Abstract
The role of NF-κB in the reactivation of human immunodeficiency virus (HIV) from latency in CD4 T lymphocytes is well documented. However, its role in driving HIV transcription in human macrophages, which contain a constitutive nuclear pool of NF-κB, is less well understood. In this study we have investigated the role that the constitutive pool of NF-κB and the NF-κB cis-acting motifs of the HIV long terminal repeat (LTR) play in regulating HIV transcription in human monocytic cells and primary macrophages. Inhibition of the constitutive nuclear pool of NF-κB (RelA and RelB) in the promonocytic U937 cell line using dominant-negative IκBα significantly decreases HIV replication. Moreover, it is demonstrated that in the differentiated monocytic cell line THP1, which contains a constitutive nuclear pool of NF-κB (RelB),an HIV provirus containing mutations of the κB cis-acting sites in the LTR is transcriptionally impaired. Reduction of the constitutive pool of NF-κB in human macrophages by an adenovirus vector expressing a dominant-negative IκBα also reduces HIV transcription. Lastly, mutation of the NF-κB cis-acting sites in the LTR of an R5 HIV provirus completely abrogates the first cycle of HIV transcription. These studies indicate that the cis-acting NF-κB motifs of the HIV LTR are critical in initiating HIV transcription in human macrophages and suggest that the constitutive nuclear pool of NF-κB is important in regulating HIV transcription in these cells.
Tissue macrophages are important targets of human immunodeficiency virus (HIV) (18, 19, 26). The ability of HIV to infect and replicate in these nondividing cells significantly contributes to the pathogenesis of AIDS. Since the viral cytopathic effects in this cell population appear to be minimal, the infected macrophage behaves as a major viral reservoir facilitating persistent HIV replication, spreading virus via cell-to cell-contact, and hence accelerating disease progression (30). How HIV transcription is regulated in macrophages is less well understood. At least two differences are noted between resting CD4 T cells and differentiated macrophages. Macrophages do not die upon HIV infection, and they have a preexisting pool of transcription factors that may allow HIV to undergo continuous gene transcription in collaboration with HIV regulatory proteins such as HIV Tat. In CD4 T cells, in contrast, HIV transcription is dependent on extracellular stimuli that lead to the translocation of transcription factors to the nuclei. This triggers HIV transcription and reactivation from latency, and ultimately cell death (reviewed in references 12 and 38).
HIV replication is tightly regulated at the transcriptional level through the specific interaction of viral regulatory proteins, namely, Tat and cellular transcription factors binding to a variety of cis-acting DNA sequences in the HIV long terminal repeat (LTR) (reviewed in reference 9.) One of the main mediators of HIV LTR transcription is NF-κB (31). NF-κB, which is an inducible transcription factor in CD4 T cells, is composed of homo- or heterodimers of Rel family proteins (reviewed in references 23 and 42). All of the NF-κB proteins contain an N-terminal Rel homology domain, which mediates DNA binding, dimerization, and interaction with the inhibitory proteins, or IκBs. In addition, c-Rel, RelA, and RelB contain a C-terminal transactivation domain (reviewed in references 23 and 42). The classical NF-κB complex (p50/RelA) is sequestered in the cytoplasm by interaction with a family of inhibitory proteins, or IκBs, including IκBα, IκBβ, IκBɛ, IκBγ, and the proto-oncogene Bcl-3 (4, 5), of which IκBα is the best-characterized inhibitor. Following cell activation by a variety of extracellular stimuli, IκBα is phosphorylated at the N-terminal residues S32 and S36 by the IκB kinase (IKK) complex, leading to ubiquitination and subsequent proteasome-mediated degradation (reviewed in reference 22), which allows NF-κB to translocate to the nucleus, where it activates gene expression. In addition to the classic inducible pool of NF-κB (p50/RelA), constitutive nuclear localization of other NF-κB heterodimers is observed in several cell populations in the absence of activating stimuli. c-Rel is present mainly in the nuclei of B and T lymphocytes (24), while constitutive RelB nuclear localization has been detected in murine dendritic cells (6), mature human dendritic cells, and differentiated human macrophages (32). In contrast to the well-characterized mechanisms regulating the inducible pool of NF-κB, the molecular mechanism regulating the constitutive NF-κB pool, especially that of RelB, remains ill defined.
Since the identification of NF-κB elements in the HIV LTR (31), multiple studies have addressed the dispensability of this family of transcription factors in the transcriptional regulation of the HIV LTR in T cells and its impact on HIV reactivation from latency (7, 8, 13–15, 17, 27, 35, 36, 39, 45, 46). Differences in the type of T cell studied, HIV-driven reporter constructs, viral stocks, and methodological approaches have yielded conflicting results. In general, in transformed CD4 T-cell lines, the HIV LTR κB sites are dispensable for viral replication (27), but they are relevant in regulating HIV transcription in primary T cells (1, 7). Study of the interaction between NF-κB and HIV in both human monocytic cells and transformed human macrophages has mainly focused on how monocyte differentiation may lead to HIV expression (20, 37, 41) and how HIV infection leads to NF-κB activation. In the promonocytic cell line U937, HIV activates the inducible pool of NF-κB (3, 21, 40, 43) as a result of enhanced IκBα degradation (10, 29) that is believed to be secondary to IKK activation (2). However, differentiated macrophages already have a constitutive pool of NF-κB in their nuclei. Therefore, apart from the HIV induction of NF-κB, it is unclear what role, if any, the preexisting pool of NF-κB or the NF-κ B cis-acting motifs play in regulating HIV transcription.
In this study, we have analyzed HIV transcription in monocytes and human macrophages expressing a constitutive nuclear pool of NF-κB. We have determined that the cis-acting motifs present in the HIV LTR are indispensable for the first cycle of HIV transcription in these primary cells.
MATERIALS AND METHODS
Antibodies.
Expression of RelA and RelB was detected with rabbit polyclonal specific antibodies (Santa Cruz Biotechnology, Santa Cruz, Calif.). Rabbit polyclonal anti p50/p105 and anti p52/p100 antibodies were generated with N-terminal peptides of p105 and p100, respectively. The gp120 envelope protein of T-tropic clones was detected with an anti-gp120 polyclonal antibody (Center for Biologics Evaluation and Research, U.S. Food and Drug Administration, Bethesda, Md.). The gp120 envelope protein of M-tropic clones was detected with serum from an HIV-infected patient (AIDS Research and Reference Reagent Repository, Rockville, Md.).
Plasmids.
cDNAs encoding human RelB, RelA, p50, and p52 were cloned into the pcDNA3 vector (Invitrogen, Carlsbad, Calif.). The HIV LTR-luciferase reporter plasmid has been described previously (34). HIV LTRΔκB-luciferase contains the ΔκB mutation described previously (1). The TK-Renilla-luciferase plasmid contains cDNA encoding Renilla luciferase under the control of an upstream herpes simplex virus thymidine kinase (TK) promoter (Promega, Madison, Wis.).
Cells and cell lines.
The U937 promonocytic cell line was purchased from the American Type Culture Collection (ATCC) and grown in RPMI 1640 supplemented with 5% heat-inactivated fetal bovine serum (FBS; Intergen, Purchase, N.Y.), 2 mM glutamine, 100 U of penicillin/ml, and 100 μg of streptomycin/ml. The generation and characterization of the spleen focus-forming virus (SFFV)- and IκBα-expressing U937 cell lines have been described previously (2). The THP1 monocytic cell line was obtained from ATCC and grown in RPMI 1640 supplemented with 10% heat inactivated FBS. Jurkat cells were obtained from ATCC and cultured in RPMI supplemented with 10% heat-inactivated FBS (Intergen), 100 U of penicillin/ml and 100 μg of streptomycin/ml, and glutamine (2 mM).
The chronically HIV type 1 (HIV-1) infected cell line ACH2 was obtained from the AIDS Research and Reference Reagent Repository and grown in RPMI 1640 supplemented with 10% heat-inactivated FBS.
Human monocyte-derived macrophages (MDM) were obtained from peripheral blood mononuclear cells from buffy coats of healthy volunteers by Ficoll-Hypaque density gradient centrifugation. A total of 40 × 106 peripheral blood mononuclear cells were incubated horizontally in RPMI plus 10% human AB serum for 5 days in T25 flasks. Thereafter, nonadherent cells were removed, and adherent cells were maintained in 10% FBS until the moment of HIV infection. In experiments using adenovirus, MDM were purified by negative depletion using magnetic beads (Stem Cell Inc., Vancouver, British Columbia, Canada). Monocyte purity was assessed by flow cytometry using an anti-CD14 antibody and was higher than 80%. The human embryonic kidney cell lines 293T and 293A were generously provided by Richard Bram and Robert Simari (Mayo Clinic, Rochester, Minn.) and grown in Dulbecco's modified Eagle medium supplemented with 10% heat inactivated FBS, 2 mM glutamine, 100 U of penicillin/ml, and 100 μg of streptomycin/ml.
Nuclear and cytosolic extracts, electrophoretic mobility shift assay, and immunoblotting.
Nuclear and cytosolic extracts from U937 cells, THP1 cells, and peripheral blood lymphocytes were prepared by a modification of the method of Dignam et al. (11) as previously described (29). Nuclear and cytosolic extracts from MDM were obtained by gentle scraping of the cells in ice-cold-phosphate-buffered saline containing 0.5 mM EDTA. Cells were then washed with buffer A (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl) and subsequently lysed for 10 min on ice in buffer A supplemented with 0.1% Nonidet P-40, 0.5 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride, and aprotinin, leupeptin, and pepstatin (all at 2 μg/ml). After centrifugation, cells were washed three times in buffer A. Nuclei were pelleted by centrifugation, and proteins were extracted by resuspension in buffer C (20 mM HEPES, 25% glycerol, 0.42 M NaCl, 1.5 mM MgCl2, 0.2 mM [each] EDTA, dithiothreitol, phenylmethylsulfonyl fluoride, aprotinin, leupeptin, and pepstatin) at 4°C for 1 h. After centrifugation the supernatants were collected and stored at −70°C or used immediately. Electromobility shift assays were performed as described previously (29). Double stranded oligonucleotide DNA probes were made by annealing sense and antisense oligonucleotides corresponding to the wild-type consensus sequences for NF-κB present in the HIV LTR (5′-AGTTGAGGGGACTTTCCCAGGC-3′) and NFAT (5′-CGCCCAAAGAGGAAAATTTGTTTCATA-3′). DNA probes with mutated binding sites were made corresponding to the sequences NF-κB mut (5′-AGTTGAGGCGACTTTCCCAGG-3′) and NFAT mut (5′-CGCCCAAAGCTT AAAATTTGTTTCATA-3′) (with mutated nucleotides italicized).
To characterize the level of expression of the Rel family members in uninfected cells, 20 μg of cytosolic and 10 μg of nuclear proteins were analyzed by sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis. Proteins were transferred to Immobilon-P membranes (Millipore, Bedford, Mass.) by standard procedures and blotted with polyclonal antibodies followed by incubation with horseradish peroxidase (Amersham, Little Chalfont, Buckinghamshire, England). Immunoreactive proteins were detected with an ECL Western blotting detection kit (Amersham).
Molecular clones of HIV.
Molecular clones HXB-wt and HXB-Δ-κB were a generous gift from David Baltimore (California Institute of Technology, Pasadena, Calif.) and were derived from molecular clone HXB-2D in proviral vector R7. The HXB-Δ-κB provirus was obtained by replacing the XhoI-SacI fragment containing the 3′ HIV-1 LTR in proviral clone R7 with a κB mutant LTR from proviral clone BH8 (7).
Macrophage-tropic viruses were derived from molecular clone pAD, kindly provided by Keith Peden (Division of Viral Products, Office of Vaccine Research and Review, Center for Biologics Evaluation and Research). To create pAD ΔκB, site-directed mutagenesis was performed by amplifying a 3-kb fragment encompassing the 3′ LTR region and subcloning the κB mutant LTR into the wild-type (wt) LTR via StuI and BamHI sites present in the 3′ end of the proviral sequence. The mutations were created with the sense primer 5′-TTTCTACTTTAAACTTTCCGCTTTAAACTTTCC-3′ and the antisense primer 5′-GGAAAGTTTAAAGCGGAAAGTTTAAAGTAGAA-3′. LTR mutations were confirmed by DNA sequencing.
Stocks of wt and mutant HIV strains were generated by transfecting 10 μg of linear viral DNA into 293T cells growing in 180-mm dishes using FuGENE6 (Roche Diagnostics Corporation, Indianapolis, Ind.). Culture supernatants from transfected cells were collected 48 h after transfection and clarified by low-speed centrifugation. The virus was then pelleted by ultracentrifugation through a cushion of sucrose buffer (20% sucrose, 20 mM Tris [pH 7.4], 100 mM NaCl) and resuspended in ice-cold RPMI with 10% FBS. The concentrated virus was filtered through a 0.2-μm-pore-size-filter and used immediately or kept frozen at −70°C until further use. To eliminate HIV proviral DNA contamination, viral stocks were treated with 10 U of RNase-free DNase per ml and 10 mM MgCl2 for 30 min at room temperature. Viral stocks were quantitated by p24 enzyme-linked immunosorbent assay (Coulter-Immunotech Immunology, Westbrook, Maine) to normalize all infections to equivalent viral input.
HIV infection.
U937 cells expressing Flag-IκBα-2N and empty vector (SFFV) U937 control cells were infected with HIV-1LAV.04, which was obtained through the AIDS Research and Reference Reagent Program, National Institutes of Health, from Malcolm Martin. Briefly, 107 exponentially growing U937 cells were sedimented by low-speed centrifugation and resuspended overnight in 10 ml of supernatant containing 360 ng of HIV-1LAV.04 p24 per ml. After 24 h, cells were extensively (six times) washed and then cultured in normal medium. For experiments addressing the first cycle of HIV replication, 107 THP1 cells were incubated with 10 ml of supernatant containing 1 μg of HIV HXB-2D p24 per ml, and MDM were cultured with 3 ml of supernatant containing 5 μg of HIV p24. After 2 h of incubation, both cell types were extensively washed and cultured in normal medium until the moment of genomic DNA or RNA extraction. Mock-infected cells were used as a control.
Detection of HIV proviral DNA.
HIV DNA was detected by PCR amplification from genomic DNA isolated from THP1 cells and MDM (Puregene DNA isolation kit; Gentra Systems, Minneapolis, Minn.). Briefly, 1 × 106 to 2 × 106 THP1 cells or 5 × 106 to 10 × 106 MDM were lysed with an anionic detergent in the presence of a DNA stabilizer. Contaminating RNA was removed by RNase digestion at 37°C for 30 min. Contaminating proteins were eliminated by salt precipitation. Thereafter, total genomic DNA was precipitated and resuspended in 20 to 30 μl of Puregene Hydration Solution (Gentra Systems). HIV DNA was amplified using the LTR sense primer (5′-GGCTAACTAGGGAACCCACTG-3′) and the Gag antisense primer (5′-TAATACTGACGCTCTCGCACC-3′). The LTR and Gag primers should form a 318-bp fragment following PCR amplification. HIV DNA was quantitated by comparison with a standard curve generated by serial dilutions of genomic DNA extracted from ACH2 cells. The human β-actin gene was amplified with the sense primer 5′-ATGGCCACGGCTGCTTCCAGC-3′ and the antisense primer 5′CATGGTGGTGCCGCCAGACAG-3′ in order to control for the integrity of cellular DNA. Amplified products were visualized by electrophoresis in a 3% agarose gel.
Detection of integrated DNA.
To detect HIV DNA integrated into the host cell chromosome, genomic DNA was extracted and amplified by PCR using the Alu sense primer (5′-GCCTCCCAAAGTGCTGGGATTA-3′) and the Gag antisense primer described above. PCR amplification products were extracted with phenol-chloroform and subsequently digested with AvaI and AvaII restriction enzymes (Roche Diagnostics Corporation). Digested products were separated by electrophoresis in a 3% agarose gel, transferred overnight onto nylon transfer membranes (Nitran, Keene, N.H.), and hybridized with a γ-32P-labeled HIV probe encompassing the sequence 5′-AGAGATTTTCCACACTGACTA-3′ in the U5 region of the HIV LTR. HIV DNA products were visualized by autoradiography. ACH2 cells containing 1 copy of integrated viral DNA per cell were used as a control.
Analysis of HIV RNA.
The first cycle of HIV transcription was detected by reverse transcription-PCR using oligonucleotide primers complementary to the flanking region of the common splice donor and acceptor sites of the env, tat, and rev genes. A 5-μl volume of total RNA extracted from an equivalent number of uninfected and HIV-infected cells was reverse transcribed with 20 U of avian myeloblastosis virus reverse transcriptase (Roche Diagnostics Corporation). Thereafter, 10 μl of this reaction product was subjected to PCR with the sense primer M669 (5′-GTGTGCCCGTCTGTTGTGTGACTCTGGTAAC-3′, nucleotides 558 to 588) and the antisense primer LA23 (5′-GCCTATTCTGCTATGTCGACACCC-3′, nucleotides 5815 to 5792) of the HXB-2D molecular clone of HIV. The M669 and LA23 primers yield a 214-bp product from spliced RNAs. In experiments using cDNA from MDM, amplification products were resolved in a 3% agarose gel, transferred overnight onto Nitran nylon transfer membranes, and hybridized with a 32P-labeled HIV probe encompassing the U5 region of the HIV LTR. The early transcripts were visualized by autoradiography. ACH2 cells and THP1 cells chronically infected with HIV HXB-wt and ΔκB were used as controls.
Adenovirus infection.
Recombinant replication-deficient adenoviral vectors encoding alkaline phosphatase (AD5AP) or the superrepressor of NF-κB activity IκBαSer32–36 (AD5IκBαSer32–36) were generously provided by Robert Simari (Mayo Clinic). Viruses were propagated in 293A cells and were purified by ultracentrifugation through two cesium chloride gradients. For experiments addressing the inhibition of NF-κB proteins, 5-day-old MDM (4 × 106 cells per flask) were infected with AD5AP or AD5IκBαSer32–36 at a multiplicity of infection of 100 in 300 μl of RPMI–1% FBS. After 2 h, cells were overlaid with RPMI containing 10% FBS and kept at 37°C until the moment of HIV infection. Uninfected cells were used as a control. The efficiency of adenovirus infection was evaluated in cells growing in 6-well plates (106 cells per well) 2 days after infection by staining for intracellular expression of alkaline phosphatase. Adenovirus infection of MDM purified from CD14+ cells resulted in 90 to 100% positivity for akaline phosphatase.
Gene transfection and reporter assays.
One million Jurkat cells per experimental point were transiently transfected by FuGENE6 (Roche Molecular Biochemicals, Indianapolis, Ind.) according to the manufacturer's protocol. Transfected Jurkat cells were lysed in the Passive Lysis Buffer supplied in the Dual-Luciferase Reporter Assay System (Promega) according to the instructions in the accompanying technical manual. Briefly, 20 μl of lysates was mixed with 100 μl of Luciferase Assay Reagent II (Promega), and luminescence was measured with a Berthold Lumat to analyze firefly luciferase levels. Then 100 μl of Stop & Glo Reagent (Promega) was added and luminescence was again measured to analyze Renilla luciferase levels. Relative luciferase units (RLU) are equivalent to firefly luciferase values normalized to Renilla luciferase values.
RESULTS
Functional relevance of the constitutive nuclear pool of NF-κB present in monocytic cells in the regulation of HIV persistence.
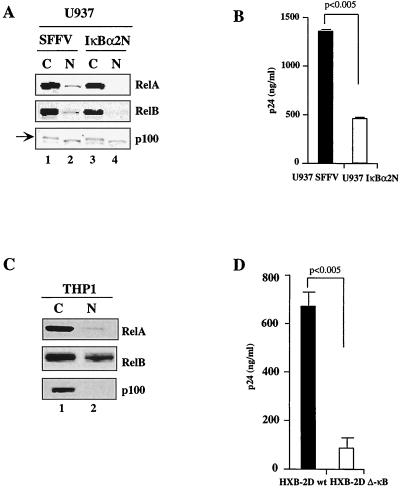
Using a promonocytic U937 cell line that expresses a constitutive pool of NF-κB in the nucleus (U937-SFFV-bis) and the control clone expressing a dominant-negative IκBα transgene (SFFV-IκBα-2N) (2), we characterized and investigated the role of the constitutive NF-κB pool in regulating HIV persistence. The molecular composition of the nuclear pool of NF-κB in the U937-SFFV and U937-IκBα-2N cell lines was first characterized. The pool of NF-κB in the nuclei of U937-SFFV cells is composed of RelA and RelB (Fig. 1A, lane 2), and stable expression of IκBα-2N abrogates this nuclear localization (lane 4). These nuclear samples were devoid of cytosolic contamination, as p100 was present only in the cytosolic compartment (lanes 1 and 3), although a faster-migrating cross-reactive band was noted in nuclear extracts of these cells. The same nuclear extracts were then analyzed by gel shift assays with an oligonucleotide containing the HIV LTR NF-κB binding sites, confirming the presence of these NF-κB components (data not shown).
FIG. 1.
Molecular characterization of the nuclear pool of NF-κB in promonocytic cells and analysis of HIV replication. (A) Immunoblotting of cytosolic (C) and nuclear (N) extracts in SFFV- and IκBα-2N-expressing U937 cells with anti-RelA, anti-RelB, and anti-p100/p52 antibodies. (B) HIV p24 values in the culture supernatants of HIV LAV-BN-infected SFFV- and IκBα-2N-expressing U937 cells at day 30 postinfection. (C) Immunoblotting of cytosolic (C) and nuclear (N) extracts from THP1 cells with anti-RelA, anti-RelB, and anti-p100 antibodies. (D) HIV p24 values in the culture supernatants of THP1 cells infected with HIV HXB2D-wt and HXB2D-ΔκB at day 14 postinfection.
To determine the role of this constitutive nuclear pool of NF-κB in regulating HIV replication, these cells were then infected with HIV strain LAV.04 and viral replication was evaluated by measuring HIV p24 levels in the cell culture supernatant at various time points after infection. As shown in Fig. 1B, there was a significant reduction in the levels of p24 in supernatants from IκBα-2N-expressing U937 cells at 30 days after infection compared to supernatants from SFFV-expressing control cells, suggesting that the nuclear presence of RelA and RelB in these promonocytic cells may impact HIV persistence.
To overcome potential differences that may have existed between the U937 SFFV and U937 IκBα-2N clones with regard to influencing HIV replication independently of abrogating the constitutive pool of NF-κB, we utilized another well-characterized promonocytic cell line known to contain a large constitutive pool of NF-κB in its nucleus, THP1. Characterization of this pool by Western blotting of the nuclear compartment demonstrated that the pool of NF-κB present in the nucleus of this monocytic cell line was composed mainly of RelB, with small amounts of RelA and no c-Rel (Fig. 1C; also data not shown).
To determine how this constitutive pool of NF-κB present in the nuclei of THP1 cells influences the replication of HIV, the molecular clone of HIV HXB-2D harboring mutations (or not)in both κB sites of the LTR was used. As shown in Fig. 1D, supernatants from THP1 cells infected with HXB-ΔκB showed a significantly lower level of replication than those from THP1 cells infected with the HXB-wt clone at 14 days postinfection, suggesting that the constitutive pool of NF-κB (RelB) present in THP1 cells may influence viral transcription.
NF-κB is essential for the first cycle of HIV transcription in THP1 cells.
To confirm that the critical step of the HIV cycle that is influenced by NF-κB occurs at the level of transcription, we characterized, in a sequential manner, viral reverse-transcription, integration, and transcription in THP1 cells infected with HXB-wt and HXB-ΔκB.
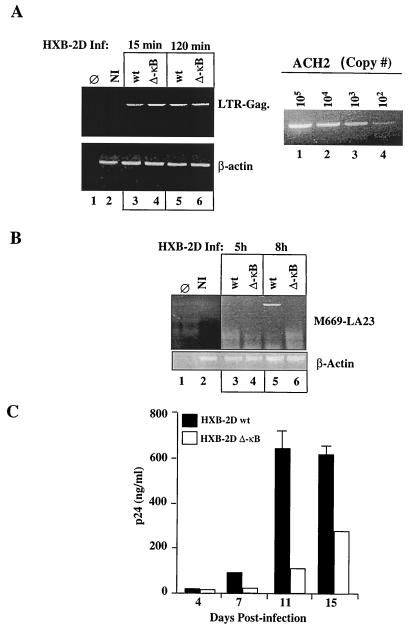
Proviral DNA was first analyzed by a semiquantitative PCR assay from total genomic DNA extracted from THP1 cells 15 and 120 min after HIV infection. As shown in Fig. 2A (left panel; lanes 3 and 4 and lanes 5 and 6, respectively), the amounts of proviral DNA amplified in cells infected with the wt and ΔκB mutant HIV clones were comparable at both time points, suggesting that equal input of each HIV strain results in comparable infection rates. The amounts of total DNA amplified within the various samples were equivalent, as shown by similar levels of β-actin amplification (Fig. 2A, lower left panel, lanes 3 to 6). The limit of detection of our assay was 102 copies of viral DNA, as detected in parallel by amplifications from serial dilutions of total genomic DNA isolated from ACH2 cells (Fig. 2A, right panel). Thus, the defect in viral replication observed with the HXB-ΔκB HIV clone in THP1 cells was not due to differences in infectivity. Since the LTR and Gag primers allow the detection of full-length or nearly full-length reverse-transcribed cDNA, these results also indicate that the NF-κB mutation in the HIV clone does not interfere with the initiation or completion of reverse transcription of HIV.
FIG. 2.
Mutation of the κB sites in the HIV LTR delays the first cycle of HIV transcription and decreases viral persistence in monocytic cells. (A) (Top left) HIV DNA amplification from uninfected (NI) and HIV-infected THP1 cells at 15 and 120 min after infection. The amplified 318-bp fragment is indicated. (Bottom left) Human β-actin amplification within the same samples. (Right) Serial dilution of known HIV-1 proviral copy numbers amplified from ACH2 cells. (B) (Top) PCR amplification of the HIV early transcripts (tat/rev, env) from uninfected (NI) and HIV-infected THP1 cells at 5 and 8 h after HIV infection. (Bottom) Human β-actin PCR amplification within the same samples. (C) HIV p24 values in the culture supernatants of THP1 cells infected with HIV-HXB-2D-wt and ΔκB at different days after viral infection. The p24 determinations were performed in duplicate. Error bars, standard errors.
We next investigated whether the first cycle of HIV transcription is modified in the HXB-ΔκB compared to the wt HIV clone. To detect viral transcription, total RNAs from uninfected and HIV-infected THP1 cells were extracted at various time points after infection. Following a first step of synthesis of cDNA, HIV early transcripts were detected by PCR amplification using primers directed to the common splice donor and acceptor sites of the tat, rev, and env regulatory genes of HIV (47). As shown in Fig. 2B, HIV-specific early transcripts were detected in THP1 cells infected with the HXB-wt HIV clone between 5 and 8 h after infection (lane 5). This transcript was sequenced and found to encompass the corresponding sequence of the molecular clone of HIV. From this, it is inferred that the delay in HIV transcription correlates with the failure of HIV-infected cells to produce progeny virions. This, in turn, results in a significant decrease in the HIV p24 levels generated in the culture supernatants of THP1 cells infected with HXB-ΔκB at different days following HIV infection (Fig. 2C).
Genetic interference with the nuclear pool of NF-κ B in human macrophages reduces HIV replication.
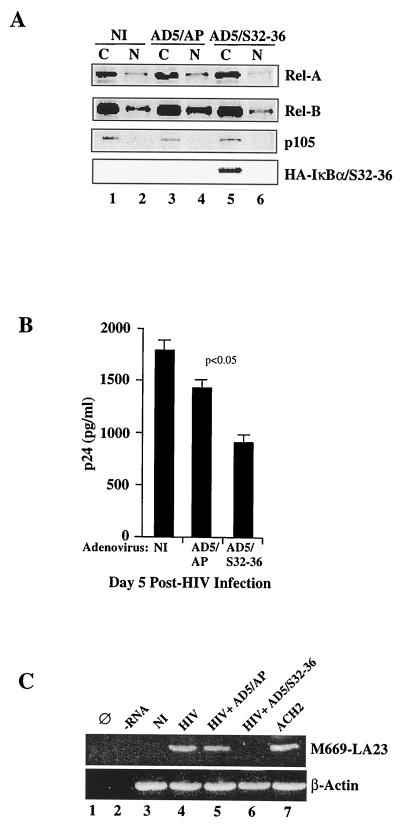
To investigate the role that the constitutive pool of nuclear NF-κB present in the nuclei of differentiated macrophages (MDM) plays in regulating HIV replication, we first attempted to inhibit such pools using a replication-deficient adenoviral vector expressing the IκBα dominant-negative protein IκBαSer32–36 (AD5/S32–36). This adenovirus vector has been shown to partially inhibit NF-κB in human macrophages (16). To confirm the function of this reagent in primary human macrophages, nuclear and cytosolic extracts were prepared from 7-day-old adherent MDM that were mock infected or adenovirus infected (AD5/S32–36 or control adenovirus vector [AD5/AP]). As shown in Fig. 3A, the major component of NF-κB in the nuclei of MDM is RelB; RelA is present to a lesser extent (lane 2). Infection with the adenovirus control vector expressing alkaline phosphatase (AD5/AP) does not inhibit, but rather increases the levels of, RelA and specifically RelB (Fig. 3A, lane 4). In contrast, infection with an adenovirus vector expressing an IκBα-negative dominant transgene (AD5/S32–36) abrogated the majority of nuclear RelA and, albeit incompletely, the majority of nuclear RelB (Fig. 3A; compare lanes 4 and 6). The expression of the hemagglutinin (HA)-tagged IκBα transgene was verified by blotting the membrane containing cytosolic extracts with anti-HA antibodies (Fig. 3A, lane 5).
FIG. 3.
Decreases in RelB and RelA nuclear levels correlate with decreased HIV-PAD-1 replication in MDM. (A) Immunoblotting of cytosolic (C) and nuclear (N) extracts from uninfected (NI) or adenovirus-infected (AD5/AP or AD5/Ser 32–36) MDM, with anti-RelA, anti-RelB, anti-p100, and anti-FLAG antibodies. (B) HIV p24 values from the culture supernatants of uninfected (NI) or adenovirus-infected (AD5/AP or AD5/S32–36) MDM. p24 determinations were performed in triplicate. Error bars, standard errors. (C) PCR amplification of early HIV transcripts (tat/rev, env) in MDM mock infected (HIV) or infected with AD5/S32–36 or the vector control (AD5/AP) followed by HIV PAD-1 wt infection in the three cases. Ø, no DNA input in the PCR; −RNA, reverse transcription in the absence of RNA. The amplified 219-bp fragment is visualized by Southern blot hybridization with a γ-32P-labeled U5 LTR internal primer.
We next asked if the partial inhibition of the constitutive pool of NF-κB in the nuclei of MDM correlates with decreased HIV replication, and if so, if this was secondary to decreased HIV transcription. To test this, 5-day-old differentiated MDM were either mock infected or infected with either AD5/S32–36 or the adenovirus control vector (AD5/AP). Two days after adenovirus infection (the same time at which NF-κB components were analyzed as described above and shown in Fig. 3A), the same MDM cultures were infected with the HIV-1AD wt molecular clone. As shown in Fig. 3B, preinfection with AD5/S32–36 decreased the replication of the R5 HIV strain HIV-1AD as measured by levels of p24 in the supernatant 3 days after HIV infection.
HIV-1AD transcription was next evaluated by amplification of HIV early transcripts from previously synthesized cDNA. As shown in Fig. 3C (upper panel, lane 6) expression of IκBαS32–36 (AD5/S32–36) specifically inhibits the appearance of HIV-1AD early transcripts. The expected 219-bp fragment was amplified exclusively from mock-adenovirus AD5/AP-infected and HIV-1AD-infected MDM (lanes 4 and 5, respectively), not from AD5/S32–36 adenovirus-infected, HIV-1AD-infected MDM (Fig. 3C, lane 6). Amplification of HIV from the HIV-infected ACH2 line was performed as a control (lane 7). The same level of β-actin amplification was achieved in all samples (Fig. 3C, lower panel, lanes 3 through 7).
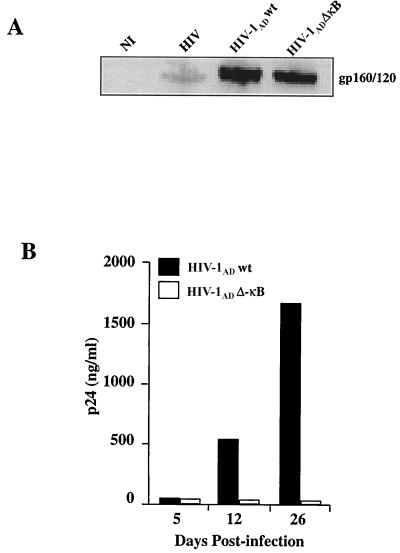
The above results suggest a partial dependence on the constitutive pool of NF-κB in regulating HIV replication in MDM. However, the adenovirus vector expressing the IκBαS32–36 transgene did not completely inhibit HIV replication in human macrophages, nor did it completely eliminate RelB from their nuclei. We therefore reasoned that to clearly confirm or disprove the role of the constitutive pool of NF-κB in HIV persistence in human macrophages, we needed to resort to the use of an R5 HIV provirus lacking (or not) functional κB cis-acting motifs in its LTR. For this purpose we mutated the two cis-acting κB sites in the HIV LTR as performed previously for the X4 strain HXB-2D (Fig. 1 and 2). The competencies of virion assembly of both pAD wt and the ΔκB mutant were confirmed to be similar when each proviral DNA was transfected into 293T cells and the resulting HIV envelope expressed was detected with anti-gp160 antibodies (Fig. 4A) (44).
FIG. 4.
Molecular characterization of the NF-κB complex and kinetics of HIV-1AD replication in MDM. (A) Assembling of HIV-1AD wt and ΔκB in 293 T cells transfected with each provirus. Cell lysates from these cells were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by immunoblotting with anti-HIV antibodies. Cell lysates from uninfected MDM (NI) or MDM infected with HIV were also analyzed. (B) HIV p24 values in the culture supernatants of MDM infected with HIV-1AD wt and ΔκB at different days after infection.
The kinetics of viral replication of these two clones were then characterized by measuring HIV p24 levels in the culture supernatants of primary macrophages infected with equal amounts of each provirus. As shown in Fig. 4B, minimal levels of HIV p24 are detected in the supernatants from MDM infected with HIV-1AD ΔκB compared to those in supernatants from MDM infected with the HIV wt R5 HIV clone at different time points postinfection. In fact, we were unable to detect any increase in HIV p24 production, even 26 days after HIV infection, in these primary macrophages. The increase of HIV-1AD wt replication in differentiated macrophages may be due to new infection of previously uninfected resting macrophages or, alternatively, to the production of cytokines, chemokines, and/or other regulatory proteins from HIV-infected macrophages that ultimately enhance HIV transcription.
Members of the NFAT family do not bind the NF-κB cis-acting motifs in human macrophages.
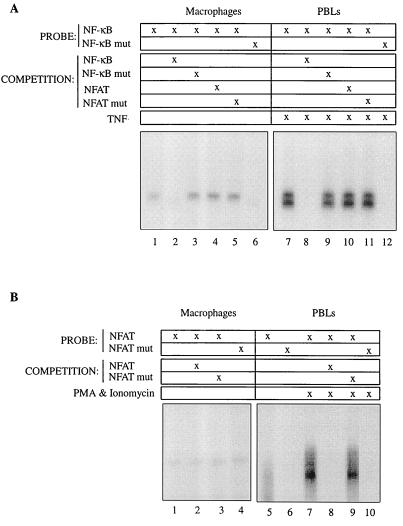
The apparent dichotomy between the partial inhibition of HIV transcription achieved by infection with the constitutive pool of NF-κB present in the nuclei of human macrophages and the near-complete lack of HIV transcription of HIV provirus containing mutations in the NF-κB cis-acting DNA binding motifs raised the question as to whether other transcription factors, separate from NF-κB members, could be binding to the NF-κB cis-acting motif. To address this, we focused on members of the NFAT family of transcription factors, which are known to interact with NF-κB members, bind to the HIV LTR, and induce its transcription in a number of transformed cells (25). Nuclear extracts from 5-day-old MDM were isolated and incubated with a 32P-labeled oligonucleotide encompassing the NF-κB cis-acting motifs present in the HIV LTR of HIV-1AD. As shown in Fig. 5A (left panel), MDM nuclear extracts contain NF-κB DNA binding activity, which is lost upon coincubation with an excess of unlabeled NF-κB oligonucleotide but not with an oligonucleotide containing the critical mutations in the cis-acting NF-κB motifs. As a control (Fig. 5A, right panel), nuclear extracts from peripheral blood lymphocytes treated with tumor necrosis factor (TNF) were incubated with the same oligonucleotide combinations, confirming the specificity of the NF-κB DNA binding activity observed in macrophages. NF-κB DNA binding activity present in the nuclei of differentiated macrophages and TNF-stimulated primary CD4 T cells was not competed with an oligonucleotide encompassing the classical NFAT consensus sequence. NFAT DNA binding activity was barely detectable in nuclear extracts from differentiated macrophages (Fig. 5B, left panel), in contrast to that observed in phorbol myristate acetate (PMA)- and ionomycin-treated CD4 T cells.
FIG. 5.
NF-κB, but not NFAT, present in the nuclei of human macrophages binds the NF-κB cis-acting motifs of the HIV LTR. (A) NF-κB DNA binding. Three micrograms of nuclear extracts either from peripheral blood lymphocytes (PBLs) treated for 30 min either with TNF (10 ng/ml) for NF-κB binding or with PMA (20 ng/ml) plus ionomycin (3.5 μg/ml) for NFAT binding or from 5-day-old plastic-adhered human macrophages was incubated with 32P-labeled DNA probes as indicated. Competition of DNA binding to the labeled oligonucleotide was analyzed in the presence of a 20-fold excess of nonlabeled DNA probe as indicated. (B) NFAT DNA binding. Extracts and experimental conditions were the same as for the experiment for which results are shown in panel A.
Therefore, these results suggest that it is the NF-κB complex, rather than other transcription factors such as NFAT family members, that interacts with the NF-κB cis-acting motif. Moreover, the NF-κB DNA binding activity was eliminated with anti-p50, anti-RelB, and anti-RelA supershifting antibodies (data not shown).
The replication defect of HIV-1AD ΔκB in primary MDM is at the level of HIV transcription.
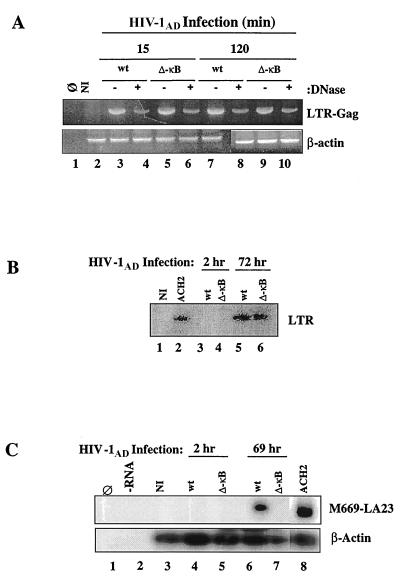
To further characterize the mechanism(s) accounting for the differences in replication between the HIV wt and ΔκB R5 HIV clones, we first excluded differences in HIV infection and integration using the same methodological approach as described for THP1 cells (Fig. 2). The HIV stocks generated from transfecting each provirus clone into 293T cells were treated with RNase-free DNase prior to their use in infection to avoid any plasmid DNA carryover. Thereafter, the proviral DNA present in the macrophages following HIV infection was analyzed. As shown in Fig. 6A, as early as 15 min after infection there was no difference in the amounts of proviral DNA amplified from macrophages infected with the wt and the ΔκB mutant virus, regardless of the presence or absence of previous DNase treatment (upper panel, lanes 3 to 6). The amounts of total DNA amplified in the various samples were equivalent, as shown by comparable levels of β-actin amplification within each sample (Fig. 6A, lower panel). Since the LTR-Gag primers are able to detect full-length or nearly full-length reverse-transcribed cDNA, these results confirm that the differences in replication between the HIV-1AD wt and ΔκB clones are not due to differences in infection or reverse transcription. Further, we determined that viral integration into the host cell was similar for each provirus (Fig. 6B, lanes 5 and 6).
FIG. 6.
The HIV-1AD ΔκB provirus does not undergo transcription in MDM. (A) (Upper panel) HIV DNA PCR amplification from uninfected (NI) and HIV-infected MDM at different times after infection. The 318-bp fragment encompassing the LTR-Gag sequence is indicated (LTR-Gag). (Lower panel) Human β-actin PCR amplification within the same samples. (B) HIV DNA integration in MDM that were infected with HIV-1AD wt or ΔκB. The viral integrated DNA LTR fragment is indicated (LTR). Genomic DNA from ACH2 cells was used as a positive control (lane 2). NI, not infected. (C) (Upper panel) PCR amplification of early HIV transcripts (tat/rev, env) in MDM that were infected with HIV-1AD wt or ΔκB. The amplified 219-bp fragment was visualized by Southern blot hybridization with a γ-32P-labeled U5 LTR internal primer. cDNA from ACH2 was used as a positive control. (Lower panel) Human β-actin hybridization in the same samples.
Based on the above observations, we speculated that the difference in viral replication in MDM between HIV-1AD wt and ΔκB clones must be due to defective transcription of the ΔκB clone. To investigate this, the kinetics of viral transcription at different times after HIV infection were studied by determining the HIV early transcripts of the tat, rev, and env genes. To increase the sensitivity of the method of detection, PCR products were then hybridized with a specific HIV U5 γ-32P-labeled probe and visualized by autoradiography. As shown in Fig. 6C (upper panel, lane 6), HIV-specific early transcripts were present by 69 h after HIV-1AD infection in only the MDM infected with the wt clone. No transcription was detected in MDM infected with the mutant virus, even after 5 days of infection (data not shown). Viral cDNA inputs in this reaction were equivalent, as shown by a comparable hybridization signal to the β-actin probe within the same experimental samples (Fig. 6C, lower panel, lanes 3 to 8). Thus, the lack of viral transcription in the HIV-1AD ΔκB clone accounts for its failure to replicate and hence to persist in human macrophages.
RelB is a trans-activator of HIV LTR via the NF-κB cis-acting motifs.
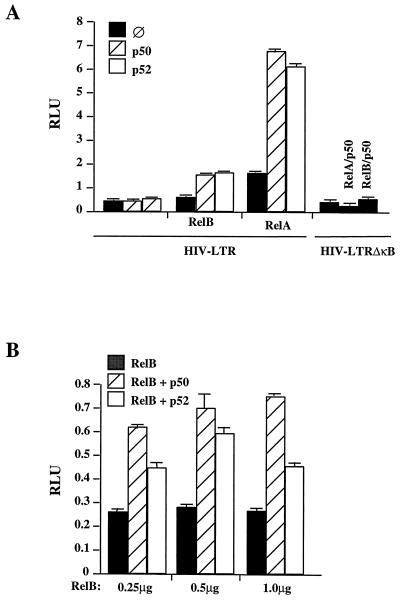
The above results imply that the constitutive pool of NF-κB present in human macrophages, which is mainly composed of RelB, is important in driving HIV LTR transcription via the NF-κB cis-acting motifs. To determine whether RelB is transcriptionally active in driving HIV LTR transcription, Jurkat cells were cotransfected with RelB expression vectors and its dimer NF-κB partners, p50 and p52, and an HIV LTR luciferase reporter gene containing NF-κB cis-acting motifs. As shown in Fig. 7, RelB, in conjunction with either p50 or p52, trans-activates HIV LTR via the NF-κB motifs in a dose-dependent manner, confirming that RelB can exert a positive regulatory activity toward the HIV LTR.
FIG. 7.
RelB is a positive trans-activator of the HIV LTR via the NF-κB cis-acting motifs. (A) Jurkat cells were transfected by FuGENE6 with 450 ng of either HIV LTR wt or an HIV LTR ΔκB-firefly luciferase reporter construct, 50 ng of a Renilla luciferase reporter construct under the control of the TK promoter, 1 μg of RelB, RelA, p50, and/or p52 expression vectors, as indicated, and empty pcDNA3 vector so that each point contained equal amounts of cDNA. Cells were lysed, and luciferase activity was determined by luminescence. Readings are expressed in RLU equivalent to expression of firefly luciferase normalized to the constitutively expressed Renilla luciferase. (B) Jurkat cells were transfected by FuGENE6 with 450 ng of an HIV LTR-firefly luciferase reporter construct, 50 ng of a Renilla luciferase reporter construct under the control of a TK promoter, 1 μg of p50 or p52 expression vector, and increasing amounts (0.25, 0.5, and 1.0 μg) of RelB expression vector, as indicated, along with empty pcDNA3 vector so that each point contained equal amounts of cDNA. Cells were lysed, and luciferase activity was determined by luminescence. Readings are expressed in RLU equivalent to expression of firefly luciferase normalized to the constitutively expressed Renilla luciferase.
DISCUSSION
Using monocytic cells and primary macrophages, we have established that the NF-κB cis-acting motifs present in the HIV LTR play an essential role in the first cycle of HIV transcription. To a lesser extent, our studies also support the important role of RelB in driving HIV transcription in human macrophages. Several mechanisms have been postulated to explain how NF-κB regulates HIV reactivation from latency, especially in quiescent T cells (17, 28). Latent viral infection is in part maintained by a lack of cellular transcription factors needed for the induction of HIV early regulatory genes. Activation of these proteins by extracellular stimuli could rescue episomal or integrated proviral DNA to generate transcriptionally active viruses. Thus, upon T-cell activation, DNA binding of specific combinations of Rel family proteins, particularly RelA/p50, that are induced to translocate to the nucleus, regulates the expression of HIV, leading to productive viral infection (33). In addition to T cells harboring latent HIV, cells of the monocyte/macrophage lineage also provide for a pool of HIV in infected individuals (19). In comparison to T cells, differentiated macrophages are unique because under resting conditions they already express a constitutive nuclear pool of NF-κB. Based on the present study, this pool of NF-κB appears to be important to allow for a basal level of HIV transcription and replication in the absence of cell stimulation.
Several experimental approaches used here have led us to conclude that NF-κB heterodimers already present in the nuclei of the host cell participate in initiating transcription upon HIV infection. Attempts at inhibiting the nuclear pool of constitutive NF-κB with a dominant-negative IκBα transgene suggests a key role of NF-κB. However, the IκBα-negative dominant transgene, when used in either promonocytic cells or human macrophages, was only partially effective in inhibiting HIV transcription. This may be secondary to the fact that the IκBα mutant only partially blocked the constitutive pool of NF-κB in the nuclei. The fact that an HIV provirus containing mutations in the κB sites is unable to transcribe in human macrophages suggests that IκBα-insensitive NF-κB members (RelB) or other transcription factors that could bind to the NF-κB motifs in the HIV LTR may be important. The former is documented in the present studies, although its functional relevance will be determined only when a RelB inhibitor is identified and functionally characterized. The second possibility was in part excluded by addressing the potential candidate role of NFAT family members. However, at this time, we cannot exclude the possibility that other, non-NFAT transcription factors may also influence HIV LTR transcription in differentiated macrophages via the NF-κB cis-acting motifs in conjunction with NF-κB.
In summary, this study extends data generated with primary resting CD4 T cells, in which HIV LTR transcription will ensue only with functional NF-κB. The main difference is that human macrophages already, in the absence of specific cellular activation, contain a pool of NF-κB in the nucleus. This pool, which is mainly composed of RelB, can, alone or in combination with other, non-NF-κB transcription factors, exert a positive transcription on the HIV gene.
ACKNOWLEDGMENTS
This work was supported by NIH grant R01-AI36076.
We thank the members of the Paya laboratory for helpful discussions and criticisms, those authors cited in Materials and Methods who provided us with invaluable reagents, and Teresa Hoff for manuscript preparation.
REFERENCES
- 1.Alcami J, Lain de Lera T, Folgueira L, Pedraza M A, Jacque J M, Bachelerie F, Noriega A R, Hay R T, Harrich D, Gaynor R B, et al. Absolute dependence on κB responsive elements for initiation and Tat-mediated amplification of HIV transcription in blood CD4 T lymphocytes. EMBO J. 1995;14:1552–1560. doi: 10.1002/j.1460-2075.1995.tb07141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asin S, Taylor J A, Trushin S, Bren G, Paya C V. Iκκ mediates NF-κB activation in human immunodeficiency virus-infected cells. J Virol. 1999;73:3893–3903. doi: 10.1128/jvi.73.5.3893-3903.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachelerie F, Alcami J, Arenzana-Seisdedos F, Virelizier J L. HIV enhancer activity perpetuated by NF-κB induction on infection of monocytes. Nature. 1991;350:709–712. doi: 10.1038/350709a0. [DOI] [PubMed] [Google Scholar]
- 4.Baldwin A S., Jr The NF-κB and IκB proteins: new discoveries and insights. Annu Rev Immunol. 1996;14:649–683. doi: 10.1146/annurev.immunol.14.1.649. [DOI] [PubMed] [Google Scholar]
- 5.Beg A A, Baldwin A S., Jr The IκB proteins: multifunctional regulators of Rel/NF-κB transcription factors. Genes Dev. 1993;7:2064–2070. doi: 10.1101/gad.7.11.2064. [DOI] [PubMed] [Google Scholar]
- 6.Carrasco D, Ryseck R P, Bravo R. Expression of relB transcripts during lymphoid organ development: specific expression in dendritic antigen-presenting cells. Development. 1993;118:1221–1231. doi: 10.1242/dev.118.4.1221. [DOI] [PubMed] [Google Scholar]
- 7.Chen B K, Feinberg M B, Baltimore D. The κB sites in the human immunodeficiency virus type 1 long terminal repeat enhance virus replication yet are not absolutely required for viral growth. J Virol. 1997;71:5495–5504. doi: 10.1128/jvi.71.7.5495-5504.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen B K, Saksela K, Andino R, Baltimore D. Distinct modes of human immunodeficiency virus type 1 proviral latency revealed by superinfection of nonproductively infected cell lines with recombinant luciferase-encoding viruses. J Virol. 1994;68:654–660. doi: 10.1128/jvi.68.2.654-660.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cullen B R. Regulation of HIV-1 gene expression. FASEB J. 1991;5:2361–2368. doi: 10.1096/fasebj.5.10.1712325. [DOI] [PubMed] [Google Scholar]
- 10.DeLuca C, Roulston A, Koromilas A, Wainberg M A, Hiscott J. Chronic human immunodeficiency virus type 1 infection of myeloid cells disrupts the autoregulatory control of the NF-κB/Rel pathway via enhanced IκBα degradation. J Virol. 1996;70:5183–5193. doi: 10.1128/jvi.70.8.5183-5193.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fauci A S. Host factors and the pathogenesis of HIV-induced disease. Nature. 1996;384:529–534. doi: 10.1038/384529a0. [DOI] [PubMed] [Google Scholar]
- 13.Finzi D, Hermankova M, Pierson T, Carruth L M, Buck C, Chaisson R E, Quinn T C, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho D D, Richman D D, Siliciano R F. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 14.Folks T, Powell D M, Lightfoote M M, Benn S, Martin M A, Fauci A S. Induction of HTLV-III/LAV from a nonvirus-producing T-cell line: implications for latency. Science. 1986;231:600–602. doi: 10.1126/science.3003906. [DOI] [PubMed] [Google Scholar]
- 15.Folks T M, Justement J, Kinter A, Dinarello C A, Fauci A S. Cytokine-induced expression of HIV-1 in a chronically infected promonocyte cell line. Science. 1987;238:800–802. doi: 10.1126/science.3313729. [DOI] [PubMed] [Google Scholar]
- 16.Foxwell B, Browne K, Bondeson J, Clarke C, de Martin R, Brennan F, Feldmann M. Efficient adenoviral infection with IκBα reveals that macrophage tumor necrosis factor α production in rheumatoid arthritis is NF-κB dependent. Proc Natl Acad Sci USA. 1998;95:8211–8215. doi: 10.1073/pnas.95.14.8211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia-Blanco M A, Cullen B R. Molecular basis of latency in pathogenic human viruses. Science. 1991;254:815–820. doi: 10.1126/science.1658933. [DOI] [PubMed] [Google Scholar]
- 18.Gartner S, Markovits P, Markovitz D M, Kaplan M H, Gallo R C, Popovic M. The role of mononuclear phagocytes in HTLV-III/LAV infection. Science. 1986;233:215–219. doi: 10.1126/science.3014648. [DOI] [PubMed] [Google Scholar]
- 19.Gendelman H E, Orenstein J M, Baca L M, Weiser B, Burger H, Kalter D C, Meltzer M S. The macrophage in the persistence and pathogenesis of HIV infection. AIDS. 1989;3:475–495. doi: 10.1097/00002030-198908000-00001. [DOI] [PubMed] [Google Scholar]
- 20.Griffin G E, Leung K, Folks T M, Kunkel S, Nabel G J. Activation of HIV gene expression during monocyte differentiation by induction of NF-κB. Nature. 1989;339:70–73. doi: 10.1038/339070a0. [DOI] [PubMed] [Google Scholar]
- 21.Jacque J M, Fernandez B, Arenzana-Seisdedos F, Thomas D, Baleux F, Virelizier J L, Bachelerie F. Permanent occupancy of the human immunodeficiency virus type 1 enhancer by NF-κB is needed for persistent viral replication in monocytes. J Virol. 1996;70:2930–2938. doi: 10.1128/jvi.70.5.2930-2938.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karin M. How NF-κB is activated: the role of the IκB kinase (IKK) complex. Oncogene. 1999;18:6867–6874. doi: 10.1038/sj.onc.1203219. [DOI] [PubMed] [Google Scholar]
- 23.Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu Rev Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 24.Kaufman P A, Weinberg J B, Greene W C. Nuclear expression of the 50- and 65-kD Rel-related subunits of nuclear factor-κB is differentially regulated in human monocytic cells. J Clin Investig. 1992;90:121–129. doi: 10.1172/JCI115824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kinoshita S, Su L, Amano M, Timmerman L A, Kaneshima H, Nolan G P. The T cell activation factor NF-ATc positively regulates HIV-1 replication and gene expression in T cells. Immunity. 1997;6:235–244. doi: 10.1016/s1074-7613(00)80326-x. [DOI] [PubMed] [Google Scholar]
- 26.Koenig S, Gendelman H E, Orenstein J M, Dal Canto M C, Pezeshkpour G H, Yungbluth M, Janotta F, Aksamit A, Martin M A, Fauci A S. Detection of AIDS virus in macrophages in brain tissue from AIDS patients with encephalopathy. Science. 1986;233:1089–1093. doi: 10.1126/science.3016903. [DOI] [PubMed] [Google Scholar]
- 27.Leonard J, Parrott C, Buckler-White A J, Turner W, Ross E K, Martin M A, Rabson A B. The NF-κB binding sites in the human immunodeficiency virus type 1 long terminal repeat are not required for virus infectivity. J Virol. 1989;63:4919–4924. doi: 10.1128/jvi.63.11.4919-4924.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCune J M. Viral latency in HIV disease. Cell. 1995;82:183–188. doi: 10.1016/0092-8674(95)90305-4. [DOI] [PubMed] [Google Scholar]
- 29.McElhinny J A, MacMorran W S, Bren G D, Ten R M, Israel A, Paya C V. Regulation of IκBα and p105 in monocytes and macrophages persistently infected with human immunodeficiency virus. J Virol. 1995;69:1500–1509. doi: 10.1128/jvi.69.3.1500-1509.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meltzer M S, Skillman D R, Hoover D L, Hanson B D, Turpin J A, Kalter D C, Gendelman H E. Macrophages and the human immunodeficiency virus. Immunol Today. 1990;11:217–223. doi: 10.1016/0167-5699(90)90086-o. [DOI] [PubMed] [Google Scholar]
- 31.Nabel G, Baltimore D. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature. 1987;326:711–713. doi: 10.1038/326711a0. . (Erratum, 344:178, 1990.) [DOI] [PubMed] [Google Scholar]
- 32.Neumann M, Fries H, Scheicher C, Keikavoussi P, Kolb-Maurer A, Brocker E, Serfling E, Kampgen E. Differential expression of Rel/NF-κB and octamer factors is a hallmark of the generation and maturation of dendritic cells. Blood. 2000;95:277–285. [PubMed] [Google Scholar]
- 33.Oakes J W, Bagasra O, Duan L, Pomerantz R J. Association of alterations in NF-κB moieties with HIV type 1 proviral latency in certain monocytic cells. AIDS Res Hum Retrovir. 1994;10:1213–1219. doi: 10.1089/aid.1994.10.1213. [DOI] [PubMed] [Google Scholar]
- 34.Paya C V, Ten R M, Bessia C, Alcami J, Hay R T, Virelizier J L. NF-κB-dependent induction of the NF-κB p50 subunit gene promoter underlies self-perpetuation of human immunodeficiency virus transcription in monocytic cells. Proc Natl Acad Sci USA. 1992;89:7826–7830. doi: 10.1073/pnas.89.16.7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perkins N D, Edwards N L, Duckett C S, Agranoff A B, Schmid R M, Nabel G J. A cooperative interaction between NF-κB and Sp1 is required for HIV-1 enhancer activation. EMBO J. 1993;12:3551–3558. doi: 10.1002/j.1460-2075.1993.tb06029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Psallidopoulos M C, Schnittman S M, Thompson L M D, Baseler M, Fauci A S, Lane H C, Salzman N P. Integrated proviral human immunodeficiency virus type 1 is present in CD4+ peripheral blood lymphocytes in healthy seropositive individuals. J Virol. 1989;63:4626–4631. doi: 10.1128/jvi.63.11.4626-4631.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raziuddin J, Mikovits A, Calvert I, Ghosh S, Kung H F, Ruscetti F W. Negative regulation of human immunodeficiency virus type 1 expression in monocytes: role of the 65-kDa plus 50-kDa NF-κB dimer. Proc Natl Acad Sci USA. 1991;88:9426–9430. doi: 10.1073/pnas.88.21.9426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rosenberg Z F, Fauci A S. Induction of expression of HIV in latently or chronically infected cells. AIDS Res Hum Retrovir. 1989;5:1–4. doi: 10.1089/aid.1989.5.1. [DOI] [PubMed] [Google Scholar]
- 39.Ross E K, Buckler-White A J, Rabson A B, Englund G, Martin M A. Contribution of NF-κB and Sp1 binding motifs to the replicative capacity of human immunodeficiency virus type 1: distinct patterns of viral growth are determined by T-cell types. J Virol. 1991;65:4350–4358. doi: 10.1128/jvi.65.8.4350-4358.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roulston A, Beauparlant P, Rice N, Hiscott J. Chronic human immunodeficiency virus type 1 infection stimulates distinct NF-κB/rel DNA binding activities in myelomonoblastic cells. J Virol. 1993;67:5235–5246. doi: 10.1128/jvi.67.9.5235-5246.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schuitemaker H, Kootstra N A, Koppelman M H, Bruisten S M, Huisman H G, Tersmette M, Miedema F. Proliferation-dependent HIV-1 infection of monocytes occurs during differentiation into macrophages. J Clin Investig. 1992;89:1154–1160. doi: 10.1172/JCI115697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sen J, Venkataraman L, Shinkai Y, Pierce J W, Alt F W, Burakoff S J, Sen R. Expression and induction of nuclear factor-κB-related proteins in thymocytes. J Immunol. 1995;154:3213–3221. [PubMed] [Google Scholar]
- 43.Suzan M, Salaun D, Neuveut C, Spire B, Hirsch I, Le Bouteiller P, Querat G, Sire J. Induction of NF-κB during monocyte differentiation by HIV type 1 infection. J Immunol. 1991;146:377–383. [PubMed] [Google Scholar]
- 44.Theodore T S, Englund G, Buckler-White A, Buckler C E, Martin M A, Peden K W. Construction and characterization of a stable full-length macrophage-tropic HIV type 1 molecular clone that directs the production of high titers of progeny virions. AIDS Res Hum Retrovir. 1996;12:191–194. doi: 10.1089/aid.1996.12.191. [DOI] [PubMed] [Google Scholar]
- 45.Wu B Y, Woffendin C, Duckett C S, Ohno T, Nabel G J. Regulation of human retroviral latency by the NF-κB/IκB family: inhibition of human immunodeficiency virus replication by IκB through a Rev-dependent mechanism. Proc Natl Acad Sci USA. 1995;92:1480–1484. doi: 10.1073/pnas.92.5.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu B Y, Woffendin C, MacLachlan I, Nabel G J. Distinct domains of IκB-α inhibit human immunodeficiency virus type 1 replication through NF-κB and Rev. J Virol. 1997;71:3161–3167. doi: 10.1128/jvi.71.4.3161-3167.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zack J A, Arrigo S J, Weitsman S R, Go A S, Haislip A, Chen I S. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell. 1990;61:213–222. doi: 10.1016/0092-8674(90)90802-l. [DOI] [PubMed] [Google Scholar]