Abstract
Introduction
Single-cell RNA sequencing (scRNA-seq) and single-nucleus RNA sequencing (snRNA-seq) provide valuable insights into the cellular states of kidney cells. However, the annotation of cell types often requires extensive domain expertise and time-consuming manual curation, limiting scalability and generalizability. To facilitate this process, we tested the performance of five supervised classification methods for automatic cell type annotation.
Results
We analyzed publicly available sc/snRNA-seq datasets from five expert-annotated studies, comprising 62,120 cells from 79 kidney biopsy samples. Datasets were integrated by harmonizing cell type annotations across studies. Five different supervised machine learning algorithms (support vector machines, random forests, multilayer perceptrons, k-nearest neighbors, and extreme gradient boosting) were applied to automatically annotate cell types using four training datasets and one testing dataset. Performance metrics, including accuracy (F1 score) and rejection rates, were evaluated. All five machine learning algorithms demonstrated high accuracies, with a median F1 score of 0.94 and a median rejection rate of 1.8 %. The algorithms performed equally well across different datasets and successfully rejected cell types that were not present in the training data. However, F1 scores were lower when models trained primarily on scRNA-seq data were tested on snRNA-seq data.
Conclusions
Despite limitations including the number of biopsy samples, our findings demonstrate that machine learning algorithms can accurately annotate a wide range of adult kidney cell types in scRNA-seq/snRNA-seq data. This approach has the potential to standardize cell type annotation and facilitate further research on cellular mechanisms underlying kidney disease.
Keywords: Kidney, RNA-seq, Machine learning, Classification, Cell identity, Annotation
1. Introduction
The human kidney is a highly complex organ composed of various cell types with distinct functions. Recent advancements in single-cell (sc) and single-nucleus (sn) RNA sequencing (RNA-seq) have provided researchers with the ability to examine the transcriptome of individual cells [1,2]. This technological breakthrough enables a detailed understanding of the components and functional processes of distinct kidney cell types and presents opportunities for targeted therapeutic interventions aimed toward these [3]. Consequently, the field of kidney medicine is poised to undergo a transformative shift towards a data-driven, precision-based approach.
Despite the improvements in the clustering of cell types made possible by these sophisticated techniques, the task of annotating the resulting data remains predominantly manual [[4], [5], [6], [7], [8], [9]]. Researchers typically rely on a combination of personally identified biomarkers to identify specific cell populations, a laborious and non-standardized process that necessitates expertise in navigating the intricate transcriptomic diversity of the human kidney [[4], [5], [6], [7], [8], [9]]. Consequently, this manual annotation introduces subjectivity into an otherwise data-driven analysis and restricts the ability of researchers to conduct cross-study and validation analyses and scale up these investigations due to inconsistent ontologies [[4], [5], [6], [7], [8], [9], [10]].
Modern machine learning tools offer a potential solution for addressing the challenge of cell type annotation. Various algorithms have been developed specifically for cell type annotation by leveraging scRNA-seq data. For instance, in one study, researchers successfully employed an extreme gradient boosting (XGBoost) algorithm as part of a machine learning pipeline to classify and predict cardiac developmental cell types [11]. Another comprehensive study conducted by Abdelaal et al. (2019) compared several supervised machine learning algorithms, such as linear discriminant analysis, nearest mean classifiers, support vector machines (SVM), random forests (RF), and k-nearest neighbors (KNN), across 27 distinct scRNA datasets encompassing brain, pancreas, and peripheral blood mononuclear cells from both human and mouse samples [4]. The results demonstrated that all the algorithms exhibited high median F1 scores and low rejection rates. Notably, the SVM classifier with a linear kernel demonstrated the most optimal performance in their analysis [4]. However, it is important to note that the study conducted by Abdelaal et al. (2019) did not specifically examine kidney cell types, leaving the applicability of machine learning algorithms for accurately predicting kidney cell types uncertain [4]. Furthermore, there are relatively fewer studies that compare machine learning methods for cell type annotations using snRNA-seq data [12].
In this study, we aimed to assess and compare the effectiveness of various machine learning algorithms for automating kidney cell type annotations. To achieve this, we utilized five publicly available scRNA-seq and snRNA-seq datasets that had been previously annotated by experts. We pooled author-identified cell types into harmonized cell types, applied five different machine learning algorithms to predict harmonized cell type annotations, and evaluated the performance of the different machine learning models using F1 scores and the rate at which models labeled cells as “unknown.” Findings from our study build on ongoing efforts focused on the development and implementation of standardized cell type ontologies, and more broadly serve to improve our understanding of kidney physiology.
2. Results
2.1. Harmonization of cell type annotations across datasets reveals 16 harmonized cell types
Our dataset encompasses a diverse collection of kidney cell-specific transcriptomic data, consisting of a total of 62,120 cells obtained from 79 kidney biopsy samples originating from 44 healthy donors across 5 different studies. We deliberately included data from donors of varying ages, spanning the cortex and medulla, and obtained through multiple sequencing technologies, as outlined in Table 1.
Table 1.
Metadata from the 5 different sc/snRNA datasets analyzed in this study.
| Study (PMID) | Number of Cells | Number of Donors | Number of Samples | Donor Age Range | Donor Sex |
Sampling Locations |
Sequencing Method | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M | F | Not Reported | Cortex | Medulla | Both | Unknown/Other | ||||||
| Menon (32107344) | 22,264 | 22 | 24 | <50 = 2 ≥50 = 13 Unknown = 7 |
7 | 6 | 9 | NA | NA | NA | 24 | (sc) 10X |
| Young (30093597) | 6197 | 5 | 17 | 49–72 | 3 | 2 | 0 | 14 | 0 | 1 | 2 | (sc) 10X |
| Liao (31896769) | 16,145 | 2 | 2 | 59–65 | 1 | 1 | 0 | NA | NA | NA | 2 | (sc) 10X |
| Wu (29980650) | 4259 | 1 | 1 | 70 | 1 | 0 | 0 | NA | NA | NA | 1 | (sn) InDrops |
| Lake (31249312) | 13,255 | 14 | 35 | <50 = 4 ≥50 = 7 Unknown = 3 |
10 | 4 | 0 | 15 | 14 | 6 | 0 | (sn) Drop-Seq |
| Total | 62,120 | 44 | 79 | 22 | 13 | 9 | 29 | 14 | 7 | 29 | ||
The age range of the donors spanned from under 30 to over 70 years old. 29 samples consisted only of cortical tissue, 14 samples consisted only of medullary tissue, 7 samples consisted of both, and the sampling location of the remaining 29 samples were either unknown (n = 28) or from ureteral tissue (n = 1). As shown in Fig. S1A, among cells of known sampling location, 8698 (44.7 %) were from the cortex alone, 7742 (39.8 %) were from the medulla alone, 2050 (10.5 %) were from the corticomedullary junction, and 962 (4.95 %) were from the ureter. Among the five datasets incorporated, three utilized the 10X single-cell technology, one used the InDrops single nucleus technology, while the remaining one employed Drop-Seq single nucleus technology as illustrated in Fig. S1B [[13], [14], [15]]. Additionally, our dataset consisted of at least 22 males and 13 females, which contributed to 31,838 (51.2 %) and 15,753 (25.4 %) cells, respectively, as shown in Fig. S1C. For comprehensive details regarding the donors, we refer readers to the original publications associated with each dataset [[5], [6], [7], [8], [9]].
To ensure the quality of our dataset, we performed quality control measures by leveraging published data and code from each study. We pre-processed each study dataset individually, including performing pertinent transformations and dropping of samples and/or cells as described in Methods. We validated the original author cell type annotations using the uniform manifold approximation and projection for dimension reduction (UMAP) visualization technique. The UMAP visualizations for each of the five datasets can be found in Fig. S2.
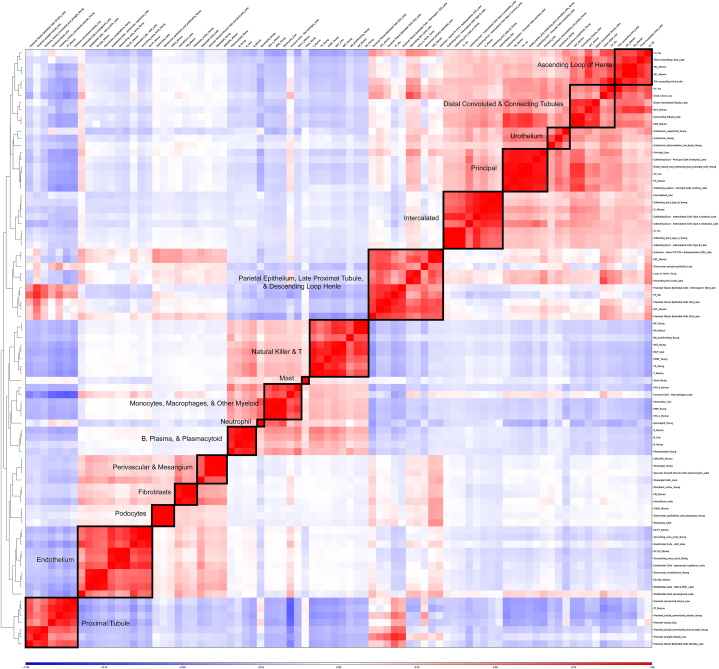
Overall, we identified a total of 84 unique cell type annotations across all five cohorts. It is important to note that all these cells were derived from healthy, adult human kidneys. To consolidate the annotations and establish a unified cell type nomenclature, we leveraged the transcriptomic data and observed high correlations between individual study annotations with respect to expression of marker genes. Consequently, cell type annotations that exhibited strong correlation patterns were grouped together into harmonized cell types. Fig. 1 illustrates the results of this analysis, revealing 16 distinct harmonized kidney cell types based on transcriptomic data. For instance, annotations from different studies that included the term “podocyte” were highly correlated with each other, leading us to assign them to a single harmonized cell type referred to as “Podocyte.” This consolidation approach was applied consistently across the remaining 15 harmonized cell types.
Fig. 1.
Heatmap of correlations between cell types across all 5 cohorts per the authors' original annotations. Axes are a symmetrical layout of annotations and annotations are grouped according to dendrogram and boxed by harmonized cell types, which are defined as groups of annotations with a high degree of correlation.
The number of individual cells included in each harmonized cell type varied across studies. As depicted in Fig. S3A the “Proximal Tubule” harmonized cell type encompassed the largest number of cells, totaling 23,177. On the other hand, the “Mast” harmonized cell type had the smallest cell count, with only 22 cells identified. Rare cell types, such as “Fibroblasts” and “B, Plasma, & Plasmacytoid” benefitted from the inclusion of multiple studies in our dataset, compensating for their low cell counts in individual studies (Fig. S3B). By combining multiple datasets, we were able to overcome the limitations of each individual study regarding the inclusion of specific cell types. This was observed even among harmonized cell types that contained a substantial number of cells. For instance, although Lake et al. had only 16 cells in the “Monocytes, Macrophages, & Other Myeloid” cell type, the inclusion of these cells from three of the other datasets compensated for this omission, resulting in 2429 “Monocytes, Macrophages, & Other Myeloid”-labeled cells in our final integrated dataset (Table S1). In a few cases, certain cell types were only present in a single study, as exemplified by the “Neutrophil,” “Mast,” and “Urothelium” harmonized cell types. This highlights the significance of incorporating multiple studies in our data to complement one another and achieve a comprehensive coverage of healthy, adult human kidneys in our training dataset.
Prior to integrating all five datasets, we examined the combined UMAP visualization of the datasets and observed the presence of batch effects, as shown in Fig. S4A. To address this issue, we employed rPCA from the Seurat package to mitigate the batch effects, resulting in a batch-corrected UMAP plot illustrated in Fig. S4B. Following batch correction, we observed a more even distribution of harmonized cell types across the different studies, as depicted in Fig. S4C. However, it's important to note that despite the batch correction, we encountered instances where certain cells did not align perfectly with the harmonized cell types based on the original authors' annotations. To address this discrepancy, we trained a support vector machine (SVM) model using all 62,120 cells. When we applied the trained model on the same data, it predicted the wrong harmonized cell type label for a subset of 4256 cells (6.9 %). Consequently, we categorized these 4256 cells as low-quality and excluded them from further analyses, as illustrated in Fig. S5. The choice of SVM for cell type classification was based on its demonstrated high performance in previous studies [4,[16], [17], [18], [19], [20]]. Our final integrated dataset included 57,864 cells that were not identified as low-quality cells. The distribution of these cells by harmonized cell type can be found in Table S1. Metadata regarding each cell and sample can be found in our Zenodo as described in Methods.
2.2. Machine learning algorithms were predictive of harmonized cell types
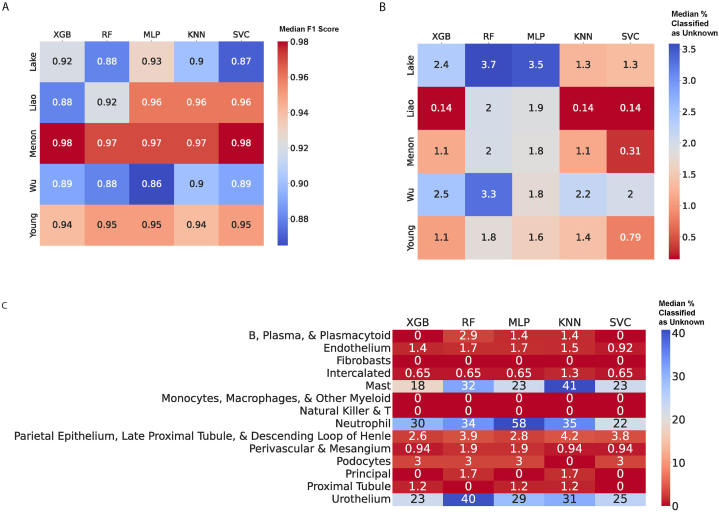
Next, we employed five distinct supervised learning methods to predict the harmonized cell type annotations in our integrated dataset. These methods included a support vector classifier (SVC), a random forest classifier (RF), a multilayer perceptron (MLP), a k-nearest neighbors classifier (KNN), and an extreme gradient boost (XGB) model. To evaluate the performance of these models, we adopted an inter-dataset evaluation scheme. This involved utilizing combinations of four out of the five datasets as the training data and using the remaining fifth dataset as the testing data. By employing this approach, we aimed to reduce the risk of overfitting by ensuring that the testing data was not used during the training process of the model. Fig. 2A demonstrates that all the employed algorithms exhibited a median F1 score of 0.92 or higher when tested on each of the individual datasets. These high median F1 scores indicate the strong performance of the algorithms in accurately identifying harmonized cell type annotations using transcriptomic data.
Fig. 2.
Heatmaps demonstrating for each testing dataset, the performances of each classification algorithm as defined by (a) median F1 score or (b) median percent of cells classified as unknowns. (c) Heatmap of each classification algorithm's rejection rate on when Young et al. was used as the testing dataset.
2.3. Feature importance analysis
In order to better understand which genes contributed most significantly to the classification of kidney cell types, we extracted the feature importance scores from all five machine learning models and generated a comparative analysis (Fig. S10). Using the Menon dataset as the testing dataset, we observed that certain genes, such as SPP11, consistently ranked highly across all five models, indicating that these genes play a key role in distinguishing specific kidney cell types. This variation in feature importance highlights the strengths of different algorithms in identifying cell type–specific markers and underlines the value of using multiple models to obtain a more comprehensive view of key gene markers.
2.4. Performance of different classifiers varied across sequencing methods and harmonized cell types
Upon comparing the performance of each algorithm across all datasets, we observed that the median F1 scores varied depending on the specific dataset used for testing. For instance, the XGB algorithm achieved the highest median F1 score across cell types when the Menon dataset was used for testing. On the other hand, the KNN algorithm achieved the highest median F1 score across cell types when the Wu dataset was used for testing and the lowest median F1 score across cell types when the Young dataset was used for testing, and the MLP algorithm attained the highest median F1 score when the Lake dataset was used for testing (Fig. 2A). However, it is noteworthy that none of the five machine learning algorithms significantly outperformed the others across the five testing sets. This observation is supported by the results of Kruskal-Wallis tests showing that the p-values for the Menon, Lake, Liao, Wu, and Young datasets were 0.62, 0.97, 0.94, 0.85, and 1, respectively (Table S2).
In some instances, certain harmonized cell types were only present in a specific study, such as “Neutrophil,” “Mast,” and “Urothelium” in the Young dataset. When the Young dataset was used as the testing dataset, models trained on the other four datasets were unable to predict these cell types, resulting in an F1 score of 0. Consequently, we also compared the rejection rates, which represent the percentage of cells labeled as “Unknown,” across the different machine learning algorithms and datasets to assess their effectiveness (Fig. 2B). Interestingly, none of the five machine learning algorithms significantly outperformed each other in terms of rejection rates across the five testing sets. The results of the Kruskal-Wallis tests yielded Holm-adjusted p-values of 0.178, 1, 0.178, 1, and 1 for the Menon, Lake, Liao, Wu, and Young datasets, respectively (Table S2). However, it is worth noting that SVC exhibited the lowest rejection rate across all five testing datasets, although this difference was not statistically significant compared to the other machine learning algorithms.
When considering the Young dataset as the testing dataset, the best model is one that accurately rejects cells in the “Neutrophil,” “Mast,” and “Urothelium” cell types as these cell types are not present in the training data. Fig. 2C demonstrates that the RF model had the highest rejection rate for cells in the “Urothelium” type, while the MLP model had the highest rejection rate for cells in the “Neutrophil” type and the KNN model had the highest rejection rate for cells in the “Mast” type when the Young dataset was used as the testing dataset.
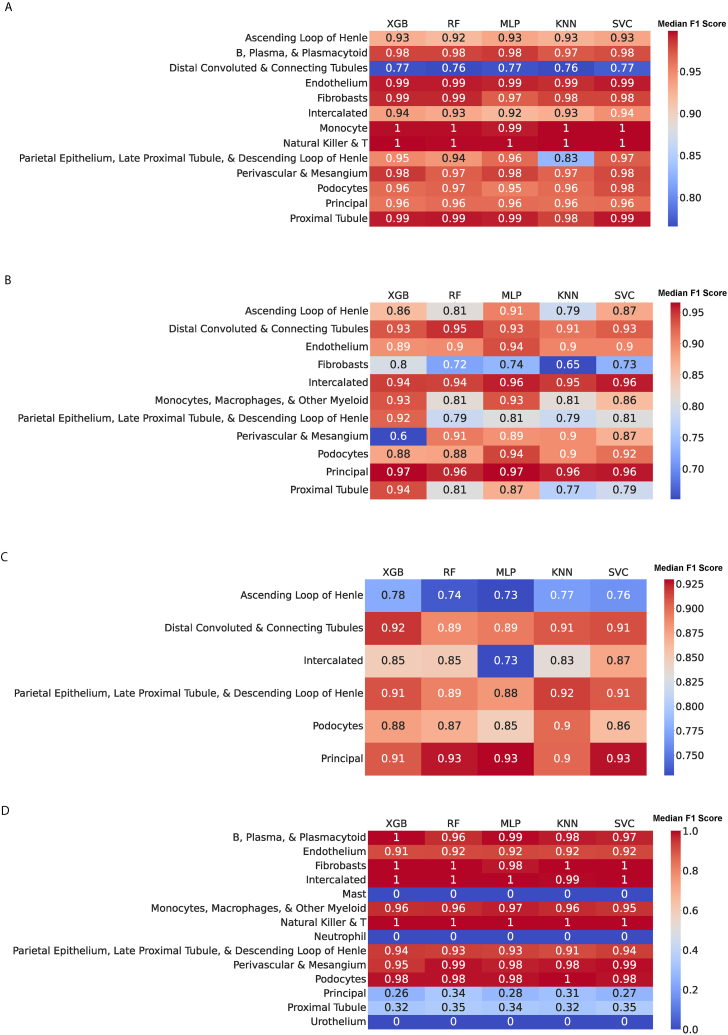
The performance of the machine learning algorithms also varied across different harmonized cell types. For instance, when Menon was used as the testing dataset, the “Distal Convoluted Tubule and Connecting Tubule” harmonized cell type exhibited lower F1 scores across the machine learning algorithms compared to harmonized cell types such as “Natural Killer & T,” “Monocytes, Macrophages, & Other Myeloid,” “Proximal Tubule,” or “Endothelium,” which had higher F1 scores across the algorithms (Fig. 3A). Specifically, the XGB model incorrectly labeled 208 out of 745 (27.9 %) cells belonging to the “Distal Convoluted and Connecting Tubule” harmonized cell type as belonging to the “Ascending Loop of Henle” harmonized cell type (Table S3). It is worth noting that the “Distal Convoluted and Connecting Tubule” and “Ascending Loop of Henle” harmonized cell types exhibited a high degree of correlation, as depicted in Fig. 1.
Fig. 3.
Heatmap of each classifier's F1 score on (a) Menon et al., (b) Lake et al., (c) Wu et al., and (d) Young et al. with respect to each harmonized cell type.
The lowest F1 scores across machine learning algorithms were observed when Lake and Wu were used as the testing datasets (Fig. 3B and. C). The harmonized cell types with the lowest F1 scores when tested on the Lake dataset were “Fibroblasts,” “Parietal Epithelium, Late Proximal Tubule, & Descending Loop of Henle,” and “Proximal Tubule” (Fig. 3B). Notably, in the Menon dataset, the F1 scores for the “Fibroblast” cell type exceeded 0.97 across machine learning algorithms (Fig. 3A), whereas in the Lake et al. dataset, the F1 scores for this cell type ranged from 0.65 to 0.8, indicating misclassification of cells of this type (Fig. 3B). For instance, the KNN model misclassified these cells as belonging to the “Perivascular & Mesangium” harmonized cell type in 18.3 % of cases and as “Endothelium” in 14.6 % of cases (Table S3). In the case of Wu, the “Ascending Loop of Henle” harmonized cell type had the lowest F1 scores across algorithms when the Wu dataset was used for testing (Fig. 3C). For example, 291 cells belonging to the “Parietal Epithelium, Late Proximal Tubule, & Descending Loop of Henle” harmonized cell type were misclassified by the MLP model as belonging to the “Ascending Loop of Henle” cell type, driving the low F1 scores for this cell type (Table S3).
When Young was used as the testing dataset, the average F1 scores for cells belonging to the “Principal” or “Proximal Tubule” harmonized cell types were below 0.35 (Fig. 3D). This can be attributed to the low precision of all the machine learning algorithms in predicting cells of the “Proximal Tubule” type and the low recall in predicting cells of the “Urothelium” type. The mislabeling of cells in “Urothelium” as cells in “Principal” instead of rejecting them and the mislabeling of “Endothelium” and “Ascending Loop of Henle” cells as “Proximal Tubule” cells were the main factors contributing to these low F1 scores. Detailed information regarding the predicted and actual labels for each cell type and classifier can be found in Table S3.
3. Discussion
In this study, we applied several machine learning algorithms including SVC, RF, MLP, KNN, and XGB to accurately classify kidney cell types using publicly available scRNA-seq and snRNA-seq datasets. Overall, the performance of the machine learning algorithms was satisfactory, with high median F1 scores and low rejection rates for most harmonized cell types across different testing datasets. This suggests that the machine learning algorithms successfully annotated the majority of cells and achieved a high level of concordance with the actual harmonized cell type annotations.
Our work is among the first to investigate how general-purpose ML models, such as XGB and SVC, perform in annotating kidney-specific cell types, which present unique challenges in the complex cellular composition and expression patterns of the kidney. Currently, there are several excellent cell-type annotation methods such as scCATCH, SCSA, SingleR, SingleCellNet, and ACTINN, as well as novel large language model applications in cell type annotations such as Cell2Sentence [[21], [22], [23], [24], [25], [26]]. Rather than propose a new algorithm for cell type annotation, our study aimed to fill a gap in knowledge by systematically evaluating the performance of established supervised ML methods in the context of kidney cell type annotation using both scRNA-seq and snRNA-seq datasets. While other comprehensive comparisons of cell type annotation methods have been published, such as the work by Ref. [4], which compared over 22 annotation tools across multiple tissues, they have not focused on kidney-specific data. Therefore, this study contributes to our understanding of which general ML methods are most suitable for kidney cell type classification and identifies areas where further improvement on these is needed.
By using inter-dataset evaluation, our study closely mimics real-world scenarios where models are trained on existing data and applied to an independent, novel study. With a sufficiently large number of cells across multiple datasets, our inter-dataset approach assessed model performance and allowed us to test model generalizability to new data across different sources. Additionally, our inter-dataset evaluation allowed cross-modality testing, where models trained on scRNA-seq data were tested on snRNA-seq data, and vice versa. Inter-dataset evaluation allowed us to explore how well models trained on one sequencing platform generalize to another, providing insights into the models' flexibility and adaptability.
No single machine learning algorithm consistently outperformed the others in our evaluation of F1 scores and rejection rates. Each had strengths and limitations. In general, XGB and SVC models performed well across most datasets, achieving high median F1 scores and relatively low rejection rates. For example, SVC models had some of the lowest rejection rates across all five datasets, suggesting that this algorithm was able to confidently label most cells. However, both XGB and SVC had relative difficulty with correctly rejecting urothelial cells, neutrophils, and mast cells when these cell types were not present in the training data, suggesting that these algorithms tended to mislabel cells of uncertain type rather than rejecting them. On the other hand, RF models had lower median F1 scores overall but the highest rejection rates for urothelial cells and some other cell types. This suggests that RF was more conservative in its labeling, better rejecting uncertain cell types rather than mislabeling them. This may be a strength for studies that prioritize precision over recall. MLP and KNN models achieved a balanced performance overall but encountered challenges with labeling cell types with low cell counts. Our results highlight the importance of carefully selecting and tuning models for specific datasets and tasks.
We observed that the overall performance of the machine learning algorithms varied in different scenarios. For example, the algorithms struggled to differentiate cell types with highly correlated transcription profiles, such as distal convoluted tubule and ascending Loop of Henle cells (Table S3). Additionally, cell types with smaller sample sizes, such as fibroblasts and principal cells, posed challenges for accurate classification. To improve the performance for cell types with limited cell numbers, we recommend investigating the correlation between clusters with regards to transcription profiles and merging highly correlated clusters into a single cluster. Notably, the performance for some small, harmonized cell type clusters, such as intercalated cells in Liao et al., showed higher accuracies, potentially due to lower correlation with other harmonized cell types.
Lower F1 scores were observed when training datasets consisted primarily of scRNA-seq data and testing datasets consisted exclusively of snRNA-seq data, as observed when Wu and Lake were used as testing datasets. This may stem from inherent differences between the two sequencing methods, as well as differences in protocols across studies. scRNA-seq tends to capture a higher proportion cytoplasmic mRNA leading to greater sensitivity for detecting highly expressed genes [[27], [28], [29]]. Previous studies have highlighted variations in cell type composition and dropout rates, including based on sample storage and processing, leading to differences in gene enrichment and subsequent cell type annotations [[27], [28], [29]]. For instance, snRNA-seq has been associated with reduced enrichment of leukocytes, including T cells, B cells, and natural killer cells, which are often indicative of underlying inflammatory states [27,28]. Notably, Wu et al. specified in their study that they were unable to detect stromal or leukocyte populations, possibly due to dissociation bias or cell frequency below the limit of detection [6]. This may contribute to reduced precision or recall for these cell types when models trained on scRNA-seq data are applied to snRNA-seq datasets. Another study comparing scRNA-seq and snRNA-seq in adult mouse kidney models reported an enrichment of specific kidney cell types, such as podocytes, mesangial cells, and endothelial cells in snRNA-seq data [30]. These differences contribute to the overall lower performance of machine learning models when tested on data obtained with a different method than the training dataset. Our results suggest that models trained primarily on a single sequencing technology may not fully capture the expression dynamics of other sequencing technologies, and therefore pursuing a multi-modal approach in future studies may improve generalizable performance.
In selecting the machine learning models for this study, we prioritized models based on several key criteria. First, we focused on widely used, open-source models from libraries such as scikit-learn [31] to ensure reproducibility and accessibility, allowing other researchers to easily expand and adapt our framework. We have also provided a detailed guide for incorporating additional datasets and ML algorithms in our GitHub repository. The models we chose—Random Forest (RF), Support Vector Machines (SVM), Multilayer Perceptrons (MLP), k-Nearest Neighbors (KN), and Extreme Gradient Boosting (XGB)—are general-purpose classifiers that are popular in bioinformatics and provide a robust baseline for kidney cell annotation. Our selection was informed by previous work comparing a wide array of ML algorithms, and this study fills a crucial gap in benchmarking the performance of these models in the context of kidney cell type annotation, laying the foundation for future work in this area [4].
Within the realm of biomarker ontologies, it is crucial to consider the diversity of the datasets analyzed in our study, which originated from distinct studies utilizing varying pipelines, ontologies, and manual annotations by experts. Despite these differences, our machine learning models, trained on standardized cell type labels, exhibited strong performance. This indicates that expert-derived annotations can be effectively harmonized across studies with several implications. First, harmonization of cell types across studies can allow for greater sample sizes in future transcriptomic analysis and allow for comparison between studies. Consequently, we believe that our approach of identifying and labeling matching cell types across studies will facilitate the adoption of standardized cell labels for identical cell populations in future research endeavors. This promotes consistency and comparability in the field of biomarker ontologies, enabling more comprehensive and cohesive analyses across diverse studies. Moreover, specifically in the field of kidney research, there are ongoing efforts to establish standardized ontologies. The Kidney Precision Medicine Project (KPMP) is actively developing the Kidney Tissue Atlas Ontology, aiming to create a unified system that incorporates clinical, pathological, imaging, and molecular data [32]. This ontology seeks to standardize labels for biomarkers, phenotypes, disease states, cell types, and anatomical structures in the kidney across both healthy and diseased conditions [32]. By utilizing scRNA-seq and snRNA-seq, KPMP aims to identify gene, metabolite, and protein biomarkers that differentiate cell types and contribute to disease pathways.
KPMP builds upon previous ontological projects in the kidney, such as the Genitourinary Development Molecular Anatomy Project and the Chronic Kidney Disease Ontology, which focused on specific disease states or cell types rather than encompassing all kidney cell types [32]. The collaboration between KPMP and the Human BioMolecular Atlas Program (HuBMAP) resulted in the publication of the Anatomical Structures, Cell Types, and Biomarkers (ASCT + B) tables in 2019 [32,33]. These tables aid in the annotation of anatomical structures, cell types, and biomarkers in the kidney. Furthermore, the HuBMAP initiative, which includes KPMP and other data consortia, is actively working on the Human Reference Atlas (HRA) which aims to develop biomarker ontologies for various organs in the human body [33]. Additionally, the Human Cell Atlas (HCA) initiative has introduced the Cell Annotation Platform (CAP), a data visualization tool intended to facilitate the visualization and integration of annotation data from multiple published studies [34]. Moreover, our work complements the exceptional work done by the Tabula Sapiens Consortium and HubMAP's Azimuth team as well as generative AI models in this space such as scGPT by utilizing general-purpose machine learning algorithms such as SVM, which were demonstrated by Abdelaal et al. to have better overall performance with faster computation time than scRNA-specific algorithms [4,[35], [36], [37]]. Our research aligns with these ongoing initiatives by providing valuable insights that can contribute to the less labor-intensive compilation of independent datasets, enhance interoperability, increase cell sample sizes, and strengthen the utilization of machine learning-derived cell type annotations using general-purpose machine learning models.
In this study, our focus was specifically on annotating healthy kidney cell types using scRNA-seq and snRNA-seq data. The decision to limit our analysis to healthy kidney cells was driven by the need to establish a robust baseline for cell type classification, free from the variability introduced by pathological states. By concentrating on healthy tissues, we aimed to assess the performance of machine learning algorithms in identifying the diverse and complex cell populations that constitute the normal kidney environment [33,38]. However, some recent studies investigating gene expression patterns in various kidney disease states have specifically focused on certain diseases, such as hypertensive or diabetic kidney disease, while others have utilized murine models to identify biomarkers and analyze cell type enrichment [[39], [40], [41]]. Lake et al. (2021) took a different approach by leveraging data from HuBMAP, KPMP, and HCA, including cells from both healthy and diseased kidneys, to characterize differential gene expression in disease states using spatial transcriptomics [42]. Their findings revealed associations between disease states, elevated cytokine production, and tubular regeneration and differentiation, as well as increased expression of inflammatory and fibrotic cell markers [42]. Machine learning models, including the general-purpose algorithms such as RF and XGB evaluated in this study, may be applied in future studies to identify key pathways involved in kidney disease. Despite not directly including patients with kidney disease, one potential application of our results in the study of kidney disease is to utilize the rejection rate of the models trained on healthy kidney cells. By identifying cells that are more likely to represent a disease state based on higher rejection rates, researchers can target those cells for further analysis of differential gene expression patterns. As databases of kidney disease continue to expand over time, similar approaches to the ones described in our study can be applied to enhance our understanding of kidney diseases.
Despite its many strengths, this study also has several limitations, including those previously acknowledged. First, we only included samples from the cortex and medulla and did not include samples from other regions of the kidney such as the renal pelvis or papilla. We focused on the regions that were most consistently available and well-represented across the publicly available datasets we used. Future studies may benefit from incorporating samples from other regions in the kidney. In addition, we included only 79 kidney biopsy samples in our study. The selection of these samples was intentional in that these were from five studies for which we could replicate the UMAP figures generated in the original publications, ensuring the integrity of the data. Moreover, the models were fed 62,120 individual cells rather than pseudobulk data. The impact of varying sample sizes on model performance was previously evaluated by Abdelaal et al., in 2019 [4]. The authors found that while using fewer than 500 cells leads to reduced accuracy, most models performed reliably when trained on 20 % of the cells or more.
While our study provides a foundational framework for the annotation of kidney cell types using machine learning, several key areas for future research could expand and enhance the utility of our findings. These include incorporating disease-specific datasets, performing cross-species analyses with animal models, and exploring sex-specific differences in kidney cell type classification. One important extension of this work involves applying machine learning models to disease-specific datasets. Identifying key genes and sequences that differentiate healthy and diseased cells could illuminate the pathways involved in the onset and progression of kidney diseases in a cell-type-specific way. Machine learning algorithms such as Random Forest (RF) and Extreme Gradient Boosting (XGB) can provide feature importance scores that highlight the genes most influential in classifying diseased states. By performing gene set enrichment analysis (GSEA) [43] and other pathway analysis methods on these key genes, researchers can connect differential gene expression in diseased cell types to specific biological pathways, such as fibrosis, inflammation, and cell death. Understanding the dysregulated molecular mechanisms driving kidney diseases could aid in the identification of novel therapeutic targets.
Another significant avenue for future exploration is the inclusion of animal sequences, particularly from murine models. Comparing human kidney cell types with those in mouse models can help evaluate how well machine learning algorithms trained on human data generalize to animal models. This cross-species analysis could reveal conserved and divergent cell types or molecular features, offering insights into kidney disease mechanisms that are relevant across species [44]. Moreover, analyzing how models trained on human data perform when applied to animal models could identify the strengths and limitations of these models in a translational research context. Additionally, exploring the impact of sex differences on kidney cell type annotation is another crucial direction for future research. By including datasets with a more balanced representation of male and female samples, future analyses can investigate how sex-specific biological pathways influence gene expression patterns, cellular composition, and disease susceptibility in the kidney. Incorporating sex as a variable in these analyses will help to understand how these differences affect the performance of machine learning algorithms in classifying kidney cell types. This understanding could lead to the development of more precise and personalized models for kidney health and disease, enabling researchers to identify pathways that are differentially regulated between male and female kidneys [[45], [46], [47]].
To ensure the reliability and consistency of our analysis, we did not perform any new cell type annotations or raw data processing for this study. Instead, we utilized pre-processed data as provided by the original authors of each dataset. Our approach was based on the principle that the original authors are experts in their respective studies and are best positioned to accurately annotate their datasets. Therefore, we focused on evaluating the performance of machine learning algorithms using the provided expert-curated annotations without introducing variability from re-annotation or re-processing steps. We successfully replicated the UMAP visualizations presented in the original publications of each study, ensuring that our analyses were consistent with the authors' original work.
To facilitate the expansion of our research by other scientists, we have made our entire pipeline available, including detailed documentation for adding new training datasets or implementing alternative machine learning algorithms. All code for our project can be found in our GitHub repository, and our data is accessible on Zenodo. By leveraging the power of machine learning algorithms and fostering collaborative efforts, we can accelerate the discovery of novel insights into kidney cell types and drive advancements in precision medicine for kidney diseases.
4. Methods
4.1. Availability of data and materials
The datasets supporting the conclusions of this article are available in Zenodo [48]. Cell and sample metadata are available on an alternate Zenodo [49]. Our results are reproducible with the code and Snakemake pipeline available in our GitHub repository (https://github.com/smadapoosi/IKCTML).
4.2. Data collection and quality control
We initially identified five studies of sc/snRNA-seq data on kidney cells from the GEO database. The selection criteria included studies with publicly available data for which we could generate UMAP visualizations that matched the original publications using the methods described in this section or the code provided on our GitHub repository. Subsequently, we filtered the data to include only normal, healthy cells with “well-annotated” cell types as described in the sections corresponding to each original study below. Our analysis pipeline was implemented using Snakemake, and a visual representation of the pipeline can be found in Fig. S8. The complete code for our analysis, including the pipeline, is available on our GitHub repository. Additionally, the data used in this study can be accessed on Zenodo, including metadata on individual cells and samples. To ensure data quality, we performed UMAP analyses, which are illustrated in Fig. S2, and compared these to the UMAPs presented in the original publications. Importantly, we utilized publicly available RDS objects or processed count matrices provided by the authors of the original studies as below and did not process raw FASTQ files.
4.2.1. Lake et al. [8]
The normalized data from Lake et al. was generously shared with us. Several of the cells in this dataset were also included in the data from Menon et al., and these duplicates were removed. Additionally, we excluded cells marked as “distressed” or “unassigned.”
4.2.2. Liao et al. [9]
The raw data from Liao et al. was downloaded from GSE131685 [9]. Our replication of the original dataset used an adaptation of the original analysis code available on Github [50]. Sample ‘kidney1’ was removed due to its uniformly high mitochondrial expression and the misalignment of cells from ‘kidney1’ with those of ‘kidney2’ and ‘kidney3,’ as visualized by UMAP (Fig. S9). The rest of the cells from this study were included.
4.2.3. Menon et al. [7]
The normalized data from this study was downloaded from GSE140989 [7]. The original annotations were recreated using the published description of their workflow from the methods section of their paper. All cells from this dataset were used.
4.2.4. Wu et al. [6]
The raw data from Wu et al. was downloaded from GSE114156. Our replication of the original dataset was based on the instructions provided in the supplementary files to the original publication. No cells were excluded from this analysis prior to the SVM quality control step. As the authors did not provide a file with marker genes to label clusters, cluster labeling was performed using the marker genes listed in Fig. 3 of the original manuscript.
4.2.5. Young et al. [5]
The raw data from this study was downloaded from the supplementary files of the study. Annotations were replicated with the provided metadata in their supplement and an adaptation of their original code, which is available on Github [51]. Samples derived from children and annotated as tumor samples were excluded using the cell manifest prior to reading the data. We then removed all cells annotated as ‘junk’, ‘private,’ or ‘nephron epithelium.’ This step resulted in the loss of several cells that clustered with proximal tubular cells, resulting in lower representation of this cell type from this particular dataset.
4.3. Batch correction
Batch Correction was performed using Seurat v4 rPCA integration [52]. The resulting integrated assay was then scaled, reduced in dimensionality, clustered, and visualized with the standard Seurat functions [53].
4.4. Harmonized cell type labeling
The cell-type annotations from the original datasets were classified into 16 different harmonized cell type classes, which were determined by the pattern of their PCA-coordinate Pearson correlations, implemented with Scanpy [54]. These categories were named based on the original, expert annotations present in each original dataset.
4.5. SVM outlier detection
We removed outlier cells from each harmonized cell type by training and testing an SVM model on the integrated dataset. Cells that were classified with low probability (<0.6) were removed. SVM was chosen for this task due to its previously shown high performance in outlier detection [4,[16], [17], [18], [19], [20]].
4.6. Supervised learning
We evaluated five different popular machine learning algorithms including a support vector classifier (SVC), a random forest classifier (RF), a multi-layer perceptron (MLP), a k-nearest neighbors classifier, and XGBoost (XGB), each implemented in the scikit-learn python library [31]. We trained the machine learning algorithms on four datasets and tested on the fifth dataset. We performed this process 5 times with unique single different testing datasets in each run. The performance of machine learning algorithms were evaluated using F1 scores and rejection rates. The overall F1 scores and rejection rates for each machine learning algorithm were calculated as the median of all individual harmonized cell types.
Ethics approval and consent to participate
Not applicable.
Funding
The KPMP is funded by the following grants from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK): U01DK133081, U01DK133091, U01DK133092, U01DK133093, U01DK133095, U01DK133097, U01DK114866, U01DK114908, U01DK133090, U01DK133113, U01DK133766, U01DK133768, U01DK114907, U01DK114920, U01DK114923, U01DK114933, U24DK114886, UH3DK114926, UH3DK114861, UH3DK114915, UH3DK11493.
CRediT authorship contribution statement
Adam Tisch: Writing – review & editing, Writing – original draft, Visualization, Software, Methodology, Formal analysis. Siddharth Madapoosi: Writing – review & editing, Validation, Software, Resources, Conceptualization. Stephen Blough: Writing – original draft, Software, Resources, Methodology. Jan Rosa: Validation, Software, Methodology. Sean Eddy: Methodology. Laura Mariani: Investigation, Conceptualization. Abhijit Naik: Software, Methodology. Christine Limonte: Writing – review & editing. Philip McCown: Writing – review & editing, Data curation. Rajasree Menon: Writing – review & editing, Data curation. Sylvia E. Rosas: Writing – review & editing. Chirag R. Parikh: Writing – review & editing. Matthias Kretzler: Writing – review & editing, Funding acquisition. Ahmed Mahfouz: Writing – review & editing, Writing – original draft, Visualization, Methodology. Fadhl Alakwaa: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Resources, Project administration, Methodology, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors would like to thank NIH-DDK and HCA for funding the studies utilized in this project, the authors of Menon et al. (2020), Lake et al. (2019), Liao et al. (2020), Wu et al. (2019), and Young et al. (2018) for generously sharing their data and code, and the kidney sample donors for their contribution to science.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e38567.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Ju W., Greene C.S., Eichinger F., Nair V., Hodgin J.B., Bitzer M., et al. Defining cell-type specificity at the transcriptional level in human disease. Genome Res. 2013;23(11):1862–1873. doi: 10.1101/gr.155697.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shen-Orr S.S., Tibshirani R., Khatri P., Bodian D.L., Staedtler F., Perry N.M., et al. Cell type-specific gene expression differences in complex tissues. Nat. Methods. 2010;7(4):287–289. doi: 10.1038/nmeth.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gawel D.R., Serra-Musach J., Lilja S., Aagesen J., Arenas A., Asking B., et al. Correction to: a validated single-cell-based strategy to identify diagnostic and therapeutic targets in complex diseases. Genome Med. 2020;12(1):37. doi: 10.1186/s13073-020-00732-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abdelaal T., Michielsen L., Cats D., Hoogduin D., Mei H., Reinders M.J.T., et al. A comparison of automatic cell identification methods for single-cell RNA sequencing data. Genome Biol. 2019;20(1):194. doi: 10.1186/s13059-019-1795-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Young M.D., Mitchell T.J., Vieira Braga F.A., Tran M.G.B., Stewart B.J., Ferdinand J.R., et al. Single-cell transcriptomes from human kidneys reveal the cellular identity of renal tumors. Science. 2018;361(6402):594–599. doi: 10.1126/science.aat1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu H., Malone A.F., Donnelly E.L., Kirita Y., Uchimura K., Ramakrishnan S.M., et al. Single-cell transcriptomics of a human kidney allograft biopsy specimen defines a diverse inflammatory response. J. Am. Soc. Nephrol. 2018;29(8):2069–2080. doi: 10.1681/ASN.2018020125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Menon R., Otto E.A., Hoover P., Eddy S., Mariani L., Godfrey B., et al. Single cell transcriptomics identifies focal segmental glomerulosclerosis remission endothelial biomarker. JCI Insight. 2020;5(6) doi: 10.1172/jci.insight.133267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lake B.B., Chen S., Hoshi M., Plongthongkum N., Salamon D., Knoten A., et al. A single-nucleus RNA-sequencing pipeline to decipher the molecular anatomy and pathophysiology of human kidneys. Nat. Commun. 2019;10(1):2832. doi: 10.1038/s41467-019-10861-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liao J., Yu Z., Chen Y., Bao M., Zou C., Zhang H., et al. Single-cell RNA sequencing of human kidney. Sci. Data. 2020;7(1):4. doi: 10.1038/s41597-019-0351-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kameneva P., Artemov A.V., Kastriti M.E., Faure L., Olsen T.K., Otte J., et al. Single-cell transcriptomics of human embryos identifies multiple sympathoblast lineages with potential implications for neuroblastoma origin. Nat. Genet. 2021;53(5):694–706. doi: 10.1038/s41588-021-00818-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galdos F.X., Xu S., Goodyer W.R., Duan L., Huang Y.V., Lee S., et al. devCellPy is a machine learning-enabled pipeline for automated annotation of complex multilayered single-cell transcriptomic data. Nat. Commun. 2022;13(1):5271. doi: 10.1038/s41467-022-33045-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Le H., Peng B., Uy J., Carrillo D., Zhang Y., Aevermann B.D., et al. Machine learning for cell type classification from single nucleus RNA sequencing data. PLoS One. 2022;17(9) doi: 10.1371/journal.pone.0275070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ziegenhain C., Vieth B., Parekh S., Reinius B., Guillaumet-Adkins A., Smets M., et al. Comparative analysis of single-cell RNA sequencing methods. Mol. Cell. 2017;65(4):631. doi: 10.1016/j.molcel.2017.01.023. 43.e4. [DOI] [PubMed] [Google Scholar]
- 14.Klein A.M., Mazutis L., Akartuna I., Tallapragada N., Veres A., Li V., et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015;161(5):1187–1201. doi: 10.1016/j.cell.2015.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lake B.B., Chen S., Sos B.C., Fan J., Kaeser G.E., Yung Y.C., et al. Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain. Nat. Biotechnol. 2018;36(1):70–80. doi: 10.1038/nbt.4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao P., Xu Z., Chen J., Ren Y., King I. Single cell self-paced clustering with transcriptome sequencing data. Int. J. Mol. Sci. 2022;23(7):3900. doi: 10.3390/ijms23073900. Published 2022 Mar 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu X., Wolfgruber T.K., Tasato A., Arisdakessian C., Garmire D.G., Garmire L.X. Granatum: a graphical single-cell RNA-Seq analysis pipeline for genomics scientists. Genome Med. 2017;9(1):108. doi: 10.1186/s13073-017-0492-3. Published 2017 Dec 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li W.V., Li J.J. An accurate and robust imputation method scImpute for single-cell RNA-seq data. Nat. Commun. 2018;9(1):997. doi: 10.1038/s41467-018-03405-7. Published 2018 Mar 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Furey T.S., Cristianini N., Duffy N., Bednarski D.W., Schummer M., Haussler D. Support vector machine classification and validation of cancer tissue samples using microarray expression data. Bioinformatics. 2000;16(10):906–914. doi: 10.1093/bioinformatics/16.10.906. [DOI] [PubMed] [Google Scholar]
- 20.Kim Bong-Hyun, Yu Kijin, Lee Peter C.W. Cancer classification of single-cell gene expression data by neural network. Bioinformatics. March 2020;36(5):1360–1366. doi: 10.1093/bioinformatics/btz772. [DOI] [PubMed] [Google Scholar]
- 21.Shao X., Liao J., Lu X., Xue R., Ai N., Fan X. scCATCH: automatic annotation on cell types of clusters from single-cell RNA sequencing data. iScience. 2020;23(3) doi: 10.1016/j.isci.2020.100882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cao Y., Wang X., Peng G. SCSA: a cell type annotation tool for single-cell RNA-seq data. Front. Genet. 2020;11:490. doi: 10.3389/fgene.2020.00490. Published 2020 May 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan Y., Cahan P. SingleCellNet: a computational tool to classify single cell RNA-seq data across platforms and across species. Cell Syst. 2019;9(2):207–213.e2. doi: 10.1016/j.cels.2019.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aran D., Looney A.P., Liu L., et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 2019;20(2):163–172. doi: 10.1038/s41590-018-0276-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ma F., Pellegrini M. ACTINN: automated identification of cell types in single cell RNA sequencing. Bioinformatics. 2020;36(2):533–538. doi: 10.1093/bioinformatics/btz592. [DOI] [PubMed] [Google Scholar]
- 26.Levine D., Rizvi S.A., Levy S., et al. Cell2Sentence: teaching large language models the language of biology. bioRxiv. 2024 doi: 10.1101/2023.09.11.557287. [Preprint] [DOI] [Google Scholar]
- 27.Denisenko E., Guo B.B., Jones M., Hou R., de Kock L., Lassmann T., et al. Systematic assessment of tissue dissociation and storage biases in single-cell and single-nucleus RNA-seq workflows. Genome Biol. 2020;21(1):130. doi: 10.1186/s13059-020-02048-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deleersnijder D., Callemeyn J., Arijs I., Naesens M., Van Craenenbroeck A.H., Lambrechts D., et al. Current methodological challenges of single-cell and single-nucleus RNA-sequencing in glomerular diseases. J. Am. Soc. Nephrol. 2021;32(8):1838–1852. doi: 10.1681/ASN.2021020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Habib N., Avraham-Davidi I., Basu A., Burks T., Shekhar K., Hofree M., et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat. Methods. 2017;14(10):955–958. doi: 10.1038/nmeth.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu H., Kirita Y., Donnelly E.L., Humphreys B.D. Advantages of single-nucleus over single-cell RNA sequencing of adult kidney: rare cell types and novel cell states revealed in fibrosis. J. Am. Soc. Nephrol. 2019;30(1):23–32. doi: 10.1681/ASN.2018090912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pedregosa F., Varoquax G., Gramfort A., Michel V., Thirion B., Grisel O., et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 32.Ong E., Wang L.L., Schaub J., O'Toole J.F., Steck B., Rosenberg A.Z., et al. Modeling kidney disease using ontology: insights from the Kidney Precision Medicine Project. Nat. Rev. Nephrol. 2020;16(11):686–696. doi: 10.1038/s41581-020-00335-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Börner K., Teichmann S.A., Quardokus E.M., Gee J.C., Browne K., Osumi-Sutherland D., et al. Anatomical structures, cell types and biomarkers of the Human Reference Atlas. Nat. Cell Biol. 2021;23(11):1117–1128. doi: 10.1038/s41556-021-00788-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hao Y., Hao S., Andersen-Nissen E., et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184(13):3573–3587.e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sapiens Consortium Tabula, Jones R.C., Karkanias J., et al. The Tabula Sapiens: a multiple-organ, single-cell transcriptomic atlas of humans. Science. 2022;376(6594) doi: 10.1126/science.abl4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Osumi-Sutherland D., Xu C., Keays M., Levine A.P., Kharchenko P.V., Regev A., et al. Cell type ontologies of the human cell atlas. Nat. Cell Biol. 2021;23(11):1129–1135. doi: 10.1038/s41556-021-00787-7. [DOI] [PubMed] [Google Scholar]
- 37.Cui H, Wang C, Maan H, Wang B. scGPT: towards building a foundation model for single-cell multi-omics using generative AI. bioRxiv. doi:10.1101/2023.04.30.538439.Preprint. [DOI] [PubMed]
- 38.Hansen J., Sealfon R., Menon R., et al. A reference tissue atlas for the human kidney. Sci. Adv. 2022;8(23) doi: 10.1126/sciadv.abn4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Obradovic A., Chowdhury N., Haake S.M., Ager C., Wang V., Vlahos L., et al. Single-cell protein activity analysis identifies recurrence-associated renal tumor macrophages. Cell. 2021;184(11):2988–3005.e16. doi: 10.1016/j.cell.2021.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Conway B.R., O'Sullivan E.D., Cairns C., O'Sullivan J., Simpson D.J., Salzano A., et al. Kidney single-cell atlas reveals myeloid heterogeneity in progression and regression of kidney disease. J. Am. Soc. Nephrol. 2020;31(12):2833–2854. doi: 10.1681/ASN.2020060806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fu J., Akat K.M., Sun Z., Zhang W., Schlondorff D., Liu Z., et al. Single-cell RNA profiling of glomerular cells shows dynamic changes in experimental diabetic kidney disease. J. Am. Soc. Nephrol. 2019;30(4):533–545. doi: 10.1681/ASN.2018090896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lake B.B., Menon R., Winfree S., et al. An atlas of healthy and injured cell states and niches in the human kidney. Nature. 2023;619(7970):585–594. doi: 10.1038/s41586-023-05769-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Subramanian A., Tamayo P., Mootha V.K., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miao J., Zhu H., Wang J., Chen J., Han F., Lin W. Experimental models for preclinical research in kidney disease. Zool. Res. 2024;45(5):1161–1174. doi: 10.24272/j.issn.2095-8137.2024.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brannon A.R., Haake S.M., Hacker K.E., et al. Meta-analysis of clear cell renal cell carcinoma gene expression defines a variant subgroup and identifies gender influences on tumor biology. Eur. Urol. 2012;61(2):258–268. doi: 10.1016/j.eururo.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu S., Wu J., Yang D., Xu J., Shi H., Xue B., Ding Z. Big data analytics for MerTK genomics reveals its double-edged sword functions in human diseases. Redox Biol. 2024 Apr;70 doi: 10.1016/j.redox.2024.103061. Epub 2024 Feb 5. PMID: 38341954; PMCID: PMC10869259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sultanova R., Schibalski R., Yankelevich I., Stadler K., Ilatovskaya D. Sex differences in renal mitochondrial function: a hormone-gous opportunity for research. Am. J. Physiol. Ren. Physiol. 2020;319(6):F1117–F1124. doi: 10.1152/ajprenal.00320.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Madapoosi Siddharth. Automatic identification of kidney cell types in scRNA-seq and snRNA-seq data using machine learning algorithms - datasets. Zenodo. 2023 doi: 10.5281/zenodo.8303415. [Data set] [DOI] [Google Scholar]
- 49.Madapoosi Siddharth. Identification of kidney cell types in scRNA-seq and snRNA-seq data using machine learning algorithms. Zenodo. 2024 doi: 10.5281/zenodo.11267675. [Data set] [DOI] [Google Scholar]
- 50.Yu Z. Lessonskit. 2019 https://github.com/lessonskit/Single-cell-RNA-sequencing-of-human-kidney [Available from: [Google Scholar]
- 51.Young M.D. constantAmateur. 2018 https://github.com/constantAmateur/scKidneyTumors [Available from: [Google Scholar]
- 52.Hao Y., Hao S., Andersen-Nissen E., Mauck W.M., Zheng S., Butler A., et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184(13):3573. doi: 10.1016/j.cell.2021.04.048. 87.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Satija R., Farrell J.A., Gennert D., Schier A.F., Regev A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015;33(5):495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wolf F.A., Angerer P., Theis F.J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19(1):15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting the conclusions of this article are available in Zenodo [48]. Cell and sample metadata are available on an alternate Zenodo [49]. Our results are reproducible with the code and Snakemake pipeline available in our GitHub repository (https://github.com/smadapoosi/IKCTML).