Abstract
Filaments in the cell commonly treadmill. Driven by energy consumption, they grow on one end while shrinking on the other, causing filaments to appear motile even though individual proteins remain static. This process is characteristic of cytoskeletal filaments and leads to collective filament self-organization. Here we show that treadmilling drives filament nematic ordering by dissolving misaligned filaments. Taking the bacterial FtsZ protein involved in cell division as an example, we show that this mechanism aligns FtsZ filaments in vitro and drives the organization of the division ring in living Bacillus subtilis cells. We find that ordering via local dissolution also allows the system to quickly respond to chemical and geometrical biases in the cell, enabling us to quantitatively explain the ring formation dynamics in vivo. Beyond FtsZ and other cytoskeletal filaments, our study identifies a mechanism for self-organization via constant birth and death of energy-consuming filaments.
Subject terms: Biological physics, Computational biophysics
Treadmilling of cytoskeletal filaments is crucial for their functional self-organization. Now the mechanism underpinning this collective organization is shown to be the dissolution of misaligned filaments.
Main
Cytoskeletal filaments are active cellular elements that continuously grow and shrink through addition and removal of subunits. One prominent example of this activity is treadmilling—a process by which protein filaments grow on one end while the other end shrinks at an equal rate. This dynamic behaviour is driven by nucleotide hydrolysis and results in the filaments seemingly moving, despite the individual filament monomers remaining in one place1. Many essential cytoskeletal filaments, such as actin, microtubules and FtsZ, exhibit this important feature2–6.
From a physics point of view, treadmilling has been often modelled as self-propulsion, where the filament centre of mass moves directionally with a certain velocity to mimic the directional filament growth7–9. While this approach can be appropriate for describing single-filament dynamics, when it comes to the assembly into higher order dynamic structures, self-propelled models might not be suitable. Such models fail to capture the characteristic constant turnover of components in treadmilling systems and by introducing propelling forces may overestimate the forces generated by filament polymerization on obstacles, which have been shown to be small or negligible10 and instead likely operate via a ratchet-like interplay between polymerization and thermal fluctuations11. The self-organization mechanisms of treadmilling polymers may hence be different from those of self-propelled filaments and remain unexplored.
Here we develop a computational model for the collective behaviour of treadmilling filaments, accounting for the kinetics of nucleation, growth and shrinkage, to study their collective dynamics. We focus on investigating the self-organization of FtsZ filaments, a highly conserved tubulin homologue12, widely present in bacteria and one of the best characterized treadmilling systems in the literature13–16. Our model allows us to explore the interplay between polymerization dynamics and collective filament organization at the cellular scale and to directly compare to in vitro and in vivo experiments. Treadmilling FtsZ filaments self-organize into a dynamic ring17 in the middle of bacterial cells (the ‘Z-ring’) that orchestrates bacterial cell division18–20. Treadmilling has been shown to be essential for cell division of Escherichia coli, Bacillus subtilis, Staphylococcus aureus and Streptococcus pneumoniae15,16,21,22, and in B. subtilis, it has been shown to be required for the formation of a coherent Z-ring23. However, how treadmilling contributes to the self-organization of individual filaments into a large-scale cytoskeletal structure remains unclear.
We first show that our model correctly reproduces single-filament treadmilling dynamics collected in vitro13 and proposed in previous theoretical kinetic studies24. We then demonstrate that treadmilling polymers collectively align due to shrinkage and ‘death’ of misaligned filaments, yielding quantitative matching with high-speed atomic force microscopy (HS-AFM) imaging of reconstituted FtsZ filaments. Finally, we find that such a system forms tight ordered dynamic rings when under external biases present in bacterial division. The model quantitatively explains the time-dependent in vivo dynamics of Z-ring condensation and maturation23,25 in B. subtilis. These results identify a mechanism for ordering of cytoskeletal filaments through dynamic growing and shrinking, which is responsible for the formation of the bacterial division ring and potentially present in a plethora of other cytoskeletal processes across the tree of life.
Model description
We model treadmilling filaments as coarse-grained polymers made of spherical beads with diameter σ (the simulation unit of length) on a two-dimensional plane with periodic boundary conditions26. Individual beads correspond to distinct monomers, with σ = 5 nm as known for FtsZ19,27,28. Filaments are formed by connecting monomers via harmonic springs and angle potentials to capture filament stiffness (see Supplementary Information for more details). We time-evolve our system in molecular dynamics simulations and impose filament nucleation, growth and shrinkage kinetics by dynamically creating and deleting monomers in the system (Fig. 1a and Supplementary Video 1; see Supplementary Information for more details).
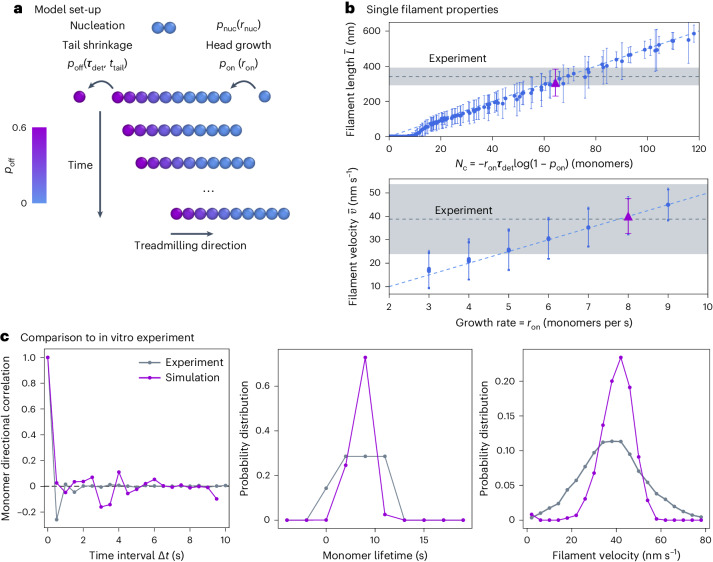
Fig. 1. Coarse-grained model for treadmilling filaments.
a, New filaments nucleate at a constant rate rnuc, grow with probability pon = rondt and shrink with probability poff, determined by the monomer detachment time and the time the monomer has been in the system. This growth and shrinkage dynamics results in the apparent directional motion of the filaments over time. b, Upper, single filament length against the corresponding intrinsic size Nc. The blue dashed line is the theoretical (σ = 5 nm). Lower, average treadmilling velocity of a filament against the corresponding growth rate ron. The blue dashed line is the theoretical . In both graphs, each point is the steady-state average over 20 replicas of single-filament simulations with error bars for the standard error of the average. The grey dashed lines correspond to the fluorescence microscopy experiments of in vitro reconstituted E. coli FtsZ treadmilling filaments on SLBs at 1.25 μM (shaded areas for error). Purple triangles correspond to ron = 8 monomers per second and s, which fit experimental measurements best and are used for the simulation data in c. c, Single-filament dynamics in simulations (in purple) quantitatively matches in vitro FtsZ dynamics (in grey). Left, directional autocorrelation of FtsZ monomers at increasing time intervals Δt decays quickly, showing the static nature of individual monomers. Middle, monomer lifetime distribution. Right, treadmilling filament velocity distribution.
Treadmilling filaments are active, dissipating energy via nucleotide hydrolysis at the monomer–monomer interface. This results in a change in the interface domains of the monomers, which then tend to dissociate when they reach the tail of the filament24,29–31. To capture these properties, we consider reactions at time intervals dtreact = 0.1 s during which individual filaments grow and shrink with probabilities pon and poff respectively, and new filaments are nucleated with probability pnuc. Treadmilling depends only on two parameters: the head polymerization rate, ron, and the typical time over which monomers become available for detachment as a result of hydrolysis, . Here ron (in monomers per second) captures both the bulk free monomer concentration, which we assume is constant, and the polymerization rate constant, such that pon = rondtreact. Monomers in solution are thus implicitly considered. Importantly, filament polymerization is allowed only if free space for monomer addition is available around the filament head. Tail depolymerization is modelled through , where (in seconds) effectively accounts for both the slow nucleotide hydrolysis at the monomer–monomer interface in the filament and the fast monomer dissociation at the filament tail, while ttail is the time for which the tail monomer has been in the system (see Supplementary Fig. 3 for a detailed exploration of the hydrolysis and dissociation reactions). Given that monomers detach into solution upon depolymerization, poff is not dependent on the environment of the filament on the surface32. To reach steady state, pon = poff must be satisfied, which defines an intrinsic size around which filaments fluctuate while treadmilling at constant velocity vc = ronσ. This also defines a typical monomer lifetime . Finally, insertion of new filament nuclei into the system (modelled as dimers) is controlled by the imposed nucleation rate rnuc (in nuclei per second), such that pnuc = rnucdtreact. Like the growth rate, rnuc captures both the nucleation rate constant and the free monomer concentration.
Single-filament dynamics
We first perform simulations of individual filaments for a range of filament growth and detachment time parameter sets . We initialize the system with a single filament nucleus and let it evolve in time (Supplementary Video 1). As expected, filaments reach a steady state where they fluctuate around a constant length and treadmilling velocity (Fig. 1b), while the individual monomers display finite lifetimes (Supplementary Figs. 4 and 5). Experimentally, FtsZ protofilaments have been measured (in vivo and in vitro) to be between 100 and 500 nm long and to treadmill at speeds between 15 and 50 nm s−1, while individual monomers turnover at lifetimes around 8 s, depending on the bacterial strain and conditions25,28,33–35, all of which are well within the range we observe in simulations (Fig. 1b).
We now turn to a direct comparison of our model with experiments. For this purpose, we reconstituted E. coli FtsZ filaments (FtsA and fluorescently labelled FtsZ) on supported lipid bilayers (SLBs) with excess adenosine triphosphate and guanosine triphosphate and used total internal reflection fluorescence microscopy to image treadmilling dynamics and filament organization (see Supplementary Information for further details). Using previously developed image analysis software36,37 we measure filament lengths and velocities (grey dashed lines and shaded regions in Fig. 1b, also reported in ref. 36) as well as individual monomer lifetimes (Supplementary Figs. 4 and 5). When choosing model parameters that match this specific experimental system (the purple triangles in Fig. 1b), the velocity and lifetime distributions also display a good agreement with the experimental data (Fig. 1c, middle and right).
Importantly, the directional autocorrelation function of individual monomers for different time intervals Δt, measured both in simulations and in in vitro experiments, presents a sharp decay to zero (Fig. 1c, left), indicating that individual monomers remain static even though the filament as a whole appears to be moving. The same is also clearly visible from the individual molecule track analysis in vitro (Supplementary Fig. 7). This lack of motility is an important feature of treadmilling filaments, which also suggests that modelling treadmilling as self-propulsion is likely unsuitable, as individual monomers do not display any directional motion. Consequently, pushing forces exerted by treadmilling filaments might be small, as previously discussed for actin bundles10. Together, these results demonstrate that our model correctly captures the main aspects of treadmilling dynamics on biologically relevant time and length scales and that it can be benchmarked for a specific protein under particular conditions and used for quantitative comparison with experiments.
Collective filament dynamics
We are now in a position to explore the collective dynamics of many treadmilling filaments and how their mortality affects the formation of higher order structures. We nucleate filaments in the system, characterized by parameters , at a constant nucleation rate rnuc. The system generally reaches steady-state surface density ρ and average filament size while the filament population undergoes continuous turnover (Supplementary Fig. 8). We observe three different collective regimes (Fig. 2a), which depend exclusively on the treadmilling kinetics or the resulting intrinsic filament size Nc (Supplementary Figs. 8–10).
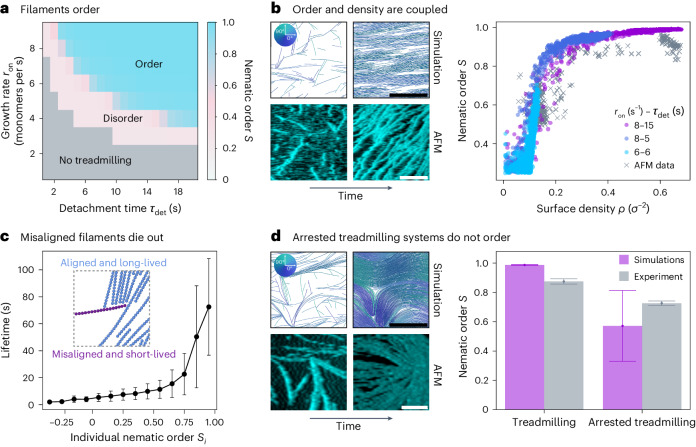
Fig. 2. Collective behaviour of treadmilling filaments.
a, Steady-state nematic order parameter S measured in simulations (average over N = 10 replicas, t > 10 min). Grey indicates the quasi-empty systems of non-treadmilling filaments, blue the nematically ordered system with polar bands and pink the disordered transitional region. b, Left, snapshots of treadmilling systems at early (leftmost column) and late (rightmost column) times in simulations and in HS-AFM experiments (filaments are coloured according to their orientation—see the colour wheel in the inset). Right, nematic order S plotted against surface density ρ at different points in time for reconstituted FtsZ on SLBs (AFM data is shown as crosses) and simulations (coloured according to kinetics and shown as circles). For experiments, data from N = 3 different videos is shown. For simulations, N = 10 different trajectories are shown for each parameter set. c, Filament lifetime for different levels of alignment (characterized by the individual nematic order Si). We consider only simulations for Nc ≥ 100 (N = 10 replicas per parameter set), which results in 226,077 data points (filaments) in total, binned in Si with width 0.1. Points correspond to the mean of the bin and the error bar to the standard deviation. Inset, snapshot of a trapped misaligned filament (purple) that dissolves over time (Supplementary Video 7). d, Left, snapshots of systems with arrested treadmilling at early (leftmost column) and late (rightmost column) times in simulations (same kinetic parameters as in b and in HS-AFM experiments). Right, steady-state nematic order S for treadmilling and non-treadmilling systems. Bars and dots indicate the average for t > 600 s (N = 10 replicas in simulations: n = 600 data points in simulations, n = 28 data points for treadmilling experiments and n = 48 for arrested treadmilling experiments), and error bars indicate the standard deviation. In experiments, ‘treadmilling’ corresponds to wild-type data and ‘arrested treadmilling’ to the L169R mutant. Unless stated otherwise, simulation data corresponds to ron = 8 s−1, and rnuc = 1 s−1 for lp = 10 μm and D = 100 nm2 s−1. Scale bars, 500 nm.
The first regime occurs for very low values of the intrinsic filament size Nc (grey area in Fig. 2a), where individual filaments die out rapidly (Supplementary Fig. 6), resulting in a quasi-zero surface density (Supplementary Figs. 8–10). As the intrinsic filament size increases (Nc ≈ 10) the system transitions into an unstable treadmilling regime, where filament size fluctuations are comparable to their size (Supplementary Fig. 6). The system is then populated by treadmilling filaments that stochastically emerge and disappear, never reaching high surface densities (pink region in Fig. 2a and Supplementary Video 2). Finally, for higher values of Nc (Nc ≫ 10), the intrinsic size fluctuations become negligible compared to the filament size (Supplementary Fig. 6), and filaments enter a highly stable and persistent treadmilling regime in which they can achieve long lifetimes. This allows the system to build up its filament population and reach higher surface densities (Supplementary Figs. 8–10).
We find that the majority of this parameter space is characterized by the emergence of a large-scale nematic order (Fig. 2a,b) and the formation of polar bands (snapshots in Fig. 2b and Supplementary Video 3). This behaviour is highly reminiscent of the nematic laning phase previously observed in self-propelled rods38,39 and is similarly characterized by high values of local and global nematic order parameter S and high local but low global values of the polar order parameter due to lanes formation (Supplementary Fig. 13). Unlike in self-propelled8,38,39 or passive40,41 nematic systems, where the system density determines the ordering, in a treadmilling system the turnover kinetics (characterized by Nc) dictates the emergent steady-state ordering and density (Fig. 2b and Supplementary Fig. 10). We find that if monomer hydrolysis and hence treadmilling are arrested and the filaments grow long, the system will evolve towards a highly populated but disordered configuration (Fig. 2d, Supplementary Fig. 15 and Supplementary Video 4). This occurs because, while thermal fluctuations and collisions can foster local alignment and bundling, the system cannot resolve the nematic defects that stochastically arise as filaments nucleate. Hence, treadmilling serves as an effective mechanism for defect healing.
We now turn to a comparison of collective ordering with experiment. As shown in Fig. 2b, HS-AFM images of E. coli FtsZ filaments on a SLB bear a striking resemblance to simulation snapshots, with an evident transition to a high nematic order as the system becomes more populated (Supplementary Video 5). These high-resolution images allow us to quantify both the orientational order and the surface density of the system at any time from vector field analysis (Supplementary Fig. 16), enabling a quantitative comparison with the simulations. Figure 2b demonstrates that both HS-AFM and simulation trajectories display the same coupling between orientational order and density, independent of the specific kinetic parameters used. Furthermore, when we perform HS-AFM with a FtsZ mutant with reduced GTPase activity and greatly inhibited depolymerization (FtsZ L169R)9, we observe that, just like in simulations, nematic defects are not resolved over time and the system remains at lower nematic order than wild type (Fig. 2d and Supplementary Video 6). These results indicate that our model correctly describes FtsZ treadmilling behaviour as observed experimentally and should therefore provide insight into the mechanisms underlying filament ordering.
In simulations, we find that the ordering transition is driven by filaments growing against each other, which eventually selects for a common alignment direction, like in systems of self-propelled rods38,39,42. However, the underlying mechanism is completely different: unlike self-propelled rods that exert force when collided with another filament43–46, treadmilling filaments stop growing when they reach another object, but still continue to depolymerize, which results in shrinkage, reorientation or even dissolution of the filaments. Ultimately, the fact that shrinkage is independent of growth causes misaligned filaments to die out, because they are more likely to grow against the neighbours than their aligned counterparts. Supplementary Video 7 exemplifies a striking example of such a filament which, by being misaligned with its surroundings, stops growing and eventually dies.
Figure 2c measures the lifetime of each filament against its average alignment, characterized by the individual nematic order Si. These two variables are indeed highly correlated: only very aligned filaments (Si ≈ 1) reach long lifetimes, up to several minutes, while strongly misaligned polymers die out after a few seconds on average and are gradually replaced with aligned ones. The aligned filaments also treadmill with a velocity that approaches their single-filament value vc = ronσ, while the misaligned filaments typically have vanishing velocities (Supplementary Fig. 11). Globally, this behaviour results in the progressive selection of the more stable filaments that align with the rest of the system, reminiscent of what was previously proposed for plant microtubules47,48. The filament death and birth mechanism and the establishment of order are not rapid but require turnover of a large number of filaments and typically take minutes (Supplementary Fig. 12).
Treadmilling drives bacterial division ring condensation
To showcase the importance of the identified self-organization mechanism in living cells, we investigate the role of treadmilling for bacterial division ring formation. Our in vivo experiments recently showed that treadmilling dynamics of FtsZ is essential for Z-ring formation in division of B. subtilis23, but the details linking the two remain elusive. We now use our coarse-grained molecular model for treadmilling filaments to explore the underlying mechanism. Bacterial FtsZ has been observed to display mild natural curvature in live cells and in vitro, with diameters similar to or smaller than typical cell diameters13,32,49,50. In addition, its membrane anchor FtsA—which makes composite polymers with FtsZ—forms intrinsically curved filaments that therefore display a preference for curved surfaces51. This suggests that the interplay between cell geometry and curvature of the (composite) filament should favour filament alignment along the circumference of the cell, similar to what has been previously shown for MreB filaments52–54. We incorporate this curvature-sensing mechanism in the model, as shown in Fig. 3a, by adding a circumference aligning force fcurv to the head and tail monomers of each filament (Supplementary Information). We find that a small force of this kind (fcurv = 5kBT/σ, where kB is the Boltzmann constant and T is the temperature) is sufficient to determine the global orientation of the filaments along the cell circumference (Fig. 3a), highlighting the importance of the interplay between filament and cellular geometry in supramolecular organization.
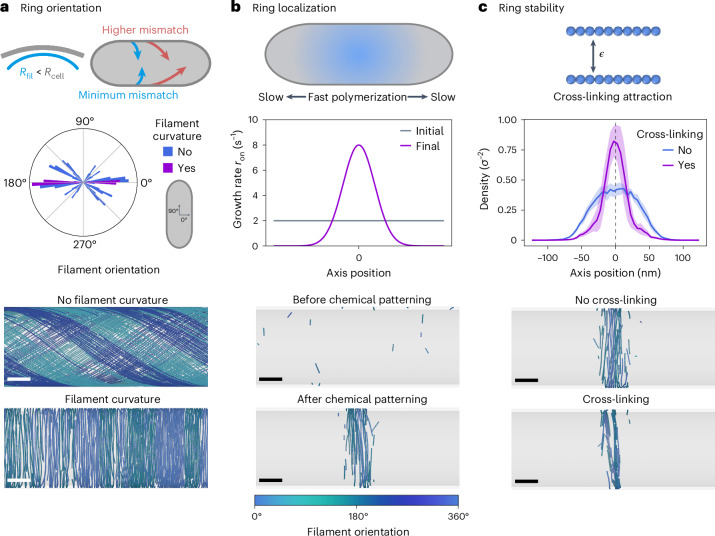
Fig. 3. Formation of the bacterial division ring.
a, Filament curvature and cell geometry drive the collective filament orientation along the cell circumference. Top, illustration of the curvature-sensing mechanism of FtsZ and FtsA filaments. Middle, measured distribution of filament orientations with and without filament curvature. Each bar plot corresponds to the average over N = 10 replicas in the nematic order region of the parameter space (ron = 8 s−1, ). Bottom, two representative snapshots of steady-state configurations of treadmilling filaments with and without filament curvature. b, Spatial modulation of FtsZ's growth and nucleation kinetics mediates its midcell localization. Top, illustration of the typical FtsZ kinetics modulation observed in vivo. Middle, example of a kinetics modulation combining an increase in the growth and nucleation rates around the midcell with a decrease at the poles. Bottom, two representative snapshots of system configurations before and after the modulation of the kinetics is switched on. c, Attractive interactions stabilize the ring and mediate tight packing. Top, illustration of the cross-linking implementation. Middle, late time (t > 8 minutes after the switch) density profiles along the cell axis with and without cross-linking interactions. The solid lines represent the mean over N = 10 replicas and the shaded region, the standard deviation. Bottom, two representative snapshots of steady-state rings with and without cross-linking interactions. In b and c we use switch parameters , and wprof = 100 nm for , where wprof defines the profile width (see Supplementary Information). In all snapshots, the filaments are coloured according to their orientation with respect to the cell circumference (see colour bar at the bottom). For all simulations, we set fcurv = 5kBT/σ, lp = 10 μm and D = 100 nm2 s−1 in a box of size L = 200 σ = 1 μm. Scale bars, 50 nm.
Bacteria have evolved a number of different molecular tools that act in complementary ways to position FtsZ filaments to the right division site—the midcell—at the right time. Such systems include dynamic chemical patterning55–60 and condensate formation61 or nucleoid occlusion62–65 and generally have the effect of spatially modulating FtsZ polymerization, resulting in higher density of filaments at the midcell and lower density around the poles (schematic in Fig. 3b). This effect can be captured by incorporating higher polymerization rates at the midcell and lower rates around the poles. Our model can incorporate this FtsZ growth modulation as an instantaneous change from a uniform distribution along the cell body to a Gaussian profile centred at the midcell for growth and nucleation rates, with a constant detachment time (Fig. 3b and Supplementary Information). We find that the resulting combination of midcell enrichment and cell pole depletion of FtsZ proteins reliably localizes the filaments to the midcell region (Fig. 3b).
Finally, we consider one more model ingredient. It has been shown that FtsZ self-interaction, as well as cross-linking by FtsZ-binding proteins such as ZapA, can promote Z-ring condensation and that cross-linker deletion can lead to axial spreading of the rings25,36,66,67. In line with this, we find that including explicit attractive interactions between filaments has an important stabilizing effect on the ring structure, further promoting filament alignment and tight packing and allowing for a sustained monomer accumulation in the ring over time (Fig. 3c and Supplementary Fig. 18). Taken together, these results show that treadmilling filaments in our model can spontaneously form stable ring-like structures when a combination of filament curvature, attractive interfilament interactions and spatio-temporal modulation of the filament kinetics are considered.
Comparison with in vivo data
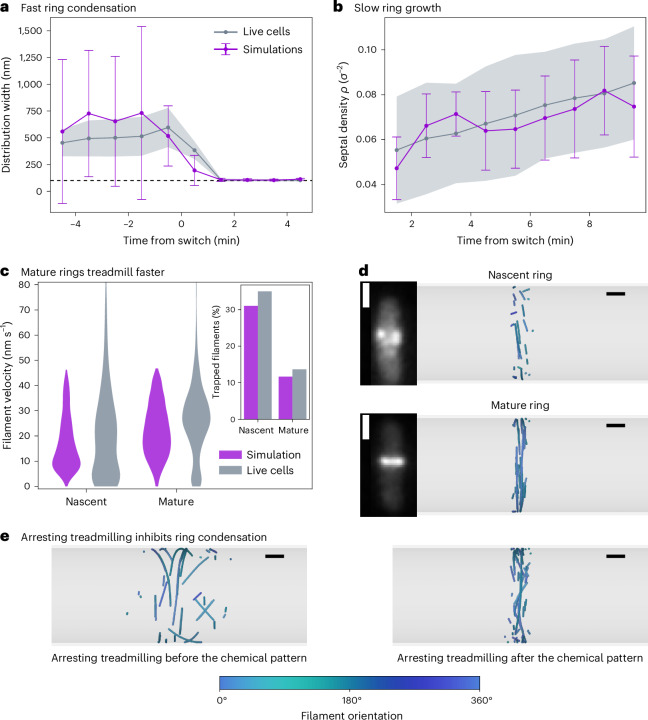
We are now in a good position to compare our model for Z-ring formation with live-cell data. Our experiments have previously directly identified a very particular dynamics of Z-ring formation in living B. subtilis cells: localization of the ring to the midcell, often referred to as ring condensation, occurs very rapidly, within one minute (Fig. 4a), while ring maturation (growth and thickening) occurs over several minutes (Fig. 4b). If treadmilling is impaired early on, the Z-ring cannot form and remains diffuse, while impairing treadmilling during maturation disrupts proper division23. Our experiments also found that the average treadmilling velocity of filaments in mature Z-rings is substantially faster than in nascent ones (Fig. 3c).
Fig. 4. Z-ring formation in vivo.
a, Z-ring condensation dynamics for live cells (grey) and simulations (purple). A rapid ring collapse around t = 0 minutes is observed. b, Ring density in time for live cells (grey) and simulations (purple). A positive average slope is observed, indicating the slow accumulation of proteins to the division site. In a and b, solid lines correspond to the average over samples (N = 10 replicas for simulations, N = 67 cells analysed from three independently prepared bacterial samples) and the shaded region or error bars to the standard deviation. Time t = 0 corresponds to the onset of the chemical pattern. c, Filament velocity distributions in nascent and mature rings for live cells (grey) and simulations (purple). Inset, fraction of trapped filaments (v < 10 nm s−1) for each distribution. In simulations we define t < 2 min for nascent and t > 8 min for mature rings. Statistics are done over n = 1,834 data points (filaments) for simulated nascent rings, n = 2,347 data points (filaments) for simulated mature rings, n = 1,066 data points for nascent in vivo rings and n = 3,126 data points for mature in vivo rings (note that this is the number of filaments imaged but the number of cells is N = 67 biological replicates). d, Representative snapshots of two successive ring configurations observed in vivo and in simulations. Microscopy images show fluorescently tagged FtsZ in live B. subtilis cells. These experiments were repeated independently with similar results. Simulations were repeated for N = 10 replicas with similar results. e, Representative snapshots of systems at t = 10 min after arresting treadmilling. Treadmilling is arrested 5 min before (left) and 1 min after (right) the onset of the chemical pattern that modulates the growth kinetics. The simulation data in a, b and c correspond to a switch with parameters , and for and wprof = 100 nm. Note that monomers in simulations are rendered with size 20 nm instead of the actual 5 nm for visualization purposes and are coloured according to their orientation with respect to the cell circumference (see the colour bar at the bottom). Scale bars, 1 μm (d, insets), 200 nm (d,e).
To explore whether our model can capture the dynamics of Z-ring formation in vivo and explain the mechanism behind it, we simulate systems on the cell scale (L = 3 μm giving R ≈ 1 μm). We limit the total amount of monomers to the estimated amount of FtsZ in B. subtilis Z-rings (Supplementary Information), resulting in a relatively low monomer surface density. Strikingly, as shown in Fig. 4a and Supplementary Video 8, our model also displays rapid ring condensation, which occurs only a few seconds after the switch of the modulation profile. This behaviour is caused by the rapid dying out of the filaments in the growth-inhibited areas of the cell, as their depolymerization rate is faster than the polymerization rate. The whole process occurs on the scale of the single monomer’s lifetime, as controlled by . Hence, treadmilling allows the system to rapidly respond to external chemical cues.
We further find, in agreement with live-cell experiments, that after rapid ring condensation at the site of division, treadmilling filaments in the model accumulate rather slowly, over a time-span of several minutes. The simulations reveal a steadily increasing surface density of monomers in the midcell region that matches well the experimental measurements (Fig. 4b). This feature is the inherent result of treadmilling-driven alignment: the ring structure transitions from an initially disordered state to an ordered state populated by several bundles of filaments tightly bound together (Fig. 4d and Supplementary Video 8) through the dissolution and replacement of misaligned filaments in a process that is analogous to what we observed in vitro for E. coli FtsZ. Mature rings are thus composed of small patches of FtsZ organized in bands that are a few filaments wide and travel together, as previously observed in live-cell imaging19,68,69 (Fig. 4d and Supplementary Video 8). These bands are tightly bound, displaying high local density of monomers, consistent with cryotomography experiments estimating an interfilament distance of approximately 6 nm in FtsZ rings70.
Ring maturation is driven by the replacement of misaligned filaments, which is at heart a trial-and-error process, rendering this process slow. The decrease in the fraction of low-velocity misaligned filaments in the mature ring also naturally leads to faster treadmilling velocities without individual filaments speeding up, which matches live-cell measurements (Fig. 4c). In other words, the surviving filaments all run parallel to each other with velocities close to maximal values. Finally, we found that arresting treadmilling in our model has similar effects to those observed in vivo. If treadmilling is arrested early on, before the onset of the chemical patterning, the filament population remains diffuse and never condenses to the midcell (Fig. 4e and Supplementary Video 9). If treadmilling is arrested after the switch of the chemical patterning, the ring forms at the midcell, but displays aberrant, ‘frozen’ configurations (Fig. 4e, Supplementary Fig. 21 and Supplementary Video 10). The crucial role of treadmilling in the positioning of the FtsZ ring that our model reveals has also been previously discussed in the context of E. coli: it has been reported that mutants that are insensitive to classical positioning systems like Min (FtsZ2, FtsZ9 and FtsZ100) all have very low or undetectable GTPase activity. This implies that the lack of treadmilling underlies their inability to correctly position the ring28.
Altogether, our treadmilling model naturally reproduces the key dynamical aspects of Z-ring formation observed in live cells. FtsZ filaments are able to localize to the midcell on timescales similar to the monomer lifetimes, driven by the underlying spatial modulation of the polymerization rates, while the tightly packed division rings grow on substantially longer timescales by virtue of misaligned filaments dying and growing again until only the aligned ones remain. This mechanism is at play irrespective of the filament density: simulations of treadmilling filaments on a surface constrained to in-vivo-like low protein concentrations show that ordering still occurs and is driven by the same mechanism of death by misalignment (Supplementary Fig. 14). More generally, our model explains the importance of treadmilling—dynamic growth and shrinkage of filaments—in localizing and timing bacterial division.
Discussion
Our treadmilling model reveals a surprisingly rich collective behaviour arising from the dissolution and replacement of misaligned filaments, characteristic of mortal polymers. These findings put forward treadmilling filaments as a paradigm for a dynamic self-ordering system that is able to quickly remodel itself in response to external cues. This suggests that both naturally occurring and artificial treadmilling polymers could be a useful tool in areas such as synthetic cell development or programmable active matter, capable of healing via local filament dissolution. For example, microfluidic devices and chemical or photo patterning could be used experimentally to generate and control dynamic treadmilling filament systems.
From a biological perspective, our model provides structural and dynamic insights into functional FtsZ assemblies. The model can further be used to investigate how the dynamics of the Z-ring affects recruitment and activation of bacterial wall synthesis machinery, resulting in the growth of the cell wall at midcell, which ultimately divides the cell into two15,16,71–73.
From a physics point of view, we find that treadmilling filaments belong to a distinct class of active matter, which consumes energy not for motility but for turnover and thus order via death of locally misaligned filaments. This mechanism is expected to have further implications for filament collective behaviour, such as the response to obstacles, perturbations and propagation of information. For instance, treadmilling filaments can be expected to evacuate the region around obstacles due to their mortality, instead of being trapped. Due to filament dissolution, such systems are also expected to locally heal upon defect introduction or external perturbation and are not expected to transduct the perturbation deep into the assembly. Overall, by revealing a distinct ordering mechanism in chemically active matter, our work highlights the importance of exploring the collective and material properties of explicitly mortal filaments.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41567-024-02597-8.
Supplementary information
Supplementary Figs. 1–21, Methods and extended results and discussion and Videos 1–10.
Acknowledgements
We thank I. Palaia (ISTA) for useful discussions and K. Lim and R. W. Wong (WPI-Nano Life Science Institute, Kanazawa University) for providing access to HS-AFM. We would like to thank B. Prats Mateu (MSD Austria, Vienna) for providing the HS-AFM data. This work was supported by the Royal Society (grant no. UF160266; C.V.-C. and A.Š.), the European Union’s Horizon 2020 Research and Innovation Programme (grant no. 802960; A.Š.), the Austrian Science Fund (FWF) Stand-Alone P34607 (M.L.) and a Wellcome Trust and Royal Society Sir Henry Dale Fellowship (grant no. 206670/Z/17/Z; S.H. and K.D.W.).
Author contributions
C.V.-C. and A.Š. designed the research aims. C.V.-C. designed the model and carried out the simulations and data analysis under supervision of A.Š. P.R. designed and performed the in vitro TIRF microscopy experiments under supervision of M.L. K.D.W. reanalysed the in vivo experimental data under supervision of S.H. A.Š. directed the research. C.V.-C. and A.Š. wrote the manuscript, and all authors commented on it.
Peer review
Peer review information
Nature Physics thanks Harold Erickson and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Data availability
The simulation data presented in this work are available from the University College London public data repository at 10.5522/04/24754527 (ref. 74). Live-cell imaging of FtsZ rings data presented in this work are from ref. 23. High-speed atomic force microscopy data presented in this work are from ref. 9. Total internal reflection fluorescence microscopy data presented in this work are from refs. 37,75,76.
Code availability
Appropriate documentation and example files to replicate the simulation results presented in this work are available from the University College London public data repository at 10.5522/04/24754527 (ref. 74). A maintained version of the code is available on GitHub77. Simulation data analysis was performed using custom Python code, which makes use of several public libraries such as NumPy, pandas or Ovito, also available at ref. 74. ImageJ (current version v.1.54) and Python (current version Jupyter Notebook v.6.5.4) were used for the in vitro data analysis. Live-cell imaging videos were analysed using Fiji (v.1.53 and v.1.54) with open-source plugins PureDenoise, StackReg, MicrobeJ (v.5.13I), ilastik (v.1.3.3post2) and custom code available via GitHub at https://github.com/HoldenLab/Ring_Analysis_IJ). Further in vivo data analysis was done using Matlab with custom code available via GitHub at https://github.com/HoldenLab/ring-fitting2; https://github.com/HoldenLab/violinplusDABEST-Matlab and https://github.com/HoldenLab/ring-simulator.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41567-024-02597-8.
References
- 1.Andreu, J. M. How protein filaments treadmill. Biophys. J.119, 717–720 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wegner, A. Head to tail polymerization of actin. J. Mol. Biol.108, 139–150 (1976). [DOI] [PubMed] [Google Scholar]
- 3.Erlenkämper, C. & Kruse, K. Treadmilling and length distributions of active polar filaments J. Chem. Phys.139, 164907 (2013). [DOI] [PubMed]
- 4.Waterman-Storer, C. M. & Salmon, E. D. Microtubule dynamics: treadmilling comes around again. Curr. Biol.7, R369–R372 (1997). [DOI] [PubMed] [Google Scholar]
- 5.Panda, D., Miller, H. P. & Wilson, L. Rapid treadmilling of brain microtubules free of microtubule-associated proteins in vitro and its suppression by tau. Proc. Natl Acad. Sci. USA96, 12459–12464 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rzadzinska, A. K., Schneider, M. E., Davies, C., Riordan, G. P. & Kachar, B. An actin molecular treadmill and myosins maintain stereocilia functional architecture and self-renewal. J. Cell Biol.164, 887–897 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denk, J., Huber, L., Reithmann, E. & Frey, E. Active curved polymers form vortex patterns on membranes. Phys. Rev. Lett.116, 178301 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Shaebani, M. R., Wysocki, A., Winkler, R. G., Gompper, G. & Rieger, H. Computational models for active matter. Nat. Rev. Phys.2, 181–199 (2020). [Google Scholar]
- 9.Dunajova, Z. et al. Chiral and nematic phases of flexible active filaments. Nat. Phys.19, 1916–1926 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Footer, M. J., Kerssemakers, J. W. J., Theriot, J. A. & Dogterom, M. Direct measurement of force generation by actin filament polymerization using an optical trap. Proc. Natl Acad. Sci. USA104, 2181–2186 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Theriot, J. A. The polymerization motor. Traffic1, 19–28 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Nogales, E., Downing, K. H., Amos, L. A. & Löwe, J. Tubulin and FtsZ form a distinct family of GTPases. Nat. Struct. Biol.5, 451–458 (1998). [DOI] [PubMed] [Google Scholar]
- 13.Loose, M. & Mitchison, T. J. The bacterial cell division proteins FtsA and FtsZ self-organize into dynamic cytoskeletal patterns. Nat. Cell Biol.16, 38–46 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramirez-Diaz, D. A. et al. Treadmilling analysis reveals new insights into dynamic FtsZ ring architecture. PLoS Biol.16, e2004845 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bisson-Filho, A. W. et al. Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science355, 739–743 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang, X. et al. GTPase activity-coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell wall synthesis. Science355, 744–747 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anderson, D. E., Gueiros-Filho, F. J. & Erickson, H. P. Assembly dynamics of FtsZ rings in Bacillus subtilis and Escherichia coli and effects of FtsZ-regulating proteins. J. Bacteriol.186, 5775–5781 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrows, J. M. & Goley, E. D. FtsZ dynamics in bacterial division: what, how, and why? Curr. Opin. Cell Biol.68, 163–172 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McQuillen, R. & Xiao, J. Insights into the structure, function, and dynamics of the bacterial cytokinetic FtsZ-ring. Annu. Rev. Biophys.49, 309–341 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahone, C. R. & Goley, E. D. Bacterial cell division at a glance. J. Cell Sci.133, jcs237057 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monteiro, J. M. et al. Peptidoglycan synthesis drives an FtsZ-treadmilling-independent step of cytokinesis. Nature554, 528–532 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perez, A. J. et al. Movement dynamics of divisome proteins and PBP2x: FtsW in cells of Streptococcus pneumoniae. Proc. Natl Acad. Sci. USA116, 3211–3220 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whitley, K. D. et al. FtsZ treadmilling is essential for Z-ring condensation and septal constriction initiation in Bacillus subtilis cell division. Nat. Commun.12, 2448 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corbin, L. C. & Erickson, H. P. A unified model for treadmilling and nucleation of single-stranded FtsZ protofilaments. Biophys. J.119, 792–805 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Squyres, G. R. et al. Single-molecule imaging reveals that Z-ring condensation is essential for cell division in Bacillus subtilis. Nat. Microbiol.6, 553–562 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hafner, A. E., Krausser, J. & Šarić, A. Minimal coarse-grained models for molecular self-organisation in biology. Curr. Opin. Struct. Biol.58, 43–52 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Löwe, J. & Amos, L. A. Crystal structure of the bacterial cell-division protein FtsZ. Nature391, 203–206 (1998). [DOI] [PubMed] [Google Scholar]
- 28.Erickson, H. P., Anderson, D. E. & Osawa, M. FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol Mol. Biol. Rev.74, 504–28 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wagstaff, J. M. et al. A polymerization-associated structural switch in FtsZ that enables treadmilling of model filaments. mBio8, e00254-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruiz, F. M. et al. FtsZ filament structures in different nucleotide states reveal the mechanism of assembly dynamics. PLoS Biol.20, e3001497 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Du, S., Pichoff, S., Kruse, K. & Lutkenhaus, J. FtsZ filaments have the opposite kinetic polarity of microtubules. Proc. Natl Acad. Sci. USA115, 10768–10773 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Radler, P. et al. In vitro reconstitution of Escherichia coli divisome activation. Nat. Commun.13, 2635 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, Z., Trimble, M. J., Brun, Y. V. & Jensen, G. J. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J.26, 4694–4708 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen, Y. & Erickson, H. P. Rapid in vitro assembly dynamics and subunit turnover of FtsZ demonstrated by fluorescence resonance energy transfer. J. Biol. Chem.280, 22549–22554 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huecas, S. et al. Energetics and geometry of FtsZ polymers: Nucleated self-assembly of single protofilaments. Biophys. J.94, 1796–1806 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caldas, P. et al. Cooperative ordering of treadmilling filaments in cytoskeletal networks of FtsZ and its crosslinker ZapA. Nat. Commun.10, 5744 (2019). [DOI] [PMC free article] [PubMed]
- 37.Caldas, P., Radler, P., Sommer, C. & Loose, M. in Methods in Cell Biology Vol. 158 (ed. Tran, P.) 145–161 (Academic, 2020). [DOI] [PubMed]
- 38.Shi, X. & Chaté, H. Self-propelled rods: linking alignment-dominated and repulsion-dominated active matter. Preprint at 10.48550/arXiv.1807.00294 (2018).
- 39.Großmann, R., Aranson, I. S. & Peruani, F. A particle-field approach bridges phase separation and collective motion in active matter. Nat. Commun.11, 5365 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Onsager, L. The effects of shape on the interaction of colloidal particles. Ann. N. Y. Acad. Sci.51, 627–659 (1949). [Google Scholar]
- 41.Tjipto-Margo, B. & Evans, G. T. The Onsager theory of the isotropic-nematic liquid crystal transition: incorporation of the higher virial coefficients. J. Chem. Phys.93, 4254–4265 (1990). [Google Scholar]
- 42.Perepelitsa, M., Timofeyev, I., Murphy, P. & Igoshin, O. A. Mean-field model for nematic alignment of self-propelled rods. Phys. Rev. E106, 34613 (2022). [DOI] [PubMed] [Google Scholar]
- 43.Schaller, V., Weber, C., Semmrich, C., Frey, E. & Bausch, A. R. Polar patterns of driven filaments. Nature467, 73–77 (2010). [DOI] [PubMed] [Google Scholar]
- 44.Sciortino, A. & Bausch, A. R. Pattern formation and polarity sorting of driven actin filaments on lipid membranes. Proc. Natl Acad. Sci. USA118, e2017047118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Henkin, G., DeCamp, S. J., Chen, D. T. N., Sanchez, T. & Dogic, Z. Tunable dynamics of microtubule-based active isotropic gels. Trans. R. Soc. A372, 20140142 (2014). [DOI] [PMC free article] [PubMed]
- 46.DeCamp, S. J., Redner, G. S., Baskaran, A., Hagan, M. F. & Dogic, Z. Orientational order of motile defects in active nematics. Nat. Mater.14, 1110–1115 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dixit, R. & Cyr, R. Encounters between dynamic cortical microtubules promote ordering of the cortical array through angle-dependent modifications of microtubule behavior. Plant Cell16, 3274–3284 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tindemans, S. H., Hawkins, R. J. & Mulder, B. M. Survival of the aligned: ordering of the plant cortical microtubule array. Phys. Rev. Lett.104, 058103 (2010). [DOI] [PubMed] [Google Scholar]
- 49.Erickson, H. P. Modeling the physics of FtsZ assembly and force generation. Proc. Natl Acad. Sci. USA106, 9238–9243 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ghosh, B. & Sain, A. Origin of contractile force during cell division of bacteria. Phys. Rev. Lett.101, 178101 (2008). [DOI] [PubMed] [Google Scholar]
- 51.Nierhaus, T. et al. Bacterial divisome protein FtsA forms curved antiparallel double filaments when binding to FtsN. Nat. Microbiol.7, 1686–1701 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ouzounov, N. et al. MreB orientation correlates with cell diameter in Escherichia coli. Biophys. J.111, 1035–1043 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hussain, S. et al. MreB filaments align along greatest principal membrane curvature to orient cell wall synthesis. eLife7, e32471 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shi, H., Quint, D. A., Grason, G. M., Gopinathan, A. & Huang, K. C. Chiral twisting in a bacterial cytoskeletal polymer affects filament size and orientation. Nat. Commun.11, 1408 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Loose, M., Fischer-Friedrich, E., Ries, J., Kruse, K. & Schwille, P. Spatial regulators for bacterial cell division self-organize. Science320, 789–792 (2008). [DOI] [PubMed] [Google Scholar]
- 56.Kiekebusch, D., Michie, K. A., Essen, L. O., Löwe, J. & Thanbichler, M. Localized dimerization and nucleoid binding drive gradient formation by the bacterial cell division inhibitor MipZ. Mol. Cell46, 245–259 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arumugam, S., Petrášek, Z. & Schwille, P. MinCDE exploits the dynamic nature of FtsZ filaments for its spatial regulation. Proc. Natl Acad. Sci. USA111, 1192–1200 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zieske, K. & Schwille, P. Reconstitution of self-organizing protein gradients as spatial cues in cell-free systems. eLife3, e03949 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Feddersen, H., Würthner, L., Frey, E. & Bramkamp, M. Dynamics of the Bacillus subtilis Min system mBio12, e00296-21 (2021). [DOI] [PMC free article] [PubMed]
- 60.Corrales-Guerrero, L. et al. MipZ caps the plus-end of FtsZ polymers to promote their rapid disassembly. Proc. Natl Acad. Sci. USA119, e2208227119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ramm, B. et al. Biomolecular condensate drives polymerization and bundling of the bacterial tubulin FtsZ to regulate cell division. Nat. Commun.14, 3825 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu, L. J. & Errington, J. Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell117, 915–925 (2004). [DOI] [PubMed] [Google Scholar]
- 63.Lutkenhaus, J. Linking DNA replication to the Z ring. Nat. Microbiol.6, 1108–1109 (2021). [DOI] [PubMed] [Google Scholar]
- 64.Männik, J. & Bailey, M. W. Spatial coordination between chromosomes and cell division proteins in Escherichia coli. Front. Microbiol.6, 306 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bernhardt, T. G. & De Boer, P. A. J. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol. Cell18, 555–564 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Buss, J. et al. In vivo organization of the FtsZ-ring by ZapA and ZapB revealed by quantitative super-resolution microscopy. Mol. Microbiol.89, 1099–1120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guan, F. et al. Lateral interactions between protofilaments of the bacterial tubulin homolog FtsZ are essential for cell division. eLife7, e35578 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Walker, B. E., Männik, J. & Männik, J. Transient membrane-linked FtsZ assemblies precede Z-ring formation in Escherichia coli. Curr. Biol.30, 499–508 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lyu, Z., Coltharp, C., Yang, X. & Xiao, J. Influence of FtsZ GTPase activity and concentration on nanoscale Z-ring structure in vivo revealed by three-dimensional superresolution imaging. Biopolymers105, 725–734 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Szwedziak, P., Wang, Q., Bharat, T. A. M., Tsim, M. & Löwe, J. Architecture of the ring formed by the tubulin homologue FtsZ in bacterial cell division. eLife3, 04601 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McCausland, J. W. et al. Treadmilling FtsZ polymers drive the directional movement of sPG-synthesis enzymes via a Brownian ratchet mechanism. Nat. Commun.12, 609 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Navarro, P. P. et al. Cell wall synthesis and remodelling dynamics determine division site architecture and cell shape in Escherichia coli. Nat. Microbiol.7, 1621–1634 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Puls, J. S., Brajtenbach, D., Schneider, T., Kubitscheck, U. & Grein, F. Inhibition of peptidoglycan synthesis is sufficient for total arrest of staphylococcal cell division. Sci. Adv.9, 9023 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vanhille-Campos, C. et al. Self-organization of mortal filaments and its role in bacterial division ring formation. University College London10.5522/04/24754527 (2024).
- 75.Baranova, N. & Loose, M. in Cytokinesis, Methods in Cell Biology Vol. 137 (ed. Echard. A.) Ch. 21 (Elsevier, 2017); 10.1016/bs.mcb.2016.03.036
- 76.Baranova, N. et al. Diffusion and capture permits dynamic coupling between treadmilling FtsZ filaments and cell division proteins. Nat. Microbiol.5, 407–417, 10.1038/s41564-019-0657-5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vanhille-Campos, C. et al. Treadmilling filaments. GitHubhttps://github.com/Saric-Group/treadmilling (2024).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figs. 1–21, Methods and extended results and discussion and Videos 1–10.
Data Availability Statement
The simulation data presented in this work are available from the University College London public data repository at 10.5522/04/24754527 (ref. 74). Live-cell imaging of FtsZ rings data presented in this work are from ref. 23. High-speed atomic force microscopy data presented in this work are from ref. 9. Total internal reflection fluorescence microscopy data presented in this work are from refs. 37,75,76.
Appropriate documentation and example files to replicate the simulation results presented in this work are available from the University College London public data repository at 10.5522/04/24754527 (ref. 74). A maintained version of the code is available on GitHub77. Simulation data analysis was performed using custom Python code, which makes use of several public libraries such as NumPy, pandas or Ovito, also available at ref. 74. ImageJ (current version v.1.54) and Python (current version Jupyter Notebook v.6.5.4) were used for the in vitro data analysis. Live-cell imaging videos were analysed using Fiji (v.1.53 and v.1.54) with open-source plugins PureDenoise, StackReg, MicrobeJ (v.5.13I), ilastik (v.1.3.3post2) and custom code available via GitHub at https://github.com/HoldenLab/Ring_Analysis_IJ). Further in vivo data analysis was done using Matlab with custom code available via GitHub at https://github.com/HoldenLab/ring-fitting2; https://github.com/HoldenLab/violinplusDABEST-Matlab and https://github.com/HoldenLab/ring-simulator.