Abstract
MYCN amplification predicts poor prognosis in childhood neuroblastoma. To identify MYCN oncogenic signal dependencies we performed N-ethyl-N-nitrosourea (ENU) mutagenesis on the germline of neuroblastoma-prone TH-MYCN transgenic mice to generate founders which had lost tumorigenesis. Sequencing of the mutant mouse genomes identified the Ring Finger Protein 121 (RNF121WT) gene mutated to RNFM158R associated with heritable loss of tumorigenicity. While the RNF121WT protein localised predominantly to the cis-Golgi Complex, the RNF121M158R mutation in Helix 4 of its transmembrane domain caused reduced RNF121 protein stability and absent Golgi localisation. RNF121WT expression markedly increased during TH-MYCN tumorigenesis, whereas hemizygous RNF121WT gene deletion reduced TH-MYCN tumorigenicity. The RNF121WT-enhanced growth of MYCN-amplified neuroblastoma cells depended on RNF121WT transmembrane Helix 5. RNF121WT directly bound MYCN protein and enhanced its stability. High RNF121 mRNA expression associated with poor prognosis in human neuroblastoma tissues and another MYC-driven malignancy, laryngeal cancer. RNF121 is thus an essential oncogenic cofactor for MYCN and a target for drug development.
Subject terms: Oncogenes, Paediatric cancer
A chemical mutagenesis screen identifies RNF121 as an oncogenic cofactor for MYCN in neuroblastoma.
Introduction
RING finger protein 121 (RNF121) consists of 327 amino acids, with an N-terminal RING domain and six transmembrane domains1. RNF121 is highly conserved from Caenorhabditis elegans to human, and, has been localized to the endoplasmic reticulum and Golgi Complex in different cell types2. RNF121 can promote ubiquitin-mediated proteasome degradation and membrane localization of voltage-gated sodium channels in zebrafish3. RNF121 can also act as a broad regulator of NF-κB signalling4.
Recent studies indicate that RING finger proteins can act as oncogenes or tumour suppressors5,6. They are involved in the regulation of cell cycle, signal transduction and metastasis of cancer cells7. One study showed that RNF121 has a negative regulatory role on renal cell carcinogenesis in vivo and in vitro8. Another study revealed the mechanism of circ (circular)-ring finger protein 121 (circ-RNF121) in colorectal cancer (CRC). Circ-RNF121 silencing repressed cell proliferation, migration, invasion and glycolysis, and induced apoptosis in CRC, which were attenuated by a miR-1224-5p inhibitor9. However, the mechanism by which RNF121 relates to cancer development remains poorly understood and is likely to be context-dependent.
MYCN amplification correlates with poor prognosis in the childhood cancer, neuroblastoma, making MYCN an ideal target for therapeutic intervention10. However, the development of inhibitors directly targeting MYC family proteins (c-Myc, MYCN and L-Myc) has proven challenging, due to the almost complete absence of secondary structure, its nuclear location, and potential side-effects in normal proliferating tissues11,12. Therefore, extensive research has focused on alternative approaches such as indirectly targeting MYCN through protein-binding partners which might serve as alternative therapeutic targets. We have defined increased MYCN protein stability as a necessary feature during MYCN-related tumorigenesis13–16. We recently identified Proliferation-Associated protein 2G4 (PA2G4) as a direct protein-binding partner of MYCN which acts in a forward feedback expression loop with MYCN to drive neuroblastoma tumourigenesis17. We also recently identified an ALYREF-MYCN coactivator complex which drives neuroblastoma tumorigenesis through effects on a deubiquitinase (USP3) and thus MYCN stability18. These studies suggest that direct competitive inhibitors of MYCN binding to these “protective” proteins will have profound anti-cancer effects in neuroblastoma and other MYCN-driven cancers.
Here we used a germline chemical mutagenesis screen to identify a hemizygous loss-of-function RNF121M158R mutation that completely blocked neuroblastoma tumorigenesis in a MYCN transgenic mouse model (TH-MYCN) which normally develops neuroblastoma in 100% of cases. Analyses demonstrated that RNF121 copy number is preferentially conserved in MYCN amplified human neuroblastoma tissues and high RNF121 gene expression associates with poor patient outcome. Mechanistic experiments revealed that RNF121 formed a protein complex with MYCN and enhanced MYCN protein stability. RNF121 expression increased human neuroblastoma cell growth via its transmembrane domain. These findings provide evidence for the future development of RNF121 inhibitors as therapeutics for neuroblastoma.
Results
RNF121M158R loss-of-function mutation blocks neuroblastoma tumorigenesis in TH-MYCN mice
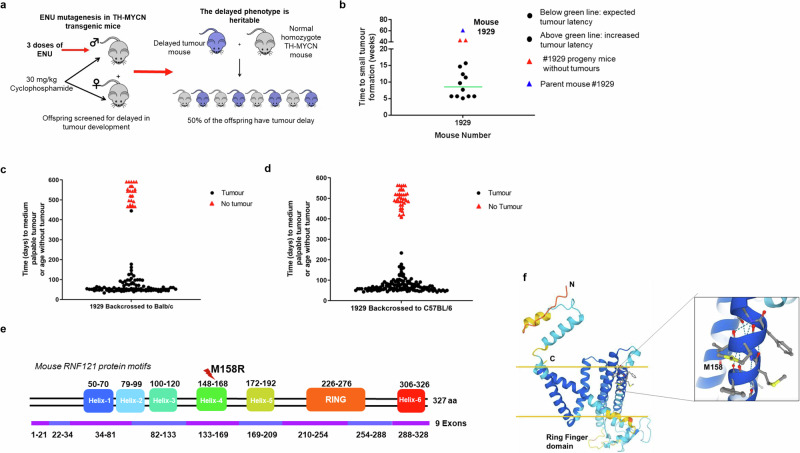
To identify genes whose loss of function suppressed MYCN-driven neuroblastoma, we subjected homozygous male TH-MYCN+/+ mice to random ENU mutagenesis. The model demonstrates a rapid tumorigenic process with 100% of homozygous TH-MYCN+/+ mice developing neuroblastoma in sympathetic ganglia tissues by 6–8 weeks postnatal. To permit ENU mutagenesis, homozygous TH-MYCN+/+ mice were treated with cyclophosphamide at 5–6 weeks of age to inhibit tumour growth as we have previously shown16. The 7–8-week-old male mice were then injected with N-Ethyl-N-Nitrosourea (ENU) to induce random mutations in the sperm genome19,20 (Fig. 1a). ENU-treated male mice were mated to cyclophosphamide-treated, female, homozygous TH-MYCN mice to generate offspring. A total of 81 pair mattings were performed to introduce the random ENU mutations into TH-MYCN+/+ offspring. Mouse #1929 showed no tumour at 61 weeks of age, and this tumour-suppressed phenotype was inherited in its offspring (Fig. 1b). To perform mapping and sequencing, suppressed mice were first backcrossed to Balb/c and C57BL/6 mice, and then crossed to TH-MYCN mice, where only those mice homozygous for the transgene were followed for tumour development. The tumour-suppressed phenotype was retained in crosses with both Balb/c (Fig. 1c) and C57BL/6 (Fig. 1d). Exome sequencing of tumours from progeny, and confirmatory PCR from #1929 mouse backcrossed with Balb/c mice, identified a missense mutation in the Ring Finger Protein 121 (Rnf121) gene as the only candidate mutation which was common to all tumour-suppressed 1929 progeny (Fig. 1e; Supplementary Fig. S1). A 3D model of RNF121 protein, built using AlphaFold221, was examined to predict the effect of the M158R mutation. Methionine 158 is located in transmembrane helix 4. The missense mutation of methionine to arginine at amino acid 158 (RNF121M158R) would introduce a charged residue into the hydrophobic core of the membrane and hence be energetically unstable due to the missense mutation (Fig. 1f).
Fig. 1. RNF121M158R mutation blocks neuroblastoma development in TH-MYCN mice.
a Homozygous TH-MYCN (TH-MYCN+/+) mice were screened for delay in tumour development after ENU chemical treatment. Homozygous TH-MYCN (TH-MYCN+/+) breeder mice were treated for 2 weeks with cyclophosphamide to prevent tumour progression, then subjected to N-ethyl-N-nitrosourea (ENU) chemical mutagenesis of their germline at 6 weeks of age to induce random mutations in their sperm genome. b Tumour latency in 13 offspring of mouse 1929. Approximately half of the progeny developed tumours by the expected 7 weeks postnatal, whereas the remainder exhibited increased tumour latency. A subset of these mice did not develop tumours. Parent mouse 1929, that did not develop a tumour, is shown as a comparison. c, d Mouse 1929 or non-ENU-treated TH-MYCN+/+ mice were crossed with BALB/c (c) and C57BL/6 (d) wild-type mice before backcrossing with Cyclophosphamide-treated TH-MYCN+/+ mice. The mice represented by red triangles were selected for exome sequencing. e An RNF121M158R mutation was identified by exome sequencing of tumours from all 14 offspring with the tumour-suppressed phenotype. f A 3D model of RNF121M158R showing the location of the mutation. The secondary structures with the light and darker blue colours are predicted with the most confidence, whereas the yellow and red colours signify the least confident predictions. The yellow horizontal bars signify the approximate locations of the membrane surfaces. A zoomed in view of the Met 158 microenvironment is shown in the box. Met 158, located in transmembrane helix 4, and surrounding residues are shown in ball-and-stick format.
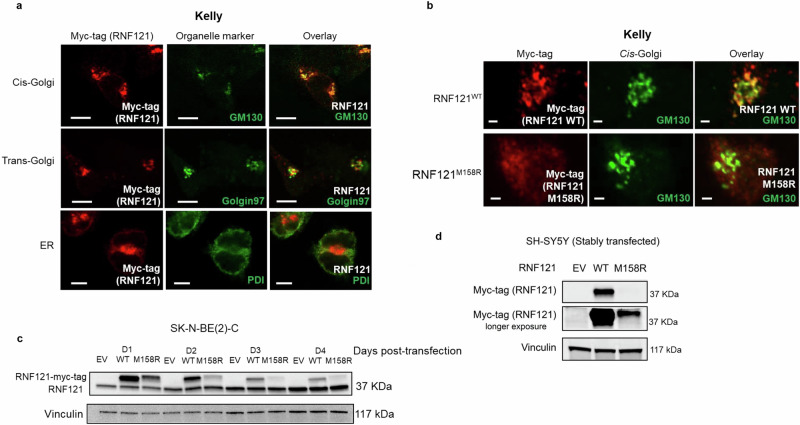
To examine the effect of the RNF121M158R mutation on wild-type RNF121 (RNF121WT) protein localization and expression, Myc-tagged RNF121WT and RNF121M158R vector constructs were overexpressed in human MYCN amplified neuroblastoma tumour cell lines, Kelly and SK-N-BE(2)-C. A previous study in Caenorhabditis elegans demonstrated that RNF121WT was an endoplasmic reticulum (ER)-localized protein and was involved in ER stress1. However, another study provided evidence that human RNF121WT was a Golgi-localized protein2. In our studies, immunofluorescence staining showed that RNF121WT co-localized preferentially with the GM130 protein in the cis-Golgi network, but minimally with Golgin97 of the trans-Golgi or with protein disulfide isomerase (PDI) of the endoplasmic reticulum (Fig. 2a and Supplementary Fig. S2a), indicating that RNF121WT protein was located on the Golgi complex in neuroblastoma cells. In contrast, RNF121M158R did not co-localize to the cis-Golgi of human neuroblastoma cells (Fig. 2b and Supplementary Fig. S2b). Instead, RNF121M158R transfected cells displayed more diffuse staining in and around the Golgi compared with RNF121WT, consistent with loss-of-function. Co-immunofluorescence using DAPI, anti-RNF121, anti-MYCN, and anti-GM130 demonstrated that MYCN and RNF121 co-localise to the cis-Golgi but MYCN is mostly found in the nucleus (Fig S2c). An immunoblot of RNF121-transfected neuroblastoma cells indicated that RNF121M158R had a much lower protein expression level than RNF121WT (Fig. 2c). Indeed, in stably transfected SH-SY5Y, a human neuroblastoma cell line with non-amplified MYCN, where transfected cells were selected in Geneticin (G418) for over 2 months, RNF121M158R was expressed at a much lower level compared to RNF121WT over time (Fig. 2d). Together, these data show that the mutant RNF121M158R protein exhibited loss-of-function features with uniformly low protein expression and stability compared to RNF121WT.
Fig. 2. The RNF121M158R mutation reduces RNF121 protein levels, stability and localization to the cis-Golgi.
a Immunofluorescent staining using a: an anti-Myc-tag antibody to identify RNF121 co-expressed with the Myc tag; GM130 antibody as a cis-Golgi marker; Golgin-97 antibody as a trans-Golgi marker; and, PDI antibody as an endoplasmic reticulum (ER) marker, in a human MYCN amplified neuroblastoma cell line, Kelly. Scale bar: 100 μm. b Kelly cells transiently transfected with Myc-tagged wild-type RNF121 (RNF121WT) or mutant RNF121M158R for 48 h. Shown here are representative images of immunofluorescent staining using an anti-Myc-tag antibody for the RNF121 protein and a GM130 antibody for the cis-Golgi. Scale bar: 20 μm. c Immunoblot analysis of RNF121 expression in human MYCN amplified neuroblastoma cell line, SK-N-BE(2)-C, across the 4 days following transfection with either RNF121WT or mutant RNF121M158R. d Immunoblot analysis of RNF121 expression in human MYCN non-amplified neuroblastoma cell line, SH-SY5Y, following transfection with either RNF121WT wild type or RNF121M158R.
RNF121WT expression is required for neuroblastoma progression in TH-MYCN mice
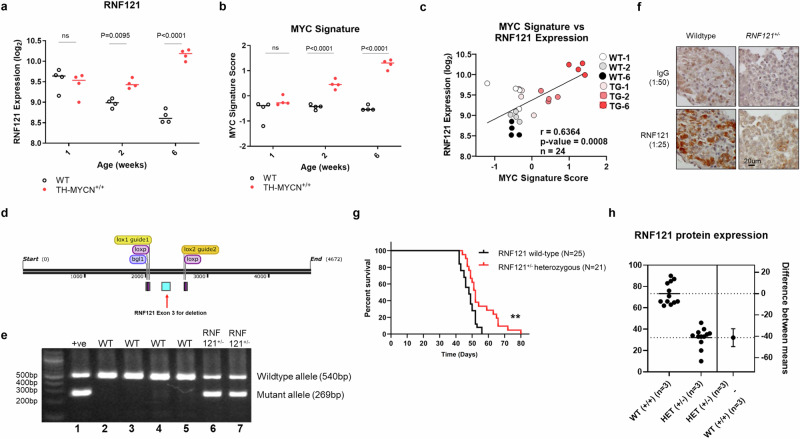
Since a mono-allelic RNF121M158R loss-of-function mutation ablated tumorigenicity in TH-MYCN mice, we mapped the pattern of RNF121WT mRNA expression throughout the course of tumorigenesis in the TH-MYCN model. When we compared TH-MYCN+/+ and non-transgenic littermate tissues for RNF121WT expression level in post-natal sympathetic ganglia and tumour tissues from 1 to 6 weeks of age, we found RNF121WT was significantly upregulated during early tumorigenesis at 2 weeks of age and the difference further increased by 6 weeks of age, at the point of palpable tumour formation22 (Fig. 3a). RNF121WT expression across the course of tumour initiation and progression also correlated with a previously established MYC-signature23, indicating a possible regulatory relationship between RNF121WT and MYCN expression (Fig. 3b, c).
Fig. 3. RNF121 is necessary for neuroblastoma development in TH-MYCN mice.
mRNA expression level of a RNF121WT, and, b a MYC gene signature23 in ganglia tissue and tumours from wild-type (WT) and TH-MYCN+/+ mice, obtained at different postnatal ages (1, 2, and 6 weeks of age; n = 4 per group). Gene expression values are represented as log2-transformed microarray probe intensities of RNA levels. c The Pearson correlation coefficient (r) and p value for RNF121WT and the MYC signature expression scores for the tissue samples in (a) and (b). d Schematic of targeted RNF121 conditional allele: Two sgRNAs (yellow) were complexed with Cas9 nuclease to generate double stranded breaks in the RNF121 locus. A targeting vector featuring exon 3 (blue) of the RNF121 gene flanked by loxP sites was used as a template for homologous recombination. The resulting RNF121 conditional mice was crossed to Cre deleter mice to delete exon 3, leading to the generation of RNF121 knockout mice. e PCR products of mouse tail genomic DNA using primers which distinguished RNF121WT (540 bp in lanes 2–5) and the successfully floxed RNF121+/− allele (269 bp in lanes 6–7). Positive control for RNF121 hemizygous mutant allele (+ ve): DNA from RNF121 targeted embryonic stem cell clone. f Immunohistochemical staining for RNF121WT protein in coeliac ganglia from 20-week-old RNF121WT or RNF121+/− mice using an anti-mouse RNF121 polyclonal antibody and a control IgG antibody. g Tumour-free survival assessed using Kaplan–Meier analyses of TH-MYCN+/+ mice back-crossed with either RNF121WT (n = 25, black line) or floxed RNF121+/− knockout (n = 21, red line) mice (**p < 0.01). h A graphical representation showing RNF121 protein expression levels in tumour tissues from TH-MYCN+/+ mice crossed with either RNF121WT or floxed RNF121+/− knockout mice, across 5 independent western Blots (each with 3 different tumour lysates per group).
To further confirm the role of the RNF121WT protein in MYCN-driven neuroblastoma tumorigenesis, RNF121 knockout mice were developed using a loxP-flanked allele of RNF121WT Exon 3 (Fig. 3d). RNF121 heterozygous knockout mice (RNF121+/−) were viable, but homozygous RNF121-/- knockout mice died in utero. Confirmation of the successful deletion of the RNF121WT gene function was evidenced by PCR using genomic DNA and immunohistochemistry for protein expression. We found the deletion mutant allele with a PCR product of 269 bp in RNF121+/− mouse tail tissue DNA (Fig. 3e) and markedly reduced RNF121WT protein expression in coeliac ganglia from RNF121+/− mice, compared to coeliac ganglia from RNF121WT mice (Fig. 3f). Hemizygous RNF121+/− knockout mice were then crossed with hemizygous TH-MYCN+/− transgenic mice over 4 generations to an estimated congenicity of 93.75%. The TH-MYCN+/+ offspring, with a single RNF121 allele deleted, exhibited a significant delay in tumour development and increased in tumour latency (Fig. 3g), supporting our previous evidence that RNF121WT plays a functional role in MYCN-driven neuroblastoma tumorigenesis. Immunoblot analysis of total protein from snap-frozen tumour tissues showed a decrease in RNF121 protein expression in TH-MYCN+/+ mice crossed with RNF121+/− mice, compared to TH-MYCN+/+ mice crossed with RNF121WT mice (Fig. 3h and Supplementary Fig. S3a).
RNF121 increases human neuroblastoma cell growth via its transmembrane domain
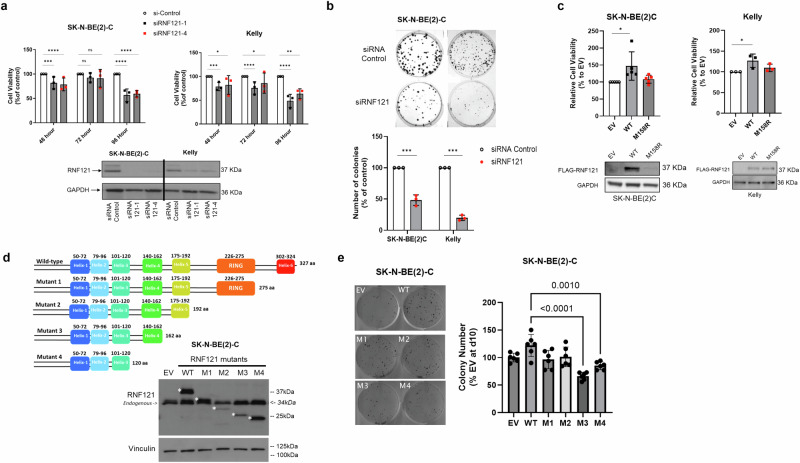
To evaluate the functional role of RNF121WT in neuroblastoma cells, we examined the expression levels of RNF121 in a panel of human neuroblastoma cell lines and found that RNF121 expression was higher in MYCN-amplified neuroblastoma cell lines, in comparison with MYCN non-amplified and normal human fibroblast cells (Supplementary Fig. S3b). We then examined the effects of RNF121WT knock-down or overexpression in two MYCN amplified human neuroblastoma cell lines, SK-N-BE(2)-C and Kelly. The siRNA-mediated repression of RNF121WT expression maximally decreased cell viability 96 h after transfection and reduced the colony forming ability of both cell lines over 10 days (Fig. 4a, b). Conversely, cell viability assays showed that an RNF121WT vector overexpressed in both cell lines increased cell viability 72 h after transfection, whereas overexpression of the RNF121M158R mutant in the two human neuroblastoma cells had no effect on viability (Fig. 4c). These data indicated an oncogenic function for RNF121WT in neuroblastoma cells, which was both MYCN-dependent and -independent, and provided further support for the loss-of-function characteristics of RNF121M158R.
Fig. 4. The RNF121 transmembrane domain 5 is required for growth and clonogenicity in vitro.
a Top panel: Cell viability analyses (n = 3) of SK-N-BE(2)-C and Kelly human neuroblastoma cells transfected with RNF121 siRNA-1, RNF121 siRNA-4 or Control siRNA for 96 h. Two-sided unpaired Student’s t-tests were performed to derive p-values. Differences in cell growth were compared to Control siRNA transfected cells. ***p < 0.001 and ****p < 0.0001. Bottom panel: Immunoblot analysis of RNF121 expression in SK-N-BE(2)-C and Kelly cells following siRNA-mediated RNF121 knockdown for 48 h. b SK-N-BE(2)-C and Kelly cells transfected with RNF121 siRNA-1 or Control siRNA were grown for 10 days in vitro and then assessed for colony formation. c Top panel: Cell viability analysis of SK-N-BE(2)-C and Kelly cells transfected with either Flag-RNF121WT, Flag-RNF121M158R or Empty vector control for 72 h. Two-sided unpaired Student’s t-tests were performed to derive p-values. *p < 0.05. Differences in cell growth were compared to Empty vector control expressing cells. Bottom panel: Immunoblot analysis of Flag-RNF121 expression in SK-N-BE(2)-C and Kelly cells following either overexpression of Flag-RNF121WT, Flag-RNF121M158R or Empty vector control for 24 h. d Top panel: Schematic representation of the Myc-tag-RNF121 deletion mutants. Bottom Panel: Representative immunoblot analysis for SK-N-BE(2)-C cells either overexpressing for 48 h empty Vector (EV), Myc-tag-RNF121WT full-length or each of the 4 Deletion Mutants of RNF121WT, probed with an anti-Myc-tag antibody. *predicated molecular weights of ectopically overexpressed Myc-RNF121 full-length and 4 Mutants with Myc-tag. e Left panel: SK-N-BE(2)C cells transfected with empty vector (EV), RNF121WT or each of the four RNF121 Deletion Mutants grown for 10 days and then assessed for colony formation. Right panel: Histogram representing the number of colonies 10 days after transfection as a percentage of control EV. **p < 0.05 for the comparison of RNF121 Mutant 3 and 4 with RNF121WT.
To map the domain of RNF121WT which was critical for the oncogenic function of RNF121WT in neuroblastoma cells, we derived 4 RNF121WT deletion mutants using a pCMV6-myc-DDK-tagged expression construct. We transiently transfected SK-N-BE(2)-C cells with constructs expressing the RNF121WT deletion mutants for 48 h, followed by immunoblotting assays confirming the expression of RNF121WT full-length (amino acid 1–327), Mutant 1 (amino acid 1–275), Mutant 2 (amino acid 1–192), Mutant 3 (amino acid 1–162) and Mutant 4 (amino acid 1–120) (Fig. 4d). Colony forming assays showed that SK-N-BE(2)-C cells transfected either with RNF121WT, Mutant 1 or Mutant 2 displayed increased colony formation, when compared to empty vector control. However, SK-N-BE(2)-C cells transfected with RNF121WT Mutant 3 and Mutant 4 exhibited reduced capacity to increase colony formation (Fig. 4e), suggesting that the putative transmembrane domain, Helix 5, of RNF121WT (amino acids 175–192) was crucial for oncogenic function of RNF121, but not the putative E3 ligase function of the RING domain.
RNF121WT forms a protein complex with MYCN and enhances MYCN protein stability
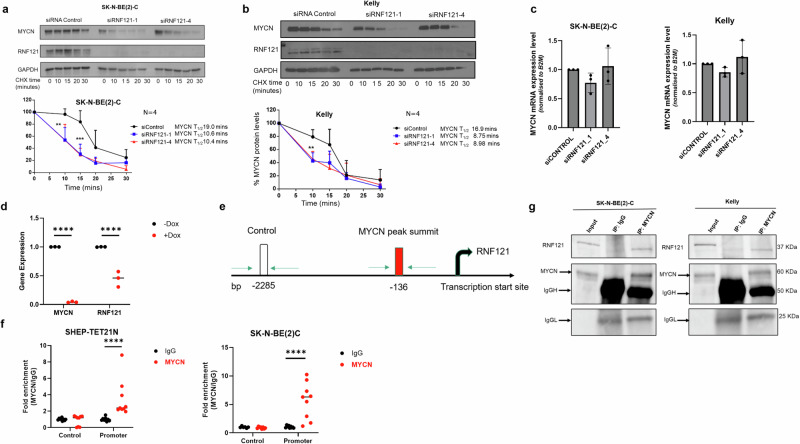
To better understand the mechanistic relationship between RNF121WT and MYCN, we performed immunoblotting using protein from the nuclear and cytosolic subcellular fractions of Kelly and SK-N-BE(2)-C cells. We found that RNF121WT was predominantly recovered in the cytoplasmic fraction, which includes organelle membranes, whereas MYCN was predominantly recovered in the nuclear fractions of MYCN-amplified neuroblastoma cells. (Supplementary Fig. S4a). We then transiently transfected Kelly and SK-N-BE(2)-C cells with two RNF121WT-specific siRNAs. We showed that the siRNA knock-down of RNF121WT resulted in a significant decrease in MYCN protein levels in both cell lines, and an almost 2-fold reduction in MYCN protein half-life in both cell lines (Fig. 5a, b). However, knocking down RNF121 did not result in reduced MYCN mRNA expression levels (Fig. 5c), suggesting that the interaction between MYCN and RNF121 was post-translation.
Fig. 5. RNF121 directly binds MYCN protein and increases its stability.
a, b Representative immunoblots from cycloheximide (CHX) chase experiments 48 h after RNF121 knockdown using control siRNA or RNF121-specific siRNAs #1 or #4 in SK-N-BE(2)-C (a) and Kelly (b) cells. Cells were then treated with 100 µg/ml CHX for up to 30 min followed by immunoblotting of total cell lysates with anti-RNF121 or anti-MYCN antibodies. The ratio of MYCN protein/Actin protein was artificially set at 1.0 for control samples and then the half-life of MYCN was calculated from the line graph. c RT-PCR of MYCN mRNA expression levels following 48 h of siRNA knockdown of RNF121. d RT-PCR of MYCN and RNF121 mRNA expression levels following 24 h of Doxycyline (Dox) treatment of Dox-inducible MYCN knockdown neuroblastoma cell line, SHEP-Tet21N. e Schematic of PCR primer sites in the RNF121 gene promoter used for ChIP-PCR, detailing the MYCN peak binding summit (−136 bp) and an upstream negative control site (−2285 bp). f ChIP-PCR assays assessing fold enrichment of MYCN protein binding at two sites within the RNF121 gene promoter: the MYCN protein binding summit (−136 bp [Promoter]) or the negative control region (−2285 bp [Control]) using an anti-IgG (control) or anti-MYCN antibody in total cell lysates from either SHEP-TET21N or SK-N-BE(2)-C neuroblastoma cells, both with basal high MYCN expression. g Representative Western blot analysis for endogenous RNF121 after immunoprecipitation of endogenous MYCN in total cell lysates from MYCN amplified Kelly and SK-N-BE(2)-C cells. 5% of the cell lysate was loaded for input.
We next used a non-MYCN amplified neuroblastoma with doxycycline (Dox)-inducible MYCN expression, the SHEP-TET21N cell line, to determine if MYCN regulated RNF121 mRNA transcription. We observed significantly decreased RNF121 mRNA levels when MYCN was switched off 24 h after adding Dox (Fig. 5d). This observation suggested that RNF121 is a transcriptional target of MYCN in neuroblastoma cells. Analysis of the RNF121 gene promoter sequence and published chromatin-immunoprecipitation (ChIP)-sequencing data24 revealed a non-canonical MYCN binding site CANNTG (at −136 bp) in close proximity to the RNF121 transcription start site (Supplementary Fig. S4b). Next, we performed ChIP with an anti-IgG or anti-MYCN antibody, followed by real-time PCR with primers targeting either a negative control region (Control, −2285 bp) or the putative MYCN binding site (−136 bp) upstream of the RNF121WT transcription start site (Fig. 5f). The MYCN antibody immunoprecipitated the MYCN binding site (−136 bp) at a level 3.5-fold higher than the negative control region in SHEP-TET21N cells, and 5.7-fold higher in SK-N-BE(2)-C cells (Fig. 5f), confirming that MYCN directly binds the RNF121 gene promoter. To further understand the regulatory interaction between RNF121 and MYCN, we examined total cellular protein extracts immunoprecipitated with a MYCN-specific antibody and found that endogenous RNF121 could directly bind the immunoprecipitated MYCN and form a protein complex in both Kelly and SK-N-BE(2)C cells (Fig. 5g). In addition, co-immunoprecipitation between MYCN and RNF121WT and four deletion mutants, confirmed that helix 5 (lacking in mutant 3 and 4) was essential for the protein complex formation between MYCN and RNF121 (Fig. S4c). Thus, high MYCN levels resulting from MYCN amplification in neuroblastoma cells stimulates RNF121WT transcription. We propose a potential oncogenic mechanism through a positive feedback loop where high levels of RNF121 protein in turn bind and “protect” MYCN from proteasomal degradation. However, this would require further mechanistic studies to establish.
The RNF121WT gene is preferentially retained in human MYCN amplified neuroblastoma tissue and high expression associates with very poor prognosis
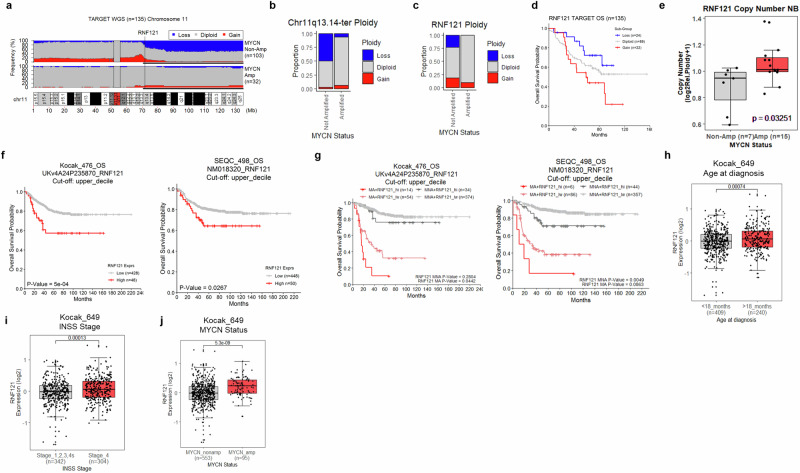
Since either a mono-allelic RNF121M158R loss-of-function mutation or RNF121WT deletion delayed or ablated tumour formation in homozygous TH-MYCN+/+ mice, we assessed RNF121WT gene copy number variation (CNV) and expression in databases of bulk mRNA and somatic DNA analyses of human neuroblastoma primary tumour tissues. In humans, RNF121 is located on Chromosome 11q13.4, a commonly deleted region in neuroblastoma which associates with poor patient outcome25–28. The deletion of Chromosome 11q and the amplification of MYCN are largely mutually exclusive events in human neuroblastoma27. Analysis of segmental CNVs across Chromosome 11 in tumour tissues from the TARGET neuroblastoma patient cohort (n = 135) revealed that 50% of MYCN non-amplified patients (n = 103) had a loss of the 11q13.14-ter region compared with only 6.25% of MYCN amplified patients (n = 32)29 (Fig. 6a, b). Moreover, focal analyses of RNF121WT gene copy number revealed that 23% of MYCN non-amplified patients had a loss of this gene compared with 0% of MYCN-amplified patients (Fig. 6c). These data support the hypothesis that MYCN amplified neuroblastoma cells rely on genes in the 11q13.14-ter region for malignant progression, such as RNF121 (Fig. 6c). RNF121 copy number-based stratification of patients in this cohort revealed a dose-dependent relationship with neuroblastoma patient prognosis, wherein patients with RNF121 loss had significantly better overall survival compared to patients with RNF121 gain (p = 0.0341) (Fig. 6d).
Fig. 6. The RNF121WT gene is preferentially conserved in MYCN amplified human neuroblastoma tumours and associates with poor patient outcome.
a Frequency of segmental copy number variation (loss [blue], gain [red]) across Chromosome 11 (10 kilobase genomic bins), from whole-genome sequencing of somatic tumour DNA (WGS) from the TARGET neuroblastoma patient cohort (n = 135)28. The 11q13.4-ter region is emphasised by a thin black bar underline. Genomic coordinates are provided in megabase measurements (Mb) below the plot. Centromeric regions were excluded from analyses (11p11.11/q11). b The proportion of patients with chromosome 11q13.4-ter segmental copy number variation (loss [blue], gain [red]) in MYCN non-amplified and amplified patient subgroups. c The proportion of patients with RNF121WT copy number variation (loss [blue], gain [red]) in MYCN non-amplified and amplified patient subgroups. d Kaplan–Meier curve of overall neuroblastoma patient survival in the TARGET cohort (n = 135) subdivided by RNF121WT ploidy. e RNF121WT copy number for 22 neuroblastoma cell lines divided into MYCN-non-amplified and amplified subgroups. Data is derived from the Dependency Map portal (DepMap; 21Q2 release). The p-value is calculated using a two-sided t-test between each subgroup. f Kaplan–Meier curves of Overall Survival probability of patients from the Kocak (n = 476) or SEQC neuroblastoma cohorts (n = 498) subgrouped above or below the upper-decile of RNF121 mRNA expression for the overall group. The p-values are calculated using a log-rank test. g Kaplan–Meier curves of Overall Survival probability for patients from the Kocak (n = 476) or SEQC neuroblastoma cohort (n = 498) subdivided by RNF121 mRNA expression and MYCN amplification status (Amplified; MA, Non-amplified; MNA). Patients were subgrouped above or below the upper decile of RNF121 mRNA expression (High; RNF121_hi, Low; RNF121_lw). The p-values were calculated using two-sided log-rank tests between each RNF121 (lw, hi) expression group after subdividing cohorts by MYCN amplification status. p values were also adjusted using the Benjamini–Hochberg method to account for multiple comparisons. h–j Comparison of RNF121 mRNA expression levels for patients subdivided by either the age at diagnosis (<18 months or >18 months), disease stage (INSS stage 1,2,3,4s vs stage 4) or MYCN amplification status (MYCN non-amplified vs amplified) in the Kocak dataset (n = 649).
Analysis of RNF121 copy number in human neuroblastoma cell lines using the Cancer Dependency Map (DepMap) revealed that MYCN amplified cell lines had significantly higher RNF121 copy number compared with MYCN non-amplified cell lines30,31 (Fig. 6e).
Analysis of bulk mRNA levels in primary human neuroblastoma tissue from the Kocak/Oberthuer (n = 476) and SEQC (n = 498) patient cohorts subdivided around the upper decile of RNF121 expression revealed an association between very high RNF121 mRNA expression and poor survival (Fig. 6f; Supplementary Fig. S5a)32,33. Further stratification of patients using both very high RNF121 expression and MYCN amplification status revealed an enhanced prognostic value, wherein patients with very high RNF121 expression and MYCN amplification had the poorest prognosis (Fig. 6g; Supplementary Fig. S5b). Comparison of RNF121 mRNA expression in patient subgroups segregated by age at diagnosis, INSS disease stage and MYCN amplification status, indicated that higher RNF121 expression associated with those subgroups with the poorest clinical outcomes (>18 months of age at diagnosis, Stage 4 disease and MYCN amplification) (Fig. 6h–j).
Collectively, this data highlighted an important link between RNF121 ploidy and MYCN amplification levels. The MYCN-amplified patient cohort showed no RNF121 loss or deletion (Fig. 6c), and this was strongly supported by the observed higher mRNA expression of RNF121 in patients with MYCN amplification (Fig. 6j). MYCN-amplified neuroblastoma may have an oncogenic dependency on RNF121 expression which is supported by the observation that when there is an 11q deletion or RNF121 loss/deletion in MYCN-amplified cells, there is significant decrease in viability (Fig. 4). Thus, RNF121WT expression patterns and ploidy define a very high-risk subgroup of neuroblastoma patients, especially if present with MYCN amplification.
Lastly, we assessed RNF121WT CNV in other cancer types using a curated dataset of 155 non-redundant studies in cBioPortal34. RNF121 was amplified in 23/95 (24.2%) of oesophageal squamous cell carcinoma and 54/523 (10.3%) of head and neck squamous cell carcinoma (HNSCC) (Supplementary Fig. S6a). High RNF121 mRNA expression (upper quartile) correlated with poor overall survival for HNSCC patients in the Cancer Genome Atlas (TCGA) database (Supplementary Fig. S6b)35. Given that c-MYC, a gene closely related to MYCN, is often amplified in HNSCC, we examined CNV for both RNF121 and c-MYC in primary tumour tissues from 66 (RNF121) and 46 (c-MYC) HNSCC patients. Fluorescence in situ hybridization (FISH) analysis of tumour tissues from the HNSCC patient cohort showed that RNF121WT and c-MYC genes were amplified in 18.2% and 41.3% of patients, respectively (Supplementary Fig. S6c, d). However, there was no direct correlation between c-MYC amplification and RNF121WT amplification in these patient samples. Our data suggests that RNF121WT amplification alone may also play a role in HNSCC tumorigenesis.
Discussion
More than half of all human cancer is driven by MYC family oncoproteins. Neuroblastoma is commonly driven by the MYCN and/or MYC oncogenic signals, and, is thus considered a model disease for studies of mechanism and therapeutics of MYC-driven cancer36. Here we used a random in vivo mutagenesis screen to define a oncogenic cofactor role for the cis-Golgi Complex protein, RNF121, in neuroblastoma, and potentially other MYC- or MYCN-driven malignancies, such as HNSCC. We show that RNF121 is a transcriptional target for MYCN, which in turn binds the MYCN protein in a “protective” manner to increase MYCN stability, possibly in the Golgi. We and others have previously shown that, even in the presence of multi-copy MYCN amplification, neuroblastoma cells still require such positive feedback mechanisms to achieve the very high levels of MYCN protein needed to sustain tumorigenesis17,18. We found RNF121 was required for MYCN-driven tumorigenesis in mouse models and human tumour cells, while neuroblastoma patients with the highest levels of both RNF121 and MYCN had the very poorest prognosis. Despite 11q deletion being common in neuroblastoma, we found, remarkably, that RNF121 (on 11q) is never lost in tumours with MYCN amplification. This supports an important role for RNF121 in MYCN oncogenesis and potentially explains the observation that the two events of MYCN amplification and 11q deletion are mutually exclusive. Thus, RNF121 represents an attractive therapeutic vulnerability in neuroblastoma, particularly as mice hemizygous for a hypo-functioning RNF121 allele appeared healthy.
In cancer cells, RING Finger proteins may behave as oncogenes or tumour suppressors dependent on the context and it is unclear which structural features determine these functions37,38. As a RING finger protein, RNF121 has been shown to promote target protein ubiquitination as an E3 ligase in some cell systems1. However, our deletion mutant studies of RNF121 in neuroblastoma cells suggest that, for its role as an oncogenic cofactor for MYCN, the RING domain of the protein was dispensable. These findings indicate that an E3 ligase function is less likely to mediate its oncogenic role. Instead, loss of transmembrane domain Helix 5 had the greatest effect on clonogenicity. This observation, coupled with our finding that the hypofunctioning RNFM158R allele induced by random chemical mutagenesis was sited in Helix 4, strongly suggests a cis-Golgi membrane-associated function for RNF121 in MYCN-driven neuroblastoma. It will be important to determine whether MYCN and RNF121 indeed bind together at this membrane and that this site of binding is necessary for its oncogenic effects.
Although RNF121 has been found in both the endoplasmic reticulum and the Golgi Complex, we show that in neuroblastoma cells RNF121WT co-localises with GM130 to the cis-Golgi. The Golgi complex is an endomembrane system with diverse functions in signalling, vesicle trafficking, and, protein and lipid processing in normal cells39,40. These functions can be disordered and quantitatively dysregulated in cancer. Golgi phosphoprotein 3 (GOLPH3) was the first described Golgi complex oncoprotein41. GOLPH3 is sited in the trans-Golgi network and is thought to stimulate aberrant glycosphingolipid biosynthesis42, mTOR signalling43 and cell growth in several cancer types44,45. However, there are now many other Golgi-sited proteins which have diverse roles in cancer cells, without a unifying explanation46. We show here that MYCN-driven tumorigenesis is completely blocked at initiation by one hypofunctioning RNF121 allele. We hypothesise that RNF121M158R, mutated in its transmembrane domain 4, is no longer incorporated into the cis-Golgi membrane where its function is essential for achieving the very high MYCN levels required at tumour initiation in its tissue of origin, the sympathetic ganglia. Post-translational modification of MYCN, through phosphorylation, for instance, has important effects on MYCN stability in normal and cancer cells47,48. RNF 121 may facilitate post-translational modification of MYCN at the cis-Golgi membrane prior to its translocation to the nucleus. This exquisite vulnerability of the MYCN oncogenic signal indicates that inhibitors of RNF121 effects on MYCN may have potent therapeutic benefits in patients.
The absence of deep pockets and the unstructured nature of the MYCN and MYC proteins has meant that designing direct drug inhibitors has been challenging. Here we show that MYCN protein stability may be a tractable target if instead competitive inhibitors are designed which block RNF121-MYCN binding. Multiple recent successes in the design of protein-protein interface inhibitors for clinical use indicate this is a feasible strategy for blocking RNF121-MYCN binding49. Indeed, our own work on another MYCN binding protein, PA2G4, showed how effective interruption of “protective” protein binding in vitro can translate into potent in vivo effectiveness17. Future drug discovery and development studies will need to address the specific mechanism by which RNF121 modifies MYCN protein half-life at a post-translational level, and the structural characteristics of RNF121-MYCN binding interface.
Our findings have important clinical implications. While MYCN amplification is detected in 20–25% of neuroblastoma patient tumours, and 11q deletion in 35–45%, the two CNVs occur as mutually exclusive cancer driver events50. Our findings suggest that when MYCN is a major driver in human neuroblastoma, RNF121 is almost always amplified or overexpressed. Furthermore, we show that patients with very high RNF121 levels and MYCN amplification together form a unique group with an exceedingly poor survival rate. This very poor prognosis group could be identified early in the treatment plan as being candidates for more experimental therapies. We also found a clinical association between RNF121WT amplification with esophageal cancer and HNSCC, both known to be driven by MYC51,52. This finding raises the exciting possibility that RNF121 may be an important drug target in a much wider range of MYC-driven cancers, a possibility supported by recent large-scale molecular characterisation of oral verrucous hyperplasia samples, a common precursor of HNSCC53.
Methods
Animal experiments
The Th-MYCN transgenic mouse model overexpresses the human MYCN gene under the control of the rat tyrosine hydroxylase promoter. All homozygote Th-MYCN mice develop neuroblastoma in sympathetic nervous tissue by 6 weeks postnatal54. The mice have been maintained by breeding hemizygous mice together. All experimental procedures were approved by the University of New South Wales Animal Care and Ethics Committee (ACEC) (Approval numbers ACEC 07/58B, 10/8B, 12/97A), according to the Animal Research Act, 1985 (New South Wales, Australia) and the Australian Code of Practice for Care and Use of Animals for Scientific Purposes (2013). The use of genetically modified mice was approved by the Institutional Biosafety Committee. We have complied with all relevant ethical regulations for animal use.
Generation of ENU mutant mice
To allow homozygous male TH-MYCN mice to reach an age where they could receive chemical mutagenesis and undergo mating prior to tumour formation, four-week-old mice were treated with 30 mg/kg Cyclophosphamide (Baxter, Illinois), daily for 5 days. As we have previously reported cyclophosphamide therapy successfully ablated tumours which had begun to form16. After a 2-week resting period, mice were treated with 3 i.p. injections of a 66 mg/kg dose of N-ethyl-N-nitrosourea (ENU) (Sigma), as previously described55,56. To generate first-generation offspring, homozygous TH-MYCN male mice were then mated with homozygote TH-MYCN female mice, which were also treated with Cyclophosphamide to ablate their early tumours. Tumour development was monitored by abdominal palpation and tumour delayed G1 offspring were mated to produce G2 offspring to test heritability. The founder 1929 mouse carrying the MYCN transgene, but which did not develop neuroblastoma, was mated, and progeny mice were used in further experiments. To identify the mutations which may be responsible for delaying tumour formation, the 1929 progeny mice were then mated to BALB/c (BALB/cJAusb) or C57BL/6 (C57BL/6JAusb) mice from Australian BioResources (ABR), Moss Vale and then backcrossed to homozygote TH-MYCN+/+ mice which had not received ENU.
Exome sequencing, confirmatory PCR and in silico modelling of membrane localized RNF121 protein
Exome sequencing of 1929 progeny was performed at the Australian Phenomics Network located at the Australian National University (Canberra, Australia) using the SureSelect XT Mouse All Exon array (Agilent). The reads were aligned and filtered. To determine the mutations responsible for the phenotype, sequence-specific assays were designed using the Primer Express™ software, Version 3.0.1 from Applied Biosystems (Thermo Fisher Scientific, Australia), based on the location of the single nucleotide variation identified by exome sequencing. Primers were synthesised by Sigma-Aldrich (Australia). To confirm the presence of the variants, PCR assays were designed as outlined in Supplementary Table 1. PCR reactions were performed using 100 ng of DNA and 1 unit of AmpliTaq Gold® DNA polymerase in a reaction mix containing 10 x PCR Buffer II (100 mM Tris-HCl, pH 8.3, 500 mM KCl), dNTPs (250 µM), primers (20 pmol) and 1.5 mM magnesium chloride in a 25 µL reaction volume. The reaction was subject to a 3-step PCR cycle for 35 cycles in a thermocycler. An initial pre-incubation step of 94 °C for 10 min was followed by 35 cycles of 94 °C for 45 s, primer annealing at 58 °C for 45 s and an extension of 90 s at 72 °C. A final extension of 72 °C for 10 min was followed by cooling to 4 °C. All PCR reactions had a primer annealing temperature of 58 °C, except for Kctd2, which was run at 55 °C. The PCR products were electrophoresed on a 12.5% acrylamide gel to confirm amplification, before being sent for sequencing.
ExoSAP-IT PCR clean up kit from Affymetrix was used for purification of PCR products for sequencing. The PCR product (10 µL) was mixed with exonuclease I (2 µL) and shrimp alkaline phosphatase (2 µL) in a PCR tube and incubated at 37 °C for 15 min to degrade primers and nucleotides, followed by 80 °C for 15 min to inactivate the ExoSAP-IT. The cleaned PCR product (7 µL) was mixed with 1 µL of forward or reverse primer and made up to 16 µL with DNase- and RNase-free water. Sequencing was performed by the Australian Genome Research Facility Westmead. A 3D model of RNF121 was constructed using the AlphaFold DB website21.
Cell lines, cell culture, and cell viability assay
Neuroblastoma cell lines, SK-N-BE(2)C, SH-EP, SH-SY5Y, LAN-1, IMR-32, NBLS, SK-N-AS, SK-N-DZ and SK-NFI were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Life technologies Australia, VIC, Australia) with 10% foetal calf serum (FCS) (Life Technologies). CHP-134 and Kelly cells were cultured in RPMI media (Life Technologies) with 10% FCS. MRC5 and WI-38 normal human fibroblasts were grown in alpha-minimum essential media (MEM) (Life Technologies) supplemented with 10% heat-inactivated FCS. Neuroblastoma cell lines, SK-N-BE(2)-C, SH-EP, and SHSY5Y cells were provided by Barbara Spengler (Fordham University, New York, NY). Neuroblastoma cell lines, IMR-32 and SK-N-FI cells were obtained from the American Type Culture Collection (ATCC) (Manassas, VA, USA). Neuroblastoma cell lines, Kelly, CHP-134, SK-N-DZ, and SK-N-AS cells were obtained from the European Collection of Cell Cultures through Sigma (Sigma, Sydney, Australia). Neuroblastoma cell line NBLS was kindly provided by Prof. Susan L. Cohn (Northwestern University, Chicago, IL, USA). Neuroblastoma cell line LAN-1 was kindly provided by Dr. John Maris (Children’s Hospital of Philadelphia, Philadelphia, USA). Non-MYCN-amplified human neuroblastoma cell line SHEP-tet21N, which is genetically modified to repress human MYCN cDNA when exposed to doxycycline, was generously supplied by Professor Jason Shohet (Texas Children’s Cancer Center, Houston). MRC5 and WI-38 normal human fibroblasts were purchased from ATCC. All cell lines used were authenticated by Cell Bank Australia (Westmead, NSW, Australia), free from mycoplasma, and cultured at 37 °C/5% CO2 in a humidifier incubator. All cells were freshly thawed from initial seeds and were not cultured for more than 2 months.
Cell viability was measured using the Alamar Blue assay (Life Technologies) according to the manufacturer’s protocol. Signals were quantitated on a VICTOR2 Multilabel counter (PerkinElmer Australia) at an excitation wavelength of 560 nm and an emission wavelength of 590 nm.
Immunoprecipitation
For endogenous MYCN immunoprecipitation assays, cells were lysed in cold NP40 buffer (Sigma) supplemented with protease inhibitor (Sigma). 5% of the cell extract was saved as the input, and the rest (750 µg total protein) was incubated with either 10 µg MYCN-specific antibody (Merck Millipore), or with 10 µg control mouse IgG antibody (Santa Cruz Biotechnology) and A/G PLUS agarose beads (Santa Cruz Biotechnology) at 4 °C overnight. After three washes with the lysis buffer, the bound proteins were eluted by boiling with SDS sample buffer. Bound proteins were resolved by SDS-PAGE.
Western blot analysis
Cell pellets were lysed with NP-40 buffer (Sigma) freshly supplemented with protease inhibitor cocktail (Sigma). Protein lysate was standardized using the BCA protein quantitation assay kit as per manufacturer’s instructions (Thermo Fisher Scientific), and 20–40 µg whole protein lysates were resolved on 10–14% Tris-HCl Criterion gels (Bio-Rad). Nitrocellulose membranes (GE Healthcare) were blocked with 5% (wt/vol) nonfat dry milk in Tris-buffered saline with Tween-20 (20 mM Tris-HCl (pH 7.6), 137 mM NaCl, 0.1% Tween-20), then incubated overnight at 4 °C with the following primary antibodies: Flag/DYKDDDDK tag (9A3; 1:1000; Cell Signaling Technologies); β-actin (AC-15; 1:10,000; Sigma), RNF121 (1:1000; Novus). Appropriate horseradish peroxidase-conjugated secondary antibodies (1:3000; Santa Cruz Biotechnologies and Merck Millipore) were diluted in Tris-buffered saline with 0.1% Tween-20 and membranes were probed at room temperature for 2 h. Immunoblots were visualized with Clarity ECL Chemiluminescence reagents (Bio-Rad). For the immunoblotting of whole cell protein lysates from mouse tumours of RNF121+/− knockout mice crossed with Th-MYCN mice, an anti-rabbit RNF121 polyclonal antibody (Sigma-Aldrich) was used in 1:500 dilution and 50 µg of protein were loaded for each sample. Anti-Vinculin antibody was used as a loading control in 1:3000 dilution.
Chromatin immunoprecipitation (ChIP)
Preparation of DNA from SK-N-BE(2)-C and SHEP-TET21N cells for ChIP assay was performed using the Chromatin-Immunoprecipitation Assay kit (Merck Millipore) according to the manufacturer’s instructions. ChIP assays were performed with an anti-MYCN antibody (sc-53993; Santa Cruz Biotechnology) or control mouse IgG antibody (#10400C; Invitrogen). DNA was purified using a MiniElute PCR Purification Kit (Qiagen). Quantification of Real-time PCR (qRT-PCR) was performed with primers designed to cover the regions of the RNF121 gene promoter containing MYCN-binding motifs or remote negative control promoter regions. qRT-PCR for gene expression was analysed using the Quantstudio™ 3 real-time PCR machine (ThermoFisher Scientific, Waltham, MA, USA). Fold enrichment of MYCN bound at the RNF121 gene promoter regions containing the MYCN binding site or a control upstream sequence using an anti-MYCN or anti-IgG antibody was calculated by dividing the PCR product from this region by the PCR product from the negative control region, relative to the input. Control-Forward primer: ACCCTGTATGCCTTCTCACT; Control-Revise primer: GTGGGAGATCATGAACATGACAA; RNF121 ChIP-Forward primer: AATGCGTTCTGGGGCTTCAA; RNF121 ChIP-Reverse primer: GCTCCCTCCACGAAAGAGTT.
Co-immunofluorescent staining of RNF121 and organelle proteins
Human neuroblastoma cells (6 × 104 for Kelly; 4 × 104 for SK-N-BE(2)-C) transfected with Myc-tag-RNF121WT or Myc-tag-RNF121M158R were seeded into each well of a 12-well plate, each containing one 13 mm2 coverslip, for 24 h before fixation with 4% paraformaldehyde for 10 min at room temperature, followed by three washes with PBS for 5 min. Cells were permeabilised with 0.1% Triton X for 15 min, followed by three washes of PBST (PBS + 0.1% Tween 20) for 5 min each. Cells were incubated for 1 h with primary antibodies, anti-Myc-tag (1:1000, Sigma-Aldrich) and organelle markers (anti-GM130 for cis-Golgi [1:200, Abcam]; anti-Golgin-97 for trans-Golgi [1:200, Abcam] or anti-PDI for ER [1:200, Thermo Fisher]), diluted in 2% BSA + PBST. Rabbit and mouse IgG antibodies were used as negative isotype controls and diluted with PBST at a ratio of 1:2500. After 3 washes of PBST for 5 min each, secondary antibodies, Alexafluor 594 anti-mouse (to detect Myc) and Alexafluor 488 anti-rabbit (to detect organelle marker), were applied at a dilution ratio of 1:500 and incubated for 45 min at room temperature. Cells were washed three times with PBST before staining with ProLong Diamond mounting medium. Fluorescent images were captured and analysed using the Leica LAS Lite software.
Generation of RNF121 knockout mice and the RNF121 knockout crossed with TH-MYCN colony
RNF121 knockout mice were generated on a C57BL/6J background by the MAGEC laboratory at the Walter & Eliza Hall Institute, as previously described57. 20 ng/μL Cas9 mRNA, 10 ng/μL single guide RNAs, and 40 ng/μL donor template were injected into fertilized one-cell stage embryos. Guides and targeting vector sequences are provided in Supplementary Table 1. Knock-in mice were backcrossed to C57BL/6J mice for 6 generations to eliminate potential off-target events. sgRNA1: AATCATTTTGCAGACCAACC; sgRNA2: TATCTGCCTATATGACCCCT. RNF121 heterozygous knockout mice were generated by crossing RNF121Flox+/− heterozygous mice with TH-Cre+/− heterozygous mice to delete RNF121 exon 3. Genomic DNA PCR primers were used for genotyping: Forward primer: AGGACTGCTGTGCATGGTG and Reverse primer: AGACAAGGCCTCTCTACAAAGC. RNF121Flox x Th-Cre x Th-MYCN homozygous neuroblastoma colony were then generated by crossing RNF121Flox+/−; Th-Cre +/−; Th-MYCN-/+ with RNF121Flox+/−; Th-Cre+/−; Th-MYCN-/+ mice.
siRNA, plasmid DNA constructions and plasmid DNA transfection
Cells were seeded on 96-well plates for cell viability assays, 6-well plates for colony assays or T25 flasks for protein lysates. Cell density was adjusted so that for all assays, 80–90% confluency would be achieved by the endpoint of the assay in the control condition. siRNA’s used were siRNF121-1: GAAAUGGUCCUCAUCCUCA (Dharmacon: J-007011-06) and siRNF121-4: CCAUAGGGUUCUACAGCGA (Dharmacon: J-007011-08) or non-targeting control siRNAs (ONTARGETplus siRNA, #D-001810-0X, Dharmacon). Expression plasmids of pCMV6-Empty Vector and pCMV6-hRNF121 wild type (WT) with Myc-DDK (FLAG) at c-terminus were purchased from Origene (Origene RC211747). pCMV6-hRNF121(M158R), pCMV6-hRNF121-M1, pCMV6-hRNF121-M2, pCMV6-hRNF121-M3 and pCMV6-hRNF121-M4 with Myc-DDK(FLAG) at c-terminus were subcloned and mutagenesis were performed by Genescript based on RC211747 RNF121 sequence (Genescript). All siRNA or plasmid transfections were conducted with Lipofectamine 2000 according to manufacturer’s instructions (Life Technologies) at 20–40 nM for siRNA or 1–2 μg of plasmid DNA. After transfection, cells were harvested for total RNA or protein at 48–72 h following transfection and analysed by quantitative PCR or western blot respectively. Stable RNF121-overexpressing cell line was created using SH-SY5Y neuroblastoma cells, and transfected with pCMV6-Empty Vector, pCMV6-hRNF121 wild type (WT) or pCMV6-hRNF121(M158R) vectors. Cells were transfected using Lipofectamine 2000 as per the transient transfection protocol, in a 100 mm cell culture dish. Selection of clones began 48 h post transfection with 500 µg/ml G418 (Life Technologies). After 30 days, visible clones were picked, expanded and screened for RNF121 protein expression.
Microarray analysis on TH-MYCN+/+ ganglia tissues
Normalised microarray data from the ganglia of TH-MYCN+/+ homozygous mice and wild-type controls collected at 2, 4, and 6 weeks of age were obtained from ArrayExpress (E-MTAB-3247) and analysed as previously described18,22. In brief, data were summarized and normalized with the vsn method, in the R statistical programming language using the “limma” package58.
Immunohistochemistry
For mouse tissue immunohistochemistry (IHC), slides were incubated with the following primary antibodies at 4 °C overnight: anti-RNF121 mouse polyclonal antibody (Novus Biologicals 1:25) or rabbit IgG (Vector Laboratories 1:50) at corresponding concentrations. Slides were then incubated with secondary goat anti-rabbit immunoglobulin/biotinylated antibody (Dako, E0432 1:500) for 1 h at room temperature. The biotinylated antibody was labelled with streptavidin-HRP (Dako, K1016) for 45 min at room temperature. The sections were developed with 3, 3’-diaminobenzidine tetrahydrochloride (Dako, K3468) at room temperature for 5 min. Slides were then washed in MilliQ water and dehydrated with 70% %, 90%, 100% ethanol sequentially.
Colony formation assays
Cells were transfected using RNF121 siRNA#1 or siControl (Dharmacon) for a period of 72 h. Kelly cells and SK-N-BE(2)-C cells were seeded into 6-well plates at a density of 80,000 or 72,000 cells per well, respectively. For mapping the domains of RNF121WT oncogenic function, SK-N-BE(2)-C cells were also transfected with either empty vector, RNF121WT, RNF121 Mutant 1, Mutant 2, Mutant 3 or Mutant 4 for 72 h. After siRNA or plasmid DNA transfection, media (10% FCS DMEM) was replaced every third day and colonies were left to grow for 10 days until visible colonies appeared. Cell colonies were stained using crystal violet (0.5% (w/v) and quantified using image J.
Cycloheximide chase assays for MYCN protein half-life
SK-N-BE(2)-C and Kelly cells were seeded at a density of 100,000 cells/well or 108,000 cells/well, respectively, in a 6-well plate before transfection using RNF121 siRNA#1, RNF121 siRNA#4, or siControl (Dharmacon) for a period of 48 h. Transfected cells were treated with 100 µg/ml of cycloheximide (Sigma, NSW, Australia) for 0, 10, 15, 20 or 30 min. Cells were washed with cold PBS after treatment and harvested. The cell pellet was resuspended in 200 µl RIPA buffer containing protease inhibitor (Sigma-Aldrich, NSW, Australia) and centrifuged at 13,000 rpm at 4 °C for 10 min. The supernatant was kept, and protein was quantified using western immunoblotting to determine the MYCN protein half-life.
Patient tumour genome/gene expression/survival analyses
Whole-genome sequencing (WGS) data, containing normalized coverage and ploidy calls for genomic segments in 135 clinically annotated primary neuroblastoma tumours (TARGET neuroblastoma cohort), were obtained through the TARGET data matrix (https://ocg.cancer.gov/programs/target/data-matrix) and further processed using the R statistical language and R Studio as previously described18. Average ploidy values were calculated in 10 kb bins across chromosome 11 using the Genomic Ranges R package (v1.40.0), wherein an average ploidy (n), n ≤ 1 was considered a loss, 1 ≤ n ≤ 3 was considered diploid and n ≥ 3 considered to be a gain29. Average ploidy across the 11q13.14-ter region was then calculated in each sample and classified according to the above strata. We utilized matched clinical annotations containing event-free and overall survival data to construct Kaplan–Meier survival curves of the different molecular subgroups using the survminer (v0.4.6) and survival R packages (v2.42.1).
RNA-seq data, expressed in reads per million (RPM) for each gene, for 498 clinically annotated primary neuroblastoma samples (SEQC neuroblastoma cohort) were obtained from the gene expression omnibus (GEO) with the accession GSE6256459. Microarray data for RNF121 expression among 649 neuroblastoma samples (of which only 476 patients have survival data) (Kocak/Oberthuer neuroblastoma cohort), were downloaded from R2 platform (http://r2.amc.nl). We utilized matched clinical annotations containing event-free and overall survival data to construct Kaplan–Meier survival curves of subgroups dichotomized by median divided by the upper decile of RNF121 gene expression values using the survival R package (v2.42.1). All statistical tests concerning Kaplan–Meier analyses were done utilizing log-rank tests, adjusted using the Bonferroni method for multiple hypothesis testing where appropriate as previously described18.
Multi-omic cancer cell line analyses
We utilized public data resources produced by the Cancer Cell Line Encyclopedia (CCLE) and Project Achilles via the Cancer Dependency Map portal (DepMap, 20Q1)31. Copy number (relative ploidy), and gene dependency (CERES) data for RNF121 were first obtained from DepMap and then filtered using R/R Studio (v1.1.456) for disease types/cell lines common to all data sources as well as those disease types that had >1 cell line present. Statistical tests were then performed between neuroblastoma cell lines and an aggregate of other cell lines, or MYCN-amplified vs non-amplified neuroblastoma cell lines, using two-sample t-tests.
RNF121 and c-MYC fluorescence in situ hybridisation (FISH)
RNF121 and c-MYC FISH assays were performed on 4 mm sections from three separate Tissue Microarray (TMA) blocks containing representative 1 mm tumour cores from head and neck squamous cell carcinomas. Slides were deparaffinized in xylene then rehydrated through graded alcohols to water. Target retrieval was performed in Heat Pre-treatment Solution (Invitrogen) in a pressure cooker at 125 °C for 2 min, followed by several washes in water, after which sections were treated with Enzyme Pre-treatment Reagent (Invitrogen) for 20 min at room temperature. Slides were then dehydrated through graded ethanols and air-dried for 15 min. The RNF121 probe (Empire Genomics) or the c-MYC probe (Vysis) was added to the slide which was coverslipped and sealed with rubber cement to prevent evaporation during hybridization. Slides were denatured for 5 min at 85 °C and then hybridized for 18 h at 37 °C on a StatSpin hybridizer (Dako). After hybridization, the coverslips were removed and slides were washed for 2 min in a 0.5x SSC stringent wash buffer at room temperature and then placed for 5 min into a 0.5x SSC stringent wash buffer at 75 °C, followed by several washes in water. Finally, sections were dried and mounted in Vectashield mounting medium with 40, 6-diamidino-2-phenylindole (DAPI; Vector), cover slipped, and sealed with nail polish. Positive (amplified) and negative (non-amplified) controls were included in each FISH run. Sections were stored at 4 °C in the dark prior to scoring. Scoring of RNF121 and c-Myc FISH was done on an Olympus BX51 fluorescence microscope. For each core, areas of tumour were identified at low magnification by using the DAPI filter. A minimum of 50 cells, typically from 2–3 fields/core, were scored for the number of RNF121 (red) and Chromosome 11 (green) signals, or for the number of c-Myc (red) and Chromosome 8 (green) signals, using a 60x water immersion lens. Raw data was entered into a Microsoft Excel spreadsheet for calculation of RNF121 and c-Myc gene amplification status. For both RNF121 and c-Myc, gene amplification was defined as >5 copies of the gene in >50% of cells or a gene/chromosome ratio >2. Gene gain was defined as an average gene copy number >2.40 or a gene/chromosome ratio between 1.30 and 1.99. Normal disomy was defined as an average gene copy number between 1.20 and 2.40. Gene loss was defined as an average gene copy number <1.20. Chromosomal disomy was defined as average gene copy number of 1.20–2.40, trisomy 2.40–3.40 and polysomy >3.40.
Statistics and reproducibility
All experiments included a minimum of three independent replicates. Statistical analysis was conducted using GraphPad Prism 8 software. P values were determined using ANOVA for multiple group comparison, or two-sided unpaired t-test for two groups. All values were expressed as mean values with 95% confidence intervals. Graphical error bars for represent mean ± SEM, and a p value < 0.05 was considered statistically significant.
Supplementary information
Description of additional supplementary files
Acknowledgements
The generation of the RNF121 knockout mice used in this study was supported by Phenomics Australia and the Australian Government through the National Collaborative Research Infrastructure Strategy (NCRIS) programme. Infrastructure support from the NHMRC Independent Research Institutes Infrastructure Support Scheme and the Victorian State Government Operational Infrastructure Support Program to St. Vincent’s Institute are gratefully acknowledged. M.W.P. is an NHMRC Leadership Fellow (APP1194263). This work was supported by program grants (G.M. Marshall, M.D. Norris, M. Haber) from the National Health and Medical Research Council (NHMRC) Australia (APP1016699), Cancer Institute NSW (10/TPG/1-13), Cancer Council NSW (PG-11-06). This work was also supported by a NHMRC Project Grant (APP1125171; G.M. Marshall, B.B. Cheung) and Cancer Council NSW Project Grants (B.B. Cheung, RG21-08 and B.B. Cheung, RG214491). This work was also supported by a grant from the Can Too Foundation in 2022 (B.B. Cheung, RG214491). This work was supported by NHMRC Fellowships APP1003021, APP1176209 and ACRF Cancer Biology Facility IMB (J.L. Stow). This work was also supported by grant from the National Cancer Institute, National Institute of Health, R01CA240323 (S. Zhu) and the V Foundation for Cancer Research. The authors thank the Steven Walter Children’s Cancer Foundation, Hyundai Help for Kids Foundation, and Neuroblastoma Australia for their financial support.
Author contributions
Conception, supervised all experiments and wrote the manuscript: B.B.C. and G.M.M. Acquisition of data: R.M., J.M., Q.W., J.A.S., M.R., I.P.L.W., D.R., A.G., A.S., S.S., L.H., P.V.F., J.T., E.S., C.M., L.L. and D.L.B. Review and revision of the manuscript: J.L.S., S.Z, R.J.Y. B.J.S., S.C., B.K., A.K., M.J.H., D.J.H., T.L., M.D.N., M.H., D.R.C. and M.W.P.
Peer review
Peer review information
Communications Biology thanks Damien Arnoult, Anthony Faber for their contribution to the peer review of this work. Primary Handling Editors: Derrick Ong and Manuel Breuer.
Data availability
The numerical source data for graphs and charts are provided in Excel files and named as Supplementary Data 1 to 6. Uncropped and unedited gel images have been included as Supplementary Fig. 7 in the Supplementary Information. Whole-genome sequencing (WGS) data, containing normalized coverage and ploidy calls for genomic segments and RNA-seq data, in the form of gene-level counts for neuroblastoma tumours (TARGET neuroblastoma cohort), were obtained through the TARGET data matrix (https://ocg.cancer.gov/programs/target/data-matrix). RNA-seq data, expressed in reads per million (RPM) for each gene, for primary neuroblastoma samples (SEQC neuroblastoma cohort) were obtained from the gene expression omnibus (GEO) with the accession GSE62564. Microarray data, expressed as normalised probe intensities (Kocak neuroblastoma cohort), were obtained from GEO with the accession GSE45547.
Competing interests
The authors declare no competing interests.
Ethics statement
The animal studies were approved by the UNSW Sydney Animal Ethics Committee (Approval numbers ACEC 07/58B, 10/8B, 12/97A).
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Belamy B. Cheung, Ritu Mittra.
Contributor Information
Belamy B. Cheung, Email: bcheung@ccia.unsw.edu.au
Glenn M. Marshall, Email: glenn.marshall@health.nsw.gov.au
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-06899-8.
References
- 1.Darom, A., Bening-Abu-Shach, U. & Broday, L. RNF-121 is an endoplasmic reticulum-membrane E3 ubiquitin ligase involved in the regulation of beta-integrin. Mol. Biol. Cell21, 1788–1798 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao, Y., Hongdu, B., Ma, D. & Chen, Y. Really interesting new gene finger protein 121 is a novel Golgi-localized membrane protein that regulates apoptosis. Acta Biochim. Biophys. Sin.46, 668–674 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Ogino, K. et al. RING finger protein 121 facilitates the degradation and membrane localization of voltage-gated sodium channels. Proc. Natl Acad. Sci. USA112, 2859–2864 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zemirli, N., Pourcelot, M., Dogan, N., Vazquez, A. & Arnoult, D. The E3 ubiquitin ligase RNF121 is a positive regulator of NF-κB activation. Cell Commun. Signal.12, 72 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeng, Y. et al. Ring finger protein 6 promotes breast cancer cell proliferation by stabilizing estrogen receptor alpha. Oncotarget8, 20103–20112 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee, N. S. et al. Ring finger protein 126 (RNF126) suppresses ionizing radiation-induced p53-binding protein 1 (53BP1) focus formation. J. Biol. Chem.293, 588–598 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao, Y. et al. Ring finger protein 43 associates with gastric cancer progression and attenuates the stemness of gastric cancer stem-like cells via the Wnt-β/catenin signaling pathway. Stem Cell Res. Ther.8, 98 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiang, P., Sun, Y., Liu, Y., Shu, Q. & Zhu, Y. Really interesting new gene finger protein 121 is a tumor suppressor of renal cell carcinoma. Gene676, 322–328 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Jiang, Z. et al. Circ-RNF121 regulates tumor progression and glucose metabolism by miR-1224-5p/FOXM1 axis in colorectal cancer. Cancer Cell Int.21, 596 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maris, J. M. Recent advances in neuroblastoma. N. Engl. J. Med.362, 2202–2211 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carabet L. A., Rennie P. S. & Cherkasov A. Therapeutic inhibition of Myc in cancer. Structural bases and computer-aided drug discovery approaches. Int. J. Mol. Sci.20, 3-32 (2018). [DOI] [PMC free article] [PubMed]
- 12.Llombart, V. & Mansour, M. R. Therapeutic targeting of “undruggable” MYC. EBioMedicine75, 103756 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marshall, G. M. et al. Transcriptional upregulation of histone deacetylase 2 promotes Myc-induced oncogenic effects. Oncogene29, 5957–5968 (2010). [DOI] [PubMed] [Google Scholar]
- 14.Marshall, G. M. et al. SIRT1 promotes N-Myc oncogenesis through a positive feedback loop involving the effects of MKP3 and ERK on N-Myc protein stability. PLoS Genet.7, e1002135 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu, P. Y. et al. The histone deacetylase SIRT2 stabilizes Myc oncoproteins. Cell Death Differ.20, 503–514 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carter, D. R. et al. Therapeutic targeting of the MYC signal by inhibition of histone chaperone FACT in neuroblastoma. Sci. Transl. Med.7, 312ra176 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koach, J. et al. Drugging MYCN oncogenic signaling through the MYCN-PA2G4 binding interface. Cancer Res.79, 5652–5667 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Nagy, Z. et al. An ALYREF-MYCN coactivator complex drives neuroblastoma tumorigenesis through effects on USP3 and MYCN stability. Nat. Commun.12, 1881 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carpinelli, M. R. et al. Suppressor screen in Mpl-/- mice: c-Myb mutation causes supraphysiological production of platelets in the absence of thrombopoietin signaling. Proc. Natl Acad. Sci. USA101, 6553–6558 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kile, B. T. & Hilton, D. J. The art and design of genetic screens: mouse. Nat. Rev. Genet.6, 557–567 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature596, 584–587 (2021). [DOI] [PMC free article] [PubMed]
- 22.Ooi, C. Y. et al. Network modeling of microRNA-mRNA interactions in neuroblastoma tumorigenesis identifies miR-204 as a direct inhibitor of MYCN. Cancer Res.78, 3122–3134 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Ji, H. et al. Cell-type independent MYC target genes reveal a primordial signature involved in biomass accumulation. PLoS ONE6, e26057–e26057 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeid, R. et al. Enhancer invasion shapes MYCN-dependent transcriptional amplification in neuroblastoma. Nat. Genet.50, 515–523 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Attiyeh, E. F. et al. Chromosome 1p and 11q deletions and outcome in neuroblastoma. N. Engl. J. Med.353, 2243–2253 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Luttikhuis, M. E. et al. Neuroblastomas with chromosome 11q loss and single copy MYCN comprise a biologically distinct group of tumours with adverse prognosis. Br. J. Cancer85, 531–537 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plantaz, D. et al. Comparative genomic hybridization (CGH) analysis of stage 4 neuroblastoma reveals high frequency of 11q deletion in tumors lacking MYCN amplification. Int. J. Cancer91, 680–686 (2001). [DOI] [PubMed] [Google Scholar]
- 28.Spitz, R., Hero, B., Simon, T. & Berthold, F. Loss in chromosome 11q identifies tumors with increased risk for metastatic relapses in localized and 4S neuroblastoma. Clin. Cancer Res.12, 3368 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Pugh, T. J. et al. The genetic landscape of high-risk neuroblastoma. Nat. Genet.45, 279–284 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ghandi, M. et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature569, 503–508 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meyers, R. M. et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nat. Genet.49, 1779–1784 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kocak, H. et al. Hox-C9 activates the intrinsic pathway of apoptosis and is associated with spontaneous regression in neuroblastoma. Cell Death Dis.4, e586 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang, C. et al. The concordance between RNA-seq and microarray data depends on chemical treatment and transcript abundance. Nat. Biotechnol.32, 926–932 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cerami, E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov.2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pérez Sayáns, M. et al. Comprehensive genomic review of TCGA head and neck squamous cell carcinomas (HNSCC). J. Clin. Med.8, 7-9 (2019). [DOI] [PMC free article] [PubMed]
- 36.Huang, M. & Weiss, W. A. Neuroblastoma and MYCN. Cold Spring Harb. Perspect. Med.3, a014415 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu, H. et al. E3 ubiquitin ligase RNF126 affects bladder cancer progression through regulation of PTEN stability. Cell Death Dis.12, 239 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lipkowitz, S. & Weissman, A. M. RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis. Nat. Rev. Cancer11, 629–643 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu, J. et al. The role of the Golgi apparatus in disease. Int. J. Mol. Med.47, 1-2 (2021). [DOI] [PMC free article] [PubMed]
- 40.Mijaljica, D., Prescott, M. & Devenish, R. J. Endoplasmic reticulum and Golgi complex: contributions to, and turnover by, autophagy. Traffic7, 1590–1595 (2006). [DOI] [PubMed] [Google Scholar]
- 41.Scott, K. L. et al. GOLPH3 modulates mTOR signalling and rapamycin sensitivity in cancer. Nature459, 1085–1090 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rizzo, R. et al. Golgi maturation-dependent glycoenzyme recycling controls glycosphingolipid biosynthesis and cell growth via GOLPH3. EMBO J.40, e107238 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frappaolo, A. et al. GOLPH3 protein controls organ growth by interacting with TOR signaling proteins in Drosophila. Cell Death Dis.13, 1003 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palm, W. GOLPH3 tunes up glycosphingolipid biosynthesis for cell growth. EMBO J.40, e108070 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang, A. et al. Golgi phosphoprotein 3 promotes colon cancer cell metastasis through STAT3 and integrin α3 pathways. Front. Mol. Biosci.9, 808152 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spano, D. & Colanzi, A. Golgi complex: a signaling hub in cancer. Cells11, 1-6 (2022). [DOI] [PMC free article] [PubMed]
- 47.Gustafson, W. C. & Weiss, W. A. Myc proteins as therapeutic targets. Oncogene29, 1249–1259 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ruiz-Pérez, M. V., Henley, A. B. & Arsenian-Henriksson, M. The MYCN protein in health and disease. Genes8, 2-13 (2017). [DOI] [PMC free article] [PubMed]
- 49.Nero, T. L., Morton, C. J., Holien, J. K., Wielens, J. & Parker, M. W. Oncogenic protein interfaces: small molecules, big challenges. Nat. Rev. Cancer14, 248–262 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Brady, S. W. et al. Pan-neuroblastoma analysis reveals age- and signature-associated driver alterations. Nat. Commun.11, 5183 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang, J. et al. Prognostic significance of c-MYC amplification in esophageal squamous cell carcinoma. Ann. Thorac. Surg.107, 436–443 (2019). [DOI] [PubMed] [Google Scholar]
- 52.Zhao, S. et al. Identification and validation of the role of c-Myc in head and neck squamous cell carcinoma. Front. Oncol.12, 820587 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hsieh, J. C. et al. Review of emerging biomarkers in head and neck squamous cell carcinoma in the era of immunotherapy and targeted therapy. Head Neck41 (Suppl 1), 19–45 (2019). [DOI] [PubMed]
- 54.Aldape, WeissW. A., Mohapatra, K., Feuerstein, G. & Bishop, B. G. JM. Targeted expression of MYCN causes neuroblastoma in transgenic mice. EMBO J.16, 2985–2995 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bode, V. C. Ethylnitrosourea mutagenesis and the isolation of mutant alleles for specific genes located in the T region of mouse chromosome 17. Genetics108, 457–470 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Salinger, A. P. & Justice, M. J. Mouse mutagenesis using N-ethyl-N-nitrosourea (ENU). CSH Protoc.2008, pdb.prot4985 (2008). [DOI] [PubMed]
- 57.Kueh, A. J. et al. An update on using CRISPR/Cas9 in the one-cell stage mouse embryo for generating complex mutant alleles. Cell Death Differ.24, 1821–1822 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res.43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Su, Z. et al. An investigation of biomarkers derived from legacy microarray data for their utility in the RNA-seq era. Genome Biol.15, 523 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of additional supplementary files
Data Availability Statement
The numerical source data for graphs and charts are provided in Excel files and named as Supplementary Data 1 to 6. Uncropped and unedited gel images have been included as Supplementary Fig. 7 in the Supplementary Information. Whole-genome sequencing (WGS) data, containing normalized coverage and ploidy calls for genomic segments and RNA-seq data, in the form of gene-level counts for neuroblastoma tumours (TARGET neuroblastoma cohort), were obtained through the TARGET data matrix (https://ocg.cancer.gov/programs/target/data-matrix). RNA-seq data, expressed in reads per million (RPM) for each gene, for primary neuroblastoma samples (SEQC neuroblastoma cohort) were obtained from the gene expression omnibus (GEO) with the accession GSE62564. Microarray data, expressed as normalised probe intensities (Kocak neuroblastoma cohort), were obtained from GEO with the accession GSE45547.