Abstract
Purpose:
The purpose of the study was to compare the effectiveness of PARP inhibitor maintenance therapy (mPARPi) in real-world practice by biomarker status [BRCA1/2 alterations (BRCAalt) and a homologous recombination deficiency signature (HRDsig)] in advanced ovarian cancer.
Experimental Design:
Patients with ovarian cancer receiving first-line platinum-based chemotherapy and either mPARPi or no maintenance were included. Patient data were obtained by a US-based de-identified ovarian cancer Clinico-Genomic Database, from ∼280 US cancer clinics (01/2015–03/2023). Real-world progression-free survival (rwPFS) and overall survival (rwOS) were compared by biomarker status using Cox models, weighted by propensity scores.
Results:
Of 673 patients, 160 received mPARPi [31.2% BRCAalt and 51.9% HRDsig(+)] and 513 no maintenance [15.6% BRCAalt and 34.1% HRDsig(+)]. BRCAalt patients receiving mPARPi versus no maintenance had favorable rwPFS [HR, 0.48; 95% confidence interval (CI), 0.26–0.87; P = 0.0154], as did BRCA wild-type (WT; HR, 0.76; 95% CI, 0.57–1.01; P = 0.0595). Favorable rwOS was not observed with mPARPi for BRCAalt or BRCA-WT. HRDsig(+) patients receiving mPARPi versus no maintenance had favorable rwPFS (HR, 0.36; 95% CI, 0.24–0.55; P < 0.001) and numerically favorable rwOS (HR, 0.46; 95% CI, 0.21–1.02; P = 0.0561). No differences were observed for HRDsig(−). mPARPi treatment interaction was observed for HRDsig(+) versus HRDsig(−) (rwPFS P < 0.001/rwOS P = 0.016) but not for BRCAalt versus BRCA-WT. Patients with BRCA-WT/HRDsig(+) receiving mPARPi had favorable rwPFS (HR, 0.40; 95% CI, 0.22–0.72; P = 0.003), whereas no difference was observed for BRCA-WT/HRDsig(−).
Conclusions:
HRDsig predicted benefit of mPARPi better than BRCAalt. Patients with HRDsig(+) status experienced favorable outcomes, even if they had BRCA-WT status. In contrast, patients with HRDsig(−) status did not show significant benefit from mPARPi treatment. HRDsig might predict benefit from mPARPi regardless of BRCAalt status.
Translational Relevance.
The use of PARP inhibitors as maintenance therapy (mPARPi) after platinum-based chemotherapy is now the standard of care for treating advanced ovarian cancer. Given the possibility of long-term administration of mPARPi, its potential benefits, adverse effects, and financial burden must be carefully evaluated. Debate remains about mPARPi use in biomarker-defined groups, especially those without BRCA1/2 alterations (BRCAalt), high genomic LOH (gLOH-high), or homologous recombination deficiency (HRD). This study shows that a novel HRD signature (HRDsig) can identify patients who benefit from mPARPi even among those with no BRCAalt. About 21% of patients had no BRCAalt and were HRDsig(+). Patients who were HRDsig(−) showed similar progression-free survival and overall survival whether they received mPARPi or no maintenance therapy, suggesting that patients who were HRDsig(−) may be spared mPARPi toxicities. HRDsig and gLOH-high were highly concordant, but HRDsig is able to be assessed on 22.4% more patients than gLOH-high, broadening its clinical application.
Introduction
The use of PARP inhibitors as maintenance therapy (mPARPi) after initial platinum-based systemic chemotherapy has become the standard of care for the treatment of advanced ovarian cancer. Due to the possibility of maintenance therapy being administered over a prolonged period of time, patients and physicians must carefully evaluate the potential benefits (disease-free interval and life extension) with the potential adverse effects of taking a PARPi. Patients may also weigh the financial considerations of this therapeutic strategy. Debate remains about the appropriate use of mPARPi in biomarker-defined groups, especially those without somatic or germline pathogenic BRCA1 or BRCA2 alterations (BRCAalt) or homologous recombination deficiency (HRD), comprising about 50% of patients.
In clinical trials, mPARPi has demonstrated the ability to delay disease progression in the BRCAalt subgroup of patients. In the SOLO1 trial, patients with BRCAalt newly diagnosed ovarian cancer with a complete or partial response (CR or PR) to chemotherapy had significantly longer progression-free survival [PFS; HR, 0.30, 95% confidence interval (CI) 0.23–0.41) and significantly longer overall survival (OS; HR, 0.55; 95% CI, 0.40–0.76) when treated with olaparib compared with the placebo (1, 2).
Looking beyond the BRCAalt population, the PRIMA, ATHENA-MONO, and PAOLA-1 studies enrolled similar patient populations with newly diagnosed advanced ovarian cancer who had CR or PR to platinum chemotherapy, regardless of BRCA status. In PRIMA, benefit in PFS with the use of niraparib maintenance versus placebo was seen in both the HRD-positive (BRCAalt or positive as tested by the Myriad MyChoice assay) population (median PFS, 21.9 vs. 10.4 months, HR, 0.43; 95% CI, 0.31–0.59) and the HRD-negative population (median PFS 8.1 vs. 5.4 months, HR, 0.68; 95% CI, 0.49–0.94) although the degree of benefit for patients who were HRD-negative was considerably less (3). Similar results were seen in ATHENA-MONO, which used the PARPi rucaparib, in which HRD positivity was defined as BRCAalt or high genomic LOH (gLOH-high) by the Foundation Medicine biomarker (HRD-positive: median PFS 28.7 versus 11.3 months, HR, 0.47; 95% CI, 0.31–0.72; HRD-negative: median PFS 12.1 versus 9.1 months, HR, 0.65; 95% CI, 0.45–0.95; ref. 4). OS data, which will significantly contextualize long-term benefit, are not mature for either trial. In the PAOLA-1 trial, benefit in PFS (the primary outcome) and OS (a secondary outcome) with the use of olaparib maintenance was more pronounced in the patients who were HRD-positive (Myriad MyChoice assay; PFS: HR, 0.33; 95% CI, 0.25–0.45; OS: HR, 0.62; 5% CI, 0.45–0.85; HRD-negative, PFS: HR, 1.00; 95% CI, 0.75–1.35; OS: HR, 1.19; 95% CI, 0.88–1.63; refs. 5, 6). However, this trial is the only maintenance trial that used an active control (bevacizumab), and there was no olaparib-alone arm, which limits the ability to interpret the relative contribution of bevacizumab. Following the use of mPARPi in these trials, questions remain about their potential effectiveness outside of clinical trial settings, their associations with OS, and the clinical benefit relative to the financial and physical costs of these drugs in different biomarker-defined subgroups.
Using real-world data from predominantly community-based practice, we aimed to compare the observed effectiveness (both PFS and OS) of mPARPi therapy in standard-of-care practice settings to that observed in clinical trials, in both BRCAalt and non-BRCAalt populations. We also seek to evaluate the performance of two genomic scar signatures, a novel HRD signature (HRDsig) and gLOH-high.
Materials and Methods
Study population and analysis overview
This study included patients with a chart-confirmed diagnosis of ovarian cancer who underwent genomic testing using Foundation Medicine tissue-based comprehensive genomic profiling (CGP) assays. Data were obtained from the US-wide Flatiron Health and Foundation Medicine Clinico-Genomic Database. Retrospective de-identified longitudinal clinical data were derived from electronic health records (EHR) from approximately 280 US cancer clinics (∼800 sites of care) between January 2015 and March 2023 and comprises patient-level structured and unstructured data, curated via technology-enabled abstraction of clinical notes and radiology/pathology reports. Clinical data included demographics, clinical and laboratory features, time of therapy exposure, and mortality. These were linked to genomic data derived from Foundation Medicine testing by de-identified, deterministic matching (7). The data are de-identified and subject to obligations to prevent re-identification and protect patient confidentiality. Predominant genetic ancestry was determined using principal component analysis of alternate allele counts at known SNP sites to quantitate the top 10 sources of genetic variation and coupled with random forest classification trained on these 10 features to distinguish between the five ancestral superpopulations in the 1,000 Genomes Project reference dataset: African, admixed American (a proxy for Hispanic), East Asian, European, and South Asian (8–10).
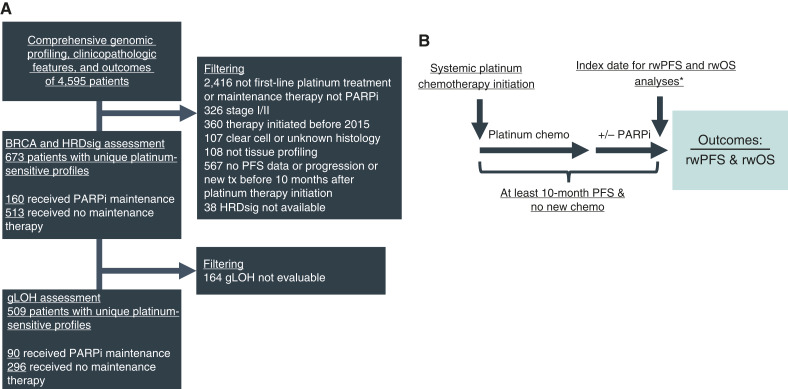
The study population comprises patients with HRDsig assessment stratified on BRCAalt (mutations or deletion) and HRDsig status that received first-line platinum-based chemotherapy with real-world PFS (rwPFS) of at least 10 months after treatment initiation and received either mPARPi (without bevacizumab) or no maintenance therapy. The inclusion criterion of rwPFS at least 10 months was used as a proxy for platinum responsiveness, due to the lack of direct chemotherapy response data in the database. The 10-month interval was chosen to allow completion of first-line chemotherapy, interval debulking, and the interval between completing chemotherapy and starting maintenance. A subpopulation of patients included in this study also had gLOH-high assessment, which requires a higher tumor purity to be assessed than HRDsig. Figure 1A shows the cohort selection, and Fig. 1B shows the temporal visualization of the analysis cohort. This study was conducted in accordance with the Declaration of Helsinki. Institutional review board approval of the study protocol was obtained before study conduct and included a waiver of informed consent based on the observational, noninterventional nature of the study (WCG IRB, Protocol No. 420180044).
Figure 1.
Cohort selection and analysis overview. Cohort selection diagram (A) and temporal visualization of the analysis cohort (B) are shown. *Four out of 160 patients receiving mPARP initiated therapy after the index date of 10 months. Chemo, chemotherapy; tx, therapy.
Comprehensive genomic profiling
Hybrid capture-based next-generation sequencing assays were performed on patient tumor specimens in a Clinical Laboratory Improvement Amendments–certified, College of American Pathologists–accredited laboratory (Foundation Medicine, Inc.). Genomic alterations were identified via CGP of >300 cancer-related genes on Foundation Medicine’s next-generation sequencing test (FoundationOneCDx or FoundationOne; refs. 11–13). gLOH-high was calculated by determining genome-wide LOH, utilizing >3,500 sequenced SNP across the genome. gLOH-high excluded whole chromosome arm losses as previously described and validated for ovarian cancer in the ARIEL2 and ARIEL3 trials (14, 15). HRDsig utilizes a broad set of copy-number features, including absolute modeled copy number, segment size, oscillation patterns, and breakpoints per chromosome arm with features examined genome-wide and specifically within the telomeric and centromeric portions of chromosome arms (16, 19, 20). The broad set of copy-number features was used as inputs into an extreme gradient boosting (XGB) machine learning model. For training, a set of samples enriched for HRD (biallelic BRCA1/2 positive cases) were labeled as HRD-positive; a set of samples depleted for HRD (no alterations in 14 HRR genes: BRCA1, BRCA2, ATM, BARD1, BRIP1, CDK12, CHEK1, CHEK2, FANCL, PALB2, RAD51B, RAD51C, RAD51D, and RAD54L) were labeled as HRD-negative. Although the labels are imperfect, these genomic correlates provide sufficient separation between groups to allow XGB modeling to identify patterns of HRD scarring. Training was performed on a subset of 282,700 pancancer samples profiled with FoundationOne or FoundationOneCDx. The XGB model outputs a score between 0 and 1, reflecting the likelihood of a sample being HRD-positive. Scores are generally bimodal. A cutoff of 0.7 was prespecified for calling a sample HRDsig(+) based on 90% sensitivity to detect biallelic BRCA1/2alt in canonically BRCA-driven diseases (ovary, prostate, pancreas, and breast cancers). Although trained using biallelic BRCA1/2 as the ground truth for HRD-positive, HRDsig also identifies BRCA1 promoter hypermethylation in ovarian cancer (17) and strongly enriches for biallelic alterations in PALB2, BARD1, BRIP1, RAD51C, and RAD51D (18). HRDsig is validated for performance down to <25% tumor purity.
Outcomes
rwPFS and rwOS were the primary endpoints. rwPFS and rwOS were indexed 10 months after the initiation of first-line platinum-based chemotherapy as a proxy for platinum sensitivity, which we used because we did not know whether a patient had a PR or CR to chemotherapy which was an inclusion criteria of the relevant clinical trials (1–4), as described previously. Thus, rwPFS was calculated from 10 months after the initiation of first-line platinum-based chemotherapy until the time of disease progression or death, and patients not yet reaching progression or death were right-censored at the date of EHR activity or CGP report. A period of 10 months was chosen to account for the usual duration of first-line platinum chemotherapy (4 months) plus 6 months without progression (time without progression to meet the definition of platinum-sensitive), as outlined in Fig. 1B. rwOS was calculated from 10 months after starting first-line platinum-based chemotherapy to death from any cause, and patients with no record of mortality were right-censored at the date of the last clinic visit or structured activity. Risk set adjustment was applied to rwOS analyses to account for immortal time caused by left truncation in the dataset, as patients cannot enter the database until a CGP report is provided (21, 22).
The mortality information in the Flatiron Health database is a composite derived from documents within the EHR, Social Security Death Index, and a commercial death dataset mining data from obituaries and funeral homes. This mortality information has been externally validated in comparison to the National Death Index with > 90% accuracy (23).
Statistical analysis
The analyses performed in this study were prespecified in a prospectively declared statistical analysis plan, with prespecified inclusion and exclusion criteria, potential biases, primary outcomes measures, and handling of missing data, consistent with Society for Pharmacoeconomics and Outcomes Research guidelines (24). The statistical analysis plan included the comparison of rwPFS and rwOS of patients with or without the biomarkers (BRCAalt, HRDsig, or gLOH-high) receiving mPARPi versus patients receiving no maintenance.
χ2 tests and Wilcoxon rank-sum tests were used to assess differences in baseline characteristics between groups of categorical and continuous variables, respectively. Baseline characteristics assessed included year of treatment initiation, ancestry, age at treatment start, practice type, Eastern Cooperative Oncology Group performance status, body mass index, disease stage at diagnosis, histology, extent of debulking surgery, residual disease, and TP53 alteration.
Differences in rwPFS and rwOS were evaluated with the log-rank test and Cox proportional hazard models, applying inverse probability of treatment weighting to adjust for potential treatment selection biases. Inverse probability of treatment weights was calculated targeting the average treatment effect in the mPARPi-treated population, implemented with R package “MatchIt.” Weights were capped at 10 equivalents to limit influence per observation. Among patients receiving no maintenance, those with characteristics most similar to patients receiving mPARPi were weighted more, and those less like patients receiving mPARPi were weighted less. These weights were included in all Kaplan–Meier visualizations and Cox PH models unless otherwise noted. Available features related to treatment assignment of mPARPi versus no maintenance included for adjustment in propensity models were stage at diagnosis (stage IV vs. III), Eastern Cooperative Oncology Group performance status (0 vs. one vs. 2), age, and BRCA status (BRCAalt vs. BRCA-WT) for the HRDsig and gLOH-high analyses. Standardized mean difference was utilized to assess balance, and within 10% was considered acceptable (25). Multiple comparison adjustments for the outcome analyses were not performed, and the P values are reported to quantify the strength of association for the treatment group and outcome not for null hypothesis significance testing. Two years of restricted mean survival times were calculated as the area under the Kaplan–Meier curves at 1-month intervals up to 24 months (26, 27). A comparison between HRDsig and Myriad MyChoice genomic instability score (GIS) results (abstracted from EHR) was conducted for patients where both tests were performed in tissue specimens collected within 90 days. R version 4.1.3 software was used for all statistical analyses.
Data availability
The data that support the findings of this study originated by Flatiron Health, Inc and Foundation Medicine, Inc. Requests for data sharing by license or by permission for the specific purpose of replicating results in this study can be submitted to PublicationsDataaccess@flatiron.com and cgdb-fmi@flatiron.com.
Results
A total of 673 patients with ovarian cancer were included in the study, with a median age of 67 (IQR, 59–73) years. Of the 673 patients, 160 received mPARPi and 513 received no maintenance therapy. Patients receiving mPARPi initiated therapy in later years (85.6% of patients receiving mPARPi vs. 27.3% of patients receiving no maintenance-initiated therapy in 2019 or later, P < 0.001) typically had tissue sample collection after mPARPi initiation (P < 0.001), had a lower prevalence of TP53-WT tumors (P = 0.039), and a higher prevalence of BRCAalt, HRDsig(+), and gLOH-high high (all P < 0.001) compared with patients not receiving maintenance therapy. No differences (P < 0.05) were observed for other baseline features (Table 1 for full cohort and Supplementary Table S1 for only BRCAalt patients). Supplementary Table S2 shows the site of tissue biopsy collection.
Table 1.
Baseline clinical and demographic characteristics of patients.
| None (N = 513) | mPARPi (N = 160) | Total (N = 673) | P value | |
|---|---|---|---|---|
| Year of therapy initiation | < 0.001 | |||
| 2015 to 2017 | 285 (55.6%) | 11 (6.8%) | 296 (43.9%) | |
| 2018 | 88 (17.2%) | 12 (7.5%) | 100 (14.9%) | |
| 2019 | 70 (13.6%) | 34 (21.2%) | 104 (15.5%) | |
| 2020 | 35 (6.8%) | 47 (29.4%) | 82 (12.2%) | |
| 2021 | 29 (5.7%) | 40 (25.0%) | 69 (10.3%) | |
| 2022 | 6 (1.2%) | 16 (10.0%) | 22 (3.3%) | |
| Ancestry | 0.283 | |||
| AFR | 40 (7.8%) | 7 (4.4%) | 47 (7.0%) | |
| AMR | 37 (7.2%) | 12 (7.5%) | 49 (7.3%) | |
| EAS | 12 (2.3%) | 8 (5.0%) | 20 (3.0%) | |
| EUR | 416 (81.1%) | 130 (81.2%) | 546 (81.1%) | |
| SAS | 8 (1.6%) | 3 (1.9%) | 11 (1.6%) | |
| Age | 0.661 | |||
| Median (Q1 and Q3) | 67.0 (58.0, 74.0) | 66.0 (60.8, 73.0) | 67.0 (59.0, 73.0) | |
| Practice type | 0.333 | |||
| Academic | 135 (26.3%) | 36 (22.5%) | 171 (25.4%) | |
| Community | 378 (73.7%) | 124 (77.5%) | 502 (74.6%) | |
| ECOG | 0.882 | |||
| 0 | 195 (38.0%) | 57 (35.6%) | 252 (37.4%) | |
| 1 | 176 (34.3%) | 58 (36.2%) | 234 (34.8%) | |
| 2+ | 41 (8.0%) | 11 (6.9%) | 52 (7.7%) | |
| Unknown | 101 (19.7%) | 34 (21.2%) | 135 (20.1%) | |
| BMI | 0.569 | |||
| Median (Q1, Q3) | 26.8 (22.4, 30.8) | 26.0 (22.0, 31.1) | 26.6 (22.3, 30.8) | |
| N-miss | 13 | 1 | 14 | |
| Stage at diagnosis | 0.043 | |||
| Stage III | 335 (65.3%) | 93 (58.1%) | 428 (63.6%) | |
| Stage IV | 108 (21.1%) | 49 (30.6%) | 157 (23.3%) | |
| Unknown/not documented | 70 (13.6%) | 18 (11.2%) | 88 (13.1%) | |
| Histology | 0.156 | |||
| Epithelial NOS | 35 (8.9%) | 15 (13.4%) | 50 (9.9%) | |
| Serous | 360 (91.1%) | 97 (86.6%) | 457 (90.1%) | |
| Extent of debulking | 0.149 | |||
| Optimal | 371 (72.3%) | 116 (72.5%) | 487 (72.4%) | |
| Suboptimal | 36 (7.0%) | 5 (3.1%) | 41 (6.1%) | |
| Unknown/not documented | 106 (20.7%) | 39 (24.4%) | 145 (21.5%) | |
| Residual disease status | 0.307 | |||
| No residual disease | 239 (46.6%) | 82 (51.2%) | 321 (47.7%) | |
| Residual disease | 147 (28.7%) | 36 (22.5%) | 183 (27.2%) | |
| Unknown/not documented | 127 (24.8%) | 42 (26.2%) | 169 (25.1%) | |
| TP53 alteration | 0.039 | |||
| Wild-type | 40 (7.8%) | 5 (3.1%) | 45 (6.7%) | |
| Positive | 473 (92.2%) | 155 (96.9%) | 628 (93.3%) | |
| BRCA group | <0.001 | |||
| BRCA1 | 52 (10.1%) | 29 (18.1%) | 81 (12.0%) | |
| BRCA2 | 28 (5.5%) | 21 (13.1%) | 49 (7.3%) | |
| Wild-type | 433 (84.4%) | 110 (68.8%) | 543 (80.7%) | |
| gLOH-high level | <0.001 | |||
| High | 136 (35.5%) | 75 (59.5%) | 211 (41.5%) | |
| Low | 247 (64.5%) | 51 (40.5%) | 298 (58.5%) | |
| N-miss | 130 | 34 | 164 | |
| HRDsig | <0.001 | |||
| (+) | 175 (34.1%) | 83 (51.9%) | 258 (38.3%) | |
| (−) | 338 (65.9%) | 77 (48.1%) | 415 (61.7%) | |
| Time of tissue collection | <0.001 | |||
| Before platinum therapy initiation | 0 (0.0%) | 11 (6.9%) | 11 (1.6%) | |
| After platinum therapy initiation | 260 (50.7%) | 42 (26.2%) | 302 (44.9%) | |
| After maintenance initiation | 253 (49.3%) | 107 (66.9%) | 360 (53.5%) | |
| PARPi used | — | |||
| Niraparib | — | 76 (47.5%) | 76 (47.5%) | |
| Olaparib | — | 55 (34.4%) | 55 (34.4%) | |
| Rucaparib | — | 13 (8.1%) | 13 (8.1%) | |
| Niraparib and olaparib | — | 10 (6.3%) | 10 (6.3%) | |
| Olaparib and rucaparib | — | 4 2.5%) | 4 2.5%) | |
| Niraparib, olaparib, and rucaparib | — | 2 (1.3%) | 2 (1.3%) | |
Abbreviations: AFR; African, AMR; admixed American, BMI; body mass index, chemo; chemotherapy, ECOG; Eastern Cooperative Oncology Group performance status, EAS; East Asian, EUR; European, NOS; not otherwise specified, SAS; South Asian,
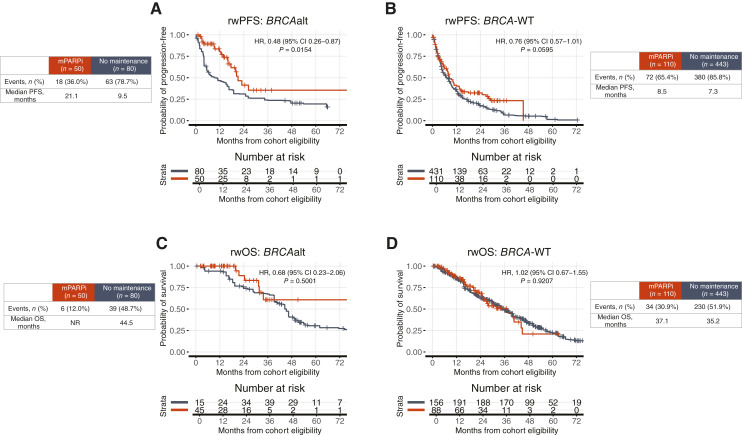
Patients with BRCAalt and BRCA-WT status had more favorable rwPFS, not rwOS, receiving mPARPi versus no maintenance
Patients with BRCAalt status receiving mPARPi versus no maintenance had more favorable rwPFS (HR, 0.48; 95% CI, 0.26–0.87; P = 0.0154; Fig. 2A). Patients with BRCA-WT status receiving mPARPi versus no maintenance also tended to have more favorable rwPFS, although to a lesser extent (HR, 0.76; 95% CI, 0.57–1.01; P = 0.0595; Fig. 2B). A significantly more favorable rwOS was not observed for either group (Fig. 2C and D). See Supplementary Fig. S1 for cohort balanced assessment using propensity weights and Supplementary Fig. S2 for unadjusted results. Supplementary Figs. S3 and S4 show rwPFS and rwOS for patients receiving mPARPi versus no maintenance in BRCA1- and BRCA2-altered groups only. The effect in the BRCA1alt group is more pronounced, but further confirmation of the individual effect of BRCA1alt versus BRCA2alt needs to be investigated in additional studies.
Figure 2.
Patients receiving maintenance PARPi had more favorable outcomes with BRCAalt but not BRCA-WT. rwPFS is shown by maintenance therapy received for (A) BRCAalt and (B) BRCA-WT. rwOS is shown by maintenance therapy received for (C) BRCAalt and (D) BRCA-WT. rwOS estimates are risk set adjusted to account for delayed entry to at-risk table (see “Materials and Methods”). Kaplan–Meier curves are adjusted by propensity weights (see Supplementary Fig. S1). Analyses unadjusted for propensity weights have similar results (see Supplementary Fig. S2). Interaction terms in interaction models (see “Materials and Methods”). tx, therapy.
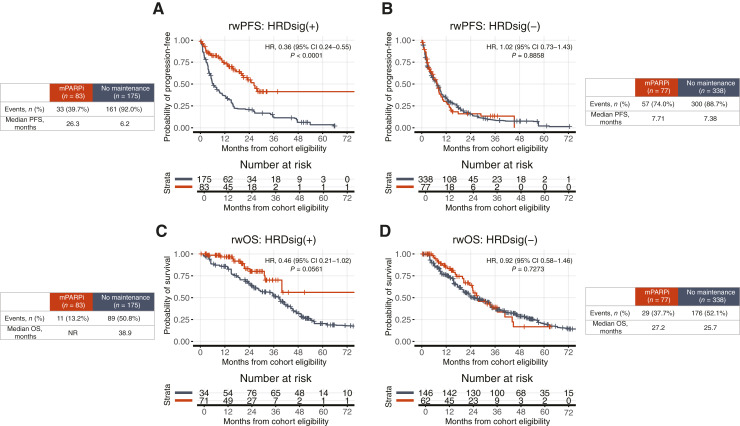
Patients with HRDsig(+) or gLOH-high-high status had better rwPFS and rwOS with mPARPi treatment, while patients with HRDsig(−) or gLOH-high-low status showed no difference in outcomes
Patients with HRDsig(+) status receiving mPARPi versus no maintenance had more favorable rwPFS (HR, 0.36; 95% CI, 0.24–0.55; P < 0.001; Fig. 3A) and tended to have more favorable rwOS (HR, 0.46; 95% CI, 0.21–1.02; P = 0.0561; Fig. 3C), whereas no significant differences were observed for patients with HRDsig(−) status (rwPFS HR, 1.02; 95% CI, 0.73–1.43; P = 0.8858; Fig. 3B; rwOS HR, 0.92; 95% CI, 0.58–1.46; P = 0.7273; Fig. 3D). See Supplementary Fig. S1 for cohort balanced assessment using propensity weights and Supplementary Fig. S5 for unadjusted results.
Figure 3.
Patients receiving maintenance PARPi had more favorable rwPFS and tended to have more favorable rwOS when HRDsig(+) but not HRDsig(−). rwPFS is shown by maintenance therapy received for (A) HRDsig(+) and (B) HRDsig(−). rwOS is shown by maintenance therapy received for (C) HRDsig(+) and (D) HRDsig(−). rwOS estimates are risk set adjusted to account for delayed entry to at-risk table (see “Materials and Methods”). Kaplan–Meier curves are adjusted by propensity weights (see Supplementary Fig. S1). Analyses unadjusted for propensity weights have similar results (see Supplementary Fig. S5). tx, therapy.
In the subcohort with gLOH-high able to be assessed (n = 509), patients with gLOH-high-high status receiving mPARPi versus no maintenance had more favorable rwPFS (HR, 0.38; 95% CI, 0.24–0.59; P < 0.001) and rwOS (HR, 0.4; CI, 0.17–0.92; P = 0.0315), and no significant differences were observed for patients with gLOH-high-low status (rwPFS HR, 0.83; 95% CI, 0.58–1.2; P = 0.3308; rwOS HR, 0.99; 95% CI, 0.58–1.7; P = 0.9841; Supplementary Fig. S6). The propensity weights used for adjusting this analysis were the same used for adjusting the HRDsig analysis (Supplementary Fig. S1), and the unadjusted results are reported in Supplementary Fig. S7.
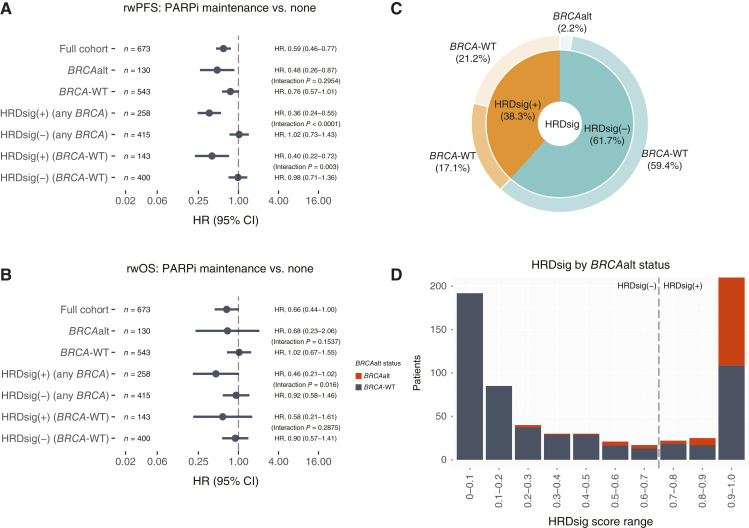
A statistically significant treatment interaction was observed for HRDsig(+) versus HRDsig(−), gLOH-high high versus gLOH-high-low, but not for BRCAalt versus BRCA-WT
In the full cohort (n = 673), a statistically significant treatment interaction was observed for HRDsig(+) versus HRDsig(−) (rwPFS P < 0.001/rwOS P = 0.016) but not for BRCAalt versus BRCA-WT (rwPFS P = 0.2954/rwOS P = 0.1537; Fig. 4A and B; Supplementary Fig. S8). A total of 143 patients were BRCA-WT and HRDsig(+). Figure 4C shows the breakdown of overlap of HRDsig(+) and BRCAalt, in which 38.3% of patients in the cohort receiving first-line platinum followed by mPARPi or no maintenance were HRDsig(+) compared with just 19.3% with BRCAalt [of which 88.6% were also HRDsig(+)]. Thus, in our cohort, HRDsig identifies a population about double the size of BRCAalt (from 19.3% to 38.3%). Figure 4D shows the bimodal distribution of HRDsig scores and the overlap of each score range and BRCAalt status.
Figure 4.
HRDsig is superior to BRCAalt alone for enrichment of favorable rwPFS and rwOS. A, rwPFS and (B) rwOS is shown by the cohort subgroup, defined by BRCAalt and/or HRDsig. Interaction P values reflect models containing the two adjacent groups. Full interaction models are available in Supplementary Fig. S8. C, HRDsig detects 21.2% patients who are BRCA-WT and might benefit from mPARPi. D, Bimodal distribution of HRDsig score and overlap with BRCAalt status. tx, therapy.
Looking specifically at BRCA-WT patients (n = 543), those who were HRDsig(+) receiving mPARPi versus no maintenance had favorable rwPFS (HR, 0.40; 95% CI, 0.0.22–0.72; median 26.8 vs. 6.2 months) and rwOS (HR, 0.58; 95% CI, 0.21–1.61; median not reached vs. 38.9 months) (Supplementary Fig. S9), whereas no difference was observed for those who were HRDsig(−) , with a statistically significant treatment interaction observed for rwPFS (P = 0.003), but not for rwOS (P = 0.2875; Fig. 4A and B; Supplementary Fig. S8).
The results were similar for the subcohort of patients with gLOH-high assessment (n = 509). These patients had a statistically significant treatment interaction for gLOH-high high versus gLOH-low (rwPFS P = 0.0054/rwOS P = 0.0072; Supplementary Figs. S10A, S10B and S11). Among patients with BRCA-WT status (n = 406), patients with gLOH-high-high status receiving mPARPi versus no maintenance also had favorable rwPFS and rwOS, whereas no difference was observed for those with gLOH-high-low status, with a statistically significant treatment interaction observed for rwPFS (P = 0.0369) but not for rwOS (P = 0.1207; Supplementary Figs. S10A, S10B, and S11). Supplementary Fig. S10C shows the breakdown of the overlap of gLOH-high high and BRCAalt for the full cohort (n = 673). Of those assessed for HRDsig, 38.3% of patients were HRDsig(+), 33.1% were gLOH-high high and 23.9% were not able to be assessed for gLOH-high. Within the subpopulation able to be assessed for gLOH-high (n = 509), 41.5% had high gLOH-high. Comparing HRDsig and gLOH-high prevalence, both biomarkers were highly concordant (Supplementary Fig. S10C). HRDsig was able to be assessed for 94.7% (673/711), whereas gLOH-high was only able to be assessed for 72.3% (514/711) because of low tumor purity issues. In the 38 patient samples that could not be assessed for HRDsig (Fig. 1A), five patients had gLOH-high data available. Consequently, HRDsig was able to be assessed in 22.4% more patients than gLOH-high.
Patients receiving mPARPi demonstrated a restricted mean survival benefit over those receiving no maintenance, among patients with BRCAalt/HRDsig(+)/gLOH-high-high, and patients with BRCA-WT and either HRDsig(+) or gLOH-high-high
Taken at face value in the full cohort, restricted mean survival times analysis showed that patients with BRCAalt had an average benefit of 7.09 months (95% CI, 3.68–10.13) of rwPFS and 4.04 (95% CI, 0.60–7.48) of rwOS in the first 24 months from mPARPi initiation, whereas those with BRCA-WT had a lower, but still positive, benefit in the first 24 months from mPARPi initiation (Supplementary Fig. S12). Patients with HRDsig(+) in the full cohort had an average benefit of 7.83 months (95% CI, 5.32–10.08) of rwPFS and 2.98 (95% CI, 1.01–5.16) of rwOS in the first 24 months from mPARPi initiation, whereas those who were HRDsig(−) had no benefit in the first 24 months from mPARPi initiation (Supplementary Fig. S12). Among patients with BRCA-WT, those with HRDsig(+) status receiving mPARPi versus no maintenance had an average benefit of 6.44 months (95% CI, 2.52–10.58) in the first 24 months from mPARPi initiation for rwPFS and 1.28 months (95% CI, -1.49–4.38) for rwOS, whereas those who were HRDsig(−) had no benefit in the first 24 months from mPARPi initiation (Supplementary Fig. S12).
In the subcohort of patients with gLOH-high available, those with gLOH-high high had an average benefit of 7.35 months (95% CI, 4.51–9.95) for rwPFS and 2.76 months (0.35–5.16) for rwOS, in the first 24 months from mPARPi initiation, whereas patients with gLOH-high-low had no benefit in the first 24 months from mPARPi initiation (Supplementary Fig. S13). Among patients with BRCA-WT status, those with gLOH-high-high status receiving mPARPi versus no maintenance had a survival benefit of 5.67 months (95% CI, 0.32–10.48) for rwPFS in the first 24 months from mPARPi initiation and 1.93 months (95% CI, −0.53–5.27) for rwOS, whereas those with gLOH-high-low status had no benefit in the first 24 months from mPARPi initiation (Supplementary Fig. S13).
HRDsig and Myriad MyChoice GIS are highly concordant
A total of 88 patients with ovarian cancer in the Clinico-Genomic Database had the Myriad MyChoice test conducted in a tissue specimen collected within 90 days of the specimen used for HRDsig assessment. We observed a 93% (82/88) agreement between HRDsig and Myriad MyChoice GIS score (Supplementary Fig. S14), and none of the six discordant cases showed the presence of biallelic BRCAalt.
Discussion
For patients with ovarian cancer, mPARPi is one of the standard-of-care options, especially for those who respond to platinum-based therapy. This present study shows that in real-world practice, a novel HRDsig can identify patients who benefit from mPARPi even among those with BRCA-WT status. Patients who were HRDsig(−) receiving mPARPi showed similar outcomes to those receiving no maintenance therapy, suggesting that patients who were HRDsig(−) may be spared the toxicities of mPARPi therapy. Similarly, gLOH-high identified patients who benefit from mPARPi, regardless of BRCAalt status, but with the caveat of being assessable in fewer patients than HRDsig.
In real-world settings, we observe similar relative benefits for patients with BRCAalt status with respect to rwPFS as seen in clinical trials for PFS (SOLO1 trial: HR, 0.30; 95% CI, 0.23–0.41; this study: HR, 0.48; 95% CI, 0.26–0.87), though in contrast to the SOLO1 trial we did not observe a statistically significant benefit in rwOS (1, 2). However, rwOS is less mature potentially attenuating any effect. The use of mPARPi in OC has been increasing over the years, as shown in Table 1. This swift change in treatment practices from nonmaintenance to mPARPi also contributes to the less mature rwOS for those patients treated with mPARPi. Similarly to the assays used in ATHENA-MONO and PRIMA, HRDsig, in this study, was able to identify patients with HRDsig(+) status with improved rwPFS from mPARPi compared with patients with HRDsig(−) status. However, in PRIMA, patients who were HRD-negative still benefited from the use of niraparib maintenance, but in our study no such benefit was observed. Interpreting results from this study in the context of PAOLA-1 should be limited because the use of bevacizumab was excluded from the study population.
With regard to the genomic signatures, we observe considerable improvement in outcome prediction using the two genomic signatures evaluated in the study, HRDsig and gLOH-high, both significantly independent to BRCAalt. Evaluating patients with BRCA-WT status specifically, both signatures identify patients of enriched observed benefit of mPARPi versus no maintenance, with an rwPFS HR of 0.40 for HRDsig and 0.42 for gLOH-high. In terms of clinical practicality, HRDsig assessment has a significant advantage over gLOH-high due to its lower requirement for tumor purity. Because gLOH-high assessment requires higher tumor purity, it can only be assessed on a limited number of patients. In contrast, HRDsig is able to be assessed on 22.4% (159) more patients than gLOH-high, enabling a broader application in clinical settings.
Although we attempted to perform a statistically rigorous analysis using validated methods, it is important to acknowledge the limitations of our study. We had to use a proxy for platinum responsiveness due to the lack of direct chemotherapy response data in the database. Although we posit that this proxy is a reasonable approximation to restrict our analysis to patients who had a PR or CR to platinum chemotherapy, we acknowledge that our inclusion criteria do not align precisely with those of prior clinical trials or approved drug indications. Comparisons of mPARPi versus no maintenance in an observational dataset are typically complicated by immortal time bias, but our landmark analysis design largely mitigated this. Nevertheless, four patients started mPARPi after the prespecified and clinically informed landmark date, contributing a small amount of residual immortal time bias. Additionally, only a subset of patients had Myriad MyChoice GIS results available for concordance analysis with HRDsig. The GIS result was abstracted from EHR, and information on the quantitative score and the biopsy site of collection for the abstracted results was unavailable. To approximate the same sample tested, we restricted the collection sample within 90 days, and 84 out of the 88 patients evaluated had specimens collected on the same date. However, we cannot confirm that the samples were from the same biopsy. This study also excluded patients who received bevacizumab maintenance with PARPi, typically used for stage IV disease, or those with residual disease after surgery, indicating that a higher-risk population was excluded. However, the difference in stage was balanced using propensity weighting (Supplementary Fig. S1), and no difference was observed in patients with residual disease between those receiving mPARPi and no maintenance. Finally, it is noteworthy that the majority of patients included in this study (81.1%) are from EUR ancestry, and the extrapolation of these results to a more diverse population warrants further confirmation.
In conclusion, this study reports on the clinical validity of a novel HRDsig biomarker to predict benefit from mPARPi over no maintenance regardless of BRCAalt status in real-world patients with ovarian cancer. Additionally, this study presents real-world evidence suggesting that those without this biomarker [i.e., patients with platinum-sensitive ovarian cancer after completion of first-line chemotherapy who are HRDsig(−)] derive minimal benefit from mPARPi and thus might be spared mPARPi therapy use, avoiding unnecessary side effects and financial toxicity, ultimately improving patient quality of life.
Supplementary Material
Supplementary Tables and Figures
Acknowledgments
We thank the patients whose data made this research possible, the clinical and laboratory staff at Foundation Medicine, and the team at Flatiron Health.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors’ Disclosures
D.L. Richardson reports personal fees from AstraZeneca, Mersana, Daiichi Sankyo, ProfoundBio, Eisai, and ImmuoGen and grants and personal fees from GlaxoSmithKline outside the submitted work. J.C.F. Quintanilha reports other support from Foundation Medicine, Inc and Roche during the conduct of the study. N. Danziger reports other support from Foundation Medicine, Inc and Roche during the conduct of the study. G. Li reports personal fees from Foundation Medicine, Inc and other support from Roche outside the submitted work. E.S. Sokol reports other support from Foundation Medicine and other support from Roche outside the submitted work. A.B. Schrock reports personal fees from Foundation Medicine and other support from Roche during the conduct of the study. E.M. Ebot reports personal fees from Foundation Medicine outside the submitted work and is a stockholder of Roche Holdings AG. N. Bhardwaj reports other support from Foundation Medicine outside the submitted work. A. Afghani reports other support from Roche during the conduct of the study, as well as other support from Roche outside the submitted work. C. Washington reports personal fees from AstraZeneca and grants from Robert A Winn Foundation STAAR outside the submitted work. J.A. Elvin reports full-time employment with Foundation Medicine, Inc. Foundation Medicine is an independent affiliate of the Roche Group, which provides equity interests as part of its employee benefit package. R.P. Graf reports other support from Foundation Medicine, Inc during the conduct of the study, as well as other support from Roche outside the submitted work. K.N. Moore reports personal fees from AstraZeneca, Aadi, Blueprint Pharma, Caris, Duality, BioNTech, Eisai, GSK, ImmunoGen, Janssen, Eli Lilly and Company, Merck, Novartis, Regeneron, Verastem, and Zentalis outside the submitted work. No disclosures were reported by the other authors.
Authors’ Contributions
D.L. Richardson: Data curation, formal analysis, investigation, visualization, methodology, writing–original draft, writing–review and editing. J.C.F. Quintanilha: Data curation, formal analysis, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. N. Danziger: Data curation, investigation, methodology, project administration, writing–review and editing. G. Li: Data curation, methodology, writing–review and editing. E. Sokol: Investigation, writing–review and editing. A.B. Schrock: Formal analysis, investigation, writing–review and editing. E. Ebot: Formal analysis, writing–review and editing. N. Bhardwaj: Writing–review and editing. T. Norris: Investigation, writing–review and editing. A. Afghani: Investigation, writing–review and editing. A. Frachioni: Investigation, writing–review and editing. C. Washington: Writing–review and editing. L. Dockery: Conceptualization, supervision, investigation, methodology, writing–review and editing. J. Elvin: Conceptualization, formal analysis, supervision, investigation, methodology, writing–review and editing. R.P. Graf: Conceptualization, formal analysis, supervision, investigation, methodology, writing–original draft, writing–review and editing. K.N. Moore: Supervision, investigation, writing–review and editing.
References
- 1. Moore K, Colombo N, Scambia G, Kim B-G, Oaknin A, Friedlander M, et al. Maintenance olaparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med 2018;379:2495–505. [DOI] [PubMed] [Google Scholar]
- 2. DiSilvestro P, Banerjee S, Colombo N, Scambia G, Kim B-G, Oaknin A, et al. Overall survival with maintenance olaparib at a 7-year follow-up in patients with newly diagnosed advanced ovarian cancer and a BRCA mutation: the SOLO1/GOG 3004 trial. J Clin Oncol 2023;41:609–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gonzalez-Martin A, Pothuri B, Vergote I, Graybill W, Lorusso D, McCormick CC, et al. Progression-free survival and safety at 3.5years of follow-up: results from the randomised phase 3 PRIMA/ENGOT-OV26/GOG-3012 trial of niraparib maintenance treatment in patients with newly diagnosed ovarian cancer. Eur J Cancer 2023;189:112908. [DOI] [PubMed] [Google Scholar]
- 4. Monk BJ, Parkinson C, Lim MC, O’Malley DM, Oaknin A, Wilson MK, et al. A randomized, phase III trial to evaluate rucaparib monotherapy as maintenance treatment in patients with newly diagnosed ovarian cancer (ATHENA-MONO/GOG-3020/ENGOT-ov45). J Clin Oncol 2022;40:3952–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ray-Coquard I, Pautier P, Pignata S, Pérol D, González-Martín A, Berger R, et al. Olaparib plus bevacizumab as first-line maintenance in ovarian cancer. N Engl J Med 2019;381:2416–28. [DOI] [PubMed] [Google Scholar]
- 6. Ray-Coquard I, Leary A, Pignata S, Cropet C, González-Martín A, Marth C, et al. Olaparib plus bevacizumab first-line maintenance in ovarian cancer: final overall survival results from the PAOLA-1/ENGOT-ov25 trial. Ann Oncol 2023;34:681–92. [DOI] [PubMed] [Google Scholar]
- 7. Singal G, Miller PG, Agarwala V, Li G, Kaushik G, Backenroth D, et al. Association of patient characteristics and tumor genomics with clinical outcomes among patients with non-small cell lung cancer using a clinicogenomic database. JAMA 2019;321:1391–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. ; 1000 Genomes Project Consortium . A global reference for human genetic variation. Nature 2015;526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Newberg J, Connelly C, Frampton G. Abstract 1599: determining patient ancestry based on targeted tumor comprehensive genomic profiling. Cancer Res 2019;79(suppl 13):1599. [Google Scholar]
- 10. Carrot-Zhang J, Chambwe N, Damrauer JS, Knijnenburg TA, Robertson AG, Yau C, et al. Comprehensive analysis of genetic ancestry and its molecular correlates in cancer. Cancer Cell 2020;37:639–54.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Frampton GM, Fichtenholtz A, Otto GA, Wang K, Downing SR, He J, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 2013;31:1023–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Woodhouse R, Li M, Hughes J, Delfosse D, Skoletsky J, Ma P, et al. Clinical and analytical validation of FoundationOne Liquid CDx, a novel 324-Gene cfDNA-based comprehensive genomic profiling assay for cancers of solid tumor origin. PLoS One 2020;15:e0237802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. He J, Abdel-Wahab O, Nahas MK, Wang K, Rampal RK, Intlekofer AM, et al. Integrated genomic DNA/RNA profiling of hematologic malignancies in the clinical setting. Blood 2016;127:3004–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Swisher EM, Lin KK, Oza AM, Scott CL, Giordano H, Sun J, et al. Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 Part 1): an international, multicentre, open-label, phase 2 trial. Lancet Oncol 2017;18:75–87. [DOI] [PubMed] [Google Scholar]
- 15. Coleman RL, Oza AM, Lorusso D, Aghajanian C, Oaknin A, Dean A, et al. Rucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ARIEL3): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2017;390:1949–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Macintyre G, Goranova TE, De Silva D, Ennis D, Piskorz AM, Eldridge M, et al. Copy number signatures and mutational processes in ovarian carcinoma. Nat Genet 2018;50:1262–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sokol ES, Madison RW, Jin DX, Chen KT, Fleischmann Z, Newberg J, et al. Abstract 966: exploration of a novel HRD signature (HRDsig) as a biomarker for rucaparib benefit in ARIEL2. Cancer Res 2023;83(suppl 7):966. [Google Scholar]
- 18. Moore JA, Chen KT, Madison R, Newberg JY, Fleischmann Z, Wang S, et al. Pan-cancer analysis of copy-number features identifies recurrent signatures and a homologous recombination deficiency biomarker to predict poly (ADP-Ribose) polymerase inhibitor response. JCO Precis Oncol 2023;7:e2300093. [DOI] [PubMed] [Google Scholar]
- 19. Antonarakis E, Moore J, Jin D, Chen T, Newberg J, Fleischmann Z, et al. Abstract 1249: Development of a pan-cancer algorithm to predict homologous recombination deficiency and sensitivity to PARPi therapy. Cancer Res 2022;82(suppl 12):1249. [Google Scholar]
- 20. Chen K-T, Madison R, Moore J, Jin D, Fleischmann Z, Newberg J, et al. A novel HRD signature is predictive of FOLFIRINOX benefit in metastatic pancreatic cancer. The Oncologist 2023;28:691–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brown S, Lavery JA, Shen R, Martin AS, Kehl KL, Sweeney SM, et al. Implications of selection bias due to delayed study entry in clinical genomic studies. JAMA Oncol 2022;8:287–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McGough SF, Incerti D, Lyalina S, Copping R, Narasimhan B, Tibshirani R. Penalized regression for left-truncated and right-censored survival data. Stat Med 2021;40:5487–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang Q, Gossai A, Monroe S, Nussbaum NC, Parrinello CM. Validation analysis of a composite real-world mortality endpoint for patients with cancer in the United States. Health Serv Res 2021;56:1281–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Berger ML, Mamdani M, Atkins D, Johnson ML. Good research practices for comparative effectiveness research: defining, reporting and interpreting nonrandomized studies of treatment effects using secondary data sources: the ISPOR Good Research Practices for Retrospective Database Analysis Task Force Report—Part I. Value Health 2009;12:1044–52. [DOI] [PubMed] [Google Scholar]
- 25. Austin PC, Stuart EA. Moving towards best practice when using inverse probability of treatment weighting (IPTW) using the propensity score to estimate causal treatment effects in observational studies. Stat Med 2015;34:3661–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liang F, Zhang S, Wang Q, Li W. Treatment effects measured by restricted mean survival time in trials of immune checkpoint inhibitors for cancer. Ann Oncol 2018;29:1320–4. [DOI] [PubMed] [Google Scholar]
- 27. Pak K, Uno H, Kim DH, Tian L, Kane RC, Takeuchi M, et al. Interpretability of cancer clinical trial results using restricted mean survival time as an alternative to the hazard ratio. JAMA Oncol 2017;3:1692–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables and Figures
Data Availability Statement
The data that support the findings of this study originated by Flatiron Health, Inc and Foundation Medicine, Inc. Requests for data sharing by license or by permission for the specific purpose of replicating results in this study can be submitted to PublicationsDataaccess@flatiron.com and cgdb-fmi@flatiron.com.