Abstract
A cyclic closed-chain dodecapeptide (cDDR5) mimicking the conformation-specific domain of CCR5 was prepared in which Gly-Asp, as a dipeptide forming a spacer arm, links the amino and carboxyl termini of the decapeptidyl linear chain (Arg168 to Thr177) derived from the undecapeptidyl arch (UPA; Arg168 to Cys178) of extracellular loop 2 (ECL2) in CCR5. Novel monoclonal antibodies were raised against cDDR5 conjugated with a multiple-antigen peptide (cDDR5-MAP), and the purified antibody [KB8C12, immunoglobulin M(κ)] reacted with cDDR5, but not with linear DDR5, in real-time biomolecular interaction analysis using surface plasmon resonance. The antibody also reacted with cells expressing CCR5, but not with cells expressing CXCR4, and the immunoreaction was competed by cDDR5-MAP. The antibody significantly interfered with chemotaxis induced by macrophage inflammatory protein, 1β, and at a concentration of 1.67 nM it almost completely inhibited infection by human immunodeficiency virus type 1 (HIV-1) R5, but not by HIV-1 X4, as observed by use of a new phenotypic assay for drug susceptibility of HIV-1 using the CCR5-expressing HeLa CD4+ cell clone 1-10 (MAGIC-5). Furthermore, cDDR5-MAP suppressed infection by HIV-1 R5 at relatively high concentrations (50 to 400 μM) in a dose-dependent manner but did not suppress infection by HIV-1 X4. Taken together, these results indicate that the antibody is conformation specific and recognizes the conformation-specific domain of the UPA of ECL2. Moreover, both the antibody and its immunogen, the cDDR5-MAP conjugate, may be useful in developing a new candidate vaccine for HIV therapy.
The chemokine receptors CCR5 and CXCR4, in addition to the CD4 molecule, are required for infection by human immunodeficiency virus type 1 (HIV-1). Moreover, the importance of CCR5 in HIV-1 transmission has been reported based on the findings that individuals homozygous for a 32-bp deletion in the CCR5-coding region have a greatly reduced susceptibility to HIV-1 infection, since the protein encoded by the defective CCR5 gene cannot be detected on the cell surface and is nonfunctional as an HIV-1 coreceptor (8, 12, 13, 21, 27). Autoantibodies against CCR5 have also been reported to be induced in the sera of HIV-seronegative individuals and to strongly block HIV infection despite multiple exposures to HIV-1; these antibodies are also confirmed to block HIV-1 infection in vitro (14). CCR5 is also considered a redundant molecule in adults, since CCR5-defective individuals have normal inflammatory and immune reactions (25). Therefore, CCR5 may become an important target for receptor antagonists, including specific antibodies against native receptors. The natural ligands for CCR5 (RANTES, macrophage inflammatory protein 1α [MIP-1α], and MIP-1β), their modified forms (Met-RANTES and aminooxypentane-RANTES), the nonpeptide CCR5 antagonist (TAK-779), and anti-CCR5 antibodies (2D7 and PA14) are known to block HIV-1 R5 infection (3, 7, 17, 19, 23, 26).
In this study, we used a different approach by developing a cyclic closed-chain dodecapeptide (cDDR5) conjugated with a multiple antigen peptide (MAP) as a new HIV-1 defense vaccine. The cDDR5-MAP, which mimics the conformation-critical domain for HIV-1 entry (the undecapeptidyl arch [UPA] of extracellular loop 2 [ECL2]), functioned as an immunogen which induced conformation-specific anti-CCR5 antibodies, and both the antibody and cDDR5-MAP inhibited infection by HIV-1 R5 in a dose-dependent manner. Therefore, we propose that cDDR5-MAP is designed for use not only as a potential ligand for defense against HIV-1 entry but also as a new vaccine in place of viral-protein-based vaccines to overcome unprecedented scientific obstacles in the development of HIV-1 vaccines.
MATERIALS AND METHODS
Preparation of cDDR5-MAP, cDDR5-Multi-Pin Block, and biotinylated cDDR5.
A CCR5-derived linear dodecapeptide (linear DDR5, H2N-D1R2S3Q4K5E6G7L8H9Y10T11G12-COOH) in which all side chain groups are protected (protected DDR5) was synthesized using an automatic peptide synthesizer and was cyclized by bond formation between the α-carboxyl group of Gly12 and the α-amino group of Asp1 after removal of the resin. The o-benzyl group of the β-carboxyl group of Asp1 in protected cDDR5 was selectively removed by reduction of palladium (0.5%) on carbon. The free β-carboxyl group of Asp1 in protected cDDR5 was separately conjugated to construct the MAP resin (Applied Biosystems Japan Ltd., Tokyo, Japan) and Multi-Pin Block according to the software manual in the Multipin peptide synthesis kit (Chiron Technologies, Clayton, Australia). It was also linked to 5-[5-(N-succinimidyloxycarbonyl)pentylamido]hexyl d-biotinamide through ethylenediamine in a similar way. cDDR5 derivatives in which all protected groups were removed by trifluoroacetic acid were used for the following purposes. Female BALB/c mice were immunized with cDDR5-MAP using the protocol of Galfre and Milstein (9). cDDR5–Multi-Pin Block was used in an enzyme-linked immunosorbent assay for screening monoclonal antibodies (MAbs) produced in hybridomas according to the experimental procedures in the Multipin peptide synthesis kit (Chiron Technologies). Biotinylated cDDR5 was used to confirm the specific binding of antibodies to cDDR5 using a BIAcore biosensor. Unless otherwise specified, all peptides used were purified by high-performance liquid chromatography (Waters), and the molecular masses of the compounds were determined by matrix-assisted laser desorption ionization–time-of-flight mass spectrometry (MALDI-TOF MS) (Burker Franzen Analytik GmbH, Bremen, Germany).
Purification of the antibody against cDDR5-MAP, KB8C12
Ten BALB/c mice were immunized intraperitoneally with cDDR5-MAP in Freund's adjuvant at 1-week intervals and administered an intravenous boost of cDDR5-MAP 3 days prior to splenectomy. We observed no adverse effects of autoantibody induction on BALB/c mice that were monitored for 3 months from the initial immunization. Thirty-three hybridomas were generated by a standard method, by which splenocytes were fused with P3U1 cells and selected in hypoxanthine-, aminopterin-, and thymidine-supplemented media. In the screening, supernatants were tested for reactivity to cDDR5–Multi-Pin Block. Hybridomas that produced the most-potent supernatants were then cloned by limiting dilution.
KB8C12, which was secreted into the culture supernatant of hybridomas, was recovered by precipitation with ammonium sulfate at 45% saturation. The immunoglobulin fraction was fractionated using a Sephadex G-150 column (Amersham Pharmacia Biotech), purified using POROS SP/H (Applied Biosystems Japan), and then eluted with 0.075 M Tris-HCl (pH 8.0) containing 1 M NaCl. The eluate was desalted with PD-10 (Amersham Pharmacia Biotech). KB8C12 was found to be monoclonal and of an immunoglobulin M(κ) [IgM(κ)] isotype.
Antiviral activities.
The antiviral activity of cDDR5-MAP or purified KB8C12 was determined using MAGIC-5 cells, which were engineered to express CCR5 in HeLa-CD4-LTR/β-Gal cells by transfection with an expression vector for CCR5 (11). MAGIC-5 cells were plated at 104 cells per well (48-well plate) and incubated overnight in a culture medium (200 μl); the medium was then replaced by a medium containing either cDDR5-MAP (50, 100, 200, or 400 μM) or a KB8C12 antibody solution (0.1, 0.2, 0.4, 0.8, or 1.5 μg/ml). Cells were then separately incubated with various HIV-1 strains (200 μl each of the R5 and X4 virus suspensions) in the presence of DEAE dextran (20 μg/ml) for 2 h, washed twice with the culture medium, and cocultured in the medium (400 μl) containing cDDR5-MAP or the antibody solution for 48 h. Cells were fixed, and HIV-1-infected cells that were stained blue were counted by conventional methods. Control experiments were carried out under identical conditions, except for the omission of cDDR5-MAP or KB8C12.
Real-time biomolecular interaction analysis using surface plasmon resonance.
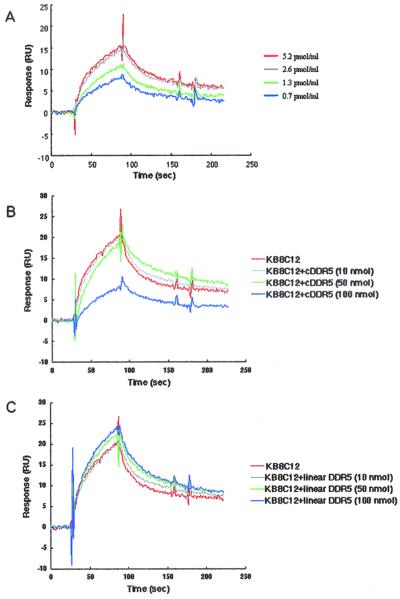
The principle of the operation of the BIAcore biosensor and its use in analyzing antigen-antibody interactions have been described previously (4, 16, 18). All interactions were analyzed in a binding buffer (0.02% KH2PO4, 0.29% Na2HPO4 · 12H2O, 0.8% NaCl, and 0.02% KCl) at a constant flow rate of 20 μl/min and a constant temperature of 25°C. Biotinylated cDDR5 was injected over a streptavidin-coated sensor chip (BIAcore) until a suitable level was achieved. Binding experiments were performed by injection of purified KB8C12 (0.7, 1.3, 2.6, and 5.2 pmol/ml) (see Fig. 2A). Competitive experiments were performed by injection of purified KB8C12 (0.52 pmol) treated with cDDR5 (10, 50, and 100 nmol) (see Fig. 2B) or linear DDR5 (10, 50, and 100 nmol) (see Fig. 2C). Bound antibody was eluted from biotinylated cDDR5 by a short pulse (20 μl) of 10 mM Gly-HCl (pH 2.0). This regeneration procedure did not, to any measurable extent, alter the ability of biotinylated cDDR5 to bind to the antibody in subsequent cycles.
FIG. 2.
Immunochemical specificity of the anti-cDDR5-MAP MAb KB8C12. The binding specificity of KB8C12 was determined by real-time biomolecular interaction analysis using surface plasmon resonance with a BIAcore biosensor. Aliquots of the deprotected cDDR5 with the free β-carboxyl group of Asp1 were used for the construction of biotinylated cDDR5 as described in Materials and Methods. Antigen-antibody binding and competitive experiments were carried out with purified KB8C12 alone (0.7, 1.3, 2.6, or 5.2 pmol/ml) (A) or with the antibody treated without (KB8C alone; 5.2 pmol/ml; black line) (B) or with cDDR5 (10, 50, or 100 nmol) (B) or linear DDR5 (10, 50, and 100 nmol) (C) using the biotinylated-cDDR5-bound BIAcore biosensor.
Flow cytometry.
The following antibodies were used: an anti-CXCR4 MAb (clone 12G5; PharMingen, San Diego, Calif.), an anti-CCR5 MAb (clone 2D7; PharMingen), purified KB8C12, an isotype-matched control antibody (Sigma Chemical Co., St. Louis, Mo.), and fluorescein isothiocyanate (FITC)-conjugated anti-mouse IgG or anti-mouse IgM.
A CD4-transduced human glioma cell line and corresponding transfectants (NP2/CD4, NP2/CD4/CXCR4, and NP2/CD4/CCR5 [24]) were incubated with 12G5, 2D7, and KB8C12 at 4°C. In addition, THP-1 cells, which are monocyte-related CCR5-positive cells (RCB1189; RIKEN Cell Bank), were exposed to KB8C12 (0.05 pmol) in the absence or presence of cDDR5-MAP (0.5 pmol). These cells were washed with a washing buffer (phosphate-buffered saline [PBS] containing 2% fetal calf serum and 0.02% NaN3), and then resuspended in the washing buffer containing FITC-conjugated anti-mouse IgG or anti-mouse IgM. After 30 min of incubation at 4°C, the cells were washed three times and then analyzed using an EPICS XL flow cytometer (Beckman Coulter).
Chemotaxis assay.
A chemotaxis assay was performed by use of the protocol of Gosling et al. (10) with THP-1 cells (106 cells) treated with or without KB8C12 (0.5 or 1.0 pmol). The assay was conducted in the presence of 20 nM MIP-1β placed in the lower chamber. Transwells (pore size, 5 μm; Corning Inc., Corning, N.Y.) were incubated for 3 h at 37°C. The cells that migrated from the upper chamber to the lower chamber were quantified by trypan blue dye exclusion.
RESULTS AND DISCUSSION
cDDR5 synthesis and peptide analysis.
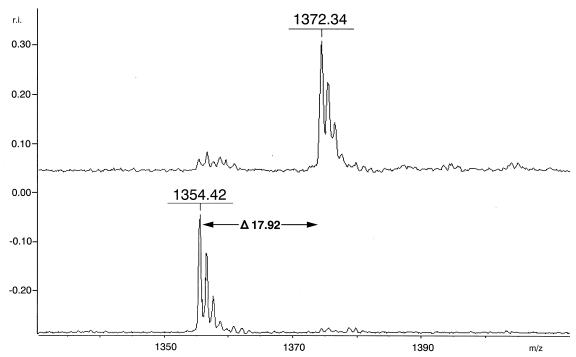
It is generally thought that the conformational B-cell epitopes involved in induction of a conformation-specific antibody would be difficult to mimic using a simple synthetic linear peptide. Therefore, a linear side chain group-blocked oligopeptide with a free-amino-terminal head and a carboxyl-terminal tail was first synthesized and then cyclized by peptidyl bond formation between the α-amino group of Asp1 and the α-carboxyl group of Gly12 on the basis of the deduced conformation of the critical domain for HIV-1 entry (UPA of ECL2) in the CCR5 structure. After removal of the resin, both the linear DDR5 (H2N-D1R2S3Q4K5E6G7L8H9Y10T11G12-COOH) and cDDR5 (cyclized at the head and tail of linear DDR5) were purified by high-performance liquid chromatography, and their molecular masses were determined by MALDI TOF-MS using α-cyano-4-hydroxy-cinnamic acid as a matrix. The spectra of purified linear DDR5 and cDDR5 exhibited major peaks at m/z 1372.34 (Fig. 1, upper spectrum) and 1354.42 (Fig. 1, lower spectrum), respectively. The difference in molecular mass between linear DDR5 and cDDR5 indicates the formation of a peptide bond. On the basis of these results, it was established that the structure of cDDR5 is cyclo(DR168S169Q170K171E172G173L174H175Y176T177G).
FIG. 1.
MALDI-TOF MS spectrum of linear DDR5 and cDDR5. The spectra exhibited two peaks at m/z 1372.34 and 1354.42: the upper peak is that of the ion derived from linear DDR5, and the lower peak is that of the ion derived from cDDR5. The matrix was a saturated solution of α-cyano-4-hydroxycinnamic acid in a solution of acetonitrile-water (1:2) containing 0.1% trifluoroacetic acid. The fraction with a molecular mass of 17.92 corresponding to H2O was deleted after cyclizing the head and tail of the peptide.
Aliquots of the protected cDDR5 with the free β-carboxyl group of Asp1 were used for construction of cDDR5-MAP (for immunization as an antigen or competitor) and cDDR5–Multi-Pin Block (antibody screening assay for enzyme-linked immunosorbent assay), and other aliquots were used for production of biotinylated cDDR5 (for determination of antigen-antibody interaction using a BIAcore biosensor) after deprotection as described in Materials and Methods.
Immunochemical specificity of the anti-cDDR5-MAP MAb, KB8C12.
Among many antibody-producing clones, one clone producing the antibody to cDDR5-MAP was effectively selected using cDDR5–Multi-Pin Block. The novel MAb [KB8C12, IgM(κ)] was purified; the immunochemical specificity of KB8C12 treated with or without cDDR5 was determined using a BIAcore biosensor bound with biotinylated cDDR5. As shown in Fig. 2A, the response of the biotinylated-cDDR5-bound biosensor increased with increasing concentration of the flowing antibody. Antibody binding was competed by cDDR5 (Fig. 2B) but not by linear DDR5 (Fig. 2C). The response curve shows a typical antibody-antigen interaction in the presence or absence of cDDR5 or linear DDR5.
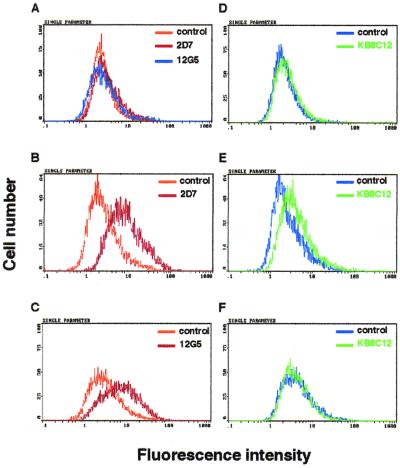
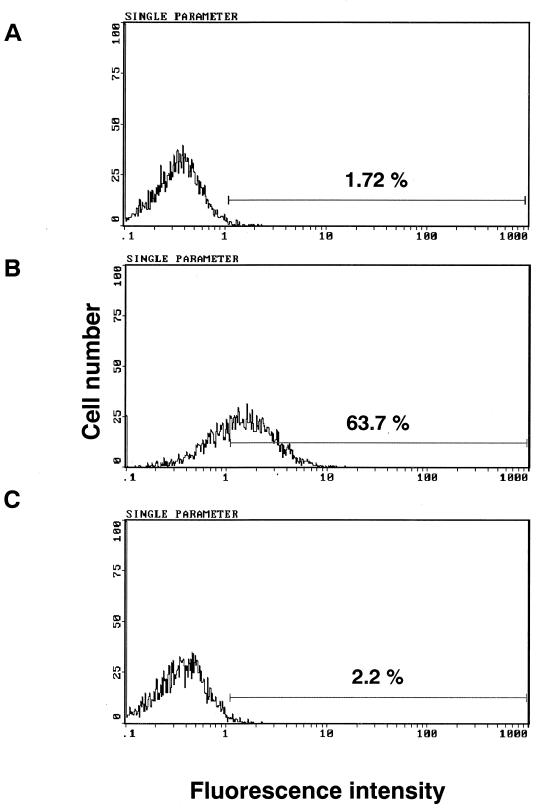
Furthermore, the binding of KB8C12 to cells expressing CCR5 was determined using a flow cytometer (Fig. 3). Anti-CCR5 and -CXCR4 MAbs (2D7 and 12G5) were used in a control experiment to confirm the expression of CCR5 and CXCR4. The antibodies reacted with CCR5- or CXCR4-expressing cells (Fig. 3B and C, respectively) but not with non-CCR5- or non-CXCR4-expressing cells (Fig. 3A). KB8C12 evidently reacted with NP2/CD4/CCR5 cells (Fig. 3E) but not with non-CCR5-expressing cells. In addition, the binding ratio of KB8C12 to CCR5 was determined with THP-1 cells expressing CCR5 at high levels. It is confirmed that 63.7% (Fig. 4B) of the total cells were stained, and this decreased to 2.2% (Fig. 4C) after treatment with cDDR5-MAP. The immunoreaction of KB8C12 to THP-1 cells was competed by cDDR5-MAP. Thus, these results taken together indicate that the MAb to cDDR5-MAP is conformation specific and that it recognizes the conformation-specific domain of UPA of ECL-2 in CCR5.
FIG. 3.
Flow cytometry of NP-2 transfectants with KB8C12. NP2/CD4 (A and D), NP2/CD4/CCR5 (B and E), and NP2/CD4/CXCR4 (C and F) were first detached by incubation in PBS containing 0.25% trypsin at 37°C for 1 min and then suspended in a cold washing buffer (PBS containing 2% fetal calf serum and 0.02% NaN3) at 106 cells/ml. These were subjected to flow cytometry as described in Materials and Methods, in which cells were separately incubated with 12G5 (1 μg; black line in Fig. 4A and C), 2D7 (1 μg; dark-gray line in Fig. 4A and black line in Fig. 4B), KB8C12 (1 μg; light-gray line in Fig. 4D, E, and F), an isotype-matched IgG antibody (control; 1 μg; light-gray line in Fig. 4A, B, and C), or an isotype-matched IgM antibody (control; 1 μ; black line in Fig. 4D, E, and F) at 4°C. Then the cells were washed with the washing buffer and resuspended in the washing buffer containing FITC-conjugated anti-mouse IgG or anti-mouse IgM.
FIG. 4.
KB8C12 binds specifically to CCR5 on the cell surface. THP-1 cells were exposed to KB8C12 (0.05 pmol) in the absence (B) or presence (C) of cDDR5-MAP (0.5 pmol), or in the presence of an isotype-matched control antibody (A) at 4°C. Positive cells were observed within the region in each histogram.
KB8C12 inhibits the chemotactic response to MIP-1β.
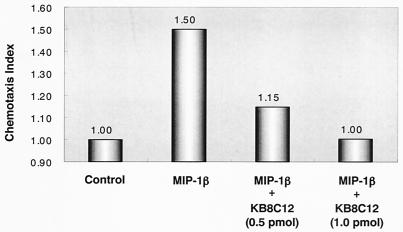
To determine whether KB8C12 affects cell functions following the blockade of the CCR5 receptor, the migration of THP-1 cells induced by a chemokine, MIP-1β, was investigated. It is known that ECL-2 of CCR5 is critical for high-affinity binding of MIP-1β (22). As expected, KB8C12 significantly interfered with chemotaxis induced by MIP-1β in a dose-dependent manner (Fig. 5). Incubation of cells with 1.0 pmol of KB8C12 was sufficient to achieve complete inhibition of THP-1 cell migration induced by MIP-1β.
FIG. 5.
KB8C12 interferes with MIP-1β-induced THP-1 chemotaxis. KB8C12 inhibits the THP-1 chemotactic response in a dose-dependent manner. Results are expressed in terms of the chemotaxis index, which represents the fold increase in the number of cells migrating in response to MIP-1β over the number migrating spontaneously in the control medium.
Antiviral activity.
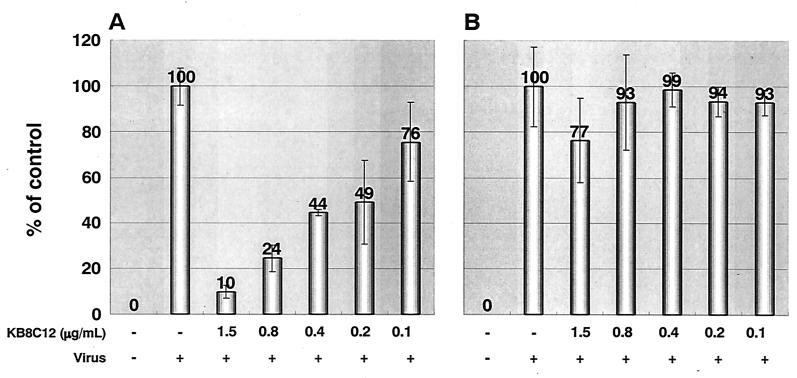
Because CCR5 is the main coreceptor for HIV-1 R5, we investigated whether KB8C12 could also inhibit HIV-1 entry via CCR5. Thus, the anti-HIV-1 activities of KB8C12 were also determined using MAGIC-5 cells that express CCR5. These cells were separately inoculated with various strains of HIV-1 (the R5 HIV-1 strain JRFL and the X4 HIV-1 strain LAV-1) in the presence of the antibody (twofold serial dilution). As expected, KB8C12 markedly suppressed infection by JRFL, but not by LAV-1, in a dose-dependent manner (Fig. 6).
FIG. 6.
Antiviral activities of KB8C12. MAGIC-5 cells were separately incubated with HIV-1 strains JRFL (amount of HIV-1 p24 antigen, 5.6 ng) (A) and LAV-1 (amount of HIV-1 p4 antigen, 56 ng) (B) and cocultured in the KB8C12-containing medium (400 μl) for 48 h as described in Materials and Methods. No significant cytotoxicity of the antibody-containing medium was observed. Values are means of triplicate determinations.
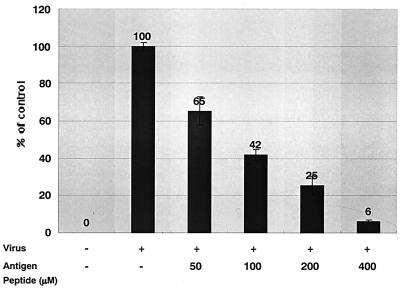
Inhibition of HIV-1 R5 infection by cDDR5-MAP.
cDDR5 was conjugated to MAP to develop a new vaccine in place of viral-protein-based vaccines. Interestingly, cDDR5-MAP also functions as a potential ligand for defense against HIV-1 entry. We evaluated the effect of cDDR5-MAP on HIV-1 replication in MAGIC-5 cells that express CCR5. Cells were inoculated with JRFL in the presence or absence of cDDR5-MAP (0, 50, 100, 200, or 400 μM). The number of cells infected (blue-stained) in the presence of cDDR5-MAP was determined microscopically and expressed as a percentage of the number of infected cells in the control. cDDR5-MAP immediately suppressed infection by JRFL in a dose-dependent manner (Fig. 7) but did not suppress infection by HIV-1 X4 (data not shown).
FIG. 7.
Antiviral activities of cDDR5-MAP. MAGIC-5 cells were separately incubated with various HIV-1 strains (JRFL; amount of HIV-1 p24 antigen, 5.6 ng) and cocultured with cDDR5-MAP (50, 100, 200, and 400 μM) for 48 h as described in Materials and Methods. No significant cytotoxicity of the cDDR5-MAP-containing medium was observed. Values are means of triplicate determinations.
Potential usefulness of antibodies raised against cDDR5-MAP.
The chemokine receptor CCR5 binds MIP-1α, MIP-1β, and RANTES and constitutes the major coreceptor that allows infection of CD4+ T lymphocytes, macrophages, and microglial cells by macrophage(M)-tropic strains of HIV (22). In particular, the second extracellular loop of CCR5 is critical for high-affinity binding of MIP-1α, MIP-1β, and RANTES and for the functional response to these chemokines. However, some studies have suggested that the multiple domains of CCR5 are used for gp120 binding (1, 2, 5, 15, 20). For example, Rucker et al. have reported that the NH2 terminus of CCR5, as well as the first extracellular loop, is important for M-tropic HIV-1 binding (20). Bieniasz et al. have indicated that the second extracellular loop is important for binding (5). Therefore, it is difficult to develop a small agonist or antagonist which by itself can inhibit HIV-1 entry via CCR5.
In this study, we provide evidence for the potential use of cDDR5-MAP, which was synthesized focusing on the potential and conformation-specific site of the chemokine receptor responsible for HIV-1 infection. cDDR5-MAP, which mimics UPA (from Arg168 to Cys178) of ECL2 in CCR5, functioned as an immunogen, which induced anti-CCR5 antibodies, and immediately inhibited infection by HIV-1 R5 in a dose-dependent manner. One antibody (KB8C12) reacted with CCR5 and blocked HIV-1 infection in the MAGIC-5 assay. With regard to strategies used for the production of AIDS vaccines, several vaccines based on native or recombinant viral materials have thus far failed to provide protection against heterologous virus infection. Furthermore, it has been reported that autoantibodies against CCR5 markedly block HIV infection despite multiple exposures to HIV-1 (14) and that autoantibodies against CCR5 could be induced in C57BL/6 mice by inoculation with recombinant papillomavirus particles representing an extracellular loop of the mouse chemokine receptor CCR5 (6). Therefore, antibodies, which may be autoantibodies, raised against cDDR5-MAP are shown to be very useful for protection or defense against HIV-1 infection and to overcome unprecedented scientific obstacles in the development of HIV-1 vaccines.
ACKNOWLEDGMENTS
We thank S. Harada and Y. Maeda (Kumamoto Medical School) for providing the NP2 cell lines. We also thank M. Tatsumi (National Institute of Infectious Diseases, Tokyo, Japan) for providing the MAGIC-5 cells.
This study was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
REFERENCES
- 1.Alkhatib G, Ahuja S S, Light D, Mummidi S, Berger E A, Ahuja S K. CC chemokine receptor 5-mediated signaling and HIV-1 co-receptor activity share common structural determinants. J Biol Chem. 1997;272:19771–19776. doi: 10.1074/jbc.272.32.19771. [DOI] [PubMed] [Google Scholar]
- 2.Atchison R E, Gosling J, Monteclaro F S, Franci C, Digilio L, Charo I F, Goldsmith M A. Multiple extracellular elements of CCR5 and HIV-1 entry: dissociation from response to chemokines. Science. 1996;274:1924–1926. doi: 10.1126/science.274.5294.1924. [DOI] [PubMed] [Google Scholar]
- 3.Baba M, Nishimura O, Kanzaki N, Okamoto M, Sawada H, Iizawa Y, Shiraishi M, Aramaki Y, Okonogi K, Ogawa Y, Meguro K, Fujino M. A small-molecule, nonpeptide CCR5 antagonist with highly potent and selective anti-HIV-1 activity. Proc Natl Acad Sci USA. 1999;96:5698–5703. doi: 10.1073/pnas.96.10.5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernard A, Bosshard H R. Real-time monitoring of antigen-antibody recognition on a metal oxide surface by an optical grating coupler sensor. Eur J Biochem. 1995;230:416–423. doi: 10.1111/j.1432-1033.1995.0416h.x. [DOI] [PubMed] [Google Scholar]
- 5.Bieniasz P D, Fridell R A, Aramori I, Ferguson S S G, Caron M G, Cullen B R. HIV-1-induced cell fusion is mediated by multiple regions within both the viral envelope and the CCR-5 coreceptor. EMBO J. 1997;16:2599–2609. doi: 10.1093/emboj/16.10.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chackerian B, Lowy D R, Schiller J T. Induction of autoantibodies to mouse CCR5 with recombinant papillomavirus particles. Proc Natl Acad Sci USA. 1999;96:2373–2378. doi: 10.1073/pnas.96.5.2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cocchi F, DeVico A L, Garzino-Demo A, Arya S K, Gallo R C, Lusso P. Identification of RANTES, MIP-1α, and MIP-1 β as the major HIV-suppressive factors produced by CD8+ T cells. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 8.Dean M, Carrington M, Winkler C, Huttley G A, Smith M W, Allikmets R, Goedert J J, Buchbinder S P, Vittinghoff E, Gomperts E, Donfield S, Vlahov D, Kaslow R, Saah A, Rinaldo C, Detels R, O'Brien S J. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 9.Galfre G, Milstein C. Preparation of monoclonal antibodies: strategies and procedures. Methods Enzymol. 1981;73:3–46. doi: 10.1016/0076-6879(81)73054-4. [DOI] [PubMed] [Google Scholar]
- 10.Gosling J, Monteclaro F S, Atchison R E, Arai H, Tsou C L, Goldsmith M A, Charo I F. Molecular uncoupling of C-C chemokine receptor 5-induced chemotaxis and signal transduction from HIV-1 coreceptor activity. Proc Natl Acad Sci USA. 1997;94:5061–5066. doi: 10.1073/pnas.94.10.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hachiya A, Aizawa-Matsuoka S, Tanaka M, Takahashi Y, Ida S, Gatanaga H, Hirabayashi Y, Kojima A, Tatsumi M, Oka S. Rapid and simple phenotypic assay for drug susceptibility of human immunodeficiency virus type 1 using CCR5-expressing HeLa CD4+ cell clone 1-10 (MAGIC-5) Antimicrob Agents Chemother. 2001;45:495–501. doi: 10.1128/AAC.45.2.495-501.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang Y, Paxton W A, Wolinsky S M, Neumann A U, Zhang L, He T, Kang S, Ceradini D, Jin Z, Yazdanbakhsh K, Kunstman K, Erickson D, Dragon E, Landau N R, Phair J, Ho D D, Koup R A. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat Med. 1996;2:1240–1243. doi: 10.1038/nm1196-1240. [DOI] [PubMed] [Google Scholar]
- 13.Liu R, Paxton W A, Choe S, Ceradini D, Martin S R, Horuk R, MacDonald M E, Stuhlmann H, Koup R A, Landau N R. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 14.Lopalco L, Barassi C, Pastori C, Longhi R, Burastero S E, Tambussi G, Mazzotta F, Lazzarin A, Clerici M, Siccardi A G. CCR5-reactive antibodies in seronegative partners of HIV-seropositive individuals down-modulate surface CCR5 in vivo and neutralize the infectivity of R5 strains of HIV-1 in vitro. J Immunol. 2000;164:3426–3433. doi: 10.4049/jimmunol.164.6.3426. [DOI] [PubMed] [Google Scholar]
- 15.Lu Z H, Berson J F, Chen Y H, Turner J D, Zhang T Y, Sharron M, Jenks M H, Wang Z X, Kim J, Rucker J, Hoxie J A, Peiper S C, Doms R W. Evolution of HIV-1 coreceptor usage through interactions with distinct CCR5 and CXCR4 domains. Proc Natl Acad Sci USA. 1997;94:6426–6431. doi: 10.1073/pnas.94.12.6426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MacKenzie C R, Hirama T, Deng S J, Bundle D R, Narang S A, Young N M. Analysis by surface plasmon resonance of the influence of valence on the ligand binding affinity and kinetics of an anti-carbohydrate antibody. J Biol Chem. 1996;271:1527–1533. doi: 10.1074/jbc.271.3.1527. [DOI] [PubMed] [Google Scholar]
- 17.Olson W C, Rabut G E, Nagashima K A, Tran D N, Anselma D J, Monard S P, Segal J P, Thompson D A, Kajumo F, Guo Y, Moore J P, Maddon P J, Dragic T. Differential inhibition of human immunodeficiency virus type 1 fusion, gp120 binding, and CC-chemokine activity by monoclonal antibodies to CCR5. J Virol. 1999;73:4145–4155. doi: 10.1128/jvi.73.5.4145-4155.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Panayotou G, Brown T, Barlow T, Pearl L H, Savva R. Direct measurement of the substrate preference of uracil-DNA glycosylase. J Biol Chem. 1998;273:45–50. doi: 10.1074/jbc.273.1.45. [DOI] [PubMed] [Google Scholar]
- 19.Proudfoot A E, Power C A, Hoogewerf A J, Montjovent M O, Borlat F, Offord R E, Wells T N. Extension of recombinant human RANTES by the retention of the initiating methionine produces a potent antagonist. J Biol Chem. 1996;271:2599–2603. doi: 10.1074/jbc.271.5.2599. [DOI] [PubMed] [Google Scholar]
- 20.Rucker J, Samson M, Doranz B J, Libert F, Berson J F, Yi Y, Smyth R J, Collman R G, Broder C C, Vassart G, Doms R W, Parmentier M. Regions in β-chemokine receptors CCR5 and CCR2b that determine HIV-1 cofactor specificity. Cell. 1996;87:437–446. doi: 10.1016/s0092-8674(00)81364-1. [DOI] [PubMed] [Google Scholar]
- 21.Samson M, Libert F, Doranz B J, Rucker J, Liesnard C, Farber C M, Saragosti S, Lapoumeroulie C, Cognaux J, Forceille C, Muyldermans G, Verhofstede C, Burtonboy G, Georges M, Imai T, Rana S, Yi Y, Smyth R J, Collman R G, Doms R W, Vassart G, Parmentier M. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 22.Samson M, LaRosa G, Libert F, Paindavoine P, Detheux M, Vassart G, Parmentier M. The second extracellular loop of CCR5 is the major determinant of ligand specificity. J Biol Chem. 1997;272:24934–24941. doi: 10.1074/jbc.272.40.24934. [DOI] [PubMed] [Google Scholar]
- 23.Simmons G, Clapham P R, Picard L, Offord R E, Rosenkilde M M, Schwartz T W, Buser R, Wells T N, Proudfoot A E. Potent inhibition of HIV-1 infectivity in macrophages and lymphocytes by a novel CCR5 antagonist. Science. 1997;276:276–279. doi: 10.1126/science.276.5310.276. [DOI] [PubMed] [Google Scholar]
- 24.Soda Y, Shimizu N, Jinno A, Liu H-Y, Kanbe K, Kitamura T, Hoshino H. Establishment of a new system for determination of coreceptor usages of HIV based on the human glioma NP-2 cell line. Biochem Biophys Res Commun. 1999;258:313–321. doi: 10.1006/bbrc.1999.0633. [DOI] [PubMed] [Google Scholar]
- 25.Stewart G. Chemokine genes—beating the odds. Nat Med. 1998;4:275–277. doi: 10.1038/nm0398-275. [DOI] [PubMed] [Google Scholar]
- 26.Wu L, LaRosa G, Kassam N, Gordon C J, Heath H, Ruffing N, Chen H, Humblias J, Samson M, Parmentier M, Moore J P, Mackay C R. Interaction of chemokine receptor CCR5 with its ligands: multiple domains for HIV-1 gp120 binding and a single domain for chemokine binding. J Exp Med. 1997;186:1373–1381. doi: 10.1084/jem.186.8.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zimmerman P A, Buckler-White A, Alkhatib G, Spalding T, Kubofcik J, Combadiere C, Weissman D, Cohen O, Rubbert A, Lam G, Vaccarezza M, Kennedy P E, Kumaraswami V, Giorgi J V, Detels R, Hunter J, Chopek M, Berger E A, Fauci A S, Nutman T B, Murphy P M. Inherited resistance to HIV-1 conferred by an inactivating mutation in CC chemokine receptor 5: studies in populations with contrasting clinical phenotypes, defined racial background, and quantified risk. Mol Med. 1997;3:23–36. [PMC free article] [PubMed] [Google Scholar]