Abstract
The vertebrate immune system has evolved to respond vigorously to microbial infection but to ignore self-antigens. Evidence has emerged that B cell responses to viruses are initiated by immune recognition of ordered arrays of antigen on the viral surface. To test whether autoantibodies against a self-antigen can be induced by placing it in a context that mimics the ordered surface of a viral particle, a peptide representing an extracellular loop of the mouse chemokine receptor CCR5 was incorporated into an immunodominant site of the bovine papillomavirus virus L1 coat protein, which self-assembles into virus-like particles. Mice inoculated with chimeric L1-CCR5 particles generated autoantibodies that bound to native mouse CCR5, inhibited binding of its ligand RANTES, and blocked HIV-1 infection of an indicator cell line expressing a human-mouse CCR5 chimera. These results suggest a general method for inducing autoantibodies against self-antigens, with diverse potential basic research and clinical applications.
The mammalian immune system is normally tolerant of its own antigens and fails to generate antibodies against circulating self-proteins or those that are expressed on the surface of circulating cells. However, cross reacting autoantibodies may be elicited in cases in which a microbial antigen mimics or incorporates a self-antigen, suggesting that B cell tolerance is not rigorous and can be broken under some circumstances. Such mechanisms have been suggested as a potential cause of human autoimmune diseases, including myasthenia gravis and autoimmune myocarditis (1, 2). Antigen arrangement may be a major determinant in inducing B cell responsiveness to self. For example, mice that were transgenic for the transmembrane envelope protein of vesicular stomatitis virus could be induced to mount an immune response against this protein. However, antibodies were elicited only when the envelope protein was presented in an ordered array on whole virions but not in animals immunized with envelope presented in a disorganized fashion, such as cell-associated or soluble envelope protein. This suggested that antigen arrangement is critical in mediating B cell responsiveness to the transgene (3). It is not known whether autoantibodies against a self-protein that has coevolved with the immune system can be induced deliberately. The ability to elicit such antibodies might have diverse applications, such as interfering with the function of a specific protein for basic research or clinical purposes.
In this report, we demonstrate that a self-protein-derived peptide, when it is presented within a highly organized context as part of the regular array of assembled viral capsomeres, can induce autoantibodies against the native protein. A self-peptide was inserted into the viral capsid (L1) protein from bovine papillomavirus type 1 (BPV-1), which has the intrinsic capacity to self-assemble into virus-like particles (VLPs) that induce high levels of neutralizing antibodies, even without adjuvant (4, 5). The self-peptide was from an extracellular (EC) loop of the mouse C-C chemokine receptor CCR5, which is expressed in numerous cell types and tissues, including memory T cells and macrophages (6). In addition to evaluating whether antibodies generated to the peptide could bind to cells expressing mouse CCR5, it was also possible to determine whether the antibodies could interfere with ligand binding to the receptor and with HIV-1 infection because macrophage-tropic (M-tropic) HIV-1 strains use human CCR5 as a coreceptor (7–11) and certain mouse-human chimeric CCR5 receptors can substitute functionally for the human receptor (12).
MATERIALS AND METHODS
Cloning and Particle Preparation.
The BPV-1 L1 gene was cloned as an EcoRI/KpnI fragment into complementary sites in the multiple cloning site of the baculovirus expression vector pFastBac1 (GIBCO/BRL). Three L1-CCR5 chimeras were generated by overlap extension PCR mutagenesis by a method adapted from Ho et al. (13). For each chimera, a portion of BPV-1 L1 sequence was replaced by a sequence predicted to encode a peptide representing the first EC loop of C57BL/6 mouse CCR5 [coding for the amino acid sequence: His Tyr Ala Ala Asn Glu Trp Val Phe Gly Asn Ile Met Cys Lys Val (14)]. The regions of BPV-1 L1 that were replaced with mCCR5 sequences were the following: L1-CCR5 chimera 1, sequence coding for L1 amino acids 130–136; L1-CCR5 chimera 2, sequence coding for L1 amino acids 275–285; and L1-CCR5 chimera 3, sequence coding for L1 amino acids 344–350. The final clones were verified by restriction digest analysis and by nucleotide sequence analysis of the PCR-amplified region.
Recombinant baculovirus stocks containing the genes coding for the chimeric L1-CCR5 proteins or wild-type BPV-1 L1 were generated by using the GIBCO/BRL baculovirus system as described by the manufacturer. Papillomavirus-like particles were purified from recombinant baculovirus-infected Sf9 cells as described (4, 5). The general morphology of the particle preparations was analyzed by mobility assay by using a fast protein liquid chromatography Superose 6 gel filtration column (Amersham Pharmacia). Eluate was collected in 1-ml fractions. The void volume of this column is 8 ml. Previously, it was determined that wild-type L1 VLPs predominantly elute in fraction 9 of the column, L1 capsomeres elute in fraction 15, and L1 monomers elute in fractions 19–21 (M. M. Okun, and J.T.S., unpublished work). Column fractions were assayed for the presence of L1 by Western blot analysis.
Inoculation of Mice.
Antisera was prepared by inoculating C57BL/6 mice with L1-CCR5 particles, wild-type BPV-1 L1 VLPs, or a synthetic CCR5 peptide representing the first EC loop of mCCR5 that was coupled to keyhole limpet hemocyanin (KLH) by using the Imject activated immunogen conjugation kit (Pierce). In some cases, mice were inoculated with particles that were denatured by boiling for 2 min in the presence of 1% SDS. Mice were inoculated intradermally with 10 μg of antigen three times at 2-week intervals. In most cases, sera were collected 2 weeks after the final boost. When adjuvant was used, antigen was prepared in Freund’s complete adjuvant for the initial injection and in Freund’s incomplete adjuvant for subsequent inoculations. All animal care was in accordance with National Institutes of Health guidelines.
ELISA.
A quantitative ELISA to detect IgG antibody against BPV-1 VLPs was performed as described (15). Antibodies that recognized the CCR5 peptide were detected as follows. A synthetic peptide representing the first EC loop of mCCR5 was generated and coupled to a BSA carrier protein by using the Imject activated immunogen conjugation kit. Anti-CCR5-specific IgG was detected by binding 300 ng of BSA-coupled CCR5 peptide in 50 μl of PBS to each well of a 96-well Immulon II microtiter plate (Dynatech) for 2 h at 37°C. After washing three times with PBS, the wells were blocked for 2 h at room temperature with 50 μl of PBS containing 0.5% nonfat dry milk plus 1% newborn calf serum and then were washed three times with PBS. Mouse serum was serially diluted in PBS plus 0.5% nonfat dry milk, and 50 μl of this diluted serum was applied to the wells after removing the last wash. The plates were incubated at room temperature for 2.5 h with gentle rocking. After five washes, 50 μl of horseradish peroxidase-conjugated goat anti-mouse IgG (Boehringer Mannheim) diluted 1:104 in 0.5% milk-PBS was added to the wells. The plate then was incubated at room temperature for 1 h with gentle rocking and then was washed three times. The peroxidase substrate ABTS (50 μl) (Boehringer Mannheim) was added to the plate, and, after 45 min of incubation at room temperature, the optical densities (ODs) were read at 405 nm in a Thermo Max microplate reader. OD405 values that were greater than twice background (usually >0.1) were considered positive.
Fluorescence-Activated Cell Sorter (FACS) Analysis.
Total IgG from pooled mouse sera was purified by affinity purification over a Protein G (Pierce) column. Peak fractions containing IgG were pooled and concentrated with a Centricon-30 spin column (Amicon). CCR5 was expressed transiently in HeLa-MAGI cells by transfection using the Lipofectamine PLUS transfection kit (GIBCO/BRL). pcDNA3-derived plasmids containing mCCR5 cloned from B6 mice and a human–mouse CCR5 chimera containing the first EC loop of mCCR5 in a background of human CCR5 were generated and described by Kuhmann et al. (12). At 48 h after transfection, the cultures were detached from the monolayer by scraping in the presence of 5 mM EDTA. Cells were washed three times in staining buffer (PBS plus 0.5% BSA). Approximately 105 cells were resuspended in 25 μl of staining buffer plus 1 μg of mouse IgG and were incubated for 45 min at 4°C. Cells then were washed three times with staining buffer, were resuspended in 25 μl of staining buffer plus 250 ng fluorescein isothiocyanate-labeled goat anti-mouse IgG (Jackson Immunoresearch), and were incubated for 30 min at 4°C. Before FACS analysis, cells were washed an additional three times with staining buffer and were resuspended in 0.5 ml of staining buffer. As a control, cells also were stained with 500 ng of fluorescein isothiocyanate-labeled mouse anti-human CCR5 mAb (mAB182) (R & D Systems) according to the manufacturer’s specifications. FACS analysis was performed on a FACSCalibur by using the cellquest software package (Becton Dickinson). Specific binding was measured relative to staining of cells transfected with pcDNA3 vector.
Chemokine Binding Assay.
HeLa-MAGI cells were transiently transfected with mouse CCR5 by using a CaPO4 mammalian transfection kit (Stratagene). Two days after transfection, the transfected cells were transferred into a 24-well culture plate, at 105 cells per well. The next day, cells were washed twice in cold PBS and then were resuspended in 150 μl of cold binding buffer [25 mM Hepes, pH 7.2/5 mM MgCl2/1 mM CaCl2/0.5% (wt/vol) BSA]. Cells were incubated for 4 h at 4°C with 0.5 nM 125I-labeled human RANTES (Amersham Pharmacia) in the absence or presence of various dilutions of mouse sera. To remove small molecules, mouse sera was buffer-exchanged to binding buffer by using Micro Bio-Spin Chromatography-6 columns (Bio-Rad) before the binding assays. As a control, some binding assays were performed in the presence of 50 nM or 500 nM cold (noniodinated) human RANTES (R & D Systems). The reactions were stopped by washing wells four times with cold binding buffer plus 0.5 M NaCl. Cells were lysed by the addition of 0.5 ml of 1% SDS. Lysates were transferred to a counting vial, and bound radioactivity was counted for 1 min in a Beckman Coulter Gamma 5500B counter.
Infectivity Assay.
HeLa-MAGI cells were transiently transfected with human/mouse CCR5 chimera (HMHH) by using CaPO4 transfection (Stratagene). Two days after transfection, and the day before infection, the indicator cells were seeded to 24-well plates at 6.5 × 104 cells per well in complete DMEM. Some infections were performed in the presence of pooled mouse sera (see above), which was buffer-exchanged to PBS by using Micro Bio-Spin Chromatography-6 columns (Bio-Rad). Before infection, cells were incubated in a total volume of 140 μl in complete DMEM with 10 μg/ml DEAE-Dextran plus dilutions of sera or antibody, for 30 min at 4°C. After this incubation, virus was added to each well to a total volume of 150 μl. Cells were incubated for 2 h at 37°C; then, 1 ml of complete DMEM was added to each well. At 3 days, cells were stained, and an infectious dose was determined by counting the number of blue nuclei in infected wells. Inhibition of viral entry was scored by comparing the average number of blue nuclei in the presence of sera with the average number of infectious centers in the absence of sera. Typically, enough infectious virions to lead to 50–75 infectious blue centers in control (no sera) wells were used in each infection. All assays were performed in duplicate on recently thawed HeLa-MAGI cells.
RESULTS
Generation of Chimeric Papillomavirus Particles.
Generation of chimeric L1-CCR5 particles required inserting the CCR5 peptide into a region of L1 that would not disrupt the ability of L1 to form particles. Although the precise structural location and function of most L1 amino acids are not known, amino acid changes that disrupt the neutralizing epitopes of various human papillomaviruses without affecting capsid assembly have been mapped to three noncontiguous regions of L1 (16–18). Because it was likely that amino acids at these sites were on the surface of the capsid, the analogous sites in BPV-1 L1 were targeted for peptide insertion. Therefore, three L1-CCR5 chimeras were constructed in which the L1 sequence at BPV-1 L1 amino acids 130–136, 275–285, or 344–350 was replaced with a sequence predicted to encode a 16-aa peptide corresponding to the first EC loop of mouse CCR5 (mCCR5) from C57BL/6 (B6) mice. These chimeras were designated L1-CCR5 chimeras 1, 2, and 3, respectively.
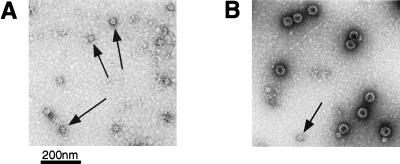
Recombinant baculoviruses containing L1-CCR5 chimeras were generated, and the resulting L1-CCR5 particles were purified by gradient centrifugation (4). To determine whether the chimeric L1-CCR5 molecules assembled into VLPs, capsomeres, or other particulate forms, Superose 6 gel filtration chromatography was performed on preparations of the three L1-CCR5 chimera. Only preparations of L1-CCR5 chimera 1 eluted in a fraction indicating an assembled particulate structure (data not shown). Therefore, further analysis was limited to this chimera. Examination of chimera 1 particles by electron microscopy revealed many particles that were smaller than wild-type L1 VLPs, ≈28 vs. 55 nm (Fig. 1A). Morphologically, the L1-CCR5 chimeric particles resemble polyomavirus 12 ICOSA shells (T = 1 particles), which are composed of a regular array of 12 pentameric capsomers of the polyomavirus major coat protein VP1 and can be generated on in vitro reassembly of VP1 capsomeres at high ionic strength (19). Small particles of a similar size to the L1-CCR5 particles often are found as a minor component of wild-type BPV-1 L1 VLP preparations (Fig. 1B, see arrow). Although the L1-CCR5 particles are smaller than wild-type VLPs, they possessed at least some characteristics of wild-type VLPs that wild-type capsomeres lack. In particular, the L1-CCR5 particles hemagglutinated mouse red blood cells and displayed ELISA reactivity to a BPV-1 neutralizing mAb (mAb 9), which specifically binds to particles but not capsomeres (data not shown; M. M. Okun, and J.T.S., unpublished work; ref. 20).
Figure 1.
Electron micrographs of L1 particles. After purification, particles were adsorbed to carbon-coated grids, were stained with 1% uranyl acetate, and were examined with a Philips electron microscope model EM 400RT at magnification ×36,000. (A) An L1-CCR5 particle preparation. Arrows identify putative 12-capsomere particles, which are ≈28 nm in diameter. (B) A preparation of wild-type L1 VLPs. Large 72-capsomere (55-nm) particles and a 12-capsomere (28-nm) particle (indicated by an arrow) are visible.
Induction of Autoantibodies.
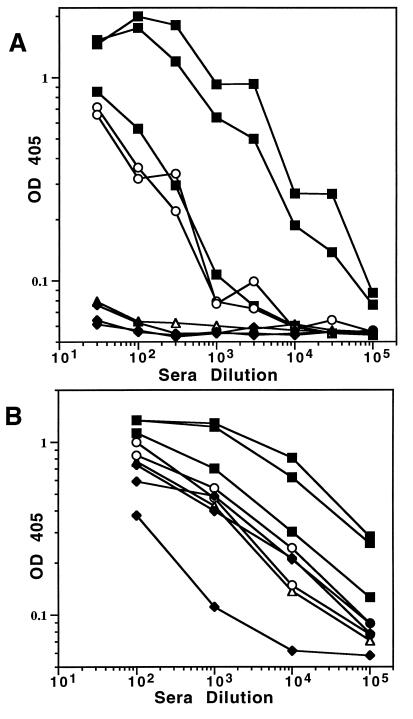
To examine whether the CCR5 chimeric particles could induce anti-CCR5 antibodies, B6 mice (a strain that encodes the identical CCR5 sequence as the insert sequence) were vaccinated with L1-CCR5 particles, denatured L1-CCR5 protein, or wild-type VLPs. Sera from these mice were tested for reactivity to CCR5 peptide and wild-type VLPs by ELISA. Sera from control mice inoculated with wild-type VLPs had no anti-CCR5 ELISA reactivity, but inoculation with L1-CCR5 particles induced sera with high anti-CCR5 ELISA titers (Fig. 2A). These titers ranged from 3 × 103 to 3 × 104 in the three animals inoculated in combination with Freund’s adjuvant and measured 3 × 103 in the two animals inoculated without adjuvant. In contrast, no CCR5-peptide-specific antibodies were detected in mice inoculated with denatured L1-CCR5 particles in combination with adjuvant. The lack of reactivity of the denatured L1-CCR5 particles was limited to the CCR5 peptide because the denatured material elicited high titers of anti-L1 antibodies (Fig. 2B).
Figure 2.
Reactivity of sera from inoculated mice by ELISA. OD 405 = OD at 405 nm. Sera from mice were inoculated with L1-CCR5 particles (■), denatured L1-CCR5 particles (♦), or BPV-1 VLPs (▵) in the presence of Freund’s adjuvant or with L1-CCR5 particles in the absence of adjuvant (○). (A) Reactivity to BSA-coupled CCR5 peptide. (B) Reactivity to BPV-1 VLPs.
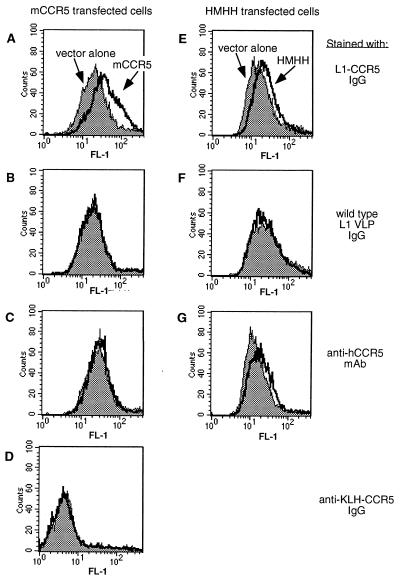
Although these results indicated that the L1-CCR5 particles elicit antibodies to the CCR5 peptide, the possibility existed that these antibodies might not recognize the peptide in its native conformation as part of membrane-associated mCCR5. To examine this question, the ability of anti-CCR5 antibodies to bind to mCCR5 on cells was tested by flow cytometric (FACS) analysis. The binding of L1-CCR5 particle sera to mCCR5 expressed on primary mouse T cells and macrophages could not be assessed because of high levels of nonspecific mouse IgG binding to these cells (data not shown). Alternatively, cloned mCCR5 from B6 mice was transiently expressed in HeLa-MAGI cells by transfection, and the binding of purified mouse IgG was measured relative to vector-transfected cells (Fig. 3). By this assay, IgG from L1-CCR5 immunized mice bound specifically to the mCCR5 transfected cells (Fig. 3A) whereas there was no significant binding with purified IgG from wild-type BPV VLP sera (Fig. 3B) or with a mAb (mAB182) that binds to the second EC loop of human CCR5 (Fig. 3C). As a control for antibody specificity, mice were inoculated with mCCR5 peptide coupled to KLH. Although these mice generated an anti-CCR5 peptide antibody response, with ELISA titers of 105 against CCR5 peptide coupled to BSA, the IgG purified from the sera of these mice failed to bind mCCR5 expressing cells (Fig. 3D). Thus, the L1-CCR5-induced antibodies, in contrast to those induced by the KLH-coupled peptide, function as true autoantibodies, in that they bind native mCCR5.
Figure 3.
Flow cytometric analysis of antibody binding to transiently transfected HeLa-MAGI cells. Constructs encoding CCR5 DNA (thick solid line) or, as a control for background staining, vector alone (shaded histogram), were transfected into the cells 2 days before staining. (A–D) Cells transfected with mouse CCR5 or vector DNA. (E–G) Cells transfected with a human/mouse CCR5 chimera (HMHH) or vector DNA. Cells were incubated with purified IgG from L1-CCR5-immunized mice (A and E), purified IgG from BPV-1 VLP-immunized mice (B and F), or purified IgG from KLH-coupled CCR5 peptide-immunized mice (D). As a control, cells also were stained with a fluorescein-labeled mAb against the second EC loop of human CCR5 (C and F).
Inhibition of Ligand and HIV-1 Binding.
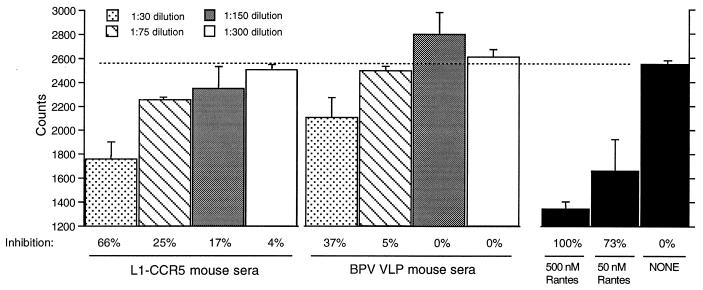
As another approach to examine the ability of the antibodies to bind native mCCR5, we examined whether the L1-CCR5 sera could compete with a chemokine ligand for mCCR5 for binding to HeLa-MAGI cells transiently transfected with mCCR5 (Fig. 4). The mouse chemokines macrophage inflammatory protein (MIP)-1α, MIP-1β, and RANTES are ligands for mCCR5. In addition, the human homologs of MIP-1β and RANTES are able to bind to mCCR5 (21, 22). In the competition assay, iodinated human RANTES was used because it is commercially available. A 1:30 dilution of L1-CCR5 sera displaced ≈66% of the iodinated human RANTES (similar to the displacement observed by using a 100-fold excess of cold RANTES), compared with 37% displacement with a 1:30 dilution of wild-type L1 VLP sera. The 1:75 and 1:150 dilutions of L1-CCR5 sera displaced 25 and 17% of the iodinated RANTES, respectively, whereas no significant displacement was observed when using control sera at these dilutions. Previous data have suggested that MIP-1α, MIP-1β, and RANTES bind to the second EC loop of hCCR5 because their binding was blocked by a mAb to this loop but not by an antibody to the amino terminus of hCCR5 (23). Our data suggest that antibodies binding to the first EC loop of mCCR5, which is located between these two sites, can partially block RANTES binding, perhaps because of the proximity of this loop to the second EC loop.
Figure 4.
Displacement of iodinated human RANTES by sera. HeLa-MAGI cells were transiently transfected with mCCR5. Three days after transfection, cells were incubated with 0.5 nM iodinated RANTES in the absence or presence of dilutions of mouse sera. Maximally bound iodinated RANTES was determined by assaying for binding in the absence of sera and corresponds to ≈2,550 cpm (indicated by the dashed line). Nonspecific binding of iodinated RANTES (≈1,300 cpm) was determined by assaying for binding in a 1,000-fold excess (500 nM) of cold (noniodinated) human RANTES. Data represents the average of duplicate wells from one experiment. This assay was repeated on two occasions to ensure reproducibility.
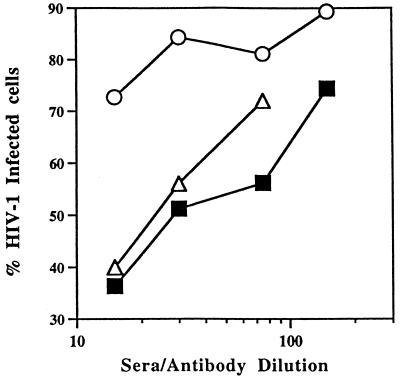
The ability of L1-CCR5-induced antibodies to block M-tropic HIV-1 infection also was tested. The interaction between HIV-1 envelope and hCCR5 is complex, likely strain-dependent, and probably involves several EC regions of CCR5. Specifically, mAb studies have implicated the second EC loop and the NH2-terminal region of hCCR5, and studies of chimeric receptors have indicated that the first and third EC loops of hCCR5 also contribute to its interaction with HIV-1 (23–28). Although mCCR5 does not function as an HIV-1 coreceptor, a human-mouse chimeric receptor (HMHH), which contains the first EC loop of mCCR5 (the B6 mouse sequence) in a background of hCCR5, has coreceptor activity (albeit at low efficiency) when expressed in human cell lines (12). We used this chimeric receptor to test whether L1-CCR5 sera could block M-tropic HIV-1 infection. To confirm that IgG purified from L1-CCR5 sera would bind HMHH, FACS analysis was performed on HeLa-MAGI cells transiently transfected with HMHH. Positive binding was obtained with IgG from L1-CCR5 mice and with a positive control mAb that binds to the second EC loop of human CCR5 whereas IgG from wild-type L1 VLP mice did not bind HMHH (Fig. 3 E–G). Based on these results, sera from L1-CCR5 mice were tested for their ability to inhibit the infection of the M-tropic BaL strain of HIV-1, in a single replication cycle assay, by using the MAGI indicator cell line (29). When indicator cells transiently transfected with HMHH were infected with HIV-1 BaL in the presence of L1-CCR5 sera, dilutions of 1:15, 1:30, and 1:75 exhibited 65, 50, and 45% neutralization, respectively, of infectivity (Fig. 5). At the same dilutions, control sera from wild-type L1 VLP mice exhibited some nonspecific neutralization, but it was only 25% at the 1:15 dilution and 15% at 1:30 and 1:75. In comparison, indicator cells infected with HIV-1 BaL in the presence of dilutions of hCCR5 binding mAb (mAB182) (at an initial concentration of 1 μg/μl) used as a positive control exhibited a similar neutralization curve (Fig. 5). The L1-CCR5 sera also were tested for neutralization activity against the T-cell tropic isolate HIV-1 LAI and, as expected, failed to show any neutralization above background levels against this isolate (data not shown).
Figure 5.
Inhibition of HIV-1 BaL infection of an indicator cell line by dilutions of L1-CCR5 sera, BPV-1 VLP sera, or a mAb against the second EC loop of human CCR5 (mAB182). Sera were pooled from three animals. HeLa-MAGI cells, an HIV-1 indicator cell line in which the nuclei of infected cells stain blue, were transiently transfected with a human-mouse CCR5 chimera (HMHH), which contains the first EC loop of mouse CCR5 in a background of the human CCR5 gene. Three days after transfection, cells were incubated with dilutions of pooled mouse sera or antibody for 30 min at 4°C. Cells then were challenged with the M-tropic isolate HIV-1 BaL. Three days after infection, infected cells were scored by counting the number of blue cells in each well. Inhibition of HIV-1 BaL infection was determined by comparing the number of blue (infected) nuclei in the presence of sera versus the number of blue nuclei in the absence of sera. Data represents the average of duplicate wells from one experiment. To ensure reproducibility, this assay was repeated on at least two other occasions, with similar results. Sera are from L1-CCR5-inoculated mice (■), BPV-1 VLP-inoculated mice (○), or mAB182 (▵).
Absence of Adverse Side Effects.
One potential concern of autoantibody induction is that these antibodies might have deleterious long term consequences for the immunized animal, possibly including uncontrolled antigenic stimulation from the native CCR5 protein. However, in three mice that were monitored over a 6-month period after L1-CCR5 particle inoculation, there was a 2- to 8-fold decrease in the titer of CCR5-specific antibodies over this period; this decline was roughly equivalent to a parallel decline in the titer of L1-specific antibodies. Two of the animals exhibited 2-fold declines in anti-CCR5 antibody titers and 3-fold declines in anti-L1 antibody titers. The third animal exhibited an 8-fold decline in its anti-CCR5 titer and a 10-fold decline in its anti-L1 antibody titer. These results suggest that continued exposure to native CCR5 does not lead to continuous B cell induction, presumably because the cellular protein remains in a context that is ignored by the immune system and, moreover, because the anti-CCR5 response depends exclusively on exposure to the CCR5 peptide on L1-CCR5 particles. The immunized mice maintained the same weight as control mice, and autopsies performed on two of the mice 6 months after the final boost did not reveal any gross pathological changes. In humans, CCR5 is expressed predominantly on memory T cells (CD3+, CD4+, CD26hi). Additionally, 1–10% of macrophages in the thymus, spleen, and lymph nodes express CCR5 (6). FACS analysis of mononuclear cells from spleen, thymus, and peripheral blood indicated that there was no decline in spleen or peripheral blood macrophage and T cell subsets that express CCR5 compared with control mice (data not shown). Thus, this limited analysis suggests that the mice immunized with L1-CCR5 particles did not suffer gross pathological changes over the period of observation.
DISCUSSION
Our studies demonstrate that incorporation of a peptide from the EC portion of a central antigen, mCCR5, into the regular array of a papillomavirus particle, followed by immunization of these particles, can induce autoantibodies that bind to the receptor and block ligand and HIV-1 binding. Autoantibodies to mCCR5 declined over time at a rate that was similar to the decline in L1-specific antibodies, suggesting that B cell stimulation by endogenous cell-surface CCR5 was not induced.
The anti-self-antibodies induced by L1-CCR5 particles efficiently bound mCCR5 expressed on the cell surface, indicating that they function as true autoantibodies. In contrast, antibodies induced by KLH-coupled CCR5 peptide failed to bind to native mCCR5. One possibility is that binding autoantibodies do not just recognize this particular amino acid sequence, but recognize this sequence in its native conformation. Moreover, the IgG from L1-CCR5-immunized mice blocks binding of a CCR5 ligand and inhibits HIV-1 infection via a chimeric CCR5 protein that contains the mouse CCR5 peptide, further demonstrating the specificity of these autoantibodies. The inhibition observed in these assays, although consistent, reproducible, specific, and similar to a control mAb against the second EC loop of human CCR5, was relatively modest. The partial inhibitory activities of the antibodies may be related to the stringency of the assays or to the indirect nature of the effect of antibody binding on viral and ligand interactions with the receptor because other regions of CCR5 have been implicated more directly in ligand binding and the precise role of the mouse CCR5 peptide in the HIV-1 infectivity assay remains unclear. The first EC loop of mCCR5 was chosen as a target because it was the only portion of mouse CCR5 that enabled us to test ligand binding as well as inhibition of HIV-1 infection. However, because other regions of CCR5 have been implicated more directly in both of these processes, it may not be surprising that only partial inhibition was obtained in the functional assays. It should be noted, however, that even partial reduction in CCR5 expression can have clinically significant effects because HIV-1-infected individuals who are heterozygous for an inactive CCR5 allele have delayed progression to AIDS (30–32). Currently, we are testing whether recombinant particles containing the homologous human/macaque CCR5 sequence induce autoantibodies in pig-tailed macaques, with the eventual goal of assessing the effects of anti-CCR5 autoantibodies on SIV infection in vivo.
We observed no adverse effects of autoantibody induction in mice that were followed for 6 months from the initial inoculation. Although we did not test for autoreactive T cells, we would not expect to break T cell tolerance to CCR5. T cells that recognize central autoantigens are strongly selected against during the development of the immune system. Presumably, the T cell help needed for Ig class switching to produce anti-CCR5 IgG is directed against the linked viral protein. Conversely, in adult animals, there is a continuous generation of antibodies with new specificities as a result of recombinase–activating gene reactivation and peripheral editing of B cell receptor genes (33–35). Our results demonstrate that adult mammals retain the capacity to produce antibodies specific for a self-antigen, provided that the antigen is presented in a context in which the host does not recognize it as self. We do not expect papillomavirus VLPs to be unique in their ability to provide this context. Indeed, it is possible that synthetic molecules may be able to provide the appropriate repetitive array necessary to stimulate autoantibody production. It remains to be determined what specific features of these arrays are critical and how the spacing of self-antigen effects autoantibody production.
The ability to induce autoantibodies has numerous potential basic and clinical applications. This technique could be used to generate mouse anti-self mAbs. Additionally, this approach could be effective as a means of modulating the activity of a soluble protein to examine its function in normal or disease processes in experimental animal models. Moreover, induction of autoantibodies may be an effective alternative to mAb therapy for human disease, such as in the treatment of breast cancer and rheumatoid arthritis with antibodies directed against ErbB-2 and tumor necrosis factor α, respectively (36, 37). The induction of potentially therapeutic autoantibodies in the individual might have some advantages over passive immunization with mAbs. For example, the anti-self antibodies might provide a longer duration of response, would not be expected to be associated with an immune response to the antibodies, and might be more effective because of the diversity of the polyclonal response. However, it would be necessary to demonstrate both long term safety and utility in experimental animals, including non-human primates, before this approach could be considered for human clinical trials.
Acknowledgments
We thank David Kabat and Shawn Kuhmann for providing CCR5 clones, Marjorie Robert-Guroff and Kris Aldrich for HIV facilities, Julie Overbaugh and Michelle Long for cell lines, Ettore Appella for peptide synthesis, Heather Greenstone and Susana Pang for assistance with electron microscopy, Jeffrey Hunt for sequencing, and Ed Berger for helpful discussions.
ABBREVIATIONS
- BPV
bovine papillomavirus
- VLP
virus-like particle
- EC
extracellular
- M-tropic
macrophage-tropic
- KLH
keyhole limpet hemocyanin
- FACS
fluorescence-activated cell sorter
- MIP
macrophage inflammatory protein
References
- 1.Rose N R. Semin Immunol. 1998;10:5–13. doi: 10.1006/smim.1997.0100. [DOI] [PubMed] [Google Scholar]
- 2.Karlsen A E, Dyrberg T. Semin Immunol. 1998;10:25–34. doi: 10.1006/smim.1997.0102. [DOI] [PubMed] [Google Scholar]
- 3.Bachmann M F, Rohrer U H, Kundig T M, Burki K, Hengartner H, Zinkernagel R M. Science. 1993;262:1448–1451. doi: 10.1126/science.8248784. [DOI] [PubMed] [Google Scholar]
- 4.Kirnbauer R, Booy F, Cheng N, Lowy D R, Schiller J T. Proc Natl Acad Sci USA. 1992;89:12180–12184. doi: 10.1073/pnas.89.24.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greenstone H L, Nieland J D, de Visser K E, De Bruijn M L H, Kirnbauer R, Roden R B S, Lowy D R, Kast W M, Schiller J T. Proc Natl Acad Sci USA. 1998;95:1800–1805. doi: 10.1073/pnas.95.4.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang L, He T, Talal A, Wang G, Frankel S S, Ho D D. J Virol. 1998;72:5035–5045. doi: 10.1128/jvi.72.6.5035-5045.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 8.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, et al. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 9.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, et al. Nature (London) 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 10.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, et al. Nature (London) 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 11.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 12.Kuhmann S E, Platt E J, Kozak S L, Kabat D. J Virol. 1997;71:8642–8656. doi: 10.1128/jvi.71.11.8642-8656.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 14.Boring L, Gosling J, Monteclaro F S, Lusis A J, Tsou C L, Charo I F. J Biol Chem. 1996;271:7551–7558. doi: 10.1074/jbc.271.13.7551. [DOI] [PubMed] [Google Scholar]
- 15.Kirnbauer R, Hubbert N L, Wheeler C M, Becker T M, Lowy D R, Schiller J T. J Natl Cancer Inst. 1994;86:494–499. doi: 10.1093/jnci/86.7.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ludmerer S W, Benincasa D, Mark G E., III J Virol. 1996;70:4791–4794. doi: 10.1128/jvi.70.7.4791-4794.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ludmerer S W, Benincasa D, Mark G E R, Christensen N D. J Virol. 1997;71:3834–3839. doi: 10.1128/jvi.71.5.3834-3839.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roden R B, Armstrong A, Haderer P, Christensen N D, Hubbert N L, Lowy D R, Schiller J T, Kirnbauer R. J Virol. 1997;71:6247–6252. doi: 10.1128/jvi.71.8.6247-6252.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salunke D, Caspar D L D, Garcea R L. Biophys J. 1989;56:887–900. doi: 10.1016/S0006-3495(89)82735-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roden R B S, Weissinger E M, Henderson D W, Booy F, Kirnbauer R, Mushinski J F, Lowy D R, Schiller J T. J Virol. 1994;68:7570–7574. doi: 10.1128/jvi.68.11.7570-7574.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyer A, Coyle A J, Proudfoot A E, Wells T N, Power C A. J Biol Chem. 1996;271:14445–14451. doi: 10.1074/jbc.271.24.14445. [DOI] [PubMed] [Google Scholar]
- 22.Nibbs R J B, Wylie S M, Pragnell I B, Graham G J. J Biol Chem. 1997;272:12495–12504. doi: 10.1074/jbc.272.19.12495. [DOI] [PubMed] [Google Scholar]
- 23.Wu L, LaRosa G, Kassam N, Gordon C J, Heath H, Ruffing N, Chen H, Humblias J, Samson M, Parmentier M, et al. J Exp Med. 1997;186:1373–1381. doi: 10.1084/jem.186.8.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rucker J, Samson M, Doranz B J, Libert F, Berson J F, Yi Y, Smyth R J, Collman R G, Broder C C, Vassart G, et al. Cell. 1996;87:437–446. doi: 10.1016/s0092-8674(00)81364-1. [DOI] [PubMed] [Google Scholar]
- 25.Atchison R E, Gosling J, Monteclaro F S, Franci C, Digilio L, Charo I F, Goldsmith M A. Science. 1996;274:1924–1926. doi: 10.1126/science.274.5294.1924. [DOI] [PubMed] [Google Scholar]
- 26.Alkhatib G, Ahuja S S, Light D, Mummidi S, Berger E A, Ahuja S K. J Biol Chem. 1997;272:19771–19776. doi: 10.1074/jbc.272.32.19771. [DOI] [PubMed] [Google Scholar]
- 27.Picard L, Simmons G, Power C A, Meyer A, Weiss R A, Clapham P R. J Virol. 1997;71:5003–5011. doi: 10.1128/jvi.71.7.5003-5011.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ross T M, Bieniasz P D, Cullen B R. J Virol. 1998;72:1918–1924. doi: 10.1128/jvi.72.3.1918-1924.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kimpton J, Emerman M. J Virol. 1992;66:2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu R, Paxton W A, Choe S, Ceradini D, Martin S R, Horuk R, MacDonald M E, Stuhlmann H, Koup R, Landau N. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 31.Samson M, Libert F, Doranz B J, Rucker J, Liesnard C, Farber C M, Saragosti S, Lapoumeroulie C, Cognaux J, Forceille C, et al. Nature (London) 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 32.Winkler C, Modi W, Smith M W, Nelson G W, Wu X, Carrington M, Dean M, Honjo T, Tashiro K, Yabe D, et al. Science. 1998;279:389–393. doi: 10.1126/science.279.5349.389. [DOI] [PubMed] [Google Scholar]
- 33.Han S, Dillon S R, Zheng B, Shimoda M, Schlissel M S, Kelsoe G. Science. 1998;278:301–305. doi: 10.1126/science.278.5336.301. [DOI] [PubMed] [Google Scholar]
- 34.Papavasiliou F, Casellas R, Suh H, Qin X F, Besmer E, Pelanda R, Nemazee D, Rajewsky K, Nussenzweig M C. Science. 1998;278:298–301. doi: 10.1126/science.278.5336.298. [DOI] [PubMed] [Google Scholar]
- 35.Hertz M, Kouskoff V, Nakamura T, Nemazee D. Nature (London) 1998;394:292–295. doi: 10.1038/28419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maini R N, Elliott M J, Brennan F M, Willians R O, Chu C Q, Paleolog E, Charles P J, Taylor P C, Feldmann M. Immunol Rev. 1995;144:195–223. doi: 10.1111/j.1600-065x.1995.tb00070.x. [DOI] [PubMed] [Google Scholar]
- 37.Baselga J, Tripathy D, Mendelsohn J, Baughman S, Benz C C, Dantis L, Sklarin N T, Seidman A D, Hudis C A, Moore J, et al. J Clin Oncol. 1996;14:737–744. doi: 10.1200/JCO.1996.14.3.737. [DOI] [PubMed] [Google Scholar]