Abstract
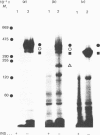
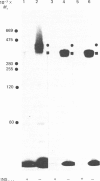
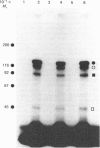
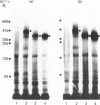
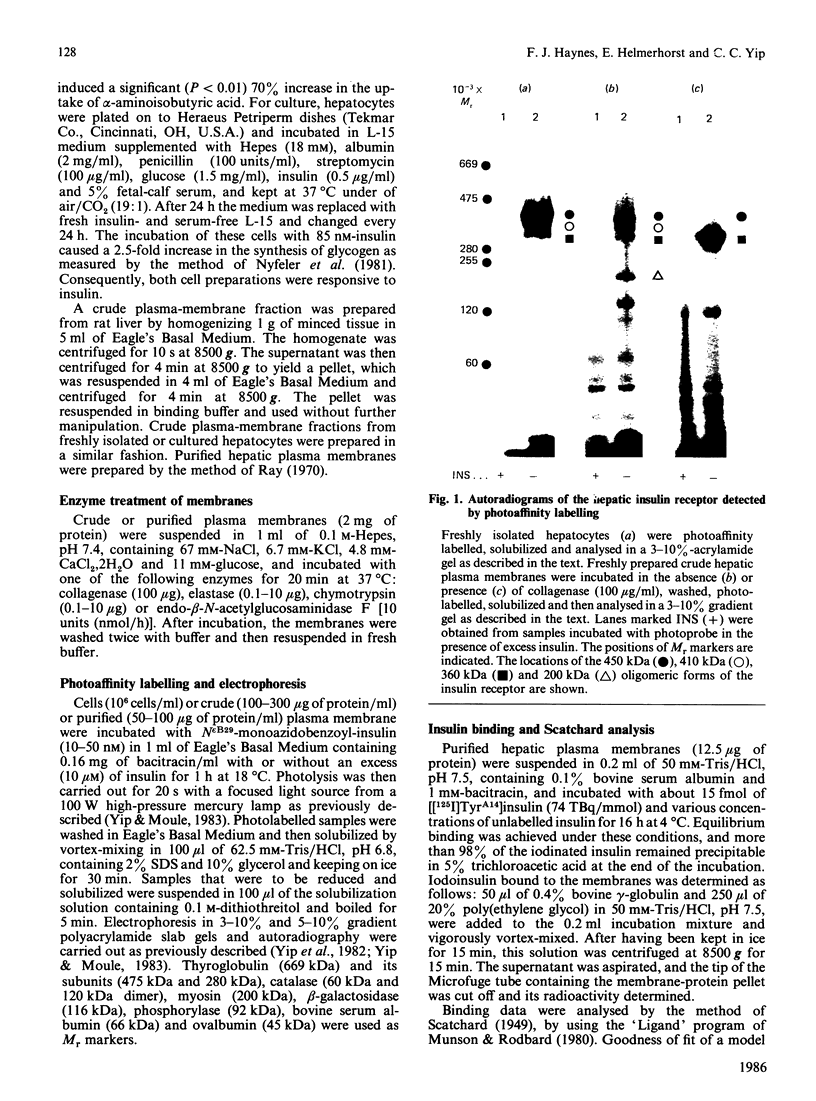
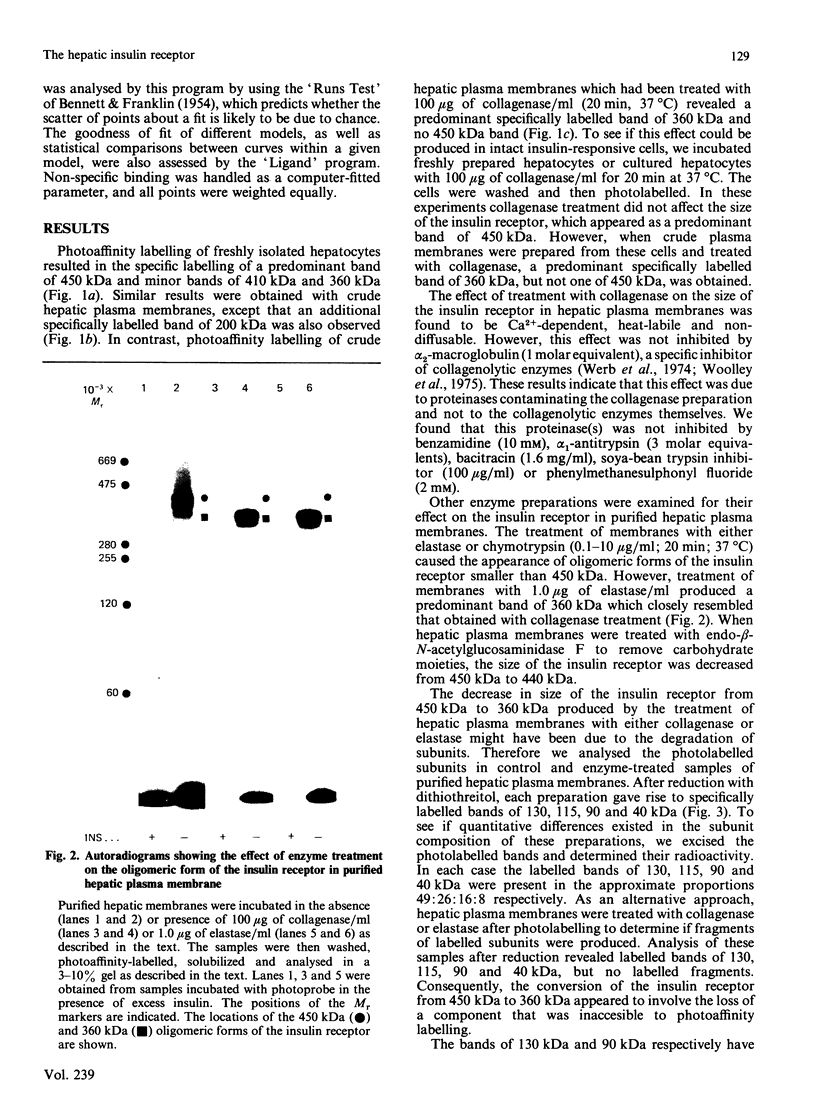
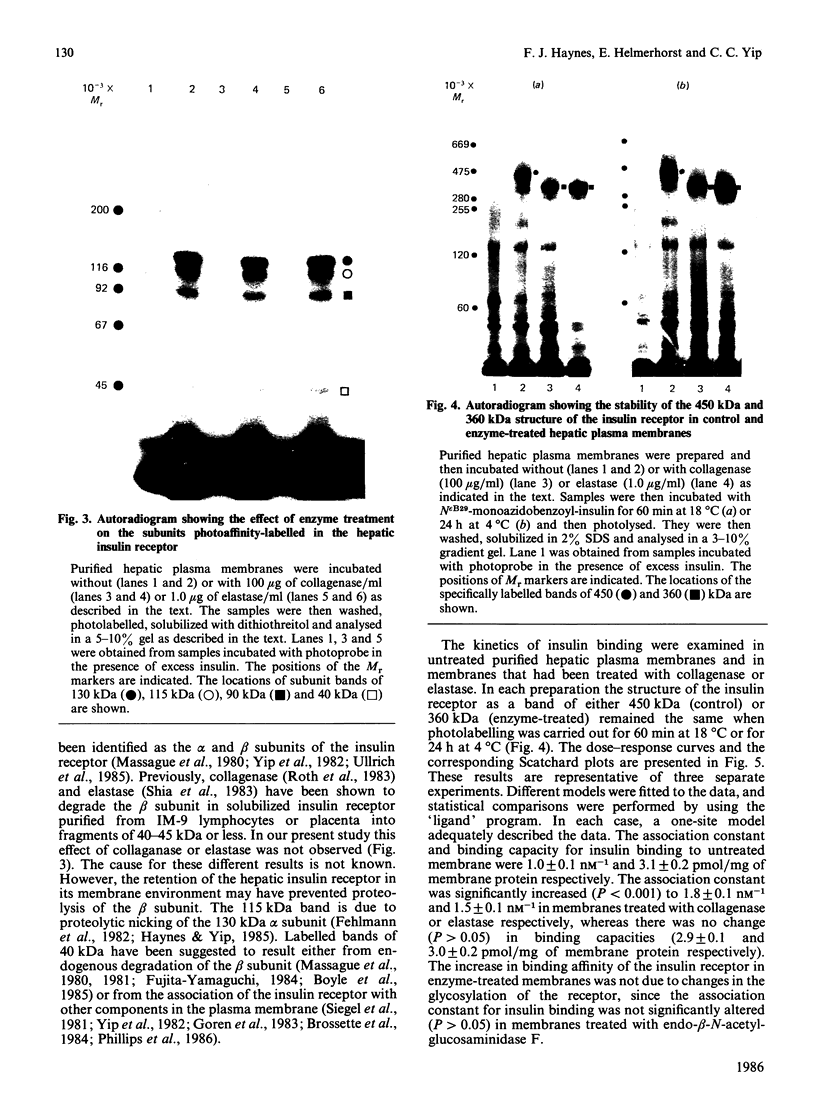
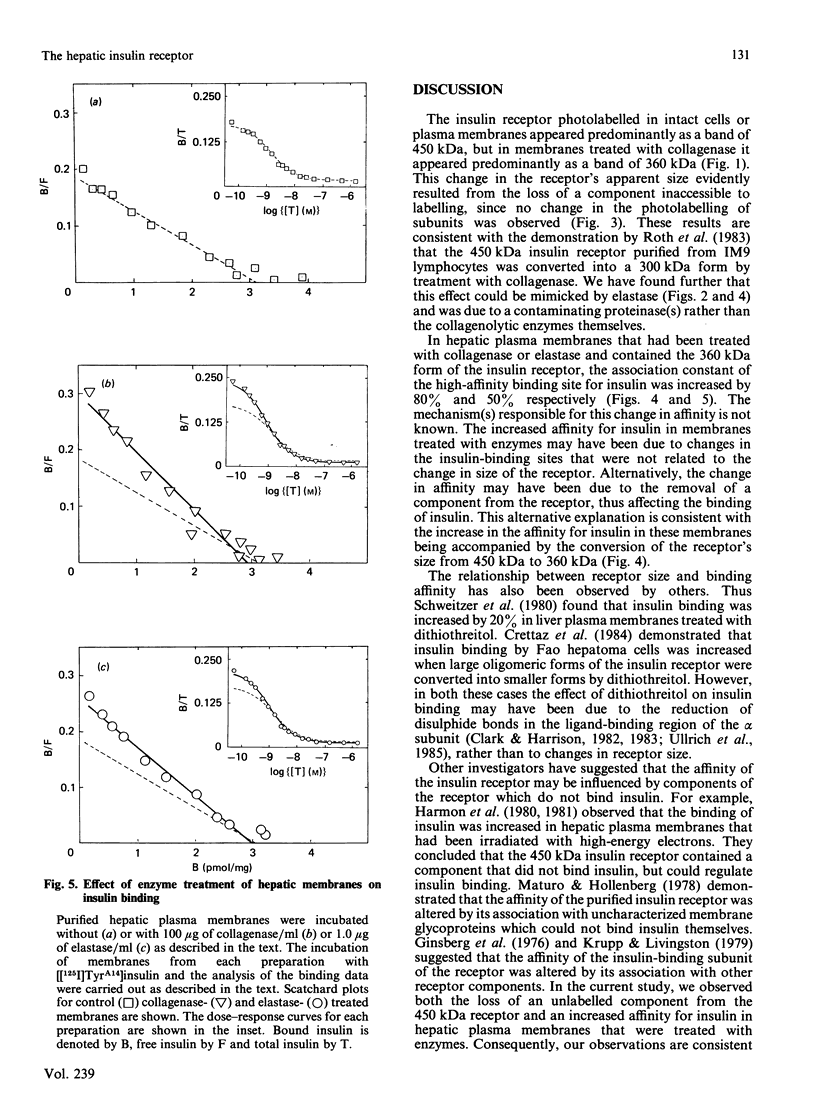
Hepatocytes or hepatic plasma membranes were photoaffinity-labelled with radioiodinated N epsilon B29-monoazidobenzoyl-insulin. Analysis of the samples by SDS/polyacrylamide-gel electrophoresis and autoradiography revealed the insulin receptor as a predominant band of 450 kDa. When hepatic plasma membranes were first treated with clostridial collagenase and then photolabelled, the insulin receptor appeared as a predominant band of 360 kDa. This effect of collagenase treatment on the insulin receptor was due to Ca2+-dependent heat-labile proteinases contaminating the preparation of collagenase, and it could be mimicked by elastase. The decrease in size of the insulin receptor to 360 kDa resulted from the loss of a receptor component that was inaccessible to photolabelling. In contrast, the size of the insulin receptor of intact cells was not affected by collagenase treatment. This suggests that the site sensitive to proteolysis was located on the cytoplasmic side of the plasma membrane. In hepatic plasma membranes that were treated with collagenase or elastase, and contained the 360 kDa form of the insulin receptor, the binding affinity for insulin was increased by up to 2-fold. These findings support the concept that a component which is either a part of, or closely associated with, the insulin receptor may regulate its affinity for insulin.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baron M. D., Sönksen P. H. Elucidation of the quaternary structure of the insulin receptor. Biochem J. 1983 Apr 15;212(1):79–84. doi: 10.1042/bj2120079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berhanu P., Olefsky J. M., Tsai P., Thamm P., Saunders D., Brandenburg D. Internalization and molecular processing of insulin receptors in isolated rat adipocytes. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4069–4073. doi: 10.1073/pnas.79.13.4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry M. N., Friend D. S. High-yield preparation of isolated rat liver parenchymal cells: a biochemical and fine structural study. J Cell Biol. 1969 Dec;43(3):506–520. doi: 10.1083/jcb.43.3.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle T. R., Campana J., Sweet L. J., Pessin J. E. Subunit structure of the purified human placental insulin receptor. Intramolecular subunit dissociation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. J Biol Chem. 1985 Jul 15;260(14):8593–8600. [PubMed] [Google Scholar]
- Brossette N., Van Obberghen E., Fehlmann M. Interaction between insulin receptors and major histocompatibility complex antigens in mouse liver membranes. Diabetologia. 1984 Jul;27 (Suppl):74–76. doi: 10.1007/BF00275651. [DOI] [PubMed] [Google Scholar]
- Clark S., Harrison L. C. Disulfide exchange between insulin and its receptor. A possible post-binding step in insulin action. J Biol Chem. 1983 Oct 10;258(19):11434–11437. [PubMed] [Google Scholar]
- Clark S., Harrison L. C. Insulin binding leads to the formation of covalent (-S-S-) hormone receptor complexes. J Biol Chem. 1982 Oct 25;257(20):12239–12244. [PubMed] [Google Scholar]
- Crettaz M., Jialal I., Kasuga M., Kahn C. R. Insulin receptor regulation and desensitization in rat hepatoma cells. The loss of the oligomeric forms of the receptor correlates with the change in receptor affinity. J Biol Chem. 1984 Sep 25;259(18):11543–11549. [PubMed] [Google Scholar]
- Deutsch P. J., Wan C. F., Rosen O. M., Rubin C. S. Latent insulin receptors and possible receptor precursors in 3T3-L1 adipocytes. Proc Natl Acad Sci U S A. 1983 Jan;80(1):133–136. doi: 10.1073/pnas.80.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo F., Elsas L. J., 2nd Structural analysis and subunit interaction of insulin receptor from membranes of cultured embryonic chick heart cells. Endocrinology. 1984 Nov;115(5):1828–1837. doi: 10.1210/endo-115-5-1828. [DOI] [PubMed] [Google Scholar]
- Fehlmann M., Le Cam A., Freychet P. Insulin and glucagon stimulation of amino acid transport in isolated rat hepatocytes. Synthesis of a high affinity component of transport. J Biol Chem. 1979 Oct 25;254(20):10431–10437. [PubMed] [Google Scholar]
- Fehlmann M., Le Marchand-Brustel Y., Van Obberghen E., Brandenburg D., Freychet P. Photoaffinity labelling of the insulin receptor in intact rat hepatocytes, mouse soleus muscle, and cultured human lymphocytes. Diabetologia. 1982 Nov;23(5):440–444. doi: 10.1007/BF00260959. [DOI] [PubMed] [Google Scholar]
- Fujita-Yamaguchi Y. Characterization of purified insulin receptor subunits. J Biol Chem. 1984 Jan 25;259(2):1206–1211. [PubMed] [Google Scholar]
- Fujita-Yamaguchi Y., Kathuria S. The monomeric alpha beta form of the insulin receptor exhibits much higher insulin-dependent tyrosine-specific protein kinase activity than the intact alpha 2 beta 2 form of the receptor. Proc Natl Acad Sci U S A. 1985 Sep;82(18):6095–6099. doi: 10.1073/pnas.82.18.6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg B. H., Kahn C. R., Roth J., De Meyts P. Insulin-induced dissociation of its receptor into subunits: possible molecular concomitant of negative cooperativity. Biochem Biophys Res Commun. 1976 Dec 20;73(4):1068–1074. doi: 10.1016/0006-291x(76)90232-1. [DOI] [PubMed] [Google Scholar]
- Goren H. J., Elliott C., Dudley R. A. Adipocyte insulin-binding species: the size and subunit composition of the larger binding species. J Cell Biochem. 1983;21(2):161–177. doi: 10.1002/jcb.240210207. [DOI] [PubMed] [Google Scholar]
- Harmon J. T., Kahn C. R., Kempner E. S., Schlegel W. Characterization of the insulin receptor in its membrane environment by radiation inactivation. J Biol Chem. 1980 Apr 25;255(8):3412–3419. [PubMed] [Google Scholar]
- Harmon J. T., Kempner E. S., Kahn C. R. Demonstration by radiation inactivation that insulin alters the structure of the insulin receptor in rat liver membranes. J Biol Chem. 1981 Aug 10;256(15):7719–7722. [PubMed] [Google Scholar]
- Haynes F. J., Yip C. C. Photoaffinity labelling of hepatic plasma membranes suggests two classes of hepatic insulin receptor. Diabetologia. 1985 Oct;28(10):786–792. doi: 10.1007/BF00265029. [DOI] [PubMed] [Google Scholar]
- Heidenreich K. A., Berhanu P., Brandenburg D., Olefsky J. M. Degradation of insulin receptors in rat adipocytes. Diabetes. 1983 Nov;32(11):1001–1009. doi: 10.2337/diab.32.11.1001. [DOI] [PubMed] [Google Scholar]
- Heidenreich K. A., Zahniser N. R., Berhanu P., Brandenburg D., Olefsky J. M. Structural differences between insulin receptors in the brain and peripheral target tissues. J Biol Chem. 1983 Jul 25;258(14):8527–8530. [PubMed] [Google Scholar]
- Helmerhorst E., Ng D. S., Moule M. L., Yip C. C. High molecular weight forms of the insulin receptor. Biochemistry. 1986 Apr 22;25(8):2060–2065. doi: 10.1021/bi00356a034. [DOI] [PubMed] [Google Scholar]
- Hofmann C., Ji T. H., Miller B., Steiner D. F. Photoaffinity labeling of the insulin receptor in H4 hepatoma cells: lack of cellular receptor processing. J Supramol Struct Cell Biochem. 1981;15(1):1–13. doi: 10.1002/jsscb.1981.380150102. [DOI] [PubMed] [Google Scholar]
- Hollenberg M. D. Hormone receptor interactions at the cell membrane. Pharmacol Rev. 1978 Dec;30(4):393–410. [PubMed] [Google Scholar]
- Jacobs S., Hazum E., Shechter Y., Cuatrecasas P. Insulin receptor: covalent labeling and identification of subunits. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4918–4921. doi: 10.1073/pnas.76.10.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga M., Karlsson F. A., Kahn C. R. Insulin stimulates the phosphorylation of the 95,000-dalton subunit of its own receptor. Science. 1982 Jan 8;215(4529):185–187. doi: 10.1126/science.7031900. [DOI] [PubMed] [Google Scholar]
- Krupp M. N., Livingston J. N. Effects of insulin on insulin-binding components extracted from rat fat cell membranes. Nature. 1979 Mar 1;278(5699):61–62. doi: 10.1038/278061a0. [DOI] [PubMed] [Google Scholar]
- Massague J., Pilch P. F., Czech M. P. A unique proteolytic cleavage site on the beta subunit of the insulin receptor. J Biol Chem. 1981 Apr 10;256(7):3182–3190. [PubMed] [Google Scholar]
- Massague J., Pilch P. F., Czech M. P. Electrophoretic resolution of three major insulin receptor structures with unique subunit stoichiometries. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7137–7141. doi: 10.1073/pnas.77.12.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J., Czech M. P. Role of disulfides in the subunit structure of the insulin receptor. Reduction of class I disulfides does not impair transmembrane signalling. J Biol Chem. 1982 Jun 25;257(12):6729–6738. [PubMed] [Google Scholar]
- Maturo J. M., 3rd, Hollenberg M. D. Insulin receptor: interaction with nonreceptor glycoprotein from liver cell membranes. Proc Natl Acad Sci U S A. 1978 Jul;75(7):3070–3074. doi: 10.1073/pnas.75.7.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munson P. J., Rodbard D. Ligand: a versatile computerized approach for characterization of ligand-binding systems. Anal Biochem. 1980 Sep 1;107(1):220–239. doi: 10.1016/0003-2697(80)90515-1. [DOI] [PubMed] [Google Scholar]
- Nyfeler F., Fasel P., Walter P. Short-term stimulation of net glycogen production by insulin in rat hepatocytes. Biochim Biophys Acta. 1981 Jun 11;675(1):17–23. doi: 10.1016/0304-4165(81)90064-7. [DOI] [PubMed] [Google Scholar]
- Phillips M. L., Moule M. L., Delovitch T. L., Yip C. C. Class I histocompatibility antigens and insulin receptors: evidence for interactions. Proc Natl Acad Sci U S A. 1986 May;83(10):3474–3478. doi: 10.1073/pnas.83.10.3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray T. K. A modified method for the isolation of the plasma membrane from rat liver. Biochim Biophys Acta. 1970 Jan 6;196(1):1–9. doi: 10.1016/0005-2736(70)90159-8. [DOI] [PubMed] [Google Scholar]
- Ronnett G. V., Knutson V. P., Kohanski R. A., Simpson T. L., Lane M. D. Role of glycosylation in the processing of newly translated insulin proreceptor in 3T3-L1 adipocytes. J Biol Chem. 1984 Apr 10;259(7):4566–4575. [PubMed] [Google Scholar]
- Rosenzweig S. A., Madison L. D., Jamieson J. D. Analysis of cholecystokinin-binding proteins using endo-beta-N-acetylglucosaminidase F. J Cell Biol. 1984 Sep;99(3):1110–1116. doi: 10.1083/jcb.99.3.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth R. A., Mesirow M. L., Cassell D. J. Preferential degradation of the beta subunit of purified insulin receptor. Effect on insulin binding and protein kinase activities of the receptor. J Biol Chem. 1983 Dec 10;258(23):14456–14460. [PubMed] [Google Scholar]
- Schweitzer J. B., Smith R. M., Jarett L. Differences in organizational structure of insulin receptor on rat adipocyte and liver plasma membranes: role of disulfide bonds. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4692–4696. doi: 10.1073/pnas.77.8.4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shia M. A., Rubin J. B., Pilch P. F. The insulin receptor protein kinase. Physicochemical requirements for activity. J Biol Chem. 1983 Dec 10;258(23):14450–14455. [PubMed] [Google Scholar]
- Siegel T. W., Ganguly S., Jacobs S., Rosen O. M., Rubin C. S. Purification and properties of the human placental insulin receptor. J Biol Chem. 1981 Sep 10;256(17):9266–9273. [PubMed] [Google Scholar]
- Ullrich A., Bell J. R., Chen E. Y., Herrera R., Petruzzelli L. M., Dull T. J., Gray A., Coussens L., Liao Y. C., Tsubokawa M. Human insulin receptor and its relationship to the tyrosine kinase family of oncogenes. 1985 Feb 28-Mar 6Nature. 313(6005):756–761. doi: 10.1038/313756a0. [DOI] [PubMed] [Google Scholar]
- Wang C. C., Hedo J. A., Kahn C. R., Saunders D. T., Thamm P., Brandenburg D. Photoreactive insulin derivatives. Comparison of biologic activity and labeling properties of three analogues in isolated rat adipocytes. Diabetes. 1982 Dec;31(12):1068–1076. doi: 10.2337/diacare.31.12.1068. [DOI] [PubMed] [Google Scholar]
- Werb Z., Burleigh M. C., Barrett A. J., Starkey P. M. The interaction of alpha2-macroglobulin with proteinases. Binding and inhibition of mammalian collagenases and other metal proteinases. Biochem J. 1974 May;139(2):359–368. doi: 10.1042/bj1390359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolley D. E., Roberts D. R., Evanson J. M. Inhibition of human collagenase activity by a small molecular weight serum protein. Biochem Biophys Res Commun. 1975 Sep 16;66(2):747–754. doi: 10.1016/0006-291x(75)90573-2. [DOI] [PubMed] [Google Scholar]
- Yip C. C., Moule M. L. Structure of the insulin receptor of rat adipocytes. The three interconvertible redox forms. Diabetes. 1983 Aug;32(8):760–767. doi: 10.2337/diab.32.8.760. [DOI] [PubMed] [Google Scholar]
- Yip C. C., Moule M. L., Yeung C. W. Subunit structure of insulin receptor of rat adipocytes as demonstrated by photoaffinity labeling. Biochemistry. 1982 Jun 8;21(12):2940–2945. doi: 10.1021/bi00541a021. [DOI] [PubMed] [Google Scholar]
- Yip C. C., Yeung C. W., Moule M. L. Photoaffinity labeling of insulin receptor of rat adiopocyte plasma membrane. J Biol Chem. 1978 Mar 25;253(6):1743–1745. [PubMed] [Google Scholar]