | 0.11 |
| Azoarcus | 0.29 | 1.3 (0.87–2.1) | 1.8 | 0.18 |
| Pseudochrobactrum | −0.35 | 0.7 (0.45–1.1) | 2.5 | 0.11 |
| Gluconobacter | −0.37 | 0.69 (0.51–0.93) | 5.9 | 0.015 |
| Parabacteroides | −0.86 | 0.42 (0.21–0.86) | 5.6 | 0.017 |
| Leptospirillum | 0.57 | 1.8 (1.2–2.6) | 8.3 | 0.0039 |
Abbreviations: KIRC, kidney renal clear cell carcinoma; Cox, Cox's proportional hazards regression model; HR, hazard ratio; CI, confidence interval.
p Values of Wilcoxon rank sum tests. The significance threshold was p<0.05.
3.2. Patients with distinct survival outcomes in KIRC possessed different intratumoral microbiotas
Data from 532 patients with KIRC were downloaded from TCGA. With regard to the OS of these patients, 96 patients (18.0 %) survived for less than 1 y, whereas 147 patients (27.6 %) survived for more than 5 y. To exclusively analyze the intratumoral microbiotas, we selected 541 cancer samples on the basis of the availability of microbiota data and categorized them into two groups according to survival outcomes: “Short” survival for patients with an OS of less than 1 y, and “Long” survival for patients with an OS of more than 5 y. In total, 201 samples fit the categories, with 53 classified as “Short” and 148 as “Long.” These two groups were chosen to explore the potential associations between microbiota composition and patient outcomes. Detailed sample characteristics and survival data are presented in Table 3.
Table 3.
Survival classification of patients with KIRC.
| Classification | Standard | Number (%) |
|---|---|---|
| Total | 613 (100) | |
| Tumor | Cancerous samples | 541 (88.3) |
| Normal | Paracancerous samples | 72 (11.7) |
| Survival analysis samples | 201 (100) | |
| Short | (OS < 1 y) and (death) | 53 (26.4) |
| Long | OS > 5 y | 148 (73.6) |
Abbreviations: Long, long survival samples; Short, short survival samples; OS, overall survival; KIRC, kidney renal clear cell carcinoma; y, year.
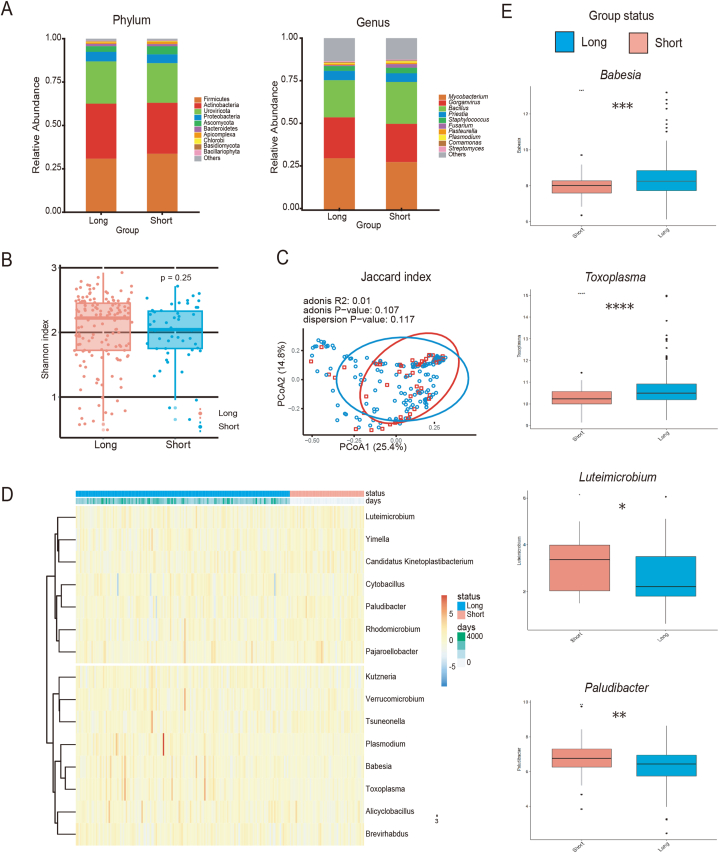
Fig. 2A displays the intratumoral microbial profiles of patients in the different survival groups. In contrast to the previous analysis that compared cancerous tissue with adjacent noncancerous samples, no significant difference in the overall composition of intratumoral microorganisms was found between patients with different survival outcomes. To gain a deeper understanding of the microbial diversity, α- and β-diversity analyses were conducted. According to the α-diversity values, which quantify the species diversity within a sample, samples in the short survival group tended to exhibit lower diversity, although the difference did not reach statistical significance (Fig. 2B). The β-diversity indices, which compare the diversity between different samples, provided additional insights into the microbial variation between the patient groups; however, still did not reach statistical significance (Fig. 2C).
Fig. 2.
Analysis of differential microbial groups between patients with different survival times
Abbreviations: Long, samples from long-survival patients; Short, samples from short-survival patients; KIRC, kidney renal clear cell carcinoma; PCoA, principal coordinate analysis; ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001; ∗∗∗∗, p < 0.0001.
(A) Overlay of the microbial compositions at the phylum and genus levels. The vertical coordinates represent the relative abundance, and colors represents the top 10 microorganisms. (B) Box plots showing the Shannon α-diversity of KIRC samples with differences in long- and short-term survival, Wilcoxon test. (C) PCoA plots showing the Jaccard index β-diversity of the KIRC samples with survival differences, Adonis test. (D) Heat map showing the differential expression of microorganisms between different survival groups. (E) Box plots showing the differential expression of microorganisms between different survival groups, Wilcoxon test.
Further analysis was conducted to identify the microbial genera that showed significant differences between the two groups. After standardization using VOOM + SNM, a Wilcoxon rank-sum test was performed on the basis of the normalized microbial signatures. Fifteen genera showed significant differences between the long and short survival groups. Of these, five genera not relevant to humans were excluded from the study. The remaining 10 genera were Plasmodium, Babesia, Toxoplasma, Luteimicrobium, Cytobacillus, Alicyclobacillus, Rhodomicrobium, Paludibacter, Verrucomicrobium, and Pajaroellobacter (Fig. 2D and E). Plasmodium, a common human pathogen transmitted by mosquitoes, causes malaria [33]. Babesia is the most common parasite associated with human babesiosis in North America and Europe, and infection by this pathogen is now classified as an emerging zoonotic disease [34]. Toxoplasma, an intracellular protozoan parasite of global importance, can infect various mammalian cells [35]. Cytobacillus is a human intestinal isolate with notable probiotic potential for promoting animal and human health [36].
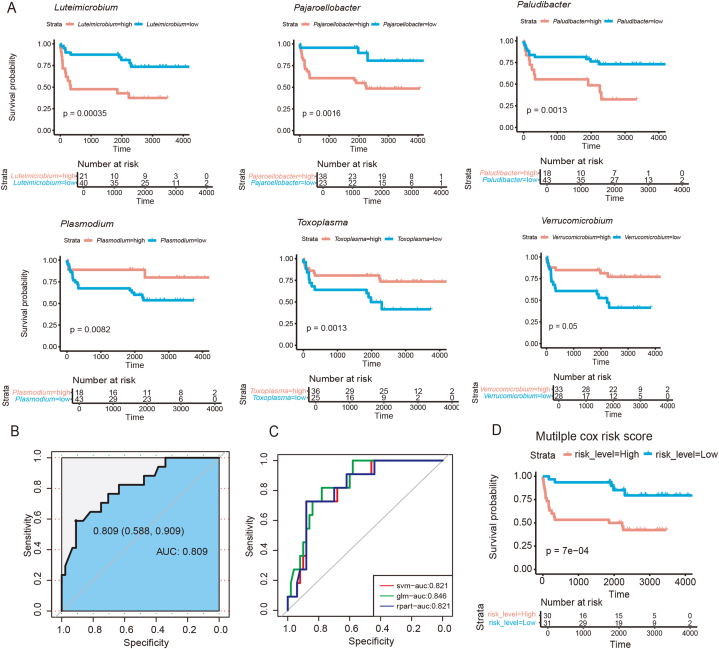
3.3. Microorganisms associated with long or short survival in patients with KIRC can act as potential prognostic factors
To investigate the impact of the 10 genera associated with long or short survival on patient outcomes, univariate Cox regression analysis was performed for each microbial genus. We performed independent prognostic predictions using univariate Cox analysis on a 7:3 randomized split dataset of the significant microbial genera. The results showed that all 10 genera significantly affected patient survival (Table 4). Therefore, the predictive value of these genera was demonstrated, reaffirming their potential as reliable prognostic indicators of patient survival in KIRC (Fig. 3A). According to this classification, the genera beneficial to patient survival were Plasmodium, Babesia, Toxoplasma, Cytobacillus, Alicyclobacillus, and Verrucomicrobium, as patients with higher levels of these microorganisms had longer survival periods. By contrast, the genera that were detrimental to patient survival were Luteimicrobium, Rhodomicrobium, Paludibacter, and Pajaroellobacter.
Table 4.
Univariate Cox statistics for the impact of long and short survival-associated microorganisms on outcomes in patients with KIRC.
| Microorganism | Beta | HR (95 % CI for HR) | Wald test | ap Value |
|---|---|---|---|---|
| Plasmodium | 0.9 | 2.5 (1.3–4.8) | 7 | 0.0082 |
| Babesia | 1.4 | 3.9 (1.4–11) | 7 | 0.0082 |
| Toxoplasma | 0.75 | 2.1 (1.3–3.4) | 10 | 0.0013 |
| Luteimicrobium | −0.83 | 0.43 (0.28–0.69) | 13 | 0.00035 |
| Cytobacillus | 0.85 | 2.3 (1.2–4.7) | 5.7 | 0.017 |
| Alicyclobacillus | 0.7 | 2 (1.2–3.3) | 7.5 | 0.0063 |
| Rhodomicrobium | −0.89 | 0.41 (0.22–0.76) | 8 | 0.0048 |
| Paludibacter | −0.75 | 0.47 (0.3–0.75) | 10 | 0.0013 |
| Verrucomicrobium | 0.46 | 1.6 (1–2.5) | 3.8 | 0.05 |
| Pajaroellobacter | −0.86 | 0.42 (0.25–0.72) | 10 | 0.0016 |
Abbreviations: KIRC, kidney renal clear cell carcinoma; Cox, Cox's proportional hazards regression model; HR, hazard ratio; CI, confidence interval.
p Values of Wilcoxon rank sum tests. The significance threshold was p<0.05.
Fig. 3.
Survival risk scores based on differentially abundant microorganisms
Abbreviations: KM curve, Kaplan–Meier survival curve; Cox analysis, Cox's proportional hazards regression model analysis; ROC, receiver operating characteristic; SVM, support vector machine; AUC, area under the ROC curve; glm, generalized linear model; rpart, decision-making tree.
(A) KM curves showing univariate Cox regression analysis results (Plasmodium, Babesia, Toxoplasma, Cytobacillus, Alicyclobacillus, and Verrucomicrobium). p values were determined using Cox proportional hazards risk models. (B) ROC curve demonstrating SVM training for 10 differential microorganisms. (C) ROC curves from three prediction methods: SVM, glm, and rpart. (D) KM curve showing multiple Cox analyses, in which the dataset was randomly divided into training and prediction sets at a 7:3 ratio.
To further validate the predictive value of these microorganisms, an SVM model was used to predict the survival of patients with KIRC. The data were divided into training and prediction sets in a 7:3 ratio and patient survival was dichotomized according to previous classification criteria. All 10 significant genera were included in the training model and the input data were replaced with previously determined cutoff values. The model was trained with data from the training set and the data from the prediction set were predicted on the basis of the training results. The AUC value was 0.809 (95 % confidence interval: 0.588–0.909), indicating that the model constructed using these 10 genera effectively predicted patient survival (Fig. 3B). Additionally, glm and rpart were used for performing predictions and the results were consistent with those of SVM (Fig. 3C). The robust performance of the 10 microbial signatures across multiple methods underscores their influence on survival outcomes.
To directly investigate the association between the microbial signatures and survival outcomes in patients with KIRC, the dataset was randomly divided into a 70 percent training set and a 30 percent prediction set and performed a multifactorial Cox regression analysis on the basis of the genera mentioned above. This analysis revealed that three genera (Luteimicrobium, Cytobacillus, and Paludibacter) were significant and mutually independent predictors of survival. The Cox regression model was used to derive a value-at-risk score that categorized patients into high- and low-risk groups. Kaplan–Meier survival curves (Fig. 3D) revealed a significant association between the risk scores and survival outcomes, with patients in the high-risk group exhibiting shorter survival times than those in the low-risk group. These findings validate the effectiveness of microbial risk scores in predicting the survival of patients with KIRC.
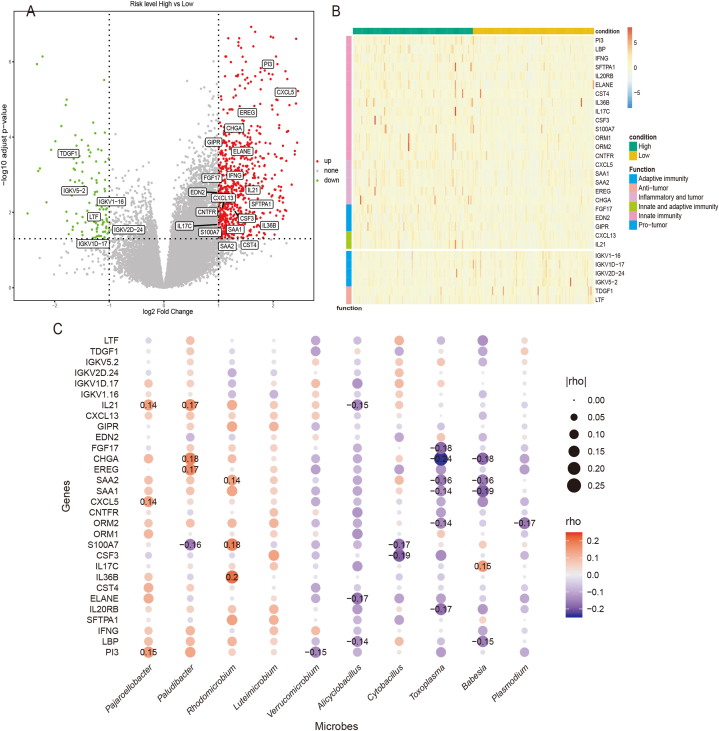
3.4. Identified prognostic-relevant microorganisms have a significant correlation with immune-related genes
To investigate the association between the risk class and gene expression, DESeq2 assessed the differential gene expression patterns between patients stratified by risk class. In total, 882 differentially expressed genes were screened, 760 of which were upregulated and 122 were downregulated. A volcano plot illustrating the differential expression of the genes is shown in Fig. 4A.
Fig. 4.
Differentially abundant microorganisms were related to immune function genes
Abbreviations: High: patients in a high-risk group; low: patients in a low-risk group; p-adj, adjusted p value.
(A) Volcano plot showing differential gene expression in patients with different risk group. Threshold: log2 fold change >1, p-adj <0.05. (B) Heat map showing 30 differential immune gene expression (24 upregulated and 6 downregulated) in patients with different risk groups. (C) Bubble plots showing the correlation between 30 differential immune gene expression and 10 differential microorganisms. Spearman correlation significant (p < 0.05) coefficients are labeled in the figure.
By specifically focusing on immune-related genes, 30 genes of interest were identified. Among these, 24 genes were upregulated in the high-risk group and were primarily associated with antimicrobial function, inflammation, and natural immunity. Examples include PI3, LBP, IFNG, and the chemokine-related genes CXCL5, SAA1, and SAA2, which may play pivotal roles in driving inflammatory responses and tumor development. Conversely, six genes were downregulated in the high-risk group and predominantly related to adaptive immunity. Notable examples are immunoglobulin family genes, such as IGKV1-16, IGKV1D-17, and IGKV2D-24, which may be involved in the adaptive immune response against tumors (Fig. 4B).
To further investigate the relationship between microorganisms and immune genes, Spearman correlation analysis was performed between the 10 differentially expressed microorganisms and 30 immune-related genes. The results were visualized using bubble plots (Fig. 4C). Notably, the correlation analysis revealed significant concordance with previous findings linking differentially expressed immune-related genes to microorganisms. Specifically, most microbial-associated genes were negatively correlated with the identified immune-related genes and were involved in antibacterial responses, inflammation induction, innate immunity, and tumor promotion. Conversely, microbial-associated genes that correlated positively with immune-related genes primarily exhibited functions related to antibody binding, adaptive immunity induction, and tumor suppression. This suggests that microorganisms with negative correlation to immune-related genes may contribute to poorer patient prognosis by promoting antibacterial responses, inflammation, and natural immunity, whereas microorganisms with positive correlation to such genes could potentially enhance adaptive immunity, leading to better patient outcomes.
3.5. Microorganism-based risk scores could stratify patients into two distinct groups characterized by different immune cell functions
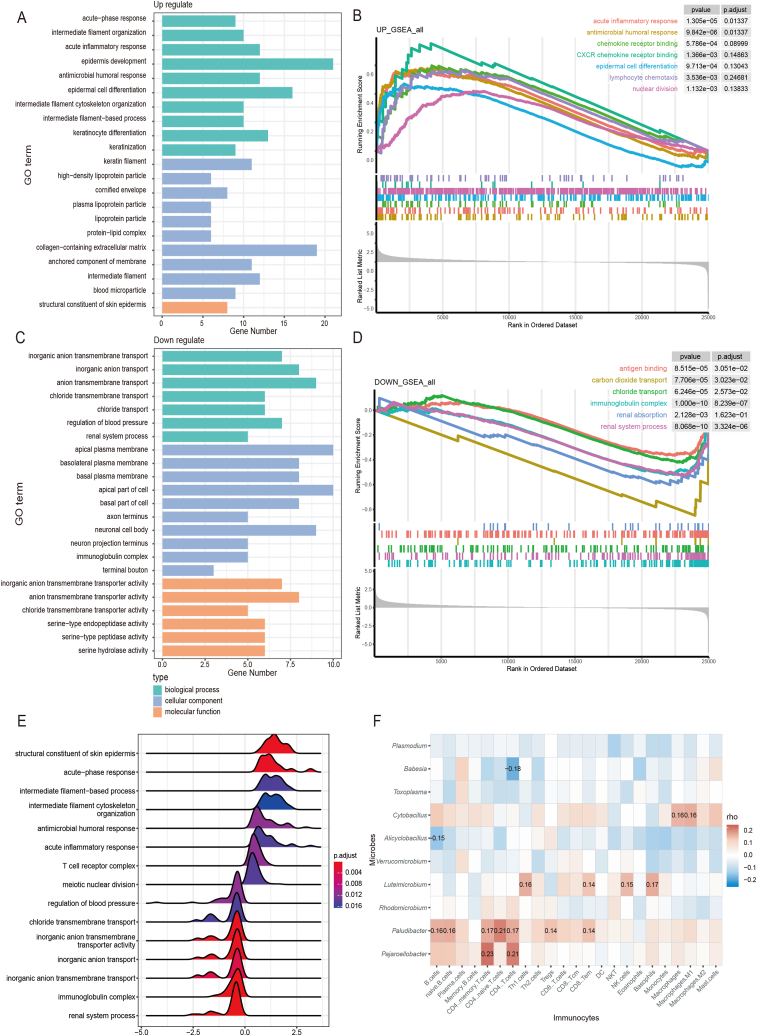
To further clarify the functions of the risk class-based differential genes, GO functional enrichment analysis was performed on 882 differentially expressed genes. The results revealed that the upregulated genes were enriched in functions related to acute response, inflammatory response, antimicrobial immunity, and epidermal cell differentiation, which are associated with tumor growth, whereas the downregulated genes were associated with immunoglobulin complexes and ion transport processes (Fig. 5A and C). After ranking all KIRC genes that underwent a log2 fold change with DESeq2 and then submitting them to GSEA for pathway enrichment, significant pathways associated with the upregulated and downregulated genes were identified. The upregulated gene pathways were those involved in epidermal growth, nuclear division, chemokines, antimicrobial responses, and acute inflammation, whereas the downregulated gene pathways were involved in antibody binding, ion transport, and adaptive immune responses (Fig. 5B and D). These GSEA results aligned with the findings of the GO enrichment analyses.
Fig. 5.
Survival risk scores distinguished high- and low-risk patients with distinct immune-related characteristics
Abbreviations: p-adj, adjusted p value; GO, Gene Ontology; GSEA, gene set enrichment analysis; NES, normalized enrichment score; FDR, false discovery rate.
(A) GO functional annotation pathways of significant upregulated genes (p < 0.05). (B) GSEA graph illustrating the results of upregulated genes in the high-risk group. Screening thresholds: |NES| > 1, p value < 0.05, and FDR (p-adj) < 0.25. (C) GO functional annotation pathways of significant downregulated genes (p < 0.05). (D) GSEA graph illustrating the results of downregulated genes in the high-risk group. (E) GSEA ridge plot with NES as the horizontal coordinate. (F) Heat map showing correlations between survival-related microorganisms and immune cells. Pearson correlation significant (p < 0.05) coefficients are labeled.
Combined with the previous correlation analyses, the microbes specific to the two risk groups were associated with different physiological pathways (Fig. 5E). Previous studies have demonstrated that microorganisms exhibiting negative survival correlation can trigger inflammation and immunosuppression, thereby promoting tumor development [14]. Microbes with negative survival correlation were associated with natural immune and inflammatory responses, potentially through the activation of antimicrobial-associated innate immunity, leading to acute inflammation and poorer prognosis. By contrast, microorganisms that correlated positively with survival may hinder carcinogenesis by modulating immune cell activation [15]. The microbes with positive survival correlation appear to be associated with adaptive immune responses, possibly through adaptive immune processes such as antibody binding, leading to a more favorable prognosis.
Given the correlation of the long- and short-term survival-associated microorganisms with specific immune-related genes and pathways, we hypothesized that these microbes may also be correlated with the corresponding immune cells. To investigate this, xcell package in R calculated the scores for 64 immune cells on the basis of the gene expression profiles of patients with KIRC. Twenty-three major immune cells were selected and Pearson correlation analysis was used to calculate their correlation scores with the 10 genera that differed between the long and short survival groups. The heat map generated from the correlation analysis revealed that the genera were correlated with immune cells (Fig. 5F). Specifically, the negative prognosis-related microorganisms were associated with monocyte, macrophage, and mast cell activities, which play crucial roles in inducing innate inflammatory responses. Conversely, the positive prognosis-related microorganisms correlated with B and T cells, which are key players in adaptive immune responses. These findings are consistent with those of previous studies and provide further insights into the complex interactions between microorganisms and the KIRC immune microenvironment.
4. Discussion
With the rapid development of high-throughput sequencing technology and the continuous updating of data analysis tools, the role of tumor microbiomes in cancer has been further revealed [37,38]. Kidney renal clear cell carcinoma, the predominant subtype of RCC, exhibits high metastatic and mortality rates, marked tumor heterogeneity, and robust immune cell infiltration [2,39]. The tumor immune microenvironment has become the focus of KIRC research [40], particularly the immunosuppressive state which poses challenges for antitumor treatment [41]. Recent decades, extensive research has revealed the potential role of microorganisms in cancer, including its diagnosis, pathogenesis, and treatment of malignant diseases [7]. As research advances, organs and tissues that were previously assumed to be sterile have been found to host diverse microbial populations [17]. Extensive research on tumor microbiomes has revealed notable insights. A recent study on patients with liver cirrhosis identified a higher abundance of Streptococcus mutans in the tumor microbiome, potentially promoting hepatocellular carcinoma through the TLR1/NF-κB/NLR family pathway and IL-3β secretion [42]. Another recent study explored fungal communities across 17 cancer types, highlighting correlations among specific fungi and tumor immune therapy responses [43]. A study in 2023 confirms the presence of a microbiota in adrenocortical carcinoma, revealing its prognostic and biological roles [44]. Consequently, characterizing the microbial communities within KIRC and understanding their relationships with the immune system has become a pivotal research area [8].
This study departed from the conventional 16S rRNA-based approach and instead used amplicon analysis to directly extract microbial information from transcriptomic data, using Kraken2 software for classification. Differentially abundant microorganisms between the cancerous and paracancerous samples were discovered via LEfSe analysis and concluded that KIRC has unique intratumoral microorganisms with a potential impact on patient survival. Subsequently, we analyzed the microbial compositions in patients stratified by long- and short-term survival and further constructed risk value scores using Cox regression and survival analyses. Multi-omics analysis between the microbiome and transcriptome was performed to thoroughly investigate the connections among the microbiota, tumor transcriptome, and tumor-infiltrating immune cells. Microorganisms that contributed to a negative prognosis were linked to natural immune activation. This activation leads to acute inflammation, which in turn is associated with tumor growth and an unfavorable prognosis. Conversely, microorganisms associated with a positive prognosis were linked to the activation of adaptive immune responses. This activation is associated with antitumor effects and a favorable prognosis. Further exploration is required to elucidate the specific mechanism by which microorganisms influence tumor growth. In subsequent studies, it would be beneficial to conduct screenings for specific microorganisms suitable for cultivation, followed by the application of quantitative polymerase chain reaction (qPCR), protein blotting, and other methodologies to validate the impact of microorganisms on tumor progression.
Currently, drugs that benefit patients with KIRC are scarce, underscoring the need for novel biomarkers [45]. This study revealed the significant impact of the tumor microbiota, suggesting valuable biomarkers for enhanced treatment and prognosis. KIRC manifests a correlation between microbiota and immune cells, hinting at two potential mechanisms. One involves activating oncogenic pathways, such as direct or indirect modulation of factors such as interleukin 6 and tumor necrosis factor-alpha, promoting tumor progression [46]. High-risk microbes, such as Luteimicrobium, were found to be associated with macrophages and inflammation, potentially promoting tumor progression [14]. The other mechanism involves reducing T cells, B cells, and natural killer cells in the TME, suppressing antitumor immune responses, regulating local immune surveillance, and weakening tumor-killing effects. Low-risk microbes, like Cytobacillus, activate adoptive immunity involving multiple T cell subsets, hindering tumor progression by modulating immune activation [15]. Although the correlation through immune microenvironment and pathway analyses has been confirmed, the specific mechanisms by which the microbiota affects tumor growth require further exploration. Future investigations could explore single-cell phenotypic changes and considering the unique metabolic features of KIRC.
Some limitations must be acknowledged. First, this study only encompassed KIRC samples. Second, contaminations were inferred from the sequencing data, which limited the reliability of the dataset. Third, the analysis was only assessed in silico. In vitro experiments are still needed to validate the interplay between tumors and their microbiotas.
In summary, microbiome-based subtyping uncovered associations between tumor microbiota, clinical prognosis, and tumor microenvironment. As an increasing number of microbiotas are being implicated in various types of malignant diseases, including liver, ovarian, and bladder cancers [[47], [48], [49]], importance of our findings may extends beyond KIRC. However, the potential challenges that lie ahead must be addressed crucially and the molecular mechanisms underlying the role of microbiotas in cancer development must be explored [16]. This study has laid the groundwork for understanding the characteristics of the RCC microbiome; however, specific mechanisms must be explored further, and clear molecular targets must be identified [50,51]. This will be the focus of future research efforts and the contribution of microbiotas to cancer biology may well become a pivotal area of cancer research in the coming decade [52]. We anticipate revealing more tumor-associated microbiotas as biomarkers, elucidating their intricate relationships with the immune microenvironment, and ultimately enabling more cancer patients to benefit from these groundbreaking findings.
5. Conclusion and future aspects
Our findings suggested several key insights. Firstly, a comparative analysis with adjacent non-tumorous tissues showed that the tumor microbiota had decreased alpha and beta diversities along with distinct microbial compositions. Furthermore, ten microbial genera that were significantly different between patients with long and short overall survival were identified and used for crafting a microbiome-based risk signature for prognosis. Worth noticing, this signature facilitated the identification of two distinct microbiome-driven subtypes that correlated with distinct clinical outcomes and immune microenvironments: four high-risk microbes were linked to pro-tumor acute inflammation while six low-risk microbes activate adoptive immunity to hinder tumor progression. In summary, associations between microbiota, prognosis and tumor immunity highlight the potential of utilizing intratumoral microbiota as a prognostic biomarker. Key future aspects include elucidating the fundamental mechanisms of intratumoral microbiota interactions with the tumor microenvironment and identifying key microbes as potential therapeutic targets. Besides, real-world clinical studies are needed to confirm the practical utility of these microbial biomarkers in predicting patient outcomes and optimizing immunotherapy responses.
Ethics statement
Not appliable.
Data availability
Datasets are available from TCGA. The following is the link to the data used in this research: https://portal.gdc.cancer.gov/projects/TCGA-KIRC.
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Consent to publish
The authors affirm that the manuscript does not contain any personal details, images, or videos of the individuals in the study. Informed consent to publish was obtained from all participants.
Funding
This research was funded by the China Postdoctoral Science Foundation (Grant No. 2023T160328 and No. 2023M731765), Wujieping Medical Foundation (Grant No. 320.6750.2021-20-2) and the University Science Research Project of Jiangsu Province (Natural Science) (Grant No.23KJB320002).
CRediT authorship contribution statement
Hengyi Xu: Writing – original draft, Supervision, Methodology, Formal analysis, Data curation. Jingze Leng: Writing – original draft, Investigation, Formal analysis. Fengshuo Liu: Writing – original draft, Methodology, Formal analysis. Tianxiang Chen: Software, Methodology. Jiangming Qu: Validation, Supervision. Yufan Yang: Validation, Supervision, Investigation. Chun Ning: Validation, Supervision, Investigation. Xindi Ke: Validation, Supervision, Investigation. Bin Xiao: Writing – review & editing, Funding acquisition, Conceptualization. Yanbin Zhang: Writing – review & editing, Funding acquisition, Conceptualization. Lejia Sun: Writing – review & editing, Project administration, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors thank Editage (https://app.editage.com) for English language editing and review services.
Contributor Information
Bin Xiao, Email: jstxiaob@163.com.
Yanbin Zhang, Email: zhangyanbin@jst-hosp.com.cn.
Lejia Sun, Email: lejiasun1881@njmu.edu.cn.
References
- 1.Xue C., Chu Q., Zheng Q., Yuan X., Su Y., Bao Z., Lu J., Li L. Current understanding of the intratumoral microbiome in various tumors. Cell Rep Med. 2023;4(1) doi: 10.1016/j.xcrm.2022.100884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer statistics. CA Cancer J Clin. 2019;69(1):7–34. doi: 10.3322/caac.21551. 2019. [DOI] [PubMed] [Google Scholar]
- 3.Powles T., Staehler M., Ljungberg B., Bensalah K., Canfield S.E., Dabestani S., Giles R.H., Hofmann F., Hora M., Kuczyk M.A., Lam T., Marconi L., Merseburger A.S., Volpe A., Bex A. European association of urology guidelines for clear cell renal cancers that are resistant to vascular endothelial growth factor receptor-targeted therapy. Eur. Urol. 2016;70(5):705–706. doi: 10.1016/j.eururo.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 4.Zhou J., Luo J., Wu K., Yun E.J., Kapur P., Pong R.C., Du Y., Wang B., Authement C., Hernandez E., Yang J., Xiao G., Cha T.L., Wu H.C., Wu D., Margulis V., Lotan Y., Brugarolas J., He D., Hsieh J.T. Loss of DAB2IP in RCC cells enhances their growth and resistance to mTOR-targeted therapies. Oncogene. 2016;35(35):4663–4674. doi: 10.1038/onc.2016.4. [DOI] [PubMed] [Google Scholar]
- 5.McDermott D.F., Regan M.M., Clark J.I., Flaherty L.E., Weiss G.R., Logan T.F., Kirkwood J.M., Gordon M.S., Sosman J.A., Ernstoff M.S., Tretter C.P., Urba W.J., Smith J.W., Margolin K.A., Mier J.W., Gollob J.A., Dutcher J.P., Atkins M.B. Randomized phase III trial of high-dose interleukin-2 versus subcutaneous interleukin-2 and interferon in patients with metastatic renal cell carcinoma. J. Clin. Oncol. 2005;23(1):133–141. doi: 10.1200/jco.2005.03.206. [DOI] [PubMed] [Google Scholar]
- 6.Huang J.J., Hsieh J.J. The therapeutic landscape of renal cell carcinoma: from the dark age to the golden age. Semin. Nephrol. 2020;40(1):28–41. doi: 10.1016/j.semnephrol.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sender R., Fuchs S., Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016;14(8) doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nejman D., Livyatan I., Fuks G., Gavert N., Zwang Y., Geller L.T., Rotter-Maskowitz A., Weiser R., Mallel G., Gigi E., Meltser A., Douglas G.M., Kamer I., Gopalakrishnan V., Dadosh T., Levin-Zaidman S., Avnet S., Atlan T., Cooper Z.A., Arora R., Cogdill A.P., Khan M.A.W., Ologun G., Bussi Y., Weinberger A., Lotan-Pompan M., Golani O., Perry G., Rokah M., Bahar-Shany K., Rozeman E.A., Blank C.U., Ronai A., Shaoul R., Amit A., Dorfman T., Kremer R., Cohen Z.R., Harnof S., Siegal T., Yehuda-Shnaidman E., Gal-Yam E.N., Shapira H., Baldini N., Langille M.G.I., Ben-Nun A., Kaufman B., Nissan A., Golan T., Dadiani M., Levanon K., Bar J., Yust-Katz S., Barshack I., Peeper D.S., Raz D.J., Segal E., Wargo J.A., Sandbank J., Shental N., Straussman R. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science. 2020;368(6494):973–980. doi: 10.1126/science.aay9189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poore G.D., Kopylova E., Zhu Q., Carpenter C., Fraraccio S., Wandro S., Kosciolek T., Janssen S., Metcalf J., Song S.J., Kanbar J., Miller-Montgomery S., Heaton R., McKay R., Patel S.P., Swafford A.D., Knight R. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature. 2020;579(7800):567–574. doi: 10.1038/s41586-020-2095-1. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Aykut B., Pushalkar S., Chen R., Li Q., Abengozar R., Kim J.I., Shadaloey S.A., Wu D., Preiss P., Verma N., Guo Y., Saxena A., Vardhan M., Diskin B., Wang W., Leinwand J., Kurz E., Kochen Rossi J.A., Hundeyin M., Zambrinis C., Li X., Saxena D., Miller G. The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature. 2019;574(7777):264–267. doi: 10.1038/s41586-019-1608-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang K.W., Alaei-Mahabadi B., Samuelsson T., Lindh M., Larsson E. The landscape of viral expression and host gene fusion and adaptation in human cancer. Nat. Commun. 2013;4:2513. doi: 10.1038/ncomms3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zitvogel L., Ma Y., Raoult D., Kroemer G., Gajewski T.F. The microbiome in cancer immunotherapy: diagnostic tools and therapeutic strategies. Science. 2018;359(6382):1366–1370. doi: 10.1126/science.aar6918. [DOI] [PubMed] [Google Scholar]
- 13.Kadosh E., Snir-Alkalay I., Venkatachalam A., May S., Lasry A., Elyada E., Zinger A., Shaham M., Vaalani G., Mernberger M., Stiewe T., Pikarsky E., Oren M., Ben-Neriah Y. The gut microbiome switches mutant p53 from tumour-suppressive to oncogenic. Nature. 2020;586(7827):133–138. doi: 10.1038/s41586-020-2541-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pushalkar S., Hundeyin M., Daley D., Zambirinis C.P., Kurz E., Mishra A., Mohan N., Aykut B., Usyk M., Torres L.E., Werba G., Zhang K., Guo Y., Li Q., Akkad N., Lall S., Wadowski B., Gutierrez J., Kochen Rossi J.A., Herzog J.W., Diskin B., Torres-Hernandez A., Leinwand J., Wang W., Taunk P.S., Savadkar S., Janal M., Saxena A., Li X., Cohen D., Sartor R.B., Saxena D., Miller G. The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discov. 2018;8(4):403–416. doi: 10.1158/2159-8290.Cd-17-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foresto-Neto O., Ghirotto B., Câmara N.O.S. Renal sensing of bacterial metabolites in the gut-kidney Axis. Kidney360. 2021;2(9):1501–1509. doi: 10.34067/kid.0000292021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang L., Li A., Wang Y., Zhang Y. Intratumoral microbiota: roles in cancer initiation, development and therapeutic efficacy. Signal Transduct Target Ther. 2023;8(1):35. doi: 10.1038/s41392-022-01304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun L., Ke X., Guan A., Jin B., Qu J., Wang Y., Xu X., Li C., Sun H., Xu H., Xu G., Sang X., Feng Y., Sun Y., Yang H., Mao Y. Intratumoural microbiome can predict the prognosis of hepatocellular carcinoma after surgery. Clin. Transl. Med. 2023;13(7) doi: 10.1002/ctm2.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Markowski M.C., Boorjian S.A., Burton J.P., Hahn N.M., Ingersoll M.A., Maleki Vareki S., Pal S.K., Sfanos K.S. The microbiome and genitourinary cancer: a collaborative review. Eur. Urol. 2019;75(4):637–646. doi: 10.1016/j.eururo.2018.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dhote R., Thiounn N., Debré B., Vidal-Trecan G. Risk factors for adult renal cell carcinoma. Urol Clin North Am. 2004;31(2):237–247. doi: 10.1016/j.ucl.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 20.Kovaleva O.V., Podlesnaya P., Sorokin M., Mochalnikova V., Kataev V., Khlopko Y.A., Plotnikov A.O., Stilidi I.S., Kushlinskii N.E., Gratchev A. Macrophage phenotype in combination with tumor microbiome composition predicts RCC patients' survival: a pilot study. Biomedicines. 2022;10(7) doi: 10.3390/biomedicines10071516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo Y., Tsai H.I., Zhang L., Zhu H. Mitochondrial DNA on tumor-associated macrophages polarization and immunity. Cancers. 2022;14(6) doi: 10.3390/cancers14061452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu K., Li Y., Ma K., Zhao W., Yao Z., Zheng Z., Sun F., Mu X., Liu Z., Zheng J. The microbiota and renal cell carcinoma. Cell. Oncol. 2023 doi: 10.1007/s13402-023-00876-9. [DOI] [PubMed] [Google Scholar]
- 23.Whiteside S.A., Razvi H., Dave S., Reid G., Burton J.P. The microbiome of the urinary tract--a role beyond infection. Nat. Rev. Urol. 2015;12(2):81–90. doi: 10.1038/nrurol.2014.361. [DOI] [PubMed] [Google Scholar]
- 24.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Colaprico A., Silva T.C., Olsen C., Garofano L., Cava C., Garolini D., Sabedot T.S., Malta T.M., Pagnotta S.M., Castiglioni I., Ceccarelli M., Bontempi G., Noushmehr H. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44(8):e71. doi: 10.1093/nar/gkv1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wood D.E., Lu J., Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20(1):257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis N.M., Proctor D.M., Holmes S.P., Relman D.A., Callahan B.J. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome. 2018;6(1):226. doi: 10.1186/s40168-018-0605-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., Feng T., Zhou L., Tang W., Zhan L., Fu X., Liu S., Bo X., Yu G. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation. 2021;2(3) doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aran D., Hu Z., Butte A.J. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18(1):220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newman A.M., Liu C.L., Green M.R., Gentles A.J., Feng W., Xu Y., Hoang C.D., Diehn M., Alizadeh A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kochar D.K., Saxena V., Singh N., Kochar S.K., Kumar S.V., Das A. Plasmodium vivax malaria. Emerg. Infect. Dis. 2005;11(1):132–134. doi: 10.3201/eid1101.040519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Herwaldt B.L., Cacciò S., Gherlinzoni F., Aspöck H., Slemenda S.B., Piccaluga P., Martinelli G., Edelhofer R., Hollenstein U., Poletti G., Pampiglione S., Löschenberger K., Tura S., Pieniazek N.J. Molecular characterization of a non-Babesia divergens organism causing zoonotic babesiosis in Europe. Emerg. Infect. Dis. 2003;9(8):942–948. doi: 10.3201/eid0908.020748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shwab E.K., Saraf P., Zhu X.Q., Zhou D.H., McFerrin B.M., Ajzenberg D., Schares G., Hammond-Aryee K., van Helden P., Higgins S.A., Gerhold R.W., Rosenthal B.M., Zhao X., Dubey J.P., Su C. Human impact on the diversity and virulence of the ubiquitous zoonotic parasite Toxoplasma gondii. Proc Natl Acad Sci U S A. 2018;115(29):E6956–e6963. doi: 10.1073/pnas.1722202115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Djukovic A., Garzón M.J., Canlet C., Cabral V., Lalaoui R., García-Garcerá M., Rechenberger J., Tremblay-Franco M., Peñaranda I., Puchades-Carrasco L., Pineda-Lucena A., González-Barberá E.M., Salavert M., López-Hontangas J.L., Sanz M., Sanz J., Kuster B., Rolain J.M., Debrauwer L., Xavier K.B., Xavier J.B., Ubeda C. Lactobacillus supports Clostridiales to restrict gut colonization by multidrug-resistant Enterobacteriaceae. Nat. Commun. 2022;13(1):5617. doi: 10.1038/s41467-022-33313-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Callahan B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J., Holmes S.P. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods. 2016;13(7):581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bolyen E., Rideout J.R., Dillon M.R., Bokulich N.A., Abnet C.C., Al-Ghalith G.A., Alexander H., Alm E.J., Arumugam M., Asnicar F., Bai Y., Bisanz J.E., Bittinger K., Brejnrod A., Brislawn C.J., Brown C.T., Callahan B.J., Caraballo-Rodríguez A.M., Chase J., Cope E.K., Da Silva R., Diener C., Dorrestein P.C., Douglas G.M., Durall D.M., Duvallet C., Edwardson C.F., Ernst M., Estaki M., Fouquier J., Gauglitz J.M., Gibbons S.M., Gibson D.L., Gonzalez A., Gorlick K., Guo J., Hillmann B., Holmes S., Holste H., Huttenhower C., Huttley G.A., Janssen S., Jarmusch A.K., Jiang L., Kaehler B.D., Kang K.B., Keefe C.R., Keim P., Kelley S.T., Knights D., Koester I., Kosciolek T., Kreps J., Langille M.G.I., Lee J., Ley R., Liu Y.X., Loftfield E., Lozupone C., Maher M., Marotz C., Martin B.D., McDonald D., McIver L.J., Melnik A.V., Metcalf J.L., Morgan S.C., Morton J.T., Naimey A.T., Navas-Molina J.A., Nothias L.F., Orchanian S.B., Pearson T., Peoples S.L., Petras D., Preuss M.L., Pruesse E., Rasmussen L.B., Rivers A., Robeson M.S., 2nd, Rosenthal P., Segata N., Shaffer M., Shiffer A., Sinha R., Song S.J., Spear J.R., Swafford A.D., Thompson L.R., Torres P.J., Trinh P., Tripathi A., Turnbaugh P.J., Ul-Hasan S., van der Hooft J.J.J., Vargas F., Vázquez-Baeza Y., Vogtmann E., von Hippel M., Walters W., Wan Y., Wang M., Warren J., Weber K.C., Williamson C.H.D., Willis A.D., Xu Z.Z., Zaneveld J.R., Zhang Y., Zhu Q., Knight R., Caporaso J.G. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019;37(8):852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang Y., An H., Xu L., Zhu Y., Yang Y., Lin Z., Xu J. Systemic inflammation score predicts postoperative prognosis of patients with clear-cell renal cell carcinoma. Br. J. Cancer. 2015;113(4):626–633. doi: 10.1038/bjc.2015.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chevrier S., Levine J.H., Zanotelli V.R.T., Silina K., Schulz D., Bacac M., Ries C.H., Ailles L., Jewett M.A.S., Moch H., van den Broek M., Beisel C., Stadler M.B., Gedye C., Reis B., Pe'er D., Bodenmiller B. An immune atlas of clear cell renal cell carcinoma. Cell. 2017;169(4):736–749.e718. doi: 10.1016/j.cell.2017.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pérez-Ruiz E., Melero I., Kopecka J., Sarmento-Ribeiro A.B., García-Aranda M., De Las Rivas J. Cancer immunotherapy resistance based on immune checkpoints inhibitors: targets, biomarkers, and remedies. Drug Resist Updat. 2020;53 doi: 10.1016/j.drup.2020.100718. [DOI] [PubMed] [Google Scholar]
- 42.Liu B., Zhou Z., Jin Y., Lu J., Feng D., Peng R., Sun H., Mu X., Li C., Chen Y. Hepatic stellate cell activation and senescence induced by intrahepatic microbiota disturbances drive progression of liver cirrhosis toward hepatocellular carcinoma. J Immunother Cancer. 2022;10(1) doi: 10.1136/jitc-2021-003069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Narunsky-Haziza L., Sepich-Poore G.D., Livyatan I., Asraf O., Martino C., Nejman D., Gavert N., Stajich J.E., Amit G., González A., Wandro S., Perry G., Ariel R., Meltser A., Shaffer J.P., Zhu Q., Balint-Lahat N., Barshack I., Dadiani M., Gal-Yam E.N., Patel S.P., Bashan A., Swafford A.D., Pilpel Y., Knight R., Straussman R. Pan-cancer analyses reveal cancer-type-specific fungal ecologies and bacteriome interactions. Cell. 2022;185(20):3789–3806.e3717. doi: 10.1016/j.cell.2022.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y., Zhang D., Wang M., Jiang H., Feng C., Li Y.X. Intratumoral microbiota is associated with prognosis in patients with adrenocortical carcinoma. iMeta. 2023:e102. doi: 10.1002/imt2.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu H., Jia Z., Liu F., Li J., Huang Y., Jiang Y., Pu P., Shang T., Tang P., Zhou Y., Yang Y., Su J., Liu J. Biomarkers and experimental models for cancer immunology investigation. MedComm. 2023;4(6):e437. doi: 10.1002/mco2.437. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garrett W.S. Cancer and the microbiota. Science. 2015;348(6230):80–86. doi: 10.1126/science.aaa4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma J., Li J., Jin C., Yang J., Zheng C., Chen K., Xie Y., Yang Y., Bo Z., Wang J., Su Q., Wang J., Chen G., Wang Y. Association of gut microbiome and primary liver cancer: a two-sample Mendelian randomization and case-control study. Liver Int. 2023;43(1):221–233. doi: 10.1111/liv.15466. [DOI] [PubMed] [Google Scholar]
- 48.Roelands J., Kuppen P.J.K., Ahmed E.I., Mall R., Masoodi T., Singh P., Monaco G., Raynaud C., de Miranda N., Ferraro L., Carneiro-Lobo T.C., Syed N., Rawat A., Awad A., Decock J., Mifsud W., Miller L.D., Sherif S., Mohamed M.G., Rinchai D., Van den Eynde M., Sayaman R.W., Ziv E., Bertucci F., Petkar M.A., Lorenz S., Mathew L.S., Wang K., Murugesan S., Chaussabel D., Vahrmeijer A.L., Wang E., Ceccarelli A., Fakhro K.A., Zoppoli G., Ballestrero A., Tollenaar R., Marincola F.M., Galon J., Khodor S.A., Ceccarelli M., Hendrickx W., Bedognetti D. An integrated tumor, immune and microbiome atlas of colon cancer. Nat Med. 2023;29(5):1273–1286. doi: 10.1038/s41591-023-02324-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sheng D., Yue K., Li H., Zhao L., Zhao G., Jin C., Zhang L. The interaction between intratumoral microbiome and immunity is related to the prognosis of ovarian cancer. Microbiol. Spectr. 2023;11(2) doi: 10.1128/spectrum.03549-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kandalai S., Li H., Zhang N., Peng H., Zheng Q. The human microbiome and cancer: a diagnostic and therapeutic perspective. Cancer Biol. Ther. 2023;24(1) doi: 10.1080/15384047.2023.2240084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.El Tekle G., Garrett W.S. Bacteria in cancer initiation, promotion and progression. Nat. Rev. Cancer. 2023;23(9):600–618. doi: 10.1038/s41568-023-00594-2. [DOI] [PubMed] [Google Scholar]
- 52.Swanton C., Bernard E., Abbosh C., André F., Auwerx J., Balmain A., Bar-Sagi D., Bernards R., Bullman S., DeGregori J., Elliott C., Erez A., Evan G., Febbraio M.A., Hidalgo A., Jamal-Hanjani M., Joyce J.A., Kaiser M., Lamia K., Locasale J.W., Loi S., Malanchi I., Merad M., Musgrave K., Patel K.J., Quezada S., Wargo J.A., Weeraratna A., White E., Winkler F., Wood J.N., Vousden K.H., Hanahan D. Embracing cancer complexity: hallmarks of systemic disease. Cell. 2024;187(7):1589–1616. doi: 10.1016/j.cell.2024.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Datasets are available from TCGA. The following is the link to the data used in this research: https://portal.gdc.cancer.gov/projects/TCGA-KIRC.